Abstract
Breast cancer progression and metastasis are driven by complex and reciprocal interactions, between epithelial cancer cells and their surrounding stromal microenvironment. We have previously shown that a loss of stromal Cav-1 expression is associated with an increased risk of early tumor recurrence, metastasis and decreased overall survival. To identify and characterize the signaling pathways that are activated in Cav-1 negative tumor stroma, we performed gene expression profiling using laser microdissected breast cancer-associated stroma. Tumor stroma was laser capture microdissected from 4 cases showing high stromal Cav-1 expression and 7 cases with loss of stromal Cav-1. Briefly, we identified 238 gene transcripts that were upregulated and 232 gene transcripts that were downregulated in the stroma of tumors showing a loss of Cav-1 expression (p ≤ 0.01 and fold-change ≥1.5). Gene set enrichment analysis (GSEA) revealed “stemness,” inflammation, DNA damage, aging, oxidative stress, hypoxia, autophagy and mitochondrial dysfunction in the tumor stroma of patients lacking stromal Cav-1. Our findings are consistent with the recently proposed “Reverse Warburg Effect” and the “Autophagic Tumor Stroma Model of Cancer Metabolism.” In these two complementary models, cancer cells induce oxidative stress in adjacent stromal cells, which then forces these stromal fibroblasts to undergo autophagy/mitophagy and aerobic glycolysis. This, in turn, produces recycled nutrients (lactate, ketones and glutamine) to feed anabolic cancer cells, which are undergoing oxidative mitochondrial metabolism. Our results are also consistent with previous biomarker studies showing that the increased expression of known autophagy markers (such as ATG16L and the cathepsins) in the tumor stroma is specifically associated with metastatic tumor progression and/or poor clinical outcome.
Key words: caveolin-1, tumor stroma, breast cancer, transcriptional profiling, clinical outcome, recurrence, metastasis, biomarkers, gene signatures, aging
Introduction
Previously, we and others identified a loss of stromal caveolin-1 (Cav-1) as a new biomarker of a “lethal” tumor microenvironment.1–5 More specifically, a loss of stromal Cav-1 in the cancer-associated fibroblast compartment of human breast cancers is a powerful single independent predictor of early tumor recurrence, lymph-node metastasis and tamoxifen-resistance.1–5 Importantly, the predictive value of a loss of stromal Cav-1 was independent of epithelial marker status, and was effective in all of the most commons sub-types of invasive ductal carcinoma, including ER(+), PR(+), HER2(+) and triple-negative tumors.2–5 In triple-negative breast cancers, loss of stromal Cav-1 was associated with a 5-year survival rate of <10%. In contrast, triple-negative patients, from the same cohort, with high stromal Cav-1, had a 12-year survival rate of >75%.4 Similarly, in DCIS patients, a loss of stromal Cav-1 was predictive of disease recurrence and progression to invasive breast cancer;3 100% of DCIS patients with a loss of stromal Cav-1 underwent recurrence and 80% of these patients progressed from DCIS to invasive disease.3 In prostate cancer, a loss of stromal Cav-1 is predictive of advanced prostate cancer, with a high Gleason score, and is associated with lymph-node or bone metastasis.6 Thus, loss of stromal Cav-1 may be a new widely applicable biomarker for various epithelial tumor types, as most solid tumors contain a significant stromal component.2 However, it remains unknown why a loss of stromal Cav-1 has such a profound effect on patient prognosis and disease progression.
To address this issue directly, here we performed laser-capture microdissection on breast cancer patient tumor tissue (i.e., frozen sections) in which the status of their stromal Cav-1 levels (positive or negative) was first determined by immuno-histochemical staining. After laser-capture, the stromal material isolated from Cav-1 (+) and Cav-1 (−) patients was then subjected to genome-wide transcriptional profiling, to mechanistically unravel which signaling pathway(s) that are activated in the microenvironment of patients which are deficient in stromal Cav-1.
We show that the new transcriptional gene signature(s) that we have defined can effectively be used to cleanly separate patients based on their stromal Cav-1 status, into Cav-1 (+) and Cav-1 (−) sub-groups, greatly facilitating the prediction of their prognosis. Our findings provide novel mechanistic insights into how the presence or absence of Cav-1 in the stroma regulates tumor progression and breast cancer metastasis.
Results
Transcriptional profiling of a Cav-1 deficient tumor microenvironment.
Loss of stromal Cav-1 in human breast cancer(s) is associated with tumor recurrence, metastasis and drug-resistance, conferring poor clinical outcome.1–5 To mechanistically understand the “lethality” of a Cav-1 negative tumor microenvironment, we performed laser capture microdissection on the tumor stroma of patients that were pre-classified as stromal Cav-1-positive(+) (N = 4) and stromal Cav-1-negative(−) (N = 7), based on immuno-histochemical (IHC) staining. Then, the RNA extracted from these samples was subjected to genome-wide transcriptional profiling.
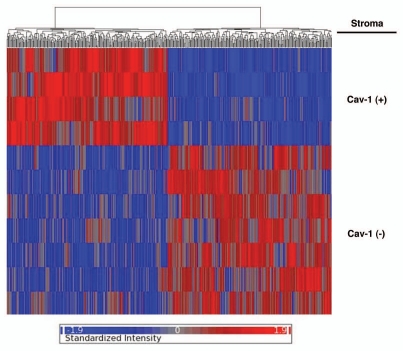
Based on this approach, we identified 238 gene transcripts that were specifically upregulated and 232 gene transcripts that were downregulated in the stroma of tumors showing a loss of Cav-1 expression [p ≤ 0.01 and fold-change (f.c.) ≥1.5] (Sup. Table 1). Using these stringent criteria, we were able to transcriptionally separate these two patient populations, in accordance with their IHC stromal Cav-1 status (negative versus positive). A HeatMap of the total patient cohort is shown in Figure 1, demonstrating that these two patient populations appear transcriptionally and “genetically” distinct.
Figure 1.
Stromal Cav-1 can be used to stratify human breast cancer patients into two transcriptionally distinct patient populations. The transcriptional profiles of Cav-1-positive (+) tumor stroma (N = 4) versus Cav-1-negative (−) tumor stroma (N = 7) were compared. We identified 238 gene transcripts that were upregulated and 232 gene transcripts that were downregulated in the stroma of tumors showing a loss of Cav-1 expression (Sup. Table 1). Note that the two patient populations are transcriptionally different. One-way ANOVA was setup to extract differentially expressed genes between Cav-1 positive and Cav-1 negative samples. The resultant p-values were further adjusted by multi-test correction (MTC) method of FDR step-up. The standardized intensity data from the stringent gene list (p-value ≤ 0.01 and fold change ≥1.5) were used in generating the hierarchical clustering HeatMap.
Gene set enrichment analysis (GSEA) of a Cav-1 deficient tumor microenvironment.
To understand what cellular processes are characteristic of a Cav-1-deficient tumor microenvironment, we next performed gene set enrichment analysis (GSEA), by comparison with other gene signatures available in various public databases. For this purpose, we focused on a wider list of gene transcripts that were upregulated in Cav-1-deficient stroma [5,424 transcripts encoding 3,459 unique genes; p ≤ 0.1 and fold-change (f.c.) ≥1.15] (Sup. Table 2), as is normally recommended for GSEA.
Table 1 shows the results of this detailed analysis. Note that we see the upregulation of cellular processes normally associated with “stemness,” inflammation, DNA damage, aging, oxidative stress, hypoxia, apoptotic signaling, autophagy and mitochondrial dysfunction in the tumor stroma of patients lacking stromal Cav-1.
Table 1.
