Abstract
Studies of the zebrafish epithalamus have provided recent insights into the development of left–right brain asymmetry, which is crucial to normal human brain function. The habenular nuclei of zebrafish are robustly asymmetric, with dense elaboration of neuropil only in the left lateral subnucleus. Because this feature is tightly correlated with asymmetric expression of K+ channel tetramerization domain-containing proteins 12.1 and 12.2 (Kctd12.1/12.2), we screened for Kctd12.1-interacting proteins to identify molecular mechanisms leading to neuropil asymmetry, and uncovered a novel interaction between Kctd12.1 and Unc-51-like kinase 2 (Ulk2). We show here that knockdown of Ulk2 or overexpression of Kctd12 proteins reduces asymmetric neuropil elaboration. Conversely, overexpression of Ulk2 or mutation of kctd12 genes causes excess neuropil elaboration. We conclude that Ulk2 activity promotes neuropil elaboration while Kctd12 proteins limit Ulk2 activity asymmetrically. This work describes a regulatory mechanism for neuronal process extension that may be conserved in other developmental contexts in addition to the epithalamus.
Introduction
Left–right (L–R) asymmetry in brain function is a feature observed throughout the vertebrate lineage (Concha and Wilson, 2001). Analysis of the behavior of many species suggests that functional asymmetries stem from an anciently derived specialization of the right side for environmentally motivated behaviors and/or pattern recognition, and the left side for self-motivated behaviors and/or detail recognition. Such specialization is thought to increase the efficiency of mental processing, and the consistent direction of asymmetry in social organisms may promote behavior that is predictable to the other members of a group (MacNeilage et al., 2009).
The zebrafish has been an important tool for understanding the molecular basis of asymmetric brain development. In particular, attention has focused on the epithalamus, which consists of the pineal organ, the parapineal organ, and the left and right habenular brain nuclei (Hb). The left-sided parapineal causes the left Hb to develop a large lateral and small medial subnucleus (Concha et al., 2000; Gamse et al., 2003, 2005; Aizawa et al., 2005), in a process that involves Notch (Aizawa et al., 2007) and Wnt signaling (Carl et al., 2007).
Hb subnuclei are asymmetric not only in size but also in anatomy and gene expression. The left lateral subnucleus exhibits a large core of dense neuropil, which consists of afferent axons from the forebrain and the Hb dendrites they contact. By contrast, the right lateral subnucleus has only a small core of neuropil (Concha et al., 2000). This neuroanatomical asymmetry correlates with the expression of the potassium channel tetramerization domain (KCTD) protein Kctd12.1 (Leftover) (Gamse et al., 2005). Kctd12.1 expression begins at 38 h postfertilization (hpf), ∼10 h before the appearance of neuropil, and is restricted to lateral Hb subnuclei; therefore, many more cells in the left Hb express Kctd12.1 than the right (Gamse et al., 2003). The medial subnucleus expresses a closely related protein, Kctd12.2, but elaborates little neuropil, similar to the right lateral subnucleus. The function of Kctd proteins in developing neurons has not been fully elucidated, but they are involved in processes as diverse as ubiquitin ligase adaption (Bayón et al., 2008) and GABAB receptor modulation (Schwenk et al., 2010).
Based on the correlation of Kctd12.1 expression with asymmetric Hb morphology, we investigated the role of Kctd12 proteins in the elaboration of asymmetric neuropil. Using cross-species two-hybrid screening, Kctd12.1 was found to physically interact with Unc-51-like 2 (Ulk2), a kinase that promotes neuronal process extension (Zhou et al., 2007). We find that antisense knockdown of zebrafish Ulk2 function causes a reduction of Hb neuropil. Overexpression of Kctd12.1 or Kctd12.2 in the Hb also results in reduced neuropil, while neuropil volume is expanded in kctd12.1 and kctd12.2 mutants. These data suggest that Kctd12 proteins are negative regulators of Ulk2, a hypothesis that is supported by our genetic interaction experiments. We conclude that Kctd12.1 and Kctd12.2 may generate asymmetry in the neuropil of the Hb nuclei through differential modulation of Ulk2 activity.
Materials and Methods
Zebrafish.
Zebrafish embryos were obtained by natural spawning of wild-type AB (Walker, 1999), Tg[cfos:gal4vp16]s1019t (referred to here as Hb:Gal) (Scott et al., 2007), Tg[UAS:kctd12.1:mt]vu260/264, Tg[UAS:kctd12.1:pA]vu302, Tg[UAS:kctd12.2:mt]vu431, kctd12.1vu442, and kctd12.2fh312 lines, by mating single male and female adult zebrafish. Embryos were raised at 28.5°C on a 14:10 light/dark cycle using standard procedures (Westerfield and Zebrafish Information Network, 2000), and staged according to hours or days postfertilization. For imaging purposes, 0.003% phenylthiourea was added to embryo media to prevent the formation of pigment.
Transgenesis.
All transgenic animals generated for this study were created using the Tol2kit (Kwan et al., 2007) and built using the Multisite Gateway (Invitrogen) cloning system in the pDestTol2CG2 transgenesis vector. For stable germline transgenics (Tg[UAS:kctd12.1:mt]vu260/264, Tg[UAS:kctd12.1:pA]vu302, and Tg[UAS:kctd12.2:mt]vu431), AB embryos were injected at late one-cell stage, screened for cardiac GFP at 3 d postfertilization (dpf), raised to adulthood, and outcrossed to AB animals. For transient scatter labeling of Hb neurons, Tg[cfos:gal4vp16]s1019t (Hb:Gal) embryos were injected with a Tol2 construct containing an upstream activating sequence (UAS) upstream of either green fluorescent protein fused to Ulk2 (gfp:ulk2) or a CaaX motif fused to GFP (memGFP) at two- to eight-cell stages with the following transmission rates: GFP:Ulk2, 1–2%; memGFP, 30–50%. Larvae with small clones of labeled Hb neurons were selected for imaging and analysis.
Mutagenesis.
The kctd12.1vu442 mutant was generated by viral insertion that interrupts the kctd12.1 coding sequence within the N-terminal oligomerization domain. Homozygous kctd12.1vu442 mutants are negative for both mRNA by in situ hybridization and protein by immunofluorescence.
The kctd12.2fh312 mutant was generated by N-ethyl-N-nitrosourea (ENU) treatment and isolated by TILLING (Draper et al., 2004). This line carries the mutation L74*. Homozygous kctd12.2fh312 mutants are negative for protein by immunofluorescence.
Immunofluorescence.
Samples for whole-mount immunohistochemistry were fixed at 96 hpf overnight at room temperature in Prefer fixative (Anatech) and processed as previously described (Snelson et al., 2008) but without proteinase K permeabilization. Primary antibodies were incubated at the following concentrations: rabbit anti-Kctd12.1 (1:500) (Gamse et al., 2005), mouse anti-acetylated tubulin (Sigma-Aldrich) (1:500), mouse anti-Myc (Calbiochem) (1:500), rabbit anti-Myc (Sigma-Aldrich) (1:500), and mouse anti-GFP (Invitrogen) (1:500). Primary antibodies were detected using goat anti-mouse or goat anti-rabbit secondary antibodies conjugated to either Alexa 488 or Alexa 568 fluorophores (Invitrogen) (1:300). To visualize cell nuclei, samples were incubated with ToPro 3 (Invitrogen; 1:1000). Samples were then cleared in glycerol and imaged with an LSM510 META (Zeiss) confocal microscope with a 40×/1.30 Plan NEOFLUAR oil-immersion objective. Z-stacks of the Hb were taken at 1 μm intervals and extended from the dorsal surface of the larva to the point at which afferents within the stria medullaris enter the Hb, a distance of 60–75 μm. All images were processed using Volocity (Improvision).
Volumetric quantification.
To accurately measure neuropil volume in each Hb subnucleus, confocal projections from the dorsal surface of the embryo to the base of the dorsal habenulae in whole-mount larvae stained with antibodies against acetylated tubulin were processed using Volocity (Improvision) software. Each Hb subnucleus (left and right, medial and lateral) was individually cropped along morphologically defined borders. Neuropil volumes were selected using the Intensity Threshold tool in the Volocity measurement software. This tool returns measurements for all individual noncontinuous objects, but only the largest continuous volume visually confirmed to correspond to Hb neuropil was selected for further measurement.
