Abstract
Legumes enter nodule symbioses with nitrogen-fixing bacteria (rhizobia), whereas most flowering plants establish symbiotic associations with arbuscular mycorrhizal (AM) fungi. Once first steps of symbiosis are initiated, nodule formation and mycorrhization in legumes is negatively controlled by a shoot-derived inhibitor (SDI), a phenomenon termed autoregulation. According to current views, autoregulation of nodulation and mycorrhization in legumes is regulated in a similar way. CLE peptides induced in response to rhizobial nodulation signals (Nod factors) have been proposed to represent the ascending long-distance signals to the shoot. Although not proven yet, these CLE peptides are likely perceived by leucine-rich repeat (LRR) autoregulation receptor kinases in the shoot. Autoregulation of mycorrhization in non-legumes is reminiscent to the phenomenon of “systemic acquired resistance” in plant-pathogen interactions.
Key words: arbuscular mycorrhiza, autoregulation, CLE peptides, mutant, nodulation, split-root system
Under natural conditions, growth of plants is often limited by the availability of nutrients such as nitrogen and phosphorous. Plants have therefore developed strategies to acquire nutrients with the help of soil microorganisms. Most land plants enter mutualistic root symbioses with arbuscular mycorrhizal (AM) fungi, whereas legumes form special root nodules containing nitrogen-fixing bacteria, so-called rhizobia.1–4 Establishment and maintenance of symbiosis requires plant resources, such as photosynthetically assimilated carbon. To minimize these costs, host plants are able to control the degree of their symbiotic interactions. Above a critical threshold level further establishment of symbiosis is restricted—a feedback phenomenon termed autoregulation of symbiosis. Autoregulation can be experimentally demonstrated in split-root systems. When legume roots are already infected by rhizobia on one side of a split-root, further nodule development is “systemically” inhibited on the other side. Similarly, prior colonization of split-roots by AM fungi on one half suppresses later fungal root colonization on the other half. Hence, important elements of the symbiotic autoregulation circuit are not only localized in roots, but also in aerial parts of the plant, implicating transport of signals in vascular bundles (Fig. 1). Whereas autoregulation of nodulation in legumes has been studied for many decades,5–9 the first publications clearly stating a shoot-controlled autoregulation of mycorrhization in split-root systems appeared in 2000 for the non-legume barley (Hordeum vulgare) and thereafter for alfalfa (Medicago sativa) and soybean (Glycine max).10–13 The data from these split-root experiments are supported by the findings that supernodulating (or hypernodulating) loss-of-autoregulation mutants displayed either an increased degree of AM colonization and/or a higher abundance of arbuscules.14–16
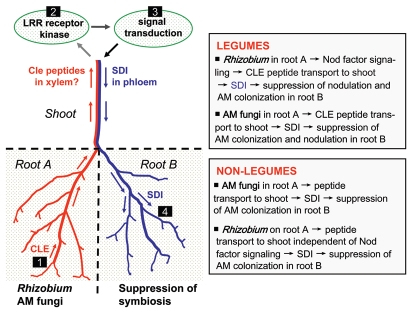
Figure 1.
Proposed model of shoot-controlled autoregulation of symbiosis in a split-root system. Prior infection of root A by rhizobia or AM fungi systemically suppresses later establishment of symbiosis in root B. Expression of specific CLE peptides (and/or other peptide hormones) is induced in response to rhizobial nodulation signals (Nod factors) and perhaps also in response to colonization by AM fungi (stage 1). The CLE peptides (and/or other signals) are then presumed to be transported in the xylem to the shoot, where they are perceived by leucine-rich repeat (LRR) autoregulation receptor kinases (stage 2). As a result of autoregulation signaling in the shoot, an unknown shoot-derived inhibitor (SDI) is produced (stage 3) and transported as a phloem-mobile signal to the root. Perception and action of the SDI signal in roots would then inhibit nodulation and root colonization by AM fungi (stage 4).
Shoot-controlled Autoregulation of Nodulation
Loss-of-autoregulation mutants of various legumes have been identified by their capacity to form significantly more nodules.5–9 Reciprocal grafting experiments with mutant and wild-type plants revealed that the supernodulating (or hypernodulating) phenotype of many mutants depended on the aerial part of the legume plants, providing genetic evidence that autoregulation is controlled by long-distance transport. Following inoculation with rhizobia, an ascending signal is transported from roots to the shoot in the xylem and a shoot-derived descending inhibitor is transported back to the roots in the phloem (Fig. 1).
