Abstract
Current macromolecule crystallization screening methods rely on the random testing of crystallization conditions, in the hope that one or more will yield positive results, crystals. Most plate outcomes are either clear or precipitated solutions, which results are routinely discarded by the experimenter. However, many of these may in fact be close to crystallization conditions, which fact is obscured by the nature of the apparent outcome. We are developing a fluorescence-based approach to the determination of crystallization conditions, which approach can also be used to assess conditions that may be close to those that would give crystals. The method uses measurements of fluorescence anisotropy and intensity. The method was first tested using model proteins, with likely outcomes as determined by fluorescence measurements where the plate data showed either clear or precipitated solutions being subjected to optimization screening. The results showed a ~83% increase in the number of crystallization conditions. The method was then tried as the sole screening method with a number of test proteins. In every case at least one or more crystallization conditions were found, and it is estimated that ~53% of these would not have been found using a plate screen.
Keywords: Crystallization screening, Fluorescence anisotropy
Introduction
Macromolecule crystallization conditions are usually determined by a screening process, which involves a search through chemical and physical precipitation parameters (or space) for those conditions that result in the most suitable crystals. The most searched parameters are in chemical space, with physical space being usually invoked by temperature changes, typically with a change from ~293K to 277K. Given the large range of chemical space, and its potential combinatorial variations, several strategies have been put forth to begin the search process. Sparse matrix screens1, which are based upon known crystallization conditions chosen to cover a broad range of crystallization space, predominate in the commercial world and practical use. Totally random screens2 avoid one of the pitfalls of sparse matrix screens by enabling the inclusion of neglected areas or components of crystallization space. Using this approach, it was estimated that ~300 trials were needed to obtain crystals having a 2% probability of success3. However, this method suffers in that one must be equipped with the ability to readily compose the array of solutions required on demand. Grid screens systematically explore regularly spaced variations in a limited number of crystallization components. Typically, lead conditions derived by other methods are refined to improved crystallization conditions using a grid screen. Incomplete factorial screens attempt to explore, as widely as possible, all the parameters of crystallization space using a screen where the various factors are present in a statistically balanced approach4-7. The data obtained is then analyzed to select those parameters most likely to result in crystallization for subsequent testing.
George and Wilson8 originally put forth the concept of there being an optimal range of values for the osmotic second virial coefficient (B22) for protein crystallization. The B22 values measured by light scattering for crystallization conditions fell within a narrow range for all proteins studied, called the crystallization slot, while those for precipitation were more negative and those for clear solutions were more positive than those for the crystallization slot. The data was interpreted to mean that crystallization is favored by mild attractive interactions between the protein molecules; too strong and more random amorphous precipitation occurs, not strong enough and the molecules stay soluble. Crystallization then involves manipulation of the solution conditions to bring about a mildly desolubilzing set of conditions, permitting the assembly of an ordered precipitate. While being within the crystallization slot has been shown to be a necessary condition, it is not in and of itself a sufficient condition. One reason for not obtaining crystals may include a surplus of interactions and thus an inability of the system to settle into a suitable consistent set of interactions. Additives would be key to obtaining crystals in this case.
The crystallization slot concept leads to an interesting thought experiment. Consider a solution of monodisperse protein in the soluble state, to which we begin incrementally changing a solution parameter. The protein goes from soluble to increasingly more desolubilized with each incremental change. A line drawn between the points in a plot of B22 vs. the variable parameter passes through the crystallization slot, which illuminates several concepts relative to obtaining crystals. First, our ability to remain within the slot is a function of the slope of the line and of how well we can control the variable parameter; steep slopes offer little room for control, while gentle slopes favor remaining within the slot. Second, there are regions immediately adjacent to and on either side of the slot, showing attractive protein-protein interactions which are close to those required for crystallization. Experimentally these points show up as either clear (more positive B22’s) or precipitated solutions (more negative B22’s). Based on crystallization plate data these outcomes are discarded, when they may in fact require only slight adjustments in the imposed conditions to bring them within the crystallization slot. This assumes that the protein can or will crystallize, and that the precipitated protein remains structurally functional. Reliance then on observed crystalline outcomes during the screening process may become counter productive to actually finding crystallization conditions.
Herein, we put forth a fluorescence anisotropy (FA) based approach to screening for macromolecule crystallization conditions. FA of appropriately labeled macromolecules is a measure of their rotational rate relative to the lifetime of the fluorescent probe employed, and thus of the volume (mass) of the rotating species. FA outcomes were first compared to those of plate assays using model proteins, then tested by subsequent crystallization screening experiments on conditions whose outcomes were similar to those of the known conditions. The data obtained led to the subsequent inclusion of fluorescence intensity data as a diagnostic, resulting in the FACTs (Fluorescence Analysis of Crystallization Technology’s) approach to crystallization screening described herein. This approach was found to substantially expand the range of crystallization conditions for model proteins. Subsequent use with test proteins, where the FACTs data was the sole screening method employed, showed this to be a useful approach to finding crystallization conditions.
Materials and Methods
Proteins
Glucose isomerase (GI) and xylanase (XLN) were obtained from Hampton Research (cat#’s HR7-100 and HR7-104, respectively). Cut canavalin (CCAN) was purified from Jack Beans (Sigma) as previously described9. Uncut, or not proteolytically cleaved, canavalin (UCAN) was purified in house by DEAE, followed by hydroxyapatite, chromatography (unpublished data). Phaseolin was purified from kidney beans using published methods10. Pv01, a ~30 kDa protein of unknown function, was purified from kidney beans by extraction at pH 5.1, chromatofocusing on cation exchange resin, followed by size exclusion chromatography. Proteins from the hyperthermophilic archaeon Thermococcus thioreducens were cloned, expressed, and purified as previously described11.
