Abstract
Pumilio proteins are a class of RNA-binding proteins harboring Puf domains (or PUM-HD; Pumilio-Homology Domain), named after the founding members, Pumilio (from Drosophila melanogaster) and FBF (Fem-3 mRNA-Binding Factor from Caenorhabditis elegans). The domains contain multiple tandem repeats each of which recognizes one RNA base and is comprised of 35–39 amino acids. Puf domain proteins have been reported in organisms ranging from single-celled yeast to higher multicellular eukaryotes, such as humans and plants. In yeast and animals, they are involved in a variety of posttranscriptional RNA metabolism including RNA decay, RNA transport, rRNA processing and translational repression. However, their roles in plants are largely unknown. Recently, we have characterized the first member of the Puf family of RNA-binding proteins, APUM23, in Arabidopsis. Here, we discuss and summarize the diverse roles and targets of Puf proteins previously reported in other organisms and then highlight the potential regulatory roles of Puf proteins in Arabidopsis, using our recent study as an example.
Key words: APUM23, Arabidopsis, nucleolus, Puf protein, Pumilio, rRNA processing
In a living cell, RNA-binding proteins interact with a variety of RNA and are involved in posttranscriptional control by regulating RNA export, splicing, stability, localization, translation initiation and decay. Although the Arabidopsis genome reportedly encodes 196 RRM (RNA recognition motif ) and 26 KH (K homology) domain proteins,1 only a few have been characterized to date. One unique class of RNA-binding protein family is the Puf family, which act mainly through the direct binding of the 3′ untranslated region (UTR).2 Puf proteins contain repeats called Puf domains, which recognize RNA in a sequence-specific manner.3 The Puf proteins are implicated in important biological processes in yeast, Drosophila, Humans and many other organisms; however, in Arabidopsis this family remains uncharacterized.
Diverse Roles of Pumilio Proteins
Puf proteins are known to play a plethora of roles in eukaryotes, including cytoplasmic de-adenylation, translational repression through RNA localization or RNA decay, maintenance of germline stem cell identity, mitochondria motility and biogenesis, translation initiation, rRNA processing and ribosome biogenesis. Recent studies on Puf protein-mediated post-transcriptional processing uncovered the role of Puf proteins as cytoplasmic deadenylation cofactors.4–7 De-adenylation of mRNA in eukaryotic cytoplasm accompanies translational repression and/or decay of mRNAs, and thus eventually regulates growth and development. In budding yeast, Puf4p and Mpt5 (Puf5p) are components of the Pop2p-Ccr4-Not de-adenylase complex, which removes the poly(A) tail of target mRNA and also recruits DExD/H-box helicase 1 (Dhh1p) and decapping enzyme 1 (Dcp1) for translational repression.
Translational repression by Puf proteins is also accomplished through the formation of the Puf-RNA complex during RNA localization. For example, the Drosophila Puf protein binds to the 3′ UTR of hunchback (hb) mRNA, thereby repressing the translation of target RNA while moving to the posterior end of the embryo.8,9 Similarly, yeast Puf6p represses the translation of Ash1 mRNA during localization to the distal tip of the budding cell.10,11 Translational repression of Ash1 mRNA by Puf6p is inhibited by eIF-5B,11 which is responsible for the conversion of 48S to the 60S ribosomal subunit.12 This suggests that RNA transport is tightly linked to ribosome maturation and assembly. In addition, Puf6p is co-purified with the 60S ribosomal subunit.13,14 This has also been shown for the yeast RNA-binding Loc1p, which has roles in rRNA processing as well as in the assembly and nucleocytoplasmic export of the 60S subunits.15–17 Consistent with these findings, Ash1 mRNA has a cis-acting binding site for Loc1p and Puf6p, and it does not properly localize to the daughter cell in Loc1 and Puf6 mutant yeast.18 Hence, ribosome biogenesis and translation machinery affect mRNA localization.16,17,19
Pumilio proteins are required to maintain germ-line stem cell identity in Drosophila, Caenorhabditis elegans and humans. In Drosophila, Pumilio acts together with Nanos to repress the translation of maternal hunchback RNA in the posterior end of the Drosophila embryo, thereby allowing abdomen formation.20 In C. elegans, double mutations in the Pumilio homologs, fbf-1 and fbf-2, result in the loss of germ-line stem cell populations.21 It has been proposed that FBF proteins control the maintenance of germ-line stem cells, at least partially, by repressing the translation of gld-1, a gene that promotes meiotic differentiation.21 Furthermore, in humans, PUM2 interacts with DAZ/DAZL proteins that function in primordial germ cell differentiation, and thus appeared to be required for maintenance of the human germ cell lineage.22