Recent findings on autoregulation of nodulation suggest that the root-derived ascending signals to the shoot are short peptides belonging to the CLE peptide family.17–19 CLE peptides have a conserved C-terminal domain (CLE domain) and are named after representative genes, CLV3 (or CLAVATA3) of Arabidopsis thaliana and ESR (endosperm-surrounding region) of Zea mays. Extracellular CLE peptides of Arabidopsis and other non-legumes possess hormone-like activities and are involved in regulation of various processes of plant development such as cell differentiation in apical shoot, floral and root meristems, as well as differentiation into vascular cells.20–23 In legumes, expression of specific CLE genes in roots is induced in response to rhizobial infection. First examples are the genes LjCLE-RS1/LjCLE-RS2 in Lotus japonicus and MtCLE12/MtCLE13 in Medicago truncatula.18,19 L. japonicus and M. truncatula mutants deficient in perception of rhizobial Nod factors (lipo-chitooligosaccharidic nodulation signals) did not show induction these genes, providing evidence that Nod factor signaling is required for expression of nodulation-related CLE genes.18,19 These findings are consistent with earlier observations that Nod factors not only initiate infection and cell division processes, but also activate the autoregulation circuit. When split-root systems of Vicia sativa subsp. nigra or alfalfa were treated on one half with active Nod factors, nodulation was significantly reduced on the other half.12,24 Similarly, grafting experiments with various Nod factor signaling mutants of pea (Pisum sativum) indicated that a functional Nod factor signal transduction cascade is required for formation (or activation) of the ascending autoregulation signal to the shoot. In the supernodulating nod3 mutant of pea, autoregulation is likely blocked at an early stage in the root, perhaps during synthesis or activation of CLE peptides.25,26 To examine the effect of symbiosis-related CLE peptides on nodule formation, transgenic L. japonicus and M. truncatula were constructed in which the Nod factor-inducible CLE genes (LjCLE-RS1/LjCLE-RS2; MtCLE12/MtCLE13) were constitutively expressed in hairy roots after transformation with Agrobacterium rhizogenes. In both legumes, overexpression of these CLE peptides under the control of the CaMV 35S promoter suppressed nodule formation in non-transgenic parts of the root system.18,19 These data provided first evidence that CLE peptides suppress nodule formation via long-distance signaling.
High nitrate levels in the soil block formation of legume root nodules. Molecular mechanisms underlying the relationship between nitrate and nodulation are poorly understood, however. Classical work on various loss-of-autoregulation mutants revealed that they not only form more nodules than wild-type plants, but also gained the ability to nodulate in the presence of high nitrate concentrations.5–9 Thus, nitrate-controlled inhibition of nodulation shares common steps with the autoregulation circuit. Interestingly, expression of LjCLE-RS2 was strongly upregulated in L. japonicus roots treated with 10 mM KNO3,18 suggesting that certain CLE peptides are not only involved in autoregulation of nodulation, but also locally inhibit nodulation when roots are exposed to high nitrate concentrations.
Future biochemical experiments are required to demonstrate that CLE peptides are indeed the ascending long-distance signals in the xylem. Alternatively, other peptides could have similar functions in systemic suppression of nodulation. In M. truncatula roots for example, rhizobial infection resulted in strongly induced expression of the genes MtRALFL1 and MtDVL1,27 which encode small peptides belonging to the RALF (“rapid alkalinization factor”) and DEVIL (ROT4) peptide family, respectively.28 When inoculated with rhizobia, transgenic M. truncatula overexpressing MtRALFL1 and MtDVL1 formed fewer nodules and exhibited an increase in the number of aborted infection threads as compared to non-transformed plants.27 Whether these peptides act locally or systemically has not been examined yet.
A first essential receptor kinase gene involved in autoregulation of nodulation has been identified by map-based cloning of supernodulating/hypernodulating mutants. The genes HAR1 in L. japonicus,29,30 SYM29 in pea,29 NARK in soybean,31 and SUNN in M. truncatula32,33 are strongly expressed in leaf tissues and encode orthologous leucine-rich repeat (LRR) receptor kinases (class XI) with sequence similarities to the CLV1 receptor kinase (CLAVATA1) of Arabidopsis. It has been shown that the CLV1 receptor biochemically interacts with the ligand CLV3, a well-investigated CLE peptide of Arabidopis.34 Although not proven yet, it is likely that root-derived CLE peptides represent the peptide ligands for the autoregulation LRR receptor kinase in legumes. Indeed, overexpression of the CLE genes LjCLE-RS1 or LjCLE-RS2 in L. japonicus showed systemic suppression effects on nodulation in wild-type plants, but not in the har1 mutant.18 Similar results were obtained in M. truncatula overexpressing MtCLE12 or MtCLE13, whereas suppressive effects on nodulation induced by MtCLE13 expression were significantly weaker in the sunn-1 mutant.19
The kinase-associated protein phosphatase genes GmKAPP1 and GmKAPP2 of soybean35 and the gene at the KLAVIER locus in L. japonicus36 encode likely additional elements of the autoregulation signal transduction pathway in the shoot. As a consequence, a shoot-derived inhibitor (SDI) is generated and transported in the phloem to the root, where it inhibits nodule formation (Fig. 1). The descending SDI signal from soybean plants has been biochemically characterized as ethanol-soluble and heat-stable molecule with a low molecular weight (<1 kDa).37–40 Grafting experiments suggested that the hypernodulating L. japonicus mutant too much love (tml) is involved in perception of the descending SDI signal.41 Accordingly, recent expression analysis with the tml mutant showed normal induction of the CLE genes LjCLE-RS1 and LjCLE-RS2 in response to rhizobial infection.42