Cellular processes that are associated with a lethal tumor micro-environment in human breast cancer (Cav-1 deficient stroma)
| Data Set | p-value | Detailed Description |
| Stem Cells, HGF/MET Signaling, the EMT and Metastasis | ||
| STEMCELL_NEURAL_UP | 1.45E-17 | Enriched in mouse neural stem cells, compared to differentiated brain and bone marrow cells |
| MORF_BMI1 | 3.74E-10 | Neighborhood of BMI1 B lymphoma Mo-MLV insertion region (mouse) in the MORF expression compendium |
| STEMCELL_EMBRYONIC_UP | 6.51E-09 | Enriched in mouse embryonic stem cells, compared to differentiated brain and bone marrow cells |
| HSC_LATEPROGENITORS_SHARED | 8.03E-09 | Upregulated in mouse hematopoietic late progenitors from both adult bone marrow and fetal liver (Cluster VI, Late Progenitors Shared) |
| STEMCELL_HEMATOPOIETIC_UP | 8.50E-09 | Enriched in mouse hematopoietic stem cells, compared to differentiated brain and bone marrow cells |
| HSC_LATEPROGENITORS_FETAL | 2.72E-08 | Up regulated in mouse hematopoietic late progenitors from fetal liver (Late Progenitors Shared + Fetal) |
| RUTELLA_HEPATGFSNDCS_UP | 6.88E-06 | Genes upregulated by HGF treatments |
| MORF_TPR | 1.59E-05 | Neighborhood of TPR translocated promoter region (to activated MET oncogene) in the MORF expression compendium |
| Inflammation, Host Defense and Graft Rejection | ||
| WIELAND_HEPATITIS_B_INDUCED | 5.92E-12 | Genes induced in the liver during hepatitis B viral clearance in chimpanzees |
| REOVIRUS_HEK293_UP | 1.56E-06 | Upregulated at any timepoint up to 24 hours following infection of HEK293 cells with reovirus strain T3Abney |
| DEFENSE_RESPONSE | 1.73E-06 | Genes annotated by the GO term GO:0006952. Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack |
| FLECHNER_KIDNEY_TRANSPLANT_ | ||
| REJECTION_UP | 3.80E-06 | Genes upreglated in acute rejection transplanted kidney biopsies relative to well functioning transplanted kidney biopsies from stable, immunosuppressed, recipients (median FDR <0.14% per comparison) |
| CARIES_PULP_UP | 2.02E-05 | Up-regulated in pulpal tissue from extracted carious teeth (cavities), compared to tissue from extracted healthy teeth |
| ER Negative Breast Cancer, BRCA1 Mutations, Cancer-Related Genes and Resistance to Chemotherapy | ||
| BRCA_ER_NEG | 6.60E-11 | Genes whose expression is consistently negatively correlated with estrogen receptor status in breast cancer] |
| MORF_DEK | 1.30E-08 | Neighborhood of DEK oncogene (DNA binding) in the MORF expression compendium |
| BRCA_BRCA1_POS | 7.38E-05 | Genes whose expression is consistently positively correlated with brca1 germline status in breast cancer |
| BRENTANI_SIGNALING | 1.19E-04 | Cancer related genes involved in the cell signaling |
| DOX_RESIST_GASTRIC_UP | 2.45E-04 | Upregulated in gastric cancer cell lines resistant to doxorubicin, compared to parent chemosensitive lines |
| MORF_SS18 | 3.32E-04 | Neighborhood of SS18 synovial sarcoma translocation, chromosome 18 in the MORF expression compendium |
| IL4/STAT6 Signaling | ||
| GNF2_STAT6 | 2.67E-10 | Neighborhood of STAT6 |
| LU_IL4BCELL | 9.16E-05 | Genes induced in peripheral B cells by 4 hours of incubation with the cytokine IL-4 |
| TNF/NFκB Signaling | ||
| GNF2_TNFRSF1B | 2.08E-08 | Neighborhood of TNFRSF1B |
| GNF2_TYK2 | 3.63E-06 | Neighborhood of TYK2 |
| GNF2_CARD15 | 6.32E-06 | Neighborhood of CARD15 |
| MORF_BUB3 | 7.64E-06 | Neighborhood of BUB3 |
| I_KAPPAB_KINASE_NF_KAPPAB_ CASCADE | 2.05E-04 | Genes annotated by the GO term GO:0007249. A series of reactions initiated by the activation of the transcription factor NF-kappaB. |
| ST_FAS_SIGNALING_PATHWAY | 2.89E-04 | The Fas receptor induces apoptosis and NFκB activation when bound to Fas ligand |
| DNA Damage and Repair | ||
| MORF_BMI1 | 3.74E-10 | Neighborhood of BMI1 B lymphoma Mo-MLV insertion region (mouse) in the MORF expression compendium |
| MORF_RAD23A | 3.80E-08 | Neighborhood of RAD23A homolog A (S. cerevisiae) in the MORF expression compendium |
| MORF_PRKDC | 3.18E-07 | Neighborhood of PRKDC protein kinase, DNA-activated, catalytic polypeptide in the MORF expression compendium |
| MORF_DDB1 | 5.42E-07 | Neighborhood of DDB1 damage-specific DNA binding protein 1, 127 kDa in the MORF expression compendium |
| DNA_METABOLIC_PROCESS | 1.01E-06 | Genes annotated by the GO term GO:0006259. The chemical reactions and pathways involving DNA, deoxyribonucleic acid, one of the two main types of nucleic acid, consisting of a long, unbranched macromolecule formed from one, or more commonly, two, strands of linked deoxyribonucleotides. |
| DNA_BINDING | 3.39E-06 | Genes annotated by the GO term GO:0003677. Interacting selectively with DNA (deoxyribonucleic acid). |
| MORF_RPA2 | 9.64E-06 | Neighborhood of RPA2 replication protein A2, 32 kDa in the MORF expression compendium |
| MORF_XRCC5 | 1.72E-05 | Neighborhood of XRCC5 X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining; Ku autoantigen, 80kDa) in the MORF expression compendium |
| MORF_RFC4 | 2.18E-05 | Neighborhood of RFC4 replication factor C (activator 1) 4, 37kDa in the MORF expression compendium |
| MORF_BUB1B | 5.69E-05 | Neighborhood of BUB1B budding uninhibited by benzimidazoles 1 homolog β (yeast) in the MORF expression compendium |
| MORF_TERF1 | 8.11E-05 | Neighborhood of TERF1 telomeric repeat binding factor (NIMA-interacting) 1 in the MORF expression compendium |
| MORF_RAD21 | 2.73E-04 | Neighborhood of RAD21 homolog (S. pombe) in the MORF expression compendium |
| Aging, Alzheimer Disease and Oxidative Stress | ||
| AGEING_KIDNEY_UP | 4.17E-07 | Upregulation is associated with increasing age in normal human kidney tissue from 74 patients |
| MORF_RAC1 | 1.88E-06 | Neighborhood of RAC1 ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) in the MORF expression compendium |
| AGED_MOUSE_NEOCORTEX_UP | 4.47E-05 | Upregulated in the neocortex of aged adult mice (30-month) vs. young adult (5-month) |
| ALZHEIMERS_DISEASE_UP | 1.81E-04 | Upregulated in correlation with overt Alzheimer's Disease, in the CA1 region of the hippocampus |
| STRESS_ARSENIC_SPECIFIC_UP | 2.12E-04 | Genes upregulated 4 hours following arsenic treatment that discriminate arsenic from other stress agents |
| Pro- and Anti-Apoptotic Signaling | ||
| GNF2_CASP1 | 1.68E-08 | Neighborhood of CASP1 |
