Abstract
Dictyostelium discoideum cells produce five dynamin family proteins. Here, we show that dynamin B is the only member of this group of proteins that is initially produced as a preprotein and requires processing by mitochondrial proteases for formation of the mature protein. Our results show that dynamin B-depletion affects many aspects of cell motility, cell-cell and cell-surface adhesion, resistance to osmotic shock, and fatty acid metabolism. The mature form of dynamin B mediates a wide range and unique combination of functions. Dynamin B affects events at the plasma membrane, peroxisomes, the contractile vacuole system, components of the actin-based cytoskeleton, and cell adhesion sites. The modulating effect of dynamin B on the activity of the contractile vacuole system is unique for the Dictyostelium system. Other functions displayed by dynamin B are commonly associated with either classical dynamins or dynamin-related proteins.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-010-0590-5) contains supplementary material, which is available to authorized users.
Keywords: Dictyostelium discoideum, Dynamin, Presequence, Peroxisome, Fission, Cell adhesion
Introduction
Dynamins are high molecular weight GTP-binding proteins with high intrinsic GTPase activity [1–3]. Classical dynamins comprise five well-defined domains: an N-terminal GTPase domain, a middle domain, a pleckstrin homology (PH) domain, a GTPases effector domain (GED), and a C-terminal proline-rich domain (PRD) [3]. Their function has been best studied in the context of the budding and scission of nascent vesicles from parent membranes [4–8]. Additional cellular functions include roles in actin-dependent membrane processes, the formation of cell surface extensions, and focal adhesion dynamics [9, 10].
Dynamin-related proteins lack a PRD and a PH domain. They have been classified into several subfamilies such as dynamin-like proteins (Dlps), OPA1/Mgm1p-like proteins, Mx proteins, mitofusins, and guanylate-binding proteins/atlastins [3]. All eukaryotic organisms studied so far possess at least a single dynamin-related protein that is involved in the division of mitochondria and peroxisomes [11–13]. It was shown for instance that mutations in yeast Dnm1 and human Drp1 strongly affect the distribution of mitochondria, but have little or no effect on membrane trafficking [11, 14, 15]. The mechanism by which these proteins control fission of mitochondria may be similar to that employed by dynamin 1 during vesicle formation [16]. Yeast Dnm1p, the plant dynamin-related proteins ADL2b, DRP3A and DRP3B, and mammalian Drp1 were shown to play a role in peroxisomal as well as mitochondrial fission [13, 17–21].
Several members of the dynamin superfamily are initially produced with an N-terminal leader sequences targeting them to mitochondria [3]. Members of this family localize to the space between the outer and inner mitochondrial membranes and play a role in fission or fusion of mitochondria, remodeling of the mitochondrial inner membrane, and cristae formation [22, 23]. The yeast dynamin-related protein Mgm1p, which is involved in mitochondrial fusion [24, 25], is an integral inner membrane protein facing the intermembrane space. Mgm1p is released into the intermembrane space after limited proteolysis by the mitochondrial rhomboid protease [26, 27]. Similarly, the human orthologue of Mgm1p, OPA1, was demonstrated to be tightly bound to the outer surface of the inner membrane [23, 28]. Downregulation of OPA1 in cultured cells leads to mitochondrial fragmentation, disruption of cristae structure, and consequent apoptosis.
Five members of the dynamin family were found in the social amoeba Dictyostelium discoideum: dynamin A (dymA), dynamin B (dymB), dynamin-like protein A (dlpA), dynamin-like protein B (dlpB), and dynamin-like protein C (dlpC). Dynamin A acts in membrane trafficking along the endo-lysosomal pathway, similar to the function of classical dynamins in higher eukaryotic cells [8]. The structure of dynamin A was studied in detail, leading to an atomic resolution model of its GTPase domain [29] and lower resolution EM-structures of the dynamin A ring complex in the presence and absence of nucleotide [30]. Dynamin-like proteins A, B, and C are closely related to one another, and resemble plant proteins with a role in cytokinesis and chloroplast division. The disruption of each individual gene results in abnormal cytokinesis [31].
Here we describe the post-translational processing of dynamin B and the cellular localization and function of the protein. Studies with cells producing altered amounts of dynamin B indicate a role in peroxisome biogenesis, maturation, and fission. Additionally, dynamin B is shown to have a modulating effect on cell adhesion, the dynamics of actin-rich structures in the cell periphery, and contractile vacuole function.
Materials and methods
Plasmid construction
Plasmids generated and used in this study are described below. The dynamin B gene contains no introns (GenBank accession no. XP_642447), and was amplified directly from genomic DNA and cloned between the SacI and XhoI sites of vector pDXA-HY and pDXA-3H [32]. The resulting plasmids pDXA-dynamin B and pC-dynamin B encode the full-length protein with N-terminal and C-terminal histidine tags, respectively. pΔNTS-dynamin B was constructed in the same way as pC-dynamin B, but using a dynamin B gene fragment lacking the first 408 base pairs.
We generated fusions with a C-terminal YFP by cloning gene fragments encoding full-length dynamin B, ΔNTS-dynamin B, and the 136 residues presequence (NTS) between the SacI and XbaI sites of pDXAmcsYFP [33]. The resulting plasmids are pDynamin B-YFP, pΔNTS-dynamin B-YFP, and pDXA/NTSYFP. Plasmid pDNeoGFP-PTS1, encoding a GFP construct with a C-terminal PTS1 tag that specifically targets GFP to peroxisomes, was a kind gift from Dr. M. Maniak.
The generation of dymB knock-out construct pE1-ΔdymB required several steps. A genomic fragment released upon digestion with EcoRI containing the first 1,466 base pairs of dynamin B coding sequence and around 1,500 base pairs of 5′UTR was cloned in pBluescript (Stratagene) giving plasmid pE1. pE1 was cut with Pst I/Xba I and religated to remove the Spe I site within the MCS (multiple cloning site). A 0.4-kb fragment, corresponding to the central part of the GTPase domain, was excised from the resulting plasmid pE1-ΔSpe I by Spe I/Nsi I digestion. The Spe I/Nsi I fragment (base pairs 613–1,020) was replaced by the 1.4-kb blasticidin S resistance cassette that had been excised by HindIII/XbaI digestion from vector pBsr2 [34]. The linearized replacement construct was transformed into AX2 cells. Transformations of D. discoideum cells with gene replacement constructs were carried out as described [35].
Antibody production and immunoblot analysis
A peptide of dynamin B containing amino acid residues from 369 to 523 was produced as hexa-histidine tagged protein in E. coli and purified by Ni–NTA chromatography (Qiagen). The peptide was used to generate polyclonal rabbit antisera against dynamin B. Antibodies were affinity purified using the purified dynamin B fragment coupled to Affi-Gel 10 (BioRad). The antiserum was diluted with TBS (PBS containing 0.05% TWEEN 20) and incubated under agitation with gel matrix overnight. The gel was washed with TBS, and antibodies were eluted with 100 mM glycine pH 2.5. The eluate was immediately neutralized with 1M Tris-HCl pH 8.0, BSA was added as stabilizer, and the solution was concentrated using Centricon 50 spin columns (Amicon).
For immunoblotting, D. discoideum proteins from wild-type and mutant strains were separated on 8% SDS-PAGE gels and transferred to a nitrocellulose membrane (Schleicher and Schuell). Membranes were blocked in TBS containing 5% non-fat dry milk powder for 1 h and incubated with 1:1,000 dilution of affinity purified anti-dynamin B antibody in the same buffer for either 1 h at room temperature or overnight at 4°C, followed by detection with an HRP-conjugated secondary antibody and ECL performed according to the manufacturer’s instructions (Pierce).
Cell culture of D. discoideum
D. discoideum AX2 cells were used throughout this work, unless otherwise indicated. Cells were grown on plates or in culture flasks stirred at 180 rpm at 21°C in HL5C media (ForMedium). Fatty acids (Sigma-Aldrich) were added to the media from ethanol stocks and mixed by vortexing followed by ultrasonication for 5 min in warm water. Identical concentrations of ethanol (usually less than 0.5%) were added as controls.
D. discoideum cells were transformed with expression constructs by electroporation, and transformants were selected in the presence of appropriate antibiotics as described [36]. Selection was performed using 5 μg/ml Blasticidin S (ICN Biomedical) or 10 μg/ml G-418 (ForMedium). In addition, dynamin B-depleted cells (dymB−) were subcloned on bacterial lawns. Deletion of dymB was verified by PCR and Southern blotting. For growth on bacterial lawns 50–100 D. discoideum cells were mixed with 0.5 ml bacterial suspension in MES-buffer (20 mM MES-NaOH pH 6.8, 2 mM MgCl2, 0.2 mM CaCl2), plated on SM agar and incubated at 21°C.
Viability of D. discoideum in HL5C (isotonic condition), 350 mM sorbitol in MES buffer (hypertonic condition), or distilled water (hypotonic condition) was determined after 1 h incubation in the respective medium with shaking at 180 rpm at 21°C. Approximately 100 cells per incubation were plated on bacterial lawns. The number of survivors was determined by colony counting after 5 days.