Autoregulation signaling affects levels and action of phytohormones, such as auxin, abscisic acid, ethylene and jasmonic acid (JA).32,43–48 In M. truncatula, an autoregulation-defective sunn mutant (mutated in the autoregulation LRR receptor kinase gene SUNN) showed increased auxin transport from the shoot to the root, suggesting that high auxin levels positively affect nodule formation.45 Following inoculation with rhizobia, the auxin-responsive gene GH3 was higher expressed in roots of the sunn mutant than in wild-type plants.32 Taken together this data suggests that auxin flow is systemically regulated by autoregulation mechanisms and that the SDI signal and auxin perhaps function antagonistically in root tissues. Furthermore, recent reports point towards a possible involvement of JA signaling in autoregulation of noduation. In soybean plants inoculated with rhizobia, genes involved in JA biosynthesis as well as JA-inducible genes were systemically regulated. This effect was abolished in nts mutants (mutated in the autoregulation LRR receptor kinase gene NARK). Furthermore, levels of JA in leaves were higher and JA-inducible genes upregulated in a non-inoculated nts mutant as compared to wild-type plants. These data suggest that NARK-mediated signaling reduces JA signaling in soybean leaves.46,48 In L. japonicus, foliar application of methyl-JA reduced nodulation.44 An inhibition effect of JA on nodulation was also seen for the tml mutant, suggesting that JA itself is not the descending SDI signal.42 Furthermore, levels of certain symbiosis-related flavonoids seem to be influenced by autoregulation signaling. When alfalfa split-root systems were infected with rhizobia, levels of formononetin and ononin were not only reduced in inoculated roots, but also systemically lowered in non-inoculated roots. Exogenous application of formononetin, and to lesser extent ononin, significantly increased nodule initiation in autoregulated roots, suggesting that these flavonoids act antagonistically to SDI-induced changes.49
Analysis of mutants revealed a close relationship beween autoregulation of nodulation and plant developmental processes. For example, most hypernodulating L. japonicus mutants show morphological changes as compared to wildtype plants. Mutations in the autoregulation LRR receptor kinase gene HAR1 resulted in plants with strongly altered root architecture,50 whereas the hypernodulating klavier mutant showed altered leaf veins, delayed flowering and a dwarf phenotype.36 Furthermore, the hypernodulating astray mutant, which is mutated in a gene encoding a basic leucine zipper protein, possesses agravitropic lateral roots.51,52 These pleiotropic effects in loss-of-autoregulation mutants indicate that autoregulation and plant morphogenesis share common mechanisms, in which CLE peptides and phytohormones possess dual functions.
Shoot-Controlled Autoregulation of Mycorrhization in Legumes and Non-Legumes
Although more information is available on autoregulation of nodulation, there is accumulating evidence that autoregulation of mycorrhization in legumes functions in a similar way. Supernodulating mutants of soybean, Lotus japonicus, M. truncatula and pea displayed increased mycorrhizal colonization, accompanied by a higher abundance of arbuscules as compared to wild-type mycorrhization.14–16 Furthermore, a mutant of M. truncatula with increased mycorrhization and low nodulation potential has been recently identified.53 These findings suggest that mycorrhization is autoregulated in legumes. Clear evidence for a shoot-controlled autoregulation circuit has been provided with mycorrhizal split-root systems. Split-roots inoculated at two different times have been found to be most suitable for analysis of systemic control of mycorrhization, as interaction between the two root sides implicates long-distance transport of signals via aerial parts of the host plant. Using such a split-root system approach, autoregulation of mycorrhization has been first identified in the non-legume barley and thereafter studied in alfalfa and soybean.10–13,49,54–56
In alfalfa, inoculation of one half of a split-root system with the fungus Glomus mosseae significantly reduced later AM colonization on the other half. A similar suppressive effect on mycorrhization was observed after inoculation with Sinorhizobium meliloti.12 Furthermore, prior addition of purified rhizobial Nod factors on one half significantly reduced mycorrhization on the other half of the split-root system. Reciprocally, prior mycorrhization on one side suppressed nodule formation on the other side of the split-root system.12 Taken these data together, they point to a common autoregulation circuit for both symbioses (Fig. 1). We suggest that Nod factor signaling as well as mycorrhizal signaling in response to unknown mycorrhizal signals (“Myc factors”) induces expression or post-translation processing of CLE peptides, which likely function as ascending long-distance signals to the shoot. Moreover, the descending SDI transported to the root seems to inhibit both, nodule formation as well as mycorrhizal root colonization. Indeed, inoculation experiments with the supernodulation mutant nts1007 provided genetic evidence for autoregulation of mycorrhization in soybean (cv. Bragg). The nts1007 mutant carries a nonsense mutation that truncates the autoregulation LRR receptor kinase NARK.31 In contrast to wild-type soybean plants, prior AM colonization of nts1007 plants on one half did not suppress later AM colonization by the AM fungus G. mosseae on the other half of the split-root system, indicating that NARK is essential for shoot-controlled autoregulation of mycorrhization.13 En6500, an allelic mutant with a similar nonsense mutation in NARK derived from cv. Enrei, retained the ability to systemically regulate AM fungi under these conditions, suggesting that the genetic background (varietal differences) influences autoregulation of mycorrhization under certain experimental conditions.56 Using reciprocal grafting experiments however, shoot-controlled autoregulation of mycorrhization has been recently demonstrated for the En6500 mutant. In this study, abundance of arbuscules was determined in roots colonized by the AM fungus Gigaspora rosea.57