| APOPTOSIS_GO | 2.01E-06 | Genes annotated by the GO term GO:0006915. A form of programmed cell death induced by external or internal signals that trigger the activity of proteolytic caspases, whose actions dismantle the cell and result in cell death. Apoptosis begins internally with condensation and subsequent fragmentation of the cell nucleus (blebbing) while the plasma membrane remains intact. Other characteristics of apoptosis include DNA fragmentation and the exposure of phosphatidyl serine on the cell surface |
| PROGRAMMED_CELL_DEATH | 2.32E-06 | Genes annotated by the GO term GO:0012501. Cell death resulting from activation of endogenous cellular processes |
| MORF_AATF | 2.84E-06 | Neighborhood of AATF apoptosis antagonizing transcription factor in the MORF expression compendium |
| GNF2_MCL1 | 4.95E-06 | Neighborhood of MCL1 |
| GNF2_BNIP2 | 6.86E-06 | Neighborhood of BNIP2 |
| GNF2_CASP8 | 5.41E-05 | Neighborhood of CASP8 |
| MORF_ANP32B | 3.34E-04 | Neighborhood of ANP32B acidic (leucine-rich) nuclear phosphoprotein 32 family, member B in the MORF expression compendium |
| Hypoxia, Mitochondrial Damage and Mitochondrial Respiration | ||
| MORF_PRKDC | 3.18E-07 | Neighborhood of PRKDC protein kinase, DNA-activated, catalytic polypeptide in the MORF expression compendium |
| RCGCANGCGY_V$NRF1_Q6 | 3.24E-07 | Genes with promoter regions [−2 kb, 2 kb] around transcription start site containing the motif RCGCANGCGY which matches annotation for NRF1: nuclear respiratory factor 1 |
| VHL_NORMAL_UP | 3.10E-05 | Upregulated in VHL-null renal carcinoma vs. normal renal cells (Fig. 2c+e) |
| V$NRF1_Q6 | 1.93E-04 | Genes with promoter regions [− 2 kb, 2 kb] around transcription start site containing the motif CGCATGCGCR which matches annotation for NRF1: nuclear respiratory factor 1 |
| MENSE_HYPOXIA_UP | 1.94E-04 | List of Hypoxia-induced genes found in both Astrocytes and HeLa Cells |
| STRESS_ARSENIC_SPECIFIC_UP | 2.12E-04 | Genes upregulated 4 hours following arsenic treatment that discriminate arsenic from other stress agents |
| Metabolic Dys-regulation: Obesity, Catabolism, Starvation and Autophagy | ||
| NADLER_OBESITY_UP | 1.90E-06 | Genes with increased expression with obesity |
| MORF_PRKAG1 | 5.94E-05 | Neighborhood of PRKAG1 protein kinase, AMP-activated, γ1 non-catalytic subunit in the MORF expression compendium |
| PENG_GLUTAMINE_UP | 5.97E-05 | Genes upregulated in response to glutamine starvation |
| BIOPOLYMER_CATABOLIC_PROCESS | 1.22E-04 | Genes annotated by the GO term GO:0043285. The chemical reactions and pathways resulting in the breakdown of biopolymers, long, repeating chains of monomers found in nature e.g., polysaccharides and proteins |
| PENG_LEUCINE_UP | 1.92E-04 | Genes upregulated in response to leucine starvation |
| Other | ||
| MGGAAGTG_V$GABP_B | 8.60E-10 | Genes with promoter regions [− 2 kb, 2 kb] around transcription start site containing the motif MGGAAGTG which matches annotation for GABPA: GA binding protein transcription factor, α subunit 60 kDa<br> GABPB2: GA binding protein transcription factor, β subunit 2 |
| SCGGAAGY_V$ELK1_02 | 1.65E-07 | Genes with promoter regions [− 2 kb, 2 kb] around transcription start site containing the motif SCGGAAGY which matches annotation for ELK1: ELK1, member of ETS oncogene family |
| GNF2_PECAM1 | 3.03E-06 | Neighborhood of PECAM1 |
| ST_GA13_PATHWAY | 2.17E-05 | G-α-13 influences the actin cytoskeleton and activates protein kinase D, PI3K and Pyk2. |
| V$E2F1_Q6 | 1.65E-04 | Genes with promoter regions [−2kb, 2kb] around transcription start site containing the motif TTTSGCGS which matches annotation for E2F1: E2F transcription factor 1 |
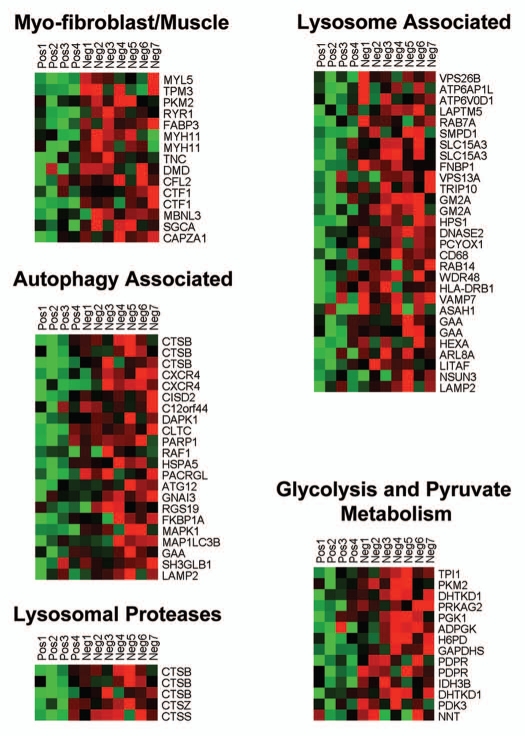
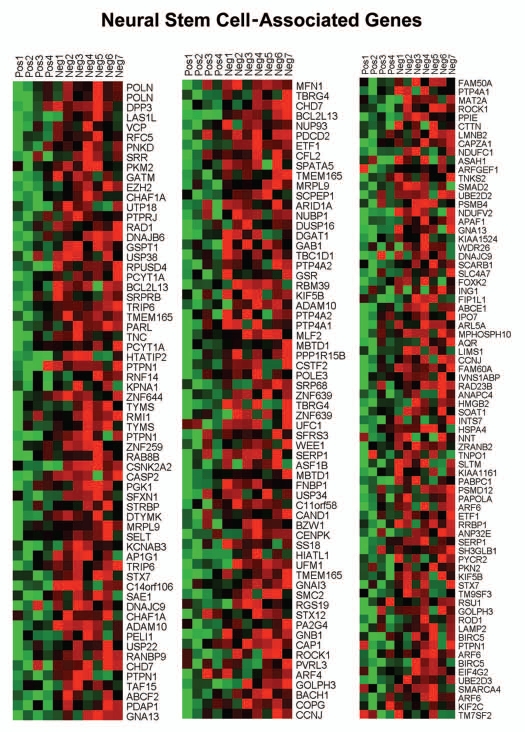
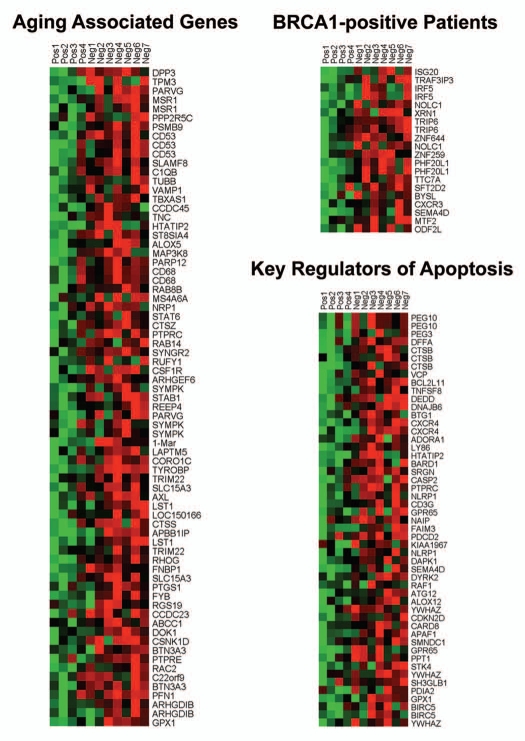
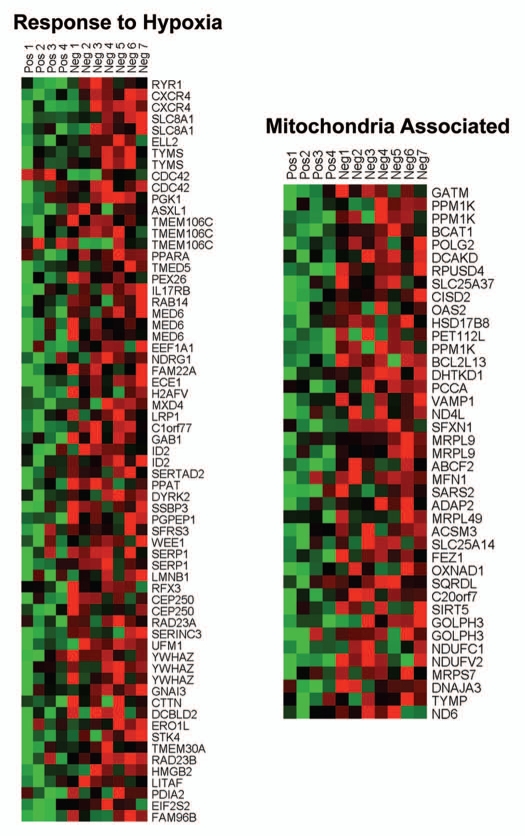
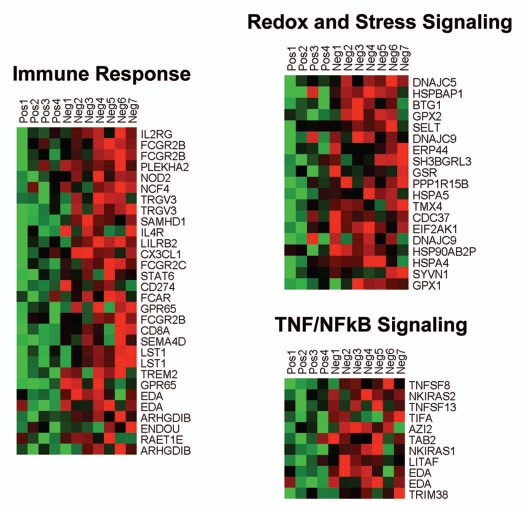
Individual HeatMaps [based on 1,819 transcripts encoding 1,297 unique genes; p ≤ 0.05 and fold-change (f.c.) ≥1.15] (Sup. Table 3) for key cellular processes are shown in Figures 2–8. These HeatMaps illustrate the upregulation of gene transcripts associated with myofibroblast differentiation, autophagy, lysosomal degradation, glycolysis, hypoxia, mitochondria, inflammation and redox signaling, DNA damage and repair, aging, BRCA1-mutation positive and ER-negative breast cancer patients, apoptosis and neural stem cells (See Sup. Tables 4–14).
Figure 2.
HeatMaps of gene transcripts associated with myofibroblast differentiation, autophagy, lysosomal degradation and glycolysis. Note that Cav-1-deficient stroma shows the upregulation of myofibroblast differentiation (15 transcripts), autophagy (22 transcripts), lysosomal proteases (5 transcripts), lysosomal proteins (29 transcripts) and glycolysis/pyruvate metabolism (15 transcripts). See Supplemental Tables 4, 5 and 10.
Figure 8.
HeatMaps of gene transcripts associated with neural stem cells. Note that Cav-1-deficient stroma shows the upregulation of genes normally associated with neural stem cells (202 transcripts). See Supplemental Table 13.
The observed association between a Cav-1-deficient tumor stroma and neural stem cells may reflect increased stromal “neurogenesis,” which has previously been implicated in an aggressive reactive stromal phenotype in a subset of prostate cancers.7,8 Similarly, “neurogenesis” was associated with increased DNA damage in the tumor stroma.7,8
In addition, the upregulation of gene transcripts or gene sets associated with GOLPH3, NRF1 (nuclear respiratory factor 1), PRKDC (protein kinase, DNA-activated, catalytic polypeptide) or arsenic treatment is highly suggestive of mitochondrial dysfunction (Table 1). NRF1 is normally upregulated during hypoxic-ischemic injury or oxidative stress, as an attempt to increase mitochondrial biogenesis.9,10 GOLPH3 (also known as MIDAS/GPP34) is a nuclear gene whose transcription is enhanced by the absence of mitochondrial DNA (mtDNA).11 MIDAS stands for mitochondrial DNA absence sensitive factor.11 PRKDC is a component of the machinery required for the proper repair and maintenance of the fidelity of mitochondrial DNA (mtDNA),12 so its upregulation is suggestive of increased mtDNA damage. Arsenic treatment is associated with increased ROS production, mtDNA damage and mitochondrial dysfunction, with reduced ATP production, driving the onset of aerobic glycolysis.13,14
Comparison with other breast cancer stromal data sets obtained by laser capture microdissection.