Crystallization Screens
Hampton Research Crystal Screen HT (high throughput), (Cat# HR2-130) was used for all 96 condition screens. Sitting drop vapor diffusion plate screens were set up using Greiner 96 well plates as previously described12. Optimization screening solutions were formulated using stock concentrated reagent solutions, prepared using commercially obtained chemicals of Reagent grade or better. Capillary counter diffusion screens were set up using 0.3 mm ID × 8 cm long borosilicate capillaries (Hilgenberg GmbH, Germany) as previously described13. Briefly, the capillary was first ~1/2 filled with protein solution, then the balance filled with precipitant solution. The ends were sealed with wax and coated in fingernail polish. The tubes were incubated at 19°C in a horizontal orientation, supported on plastic strips inside CD cases.
FA Screen
The FA screen was prepared in black 1536 well plates (Matrical, Cat# MP111-1-PS). Each precipitant solution was tested using seven assay protein concentrations of 0.2, 0.4, 0.8, 1.6, 2.4, 3.2, and 4.0 mg/mL, plus one buffer blank having no protein. Each solution was composed of 0.6 μL of 5X concentrated protein stock solution plus 2.4 μL of precipitant solution. Plates were set up by first dispensing the protein solution with multichannel pipetters, oriented (parallel to the plate Y axis) such that the first four wells in the first column had buffer solution, the second four had the lowest protein concentration, etc., to the last four wells which has the highest protein concentration. Protein was dispensed into every other column of wells. Precipitant solution was dispensed with a 12 channel pipetter oriented perpendicular (parallel to the plate X axis) to that of the protein dispensing pipette. Solutions were dispensed such that the first four wells of the first column had precipitant solutions A1, B1, C1, and D1 respectively. This pattern is repeated down the column. The E, F, G, and H solutions are dispensed by offsetting the position of the first channel of the pipetter by two wells in the X direction and repeating the same pattern. Thus one half of the wells are filled, with an empty column between each column of filled wells. After all solutions were dispensed the plate is covered with a clear film (Greiner, cat# 676070), then briefly centrifuged (2 minutes, 800 × g) to remove any air bubbles in the wells and ensure that all solutions were centered in the wells and not adhering to the sides.
Fluorescent labeling
Protein was covalently labeled with the fluorescent probe ruthenium bis(2,2′-bipyridine)-4,4′-dicarboxybipyridine disuccinimidyl ester (Ru(bpy)2dcbpy, Sigma cat# 96632). A 5 mg bottle of the probe was dissolved in 1.0 mL of acetonitrile, giving a solution of 4.34 × 10−6 moles/mL. From 1 to 1.5 mg of protein solution was buffer exchanged using a centrifugal desalting column (Pierce) into 0.05 M sodium borate, pH 8.8, and brought to 4 mLs volume with the same buffer. The total moles of protein present was determined, and probe solution added in aliquots over a 2 hr period to a 4:1 (probe:protein) molar ratio. The reaction was allowed to sit for another 2 hrs to overnight at 277K, then terminated by adding 20 μL of 0.5 M glycine. The resulting solution was centrifugally concentrated (Vivascience) using appropriate MWCO membranes to ~100 μL, then buffer exchanged as above into 0.05 M sodium hepes, 0.1 M NaCl, pH 7.5. Protein and dye concentrations were spectrophotometrically determined (Ru(bpy)2dcbpy ε460 = 14,500 M−1,)14. The measured protein absorption at 280 nm was corrected for the absorption of the dye, based upon an experimentally determined dye 280/460 absorption ratio of 4.52. The molar concentrations of each were determined and used to calculate the percentage of protein that was covalently modified with dye. Protein dilution tables to prepare stock solutions for setting up the FA assay were calculated using an Excel spreadsheet, with the inputs being the labeled protein concentration (mg/mL) and percentage labeled, the unlabeled protein concentration, protein MW, desired final probe concentration in the assay, and desired final volume of each solution to be made.
FA measurements
The FA measurements were made using an in-house built instrument having the optical layout of an epifluorescence microscope. The optical components were for the most part obtained as commercial off the shelf components (Edmund Optics), with some parts (plate holder) being made by a local machine shop or, where noted below, made in-house. An estimated total cost for the system is ~$30K. Excitation light comes from a pulsed blue light emitting diode (Superbrightleds.com, Cat# RL5-B5515), having an output peak at 470 nm. The excitation pulse was collected by a lens and then passed through a polarizer (Casix, cat# PGM5310) to give vertically polarized light. The light then passed through a 460 nm high pass filter (Omega, cat# 3RD460LP) and reflected from a dichroic mirror (Omega, cat# XF2009) to a lens that focused the light onto the sample and also collects the emitted fluorescent light. The fluorescence emission passes through the dichroic mirror, then through 590 nm long pass and 700 nm low pass filters (Omega, cat# 3RD590LP and 3RD700SP, respectively), followed by a calcite polarizer (Casix., cat# PGM5310) mounted such that it could be rotated 90° by a stepper motor to select for either vertically or horizontally polarized emitted light. The emitted light is then focused onto a photomultiplier tube (PMT) module (Hamamatsu, P/N H8249-102), which operates in photon counting mode.
Data collection is initiated by first placing the emission polarizer in the vertical orientation, then sending a pulse from the controlling PC, which triggers a ~300 ns pulse of the light emitting diode (LED). Emission pulses from the PMT are inhibited from reaching the counter circuit by a logic gate which blocks the signal until ~60 ns after the end of the LED trigger pulse. At this point the signal pulses are allowed to reach the counter, and counting is continued for a period of 10 μs, after which it is stopped and the counts transferred to the PC through a parallel IO port. This process is repeated 10 × 4000 times, with the data summed in 4000 pulse bins, after which the emission polarizer is rotated 90° to the horizontal orientation and the process repeated. After collection of the vertical-vertical and vertical-horizontal data for a well the plate is positioned to the next well using a stepper motor driven X-Y stage and the process repeated. Data is sorted and stored to a PC file after collection from each column of 32 wells, with a new file used for each ¼ plate of data (6 columns). All data collection and LED pulsing control circuits were assembled in house. Control and data acquisition programs were written in-house in C++.
Data analysis is carried out by importing the raw collected data into Excel template files. These carry out the blank subtraction for each point of the data set, calculate the anisotropy and total intensity values for each point, and plot the anisotropy data vs. assay protein concentration. The calculated slopes of the anisotropy and intensity values vs. concentration are determined and collected for use in subsequent analysis. Three data collection runs are made for each plate set up, at ~1 hr, ~4 hrs, and ~24 hrs after set up.