In yeast, Puf3p is involved in mitochondrial biogenesis as it preferentially binds to the mRNAs of nuclear-encoded mitochondrial proteins.23 It also contributes to mitochondrial motility during budding from the mother cell by linking the mitochore, a protein complex implicated in linking mitochondria to actin filaments, with the Arp2/3 complex, which generates the force needed for actin dependent mitochondria movement.24 A similar role was also proposed for yeast Puf1p/Jsn1p in recruiting the Arp2/3 complex required for mitochondrial motility.25
Usually, Puf proteins affect translation by directly controlling the abundance of transcript encoding translation initiation factor through 3′UTR mediated RNA decay pathways. For example, Drosophila Pum and mammalian Pumilio 2 proteins control the abundance of eIF-4E mRNA by binding its 3′UTR and mediating its decay.26,27 However, recently mammalian Pumilio 2 was shown to interfere with translation control through a non-decay pathway by competing with eIF-4E protein for access to the 7-methyl guanosine cap and repressing the initiation of translation by outcompeting eIF-4E, thereby preventing formation of the initiation complex.28 Hence, certain Pumilio proteins appear to play a role in the initiation of translation by controlling eIF-4E.
Puf proteins, such as Trypanosome TbPUF7 and yeast ScNop9, have characterized roles in rRNA processing and ribosome biogenesis.29,30 Both are localized in the nucleoli and affect early steps of rRNA processing. ScNop9 is associated with the pre-40S ribosome subunit.
Target RNAs for Pumilio Proteins: Single or Multiple?
Wang et al.31 determined the structure of the RNA-binding domain of the human Puf protein, Pumilio 1, and suggested that PUM-HD forms a concave structure, interacting with RNA on its inner face. Moreover, the crystal structure of yeast Puf4p showed that the UGUR core sequence in target RNA is sufficient for its binding.32 Individual target RNAs have been reported for specific Puf proteins; e.g., hb, cyclin B, HO, fem-3, COX17 and gld-1 mRNA.33 In contrast to these reports, genome-wide RNA immunoprecipitation (RIP)-coupled microarray analyses showed that a single Puf protein is able to bind to multiple target RNAs.23,34 For example, each of the five Puf proteins in yeast binds different subsets of mRNAs.23 The basis for this selectivity lies in the nucleotides at positions 4–6 of the target mRNA.35 Further complicating the issue, Puf proteins can act alone or in combination with other Puf proteins or protein partners to recognize their target RNAs.9,36–38
Puf Proteins in Arabidopsis
Although a wealth of information about the role of Puf proteins is available for other organisms, little is known about their role in Arabidopsis. As some of Arabidopsis Puf proteins show similar primary structures to those of other organisms, while others have distinct types that are found only in plants, some may maintain conserved roles among organisms and many others may have plant-specific functions. Additionally, the characterization of Puf proteins in rice will serve as a model for the role of Puf proteins in monocot plants.
Arabidopsis contains 25 Puf proteins, many of which represent examples of gene duplication events, such as APUM1 (At2g29200)/APUM2 (At2g29190)/APUM3 (At2g29140), APUM9 (At1g35730)/APUM10 (At1g35750) and APUM18 (At5g60110)/APUM19 (At5g60180). Duplicated genes frequently have redundant functions, suggesting that single gene mutants defective in one of the duplicated genes might not show noticeable phenotypic changes, enabling easy identification of gene functions by a forward genetic approach. The developmental roles of Arabidopsis Puf proteins are just beginning to emerge. For example, APUM1, APUM2 and APUM3 appear to be involved in stem cell maintenance and differentiation, directly or indirectly. This possibility was proposed because FASCIATA-2, WUSCHEL, CLAVATA-1 and ZWILLE/PINHEAD RNAs all contain a putative Nanos response element (NRE) sequence in their 3′UTR, and indeed, interact with APUM2 protein when analyzed using a yeast three-hybrid assay system.39 However, the biological implication of such an interaction remains unverified in planta. In another example, APUM10 was proposed as a reliable stem cell niche marker because of its specific expression in the apical cell layers of the heart stages of embryos.40 Furthermore, APUM16 (At5g59280) and APUM17 (At1g35850) are specifically expressed in sperm cells41 and probably have roles in the polarized nuclear division and/or speciation of sperm cells. However, these studies provide only limited information, and thus like other Puf proteins, they await detailed characterization.