Next, we compared our new results with an independent previously published transcriptional data set generated by Morag Park and colleagues,15 via laser capture of the tumor stroma of human breast cancers. However, these previously published data were not stratified based on stromal Cav-1 status.15
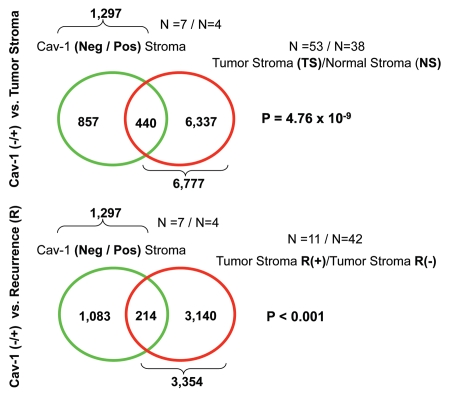
More specifically, we compared the gene transcripts upregulated in Cav-1-deficient tumor stroma (1,819 transcripts encoding 1,297 unique genes; p ≤ 0.05 and fold-change (f.c.) ≥1.15) (Sup. Table 3) with (1) tumor stroma and (2) recurrence stroma gene lists, as defined below:
Tumor Stroma vs. Normal Stroma List—Compares the transcriptional profiles of tumor stroma obtained from 53 patients to normal stroma obtained from 38 patients. Genes transcripts that were consistently upregulated in the tumor stroma were selected and assigned a p-value, with a cut-off of p ≤ 0.05 (this set contains 6,777 unique genes).16
Recurrence Stroma List—Compares the transcriptional profiles of tumor stroma obtained from 11 patients with tumor recurrence to the tumor stroma of 42 patients without tumor recurrence. Genes transcripts that were consistently upregulated in the tumor stroma of patients with recurrence were selected and assigned a p-value, with a cut-off of p ≤ 0.05 (this set contains 3,354 unique genes).16
Figure 9 shows that the gene transcripts upregulated in Cav-1-deficient tumor stroma show significant overlap with tumor stroma (a 440 transcript overlap; p = 4.76 × 10−9) and recurrence st'roma (a 214 transcript overlap; p < 0.001). Thus, the gene transcripts upregulated in Cav-1-deficient tumor stroma show a significant association with tumor recurrence, and are in accordance with the results of Morag Park and colleagues.15
Figure 9.
Venn diagrams for the intersection of the Cav-1-deficient stromal gene signature with other breast cancer tumor stromal gene sets. We compared the gene transcripts upregulated in Cav-1-deficient tumor stroma (1,819 transcripts encoding 1,297 unique genes; p ≤ 0.05 and fold-change (f.c.) ≥ 1.15; Sup. Table 3) with (upper) tumor stromal and (lower) recurrence stromal gene lists, as defined in the text of the manuscript. Note that the gene transcripts upregulated in Cav-1-deficient tumor stroma show significant overlap with tumor stroma (a 440 transcript overlap; p = 4.76 × 10−9) and recurrence stroma (a 214 transcript overlap; p < 0.001).
Comparison with the transcriptional profiles of breast cancers that were not subjected to laser capture microdissection.
Most of the published and publicly available transcriptional profiles for breast cancer patients are derived from the analysis of whole tumors, which are not separated into stromal and epithelial compartments prior to analysis. Thus, we compared the gene transcripts upregulated in Cav-1-deficient tumor stroma (Sup. Table 1), with the transcriptional profiles of normal breast tissue and whole breast tumors.17,18
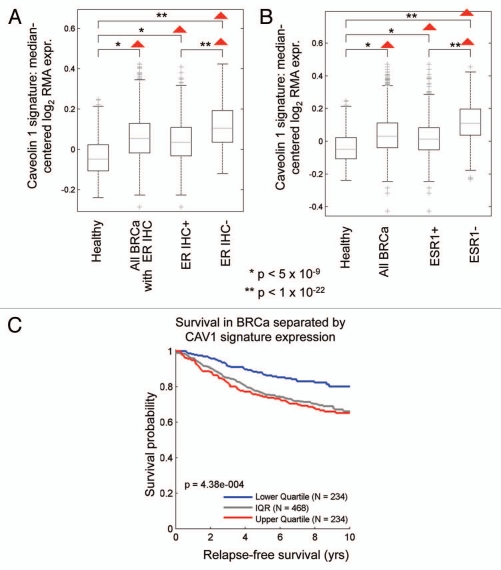
Figure 10A and B shows that the Cav-1-deficient tumor stromal signature is upregulated in breast cancer(s), both ER(+) and ER(−) sub-types. However, a more significant association was observed with ER(−) breast cancers. In addition, the Cav-1-deficient stromal signature was also associated with increased recurrence in breast cancer patients (Fig. 10C).
Figure 10.
The Cav-1-deficient stromal gene signature is upregulated in breast cancer and is associated with tumor recurrence. In (A and B), box-plots show that the Cav-1-deficient stromal signature is upregulated in all breast cancers, both ER(+) and ER(−) sub-types, relative to normal healthy breast tissue. In (A), ER status was determined by immuno-histochemistry, while in (B), ER status was inferred from ESR1 transcript expression. In (C), the Cav-1-deficient stromal signature was associated with increased recurrence in breast cancer patients. In (A–C), we used the Cav-1-deficient stromal signature included in Supplemental Table 1 (238 transcripts that were specifically upregulated; p ≤ 0.01 and fold-change (f.c.) ≥ 1.5). Qualitatively similar results were also obtained with the longer signature included in Supplemental Table 3. IQR, inter-quartile
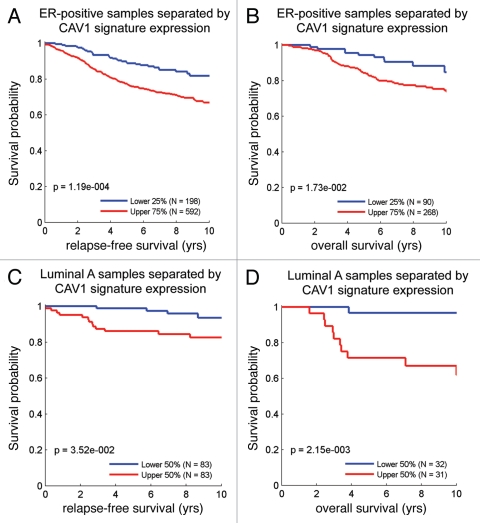
Similarly, we also looked at the prognostic value of the Cav-1-deficient stromal signature in ER(+) and luminal A breast cancer patients, which account for nearly 60% of all breast cancer patients.17,18 Interestingly, Figure 11 shows that the Cav-1-deficient stromal signature is clearly associated with increased recurrence and decreased overall survival, despite the fact that these breast cancer-derived tumors were not subjected to laser capture microdissection.
Figure 11.
The Cav-1-deficient stromal gene signature is associated with tumor recurrence and poor survival in ER(+) and luminal A breast cancer patients. Note that the Cav-1-deficient stromal signature (238 transcripts that were specifically upregulated; p ≤ 0.01 and fold-change (f.c.) ≥ 1.5; Sup. Table 1) is clearly associated with increased recurrence (A and C) and decreased overall survival (B and D), despite the fact that these breast cancer-derived tumors were not subjected to laser capture microdissection. (A and B) are ER(+) breast cancer patients, while (C and D) are the luminal A subset of ER(+) breast cancer patients. Qualitatively similar results were also obtained with the longer signature included in Supplemental Table 3.
Discussion
Given the powerful predictive value of a loss of stromal Cav-1 and its close association with tumor recurrence and metastasis in breast cancer patients,1–5 we wanted to determine what patho-physiological signaling processes are activated in this tumor microenvironment, driving poor clinical outcome. To achieve this goal, we took several independent approaches. In one approach, we developed a new co-culture system in which we could show that breast cancer cells (MCF7) drive the loss of stromal Cav-1 in adjacent cancer-associated fibroblasts.19 Under these conditions, the loss of stromal Cav-1 could be effectively blocked by the treatment of these co-cultures with anti-oxidants or agents that prevent lysosomal degradation.19–21 Thus, the Cav-1 protein is destroyed in fibroblasts in response to oxidative stress, via lysosomal degradation, directly implicating autophagy in this process.22–24 Cav-1 degradation in fibroblasts could also be induced by hypoxia, via the activation of autophagy.21 As such, loss of stromal Cav-1 is a functional marker of oxidative stress (pseudo-hypoxia), hypoxia and autophagy in the tumor microenvironment.21
Based on these, and other validating experiments, we demonstrated that cancer cells induce oxidative stress (pseudo-hypoxia) in adjacent fibroblasts, which activates certain key transcription factors (namely HIF-1α and NFκB), that then drive the onset of autophagy, leading to the cancer-associated fibroblast phenotype.20,21 HIF-1α and NFκB are the master regulator(s) of aerobic glycolysis (the response to hypoxia and oxidative stress) and inflammation, respectively.25–27 In turn, these autophagic fibroblasts underwent mitophagy, forcing them to use glycolysis as their main energy source.20,21 Interestingly, these autophagic stromal fibroblasts produced high-energy nutrients (lactate, ketones and glutamine) that could then be used as “fuel” for oxidative mitochondrial metabolism in epithelial breast cancer cells.28–30 Also, the ROS that was generated in cancer-associated fibroblasts was associated with DNA-damage (double-strand breaks) in both cancer cells and fibroblasts.19–21 We have termed this new paradigm for understanding tumor-stroma co-evolution as “The Autophagic Tumor Stroma Model of Cancer.”30
Many of the predictions of this new model were also validated using Cav-1 (−/−) null mice and stromal cells derived from these animals.16,30–32 Independently, knock-down of Cav-1 in human immortalized fibroblasts, using a targeted siRNA-approach, was indeed sufficient to generate high ROS levels, with activation of HIF1- and NFκB, as well as the onset of autophagy and mitochondrial dys-function.20,21 These Cav-1-deficient fibroblasts promoted tumor growth in vivo, when co-injected with triple-negative breast cancer cells (MDA-MB-231) in murine xenografts.33,34 Notably, the tumor growth-promoting effects of Cav-1 knock-down fibroblasts could be genetically dampened by recombinant overexpression of the anti-oxidant SOD2 (MnSOD),33 thereby relieving mitochondrial oxidative stress in Cav-1 deficient fibroblasts.