To estimate the average dendrite volume per Hb neuron, confocal images of transient scatter-labeled Hb:Gal>memGFP were processed using Volocity (Improvision) software. Regions containing GFP+ clones were cropped and the Intensity Threshold tool was used to determine the continuous volume of GFP signal. These measurements (Vtotal) represent the volume of all dendrites and the soma of labeled cells, but because Hb neurons are unipolar (Bianco et al., 2008), signal from Hb axons is not included. Estimates for average soma volume (Vsom avg = 268.9 ± 7.29 μm3/soma; N = 26) were made by carefully outlining isolated soma labeled with memGFP and measuring their volume with the Intensity Threshold tool. With these measurements in hand, the dendrite volume per cell (Vden) was estimated as follows: Vden = [Vtotal − (Vsom avg*N)]/N, where N is the number of neurons in the labeled clone.
Statistical analysis consisted of two-tailed Student's t tests using Excel (Microsoft).
In situ hybridization.
Samples for whole-mount in situ hybridization were fixed overnight at 4°C in 4% paraformaldehyde and then dehydrated in methanol at −20°C. Samples were processed as previously described (Thisse and Thisse, 2008). For colorimetric precipitate, samples were developed in a solution of NBT (4-nitroblue tetrazolium) and BCIP (5-bromo-4-chloro-3-indolyl-phosphate). Bright-field images of glycerol-cleared samples were captured with a Leica 6000M compound microscope. When combining fluorescent precipitate with immunofluorescent labeling, samples were developed using Fast Red TR/Naphthol (Sigma-Aldrich) and subsequently processed for immunofluorescence.
Yeast two-hybrid.
Potential Kctd12.1 interactors were screened using a commercially available human fetal brain cDNA library (Clontech) fused to the Gal4 activation domain pretransformed into Y187 α-type yeast. A plasmid containing human Kctd12 fused to the Gal4 DNA binding domain was transformed into AH109 α-type yeast. Mating of yeast strains and isolation of interaction-positive colonies were performed according to manufacturer's instructions. Subsequent yeast two-hybrid assays were performed by cotransformation of DNA binding and activation domain fusion plasmids and plated on interaction-selective media (−ADE, −HIS). The zebrafish homolog of Ulk2 was cloned from cDNA using the following primers: forward (F), 5′-ATGGAGACGGTGGGAGATTT-3′, and reverse (R), 5′-TTCGTACAGGGTGACGGTG-3′, and tested in subsequent yeast two-hybrid assays for interaction with zebrafish Kctd12.1.
Coimmunoprecipitation.
MYC:Ulk2 and HA:Kctd12.1 fusion proteins were generated in vitro by coupled transcription and translation using the TNT quick coupled transcription translation kit (Promega) according to manufacturer's instructions. Fusion proteins were then incubated to allow binding without further purification. Fusion protein mixtures were then incubated with antibodies against either Myc or HA epitope tags and subsequently bound to protein A-coated Sepharose beads (Pierce). After extensive washing, complexes were released from beads by incubation at 90°C in SDS buffer. Samples were then detected using standard Western blotting on a 4–20% gradient polyacrylamide gel.
Morpholino knockdown of Ulk2.
Morpholino antisense oligonucleotides (Gene Tools) were designed to hinder Ulk2 translation by binding to the start site (ulk2 MOATG, 5′-ATTCAAAATCTCCCACCGTCTCCAT) or to a splice acceptor site at the beginning of exon 7 (ulk2 MOspl, 5′-TCGGCTGTTAAACAAAGAGAGCGCC) resulting in deletion of exon 7 and a frameshift-induced stop codon in exon 8. Morpholinos were resuspended in distilled, deionized water and stored at room temperature. Morpholinos were pressure injected into the yolk of one-cell stage embryos.
Semiquantitative RT-PCR.
RT-PCRs were performed on total RNA isolated from 72 hpf embryos with Trizol (Invitrogen). Reverse transcription was performed with random hexameric primers, followed by PCR amplification using primers to amplify sequence from β-actin (F, 5′-CCATGGATGGGAAAATCGCTGC-3′; R, 5′-GTCACACCATCACCAGAGTCC-3′), ulk2 (F, 5′-CCTTAACAGCAAGGGGATCA-3′; R, 5′-ATGCTCCACAGGTCAGCTTT-3′), or kctd12.1 (F, 5′-CGAACTTAACGTGGGAGGAC-3′; R, 5′-CGTTCAATGTGTCTCCAAACACCTC-3′). Band intensity quantification was performed with Quantity One (Bio-Rad) software.
Overexpression of Ulk2.
ulk2 mRNA was transcribed in vitro using the mMessage mMachine transcription kit (Ambion) and pressure injected into one-cell stage embryos.
Results
Quantitative description of Hb neuropil asymmetry
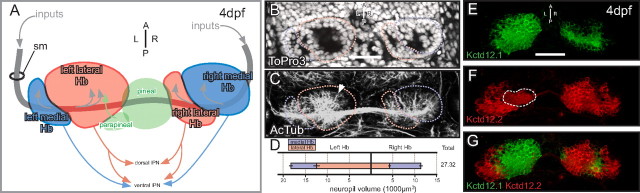
Although asymmetric neurite development has been described qualitatively in several reports (Concha et al., 2000, 2003; Bianco et al., 2008), we developed metrics to quantitatively measure Hb asymmetry. In the zebrafish epithalamus (schematized in Fig. 1A), the L–R asymmetric size ratio of medial and lateral habenular subnuclei can be distinguished based on soma distribution (Fig. 1B). During late development, the lateral Hb subnuclei shift dorsal and lateral to the medial subnuclei. Thus, the designation of “medial” and “lateral” Hb subnuclei refers to their relative positions in the adult brain, which is the opposite of their positions in the 96 hpf larva. Antibodies against acetylated tubulin mark all axons and dendrites, collectively referred to as neuropil (Fig. 1C). Distribution of neuropil volume is robustly asymmetric in the zebrafish habenulae, with the greatest volume of neuropil contained in a large soma-free region at the core of the left lateral subnucleus (Fig. 1C, arrow). To quantitatively describe Hb neurite development, we used image analysis software (Volocity) to precisely determine the neuropil volume in each individual subnucleus. The left Hb contains significantly more neuropil than the right (p = 1.34E-7; n = 12), owing largely to the contribution of the left lateral subnucleus.
Figure 1.
Asymmetry in the 96 hpf zebrafish habenular nuclei. A, The paired Hb receive input via the stria medullaris (sm) and send efferents to the midbrain target, the interpeduncular nucleus (IPN). The asymmetric parapineal organ innervates the left lateral subnucleus (green arrow) and is instrumental in establishing Hb laterality. Each subnucleus is asymmetrically subdivided into medial (blue) and lateral (red) subnuclei, with the lateral subnucleus much larger on the left and the medial subnucleus larger on the right. The nomenclature distinguishing the medial from lateral subnuclei is based on their position in the adult brain, which is the opposite of their position in the 96 hpf larvae shown here. B, An optical slice through a ToPro3 (nuclear marker)-stained larvae demonstrates asymmetrical subnucleus organization. C, The soma-free regions inside each Hb subnucleus are filled with neuronal processes [acetylated tubulin (AcTub) immunofluorescence] including afferent axons and Hb dendrites. The greatest volume of neuropil is found in the large left lateral subnucleus (arrow). D, Volumetric quantification of neuropil volume in each Hb subnuclei. The left lateral subnucleus contains much more neuropil volume than the right, so the total neuropil volume is greater in the left Hb. E, Kctd12.1 protein (green) is primarily expressed in lateral Hb subnuclei. F, G, Kctd12.2 (red) is expressed in most neurons of the medial Hb subnuclei (white dashed line indicates signal from dendritic processes, not Kctd12.2-positive soma) (F) in a largely complementary pattern to Kctd12.1 (G). Scale bars, 50 μm.
Asymmetry in subnucleus and neuropil organization is tightly correlated with asymmetric expression of Kctd12 proteins. Kctd12.1 (Fig. 1E) expression is restricted to neurons of the lateral subnuclei, while Kctd12.2 (Fig. 1F) is expressed primarily in medial Hb neurons in a pattern largely complementary to Kctd12.1 (Fig. 1G). The correlation between asymmetric morphological organization of the Hb and asymmetric expression of Kctd12 proteins lead us to investigate their role in Hb development.
Identification and confirmation of Ulk2 as Kctd12.1 interactor
To assess the function of Kctd12.1 protein in Hb neurons, we used the yeast two-hybrid assay to screen a brain cDNA library to identify protein–protein interactions with Kctd12. Because a high-quality zebrafish brain cDNA library is not available, we used the single human homolog of Kctd12.1 and Kctd12.2, HhKCTD12, as bait (fused to the Gal4 DNA binding domain) and screened a commercially available human fetal brain cDNA library (Clontech). We isolated 66 interaction-positive clones after screening through ∼2.3 × 106 library clones. Interacting clones included AKIP1 (A-kinase-interacting protein 1), UBE2NL (ubiquitin-conjugating enzyme E2N-like), and several Golgins. Sequence corresponding to Ulk2 was isolated from three independent clones.