FACTs lead optimization
Two methods were used for testing or optimizing leads found by FACTs measurements. The first used a 4 × 4 sitting drop vapor diffusion grid screen about the condition of interest. Concentrations of the screen solutions were calculated using an Excel spreadsheet template, which output a pipetting table for preparing each solution from the available stock solutions. The second method was capillary counter diffusion, which was set up as previously described13. Each lead was tested using a four solution grid, composed of; solution 1, each precipitant at ½ concentration; solution 2, precipitant A at ½ concentration, B at full concentration; solution 3, precipitant A at full concentration, B at ½ concentration; solution 4, precipitants A and B at full concentration. The buffer is at full screen concentration (0.1 M) in all cases. For conditions where there was no buffer, 0.1 M hepes, pH 7.5 was used. If only one precipitant is given then the ionic liquid 1-butyl-3-methyl imidazolium HCl (Bmim-Cl) was used15 at 0 and 0.2 M concentrations respectively. All optimization experiments were incubated at 291K for at least 4 weeks before being transferred to 286K. The incubator was a commercially available wine cooler having two temperature ranges. Lowering the temperature by just 5K was done to facilitate subsequent observations at room temperature while minimizing concerns about crystals dissolving upon being warmed to the higher temperatures.
Fluorescence anisotropy
Fluorescing species absorb maximally when their absorption vectors are aligned parallel to the polarization of the exciting light. Emission is along a separate vector, and the angular difference between these two vectors is a property of the structure of the fluorescing species. The fundamental anisotropy, r0, defines the angular difference between the absorption and emission dipoles in the absence of any other depolarizing processes. This term can vary between 0.4 (angular difference between the absorption and emission dipoles = 0°) and −0.2 (angular difference = 90°). For a more comprehensive discussion, see Lakowicz16.
Fluorescence depolarization can occur for several reasons, the most important being rotation of the attached fluorescing species due to the rotation of its substrate. The Perrin equation describes the case for a spherical rotator
| (1) |
where r0 is the fundamental anisotropy of the fluorescing species, r is the measured anisotropy, τ is the fluorescence lifetime, θ is the rotational correlation time, and D the rotational diffusion coefficient. The rotational correlation time is related to the molecular mass (M) by
| (2) |
where is η the viscosity, V the volume of the rotating species of mass M, R the gas constant, T the temperature, the specific volume of the rotator and h the hydration, grams of H2O per gram of protein. The anisotropy is classically measured by exciting the sample with vertically polarized light, then measuring the intensities of the vertically polarized (IVV) and horizontally polarized (IVH) emission light. The anisotropy is calculated from these intensities by
| (3) |
Where G is a correction factor for the sensitivity of the detection system to the different optical paths and the denominator of eqn. 3 is the total light intensity.
Results
FACTs data collection
Model proteins, defined as those for which crystallization plate data was used to interpret the FACTs results, were initially used to establish plate set-up, data collection, and data analysis procedures. Table I lists the model proteins used, the numbers of conditions that gave crystals in a plate assay, and the number of additional hits obtained using the FA data. When setting up the FACTs experiment, one has to decide what protein concentration range to employ and the final precipitant:protein ratio. Due to dispensing difficulties the latter was quickly established at a 80:20 ratio. Attempts at using a 90:10 ratio with a 3.0 uL final assay solution volume consistently failed.
Table I.
Model Protein results
| Found by | |||
|---|---|---|---|
| Protein | By Plate | FA1 | % Increase |
| Glucose Isomerase | 33 | 5 / 3 | 27.3 |
| Xylanase | 12 | 02 / 17 | 141.7 |
| Cut Canavalin | 15 | 7 / 7 | 93.3 |
| Uncut Canavalin | 6 | 10 / 11 | 350.0 |
| Phaseolin | 10 | 6 / 14 | 200.0 |
| Tt141 Pyrophosphatase | 25 | 6 / 11 | 68.0 |
| Tt104 Amino Peptidase | 16 | 0 / 10 | 62.5 |
| Tt36 ATP dep. DNA ligase | 6 | 1 / 4 | 83.3 |
| Tt186 Alcohol dehydrogenase | 17 | 1 / 4 | 29.4 |
| Total | 140 | 117 | 83.6 |
Data presented as C / P, where C is the number of conditions where the plate results were clear solutions and P those where the results were precipitated solutions.
All clear solutions at 1:1 had precipitate at 2:1 & 4:1 protein:precipitate ratio’s.
The FACTs measurements, similar to light scattering8, are used as a dilute solution indicator of protein-protein interactions. The starting approach was to determine the trend in FA with protein concentration as our diagnostic, and the concentration range chosen needed to go sufficiently high to show a sufficient change in the anisotropy due to protein self interaction, presumably due to the events leading to crystal nucleation. Strong self interactions would be expected in the case of amorphous precipitate formation, and are expected to show up as high anisotropy at low protein concentrations. Such is also expected to be the case when the protein is wholly or partially unfolded. Initial measurements were made using stock protein concentration ranges from 1 to 10 mg/mL, corresponding to assay concentration ranges of 0.2 to 2.0 mg/mL. Stock concentration ranges as high as 4 to 40 mg/mL were tested with dilution ratios to 90:10. Higher protein concentration ranges did not result in appreciably better data, although they do require proportionately more protein per screen. No definitive conclusions can yet be made concerning precipitant:protein ratios, due to pipetting difficulties discussed below.
It takes 15-20 min to manually set up a FACTs assay plate, taking care to ensure that all solutions are dispensed to the correct wells. The order of dispensing was protein followed by precipitant solution, to minimize carry-over from well to well of precipitant. The accuracy with which the multichannel pipetters could dispense volumes < 1.0 μL limited the use of the FACTs assay to total assay volumes of 3.0 μL, using an 80:20 (precipitant:protein) ratio. The major difficulty is apparently failure of the dispensed volumes, specifically the smaller protein volume, to separate from the tip within the wells of the plate, resulting in some wells having no protein and others to much. Care in ensuring that the pipette tips touched the bottom surface of the wells helped to reduce this problem, but even at 0.6 μL a number of mis-dispensed wells are obtained in every plate.