Fluorescence microscopy
A total of 2–4 × 106 cells were placed on 22 × 22 mm cover slips in media and allowed to attach for 30 min. Cells were washed twice with 10 mM MES-NaOH, pH 6.5, fixed with 3% paraformaldehyde in 10 mM PIPES buffer pH 6.0 for 30 min, and washed with PBS. Unreacted paraformaldehyde was quenched with 100 mM glycine in PBS for 5 min and permeabilized by washing with 70% ethanol or in case of tubulin staining by incubation with 0.02% Triton X-100 for 5 min followed by three washes with PBS. Cells were blocked with 0.045% fish gelatin (Sigma-Aldrich) and 0.5% BSA in PBS (PBG) for 1 h at room temperature followed by incubation in primary antibody diluted in PBG overnight at 4°C unless otherwise stated. Mitochondria were stained with 1:100 diluted mouse monoclonal anti-mitoporin antibody 70-100-1 [37], tubulin with 1:150 diluted bovine α-tubulin antiserum T9026 (Sigma–Aldrich), and all dynamin B-YFP fusions for 3 h at room temperature with 1:200 diluted rabbit polyclonal anti-GFP antibody (AB3080 Millipore). For myosin 2 and α-actinin staining mouse monoclonal myosin 2 56-396-5 [38] and mouse monoclonal α-actinin 47-60-8 [39] were used in 1:150 dilutions. After extensive washing with PBS, cells were labeled for 1 h at room temperature with 1:250 dilutions of the appropriate secondary antibodies conjugated with Alexa Fluor 488 or Alexa Fluor 555 or Alexa Fluor 594 (Invitrogen). The F-actin cytoskeleton was stained with Alexa Fluor 633-phalloidin (Invitrogen) for 30 min at room temperature. After extensive washing with PBS, cover slips were mounted on glass slides with SlowFade Gold antifade reagent (Invitrogen). Images were recorded using a Leica TCS SP2 inverted confocal microscope, 63 × 1.4 NA oil immersion objective; identical laser intensity and photo-detector gain were applied for all image acquisition. 3D reconstitutions were obtained by the superimposition of 30 overlapping Z-sections using the Leica software. Images from all focal planes were rendered as a single maximum-intensity projection using Leica software. Peroxisome numbers were counted for 110 cells from three independent experiments. Statistical significance was calculated by paired t test (N = 110, P < 0.0001). Peroxisome velocity was tracked by live time-lapse epi-fluorescence imaging, and DiaTrack 3.01 (Semasopht) was used for image analysis.
For the imaging of the peripheral actin cytoskeleton by TIRF microscopy, cells were placed in 35-mm glass-bottom petri plates (MatTek), allowed to adhere for 5 min, and fixed. Cells were stained with Alexa Fluor 633 phallodin (Invitrogen) and subsequently kept in antibleach mix containing 0.1 mg/ml glucose oxidase, 0.018 mg/ml catalase, and 3 mg/ml glucose. For live cell TIRF microscopy, cells expressing GFP-actin were used. Cells were washed three times with MES buffer and kept in MES buffer for 1 h prior to imaging.
For dynamin B TIRF microscopy, AX2 cells producing ΔNTS-dynamin B fused to YFP were used. Cells were washed three times with MES buffer prior to image acquisition. All TIRF images were taken with an Olympus 1X81 inverted microscope equipped with a TIRF module, a 60×/1.49 NA oil immersion objective, and a Hamamatsu C10600 ORCA-R2 CCD camera. A 1.6× Optovar lens was used to enlarge the image on the camera chip. Images were processed using Adobe Photoshop, and figures were compiled using Adobe Illustrator.
Electron microscopy
Cells were prepared for transmission electron microscopy as previously described [40]. Labeling of cells with colloidal gold was done according to Griffiths and Consigili [41]. AX2 cells over-producing dynamin B-YFP were fixed with 4% paraformaldehyde (Merck) and 0.2% glutaraldehyde (Sigma-Aldrich) in 200 mM HEPES-NaOH, pH 7.3, for 1 h at room temperature, followed by 1 h in 4% paraformaldehyde. Fixed cells were washed with PBS, pelleted at low speed, embedded in 2% low melting point agarose in PBS, and chilled. Small blocks were cut and incubated overnight with 2.3 M sucrose in PBS and then frozen in liquid nitrogen. Ultrathin cryo-sections were cut at −105°C with a Leica FCS cryo-microtome using a Drukeer diamond knife and transferred to coated grids. Grids were blocked for 15 min with 0.5% fish gelatin and 20 mM glycine in PBS. Sections were labeled with rabbit polyclonal anti-GFP antibody (Clontech Laboratories, Inc.) 1:10 diluted blocking buffer and detected with protein A-colloidal gold. Grids were then embedded in aqueous 2% methylcellulose (Sigma-Aldrich) supplemented with 0.3% uranyl acetate (SERVA Electrophoresis GmbH). Images were recorded with a Zeiss EM-10 electron microscope.
RIC/DIC double-view microscopy
Live cells were viewed in glass-bottom petri plates (MatTek, Corp.). Cells were washed and kept in MES buffer. Images were taken with an Olympus FV1000 microscope. An UPLSAPO 60× 1.35N.A. objective, a Chroma HQ filter set, and a 635-nm HeNe laser were used for imaging. Double-view microscopy was performed as described earlier [42]. Cell size and adhesion areas detected by DIC and RIC microscopy were quantified using ImageJ software [43]. The RIC/DIC ratio is used as an indicator of the relative size of the adhesion area per cell. Every fifth frame of 50 frames was measured; data from different measurement were pooled and plotted using Origin 7 software. Statistical significance was calculated by paired t test (N = 132, P < 10−15).
Functional analysis of mitochondria and peroxisomes
The activity of the mitochondrial marker succinate dehydrogenase was measured in whole-cell lysates as described previously [8]. Total catalase was measured using the Amplex catalase kit (Invitrogen) following the manufacturer’s protocol. Whole-cell lysates were used for measurements, and total catalase activity normalized for protein content was determined. Statistical significance was calculated by paired t test (N = 9, P < 0.005).
Protease accessibility assay
Mitochondria from AX2 cells overproducing dynamin B-YFP were prepared as described [37]. Purified mitochondria were incubated on ice for 5 or 30 min in the presence or absence of up to 0.1 mg/ml trypsin. Reactions were stopped by the addition of 10 mM PMSF; equal amounts of protein were separated by SDS-PAGE and analyzed by immunoblotting on nitrocellulose membranes. Dynamin B was detected as described above, and mitochondrial nucleoside diphosphate kinase (mNDP kinase) was detected using the rabbit polyclonal antibody described by Troll and coworkers [44].
Adhesion assays
Cell-to-substrate adhesion was analyzed in HL5C media in the presence and absence of 10 mM EDTA; 4 ml cell suspension containing 1 × 106 cells/ml was placed in 50-ml conical flasks and incubated for 45 min without agitation, followed by 3 min incubation on a rotary shaker at 80 rpm. The number of non-adherent cells was determined by counting cells in the supernatant.
For cell-to-cell adhesion, a suspension with a final density of 1 × 105 cells/ml was incubated on a rotary shaker at 180 rpm for 3 days. At intervals, samples were taken, and the number of non-aggregated cells was determined by counting single cells only. The total number of cells was determined after breaking down all aggregates by passing the suspension 20 times through a narrow-bore plastic pipette tip. The fraction of single cells was used as the measure of cell-to-cell adhesion. Measurements were performed each time in triplicate, and the results shown represent several independent experiments.
Pinocytosis and phagocytosis assays
Phagocytosis and pinocytosis were measured as described previously [8].