First, we wanted to confirm the interaction by testing the zebrafish homologs of Kctd12.1 and Ulk2. After cloning zebrafish Ulk2, we found that it is able to activate interaction-selection cassettes when cotransformed along with Kctd12.1 (Fig. 2A). To gain an initial understanding of structure–function relationships, we tested deletions of the two primary domains of Kctd12.1 for their ability to interact with full-length Ulk2. The N-terminal domain is thought to promote oligomerization of Kctd monomers (Dementieva et al., 2009), and the C-terminal domain has no described function. We found that deletion of either the N-terminal or C-terminal domain of Kctd12.1 abolishes all interaction with full-length Ulk2.
Figure 2.
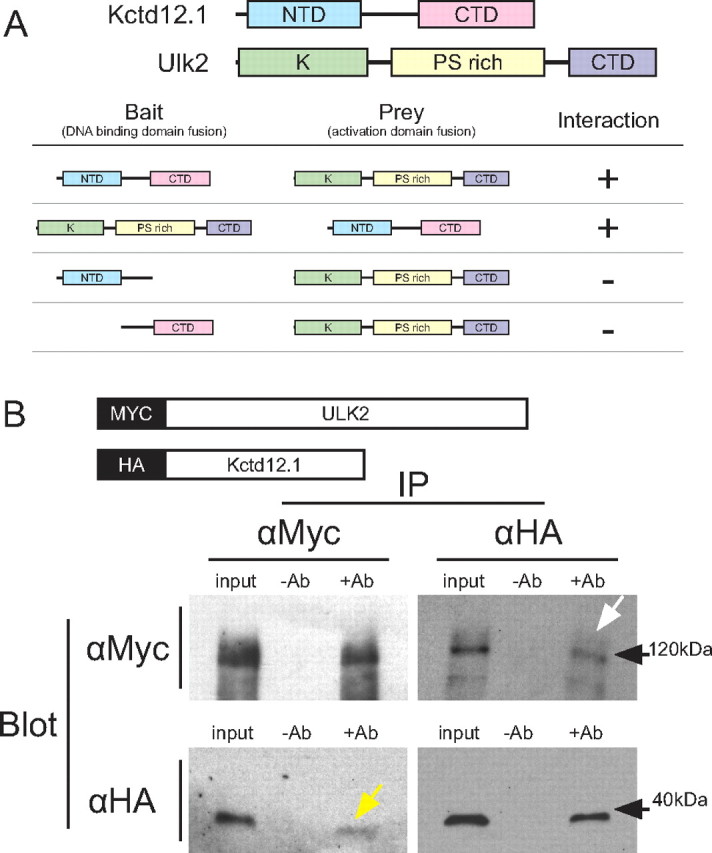
Kctd12.1 interacts with Ulk2. A, Kctd12.1 and Ulk2 interact in a yeast two-hybrid assay. Kctd12.1 is composed of two domains: an N-terminal domain that promotes oligomerization, and a C-terminal domain of undefined function. Ulk2 is composed of three domains: an N-terminal serine-threonine kinase domain (K), an internal proline-serine-rich region (PS rich), and a CTD involved in protein–protein interactions. Neither N- nor C-terminal deletions of Kctd12.1 are able to interact with full-length Ulk2. B, The interaction between Ulk2 and Kctd12.1 is confirmed by in vitro coimmunoprecipitation. After incubation of fusion proteins to allow binding, MYC:Ulk2 could be detected via Western blotting in samples immunoprecipitated with antibodies against HA (white arrow), and likewise, HA:Kctd12.1 was detected upon immunoprecipitation with antibodies against MYC (yellow arrow).
To independently verify this interaction, we generated HA:Kctd12.1 and MYC:Ulk2 fusion proteins by in vitro coupled transcription and translation (Fig. 2B). In coimmunoprecipitation experiments, we found that MYC:Ulk2 can be pulled down with HA:Kctd12.1 and that HA:Kctd12.1 can likewise be pulled down along with MYC:Ulk2.
Kctd12.1 and Ulk2 colocalize in Hb processes
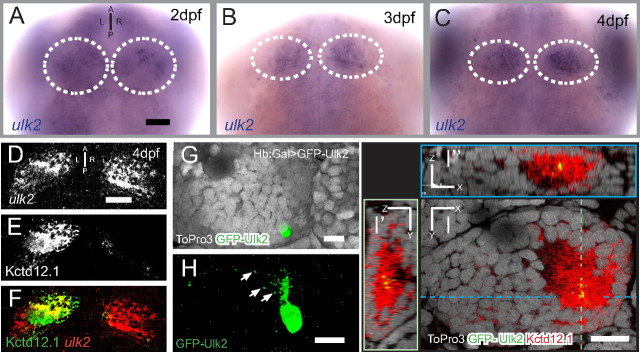
Kctd12 proteins and Ulk2 must be coexpressed in Hb neurons to be considered relevant to Hb neuropil development. By in situ hybridization (Fig. 3A–C), we detect ulk2 mRNA in most neurons of the CNS, consistent with previous reports of broad brain expression patterns in other organisms (Yan et al., 1999). Importantly, we find ulk2 mRNA bilaterally enriched in Hb neurons as early as 48 hpf, and this expression continues until at least 96 hpf. A combination of fluorescent in situ hybridization for ulk2 and immunofluorescence for Kctd12.1 protein confirms that, at the level of tissue organization, ulk2 and Kctd12.1 are coexpressed in neurons of the lateral subnucleus of the left Hb (Fig. 3D–F). We also analyzed the expression pattern of the related genes ulk1a and ulk1b. These two genes are expressed broadly in neurons of the brain, but unlike ulk2, ulk1a and ulk1b are not enriched in the Hb between 48 and 96 hpf (data not shown).
Figure 3.
Ulk2 is expressed in the Hb and colocalizes with Kctd12.1. A–C, ulk2 transcript is detected by in situ hybridization in many neurons of the CNS, but is enriched in the paired Hb (white outlines) at 48, 72, and 96 h post fertilization. D–F, A combination of fluorescent in situ for ulk2 transcript and Kctd12.1 immunofluorescence reveals coexpression in the left lateral Hb. G, H, Transgenic expression of GFP:Ulk2 fusion protein in a single neuron of the left lateral Hb shows localization of GFP:Ulk2 to puncta in Hb dendrites (arrows). I–I″, GFP:Ulk2 colocalizes with Kctd12.1. The image in I is an optical section through the left Hb in G and H with medial to the right and dorsal to the top; dashed lines in I indicate the relative orientation of optical sections shown in I′ and I″. Scale bars: A, D, 50 μm; G–I, 15 μm.
We next wanted to determine the localization of Kctd12.1 and Ulk2 proteins at the subcellular level in Hb neurons. Ulk2 has previously been reported to localize to cytoplasmic puncta in neuronal processes (Zhou et al., 2007). Because no suitable antibody against zebrafish Ulk2 is available, we expressed a GFP:Ulk2 fusion protein in small numbers of Hb neurons by Gal4:UAS-based scatter labeling (Fig. 3G) in the Hb>Gal background. Consistent with previous reports, this fusion protein is present in a punctate pattern in dendritic (Fig. 3H, arrows) and axonal (data not shown) processes of Hb neurons. Counterstaining with antibodies against Kctd12.1 shows that GFP:Ulk2 and Kctd12.1 colocalize at the subcellular level (Fig. 3I) (N = 7). According to these findings, Ulk2 and Kctd12.1 are present together at a relevant developmental time to affect Hb process development.
Morpholino knockdown of Ulk2 inhibits elaboration of Hb neuropil
Based on previous reports of Ulk protein abrogation in developing neurons, we hypothesized that knockdown of Ulk2 in zebrafish would lead to inhibition of neurite elaboration in the developing habenulae. To test the role of Ulk2 in Hb neuron development, we adopted a knockdown strategy using antisense morpholino oligonucleotide (MO) injection. Although Ulk2 morphants have slightly reduced head size and body length (Fig. 4A,B), general neurogenesis is not perturbed, as both brain organization and axonal extension by motor neurons indicate no overall defects in the development of the CNS (Fig. 4A,B, insets). The slight morphological defects associated with morpholino injection can be rescued by coinjection of 150 pg of ulk2 mRNA (Fig. 4C).
Figure 4.