The free fluorescent probe, Ru(bpy)2dcbpy has a lifetime of 375 ns17 and a quantum efficiency of ~0.0514. The probe also has a relatively low absorptivity14, 14,500 M−1. Advantages include the long lifetime and a very large Stokes shift between the excitation (Exmax) and emission maxima (Emmax) (460 and 650 nm, respectively). The Exmax makes the use of blue light emitting diodes, having peak emission wavelengths in the 460-470 nm region, very good sources of excitation light. However, this advantage is offset by the wavelength dependence of the fundamental anisotropy r0, which peaks at r0 ~ 0.28 at ~485 nm. At 470 nm r0 ~ 0.1814, reducing the ’operating range’ over which measurements can be made.
FACTs data analysis
Initial data analysis consisted of visual examination of graphs of anisotropy vs. concentration data. While highly subjective, this approach was primarily used for the data collected with the proteins XLN, GI, CCAN, and UCAN, as well as for initial experiments with T. thioreducens proteins. This approach worked satisfactorily, as indicated by the results shown in Table I. However, the data were often very irregular, attributed to the pipetting problems at low solution volumes, and the subjective nature of this approach did not help in better defining FA outcomes that were most likely to result in crystallization conditions. The results for XLN, GI, CCAN, and UCAN in Table I are for visual examination of the FA data, while those for the balance of the proteins reflects analysis based upon calculated slopes as described below.
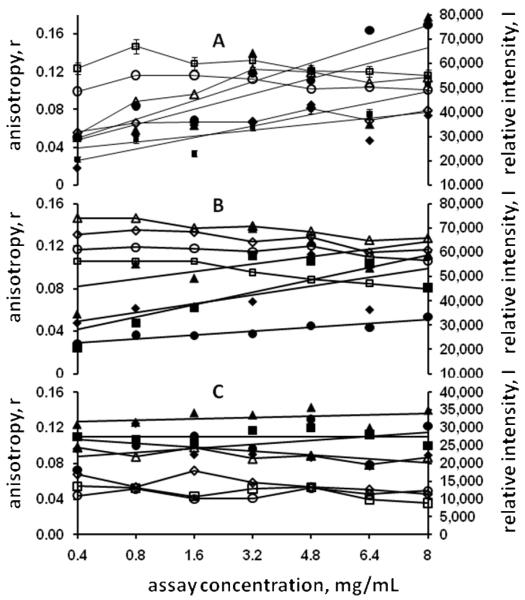
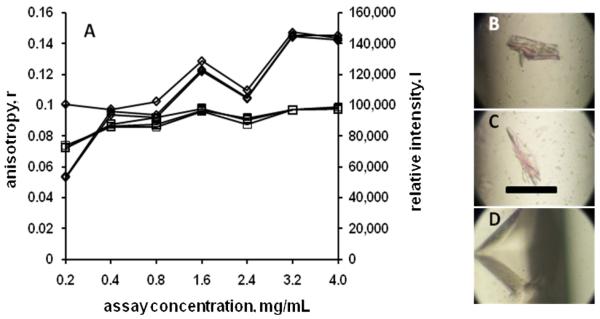
As the probe concentration was kept constant, independent of the changing protein concentration, the total intensity, the denominator in Eqn. 3, was added to the data analysis as a check of pipetting fidelity. It was initially assumed that the total intensity would remain constant if all dispensing operations were satisfactory. However, it was quickly noted that the total intensity values also changed with the protein concentration, and that the changes were apparently positive in those conditions judged to be possible leads based on the anisotropy data and neutral or even negative in those cases resulting in negative plate screening outcomes. Examples of this are shown in Figure 1, for the model protein T.t. 186, with Panel A showing FA data for known crystallization conditions, based upon crystallization plate assays, and Panels B and C showing the data obtained for precipitated and clear solution outcomes, respectively. The FA data in Panel A shows two different outcomes for the known conditions. The data for condition D1 (Panel A, open and closed squares) includes 5% error bars for both the anisotropy and intensity data points. In actuality the percentage errors ranged from 0.287-0.516 (intensity) and 1.06-4.18 (anisotropy). These values are representative of the variation in all data obtained. Regression fits to the anisotropy data for conditions D1 and A10 gave negative slopes, while that for conditions A1 and H7 gave positive slopes. However, in all cases the intensity values have positive slopes. From these and similar results it was concluded that the intensity data was also a strong indicator of likely crystallization conditions. For the known precipitation conditions the anisotropy data slopes were all negative, while the intensity data slopes were mixed. Known clear solution outcomes gave all negative anisotropy slopes and ~level or negative intensity slopes.
Figure 1.
Measured anisotropy (open symbols) and intensity values (closed symbols) for model protein T.t. 186. Panel A, known crystallization conditions. Legend: ◇, condition A1; □, condition D1; △, condition H7; ○, condition A10. Note that for conditions A1 and H7 both the anisotropy and intensity data have positive slopes, while for conditions D1 and A10 only the intensity slopes are positive. Panel B, known precipitation conditions. Legend: ◇, condition B8; □, condition E4; △, condition F1; ○, condition G6. Panel C, known clear solution conditions. Legend: ◇, condition C1; □, condition B2; △, condition C2; ○, condition A5.
The extrapolated anisotropy values in Figure 1 Panel B are somewhat higher than those in Panel C, suggesting that the solutions are aggregated even at low protein concentrations. However, another reason for this would be the viscosity of the precipitant solution as well as the low concentration aggregation state of the protein in the precipitant. The different intensity levels are due to the effects of precipitant solution components on the fluorescent probe.
Data analysis was subsequently modified to take both the slopes of the anisotropy (r) and intensity (I) vs. concentration curves into account. The slopes for each are calculated for each data set and their product, the value of r × I, is determined as part of a spreadsheet-based analysis process. A logical operation is used to assign a negative value to the product when both r and I are negative. Currently analysis of the FACTs data results in three sets of data, the r × I, r, and I, vs. concentration data. Each is ranked as a percentile of the highest positive value within its set, and the top 20 of each, excluding duplicates with other sets, are used as the basis for subsequent optimization trials.