Results
Domain organization of D. discoideum dynamin B
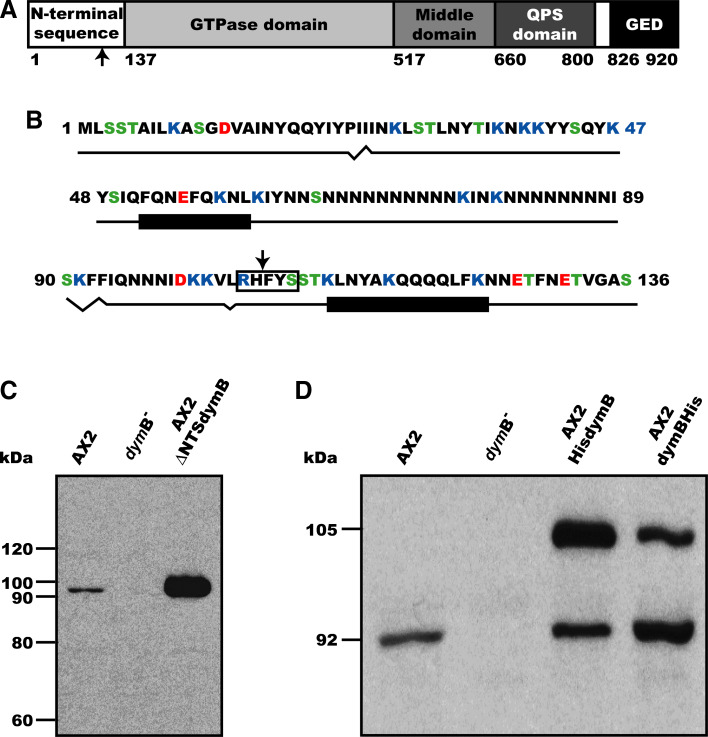
The coding sequence of dynamin B gene (GenBank AJ251163) consists of 2,760 base pairs and contains no introns. With a calculated molecular mass of 105.3 kDa, dynamin B is one of the largest members of the dynamin family. Dynamin B contains four conserved domains: a consensus GTPase domain, a middle domain, an unstructured QPS domain that is rich in glutamine, proline, and serine residues, and a carboxy-terminal GTPase effector domain (GED) (Fig. 1a). The maximum-likelihood tree constructed by Urushihara and co-workers, which is based on the alignment of 78 dynamin family proteins, indicates that dynamin B is most closely related to Vps1p [31]. In contrast to Vps1p, dynamin B is produced as a preprotein with an unusually long presequence consisting of 136 amino acids. Typically presequences consist of 15–50 amino acids [45]. Presequences serving as mitochondrial targeting sequence are a feature of members of the highly conserved OPA1/Mgm1p family of dynamin-related proteins that control mitochondrial integrity and dynamics [3]. The dynamin B presequence is basic with a pI of 9.8 and rich in Asn (26%), Gln (8%), Ile (10%), Lys (12%), and Tyr (9%) and Ser (8%) residues (Fig. 1b). The GTPase domain of dynamin B (residues 137–516) shows the highest sequence identity for this region with Dictyostelium dynamin A (53%), rat Drp1 (52%), yeast Dnm1p and Vps1p (49%), and human dynamin 1 (48%). The middle domain (residues 517–659) shares the highest degree of sequence identity with dynamin A (32%), Dnm1p (30%), and Drp1 (28%). Residues 660–800 form the QPS-domain. A similar region is present in dynamin A, but not in any other dynamin-like GTPases. It consists of many polar residues (63%), but only a few charged amino acids (12%). This region is especially rich in glutamine (31%), serine (21%), and proline residues (10%), which occur in stretches of up to 12 amino acids. Proline residues are clustered in the center of the QPS domain and may serve a similar function in mediating protein-protein interactions as in the PRD of mammalian dynamin 1. The extreme C-terminus of dynamin B (residues 826–920) shows higher sequence homology to the corresponding domains of other dynamins Drp1 (32%), Dnm1p (30%), dynamin A (29%) and dynamin 1 (21%). This region is known as the GTPase-effector domain (GED) in dynamin 1 because it interacts with the GTPase domain and stimulates its enzymatic activity. Common to GEDs is their strong α-helical character. Based on sequence composition, this is also true for the C-terminal region of dynamin B.
Fig. 1.
Domain organization and post-translational processing of dynamin B. a Domain organization of dynamin B. Domains are represented as boxes. The numbers indicate the first amino acid residue for each domain; arrow indicates the predicted mitochondrial processing protease cleavage site. b Dynamin B presequence that acts as efficient mitochondrion-targeting sequence (MTS). Positively charged amino acid residues are shown in blue, negatively charged residues in red, hydroxyl-containing residues in green. A predicted R-2 motif is boxed, and the arrow indicates the proteolytic cleavage site. Predicted secondary structure is shown under the sequence. Black rectangles represent α-helices, broken line β-sheets, and straight line unstructured regions. c Molecular weight of native dynamin B. Equal amounts of whole-cell lysates from AX2, AX2 dymB−, and AX2 overproducing ∆NTS-dynamin B cells were separated on 8% SDS-PAGE, blotted, and probed with affinity-purified anti-dynamin B rabbit antibody. d Dynamin B is produced as a longer precursor that is subsequently proteolytically processed. Equal amounts of whole-cell lysates from AX2, dymB− cells, and AX2 cells overproducing dynamin B carrying a His-tag at either the N- or C-terminus were loaded
Processing and subcellular localization of dynamin B
Dynamin B isolated from wild-type cells migrates on SDS-PAGE with an apparent molecular weight of 92 kDa, which corresponds to the predicted size of dynamin B after truncation of the predicted presequence (Fig. 1c). Over-production of recombinant dynamin B preprotein and amino- or carboxy-terminal histidine tagged versions of the protein result in the appearance of a second band corresponding to the unprocessed preprotein. In addition to a partial overloading of the processing machinery that is apparent when the untagged protein is overproduced, the presence of sequence tags appears to interfere with effective processing. The presence of an amino-terminal histidine tag results in stronger interference with protein processing than C-terminal tagging (Fig. 1d). Amino-terminal YFP-tagging blocks proteolytic processing of the preprotein completely (data not shown).
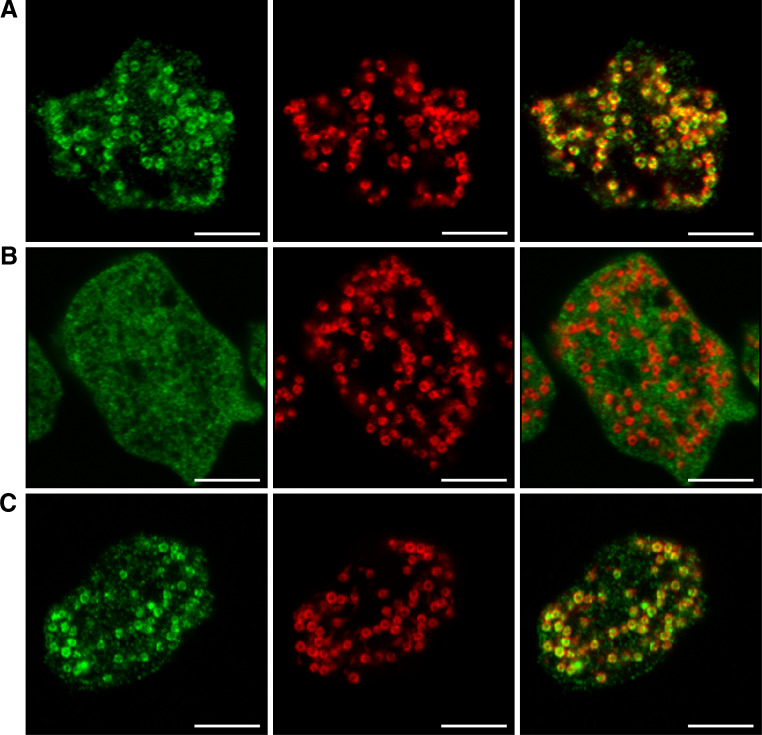
Since the anti-dynamin B antibody used in immunoblotting experiments gives a strong background signal in cellular localization experiments, we used a fluorescent tag to probe dynamin B localization. We transformed wild-type cells with a plasmid carrying a gene cassette under the control of actin-15 promoter that encodes dynamin B preprotein carrying a carboxy-terminal yellow fluorescent protein tag (dynamin B-YFP) [33]. We observed dynamin B-YFP predominantly in association with mitochondria (Fig. 2a). The MitoProt program [46] predicts, albeit with relatively low probability, that the dynamin B presequence can serve as a mitochondrial targeting signal. The presequence contains a potential cleavage site for the mitochondrial processing peptidase (R-2 motif) at position 106 [47]. The presequence is, however, unusually large compared to most mitochondrial targeting sequences and has an amino acid composition with a higher ratio of uncharged hydrophilic to hydrophobic amino acids than commonly observed in presequences [48]. To further investigate the role of the presequence, we generated fusion construct ΔNTS-dynamin B-YFP, lacking the presequence but with a carboxy-terminal YFP tag, and the expression construct NTS-YFP with the dynamin B presequence directly fused to the amino-terminus of YFP. ΔNTS-dynamin B-YFP displays a cytoplasmic distribution (Fig. 2b), whereas the presequence-tagged version of YFP is efficiently targeted to mitochondria (Fig. 2c). Dynamin B-preprotein carrying an amino-terminal YFP-tag is found in the cytosol (data not shown). These experiments demonstrate that the dynamin B-presequence is both necessary and sufficient for mitochondrial targeting, if it is positioned at the amino-terminus of a protein.
Fig. 2.