To gain information about the tissue-specific expression pattern of eight Puf proteins, we generated Arabidopsis transgenic lines expressing the β-glucuronidase (GUS) gene under the control of the promoters of specific Puf genes. While APUM4 (At3g10360) and APUM9 showed constitutive expression with higher levels of expression in the leaf vasculature, some Puf proteins such as APUM1, APUM2, APUM5 (At3g20250) and APUM6 (At4g25880) showed higher expression in the shoot meristem and newly emerging leaves (Fig. 1), suggesting that these Puf proteins are required for active cell division. We also observed GUS expression in developing pollens for APUM4 and APUM17 (At1g35850) transgenic lines. Unlike APUM4 transcript, which was amplified in entire seedling as well as anther containing pollen granules, APUM17 transcript was detected preferentially in pollen (data not shown). Probably, APUM17 is transiently expressed during pollen development as shown by GUS staining of pollen grains in PAPUM17:GUS transgenic plants. Further, PAPUM6:GUS and PAPUM18:GUS transgenic plants showed GUS expression in the guard cells of the cotyledons and rosette leaves (Fig. 1).
Figure 1.

Histochemical GUS staining in the transgenic plants expressing β-glucuronidase gene under the control of APUM gene promoters.
Some of Arabidopsis Puf proteins may be localized to the chloroplasts, which do not exist in non-photosynthetic organisms. Six Puf proteins appear to have a putative transit peptide sequence when analyzed by PSORT42 and ChloroP,43 suggesting that they may have chloroplast-related functions, such as posttranscriptional regulation of chloroplast RNA. The data on cellular localization demonstrated that cytoplasmic punctuate structures are formed for many of the Arabidopsis Puf proteins, such as APUM7 (At1g78160), APUM8 (At1g22240), APUM9, APUM10, APUM12 (At5g56510), APUM14 (At5g43110) and APUM18.44 However, others, i.e., APUM23 (At1g72320) and APUM24 (At3g16810), localize to the nucleolus,44,45 suggesting that certain Puf proteins are involved in organelle-specific functions.
APUM23 is Required for Normal Growth Patterning and Development in Arabidopsis
In an attempt to characterize Puf proteins in Arabidopsis, we analyzed in depth T-DNA mutants defective in APUM23. Both transient and constitutive assays showed that APUM23 is exclusively localized in the nucleolus, distinct from any other nucleolar body, such as the Cajal body. APUM23 transcripts showed upregulation by glucose and sucrose, suggesting a role of responding to the nutrient supply that is required for active growth. Consistently, APUM23 was expressed in rapidly dividing cells, such as newly emerging leaves and pollen, as evident in the GUS stains of PAPUM23:GUS transgenics. APUM23 is involved in rRNA processing and consequently affects ribosome biogenesis. Thus, in the T-DNA insertion mutants of APUM23, 35S pre-rRNA and polyadenylated transcripts of rRNA processing by-products were accumulated (Fig. 2). In spite of the phenotypic similarity of apum23 mutant to the “loss of function” mutants of nucleolin 1 (AtNuc-L1)46–48 and ribosomal protein (RP)49–52 genes, our microarray data showed the transcriptional upregulation of a portion of the RP genes, although the expression level of the AtNuc-L1 transcript did not change. However, AtNuc-L2, which is not expressed under normal growth conditions in wild-type plants, was greatly upregulated. Similar to the increased expression of AtNuc-L2 in apum23 mutant, atNuc-L1 mutants also accumulated AtNuc-L2. AtNuc-L1 is also required in pre-rRNA processing, similar to APUM23.46,48 Hence, AtNuc-L2 appears to be upregulated to compensate for the reduced supply rate of rRNA. Upregulation of RP gene expression in the apum23 mutant also may be a feed-back mechanism to compensate either the reduced rate of ribosome assembly or the imbalance of ribosome subunits due to a limited supply of integral rRNAs.
Figure 2.