The initial impetus for performing data collection at ~1, 4, and 24 hrs after plate set-up was the possibility of a kinetic component to the data and to determine the optimum time after set-up for data collection. If present, it was expected to manifest as a positive increase in the slope in the anisotropy or intensity vs. concentration curves over time. Evaluation of the hits data to identify a kinetic component for the test proteins was somewhat ambiguous, with ~61% of the r and I vs. concentration slopes showing a positive change with time and the balance showing a negative slope. This suggests that if only one data collection was made then it should be the last, at 24 hrs. However, more extensive data collection and analysis will be needed to definitively identify the optimal data collection time.
Optimization screening
All optimization trials were carried out on leads derived from the FA screening assay. Optimization screening outcomes were considered positive for any crystalline outcomes, from large 3D crystals to fine needles, “urchins”, or dendritic structures. Positive outcomes here are not a guarantee that the crystals are useable for X-ray diffraction data collection. In many cases the results indicated that further optimization trials were warranted; however these were not counted as positive outcomes unless those subsequent trials actually resulted in crystals. Multiple optimization rounds were only carried out for one protein, CCAN, where up to three optimization rounds were performed for some conditions. This is reflected in the high success rate for this protein, and it is anticipated that overall success rates, as shown in Table I (and II) would be higher with more aggressive pursuit of the leads obtained.
Table II.
Test Protein Results.
| Protein | Test Round | Crystals | # w/no Cond 4* |
Notes |
|---|---|---|---|---|
| T.t. 46 SS DNA-spec. Exonuclease (m + r) | 1 | 3 | 3 | |
| T.t. 71 Intracellular protease/amidase (m + r) | 1 | 22 | 5 | |
| T.t. 75 Prolyl endopeptidase (m) | 1 | 4 | 1 | |
| T.t. 97 Aspartate Racemase (m + r) | 1 | 11 | 5 | |
| T.t. 101 Amino Peptidase ( r ) | 1 | 1 | ||
| T.t. 103 Acid Phosphatase (m) | 1 | 6 | 2 | 1 |
| 2 | 5 | 2 | 2 | |
| T.t. 105 2-hydroxyacid Dehydrogenase (m) | 1 | 9 | 4 | |
| T.t. 169 RNAse (m) | 1 | 5 | 0 | |
| T.t. 185 Nucleotide Kinase (m) | 1 | 4 | 2 | |
| T.t. 187 Mg-dependent DNAse (m) | 1 | 5 | 3 | |
| T.t. 189 Nucleoside Diphosphate Kinase (m) | 1 | 16 | 5 | |
| P.v. 01 unknown protein (m) | 1 | 6 | 3 | |
| ProtX - provided by J. Gavira, Granada ( r ) | 1 | 7 | 3 | 3 |
Optimizations where crystals were not obtained in the 4th (stock conditions) capillaries.
3 unique to round 1, temp. reduced to 13°C
2 unique to round 2
only 12 leads tested
m = manual & r = robotic plate setup.
Optimization screening was initially carried out by setting up a 4×4 grid screen around the found lead conditions. This approach was primarily used for the four model proteins GI, XLN, UCAN, and CCAN. While successful, the method proved to be overly labor intensive and led to a switch to the capillary counter diffusion-based approach, which has been shown to result in a higher success rate compared to sitting or handing drop vapor diffusion13. Using a ‘standard’ grid of four solutions as described in the Methods section, a capillary counter diffusion optimization screen of 30 conditions could be set up in ~1-2 hrs. The approach taken, of four different ratios of two precipitants, was found to be very useful. Many of the optimized conditions only have crystals in one or two capillaries, or had crystals with markedly improved visual appearance in only one of the capillaries. As shown in the test protein data (Table II), ~37% of the found lead conditions did not have crystals in the 4th capillary, corresponding to the stock screen solution conditions. We can tentatively assign these as conditions where crystals would not have occurred in plate screening assays, and also would not have occurred in counter diffusion experiments where only the stock solution conditions were employed. We estimate that the FACTs screening approach, coupled with the described capillary counter diffusion approach, resulted in at least a ~53% increase in the number of crystallization conditions obtained.
Model proteins results
Model proteins, defined as those for which crystallization plate data was used to interpret the FACTs results, were initially used to establish plate set-up, data collection, and starting data analysis procedures. Table I lists the model proteins used, the numbers of conditions that gave crystals in a plate assay, and the number of additional conditions found using the FA method. In all cases FACTs-based screening resulted in a substantial increase in the number of crystallization conditions found.