Similar to autoregulation of nodulation, the SDI signal seems to induce physiological changes in the root that limit AM root colonization. Analysis of phytohormonal changes in soybean points to a possible role of auxin in autoregulation of mycorrhization.13 Levels of isoflavonoids such as formononetin and ononin were systemically reduced in non-infected parts, when alfalfa split-roots were infected with AM fungi on one half. On the other hand, exogenous application of ononin to autoregulated root parts stimulated AM colonization, indicating that certain flavonoids thwart SDI-induced inhibition effects on mycorrhization.49 Proteomic analysis of the mycorrhized autoregulation-defective sunn mutant (mutated in the autoregulation LRR receptor kinase gene SUNN of M. truncatula) revealed increased accumulation of proteins involved in plant defense reactions, cytoskeleton rearrangements and auxin signaling, which perhaps reflect the higher number of formed arbuscules in this mutant as compared to wild-type plants.58
Data from classical split-root experiments indicate that autoregulation of mycorrhization also exists in non-legume plants. In barley, mycorrhization was significantly reduced when other parts of the root system were already colonized by AM fungi.10,11 The feedback inhibition effect depended on the degree of AM colonization in the first half of the split-root system, suggesting a dose-dependent effect.54 We suggest that autoregulation processes of mycorrhization in legumes and non-legumes are controlled by similar molecules (Fig. 1). It would be interesting to find out whether CLE peptides17–23 or other plant bio-active peptides28,59 represent the ascending signals in non-legumes.
Using split-root systems, a recent study demonstrated that the broad-host range strain Rhizobium sp. NGR234 systemically suppressed mycorrhization of barley roots.60 Rhizobia cannot infect barley, but bacterial rhizophere colonization in one half of the split-root system systemically affected AM root colonization on the other half. The observed suppression effect was independent of Nod factors, as a mutant strain of NGR234 deficient in Nod factor synthesis (strain NGRΔnodABC) suppressed mycorrhization in a similar way. These data suggest that barley roots can perceive the rhizobia in the rhizophere. We postulate that there are rhizobial elicitors (microbe-associated molecular patterns), which are recognized by corresponding pattern recognition receptors and trigger activation of plant defense reactions in barley roots. Indeed, roots challenged with NGR234 showed increased levels of free salicylic acid, a typical defense response against pathogens.60
Work on cucumber (Cucumis sativus) revealed that application of root exudates from mycorrhizal plants reduced the degree of AM root colonization, whereas root exudates from non-infected plants stimulated mycorrhization. Root exudates from the nonmycorrhizal half of a split-root system (with mycorrhizal roots on the other half) partially inhibited mycorrhization of cucumber plants.61 These data indicate that the composition of root exudates is systemically regulated and suggest a systemic plant defense response against AM fungi. This is reminiscent to the effects of the “endogenous elicitor” systemin, a peptide hormone of tomato (Lycopersicon peruvianum) involved in systemic activation of plant defense reactions.62
Interestingly, prior mycorrhization in barley split-root systems not only suppressed later mycorrhization, but also systemically reduced subsequent infection of the pathogenic fungus Gaeumannomyces graminis.63 Hence, autoregulation of mycorrhization and the systemic biocontrol effect of AM fungi (or rhizobia) on pathogenic fungi could be regulated by a similar SDI signal.64 Apparently, autoregulation of mycorrhization possesses certain parallels with the phenomenon of “systemic acquired resistance” in plant-pathogen interactions, where prior infection by a pathogen systemically induces plant defense reactions in the host plant.65 It is worth mentioning in this context that autoregulation signaling affected root-knot nematode infection in L. japonicus roots. The har1 mutant (mutated in the autoregulation LRR receptor kinase gene HAR1) was hyperinfected by Meloidogyne incognita and formed significantly more galls than wild-type plants.66 Similarly, as compared to the parent cv. Enrei, the supernodulating soybean line Sakukei4 was more damaged by red crown rot, which is caused by Calonectria ilicilola.67 These differences could be due to reduced expression of disease resistance genes in loss-of-autoregulation mutants and point to a possible crosstalk between autoregulation and defense signaling pathways.46
Concluding Remarks
Whereas knowledge on autoregulation of nodulation considerably increased during the recent years, studies on autoregulation of mycorrhization are still in their infancy. In legumes, autoregulation of nodulation and mycorrhization seem to be regulated by the same signaling pathway in the shoot. Autoregulation of mycorrhization in non-legumes is reminiscent to “systemic acquired resistance” in plant-pathogen interactions. CLE peptides are putative ligands for the autoregulation LRR receptor kinases HAR1/SYM29/NARK/SUNN. It will be of interest to investigate these receptor-ligand interactions biochemically in order to characterize their specificity. Further research on loss-ofautoregulation mutants in legumes and identification of mutants from non-legumes will provide a way to understand autoregulation signaling in more detail.