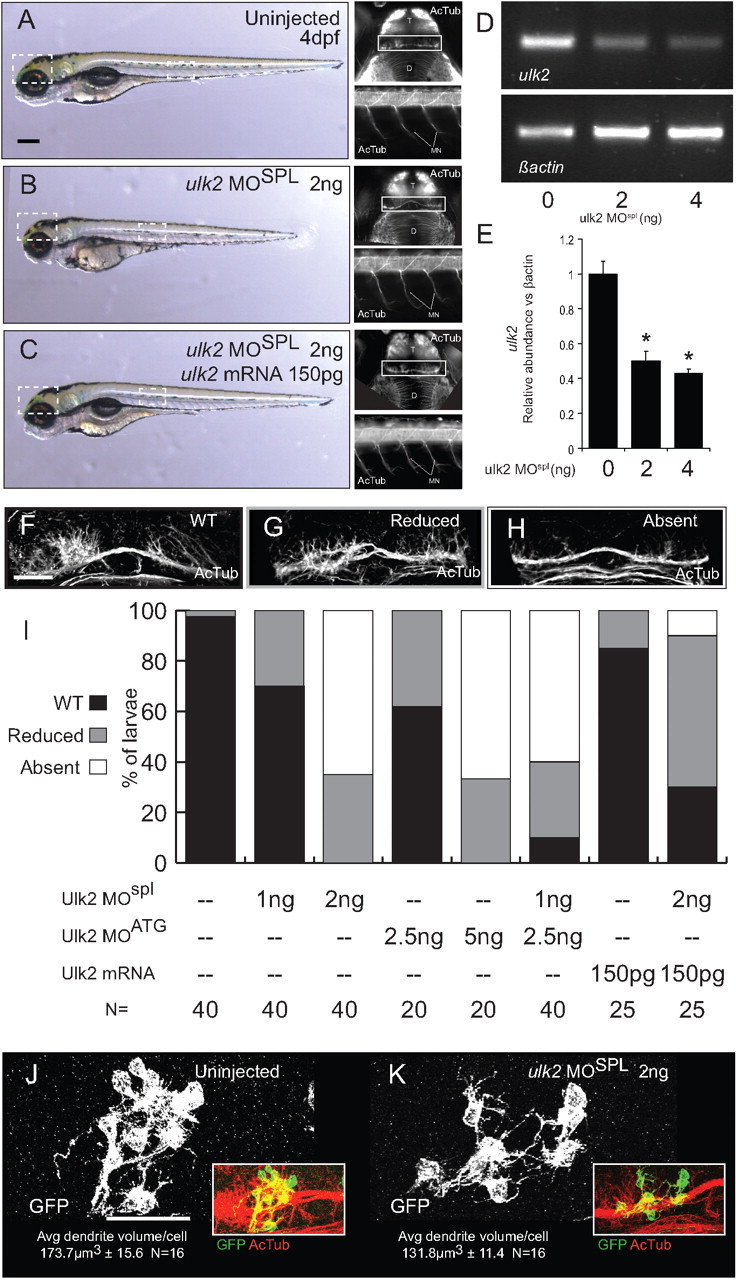
Antisense knockdown of Ulk2 inhibits the formation of Hb neuropil. A, B, Injection of 2 ng of ulk2MOspl at the one-cell stage produces a mild decrease in head size and body length at 4 dpf. The general organization of the central (top insets) and peripheral (bottom insets) nervous system as revealed by acetylated tubulin immunofluorescence is intact in morphant larvae (D, diencephalon; T, telencephalon; MN, motor neurons), but Hb neuropil development (white boxes) is disrupted. C, Injection of prespliced Ulk2 mRNA at the one-cell stage rescues all the phenotypes of Ulk2 morpholino treatment. D, The ulk2 transcript is depleted by morpholino injection. RT-PCR with ulk2 primers (top panel) shows reduction of ulk2 mRNA relative to β-actin (bottom panel). E, Quantification of band intensity in RT-PCR replicates (0 ng = 1 ± 0.07 A.U., 2 ng = 0.5 ± 0.05 A.U., 4 ng = 0.43 ± 0.02 A.U., N = 3). F–H, Morphant larvae can be assigned to one of three categories based on the degree of Hb neuropil elaboration. I, When injected with either 2 ng of ulk2MOspl or 5 ng of ulk2MOATG, the formation of neuropil in the Hb is inhibited in most larvae (white bars), a phenotype not observed in injections of half-maximal dosage (1 and 2.5 ng, respectively). Hb neuropil inhibition was also observed when half-maximal doses of each MO were combined, indicating that both morpholinos target the same transcript. Injection of 150 pg of in vitro-transcribed ulk2 mRNA was able to rescue Hb neurite formation when coinjected with 2 ng of ulk2MOspl. J, K, Mosaic scatter labeling of Hb neurons with memGFP, followed by average dendrite volume quantification highlights changes in average dendrite volume between uninjected (J) and ulk2MO-treated (K) larvae. Scale bars: A, 100 μm; F, J, 50 μm.
To assay the effectiveness of morpholino injection at depleting ulk2 mRNA, we performed RT-PCR using primers within either the ulk2 or β-actin coding region on groups of larvae injected with either 0, 2, or 4 ng of ulk2MOSPL and collected at 3 dpf (Fig. 4D). We found that 2 ng of ulk2MOSPL is sufficient to knock down ulk2 mRNA to ∼50% of the levels found in WT larvae (2 ng injection, 50.4 ± 0.05% of WT level; N = 3), probably through the process of nonsense-mediated decay.
To gauge habenular defects in populations of morphant larvae, we separated larvae into three groups based on the morphology of Hb neuropil: WT (Fig. 4F), reduced (Fig. 4G), and absent (Fig. 4H) for population frequency analysis. Samples were categorized without knowledge of treatment group.
Following treatment with either of two morpholinos complementary to different regions of the ulk2 mRNA (5 ng of ulk2MOATG or 2 ng of ulk2MOSPL), Hb neuropil is absent (Fig. 4I, white bars) in many larvae (Spl: 65%, N = 40; ATG: 66.67%, N = 21). This effect was not elicited by injection of half-maximal doses of either morpholino (2.5 ng of ulk2MOATG or 1 ng of ulk2MOSPL, respectively), but coinjection of a mixture of suboptimal morpholino concentrations (2.5 ng of ulk2MOATG plus 1 ng of ulk2MOSPL) was able to effectively inhibit development of Hb neuropil in most larvae (60%; N = 20), suggesting that both morpholinos target the same transcript.
To confirm that targeting of the ulk2 transcript is responsible for the Hb neuropil phenotype, we injected prespliced ulk2 mRNA along with 2 ng of ulk2MOSPL. We found that injection of 150 pg of ulk2 mRNA is able to rescue neuropil development in many 2 ng ulk2MOSPL larvae.
To discount the possibility that reduction in Hb neuropil in Ulk2 morphants is caused by a loss of Hb neurons, we counted Kctd12.1-positive neurons in larvae treated with 2 ng of ulk2MOSPL. Morphants and uninjected larvae do not have significantly different numbers of Kctd12.1-positive neurons (uninjected, 260 ± 7.76; 2 ng of ulk2MOSPL, 234 ± 8.53; p = 0.12; n = 12), indicating that loss of Hb neurons is not responsible for the morphant phenotype (data not shown).
Because measurements of neuropil volume unavoidably include signal from the axons of Hb afferents, we verified that Hb dendrites were specifically affected by Ulk2 knockdown. Using membrane-localized GFP (memGFP) to label small clones of cells in control and ulk2MOSPL-injected embryos, we measured average dendrite volume per Hb neuron (Fig. 4J,K) (see Materials and Methods). Average dendritic volume was significantly reduced in ulk2 morphants (uninjected: 173.7 ± 15.6 μm3, N = 16; 2 ng of ulk2MOSPL: 131.8 ± 11.4 μm3, N = 16; p = 0.028).