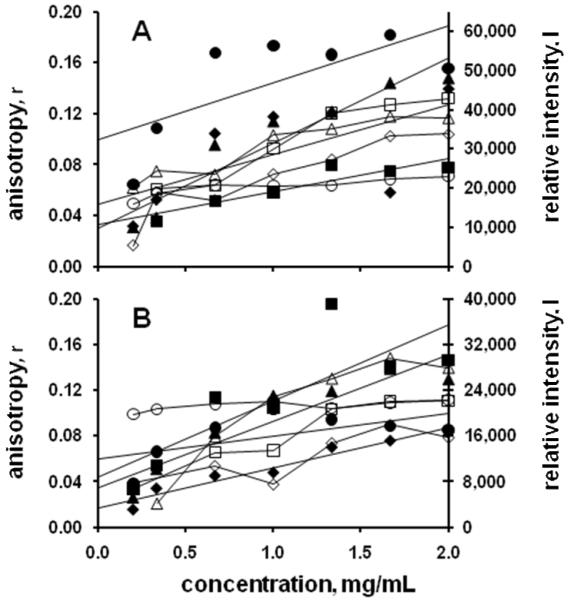
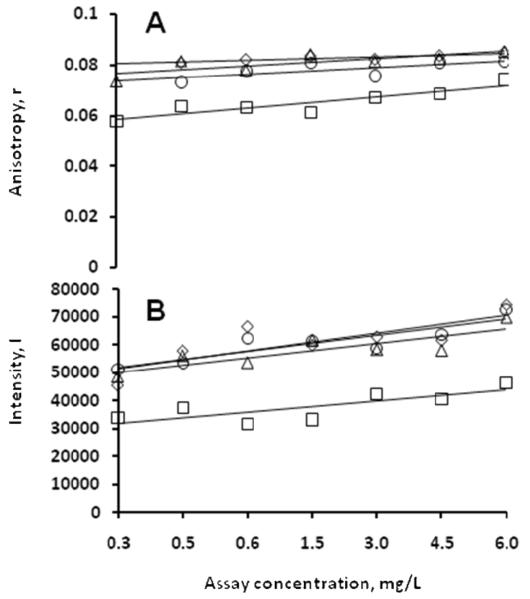
A number of model proteins had measured FACTs data interpreted to correspond to likely crystallization conditions where the plate data showed either precipitated or clear solutions, as shown in Figure 2. Optimization for the clear solution conditions was by increasing the protein concentration 4 to 5 fold and repeating the plate screen. For capillary screens the protein concentration was similarly increased and the 1st and 4th solutions used to repeat the screen. Optimization success with these FACTs-derived outcomes indicate that the 10 mg/mL protein concentrations used for the plate screen were not sufficiently high for those conditions. As shown in Figure 3, the concentration increase was not always sufficient to result in crystallization. Two of the conditions shown (A5 and B9) resulted in crystals in this case when the protein concentration was raised to 85 mg/mL using a plate-based optimization screen. The other two, which have essentially similar data trends, remained clear, suggesting that possibly still higher protein concentrations and/or adjustment to the solution conditions were needed. The advantage of this approach is that in both instances the optimization screening process can be rapidly carried out. However, clear solution-based leads were usually not as productive as precipitated solution-based leads; most of the FACTs-derived leads for the model proteins which were subsequently converted to hits came from conditions which yielded precipitated protein in the plate assays.
Figure 2.
Measured anisotropy (open symbols) and intensity values (closed symbols) for the model protein canavalin. Panel A shows data for cases where the plate outcome was a precipitate, but where the conditions were optimized to obtain crystals on the basis of the fluorescence data. Legend: ◇, condition A9; □, condition B3; △, condition D10, ○, condition H3. Panel B shows data for cases where the plate outcome was a clear solution, but crystals were obtained by increasing the protein concentration. Legend: ◇, condition A12; □, condition C2; △, condition D2; ○, condition G7.
Figure 3.
Model protein (phaseolin) fluorescence data where plate screen outcomes were clear solutions. Panel A shows the measured anisotropy data. Panel B shows the measured intensity data. Legend: ○, condition A5; △, condition B9; ◇, condition A1; □, condition E11.
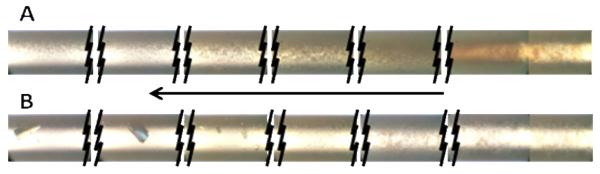
As shown in Table II the FACTs method successfully identified one or more crystallization conditions for every test protein. However, the FACTs data is only an indication of the proximity to potential crystallization conditions, and obtaining crystals may require manipulations beyond simple variations in the stock solution conditions, such as additives. In one case, protein T.t 103, a second round of optimization trials was implemented based upon the outcomes observed in the first round of trials. A common outcome for many of the first round trials was a precipitation pattern that started as a brown precipitate, becoming progressively more granular along the length of the capillary, then abruptly changing to small clear spheroids, as shown in Figure 4, panel A. Ten of the T.t. 103 precipitated outcomes were retested after making the protein solution 0.1 M arginine as an aggregation inhibitor18, 19 and reducing the protein concentration to 20 mg/mL. Figure 4, panel B, shows the crystals obtained in one of the conditions where this approach succeeded. While three of the crystallization conditions for T.t.103 were common to the two trials, three of the original trial series and two of the second (+ arginine) series were unique. The second round crystals were all obtained from conditions which had not resulted in crystals during the first round, and all grew within 2 weeks of incubation at 292K, while most of the first round crystals were obtained only after reducing the temperature to 286K. Many of the other test proteins had precipitation outcomes similar to those shown in Fig. 3A, and it is likely that additional crystals could have been obtained from them using this approach.
Figure 4.
Test protein T.t.103. Panel A (upper) showing capillary precipitation results as set-up. Panel B (lower) showing the capillary outcome with protein solution made 20 mg/mL + 0.1 M arginine. The arrow shows the direction of precipitant diffusion. Capillaries are 0.3 mm ID, 0.4 mm OD.
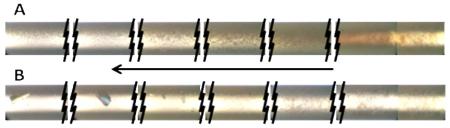
The four capillary approach to optimization trials was found to be considerably more productive than the use of just a single capillary, as shown in Figure 5. This approach was implemented as it was realized that different precipitant solution components will diffuse at different rates, and that varying profiles could be obtained by mixing their ratios. In this case all three anisotropy and intensity data sets for the lead condition are shown. Note that there is a distinct change in the visual appearance of the crystals between the three capillaries. The fourth capillary, having stock precipitant solution concentrations, had no crystals in this case.
Figure 5.
Test protein T.t. 189 fluorescence data and optimization screen outcomes for screen condition F1. Panel A shows the measured anisotropy (□) and intensity (◇) data. Panels B, C, and D show the optimization screen outcomes for capillary’s 1, 2, and 3 respective. Capillary 4 had no crystals.