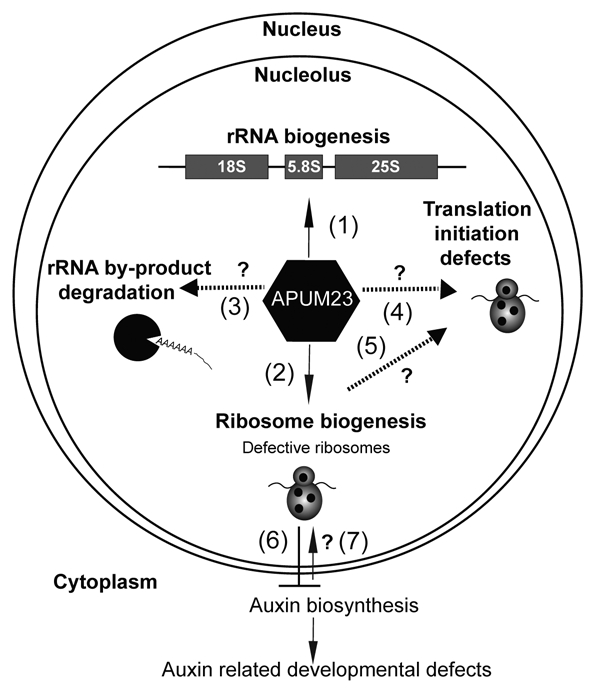
Possible nucleolar functions of APUM23. APUM23 is required for rRNA processing (1) and affects ribosome biogenesis (2). Because APUM23 is a nucleolar protein, it is possible that it has also other nucleolar functions such as rRNA byproduct degradation by exosome (3), a direct role in translation (4) or an indirect role via its role in ribosome biogenesis (5). Further, auxin and ribosome biogenesis are linked with each other such that defective ribosomes inhibit auxin biosynthesis54–56 (6) and a local auxin accumulation can translationally repress ribosomal genes (7).57
Other Possible Nucleolar Functions of APUM23
Nucleolar localized Puf proteins have also been reported in Saccharomyces cerevisae (ScPuf6 and ScNop9),10,11,30 and Trypanosoma brucei (TbPUF7).29 Most of the nucleolar Puf proteins are associated with rRNA processing and ribosome biogenesis. Our data on the nucleolar-localized Puf protein, APUM23, provide another example of nucleolar Pufs that are involved in rRNA processing. However, because the nucleolus is a multifunctional organelle and APUM23 is resident in nucleolus, APUM23 may participate in other nucleolar functions as well. As shown in Figure 2, it is possible that APUM23 associates with the nuclear exosome and mediates the degradation of the by-products of rRNA processing. Like cytoplasmic yeast Puf5p, APUM23 may interact with a nuclear deadenylation complex and mediate RNA decay in the nucleus. It is also possible that APUM23 interacts with translation initiation factors, such as eIF-4E, and mediates translation control, as do mammalian Puf proteins,28 or it may bind to the 3′UTR of eIF-4E mRNA to control its translation.26,27 The increased sensitivity of apum23-1 to cycloheximide, a eukaryotic protein synthesis inhibitor and its resistance to the antibiotic streptomycin, argue in favor of the involvement of APUM23 in the control of translation and ribosome biogenesis.
Ribosomal proteins and rRNA together constitute the structural components of the ribosomes. Mutants that are defective in ribosome biogenesis, such as mutants for RP genes and genes involved in rRNA biogenesis, often show phenotypic similarities and a reduced auxin response. Auxin and ribosomes share a reciprocal relationship. Any mutation in the ribosomal subunit genes affects auxin biosynthesis due to defective ribosomes, which translationally repress the auxin responsive factors, such as HD-ZIPIII, KANADI and ARF3/ARF4, and thereby causes auxin related developmental defects.53–55 Although apum23 mutants show phenotypes and auxin distributions similar to those of RP gene mutants, further biochemical and molecular studies are needed to explain defective auxin homeostasis.
Possible Protein Interaction Partners and RNA Targets of APUM23
APUM23 may associate with a variety of nucleolar proteins involved in rRNA processing and ribosome biogenesis, like ScNOP9.29,30 To investigate the possible protein partners of APUM23, we used Arabidopsis Interaction Viewer (http://bar.utoronto.ca/interactions/cgi-bin/arabidopsis_interactions_viewer.cgi). The potential interacting partners were mainly those proteins that are involved in rRNA processing, ribosome biogenesis and nucleic acid binding. Because Puf proteins in other organisms are known to interact with multiple partners, it is possible that APUM23 may interact with a variety of nucleolar proteins transiently over time. It is also possible that APUM23 interacts with other nucleolar-localized Puf proteins, including APUM24, to mediate its action.