The overlaps between autoregulation of symbiosis and plant developmental processes might complicate the molecular analysis of symbiotic autoregulation circuits. Redundancy in expression and multiple functions of CLE peptides likely contribute to a high level of complexity. CLE peptides are processed from longer polypeptide chains and certain CLE peptides undergo post-translational modifications, such as arbinosylation to gain their biological activity.20–23,68 Furthermore, CLE peptides seem to affect nodule inhibition not only via shoot-controlled autoregulation, but also directly within roots. Thus, short-distance transport of CLE peptides within roots could interfere with shoot-controlled autoregulation of symbiosis. We hypothesize that expression and short-distance transport of CLE peptides is particularly important for nitrate-mediated inhibition of nodulation as well as for phosphate-mediated inhibition of mycorrhization. Indeed, recent expression studies indicated that LjCLE-RS2 in L. japonicus roots is induced by KNO3, whereas expression levels of LjCLE19/LjCLE20 were stimulated when plants were grown at high phosphate concentrations.18,69 It is tempting to speculate that autoregulation LRR receptor kinases expressed in roots are receptors for CLE peptides induced by high nitrate or phosphate levels. Consequently, an inhibitor identical or related to the SDI signal would be also locally synthesized in roots. In other words, mechanisms of nutrient-mediated inhibition of symbiosis and autoregulation of symbiosis controlled by long-distance signaling would represent variations of a common theme.
Acknowledgements
This work was funded by the National Basic Research Program of China (973 program, No. 2010CB126501; grant to C.S. and Z.P. Xie) and by the Comisión Interministerial de Ciencia y Teconlogía (CICYT) and Fondos Europeos de Desarrollo Regional (FEDER) through the Ministerio de Ciencia e Innovación, Spain (AGL2008-00742).
Abbreviations
- AM
arbuscular mycorrhizal
- JA
jasmonic acid
- LRR
leucine-rich repeat
- SDI
shoot-derived inhibitor
References
- 1.Perret X, Staehelin C, Broughton WJ. Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Rev. 2000;64:180–201. doi: 10.1128/mmbr.64.1.180-201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibson KE, Kobayashi H, Walker GC. Molecular determinants of a symbiotic chronic infection. Annu Rev Genet. 2008;42:413–441. doi: 10.1146/annurev.genet.42.110807.091427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reinhardt D. Programming good relations—development of the arbuscular mycorrhizal symbiosis. Curr Opin Plant Biol. 2007;10:98–105. doi: 10.1016/j.pbi.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Parniske M. Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol. 2008;6:763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- 5.Caetano-Anolés G, Gresshoff PM. Plant genetic control of nodulation. Annu Rev Microbiol. 1991;45:345–382. doi: 10.1146/annurev.mi.45.100191.002021. [DOI] [PubMed] [Google Scholar]
- 6.Kinkema M, Scott PT, Gresshoff PM. Legume nodulation: successful symbiosis through short- and long-distance signalling. Funct Plant Biol. 2006;33:707–721. doi: 10.1071/FP06056. [DOI] [PubMed] [Google Scholar]
- 7.Oka-Kira E, Kawaguchi M. Long-distance signaling to control root nodule number. Curr Opin Plant Biol. 2006;9:496–502. doi: 10.1016/j.pbi.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 8.Magori S, Kawaguchi M. Long-distance control of nodulation: molecules and models. Mol Cells. 2009;27:129–134. doi: 10.1007/s10059-009-0016-0. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson BJ, Indrasumunar A, Hayashi S, Lin MH, Lin YH, Reid DE, et al. Molecular analysis of legume nodule development and autoregulation. J Integr Plant Biol. 2010;52:61–76. doi: 10.1111/j.1744-7909.2010.00899.x. [DOI] [PubMed] [Google Scholar]
- 10.Vierheilig H, Garcia-Garrido JM, Wyss U, Piché Y. Systemic suppression of mycorrhizal colonization of barley roots already colonized by AM fungi. Soil Biol Biochem. 2000;32:589–595. [Google Scholar]
- 11.Vierheilig H, Maier W, Wyss U, Samson J, Strack D, Piché Y. Cyclohexenone derivative- and phosphate-levels in split-root systems and their role in the systemic suppression of mycorrhization in precolonized barley plants. J Plant Physiol. 2000;157:593–599. [Google Scholar]
- 12.Catford JG, Staehelin C, Lerat S, Piché Y, Vierheilig H. Suppression of arbuscular mycorrhizal colonization and nodulation in split-root systems of alfalfa after pre-inoculation and treatment with Nod factors. J Exp Bot. 2003;54:1481–1487. doi: 10.1093/jxb/erg156. [DOI] [PubMed] [Google Scholar]