The presequence of dynamin B is necessary and sufficient for targeting proteins to mitochondria. a Dynamin B-preprotein carrying a C-terminal YFP-tag localizes to mitochondria. b YFP-tagged dynamin B lacking the presequence is not targeted to mitochondria. c A YFP construct carrying the dynamin B-presequence is efficiently targeted to mitochondria. Cells were fixed, permeabilized, and immunostained with anti-GFP (left panels, green). Anti-mitoporin was used for staining mitochondrial outer membranes (middle panels, red). Merged images are shown in the right panels. Single confocal planes are shown. Scale bars 5 μm
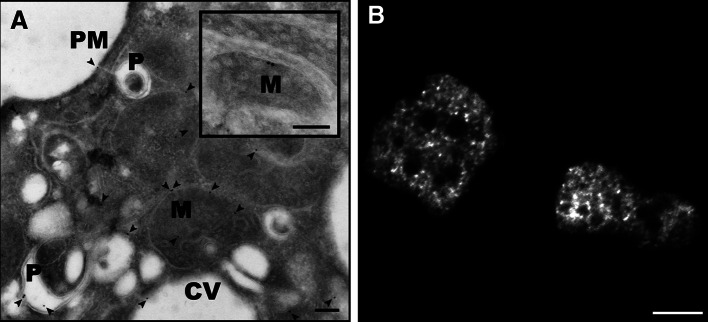
The doughnut-like structures observed in fluorescence images of dynamin B-YFP-producing cells (Fig. 2a) suggest that dynamin B is predominantly associated with the outer mitochondrial membrane. To reveal the exact localization of dynamin B-YFP, we performed immunogold electron microscopy. Ultra-thin sections of dynamin B-YFP-producing cells were stained with a polyclonal anti-GFP antibody. The resulting electron micrographs show that dynamin B-YFP is in part associated with mitochondria (Fig. 3a). Additionally, the immunogold labeling indicated the presence of dynamin B on other membranous organelles and at the plasma membrane. Localization to the plasma membrane or structures in close association with it was confirmed by evanescent field microscopy (Fig. 3b).
Fig. 3.
Cellular localization of dynamin B. a Electron micrograph showing colloidal gold immunostaining for dynamin B (black dots). YFP-tagged dynamin B is found in association with mitochondria (M), phagosomes (P), the contractile vacuole (CV), and the plasma membrane (PM). Arrows show gold particles. The inset shows an enlarged cross-section of a mitochondrion. Scale bars 0.2 μm. b Evanescent field fluorescence micrograph showing the presence of dynamin B-rich structures close to the plasma membrane. Scale bar 5 μm
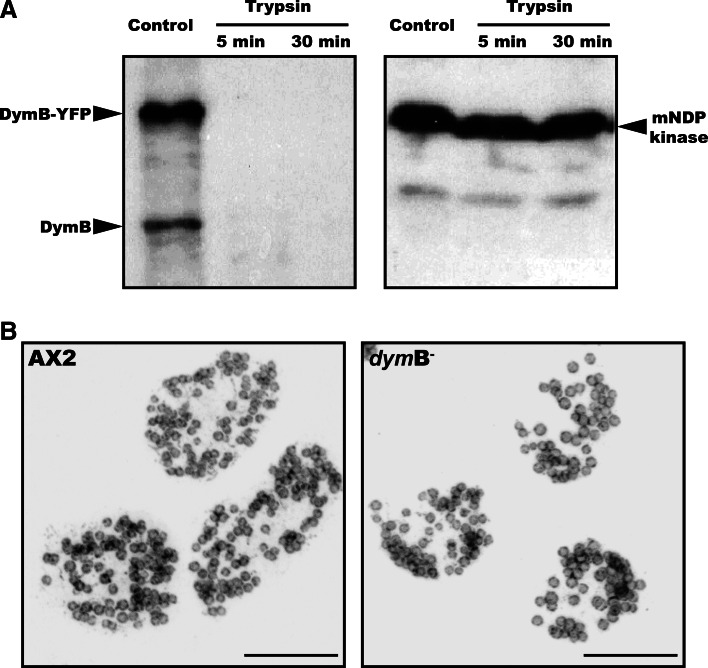
To address the question whether dynamin B-YFP localizes to the outside or inside of the outer mitochondrial membrane, the sensitivity of the fusion protein to trypsin exposure was tested in a protease accessibility assay. Fractions of purified mitochondria were incubated with and without trypsin, followed by separation of the mitochondrial proteins by SDS-PAGE and analysis by immunoblotting using polyclonal antibodies raised against dynamin B or a polyclonal anti-GFP antibody. We used the mitochondrial intermembrane space protein mNDP-kinase as a control [44]. In agreement with a predominant localization of the dynamin B construct at the outside of the outer mitochondrial membrane, dynamin B was completely digested upon trypsin treatment, and mNDP-kinase remained largely intact (Fig. 4a).
Fig. 4.
Dynamin B localizes to the outer mitochondrial membrane. a Dynamin B preprotein localizes to the outside of the outer mitochondrial membrane. Intact mitochondria from AX2 cells overproducing dynamin B-preprotein carrying a C-terminal YFP-tag were purified, treated with trypsin on ice, separated on 8% SDS-PAGE, blotted, and probed with affinity purified rabbit anti-dynamin B antibody or rabbit anti-mNDP kinase. The intermembrane space protein mNDP kinase is not affected, excluding the presence of damaged organelles. The arrows point to the bands for YFP-tagged dynamin B-preprotein, endogenous dynamin B, and mNDP kinase, respectively. b Depletion of dynamin B does not affect mitochondrial morphology. AX2 and dymB− cells were fixed and probed with anti-mitoporin antibody as described above. The superimposition of 30 confocal sections covering the entire cell volume is shown. Scale bars 10 μm
As it is well established that dynamins play an important role in mitochondrial fusion and fission [49], we generated dymB deletion mutants by homologous recombination to test for changes in mitochondrial morphology and function. The resulting dymB− cells did not display obvious differences in mitochondrial morphology (Fig. 4b). An apparent reduction in the number of mitochondria correlates with the smaller cell size of dymB− cells. Succinate dehydrogenase activity normalized for protein content is slightly reduced in dymB− cells (−11 ± 6%).
Cellular morphology of dynamin B-depleted cells
We measured cell-spreading areas of 113 ± 27 μm2 and 150 ± 40 μm2 for dymB− and wild-type cells, respectively (N = 200, P value <0.0005). In addition to their smaller size, dymB− cells show a number of subtle changes in their overall morphology. Deletion of the genes encoding dynamin A, DlpA, DlpB, or DlpC was shown to interfere with normal cytokinesis and to induce the formation of large multinucleated cells [8, 31]. In contrast, dymB− cells show no cytokinesis defect. Similarly, development and differentiation are not affected by dynamin B-depletion, and spore viability is normal in dymB− cells.
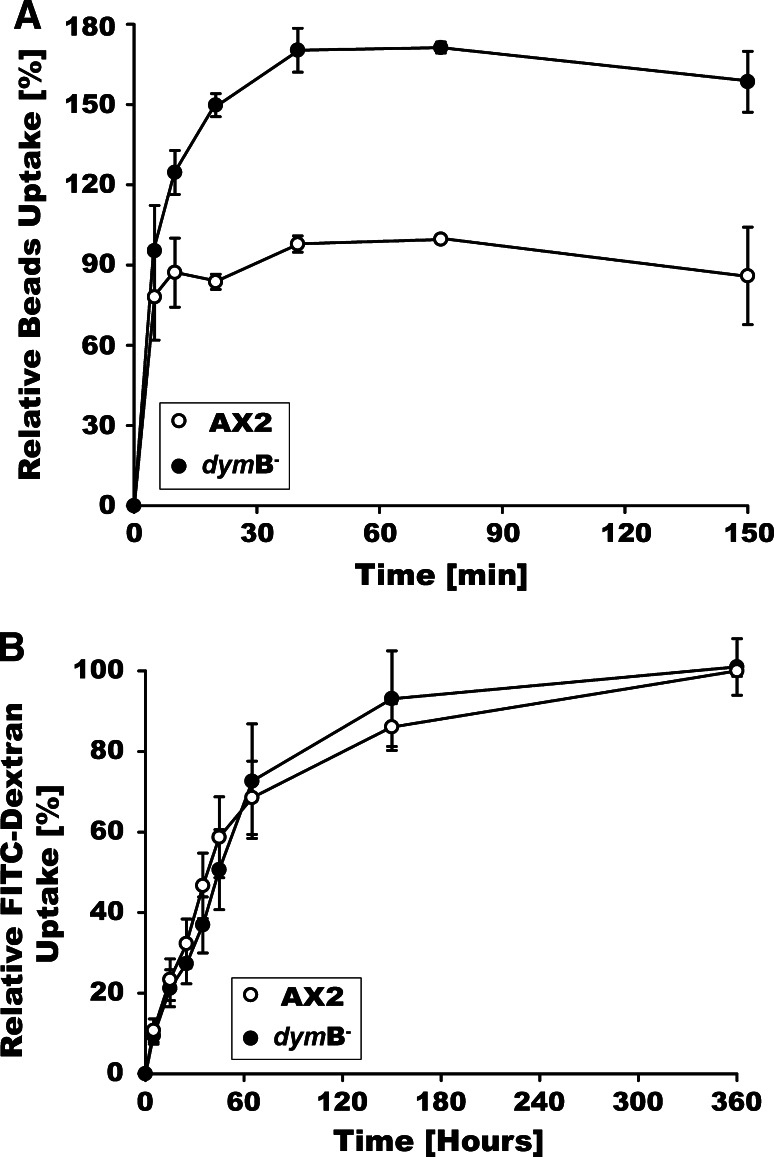
Dynamin B-depleted cells grow slower in suspension culture, with doubling times of 11.1 instead of 9.2 h (Supplementary Fig. 1A). However, growth on bacterial lawns as measured by the increase in plaque diameter is 40 ± 8% faster with dymB− cells (Supplementary Fig. 1B). The enhanced growth rate in the presence of a bacterial food source appears to be linked to an increased capacity for particle uptake by phagocytosis. Dynamin B-depleted cells display a 75% increase in total particle uptake, as measured by the rate of ingestion of fluorescent polystyrene beads of 1 μm diameter (Fig. 5a). In contrast, the rate at which fluid phase endocytosis proceeds in dymB− cells appears unaffected (Fig. 5b).