Different Puf proteins interact with different RNA substrates, suggesting that Puf proteins exert specificity towards their RNA targets despite recognizing the same UGUR sequence.23,34 Recently in Arabidopsis, Francischini and Quaggio39 showed that APUM1 to APUM6 are capable of recognizing the Drosophila hunchback Nanos Response Element (NRE) sequence.8 However, the specificity of the remaining 20 APUMs in Arabidopsis remains unexplored. As many Puf proteins in Arabidopsis appear to be plant-specific, their targets accordingly should have plant-specific functions.
Conclusions and Perspectives
Here, we introduced Arabidopsis Puf proteins by presenting examples of Puf proteins from other organisms and summarized our recent results on the role of APUM23 in ribosome biogenesis. Future work is necessary to elucidate the biological roles, spatial and temporal regulation, interacting partners and target RNAs of Puf proteins. Because Arabidopsis contains the highest number of Puf proteins, far more than in any species studied thus far, it is tempting to speculate that Puf proteins perform some major regulatory roles in plants and these roles may be even more complex than those seen in other organisms.
Acknowledgements
The referenced study on Arabidopsis and rice Puf proteins was supported by grants from the Crop Functional Genomics Center (CG2151) funded by the Ministry of Education, Science and Technology, and from Agenda Program (PJ006680), Rural Development Administration, Korea to S.B.C. N.A. was supported by the Brain Korea 21 (BK21) program funded by the Ministry of Education, Science and Technology, Korea.
References
- 1.Lorkovic ZJ, Barta A. Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucl Acids Res. 2002;30:623–635. doi: 10.1093/nar/30.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spassov DS, Jurecic R. The PUF family of RNAbinding proteins: does evolutionarily conserved structure equal conserved function? IUBMB Life. 2003;55:359–366. doi: 10.1080/15216540310001603093. [DOI] [PubMed] [Google Scholar]
- 3.Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- 4.Chritton JJ, Wickens M. Translational repression by PUF proteins in vitro. RNA. 2010;16:1217–1225. doi: 10.1261/rna.2070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- 6.Goldstrohm AC, Seay DJ, Hook BA, Wickens M. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J Biol Chem. 2007;282:109–114. doi: 10.1074/jbc.M609413200. [DOI] [PubMed] [Google Scholar]
- 7.Hook BA, Goldstrohm AC, Seay DJ, Wickens M. Two yeast PUF proteins negatively regulate a single mRNA. J Biol Chem. 2007;282:15430–15438. doi: 10.1074/jbc.M611253200. [DOI] [PubMed] [Google Scholar]
- 8.Murata Y, Wharton RP. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell. 1995;80:747–756. doi: 10.1016/0092-8674(95)90353-4. [DOI] [PubMed] [Google Scholar]
- 9.Sonoda J, Wharton RP. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 1999;13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng Y, Singer RH, Gu W. Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev. 2008;22:1037–1050. doi: 10.1101/gad.1611308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu W, Deng Y, Zenklusen D, Singer RH. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 2004;18:1452–1465. doi: 10.1101/gad.1189004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CU. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- 13.Lee I, Li Z, Marcotte EM. An improved, bias-reduced probabilistic functional gene network of baker's yeast, Saccharomyces cerevisiae. PLoS ONE. 2007;2:988. doi: 10.1371/journal.pone.0000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harnpicharnchai P, Jakovljevic J, Horsey E, Miles T, Roman J, Rout M, et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- 16.Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urbinati CR, Gonsalvez GB, Aris JP, Long RM. Loc1p is required for efficient assembly and nuclear export of the 60S ribosomal subunit. Mol Genet Genomics. 2006;276:369–377. doi: 10.1007/s00438-006-0151-7. [DOI] [PubMed] [Google Scholar]
- 18.Long H, Zeng XL, Hu B, Sun HJ, Liu ZL, Hao S. Subnucleolar distribution and transportation of U3 snoRNA in the nucleolus of Pisum sativum. Acta Botanica Sinica. 2003;45:317–321. [Google Scholar]