Discussion
The FACTs approach is based upon splitting the macromolecule crystallization process into two steps. The first step is to identify those parameters most likely to yield crystals, and the second is to focus on those parameters and optimize them to obtain the best diffracting crystal(s). The development process described herein is focused primarily on the first step, for which it is found that the FACTs approach has proven to be a powerful approach to screening for protein crystallization conditions. The method has the potential to be a very rapid screening procedure, with lead conditions being found and subjected to optimization screening potentially within 30 hrs after pure protein is ‘in hand’. Still shorter times are possible if the number of measurement runs is reduced.
Use of the FACTs method in the presence of detergents and integral membrane proteins is anticipated to be similar to its use with soluble proteins. The major difference will be the need for extra care in post derivatization processing of the labeled protein to ensure the removal of free detergent. Some benefits to the signal intensity may be found as well from the probes being attached in close proximity to a hydrophobic environment. Fluorescence anisotropy-based screening has previously been reported for detergent optimization in the crystallization of an integral membrane protein20, following the approach of Jullian and Crosio21 for the use of anisotropy in estimating the second virial coefficient and molecular interactions.
The current major weakness of the FACTs approach is the relatively large amount of protein required. At a top assay concentration of 20 mg/mL, using an 80:20 ratio (precipitant:protein), and a 3.0 μL volume per assay solution, a 96 condition screen requires 3.63 mg of protein. Use of a smaller assay solution volume can significantly reduce protein requirements for assay. Current plans are to reduce these volumes, with a final target assay solution volume of 10-20 nL. These reduced volume ranges are well beyond the ability of manual pipetting methods and will require use of specialized dispensing systems. Current syringe-based robotic dispensing systems can accommodate total assay volumes down to ~0.25 μL, requiring only ~0.3 mg of protein for a 96 condition screen. The 1536 well plates employed have a recommended minimum volume of 0.5 μL; volumes below this will require a new plate design.
Considerably more protein is used under the current optimization protocols; 30 conditions at 4 capillaries/condition requires ~12 mg of protein at a concentration of 25 mg/mL. Two approaches can be used to reduce this burden. First is to reduce the amount of protein used in capillary counter diffusion screens by, for example, using microfluidics-based capillary columns. Use of a 0.1 mm × 2 cm capillary, such as that described by Ng et al.22, would reduce the total protein for 30 condition optimizations as described to 0.6 mg. The second approach lies in better selection of the conditions for subsequent optimization. We are currently collecting FACTs data from a wide range of proteins, both prepared in-house and obtained from outside laboratories. The scoring system currently employed facilitates summarizing the results obtained, such that the scoring percentile ranges having the highest likelihood of giving crystals can be identified. It is likely once this has been achieved that only 8-10 of the FACTs-defined lead conditions from a given screen will need to be optimized to find crystallization conditions. While success in the high likelihood leads is not guaranteed, there are still additional found leads that can be subsequently explored.
Implementation of the above improvements would lead to a crystallization screen with first round optimization requiring ~0.23 mg of protein for a 96 condition screen. These amounts are in line with current low screening volume practices, with several added benefits. First, more lead conditions are likely from a given screen, reducing the number of screens that need to be carried out and thus the time and effort required to obtain crystals. Second, there are no ‘scale up’ issues in converting results obtained from low nL volume screens to obtaining crystals at higher solution volumes. Third, crystals obtained in optimization can be directly analyzed by X-ray diffraction, whether obtained in a capillary13 or microfluidics-type capillary22. Fourth, the initial screening data are obtained as numerical values, not images that need to be viewed and analyzed for the presence of crystals. This data, dependent upon the behavior of the labeled protein, should not be affected by the presence or absence of salt crystals in the solution. With these benefits, it must be borne in mind that, similar to the crystallization slot hypothesis of Wilson8, FACTs-derived lead conditions are not a guarantee of crystals or of their diffraction quality, but rather an indication of where crystallization conditions are most likely to be found. In this case, knowing that the conditions are highly favorable to crystallization makes them worth the additional effort to obtain crystals. Also, the FACTs-derived lead conditions would be the best candidates for subsequent additive screening.
The FACTs approach to screening for likely crystallization conditions could also be taken using light scattering, where one would not have to covalently label the protein. However, there are advantages to the FACTs method. Light scattering data interpretation is dependent upon the angular relationship between the incident and scattered light. FACTs data is not angularly dependent, and the measurements can be made, as done here, with an epifluorescence-type instrument. Light scattering measures the immediate signal, and is sensitive to the light path and any interfaces between the light source and the detector. By using long lifetime fluorescent probes we can gate out the prompt signals, collecting only those resulting from sample fluorescence.
The percentage of protein having covalently bound label varies with the protein concentration, typically ranging from ~20% at the lowest protein assay concentration to ~1-2% at the highest. Covalently labeled protein is only used in the FACTs assay. Subsequent optimization trials used unlabelled protein, and as a result we cannot determine what effects the presence of the probe may or may not have on the FACTs data obtained or on the crystallization process. The fluorescent label may affect or be affected by small molecule ligands in crystallization binding structures. In this case we would suggest that a course of action would be to first identify likely crystallization conditions in the absence of the ligand, then test the lead conditions for crystallization in the absence of the probe, similar to the approach taken for using arginine as an additive.
The current screening paradigm is to test as many crystallization conditions as possible while using the smallest amount of protein, in the hopes that one ‘gets lucky’ with one or more of the conditions tested. The FACTs approach does not well lend itself to this paradigm, as it is focused on not simply finding only clear hits but also identifying those conditions that are sufficiently close that they can be potentially optimized to hits. Optimization may take more than a single round of trials; early work with some model proteins required up to three rounds before crystals were obtained (data not shown). An advantage of this approach is that with careful analysis one can potentially derive those solution components that are most likely to yield crystallization conditions from the FACTs-derived lead conditions. This suggests that the FACTs method would work even better if coupled to a method such as the incomplete factorial approach4-7.