Overexpression of Kctd12 proteins inhibits Hb neuropil development
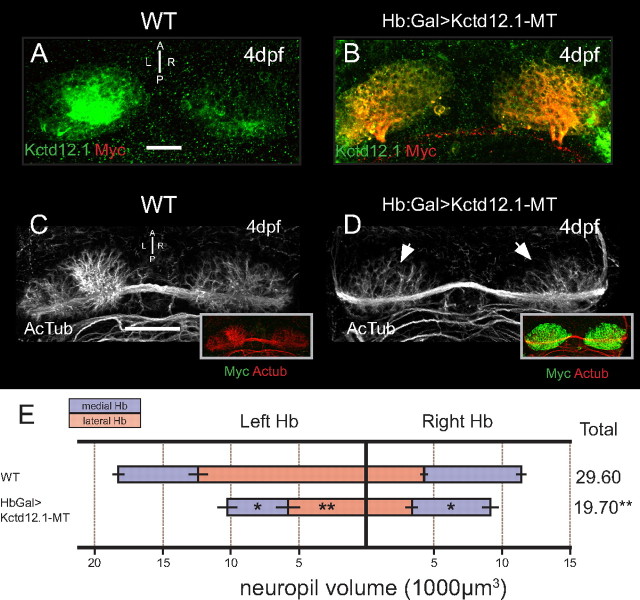
To investigate the impact of Kctd12.1 expression on developing Hb neurons, we used the Gal4/UAS expression system to overexpress Kctd12.1 in the Hb of larvae. Hb:Gal drives expression of Gal4 transcription factor bilaterally in almost all Hb neurons by 4 dpf (Scott et al., 2007). We generated response lines with transgenes containing a UAS element upstream of a Kctd12.1:Myc tag (MT) fusion protein (Tg[UAS:kctd12.1-mt]). In Hb:Gal>Kctd12.1-MT larvae, we detected bilateral kctd12.1 mRNA (data not shown) and overlapping Myc and Kctd12.1 immunofluorescence (Fig. 5B). Semiquantitative RT-PCR revealed a doubling in the level of kctd12.1 mRNA present in double-transgenic samples (WT, 1.0 ± 0.11 A.U.; Hb:Gal>Kctd12.1-MT, 2.11 ± 0.17 A.U.; N = 3). At 4 dpf, fusion protein was present in almost all Hb neurons. The pattern of Hb neuropil extension in Kctd12.1 overexpression larvae was then assessed by acetylated tubulin immunofluorescence.
Figure 5.
Overexpression of Kctd12.1 inhibits elaboration of Hb neuropil. A, Kctd12.1 (green) is normally expressed only in Hb neurons of the lateral subnuclei. B, In Hb:Gal>Kctd12.1-MT larvae, Kctd12.1-MT fusion protein (red) is expressed at high levels in nearly all Hb neurons. C, D, WT larvae have an elaborate network of neuropil that segregates within each Hb subnucleus (C), but the presence of high levels of ectopic Kctd12.1-MT fusion inhibits the elaboration of Hb neuropil (D). E, Volumetric quantification of Hb neuropil reduction. Overexpression of Kctd12.1-MT causes significant reduction of total Hb neuropil volume compared with WT (19,693 ± 1664 vs 29,602 ± 1426 μm3; p = 0.002; n = 16). All subnuclei are significantly affected with the exception of the right lateral subnucleus (asterisks indicate statistical difference compared with WT). Scale bars, 50 μm. *p < 0.05, **p < 0.01, two-tailed t test.
When Kctd12.1 is overexpressed in all Hb neurons, the large region of dense neuropil in the left Hb is dramatically reduced (Fig. 5C,D). Larvae with normal levels of Kctd12.1 expression have nearly twice the process volume in the left Hb compared with the right, but in Hb:Gal>Kctd12.1:MT larvae, only a low volume of neuropil is detected in both habenulae (Fig. 5D, arrows). Quantification of neuropil volumes in each subnucleus revealed a significant decrease in neuronal processes in the left lateral (p = 0.0013; n = 16) and both left (p = 0.043; n = 16) and right (p = 0.046; n = 16) medial subnuclei.
Hb neuropil defects are not due to an effect of the Myc epitope tag, as overexpression of untagged Kctd12.1 produces an identical phenotype (data not shown). This phenotype is also not due to parapineal asymmetry defects, as parapineal placement and morphology are unaffected (data not shown).
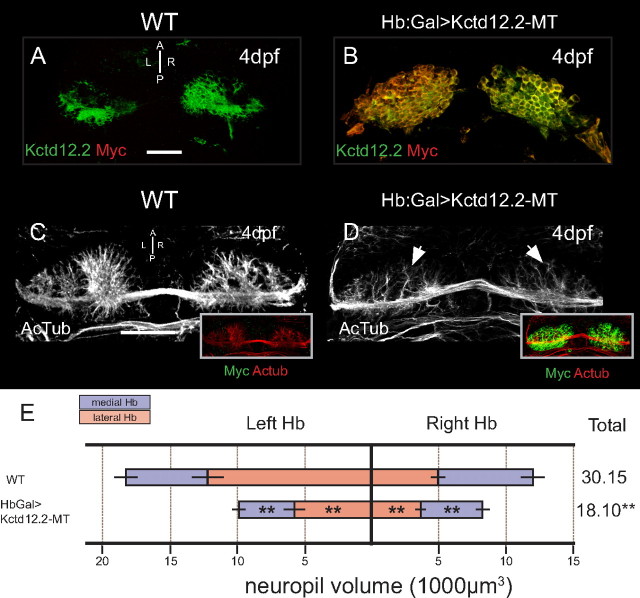
We also tested the effect of overexpression of Kctd12.2 (Fig. 6). Based on the high level of conservation between Kctd12.1 and Kctd12.2, we hypothesized that Kctd12.2-MT fusion protein would similarly inhibit the extension of Hb neuropil when overexpressed. Indeed, neuropil volume is dramatically reduced in larvae that overexpress Kctd12.2-MT (Fig. 6D, arrows). Volumetric quantification of neuropil in Hb:Gal>Kctd12.2-MT reveals significant reductions in neuropil volume in all subnuclei when compared with WT larvae (left medial: p = 1.25E-5, n = 16; left lateral: p = 8.21E-8, n = 16; right lateral: p = 1.8E-4, n = 16; right medial: p = 1.8E-6, n = 16).
Figure 6.
Overexpression of Kctd12.2 inhibits elaboration of Hb neuropil. A, Kctd12.2 (green) is normally expressed in neurons of the medial subnuclei. B, In Hb:Gal>Kctd12.2-MT larvae, expression of Kctd12.2-MT fusion protein is driven at high levels in nearly all Hb neurons. C, D, WT larvae have an elaborate network of neuropil that segregates within each Hb subnucleus (C), but the presence of high levels of ectopic Kctd12.2-MT fusion inhibits the elaboration of Hb neuropil (D). E, Volumetric quantification of Hb neuropil reduction. Overexpression of Kctd12.2-MT causes a significant reduction of total Hb neuropil volume compared with WT (18,103 ± 789 vs 30,147 ± 2588 μm3; p = 7.7E-8; n = 16). All subnuclei are significantly affected (asterisks indicate statistical difference compared with WT). Scale bars, 50 μm. *p < 0.05, **p < 0.01, two-tailed t test.
Mutation of Kctd12 gene leads to excess Hb neuropil
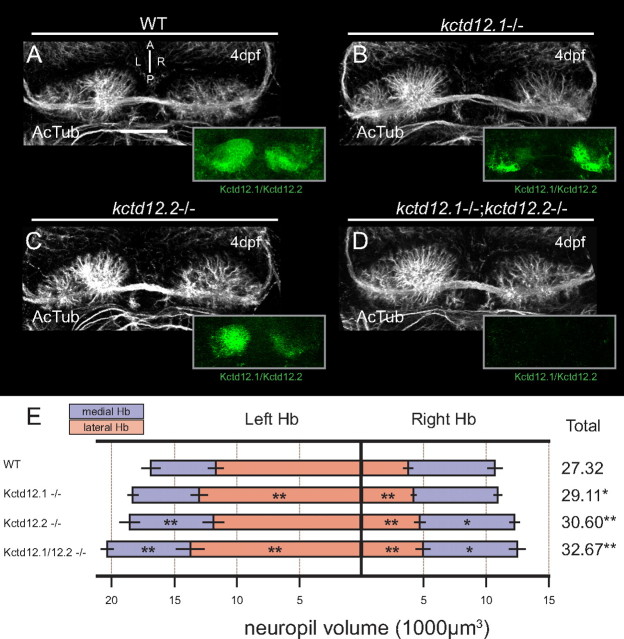
Based on the dramatic neuropil reduction when Hb neurons overexpress Kctd12 proteins, we hypothesized that loss of Kctd12 expression through mutation may lead to an excess of Hb neuropil. To test this hypothesis, we made use of a Kctd12.1-null mutant allele, kctd12.1vu442, which carries a large viral insertion interrupting the kctd12.1 locus, and a Kctd12.2 mutant allele, kctd12.2fh312, which carries an ENU-induced stop codon (L74*). Both mutations are null, with no protein detected by immunofluorescence staining in homozygous mutants (Fig. 7A–D, insets), and are homozygous viable. We analyzed neuropil volume in single- and double-homozygous mutant larvae to uncover any subtle differences in neurite volume extension in the absence of Kctd12 proteins. In kctd12.1vu442 homozygotes, we detected a significant increase in neuropil volume in both the left (p = 0.0054; N = 16) and right (p = 0.0039; N = 16) lateral Hb subnuclei, compared with wild type (Fig. 7E). The affected subnuclei are those that express Kctd12.1 in wild-type larvae (Fig. 1E–G). Similarly, in homozygous kctd12.2fh312 mutants, quantification reveals excess Hb neuropil in the left medial, right lateral, and right medial subnuclei (p = 0.0023, n = 16; p = 0.000051, n = 16; p = 0.048, n = 16, respectively) (Fig. 7E). Indeed, the only subnucleus unaffected by loss of Kctd12.2 is the left lateral subnucleus, from which Kctd12.2 is normally excluded. Comparison of neuropil between kctd12.1vu442 and kctd12.2fh312 single mutants reveals a significantly greater increase in neuropil in kctd12.2fh312 mutants (p = 0.00912; N = 16).