- 19.Paquin N, Chartrand P. Local regulation of mRNA translation: new insights from the bud. Trends Cell Biol. 2008;18:105–111. doi: 10.1016/j.tcb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 20.Forbes A, Lehmann R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 1998;125:679–690. doi: 10.1242/dev.125.4.679. [DOI] [PubMed] [Google Scholar]
- 21.Crittenden SL, Bernstein DS, Bachorik JL, Thompson BE, Gallegos M, Petcherski AG, et al. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002;417:660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- 22.Moore FL, Jaruzelska J, Fox MS, Urano J, Firpo MT, Turek PJ, et al. Human Pumilio-2 is expressed in embryonic stem cells and germ cells and interacts with DAZ (Deleted in Azoospermia) and DAZ-like proteins. Proc Natl Acad Sci USA. 2003;100:538–543. doi: 10.1073/pnas.0234478100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Rodriguez LJ, Gay AC, Pon LA. Puf3p, a Pumilio family RNA binding protein, localizes to mitochondria and regulates mitochondrial biogenesis and motility in budding yeast. J Cell Biol. 2007;176:197–207. doi: 10.1083/jcb.200606054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fehrenbacher KL, Boldogh IR, Pon LA. A role for Jsn1p in recruiting the Arp2/3 complex to mitochondria in budding yeast. Mol Biol Cell. 2005;16:5094–5102. doi: 10.1091/mbc.E05-06-0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menon KP, Sanyal S, Habara Y, Sanchez R, Wharton RP, Ramaswami M, et al. The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron. 2004;44:663–676. doi: 10.1016/j.neuron.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 27.Vessey JP, Schoderboeck L, Gingl E, Luzi E, Riefler J, Di Leva F, et al. Mammalian Pumilio 2 regulates dendrite morphogenesis and synaptic function. Proc Natl Acad Sci USA. 2010;107:3222–3227. doi: 10.1073/pnas.0907128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao Q, Padmanabhan K, Richter JD. Pumilio 2 controls translation by competing with eIF4E for 7-methyl guanosine cap recognition. RNA. 2010;16:221–227. doi: 10.1261/rna.1884610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Droll D, Archer S, Fenn K, Delhi P, Matthews K, Clayton C. The trypanosome Pumilio-domain protein PUF7 associates with a nuclear cyclophilin and is involved in ribosomal RNA maturation. FEBS Lett. 2010;584:1156–1162. doi: 10.1016/j.febslet.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomson E, Rappsilber J, Tollervey D. Nop9 is an RNA binding protein present in pre-40S ribosomes and required for 18S rRNA synthesis in yeast. RNA. 2007;13:2165–2174. doi: 10.1261/rna.747607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Zamore PD, Hall TM. Crystal structure of a Pumilio homology domain. Mol Cell. 2001;7:855–865. doi: 10.1016/s1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- 32.Miller MT, Higgin JJ, Hall TM. Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p. Nat Struct Mol Biol. 2008;15:397–402. doi: 10.1038/nsmb.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernstein D, Hook B, Hajarnavis A, Opperman L, Wickens M. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA. 2005;11:447–458. doi: 10.1261/rna.7255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cridge AG, Castelli LM, Smirnova JB, Selley JN, Rowe W, Hubbard SJ, et al. Identifying eIF4E-binding protein translationally-controlled transcripts reveals links to mRNAs bound by specific PUF proteins. Nucleic Acids Res. 2010;38:8039–8050. doi: 10.1093/nar/gkq686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Opperman L, Wickens M, Hall TM. Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein. Proc Natl Acad Sci USA. 2009;106:20186–20191. doi: 10.1073/pnas.0812076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piqué M, López JM, Foissac S, Guigó R, Méndez R. A combinatorial code for CPE-mediated translational control. Cell. 2008;132:434–448. doi: 10.1016/j.cell.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 37.Ulbricht RJ, Olivas WM. Puf1p acts in combination with other yeast Puf proteins to control mRNA stability. RNA. 2008;14:246–262. doi: 10.1261/rna.847408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wharton RP, Aggarwal AK. mRNA regulation by Puf domain proteins. Sci STKE. 2006;2006:37. doi: 10.1126/stke.3542006pe37. [DOI] [PubMed] [Google Scholar]