Finally, we could pharmacologcally pheno-copy the effects of Cav-1-deficient fibroblasts on cancer cells, simply by providing cancer cells with L-lactate. Under these conditions, lactate (a high-energy metabolite) treatment was sufficient to drive mitochondrial biogenesis in MCF7 cells,21,23 and promoted the metastasis of MDA-MB-231 cells in vivo.35 We could also genetically pheno-copy the effects of loss of Cav-1 in fibroblasts by stably-expressing activated HIF1 or activated NFκB, in cancer-associated fibroblasts.28 Under these conditions, activated HIF1 and/or NFκB drove autophagy/mitophagy in stromal fibroblasts, increased lactate production in vitro and promoted the growth of MDA-MB-231 triple-negative tumor xenografts in vivo.28
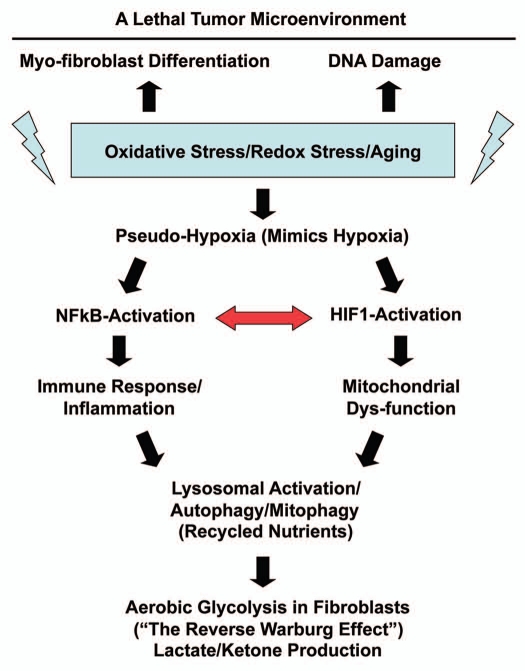
Here, we further attempted to validate this new model in vivo, specifically in human breast cancers that lack stromal Cav-1. Toward this end, we isolated the tumor stroma from breast cancer patients with a loss of stromal Cav-1 and patients with high stromal Cav-1, for direct comparison. Importantly, we demonstrate that the Cav-1 deficient tumor stroma from human breast cancer patients shows transcriptional evidence for all the same biological processes that we have implicated in the “Autophagic Tumor Stroma Model of Cancer.” These biological processes include oxidative stress/aging, hypoxia, a compensatory increase in mitochondria-associated genes, NFκB-activation, inflammation, DNA damage, aerobic glycolysis, autophagy/apoptosis, lysosomal degradation and “stemness” (summarized in Fig. 12). Previously, we have demonstrated that exactly the same biological processes are transcriptionally activated in mesenchymal stromal cells derived from the bone marrow of Cav-1 (−/−) null mice,16,31,32 which reflects oxidative stress and mitochondrial dys-function, leading to a glycolytic phenotype.
Figure 12.
Understanding the hierarchy of the cellular processes associated with a Cav-1-deficient tumor microenvironment. Oxidative stress in fibroblasts is known to be sufficient to induce (1) myo-fibroblast differentiation, (2) DNA damage and (3) a pseudo-hypoxic state.16,19–21,31,32 This pseudohypoxic state in fibroblasts then leads to the activation of NFκB and HIF-1α, which are master regulators of the immune response and mitochondrial function, as well as autophagy.19–21 The autophagic destruction of mitochnodria then drives aerobic glycolysis.22 We have previously shown that transient knock-down of Cav-1 in fibroblasts, using a targeted siRNA-approach, is sufficient to induce myo-fibroblast differentation, DNA damage and ROS production, leading to a pseudo-hypoxic state.19–21 Similarly, knock down of Cav-1 in fibroblasts is sufficient to drive NFκB- and HIF1-activation, as well as mitochondrial dys-function, autophagy and the induction of glycolytic enzymes.19–21 Interestingly, transcriptional profiling of a Cav-1-deficent tumor microenvironment provides direct evidence to support the involvement of all of these biological processes (See HeatMaps in Figs. 2–6). The red arrow denotes that NFκB-activation is known to augment HIF1-activation and visa versa, indicating that they act synergistically.28
We also observed an association between Cav-1 deficient tumor stroma and ER(−) breast cancer. For example, gene profiles from Cav-1 deficient tumor stroma were elevated in all breast cancers, relative to normal healthy breast tissue. However, there was a closer association with ER(−) negative breast cancers, that was identified either by ER IHC or via ESR1 transcriptional analysis (Fig. 10). Furthermore, we also observed an association with an ER(−) gene signature via gene set enrichment analysis (GSEA) (Fig. 7), as well as hereditary breast cancer patients that harbor BRCA1-mutations (Fig. 6). Importantly, these associations were all made with existing gene set data that were derived from whole breast tumors that were not laser-captured to isolate their stroma. This indicates that these stromal signatures can still be used effectively in conjunction with gene profiling data obtained from whole breast tumors.
Figure 7.
HeatMaps of gene transcripts associated with ER-negative breast cancers. Note that Cav-1-deficient stroma shows the upregulation of genes associated with ER-negative breast cancers (96 transcripts). See Supplemental Table 8.
Figure 6.
HeatMaps of gene transcripts associated with aging, apoptosis and BRCA1 mutation-positive breast cancer patients. Note that Cav-1-deficient stroma shows the upregulation of aging (73 transcripts), apoptosis (51 transcripts) and BRCA1-mutation associated genes (20 transcripts). See Supplemental Tables 7, 9 and 14.
Also, we showed that the stromal Cav-1 deficient gene signature could be used to predict outcome in breast cancer patients. For example, in ER(+) and luminal A breast cancer patients, the Cav-1-deficient stromal signature is clearly associated with increased recurrence and decreased overall survival, despite the fact that these breast cancer-derived tumors were not subjected to laser capture microdissection.
Interestingly, tumors with a “fibrotic focus” or “central scar” are usually associated with a poor prognosis.36,37 In accordance with this notion, keloid fibroblasts (derived from non-tumorous “scarred skin”) show many of the same characteristics as Cav-1 deficient tumor stroma, with activation of HIF1-α and NFκB, as well as a shift towards aerobic glycolysis.27,38,39 Thus, there may be a mechanistic connection between loss of Cav-1 as a driver of tissue fibrosis, tumor progression and metastasis.40 Consistent with this notion, a fibrotic focus in breast cancers is thought to be a surrogate marker for hypoxia.36,37 Similarly, a loss of Cav-1 drives activation of TGFβ signaling in fibroblasts,19,41–43 and treatment of fibroblasts with the TGFβ ligand is sufficient to induce autophagy44 and aerobic glycolysis,45 possibly explaining the association of tumor fibrosis with a poor prognosis in cancer patients.
Charis Eng and colleagues have previously reported that genomic instability (loss of heterozygosity/allelic imbalance) in human breast cancer tumor stroma is specifically associated with increased tumor grade and lymph-node metastasis.46,47 Similarly, they demonstrated increased genomic instability in the stroma of hereditary breast cancer patients that harbor BRCA1/2 mutations.48 Here, we arrived at a similar conclusion. Gene set enrichment analysis (GSEA) of the tumor stroma of breast cancer patients with a loss of stromal Cav-1 showed the dramatic upregulation of gene transcripts normally associated with DNA damage and repair (Fig. 5) and breast tumors with BRCA1-mutations (Fig. 6). Thus, stromal ROS production, resulting in stromal DNA damage, may be an underestimated driver of tumor progression and metastasis. These findings may also have relevance for guiding the use of DNA damaging agents and/or PARP inhibitors, as potential treatments for breast cancer patients with Cav-1-deficient stroma.
Figure 5.

HeatMaps of gene transcripts associated with DNA damage and repair. Note that Cav-1-deficient stroma shows the upregulation of the DNA damage response (67 transcripts). See Supplemental Table 11.
Similarly, in accordance with our current findings, the overexpression of several different markers of autophagy or oxidative stress in the tumor stroma has been shown to be associated with increased lympho-vascular invasion, increased lymph-node metastasis or poor clinical outcome, in a variety of different epithelial cancer subtypes. These markers include ATG16L1 and the cathepsins (K and D) for autophagy, as well as carbonic anhydrase IX (CAIX) and HIF1-α for oxidative stress.49–56
Furthermore, re-analysis of the published transcriptional profiles15 of laser-captured tumor stroma, isolated from human breast cancers, reveals strong evidence for the enrichment of autophagy genes, lysosomal markers, as well as indicators of oxidative stress (such as peroxisomes), which were associated with tumor recurrence and metastasis.16,30 These tumor stroma transcriptional profiles also showed significant overlap with the transcriptional profiles of Cav-1 (−/−) null stromal cells,16,30,31 and Alzheimer disease brain (known to be associated with oxidative stress).16,31 In fact the transcriptional profile of Alzheimer disease brain was most closely related to the tumor stroma of human breast cancer patients that had undergone metastasis,16 implicating stromal oxidative stress in promoting cancer cell metastasis.
Other laboratories have also clearly established that oxidative stress in the tumor microenvironment is a key driver of tumor recurrence and metastasis. For example, simple injection of purified protein anti-oxidant enzymes (such as catalase or SOD2, which detoxify hydrogen peroxide and super-oxide anions) is indeed sufficient to block both tumor recurrence and metastasis, in numerous studies.57–64 This premise has now been further validated using genetic mouse animal models of oxidative stress, using SOD2, CAV1 and JUN-D, as targets.33,65–67
In summary, we show here that simple immuno-staining with antibodies directed against Cav-1 can be used effectively to detect two different populations of breast cancer patients (high-risk vs. low-risk), based on their stromal Cav-1 status. Genome-wide transcriptional profiling of these two different stromal populations shows that they are dramatically different, each with a unique transcriptional profile. We focused on the stromal Cav-1-deficient population, as they have been associated with early tumor recurrence and metastasis. Gene set enrichment analysis (GSEA) of this population provides in vivo human data to directly support the “Autophagic Tumor Stroma Model of Cancer,” which is largely focused on oxidative stress, hypoxia, autophagy and DNA damage in the tumor microenvironment, as well as the pro-inflammatory response (Fig. 12).
Experimental Procedures
Laser capture microdissection and genome-wide expression profiling.