Fig. 5.
Dynamin B-depleted cells display increased phagocytosis, while pinocytosis is unaffected. a Phagocytosis is increased significantly after dynamin B-depletion. AX2 and dymB− cells were mixed with fluorescently labeled polystyrene-beads. At the indicated time points, beads were removed from the samples, cells were lysed, and phagocytosis was determined. Maximum uptake by AX2 cells was set as 100%. Error bars represent the standard deviations for three independent experiments. b Pinocystosis is independent of dynamin B. AX2 and dymB− cells were incubated in the presence of media containing FITC-dextran. At indicated times cells were lysed, and FITC-dextran uptake was determined by fluorescence. Maximum uptake by AX2 cells was set as 100%. The values represent averages from five independent experiments. Error bars indicate standard error of the mean
Dynamin B-depletion affects peroxisome morphology and function
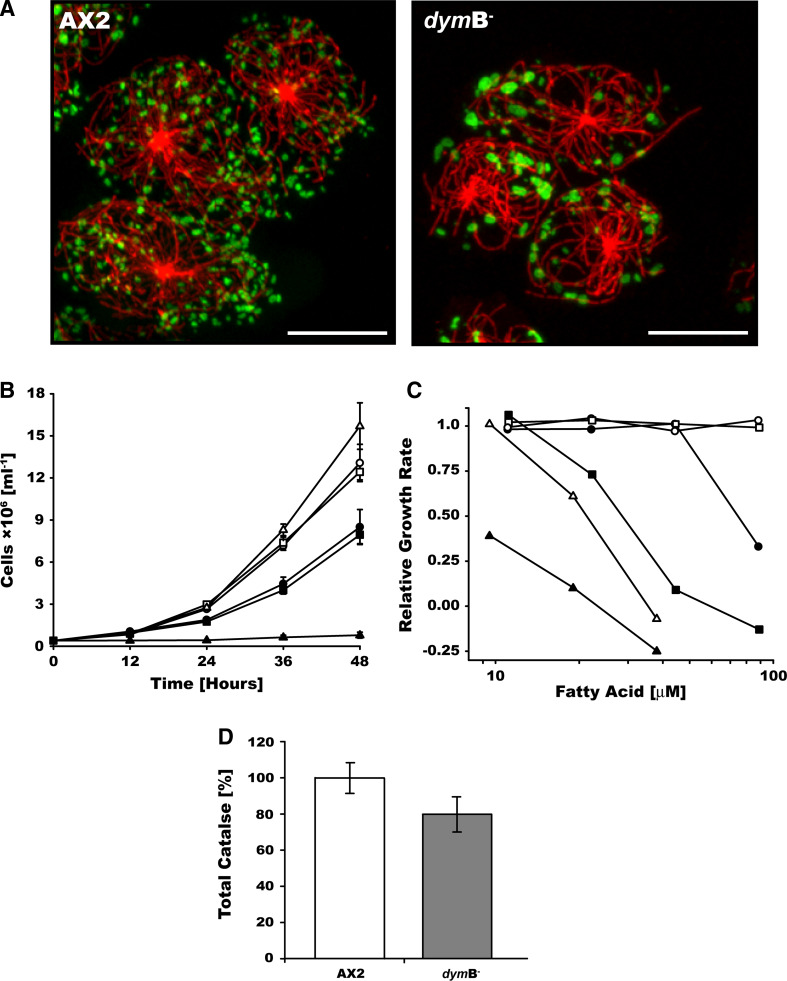
Peroxisomes are pleomorphic, single-membrane-bound organelles that house a wide variety of critical metabolic pathways. Their proper functioning is vital for lipid metabolism and free radical detoxication [50]. Similar to the morphological and functional changes induced by the depletion of the dynamin-like proteins Vps1p in yeast and Drp1 in mammalian cells [21, 51], depletion of dynamin B leads to a marked increase in the size of peroxisomes, a more than threefold reduction in their number, and a shift in their localization towards the cell periphery (Fig. 6a, supplementary movie 1 and 2). Time lapse live confocal microscopy of dymB− cells producing GFP-PTS1 shows fewer peroxisomes and more restricted movements of the organelle compared to wild-type cells (Supplementary movie 3 and 4). The saltatory movement of peroxisomes is unchanged and was determined as 0.8 ± 0.01 μm/s in both wild-type and dymB− cells.
Fig. 6.
Dynamin B-depletion affects the peroxisomal morphology and number, and alters the metabolism of fatty acids. a The number of peroxisomes is reduced and their size increased in dymB− cells. AX2 and dymB− cells producing construct GFP-PTS1 with type 1 peroxisomal targeting signal (green) were fixed, permeabilized, and immunostained for α-tubulin (red). The superimposition of 30 confocal sections covering the entire cell volume is shown. A threefold reduction in the number of peroxisomes was determined for dymB− cells. Scale bars 10 μm. b Dynamin B deletion increases the sensitivity of cells towards the supplementation of growth media with fatty acid. Growth curve of AX2 and dymB− cells in the presence of fatty acid. AX2 (open symbols) and dymB− cells (filled symbols) were grown in suspension on HL5C media in absence (circles) or presence of 12.5 μg/ml (squares) or 25 μg/ml (triangles) oleic acid. Error bars represent the standard deviations for three independent cultures grown in parallel. c Dynamin B null cells are more sensitive to long chain unsaturated fatty acid. Relative growth rates of AX2 (open symbols) and dymB− cells (filled symbols) in the presence of increasing concentrations of oleic (circles), linoleic (squares) and cis-4,7,10,13,16,19-docosahexanoic (triangles) acids are shown. Growth rates are calculated from the growth curves obtained from three independent cultures for each fatty acid concentration. d Total catalase activity normalized for protein content (N = 9, P < 0.005)
Mitochondria and peroxisomes are metabolically linked organelles, with the β-oxidation of very long chain fatty acid being restricted to peroxisomes [52, 53]. To test the functional competence of peroxisomes in dymB− cells, we grew cells in media supplemented with different concentrations of oleic, linoleic, and cis-4,7,10,13,16,19-docosahexanoic acid. Unsaturated long-chain fatty acids have no effect on cell growth at concentrations below 10 μM. However, the reduced tolerances of dymB− cells to media supplementation with fatty acids concentrations above 10 μM suggest that dynamin B-depletion affects the metabolic competence of peroxisomes (Fig. 6b, c).
Peroxisomes play a key role in governing H2O2 metabolism [54]. To examine the extent to which the reduced number and altered morphology of peroxisomes in dymB− cells affect their function, we compared catalase activity levels in total cell lysates from AX2 and dymB− cells. Normalized for protein content, dymB− cells displayed a 20% reduction in catalase activity (Fig. 6d). Therefore, our results indicate that peroxisome morphology and function are compromised in dymB− cells.