Figure 7.
Mutation of Kctd12 proteins leads to excess Hb neuropil. A, By 4 dpf, WT larvae display elaborate extension of neuropil in both Hb and express both Kctd12.1 and 12.2 (green, inset). B, The kctd12.1 coding sequence is disrupted by a large viral insertion in kctd12.1vu442 mutants (note lack of Kctd12.1 staining in lateral subnuclei in inset). Hb neuropil in Kctd12.1-negative larvae is slightly expanded in lateral subnuclei. C, An ENU-induced stop codon in the coding sequence of kctd12.2 in kctd12.2fh312 mutants leads to loss of Kctd12.2 protein expression (note lack of Kctd12.2 staining in medial subnuclei in inset). Kctd12.2-negative larvae also display excess elaboration of Hb neuropil, particularly in the medial subnuclei. D, kctd12.1vu442; kctd12.2fh312 double mutants are negative for both Kctd12 proteins (green, inset), and Hb neuropil is expanded in both the lateral and medial subnuclei. E, Volumetric quantification of neuropil expansion in kctd12 mutants. Neuropil expansion is restricted to lateral subnuclei in Kctd12.1-negative larvae, consistent with the expression pattern of Kctd12.1. Neuropil expansion in Kctd12.2-negative larvae affects medial subnuclei as well as the right lateral subnucleus. Double mutant larvae display neuropil expansion in all subnuclei (asterisks indicate statistical difference compared with WT). Scale bars, 50 μm. *p < 0.05, **p < 0.01, two-tailed test.
Consistent with the hypothesis that Kctd12.1 and Kctd12.2 proteins negatively regulate neuropil formation in different subnuclei, we found neuropil volumes in kctd12.1vu442; kctd12.2fh312 double mutants to be additively greater than either single mutant alone, and this increase in neuropil volume affects all Hb subnuclei (Fig. 7E).
Overexpression of both Kctd12.1 and Ulk2 can restore normal habenular development
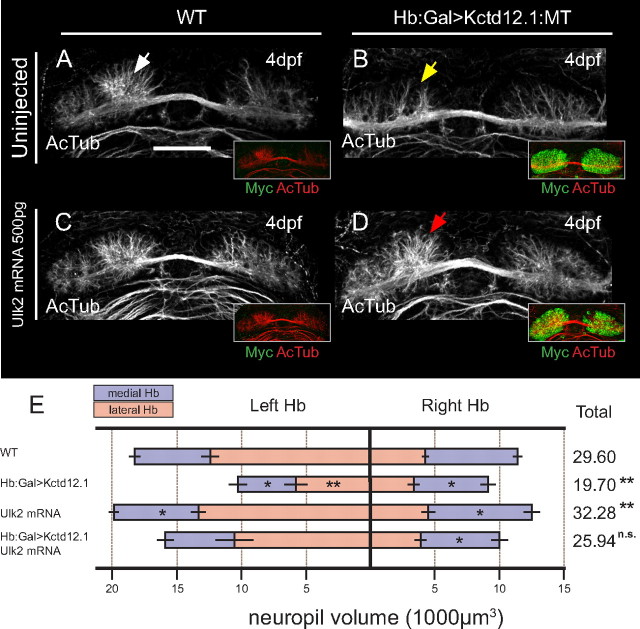
If Kctd12 proteins and Ulk2 operate in the same pathway, as their interaction suggests, we reasoned that overexpression of Ulk2 should be able to rescue Hb neuropil defects in Hb:Gal>Kctd12.1:MT larvae by restoration of correct relative levels of both proteins in Hb neurons. To test this hypothesis, we overexpressed Ulk2 by exogenous mRNA injection in the background of Kctd12.1 overexpression (Fig. 8). As in previous experiments, overexpression of Kctd12.1-MT leads to dramatic loss of Hb neuropil volume compared with controls (Fig. 8A,B). Conversely, we find that injection of 500 pg of ulk2 mRNA is able to increase Hb neuropil volumes, particularly in medial subnuclei (Fig. 8C), which supports the idea that Ulk2 acts to promote the extension of neurites.
Figure 8.
Overexpression of Ulk2 by mRNA injection rescues neuropil reduction caused by Kctd12.1 overexpression. A–C, The large volume of neuropil present in the left Hb of wild-type larvae (white arrow) (A) is reduced (yellow arrow) by overexpression of Kctd12.1-MT (green in insets) (B) and is slightly increased by injection of 500 pg of ulk2 mRNA alone (C). D, Injection of 500 pg of ulk2 mRNA restores relatively normal neuropil volume in the left Hb (red arrow) of larvae overexpressing Kctd12.1-MT. E, Volumetric quantification of neuropil phenotype. Overexpression of Kctd12.1-MT significantly reduces neuropil volume in the left lateral and both medial Hb subnuclei. Injection of ulk2 mRNA slightly increases neuropil volume in the medial subnuclei. Injection of ulk2 mRNA into Kctd12.1-MT overexpression larvae restores wild-type neuropil volumes to all subnuclei with the exception of the right medial subnucleus (asterisks indicate statistical difference compared with WT). Scale bar, 50 μm. *p < 0.05, **p < 0.01, two-tailed test.
When exogenous ulk2 mRNA is administered to Hb:Gal>Kctd12.1:MT embryos, we find that total Hb neuropil volume is restored to levels statistically indistinguishable from WT (p = 0.072; n = 8) (Fig. 8D). In these experiments, overexpression of Kctd12.1 inhibits endogenous Ulk2 activity to an inappropriate degree, but providing high levels of exogenous Ulk2 can overcome this inhibition and reestablish normal Ulk2-dependent dendrite outgrowth.
Ulk2 depletion is epistatic to Kctd12.1 mutation
Based on reported roles of Ulk2 as a positive regulator of neurite outgrowth and the excessive neurites in kctd12 mutations, we hypothesized that Kctd12 proteins negatively regulate the activity of Ulk2 kinase, which positively regulates neuropil formation. To test this hypothesis, we examined the phenotype of kctd12.1vu442 treated with an Ulk2 morpholino (Fig. 9). If Kctd12 activity is upstream of Ulk2, we expect the Hb neuropil of mutant/morphant larvae to resemble that of Ulk2 knockdown alone. Mutation of Kctd12.1 in the context of Ulk2 morpholino treatment (2 ng of ulk2 MOspl) results in severely reduced neuropil elaboration relative to WT or mutation of Kctd12.1 alone (Fig. 9A–E).
Figure 9.
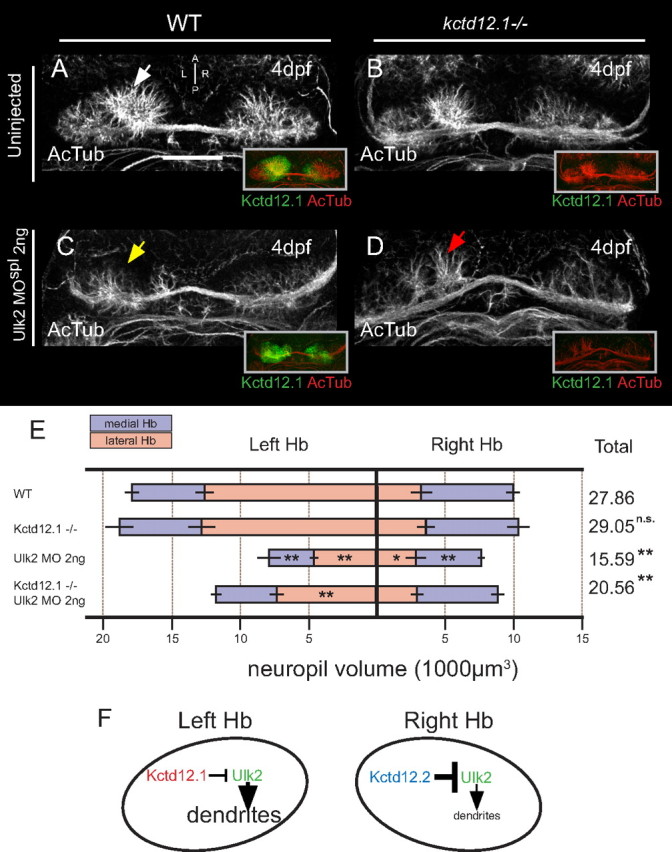
Ulk2 depletion is epistatic to Kctd12.1 mutation. A, B, Compared with normal Hb (A), neuropil development (white arrow) is slightly expanded in homozygous null kctd12.1 mutant animals [note absence of Kctd12.1 (green) in insets at right] (B). C, D, Reduced neuropil (yellow arrow) in the left Hb as a result of Ulk2 antisense depletion (C) also occurs (red arrow) when Ulk2 is depleted in a Kctd12.1 homozygous mutant, indicating that Ulk2 depletion is largely epistatic to mutation of Kctd12.1 (D). E, Volumetric quantification of Hb neuropil phenotype. Total neuropil volume is significantly reduced following injection of ulk2 MOspl in either a wild-type or Kctd12.1 mutant background (asterisks indicate statistical difference compared with WT). Scale bar, 50 μm. *p < 0.05, **p < 0.01, two-tailed test. F, Model of proposed regulatory system resulting in asymmetric habenular neuropil extension. Ulk2 kinase acts bilaterally to promote elaboration of Hb neuropil but is negatively regulated by Kctd12 proteins. Asymmetry in neuropil arises from the greater relative potency of Kctd12.2 acting in medial subnuclei.