Tumor stroma was laser capture microdissected using a Leica LCM system from 4 cases showing high stromal Cav-1 expression and 7 cases with loss of stromal Cav-1. Total RNA was amplified using the NuGEN™ WT-Ovation™ FFPE RNA Amplification System V2 and cDNA was hybridized to Affymetrix GeneChip® arrays. One-way ANOVA was setup to extract differentially expressed genes between Cav-1 positive and negative stromal samples. Stromal tissue was laser capture micro-dissected (LCM) from fresh frozen (FF) tumor tissue samples collected from the patients pre-treatment and samples included were closely matched for age, race, stage and grade.
All eukaryotic target preparations (ALMAC Diagnostics, Inc.) using the project-appropriate NuGEN™ RNA Amplification System in combination with the Encore™ Biotin Module were performed in accordance with the guidelines detailed in the corresponding NuGEN™ technical manual. Total RNA was amplified using the NuGEN™ Ovation™ Pico WTA System. First-strand synthesis of cDNA was performed using a unique first-strand DNA/RNA chimeric primer mix, resulting in cDNA/mRNA hybrid molecules. Following fragmentation of the mRNA component of the cDNA/mRNA molecules, second-strand synthesis was performed and double-stranded cDNA was formed with a unique DNA/RNA heteroduplex at one end. In the final amplification step, RNA within the heteroduplex was degraded using RNaseH and replication of the resultant single-stranded cDNA was achieved through DNA/RNA chimeric primer binding and DNA polymerase enzymatic activity. The amplified single-stranded cDNA was purified for accurate quantitation of the cDNA and to ensure optimal performance during the fragmentation and labeling process. The single stranded cDNA was assessed using spectrophotometric methods in combination with the Agilent Bioanalyzer. The appropriate amount of amplified single-stranded cDNA was fragmented and labeled using the Encore™ Biotin Module. The enzymatically and chemically fragmented product (50–100 nt) was labeled via the attachment of biotinylated nucleotides onto the 3′-end of the fragmented cDNA. The resultant fragmented and labeled cDNA was added to the hybridization cocktail in accordance with the NuGEN™ guidelines for hybridization onto Affymetrix GeneChip® arrays. Following the hybridization for 18 hours at 45°C in an Affymetrix GeneChip® Hybridization Oven 640, the array was washed and stained on the GeneChip® Fluidics Station 450 using the appropriate fluidics script, before being inserted into the Affymetrix autoloader carousel and scanned using the GeneChip® Scanner 3000.
Gene set enrichment analysis (GSEA).
For the list of genes that are upregulated in Cav-1-negative patients compared to Cav-1-positive patients (fold change ≥1.15 and p-value ≤ 0.1) (Sup. Table 2), we computed enrichments in gene sets contained in the latest release of Molecular Signatures Database (MSigDB v2.5, April 2008, reviewed in ref. 68). MSigDB is a database of gene sets:
collected from various sources, such as online pathway databases, publications and knowledge of domain experts,
comprising genes that share a conserved cis-regulatory motif across the human, mouse, rat and dog genomes,
identified as co-regulated gene clusters by mining large collections of cancer-oriented microarray data and
annotated by a common Gene Ontology (GO) term.
The enrichment analysis consisted of computing p-values for the intersections between a gene list of interest X and each gene set Y in MSigDB. First, we computed the overlap between X and Y. Then, we computed the probability (p-value) that the observed overlap between sets X and Y is less than or equal to the overlap between set X and a randomly-chosen set of size equal to the size of set Y. This probability was calculated by applying the cumulative density function of the hypergeometric distribution on the size of set X, the size of set Y, the observed overlap between X and Y and the total number of available genes. In order to account for multiple hypothesis testing, we used randomly selected gene lists of the same size as the list under consideration to estimate the false discovery rate at a given p-value threshold. The final list of significant MSigDB gene sets was determined by setting a p-value threshold which ensures that the false discovery rate does not exceed 5%.
Analysis of clinical outcome in human breast cancer patients.
A microarray dataset that was previously compiled from the public repositories Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo),69 and ArrayExpress (www.ebi.ac.uk/arrayexpress),70 was used to evaluate the stromal Cav-1 deficient gene signature in the context of clinical samples.17,18 Samples were first analyzed in subsets based on their ER status. Additional subsets were defined by classifying samples among five canonical breast cancer subtypes, including luminal A, luminal B, normal-like, basal and Her-2-overexpressing disease. Samples were classified by computing their correlation against five expression profile centroids representing these breast cancer subtypes and assigning them to the subtype with the highest corresponding correlation coefficient.71 Samples with a maximum correlation coefficient below 0.3 were considered unclassified. Differential expression of the averaged gene signature magnitude among these sample subsets was evaluated using a two-tailed t-test. Kaplan-meier analysis was used to evaluate survival trends within sample subsets. The Log-rank test was used to evaluate differences in survival curves for high vs. low signature-expressing populations. In the BoxPlots (Fig. 10A), samples with ER immuno-histochemistry (IHC) data were selected for analysis, including 959 ER-positive and 323 ER-negative, for a total of 1,282 samples; in addition, 102 normal healthy breast tissue controls were used for comparison purposes. In the BoxPlots (Fig. 10B), samples with information on ESR1 mRNA transcript levels were selected for analysis, including 1,674 ESR1-positive and 478 ESR1-negative, for a total of 2,152 samples; in addition, 102 normal healthy breast tissue controls were used for comparison purposes. The ESR1 high/low expression cutoff was defined at the RMA-normalized expression value of 7.5. This was determined from the bimodal expression transcript profile of ESR1, previously described in Ertel et al. 2010.17,18
Figure 3.
HeatMaps of gene transcripts associated with the response to hypoxia and mitochondria. Note that Cav-1-deficient stroma shows the upregulation of hypoxia target genes (65 transcripts) and mitochondrial-associated proteins (41 transcripts). See Supplemental Tables 10 and 12.
Figure 4.
HeatMaps of gene transcripts associated with inflammation and redox/stress signaling. Note that Cav-1-deficient stroma shows the upregulation of TNF/NFκB signaling (11 transcripts), the immune response (31 transcripts) and redox/stress signaling (19 transcripts). See Supplemental Tables 6 and 10.
Acknowledgments
M.P.L. and his laboratory were supported by grants from the NIH/NCI (R01-CA-080250; R01-CA-098779; R01-CA-120876; R01-AR-055660) and the Susan G. Komen Breast Cancer Foundation. A.K.W. was supported by a Young Investigator Award from Breast Cancer Alliance, Inc., and a Susan G. Komen Career Catalyst Grant. F.S. was supported by grants from the W.W. Smith Charitable Trust, the Breast Cancer Alliance (BCA) and a Research Scholar Grant from the American Cancer Society (ACS). Funds were also contributed by the Margaret Q. Landenberger Research Foundation (to M.P.L.).
This project is funded, in part, under a grant with the Pennsylvania Department of Health (to M.P.L.). The Department specifically disclaims responsibility for any analyses, interpretations or conclusions.
We also thank ALMAC Diagnostics, Inc., for their expert technical assistance with RNA preparation and gene transcriptional profiling.