Dynamin B affects cell adhesion, cytoskeletal structures, and osmo-regulation
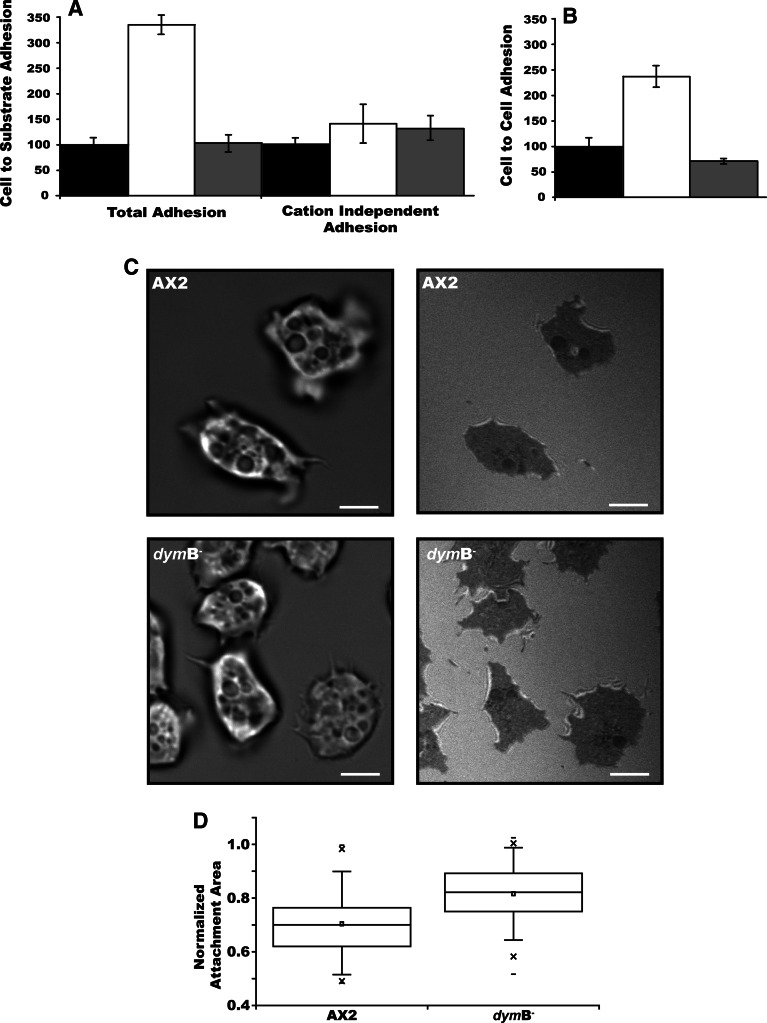
Dynamin B deletion mutants adhere to the surface more than threefold stronger than wild-type cells. It is known that D. discoideum has two types of adhesion sites that differ in their sensitivity to bivalent cations. Contact sites of type A (CsA) are stable in the presence of EDTA concentrations above 10 mM and are produced by the cells during the aggregation stage of development. Contact sites of type B (CsB) appear during early development and are easily disrupted in the presence of 1–2 mM EDTA [55]. In the presence of EDTA dymB− cells displayed only marginally better adhesion, indicating that the observed effect is CsB dependent. Transformation of dymB− cells with an expression vector carrying a functional copy of the dymB gene leads to the complete rescue of the adhesion phenotype (Fig. 7a).
Fig. 7.
Changes in dymB expression affect adhesion. Adhesion levels for wild-type (black bars), dymB− (white bars), and dymB+ cells (grey bars) are shown normalized for control measurements with wild-type cells. a Substrate adhesion of wild-type, dymB−, and dymB+ cells was measured in the absence (total adhesion) and in the presence (cation-dependent adhesion) of EDTA. b Cell-to-cell adhesion is increased in dymB− and decreased in dymB+ cells. Adhesion of AX2 cells was set as 100%. dymB+ represents dymB− cells overproducing dynamin B protein. Error bars represent the standard deviations for five independent measurements. c AX2 and dymB− cells were analyzed by RICM/DIC double-view live-cell confocal microscopy. Dynamin B-depleted cells are flatter than wild-type cells and display an increased contact surface. RICM images are shown in the right panel and the corresponding bright-field images in the left panel. Scale bar 5 μm. d Depletion of dynamin B leads to an increase in the cell-attachment area (N = 132, P value <10−15). The normalized attachment area per cell was calculated as ratio of the contact areas observed in RIC and DIC images
Compared to wild-type cells, dymB− cells display a stronger tendency to form cellular aggregates. We observed large cell aggregates when dymB− cells were cultivated in suspension. A suspension of dymB− cells with a final density of 1 × 105 cells/ml was grown for 3 days on a rotary shaker. At intervals, samples were taken, and the number of non-aggregating single cells was counted. In parallel a second sample was taken, and aggregates were disrupted by repeated pipetting before counting. The number of cells in aggregates, calculated as the difference in cell counts of both samples, was significantly higher for the dymB null cells 66% (±6%) in comparison to the wild-type cells 27% (±5%). This means that under these conditions, the apparent cell-cell adhesion of dymB null cells is on average 2.4-fold greater than for wild-type cells. Cells over-producing dynamin B sixfold form significantly fewer aggregates. The number of cells in aggregates was 18% (±6%) corresponding to a 1.5-fold reduction in their case (Fig. 7b). These results indicate that dynamin B negatively regulates bivalent cation-sensitive cell-to-cell and cell-to-surface adhesion.
Relative to their size, dynamin B null cells form larger contact areas with the glass surface than control cells (Fig. 7c). This can be visualized by reflection interference contrast microscopy (RICM) when applied in combination with differential interference contrast microscopy (DICM) [42]. Compared to wild-type cells, migrating dymB− cells display similar activity in the extension and retraction of pseudopodial structures, but form more stable contact areas and display slower and less directed movements of the cell centroid (Supplementary movie 5 and 6). The ratio of the RICM-to-DICM areas gives the normalized attachment area. Our results show that the deletion of dynamin B significantly (P < 10−15) changes cell attachment; dymB− cells are more adherent with a mean ratio of 0.82 ± 0.1 compared to 0.70 ± 0.1 for AX2 (Fig. 7d).
In addition to revealing the surface areas that are in close contact with the substratum, RIC microscopy is ideally suited for imaging membranous organelles that are in very close proximity to the plasma membrane and near to the cover glass surface. The contractile vacuole system of D. discoideum is an osmoregulatory organelle formed by a circumferential network that directly underlies the plasma membrane [56, 57]. The activity of the organelle allows D. discoideum to survive under hypotonic stress by accumulating and expelling excess water out of the cell. An association of dynamin B with the contractile vacuole system was first suggested by a partial co-localization in immunogold-labeled electron micrographs (Fig. 3a). A role of dynamin B in the regulation or maintenance of this organelle was further indicated by the twofold increase in viability of dymB− cells following exposure to hypotonic conditions (Supplementary Fig. 1C). The cisternae and interconnecting ducts of the contractile vacuole system are well visible in RIC images as dark vesiculotubular structures. Movie sequences assembled from RIC images show that the morphology and dynamics of the contractile vacuole system is altered in dymB− cells in favor of increased tubulation and decreased contractile activity of large cisternae (Supplementary movie 5 and 6).
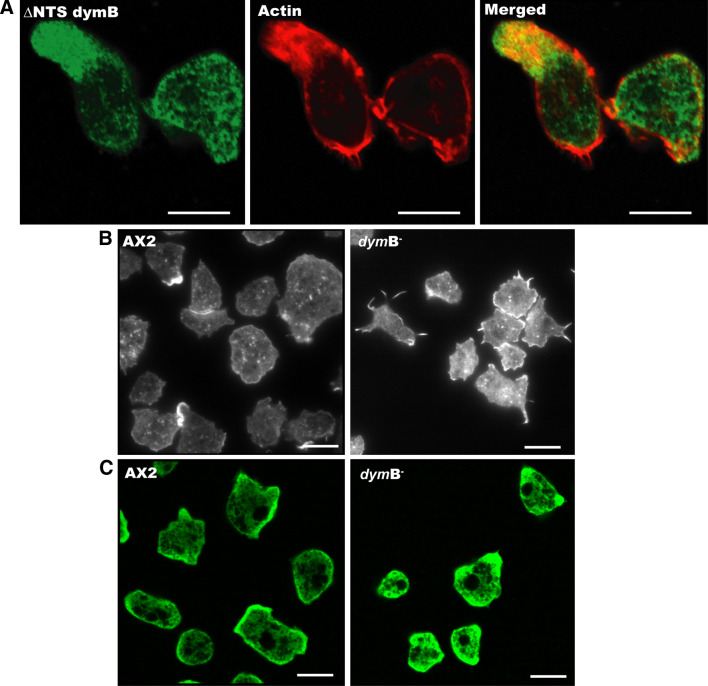
Next, we checked whether the actin cytoskeleton is affected by dynamin B-depletion. Evanescent field and confocal micrographs indicate the presence of dynamin B in the cell periphery with a particular enrichment in highly dynamic actin-rich structures at the leading edge of cells (Figs. 3b, 8a). Depletion of dynamin B leads to increased F-actin accumulation at the cell periphery along with a more polarized distribution of actin-rich structures. Evanescent field imaging of endogenous F-actin shows that wild-type cells contain more actin foci, whereas dynamin B null cells have longer cell surface extensions at the adhesion sites and show increased accumulation of F-actin near the cell surface (Fig. 8b). Evanescent field imaging of Dictyostelium cells producing GFP-actin reveals differences in motile activity and confirms the results obtained by RICM/DICM double imaging showing that dymB− cells form larger contact areas with the surface during active migration (Supplementary movie 7 and 8). To further investigate the effects of dynamin B-depletion on cytoskeletal and adhesion structures, we examined changes in the cellular localization of α-actinin and myosin 2. The concentration of α-actinin and myosin 2 appears to be significantly increased in dymB− cells, and the protein’s localization is less restricted to the cell periphery (Fig. 8c and supplementary Fig. 2).
Fig. 8.