Discussion
The results of the present study indicate that Ulk2 and Kctd12 proteins coordinately regulate the development of Hb neuronal processes, specifically that Ulk2 promotes neuropil outgrowth and that Kctd12.1 is a negative regulator of Ulk2 activity. We have shown that (1) Kctd12.1 physically interacts with Ulk2, (2) Ulk2 knockdown and Kctd12 overexpression can both inhibit neuropil formation, (3) loss of Kctd12 expression and overexpression of Ulk2 lead to enhanced elaboration of Hb neuropil, (4) Ulk2 and Kctd12 proteins are in the same pathway, and (5) Ulk2 depletion is epistatic to Kctd12 mutation. These novel findings reveal a previously uncharacterized interaction and may open a new line of inquiry into the fine control of neurite extension in the developing vertebrate nervous system.
The Ulk2–Kctd12 interaction may define a novel Ulk regulatory system
The interaction between Kctd12.1 and Ulk2 is previously unreported. We find that it is conserved between humans and zebrafish; as fruit flies and nematodes also have Kctd12 and Ulk2 orthologs, it will be interesting to see whether this mechanism for regulating dendritogenesis is conserved in invertebrates.
The structure of Kctd proteins is still poorly understood. Our finding that Kctd12.1–Ulk2 interaction is abolished by deletion of either Kctd12.1 domain raises the possibility that either the Ulk2 binding site spans both domains, or that one domain is required for the correct coordination of the binding site in the other domain.
Ulk2 is thought to promote process extension through stimulation of early endosome trafficking at growing neurite tips. Ulk2 has been shown to increase early endosome formation by activation of the small GTPase Rab5 (Tomoda et al., 2004). In axons, early endosomes that contain activated growth factor receptors (e.g., TrkA bound to NGF) move in a retrograde fashion to the cell body, and this is thought to bring them close enough to the nucleus to allow intracellular signaling to affect transcription and axon extension (Delcroix et al., 2003). A similar process is thought to occur in dendrites, although the receptors and ligands involved remain to be identified (Satoh et al., 2008). We speculate that Kctd12 proteins could regulate this process at a number of steps, but the most likely scenarios involve either a regulation of Ulk2 activity by preventing autophosphorylation or a sequestration model in which Kctd12 proteins inhibit localization of Ulk2 proteins to early endosome signaling centers.
To our knowledge, this is the first reported whole-organism knockdown of Ulk2 in a vertebrate model system. Ulk2 is a highly conserved protein from yeast (Atg1) to humans and acts in autophagy as well as early endosome formation. Given the multiple roles of this protein, why do Ulk2 morphants have a relatively mild dendritic outgrowth phenotype in the Hb? First, it is likely that we are only reducing, not eliminating, Ulk2 function in our morpholino-treated embryos, as evidenced by RT-PCR quantification. Second, there is likely some redundancy with the closely related Ulk1a/1b proteins. The selective appearance of an Hb phenotype may be due to an elevated requirement for Ulk2 in Hb neurons, a conclusion supported by the relatively high levels of ulk2 transcripts found in these cells.
We have attempted to order the Kctd12/Ulk2 pathway via genetic epistasis. The phenotype of double mutant/morphants is similar to Ulk2 single morphants, and therefore the most parsimonious explanation is that Ulk2 is downstream of Kctd12.1. However, double mutant/morphants do exhibit some increased neuropil relative to Ulk2 morphants, likely because Ulk2 morpholino treatment cannot completely eliminate Ulk2 protein. As higher levels of morpholino are toxic to embryos (R. W. Taylor, unpublished observation), we cannot exclude the possibility that Kctd12.1 is downstream of Ulk2 until an ulk2-null mutation is isolated.
Additionally, although this study finds that Kctd12 proteins regulate Ulk2, it is yet unclear how this regulation is accomplished on a molecular scale. Kctd12 proteins are unlikely to regulate Ulk2 at the transcription or translation level, as Kctds are not considered transcription factors, nor are they known to be involved in ribosomal regulation. We think it most likely that by binding to Ulk2, Kctd12 proteins either inhibit the kinase activity of Ulk2 or alter the subcellular localization of Ulk2 such as to prevent their association with early endosomes.
Relative potency of Kctd12s as Ulk2 regulators may underlie habenular asymmetry
It initially appears counterintuitive that Hb neurons should express negative regulators of neuropil extension (Kctd12 proteins) given their elaborate dendritic processes. We propose that differential neurite extension among Hb neurons may be controlled by different potencies of Kctd12.1 and Kctd12.2 in the downregulation of Ulk2 activity. The bilateral expression of ulk2 mRNA suggests that an Ulk2-dependent process is active in all Hb neurons. Asymmetric Kctd12 expression overlaid on symmetric expression of the proneurite factor Ulk2 could explain how differential process extension occurs. This model predicts that Kctd12.2 more effectively downregulates Ulk2-dependent neuropil outgrowth than Kctd12.1, resulting in the characteristic asymmetric neuropil observed in the zebrafish habenulae (Fig. 9F). Our data support this model, as overexpression of Kctd12.2 but not Kctd12.1 is able to reduce neuropil volume in all subnuclei, and mutation of Kctd12.2 results in a greater neuropil overelaboration phenotype than mutation of Kctd12.1.
Since Kctd12.1 and 12.2 are closely related proteins, with ∼80% similarity at the amino acid level (Gamse et al., 2005), differential ability to affect neuropil formation in the habenular nuclei was unexpected. However, closer inspection of the protein sequences reveals 5 aa differences between Kctd12.1 and 12.2 in the C-terminal domain (CTD). As the CTD is essential for Kctd12.1 interaction with Ulk2, these differences may affect the strength of the interaction. Determination of CTD structure at high resolution is underway to identify the residues that contact Ulk2 and reveal the significance of amino acid differences between Kctd12.1 and 12.2.
Kctd12.1–Ulk2 interaction may intersect with GABAB receptor complexes
Kctd12 (the single mammalian homolog of zebrafish Kctd12.1/12.2) was recently shown to interact with the metabotropic GABAB G-protein-coupled receptor. This interaction affects ligand sensitivity, desensitization, and kinetics of this important regulator of synaptic transmission and signal propagation (Schwenk et al., 2010). This result can be interpreted in two possible ways. First, Kctd12 may have distinct roles during the life of a neuron. During embryonic development, Kctd12 may modulate Ulk2-dependent mechanisms of neurite outgrowth, and then regulate electrical activity in mature neurons by an independent interaction with GABAB receptors. Second, and more intriguingly, we can speculate that the ability of Kctd12 proteins to interact with both Ulk2 and GABAB receptors may reflect a role in GABA-mediated neurite outgrowth. During embryonic development, GABA is known to regulate axon and dendrite formation before synapse formation, by activation of GABA receptors, including GABAB receptors (Sernagor et al., 2010). By analogy with NGF/TrkA, internalization of GABA/GABAB receptor complexes into endosomes may affect their signaling properties. Indeed, GABAB receptors in cultured neurons are internalized and recycled via an endosomal pathway, which appears to be dendrite specific (González-Maeso et al., 2003; Grampp et al., 2008; Vargas et al., 2008). The presence of GABAB receptor-associated Kctd12 could prevent Ulk2-stimulated endocytosis of the receptor, or affect the recycling of GABAB receptor-containing endosomes to the cell surface versus targeting to the proteasome.