- 13.Meixner C, Ludwig-Müller J, Miersch O, Gresshoff P, Staehelin C, Vierheilig H. Lack of mycorrhizal autoregulation and phytohormonal changes in the supernodulating soybean mutant nts1007. Planta. 2005;222:709–715. doi: 10.1007/s00425-005-0003-4. [DOI] [PubMed] [Google Scholar]
- 14.Shrihari PC, Sakamoto K, Inubushi K, Akao S. Interaction between supernodulating or non-nodulating mutants of soybean and two arbuscular mycorrhizal fungi. Mycorrhiza. 2000;10:101–106. [Google Scholar]
- 15.Morandi D, Sagan M, Prado-Vivant E, Duc G. Influence of genes determining supernodulation on root colonization by the mycorrhizal fungus Glomus mosseae in Pisum sativum and Medicago truncatula mutants. Mycorrhiza. 2000;10:37–42. [Google Scholar]
- 16.Solaiman MZ, Senoo K, Kawaguchi M, lmaizumi-Anraku H, Akao S, Tanaka A, et al. Characterization of mycorrhizas formed by Glomus sp. on roots of hypernodulating mutants of Lotus japonicus. J Plant Res. 2000;113:443–448. [Google Scholar]
- 17.Oelkers K, Goffard N, Weiller GF, Gresshoff PM, Mathesius U, Frickey T. Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biol. 2008;8:1–15. doi: 10.1186/1471-2229-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okamoto S, Ohnishi E, Sato S, Takahashi H, Nakazono M, Tabata S, et al. Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol. 2009;50:67–77. doi: 10.1093/pcp/pcn194. [DOI] [PubMed] [Google Scholar]
- 19.Mortier V, Den Herder G, Whitford R, Van de Velde W, Rombauts S, D'haeseleer K, et al. CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol. 2010;153:222–237. doi: 10.1104/pp.110.153718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jun JH, Fiume E, Fletcher JC. The CLE family of plant polypeptide signaling molecules. Cell Mol Life Sci. 2008;65:743–755. doi: 10.1007/s00018-007-7411-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchum MG, Wang XH, Davis EL. Diverse and conserved roles of CLE peptides. Curr Opin Plant Biol. 2008;11:75–81. doi: 10.1016/j.pbi.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 22.Miwa H, Kinoshita A, Fukuda H, Sawa S. Plant meristems: CLAVATA3/ESR-related signaling in the shoot apical meristem and the root apical meristem. J Plant Res. 2009;122:31–39. doi: 10.1007/s10265-008-0207-3. [DOI] [PubMed] [Google Scholar]
- 23.Wang G, Fiers M. CLE peptide signaling during plant development. Protoplasma. 2010;240:33–43. doi: 10.1007/s00709-009-0095-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Brussel AAN, Tak T, Boot KJM, Kijne JW. Autoregulation of root nodule formation: signals of both symbiotic partners studied in a split-root system of Vicia sativa subsp. nigra. Mol Plant Microbe Interact. 2002;15:341–349. doi: 10.1094/MPMI.2002.15.4.341. [DOI] [PubMed] [Google Scholar]
- 25.Li DX, Kinkema M, Gresshoff PM. Autoregulation of nodulation (AON) in Pisum sativum (pea) involves signalling events associated with both nodule primordia development and nitrogen fixation. J Plant Physiol. 2009;166:955–967. doi: 10.1016/j.jplph.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Novák K. Early action of pea symbiotic gene NOD3 is confirmed by adventitious root phenotype. Plant Sci. 2010;179:472–478. doi: 10.1016/j.plantsci.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Combier JP, Küster H, Journet EP, Hohnjec N, Gamas P, Niebel A. Evidence for the involvement in nodulation of the two small putative regulatory peptide-encoding genes MtRALFL1 and MtDVL1. Mol Plant Microbe Interact. 2008;21:1118–1127. doi: 10.1094/MPMI-21-8-1118. [DOI] [PubMed] [Google Scholar]
- 28.Butenko MA, Vie AK, Brembu T, Aalen RB, Bones AM. Plant peptides in signalling: looking for new partners. Trends Plant Sci. 2009;14:255–263. doi: 10.1016/j.tplants.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 29.Krusell L, Madsen LH, Sato S, Aubert G, Genua A, Szczyglowski K, et al. Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature. 2002;420:422–426. doi: 10.1038/nature01207. [DOI] [PubMed] [Google Scholar]
- 30.Nishimura R, Hayashi M, Wu GJ, Kouchi H, Imaizumi-Anraku H, Murakami Y, et al. HAR1 mediates systemic regulation of symbiotic organ development. Nature. 2002;420:426–429. doi: 10.1038/nature01231. [DOI] [PubMed] [Google Scholar]
- 31.Searle IR, Men AE, Laniya TS, Buzas DM, Iturbe-Ormaetxe I, Carroll BJ, et al. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science. 2003;299:109–112. doi: 10.1126/science.1077937. [DOI] [PubMed] [Google Scholar]