Cellular localization of dynamin B and effect of dynamin B-depletion on the actin cytoskeleton. a Dynamin B colocalizes with actin-rich structures and is highly enriched in pseudopodia. ∆NTS dynamin B-YFP is shown in green, and F-actin stained with Alexa Fluor 633-phalloidin is shown in red. b Effects of dynamin B-depletion on F-actin organization near the cell surface. Dynamin B-depleted cells show a marked increase in F-actin accumulation near cell-substrate contact sites and an increase in the number of large spike-like extensions that are in contact with the cover glass surface, as indicated by TIRF microscopy images of dymB− cells immunostained with Alexa Fluor 633-conjugated phalloidin. c Distribution of α-actinin in AX2 and dymB− cells. In AX2 cells immunostaining for α-actinin is most prominent in the cell periphery. Dynamin B-depleted cells display stronger α-actinin staining that is less restricted to the cell periphery. A single confocal plane close the ventral surface is shown. Scale bars 10 μm
Discussion
Here we describe the post-translational processing and the cellular functions of D. discoideum dynamin B. Dynamin B is a nuclear-encoded protein, which is synthesized as preprotein in the cytosol, shuttled for processing to mitochondria, and returned in mature form to the cytosol, where it interacts with a distinct subset of organelle membranes, the plasma membrane, and cytoskeletal structures. Dynamin B contains a QPS-rich domain that appears to mediate many of the interactions involved. The presence of one or more regions that are enriched for glutamine, proline, serine, and asparagine residues or contain repeats of these residues is a frequent feature of Dictyostelium proteins. These regions are thought to mediate protein-protein interactions [58], and they are frequently found at positions where protein interaction domains are present in the metazoan orthologs. In the case of dynamin A, which contains a QNS-rich domain but lacks a PH domain, we were able to show direct binding to lipid membranes containing phosphatidic acid or phosphatidylinositol phosphates [30].
Cellular localization studies with YFP fused to the dynamin B presequence and YFP-tagged dynamin B preprotein show that the 136-residue preprotein acts as an efficient mitochondrial targeting sequence. The protease accessibility assay indicates that the protein remains mostly exposed at the outside of mitochondria while processing occurs. Therefore, the unusually long presequence appears to enter the mitochondrial intermembrane space and the matrix, where a yet unidentified protease converts the 105-kDa preprotein into the mature 92-kDa form. The processing of dynamin B thus differs from that of Mgm1p, the yeast homologue of the mammalian protein OPA1. Mgm1p functions specifically in membrane fusion. It is the only one of the three dynamin family GTPases regulating mitochondrial membrane dynamics in yeast that has an amino-terminal signal peptide. Mgm1p is found in the full-length and rhomboid (Rbd1p) processed forms, and both forms are required for efficient fusion [26]. Dynamin B lacks transmembrane domains, but contains an R-2 sequence motif that can be recognized by matrix processing proteases involved in preprotein processing [47]. This suggests that matrix processing proteases are responsible for the cleavage of dynamin B. The mature protein is released into the cytosol. A transient association of the preprotein with mitochondria is supported by the protease accessibility assay, whereas the fluorescence pattern of the YFP-tagged preprotein appears to indicate a more stable association of dynamin B with mitochondria. However, this can be explained by the combined effects of the YFP-tag interfering with effective processing and the high level production of the recombinant protein overloading the mitochondrial processing machinery. Dynamin B is produced in low abundance in vegetatively growing cells (data not shown). The processing and stable mitochondrial association of the presequence-tagged YFP construct demonstrates that the dynamin B presequence is sufficient to direct proteins to mitochondria.
Despite being targeted to and processed in association with mitochondria, dynamin B plays no obvious role in the division and maintenance of mitochondria. The mitochondrial fission mechanism employed by D. discoideum involves the extra-mitochondrial dynamin-based system used in plants and fungi and the ancient FtsZ-based intra-mitochondrial fission process inherited from the bacterial ancestor [59]. Deletion of D. discoideum dynamin A or the FtsZ orthologues FszA and FszB was shown to result in the formation of large tubular mitochondria [8, 60]. FtsZ proteins were functionally replaced by dynamin-like proteins in most eukaryotes, but they still retain this function in some lower eukaryotic organisms [61]. Experiments designed to knock out the genes encoding dynamin A, FszA, or FszB in a dymB− background produced no viable transformants (Nöthe, Rai, Manstein, unpublished results).
Several dynamin family members were shown to represent evolutionarily conserved components of the peroxisomes division/proliferation machinery. Consistent with the close relationship of dynamin B and yeast Vps1p indicated by phylogenetic analysis [31], the phenotypic changes displayed by dynamin B-depleted cells indicate a role of the protein in peroxisome biogenesis, maturation, and fission. In particular, a cargo-selected vesicular transport pathway from mitochondria to peroxisomes has been reported [62]. Some dynamin family members, such as plant DRP3A and DRP3B, yeast Dnm1p, and mammalian Drp1, participate in the division of peroxisomes and mitochondria, possibly coordinating the division of these metabolically linked organelles [13, 19, 63]. Our results show that similar to the situation following depletion of the dynamin-like GTPases Vps1p and Dnm1p in yeast and Drp1 in mammalian cells [13, 21, 51], depletion of dynamin B severely affects peroxisome morphology and abundance. The increased sensitivity to media supplementation with long chain fatty acids and decreased amount of catalase indicates that peroxisome function is affected by the depletion of dynamin B.
Mitochondria and peroxisomes are metabolically linked organelles. They cooperate in the β-oxidation of fatty acids and metabolism of reactive oxygen species (ROS). The preferred initial substrates for peroxisomal β-oxidation are the CoA esters of long chain fatty acids including (poly-) unsaturated fatty acids. Peroxisomal β-oxidative chain shortening generates H2O2 and ceases at the stage of octanyl CoA [64]. The observed correlation between the chain length of the supplemented fatty acids and the reduction in cell growth in combination with reduced levels of catalase activity suggests that dynamin B-depleted cells are less efficient in degrading ROS or less able to cope with ROS-induced damage. Moreover, reduced peroxisomal β-oxidation can increase the intracellular concentrations of free unsaturated long chain fatty acids. This may further contribute to the observed phenotype because of interference with calcineurin signaling pathways [65].
Our analysis of dynamin B-depleted cells indicates extensive changes in the morphology and function of the contractile vacuole system, a highly specialized osmoregulatory organelle, which is driven by actomyosin in cooperation with microtubule-associated motors. Increased tubulation of the contractile vacuole system observed in dynamin B-deficient cells correlates with F-actin enrichment at the cell periphery and can be a secondary effect resulting from changes in actin organization. However, the observed association of dynamin B with contractile vacuole membranes suggests a more direct involvement of the protein in the dynamics of the contractile vacuole system.
Our results indicate that dynamin B plays yet another role as a modulator of cell-surface interactions and cytoskeletal dynamics. The observed increase in capacity for the uptake of particles and the faster growth rate displayed by dymB− cells on bacterial lawns appears to be directly related to the formation of stronger cell-surface contacts. Similarly, classical dynamins have been implicated in controlling cell adhesion in cells forming focal contacts; planar assemblies of proteins formed at the sites where clustered integrins interact with immobilized extracellular matrix [66]. The dominant negative K44A mutation of mammalian dynamin 1 affects adhesion and associated changes in the organization of the actin cytoskeleton [67]. Dynamin 2-depleted U2-OS cells display increased levels of α-actinin-induced actin crosslinking, while depletion of dynamin 2 in fibroblasts inhibits migration of adherent cells [68]. These findings link the function of dynamin B to the regulation of cell adhesion and actin-dependent processes. Thus, they strengthen the functional parallel between focal adhesion sites of mammalian cells and actin foci in D. discoideum. Actin foci, which are formed in migrating D. discoideum cells at the sites where the ventral cell membrane is closely opposed to the substratum, are considered to be functionally similar to the focal adhesions or podosomes of higher eukaryotes [69, 70].