Supplementary Material
References
- 1.Sloan EK, Ciocca D, Pouliot N, Natoli A, Restall C, Henderson M, et al. Stromal cell expression of caveolin-1 predicts outcome in breast cancer. Am J Pathol. 2009;174:2035–2043. doi: 10.2353/ajpath.2009.080924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Witkiewicz AK, Casimiro MC, Dasgupta A, Mercier I, Wang C, Bonuccelli G, et al. Towards a new “stromal-based” classification system for human breast cancer prognosis and therapy. Cell Cycle. 2009;8:1654–1658. doi: 10.4161/cc.8.11.8544. [DOI] [PubMed] [Google Scholar]
- 3.Witkiewicz AK, Dasgupta A, Nguyen KH, Liu C, Kovatich AJ, Schwartz GF, et al. Stromal caveolin-1 levels predict early DCIS progression to invasive breast cancer. Cancer Biol Ther. 2009;8:1167–1175. doi: 10.4161/cbt.8.11.8874. [DOI] [PubMed] [Google Scholar]
- 4.Witkiewicz AK, Dasgupta A, Sammons S, Er O, Potoczek MB, Guiles F, et al. Loss of stromal caveolin-1 expression predicts poor cinical outcome in triple negative and basal-like breast cancers. Cancer Biol Ther. 2010;10:135–143. doi: 10.4161/cbt.10.2.11983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Witkiewicz AK, Dasgupta A, Sotgia F, Mercier I, Pestell RG, Sabel M, et al. An absence of stromal caveolin-1 expression predicts early tumor recurrence and poor clinical outcome in human breast cancers. Am J Pathol. 2009;174:2023–2034. doi: 10.2353/ajpath.2009.080873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Vizio D, Morello M, Sotgia F, Pestell RG, Freeman MR, Lisanti MP. An absence of stromal caveolin-1 is associated with advanced prostate cancer, metastatic disease and epithelial Akt activation. Cell Cycle. 2009;8:2420–2424. doi: 10.4161/cc.8.15.9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dakhova O, Ozen M, Creighton CJ, Li R, Ayala G, Rowley D, Ittmann M. Global gene expression analysis of reactive stroma in prostate cancer. Clin Cancer Res. 2009;15:3979–3989. doi: 10.1158/1078-0432.CCR-08-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayala GE, Dai H, Powell M, Li R, Ding Y, Wheeler TM, et al. Cancer-related axonogenesis and neurogenesis in prostate cancer. Clin Cancer Res. 2008;14:7593–7603. doi: 10.1158/1078-0432.CCR-08-1164. [DOI] [PubMed] [Google Scholar]
- 9.Yin W, Signore AP, Iwai M, Cao G, Gao Y, Chen J. Rapidly increased neuronal mitochondrial biogenesis after hypoxic-ischemic brain injury. Stroke. 2008;39:3057–3063. doi: 10.1161/STROKEAHA.108.520114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee HM, Greeley G, Jr, Englander EW. Sustained hypoxia modulates mitochondrial DNA content in the neonatal rat brain. Free Radic Biol Med. 2008;44:807–814. doi: 10.1016/j.freeradbiomed.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakashima-Kamimura N, Asoh S, Ishibashi Y, Mukai Y, Shidara Y, Oda H, et al. MIDAS/GPP34, a nuclear gene product, regulates total mitochondrial mass in response to mitochondrial dysfunction. J Cell Sci. 2005;118:5357–5367. doi: 10.1242/jcs.02645. [DOI] [PubMed] [Google Scholar]
- 12.Papeta N, Zheng Z, Schon EA, Brosel S, Altintas MM, Nasr SH, et al. Prkdc participates in mitochondrial genome maintenance and prevents Adriamycin-induced nephropathy in mice. J Clin Invest. 2010;120:4055–4064. doi: 10.1172/JCI43721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jomova K, Jenisova Z, Feszterova M, Baros S, Liska J, Hudecova D, et al. Arsenic: Toxicity, oxidative stress and human disease. J Appl Toxicol. 2011;31:95–107. doi: 10.1002/jat.1649. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Liu Y, An Z, Huang D, Qi Y, Zhang Y. Mediating effect of ROS on mtDNA damage and low ATP content induced by arsenic trioxide in mouse oocytes. Toxicol In Vitro. 2011;25:979–984. doi: 10.1016/j.tiv.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 15.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 16.Pavlides S, Tsirigos A, Vera I, Flomenberg N, Frank PG, Casimiro MC, et al. Transcriptional evidence for the “reverse Warburg effect” in human breast cancer tumor stroma and metastasis: Similarities with oxidative stress, inflammation, Alzheimer's disease and “neuron-glia metabolic coupling”. Aging. 2010;2:185–199. doi: 10.18632/aging.100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ertel A. Bimodal gene expression and biomarker discovery. Cancer Inform. 2010;9:11–14. doi: 10.4137/cin.s3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ertel A, Dean JL, Rui H, Liu C, Witkiewicz AK, Knudsen KE, et al. RB-pathway disruption in breast cancer: Differential association with disease subtypes, disease-specific prognosis and therapeutic response. Cell Cycle. 2010;9:4153–4163. doi: 10.4161/cc.9.20.13454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Outschoorn UE, Pavlides S, Whitaker-Menezes D, Daumer KM, Milliman JN, Chiavarina B, et al. Tumor cells induce the cancer-associated fibroblast phenotype via caveolin-1 degradation: Implications for breast cancer and DCIS therapy with autophagy inhibitors. Cell Cycle. 2010;9:2423–2433. doi: 10.4161/cc.9.12.12048. [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Outschoorn UE, Balliet RM, Rivadeneira DB, Chiavarina B, Pavlides S, Wang C, et al. Oxidative stress in cancer-associated fibroblasts drives tumor-stroma co-evolution: A new paradigm for understanding tumor metabolism, the field effect and genomic instability in cancer cells. Cell Cycle. 2010;9:3256–3276. doi: 10.4161/cc.9.16.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez-Outschoorn UE, Trimmer C, Lin Z, Whitaker-Menezes D, Chiavarina B, Zhou J, et al. Autophagy in cancer associated fibroblasts promotes tumor cell survival: Role of hypoxia, HIF1 induction and NFκB activation in the tumor stromal microenvironment. Cell Cycle. 2010;9:3515–3533. doi: 10.4161/cc.9.17.12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez-Outschoorn UE, Whitaker-Menezes D, Pavlides S, Chiavarina B, Bonuccelli G, Casey T, et al. The autophagic tumor stroma model of cancer or “battery-operated tumor growth”: A simple solution to the autophagy paradox. Cell Cycle. 2010;9:4297–4306. doi: 10.4161/cc.9.21.13817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez-Outschoorn UE, Pavlides S, Howell A, Pestell RG, Tanowitz HB, Sotgia F, et al. Stromal-epithelial metabolic coupling in cancer: Integrating autophagy and metabolism in the tumor microenvironment. Int J Biochem Cell Biol. 2011 doi: 10.1016/j.biocel.2011.01.023. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lisanti MP, Martinez-Outschoorn UE, Chiavarina B, Pavlides S, Whitaker-Menezes D, Tsirigos A, et al. Understanding the “lethal” drivers of tumor-stroma co-evolution: Emerging role(s) for hypoxia, oxidative stress and autophagy/mitophagy in the tumor microenvironment. Cancer Biol Ther. 2010;10:537–542. doi: 10.4161/cbt.10.6.13370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazure NM, Pouyssegur J. Atypical BH3-domains of BNIP3 and BNIP3L lead to autophagy in hypoxia. Autophagy. 2009;5:868–869. doi: 10.4161/auto.9042. [DOI] [PubMed] [Google Scholar]
- 26.Mazure NM, Pouyssegur J. Hypoxia-induced autophagy: Cell death or cell survival? Curr Opin Cell Biol. 2010;22:177–180. doi: 10.1016/j.ceb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 27.Messadi DV, Doung HS, Zhang Q, Kelly AP, Tuan TL, Reichenberger E, et al. Activation of NFkappaB signal pathways in keloid fibroblasts. Arch Dermatol Res. 2004;296:125–133. doi: 10.1007/s00403-004-0487-y. [DOI] [PubMed] [Google Scholar]
- 28.Chiavarina B, Whitaker-Menezes D, Migneco G, Martinez-Outschoorn UE, Pavlides S, Howell A, et al. HIF-1alpha functions as a tumor promoter in cancer-associated fibroblasts and as a tumor suppressor in breast cancer cells: Autophagy drives compartment-specific oncogenesis. Cell Cycle. 2010;9:3534–3551. doi: 10.4161/cc.9.17.12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Migneco G, Whitaker-Menezes D, Chiavarina B, Castello-Cros R, Pavlides S, Pestell RG, et al. Glycolytic cancer-associated fibroblasts promote breast cancer tumor growth, without a measurable increase in angiogenesis: Evidence for stromal-epithelial metabolic coupling. Cell Cycle. 2010;9:2412–2422. doi: 10.4161/cc.9.12.11989. [DOI] [PubMed] [Google Scholar]
- 30.Pavlides S, Tsirigos A, Migneco G, Whitaker-Menezes D, Chiavarina B, Flomenberg N, et al. The autophagic tumor stroma model of cancer: Role of oxidative stress and ketone production in fueling tumor cell metabolism. Cell Cycle. 2010;9:3485–3505. doi: 10.4161/cc.9.17.12721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pavlides S, Tsirigos A, Vera I, Flomenberg N, Frank PG, Casimiro MC, et al. Loss of stromal caveolin-1 leads to oxidative stress, mimics hypoxia and drives inflammation in the tumor microenvironment, conferring the “reverse Warburg effect”: A transcriptional informatics analysis with validation. Cell Cycle. 2010;9:2201–2219. doi: 10.4161/cc.9.11.11848. [DOI] [PubMed] [Google Scholar]
- 32.Pavlides S, Whitaker-Menezes D, Castello-Cros R, Flomenberg N, Witkiewicz AK, Frank PG, et al. The reverse Warburg effect: Aerobic glycolysis in cancer-associated fibroblasts and the tumor stroma. Cell Cycle. 2009;8:3984–4001. doi: 10.4161/cc.8.23.10238. [DOI] [PubMed] [Google Scholar]
- 33.Trimmer C, Sotgia F, Whitaker-Menezes D, Balliet RM, Eaton G, Martinez-Outschoorn UE, et al. Caveolin-1 and mitochondrial SOD2 (MnSOD) function as tumor suppressors in the stromal microenvironment: A new genetically tractable model for human cancer-associated fibroblasts. Cancer Biol Ther. 2011;11:383–394. doi: 10.4161/cbt.11.4.14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonuccelli G, Whitaker-Menezes D, Castello-Cros R, Pavlides S, Pestell RG, Fatatis A, et al. The reverse Warburg effect: Glycolysis inhibitors prevent the tumor promoting effects of caveolin-1-deficient cancer-associated fibroblasts. Cell Cycle. 2010;9:1960–1971. doi: 10.4161/cc.9.10.11601. [DOI] [PubMed] [Google Scholar]