- 32.Penmetsa RV, Frugoli JA, Smith LS, Long SR, Cook DR. Dual genetic pathways controlling nodule number in Medicago truncatula. Plant Physiol. 2003;131:998–1008. doi: 10.1104/pp.015677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schnabel E, Journet EP, Carvalho-Niebel F, Duc G, Frugoli J. The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol Biol. 2005;58:809–822. doi: 10.1007/s11103-005-8102-y. [DOI] [PubMed] [Google Scholar]
- 34.Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science. 2008;319:294. doi: 10.1126/science.1150083. [DOI] [PubMed] [Google Scholar]
- 35.Miyahara A, Hirani TA, Oakes M, Kereszt A, Kobe B, Djordjevic MA, et al. Soybean nodule autoregulation receptor kinase phosphorylates two kinase-associated protein phosphatases in vitro. J Biol Chem. 2008;283:25381–25391. doi: 10.1074/jbc.M800400200. [DOI] [PubMed] [Google Scholar]
- 36.Oka-Kira E, Tateno K, Miura Ki, Haga T, Hayashi M, Harada K, et al. klavier (klv), a novel hypernodulation mutant of Lotus japonicus affected in vascular tissue organization and floral induction. Plant J. 2005;44:505–515. doi: 10.1111/j.1365-313X.2005.02543.x. [DOI] [PubMed] [Google Scholar]
- 37.Lin YH, Ferguson BJ, Kereszt A, Gresshoff PM. Suppression of hypernodulation in soybean by a leaf-extracted, NARK- and Nod factor-dependent, low molecular mass fraction. New Phytol. 2010;185:1074–1086. doi: 10.1111/j.1469-8137.2009.03163.x. [DOI] [PubMed] [Google Scholar]
- 38.Yamaya H, Arima Y. Evidence that a shoot-derived substance is involved in regulation of the super-nodulation trait in soybean. Soil Sci Plant Nutr. 2010;56:115–122. [Google Scholar]
- 39.Yamaya H, Arima Y. Shoot-synthesized nodulation-restricting substances are present in the medium-polarity fraction of shoot extracts from wild-type soybean plants. Soil Sci Plant Nutr. 2010;56:418–421. [Google Scholar]
- 40.Kenjo T, Yamaya H, Arima Y. Shoot-synthesized nodulation-restricting substances of wild-type soybean present in two different high-performance liquid chromatography peaks of the ethanol-soluble medium-polarity fraction. Soil Sci Plant Nutr. 2010;56:399–406. [Google Scholar]
- 41.Magori S, Oka-Kira E, Shibata S, Umehara Y, Kouchi H, Hase Y, et al. TOO MUCH LOVE, a root regulator associated with the long-distance control of nodulation in Lotus japonicus. Mol Plant Microbe Interact. 2009;22:259–268. doi: 10.1094/MPMI-22-3-0259. [DOI] [PubMed] [Google Scholar]
- 42.Magori S, Kawaguch M. Analysis of two potential long-distance signaling molecules, LjCLE-RS1/2 and jasmonic acid, in a hypernodulating mutant too much love. Plant Signal Behav. 2010;5:403–405. doi: 10.4161/psb.5.4.10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caba JM, Luz Centeno M, Fernández B, Gresshoff PM, Ligero F. Inoculation and nitrate alter phytohormone levels in soybean roots: differences between a supernodulating mutant and the wild type. Planta. 2000;211:98–104. doi: 10.1007/s004250000265. [DOI] [PubMed] [Google Scholar]
- 44.Nakagawa T, Kawaguchi M. Shoot-applied MeJA suppresses root nodulation in Lotus japonicus. Plant Cell Physiol. 2006;47:176–180. doi: 10.1093/pcp/pci222. [DOI] [PubMed] [Google Scholar]
- 45.van Noorden GE, Ross JJ, Reid JB, Rolfe BG, Mathesius U. Defective long-distance auxin transport regulation in the Medicago truncatula super numeric nodules mutant. Plant Physiol. 2006;140:1494–1506. doi: 10.1104/pp.105.075879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kinkema M, Gresshoff PM. Investigation of downstream signals of the soybean autoregulation of nodulation receptor kinase GmNARK. Mol Plant Microbe Interact. 2008;21:1337–1348. doi: 10.1094/MPMI-21-10-1337. [DOI] [PubMed] [Google Scholar]
- 47.Gresshoff PM, Lohar D, Chan PK, Biswas B, Jiang Q, Reid D, et al. Genetic analysis of ethylene regulation of legume nodulation. Plant Signal Behav. 2009;4:818–823. doi: 10.4161/psb.4.9.9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seo HS, Li J, Lee SY, Yu JW, Kim KH, Lee SH, et al. The hypernodulating nts mutation induces jasmonate synthetic pathway in soybean leaves. Mol Cells. 2007;24:185–193. [PubMed] [Google Scholar]
- 49.Catford J, Staehelin C, Larose G, Piché Y, Vierheilig H. Systemically suppressed isoflavonoids and their stimulating effects on nodulation and mycorrhization in alfalfa split-root systems. Plant Soil. 2006;285:257–266. [Google Scholar]