Footnotes
This work was supported by a Vanderbilt University Discovery Grant (J.T.G.) and by NIH Grants HG002995 (C.B.M.), GM53396 (D.J.K.), and HC054534 (J.T.G.). C.B.M. is an investigator with HHMI. We thank Erin Booton, Gena Gustin, and Qiang Guan for expert fish care.
References
- Aizawa H, Bianco IH, Hamaoka T, Miyashita T, Uemura O, Concha ML, Russell C, Wilson SW, Okamoto H. Laterotopic representation of left-right information onto the dorso-ventral axis of a zebrafish midbrain target nucleus. Curr Biol. 2005;15:238–243. doi: 10.1016/j.cub.2005.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa H, Goto M, Sato T, Okamoto H. Temporally regulated asymmetric neurogenesis causes left-right difference in the zebrafish habenular structures. Dev Cell. 2007;12:87–98. doi: 10.1016/j.devcel.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Bayón Y, Trinidad AG, de la Puerta ML, Del Carmen Rodríguez M, Bogetz J, Rojas A, De Pereda JM, Rahmouni S, Williams S, Matsuzawa S, Reed JC, Crespo MS, Mustelin T, Alonso A. KCTD5, a putative substrate adaptor for cullin3 ubiquitin ligases. FEBS J. 2008;275:3900–3910. doi: 10.1111/j.1742-4658.2008.06537.x. [DOI] [PubMed] [Google Scholar]
- Bianco IH, Carl M, Russell C, Clarke JD, Wilson SW. Brain asymmetry is encoded at the level of axon terminal morphology. Neural Dev. 2008;3:9. doi: 10.1186/1749-8104-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carl M, Bianco IH, Bajoghli B, Aghaallaei N, Czerny T, Wilson SW. Wnt/Axin1/beta-catenin signaling regulates asymmetric nodal activation, elaboration, and concordance of CNS asymmetries. Neuron. 2007;55:393–405. doi: 10.1016/j.neuron.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha ML, Wilson SW. Asymmetry in the epithalamus of vertebrates. J Anat. 2001;199:63–84. doi: 10.1046/j.1469-7580.2001.19910063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha ML, Burdine RD, Russell C, Schier AF, Wilson SW. A nodal signaling pathway regulates the laterality of neuroanatomical asymmetries in the zebrafish forebrain. Neuron. 2000;28:399–409. doi: 10.1016/s0896-6273(00)00120-3. [DOI] [PubMed] [Google Scholar]
- Concha ML, Russell C, Regan JC, Tawk M, Sidi S, Gilmour DT, Kapsimali M, Sumoy L, Goldstone K, Amaya E, Kimelman D, Nicolson T, Gründer S, Gomperts M, Clarke JD, Wilson SW. Local tissue interactions across the dorsal midline of the forebrain establish CNS laterality. Neuron. 2003;39:423–438. doi: 10.1016/s0896-6273(03)00437-9. [DOI] [PubMed] [Google Scholar]
- Delcroix JD, Valletta JS, Wu C, Hunt SJ, Kowal AS, Mobley WC. NGF signaling in sensory neurons: evidence that early endosomes carry NGF retrograde signals. Neuron. 2003;39:69–84. doi: 10.1016/s0896-6273(03)00397-0. [DOI] [PubMed] [Google Scholar]
- Dementieva IS, Tereshko V, McCrossan ZA, Solomaha E, Araki D, Xu C, Grigorieff N, Goldstein SA. Pentameric assembly of potassium channel tetramerization domain-containing protein 5. J Mol Biol. 2009;387:175–191. doi: 10.1016/j.jmb.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, McCallum CM, Stout JL, Slade AJ, Moens CB. A high-throughput method for identifying N-ethyl-N-nitrosourea (ENU)-induced point mutations in zebrafish. Methods Cell Biol. 2004;77:91–112. doi: 10.1016/s0091-679x(04)77005-3. [DOI] [PubMed] [Google Scholar]
- Gamse JT, Thisse C, Thisse B, Halpern ME. The parapineal mediates left-right asymmetry in the zebrafish diencephalon. Development. 2003;130:1059–1068. doi: 10.1242/dev.00270. [DOI] [PubMed] [Google Scholar]
- Gamse JT, Kuan YS, Macurak M, Brösamle C, Thisse B, Thisse C, Halpern ME. Directional asymmetry of the zebrafish epithalamus guides dorsoventral innervation of the midbrain target. Development. 2005;132:4869–4881. doi: 10.1242/dev.02046. [DOI] [PubMed] [Google Scholar]
- González-Maeso J, Wise A, Green A, Koenig JA. Agonist-induced desensitization and endocytosis of heterodimeric GABAB receptors in CHO-K1 cells. Eur J Pharmacol. 2003;481:15–23. doi: 10.1016/j.ejphar.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Grampp T, Notz V, Broll I, Fischer N, Benke D. Constitutive, agonist-accelerated, recycling and lysosomal degradation of GABAB receptors in cortical neurons. Mol Cell Neurosci. 2008;39:628–637. doi: 10.1016/j.mcn.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- MacNeilage PF, Rogers LJ, Vallortigara G. Origins of the left and right brain. Sci Am. 2009;301:60–67. doi: 10.1038/scientificamerican0709-60. [DOI] [PubMed] [Google Scholar]
- Satoh D, Sato D, Tsuyama T, Saito M, Ohkura H, Rolls MM, Ishikawa F, Uemura T. Spatial control of branching within dendritic arbors by dynein-dependent transport of Rab5-endosomes. Nat Cell Biol. 2008;10:1164–1171. doi: 10.1038/ncb1776. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Metz M, Zolles G, Turecek R, Fritzius T, Bildl W, Tarusawa E, Kulik A, Unger A, Ivankova K, Seddik R, Tiao JY, Rajalu M, Trojanova J, Rohde V, Gassmann M, Schulte U, Fakler B, Bettler B. Native GABAB receptors are heteromultimers with a family of auxiliary subunits. Nature. 2010;465:231–235. doi: 10.1038/nature08964. [DOI] [PubMed] [Google Scholar]
- Scott EK, Mason L, Arrenberg AB, Ziv L, Gosse NJ, Xiao T, Chi NC, Asakawa K, Kawakami K, Baier H. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat Methods. 2007;4:323–326. doi: 10.1038/nmeth1033. [DOI] [PubMed] [Google Scholar]
- Sernagor E, Chabrol F, Bony G, Cancedda L. GABAergic control of neurite outgrowth and remodeling during development and adult neurogenesis: general rules and differences in diverse systems. Front Cell Neurosci. 2010;4:11. doi: 10.3389/fncel.2010.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelson CD, Santhakumar K, Halpern ME, Gamse JT. Tbx2b is required for the development of the parapineal organ. Development. 2008;135:1693–1702. doi: 10.1242/dev.016576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Tomoda T, Kim JH, Zhan C, Hatten ME. Role of Unc51.1 and its binding partners in CNS axon outgrowth. Genes Dev. 2004;18:541–558. doi: 10.1101/gad.1151204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas KJ, Terunuma M, Tello JA, Pangalos MN, Moss SJ, Couve A. The availability of surface GABAB receptors is independent of gamma-aminobutyric acid but controlled by glutamate in central neurons. J Biol Chem. 2008;283:24641–24648. doi: 10.1074/jbc.M802419200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C. Haploid screens and gamma-ray mutagenesis. Methods Cell Biol. 1999;60:43–70. doi: 10.1016/s0091-679x(08)61893-2. [DOI] [PubMed] [Google Scholar]
- Westerfield M, Zebrafish Information Network . Eugene, OR: Zebrafish Information Network; 2000. The zebrafish book: a guide for the laboratory use of zebrafish Danio (Brachydanio) rerio. [Google Scholar]
- Yan J, Kuroyanagi H, Tomemori T, Okazaki N, Asato K, Matsuda Y, Suzuki Y, Ohshima Y, Mitani S, Masuho Y, Shirasawa T, Muramatsu M. Mouse ULK2, a novel member of the UNC-51-like protein kinases: unique features of functional domains. Oncogene. 1999;18:5850–5859. doi: 10.1038/sj.onc.1202988. [DOI] [PubMed] [Google Scholar]
- Zhou X, Babu JR, da Silva S, Shu Q, Graef IA, Oliver T, Tomoda T, Tani T, Wooten MW, Wang F. Unc-51-like kinase 1/2-mediated endocytic processes regulate filopodia extension and branching of sensory axons. Proc Natl Acad Sci U S A. 2007;104:5842–5847. doi: 10.1073/pnas.0701402104. [DOI] [PMC free article] [PubMed] [Google Scholar]