The test protein data indicates that I vs. concentration data may be a better diagnostic for crystallization conditions than the r vs. concentration data. The r vs. concentration data still has a powerful utility in the determination of crystallization conditions, and in fact can be used to better direct the subsequent optimization approach. While a number of hits are found, there are still more conditions where the FACTs data indicated leads but no crystals were obtained. For a number of these the tubes remained clear, suggesting that an increase in protein concentration, possibly followed by additional optimization, may be sufficient. For those cases where the outcome was a precipitate, the answer may be given by a linear extrapolation of r to low concentration. Examination of the data shows that in many cases the extrapolated data gives rather large values of r, suggesting that the protein is already substantially aggregated even at low concentration. In some instances, particularly where the protein was known to form multimers based on gel filtration data, the extrapolated r values may be smaller than expected, suggesting that the precipitants caused dissociation of the multimers. We have now started to routinely include a reference data set during the plate set-up process, whereby one of the extra columns of wells in the 1536-well plate is filled with protein diluted into buffer, not precipitant. This data (not shown) indicates the solution MW of the protein. Inclusion of the precipitant viscosity into the data analysis will enable referencing the experimental results to the proteins solution mass (Eqn. 2), providing an external metric and enabling estimation of the low concentration aggregation state of the protein in the precipitant conditions.
Improved interpretation of the FACTs data comes from placing the results in their proper context with respect to solution phase events. The rationale for increasing r as a result of self association was the logical starting point for this approach. A working hypothesis for increasing I is that more structured, thus more compact, assembly’s may better shield the fluorescent probe and result in a more consistent environment than would be found in an amorphous precipitate, however more experiments are needed to more definitively address this finding. Again, very strong aggregation may result in probe being buried sufficiently deeply that an increased intensity is observed. For these reasons I and r vs. concentration data cannot be used independently in analyzing the FACTs screen results, but each informs the analysis of the other.
Synopsis.
A fluorescence anisotropy based approach, which has been shown to effectively find crystallization conditions, is being developed as an alternative to plate screening for the determination of protein crystallization conditions.
Acknowledgements
Funding support for this work came from NIH/NIGMS SBIR Grant 1R43 GM084488-01 and -02. The Author also gratefully acknowledges the support provided by Dr. Joseph Ng and his laboratory at the Univ. of Alabama in Huntsville. The Author also gratefully thanks Dr. J. Gavira, Univ. of Granada, Spain, for providing one of the proteins (ProtX) used in this work.
References
- (1).Jancarik J, Kim S-H. J. Appl. Cryst. 1991;24:409–411. [Google Scholar]
- (2).Rupp B, Segelke BW, Krupka HI, Lekin TP, Schafer J, Zemla A, Toppani D, Snell G, Earnest T. Acta Cryst. D. 2002;58:1514–1518. doi: 10.1107/s0907444902014282. [DOI] [PubMed] [Google Scholar]
- (3).Segelke BW. J. Cryst. Growth. 2001;232:553–562. [Google Scholar]
- (4).Carter CW, Carter CW., Jr. J. Biol. Chem. 1979;254:12219–12223. [PubMed] [Google Scholar]
- (5).Sedzik J. Arch. Biochem. Biophys. 1994;308:342–348. doi: 10.1006/abbi.1994.1049. [DOI] [PubMed] [Google Scholar]
- (6).Sedzik J. Biochem. Biophys. Acta. 1995;1251:177–185. doi: 10.1016/0167-4838(95)00094-b. [DOI] [PubMed] [Google Scholar]
- (7).DeLucas LJ, Bray TL, Nagy L, McCombs D, Chernov N, Hamrick D, Cosenza L, Belgovskiy A, Stoops B, Chait A. J. Struct. Biol. 2003;142:188–206. doi: 10.1016/s1047-8477(03)00050-9. [DOI] [PubMed] [Google Scholar]
- (8).George A, Wilson WW. Acta Cryst. D. 1994;50:361–365. doi: 10.1107/S0907444994001216. [DOI] [PubMed] [Google Scholar]
- (9).Sumner JB, Howell SF. J. Biol. Chem. 1936;113:607–610. [Google Scholar]
- (10).Suzuki E, Van Dankelaar A, Varghese JN, Lilley GG, Blagrove RJ, Colman PM. J. Biol. Chem. 1982;258:2634–2636. [PubMed] [Google Scholar]
- (11).Hughes RC, Ng JD. Crystal Growth and Design. 2007;7:2226–2238. [Google Scholar]
- (12).Forsythe E, Achari A, Pusey ML. Acta Cryst. D. 2006;62:339–346. doi: 10.1107/S0907444906000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Ng JD, Gavira JA, Garcia-Ruiz JM. J. Struct. Biol. 2003;142:218–231. doi: 10.1016/s1047-8477(03)00052-2. [DOI] [PubMed] [Google Scholar]
- (14).Terpetschnig E, Szmacinski H, Lakowicz JR. Anal. Biochem. 1995b;227:140–147. doi: 10.1006/abio.1995.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Pusey ML, Paley MS, Turner MB, Rogers RD. Crystal Growth and Design. 2007;7:787–793. [Google Scholar]
- (16).Lakowicz JR. Principles of Fluorescence Spectroscopy. second edition. Kluwer Academic / Plenum Publishers; NY: 1999. p. 302. NY. [Google Scholar]
- (17).Terpetschnig E, Szmacinski H, Malak H, Lakowicz JR. Biophys. J. 1995a;68:342–350. doi: 10.1016/S0006-3495(95)80193-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Ito L, Kobayashi T, Shiraki K, Yamaguchi H. J. Sync. Rad. 2008;15:316–318. doi: 10.1107/S0909049507068598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).McPherson A, Cudney RJ. Struct. Biol. 2006;156:387–406. doi: 10.1016/j.jsb.2006.09.006. [DOI] [PubMed] [Google Scholar]
- (20).Furuichi M, Nishimoto E, Koga T, Takase A, Yamashita S. Biochem. Biophys. Res. Comm. 1997;233:555–558. doi: 10.1006/bbrc.1997.6429. [DOI] [PubMed] [Google Scholar]
- (21).Jullian M, Crosio MJ. Cryst. Growth. 1991;110:182–187. [Google Scholar]
- (22).Ng JD, Clark PJ, Stevens RC, Kuhn P. Acta Cryst. D. 2008;64:189–197. doi: 10.1107/S0907444907060064. [DOI] [PubMed] [Google Scholar]