- 39.Francischini CW, Quaggio RB. Molecular characterization of Arabidopsis thaliana PUF proteins—binding specificity and target candidates. FEBS J. 2009;276:5456–5470. doi: 10.1111/j.1742-4658.2009.07230.x. [DOI] [PubMed] [Google Scholar]
- 40.Aggarwal P, Yadav RK, Reddy GV. Identification of novel markers for stem-cell niche of Arabidopsis shoot apex. Gene Expr Patterns. 2010;10:259–264. doi: 10.1016/j.gep.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 41.Borges F, Gomes G, Gardner R, Moreno N, McCormick S, Feijo JA, et al. Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol. 2008;148:1168–1181. doi: 10.1104/pp.108.125229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakai K, Horton P. PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 43.Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tam PP, Barrette-Ng IH, Simon DM, Tam MW, Ang AL, Muench DG. The Puf family of RNA-binding proteins in plants: phylogeny, structural modeling, activity and subcellular localization. BMC Plant Biol. 2010;10:44. doi: 10.1186/1471-2229-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abbasi N, Kim HB, Park Ni, Kim YK, Park YI, Choi SB. APUM23, a Nucleolar PUF domain RNA-binding protein, is required for pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 2011;64:960–976. doi: 10.1111/j.1365-313X.2010.04393.x. 65. [DOI] [PubMed] [Google Scholar]
- 46.Kojima H, Suzuki T, Kato T, Enomoto KI, Sato S, Kato T, et al. Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J. 2007;49:1053–1063. doi: 10.1111/j.1365-313X.2006.03016.x. [DOI] [PubMed] [Google Scholar]
- 47.Petricka JJ, Nelson TM. Arabidopsis nucleolin affects plant development and patterning. Plant Physiol. 2007;144:173–186. doi: 10.1104/pp.106.093575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pontvianne F, Matia I, Douet J, Tourmente S, Medina FJ, Echeverria M, et al. Characterization of AtNUC-L1 reveals a central role of nucleolin in nucleolus organization and silencing of AtNUC-L2 gene in Arabidopsis. Mol Biol Cell. 2007;18:369–379. doi: 10.1091/mbc.E06-08-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Degenhardt RF, Bonham-Smith PC. Arabidopsis ribosomal proteins RPL23aA and RPL23aB are differentially targeted to the nucleolus and are desperately required for normal development. Plant Physiol. 2008;147:128–142. doi: 10.1104/pp.107.111799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ito T, Kim GT, Shinozaki K. Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J. 2000;22:257–264. doi: 10.1046/j.1365-313x.2000.00728.x. [DOI] [PubMed] [Google Scholar]
- 51.van Lijsebettens M, Vanderhaeghen R, De Block M, Bauw G, Villarroel R, van Montagu M. An S18 ribosomal protein gene copy at the Arabidopsis PFL locus affects plant development by its specific expression in meristems. EMBO J. 1994;13:3378–3388. doi: 10.1002/j.1460-2075.1994.tb06640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weijers D, Franke-van Dijk M, Vencken RJ, Quint A, Hooykaas P, Offringa R. An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development. 2001;128:4289–4299. doi: 10.1242/dev.128.21.4289. [DOI] [PubMed] [Google Scholar]
- 53.Nishimura T, Wada T, Yamamoto KT, Okada K. The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell. 2005;17:2940–2953. doi: 10.1105/tpc.105.036533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pinon V, Etchells JP, Rossignol P, Collier SA, Arroyo JM, Martienssen RA, et al. Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development. 2008;135:1315–1324. doi: 10.1242/dev.016469. [DOI] [PubMed] [Google Scholar]
- 55.Yao Y, Ling Q, Wang H, Huang H. Ribosomal proteins promote leaf adaxial identity. Development. 2008;135:1325–1334. doi: 10.1242/dev.017913. [DOI] [PubMed] [Google Scholar]
- 56.Nishimura N, Kitahata N, Seki M, Narusaka Y, Narusaka M, Kuromori T, et al. Analysis of ABA hypersensitive germination2 revealed the pivotal functions of PARN in stress response in Arabidopsis. Plant J. 2005;44:972–984. doi: 10.1111/j.1365-313X.2005.02589.x. [DOI] [PubMed] [Google Scholar]
- 57.Rosado A, Sohn EJ, Drakakaki G, Pan S, Swidergal A, Xiong Y, et al. Auxin-mediated ribosomal biogenesis regulates vacuolar trafficking in Arabidopsis. Plant Cell. 2010;22:143–158. doi: 10.1105/tpc.109.068320. [DOI] [PMC free article] [PubMed] [Google Scholar]