- 35.Bonuccelli G, Tsirigos A, Whitaker-Menezes D, Pavlides S, Pestell RG, Chiavarina B, et al. Ketones and lactate “fuel” tumor growth and metastasis: Evidence that epithelial cancer cells use oxidative mitochondrial metabolism. Cell Cycle. 2010;9:3506–3514. doi: 10.4161/cc.9.17.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van den Eynden GG, Smid M, Van Laere SJ, Colpaert CG, Van der Auwera I, Bich TX, et al. Gene expression profiles associated with the presence of a fibrotic focus and the growth pattern in lymph node-negative breast cancer. Clin Cancer Res. 2008;14:2944–2952. doi: 10.1158/1078-0432.CCR-07-4397. [DOI] [PubMed] [Google Scholar]
- 37.Van den Eynden GG, Colpaert CG, Couvelard A, Pezzella F, Dirix LY, Vermeulen PB, et al. A fibrotic focus is a prognostic factor and a surrogate marker for hypoxia and (lymph)angiogenesis in breast cancer: Review of the literature and proposal on the criteria of evaluation. Histopathology. 2007;51:440–451. doi: 10.1111/j.1365-2559.2007.02761.x. [DOI] [PubMed] [Google Scholar]
- 38.Vincent AS, Phan TT, Mukhopadhyay A, Lim HY, Halliwell B, Wong KP. Human skin keloid fibroblasts display bioenergetics of cancer cells. J Invest Dermatol. 2008;128:702–709. doi: 10.1038/sj.jid.5701107. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Q, Oh CK, Messadi DV, Duong HS, Kelly AP, Soo C, et al. Hypoxia-induced HIF-1 alpha accumulation is augmented in a co-culture of keloid fibroblasts and human mast cells: Involvement of ERK1/2 and PI-3K/Akt. Exp Cell Res. 2006;312:145–155. doi: 10.1016/j.yexcr.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 40.Qian N, Ueno T. Is dysfunction of caveolin-1 a link between systemic sclerosis and breast cancer, opening a window on both etiologies? Arch Med Res. 2010;41:297–301. doi: 10.1016/j.arcmed.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 41.Razani B, Zhang XL, Bitzer M, von Gersdorff G, Bottinger EP, Lisanti MP. Caveolin-1 regulates transforming growth factor (TGF)beta/SMAD signaling through an interaction with the TGFbeta type I receptor. J Biol Chem. 2001;276:6727–6738. doi: 10.1074/jbc.M008340200. [DOI] [PubMed] [Google Scholar]
- 42.Del Galdo F, Lisanti MP, Jimenez SA. Caveolin-1, transforming growth factorbeta receptor internalization and the pathogenesis of systemic sclerosis. Curr Opin Rheumatol. 2008;20:713–719. doi: 10.1097/bor.0b013e3283103d27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Del Galdo F, Sotgia F, de Almeida CJ, Jasmin JF, Musick M, Lisanti MP, et al. Decreased expression of caveolin-1 in patients with systemic sclerosis: Crucial role in the pathogenesis of tissue fibrosis. Arthritis Rheum. 2008;58:2854–2865. doi: 10.1002/art.23791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiyono K, Suzuki HI, Matsuyama H, Morishita Y, Komuro A, Kano MR, et al. Autophagy is activated by TGFbeta and potentiates TGFbeta-mediated growth inhibition in human hepatocellular carcinoma cells. Cancer Res. 2009;69:8844–8852. doi: 10.1158/0008-5472.CAN-08-4401. [DOI] [PubMed] [Google Scholar]
- 45.Racker E, Resnick RJ, Feldman R. Glycolysis and methylaminoisobutyrate uptake in rat-1 cells transfected with ras or myc oncogenes. Proc Natl Acad Sci USA. 1985;82:3535–3538. doi: 10.1073/pnas.82.11.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fukino K, Shen L, Patocs A, Mutter GL, Eng C. Genomic instability within tumor stroma and clinicopathological characteristics of sporadic primary invasive breast carcinoma. Jama. 2007;297:2103–2111. doi: 10.1001/jama.297.19.2103. [DOI] [PubMed] [Google Scholar]
- 47.Eng C, Leone G, Orloff MS, Ostrowski MC. Genomic alterations in tumor stroma. Cancer Res. 2009;69:6759–6764. doi: 10.1158/0008-5472.CAN-09-0985. [DOI] [PubMed] [Google Scholar]
- 48.Weber F, Shen L, Fukino K, Patocs A, Mutter GL, Caldes T, et al. Total-genome analysis of BRCA1/2-related invasive carcinomas of the breast identifies tumor stroma as potential landscaper for neoplastic initiation. Am J Hum Genet. 2006;78:961–972. doi: 10.1086/504090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nomura H, Uzawa K, Yamano Y, Fushimi K, Ishigami T, Kouzu Y, et al. Overexpression and altered subcellular localization of autophagy-related 16-like 1 in human oral squamous-cell carcinoma: correlation with lymphovascular invasion and lymph-node metastasis. Hum Pathol. 2009;40:83–91. doi: 10.1016/j.humpath.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 50.Kleer CG, Bloushtain-Qimron N, Chen YH, Carrasco D, Hu M, Yao J, et al. Epithelial and stromal cathepsin K and CXCL14 expression in breast tumor progression. Clin Cancer Res. 2008;14:5357–5367. doi: 10.1158/1078-0432.CCR-08-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nadji M, Fresno M, Nassiri M, Conner G, Herrero A, Morales AR. Cathepsin D in host stromal cells, but not in tumor cells, is associated with aggressive behavior in node-negative breast cancer. Hum Pathol. 1996;27:890–895. doi: 10.1016/s0046-8177(96)90214-2. [DOI] [PubMed] [Google Scholar]
- 52.Colpaert CG, Vermeulen PB, Fox SB, Harris AL, Dirix LY, Van Marck EA. The presence of a fibrotic focus in invasive breast carcinoma correlates with the expression of carbonic anhydrase IX and is a marker of hypoxia and poor prognosis. Breast Cancer Res Treat. 2003;81:137–147. doi: 10.1023/A:1025702330207. [DOI] [PubMed] [Google Scholar]
- 53.Kuijper A, van der Groep P, van der Wall E, van Diest PJ. Expression of hypoxia-inducible factor 1alpha and its downstream targets in fibroepithelial tumors of the breast. Breast Cancer Res. 2005;7:808–818. doi: 10.1186/bcr1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakao M, Ishii G, Nagai K, Kawase A, Kenmotsu H, Kon-No H, et al. Prognostic significance of carbonic anhydrase IX expression by cancer-associated fibroblasts in lung adenocarcinoma. Cancer. 2009;115:2732–2743. doi: 10.1002/cncr.24303. [DOI] [PubMed] [Google Scholar]
- 55.Cleven AH, van Engeland M, Wouters BG, de Bruine AP. Stromal expression of hypoxia regulated proteins is an adverse prognostic factor in colorectal carcinomas. Cell Oncol. 2007;29:229–240. doi: 10.1155/2007/945802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.von Bonsdorf CH, Fuller S, Simons K. Apical and basolateral endocytosis in Madin-Darby canine kidney (MDCK) cells grown on nitrocellulose filters. EMBO J. 1985;11:2781–2792. doi: 10.1002/j.1460-2075.1985.tb04004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hyoudou K, Nishikawa M, Umeyama Y, Kobayashi Y, Yamashita F, Hashida M. Inhibition of metastatic tumor growth in mouse lung by repeated administration of polyethylene glycol-conjugated catalase: Quantitative analysis with firefly luciferase-expressing melanoma cells. Clin Cancer Res. 2004;10:7685–7691. doi: 10.1158/1078-0432.CCR-04-1020. [DOI] [PubMed] [Google Scholar]
- 58.Nishikawa M, Hyoudou K, Kobayashi Y, Umeyama Y, Takakura Y, Hashida M. Inhibition of metastatic tumor growth by targeted delivery of antioxidant enzymes. J Control Release. 2005;109:101–107. doi: 10.1016/j.jconrel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 59.Hyoudou K, Nishikawa M, Kobayashi Y, Umeyama Y, Yamashita F, Hashida M. PEGylated catalase prevents metastatic tumor growth aggravated by tumor removal. Free Radic Biol Med. 2006;41:1449–1458. doi: 10.1016/j.freeradbiomed.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 60.Nishikawa M, Hashida M, Takakura Y. Catalase delivery for inhibiting ROS-mediated tissue injury and tumor metastasis. Adv Drug Deliv Rev. 2009;61:319–326. doi: 10.1016/j.addr.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 61.Hyoudou K, Nishikawa M, Ikemura M, Kobayashi Y, Mendelsohn A, Miyazaki N, et al. Prevention of pulmonary metastasis from subcutaneous tumors by binary system-based sustained delivery of catalase. J Control Release. 2009;137:110–115. doi: 10.1016/j.jconrel.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 62.Nishikawa M, Tamada A, Hyoudou K, Umeyama Y, Takahashi Y, Kobayashi Y, et al. Inhibition of experimental hepatic metastasis by targeted delivery of catalase in mice. Clin Exp Metastasis. 2004;21:213–221. doi: 10.1023/b:clin.0000037706.13747.5e. [DOI] [PubMed] [Google Scholar]
- 63.Hyoudou K, Nishikawa M, Kobayashi Y, Ikemura M, Yamashita F, Hashida M. SOD derivatives prevent metastatic tumor growth aggravated by tumor removal. Clin Exp Metastasis. 2008;25:531–536. doi: 10.1007/s10585-008-9165-3. [DOI] [PubMed] [Google Scholar]
- 64.van Rossen ME, Sluiter W, Bonthuis F, Jeekel H, Marquet RL, van Eijck CH. Scavenging of reactive oxygen species leads to diminished peritoneal tumor recurrence. Cancer Res. 2000;60:5625–5629. [PubMed] [Google Scholar]
- 65.Zhao Y, Xue Y, Oberley TD, Kiningham KK, Lin SM, Yen HC, et al. Overexpression of manganese superoxide dismutase suppresses tumor formation by modulation of activator protein-1 signaling in a multistage skin carcinogenesis model. Cancer Res. 2001;61:6082–6088. [PubMed] [Google Scholar]
- 66.Vozenin-Brotons MC, Sivan V, Gault N, Renard C, Geffrotin C, Delanian S, et al. Antifibrotic action of Cu/Zn SOD is mediated by TGFbeta1 repression and phenotypic reversion of myofibroblasts. Free Radic Biol Med. 2001;30:30–42. doi: 10.1016/s0891-5849(00)00431-7. [DOI] [PubMed] [Google Scholar]
- 67.Toullec A, Gerald D, Despouy G, Bourachot B, Cardon M, Lefort S, et al. Oxidative stress promotes myofibroblast differentiation and tumour spreading. EMBO Mol Med. 2010;2:211–230. doi: 10.1002/emmm.201000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, et al. NCBI GEO: Mining tens of millions of expression profiles—database and tools update. Nucl Acids Res. 2007;35:760–765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brazma A, Parkinson H, Sarkans U, Shojatalab M, Vilo J, Abeygunawardena N, et al. ArrayExpress—a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003;31:68–71. doi: 10.1093/nar/gkg091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.