- 50.Wopereis J, Pajuelo E, Dazzo FB, Jiang Q, Gresshoff PM, De Bruijn FJ, et al. Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. Plant J. 2000;23:97–114. doi: 10.1046/j.1365-313x.2000.00799.x. [DOI] [PubMed] [Google Scholar]
- 51.Nishimura R, Ohmori M, Kawaguchi M. The novel symbiotic phenotype of enhanced-nodulating mutant of Lotus japonicus: astray mutant is an early nodulating mutant with wider nodulation zone. Plant Cell Physiol. 2002;43:853–859. doi: 10.1093/pcp/pcf098. [DOI] [PubMed] [Google Scholar]
- 52.Nishimura R, Ohmori M, Fujita H, Kawaguchi M. A Lotus basic leucine zipper protein with a ring-finger motif negatively regulates the developmental program of nodulation. Proc Natl Acad Sci USA. 2002;99:15206–15210. doi: 10.1073/pnas.222302699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morandi D, le Signor C, Gianinazzi-Pearson V, Duc G. A Medicago truncatula mutant hyper-responsive to mycorrhiza and defective for nodulation. Mycorrhiza. 2009;19:435–441. doi: 10.1007/s00572-009-0242-5. [DOI] [PubMed] [Google Scholar]
- 54.Vierheilig H. Further root colonization by arbuscular mycorrhizal fungi in already mycorrhizal plants is suppressed after a critical level of root colonization. J Plant Physiol. 2004;161:339–341. doi: 10.1078/0176-1617-01097. [DOI] [PubMed] [Google Scholar]
- 55.Vierheilig H. Regulatory mechanisms during the plant-arbuscular mycorrhizal fungus interaction. Can J Bot. 2004;82:1166–1176. [Google Scholar]
- 56.Meixner C, Vegvari G, Ludwig-Müller J, Gagnon H, Steinkellner S, Staehelin C, et al. Two defined alleles of the LRR receptor kinase GmNARK in supernodulating soybean govern differing autoregulation of mycorrhization. Physiol Plant. 2007;130:261–270. [Google Scholar]
- 57.Sakamoto K, Nohara Y. Soybean (Glycine max [L.] Merr.) shoots systemically control arbuscule formation in mycorrhizal symbiosis. Soil Sci Plant Nutr. 2009;55:252–257. [Google Scholar]
- 58.Amiour N, Recorbet G, Robert F, Gianinazzi S, Dumas-Gaudot E. Mutations in DMI3 and SUNN modify the appressorium-responsive root proteome in arbuscular mycorrhiza. Mol Plant Microbe Interact. 2006;19:988–997. doi: 10.1094/MPMI-19-0988. [DOI] [PubMed] [Google Scholar]
- 59.Germain H, Chevalier E, Matton DP. Plant bioactive peptides: an expanding class of signaling molecules. Can J Bot. 2006;84:1–19. [Google Scholar]
- 60.Khaosaad T, Staehelin C, Steinkellner S, Hage-Ahmed K, Ocampo JA, Garcia-Garrido JM, et al. The Rhizobium sp. strain NGR234 systemically suppresses arbuscular mycorrhizal root colonization in a split-root system of barley (Hordeum vulgare) Physiol Plant. 2010;140:238–245. doi: 10.1111/j.1399-3054.2010.01396.x. [DOI] [PubMed] [Google Scholar]
- 61.Vierheilig H, Lerat S, Piché Y. Systemic inhibition of arbuscular mycorrhiza development by root exudates of cucumber plants colonized by Glomus mosseae. Mycorrhiza. 2003;13:167–170. doi: 10.1007/s00572-002-0219-0. [DOI] [PubMed] [Google Scholar]
- 62.Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 63.Khaosaad T, Garcia-Garrido JM, Steinkellner S, Vierheilig H. Take-all disease is systemically reduced in roots of mycorrhizal barley plants. Soil Biol Biochem. 2007;39:727–734. [Google Scholar]
- 64.Vierheilig H, Steinkellner S, Khaosaad T, Garcia-Garrido JM. The biocontrol effect of mycorrhization on soil-borne fungal pathogens and the autoregulation of the AM symbiosis: one mechanism, two effects? In: Varma A, editor. Mycorrhiza: Genetics and Molecular Biology, Eco-Function, Biotechnology, Eco-Physiology, Structure and Systematics. Heidelberg: Springer; 2008. pp. 307–320. [Google Scholar]
- 65.Heil M, Ton J. Long-distance signalling in plant defence. Trends Plant Sci. 2008;13:264–272. doi: 10.1016/j.tplants.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 66.Lohar DP, Bird DM. Lotus japonicus: A new model to study root-parasitic nematodes. Plant Cell Physiol. 2003;44:1176–1184. doi: 10.1093/pcp/pcg146. [DOI] [PubMed] [Google Scholar]
- 67.Tazawa J, Takahashi M, Usuki K, Yamamoto H. Nodulation during vegetative growth of soybean stage does not affect the susceptibility to red crown rot caused by Calonectria ilicicola. J Gen Plant Pathol. 2007;73:180–184. [Google Scholar]
- 68.Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol. 2009;5:578–580. doi: 10.1038/nchembio.182. [DOI] [PubMed] [Google Scholar]
- 69.Funayama-Noguchi S, Noguchi K, Yoshida C, Kawaguchi M. Two CLE genes are induced by phosphate in roots of Lotus japonicus. J Plant Res. 2010;124:155–163. doi: 10.1007/s10265-010-0342-5. [DOI] [PubMed] [Google Scholar]