In summary, our data show that the low abundance protein dynamin B interacts with multiple target structures and plays a critical role in a wide range of cellular processes, which require the action of distinct classical and non-classical dynamin family members in other organisms. Dynamin B is initially produced as a preprotein with an unusually long 136-residue presequence that is efficiently targeted to mitochondria for processing. Processing involves a transient association with the outer mitochondrial membrane and goes to completion. The mature 92-kDa form of the protein functions in the context of actin-rich structures and multiple target membranes, including peroxisomes, the osmoregulatory contractile vacuole system, phagosomes, and the plasma membrane.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 5 (MOV 10,014 kb)
Acknowledgments
We thank S. Eschenburg, T.F. Reubold, G. Griffiths, and H. Horstmann for their help and discussions; M. Maniak for providing GFP-PTS1. The monoclonal anti-mitoporin 70-100-1, monoclonal anti-myosin 2 56-396-5, and α-actinin antibody developed by G. Gerisch and co-workers were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA, 52242. The work was supported by Deutsche Forschungsgemeinschaft Grant MA 1081/7-2 to D.J.M.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- DICM
Differential interference contrast microscopy
- Drp
Dynamin-related protein
- GED
GTPase effector domain
- GFP
Green fluorescent protein
- MTS
Mitochondrial targeting sequence
- NTS
N-terminal sequence or presequence
- ΔNTS
Truncated presequence
- PH
Pleckstrin homology
- PRD
Proline rich domain
- PTS1
Peroxisomal targeting signal of type 1
- QNS-rich
Glutamine-asparagine-serine-rich
- RICM
Reflection interference contrast microscopy
- TIRF
Total internal reflection fluorescence
- YFP
Yellow fluorescent protein
References
- 1.Schmid SL, McNiven MA, De Camilli P. Dynamin and its partners: a progress report. Curr Opin Cell Biol. 1998;10:504–512. doi: 10.1016/S0955-0674(98)80066-5. [DOI] [PubMed] [Google Scholar]
- 2.Danino D, Hinshaw JE. Dynamin family of mechanoenzymes. Curr Opin Cell Biol. 2001;13:454–460. doi: 10.1016/S0955-0674(00)00236-2. [DOI] [PubMed] [Google Scholar]
- 3.Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 4.Schlunck G, Damke H, Kiosses WB, Rusk N, Symons MH, Waterman-Storer CM, Schmid SL, Schwartz MA. Modulation of Rac localization and function by dynamin. Mol Biol Cell. 2004;15:256–267. doi: 10.1091/mbc.E03-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Bliek AM, Redelmeier TE, Damke H, Tisdale EJ, Meyerowitz EM, Schmid SL. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pohl J, Ring A, Korkmaz U, Ehehalt R, Stremmel W. FAT/CD36-mediated long-chain fatty acid uptake in adipocytes requires plasma membrane rafts. Mol Biol Cell. 2005;16:24–31. doi: 10.1091/mbc.E04-07-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu X, Verma DP. Dynamics of phragmoplastin in living cells during cell plate formation and uncoupling of cell elongation from the plane of cell division. Plant Cell. 1997;9:157–169. doi: 10.1105/tpc.9.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wienke DC, Knetsch ML, Neuhaus EM, Reedy MC, Manstein DJ. Disruption of a dynamin homologue affects endocytosis, organelle morphology, and cytokinesis in Dictyostelium discoideum . Mol Biol Cell. 1999;10:225–243. doi: 10.1091/mbc.10.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buccione R, Orth JD, McNiven MA. Foot and mouth: podosomes, invadopodia and circular dorsal ruffles. Nat Rev Mol Cell Biol. 2004;5:647–657. doi: 10.1038/nrm1436. [DOI] [PubMed] [Google Scholar]
- 10.Kruchten AE, McNiven MA. Dynamin as a mover and pincher during cell migration and invasion. J Cell Sci. 2006;119:1683–1690. doi: 10.1242/jcs.02963. [DOI] [PubMed] [Google Scholar]
- 11.Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell. 1999;4:815–826. doi: 10.1016/S1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- 13.Kuravi K, Nagotu S, Krikken AM, Sjollema K, Deckers M, Erdmann R, Veenhuis M, van der Klei IJ. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae . J Cell Sci. 2006;119:3994–4001. doi: 10.1242/jcs.03166. [DOI] [PubMed] [Google Scholar]
- 14.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tieu Q, Nunnari J. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J Cell Biol. 2000;151:353–366. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Vos KJ, Sheetz MP. Visualization and quantification of mitochondrial dynamics in living animal cells. Methods Cell Biol. 2007;80:627–682. doi: 10.1016/S0091-679X(06)80030-0. [DOI] [PubMed] [Google Scholar]
- 17.Motley AM, Ward GP, Hettema EH. Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J Cell Sci. 2008;121:1633–1640. doi: 10.1242/jcs.026344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arimura S, Aida GP, Fujimoto M, Nakazono M, Tsutsumi N. Arabidopsis dynamin-like protein 2a (ADL2a), like ADL2b, is involved in plant mitochondrial division. Plant Cell Physiol. 2004;45:236–242. doi: 10.1093/pcp/pch024. [DOI] [PubMed] [Google Scholar]
- 19.Fujimoto M, Arimura S, Mano S, Kondo M, Saito C, Ueda T, Nakazono M, Nakano A, Nishimura M, Tsutsumi N. Arabidopsis dynamin-related proteins DRP3A and DRP3B are functionally redundant in mitochondrial fission, but have distinct roles in peroxisomal fission. Plant J. 2009;58:388–400. doi: 10.1111/j.1365-313X.2009.03786.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Hu J. Two small protein families, DYNAMIN-RELATED PROTEIN3 and FISSION1, are required for peroxisome fission in Arabidopsis . Plant J. 2009;57:146–159. doi: 10.1111/j.1365-313X.2008.03677.x. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Gould SJ. The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J Biol Chem. 2003;278:17012–17020. doi: 10.1074/jbc.M212031200. [DOI] [PubMed] [Google Scholar]
- 22.Amutha B, Gordon DM, Gu Y, Pain D. A novel role of Mgm1p, a dynamin-related GTPase, in ATP synthase assembly and cristae formation/maintenance. Biochem J. 2004;381:19–23. doi: 10.1042/BJ20040566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griparic L, van der Wel NN, Orozco IJ, Peters PJ, van der Bliek AM. Loss of the intermembrane space protein Mgm1/OPA1 induces swelling and localized constrictions along the lengths of mitochondria. J Biol Chem. 2004;279:18792–18798. doi: 10.1074/jbc.M400920200. [DOI] [PubMed] [Google Scholar]
- 24.Sesaki H, Southard SM, Yaffe MP, Jensen RE. Mgm1p, a dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane. Mol Biol Cell. 2003;14:2342–2356. doi: 10.1091/mbc.E02-12-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shepard KA, Yaffe MP. The yeast dynamin-like protein, Mgm1p, functions on the mitochondrial outer membrane to mediate mitochondrial inheritance. J Cell Biol. 1999;144:711–720. doi: 10.1083/jcb.144.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herlan M, Vogel F, Bornhovd C, Neupert W, Reichert AS. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J Biol Chem. 2003;278:27781–27788. doi: 10.1074/jbc.M211311200. [DOI] [PubMed] [Google Scholar]
- 27.Wong ED, Wagner JA, Scott SV, Okreglak V, Holewinske TJ, Cassidy-Stone A, Nunnari J. The intramitochondrial dynamin-related GTPase, Mgm1p, is a component of a protein complex that mediates mitochondrial fusion. J Cell Biol. 2003;160:303–311. doi: 10.1083/jcb.200209015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olichon A, Emorine LJ, Descoins E, Pelloquin L, Brichese L, Gas N, Guillou E, Delettre C, Valette A, Hamel CP, Ducommun B, Lenaers G, Belenguer P. The human dynamin-related protein OPA1 is anchored to the mitochondrial inner membrane facing the inter-membrane space. FEBS Lett. 2002;523:171–176. doi: 10.1016/S0014-5793(02)02985-X. [DOI] [PubMed] [Google Scholar]
- 29.Niemann HH, Knetsch ML, Scherer A, Manstein DJ, Kull FJ. Crystal structure of a dynamin GTPase domain in both nucleotide-free and GDP-bound forms. EMBO J. 2001;20:5813–5821. doi: 10.1093/emboj/20.21.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klockow B, Tichelaar W, Madden DR, Niemann HH, Akiba T, Hirose K, Manstein DJ. The dynamin A ring complex: molecular organization and nucleotide-dependent conformational changes. EMBO J. 2002;21:240–250. doi: 10.1093/emboj/21.3.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyagishima SY, Kuwayama H, Urushihara H, Nakanishi H. Evolutionary linkage between eukaryotic cytokinesis and chloroplast division by dynamin proteins. Proc Natl Acad Sci USA. 2008;105:15202–15207. doi: 10.1073/pnas.0802412105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manstein DJ, Schuster HP, Morandini P, Hunt DM. Cloning vectors for the production of proteins in Dictyostelium discoideum . Gene. 1995;162:129–134. doi: 10.1016/0378-1119(95)00351-6. [DOI] [PubMed] [Google Scholar]
- 33.Knetsch ML, Tsiavaliaris G, Zimmermann S, Rühl U, Manstein DJ. Expression vectors for studying cytoskeletal proteins in Dictyostelium discoideum . J Muscle Res Cell Motil. 2002;23:605–611. doi: 10.1023/A:1024498805771. [DOI] [PubMed] [Google Scholar]
- 34.Sutoh K. A transformation vector for Dictyostelium discoideum with a new selectable marker bsr. Plasmid. 1993;30:150–154. doi: 10.1006/plas.1993.1042. [DOI] [PubMed] [Google Scholar]
- 35.de Hostos EL, Bradtke B, Lottspeich F, Guggenheim R, Gerisch G. Coronin, an actin binding protein of Dictyostelium discoideum localized to cell surface projections, has sequence similarities to G protein beta subunits. EMBO J. 1991;10:4097–4104. doi: 10.1002/j.1460-2075.1991.tb04986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Egelhoff TT, Brown SS, Manstein DJ, Spudich JA. Hygromycin resistance as a selectable marker in Dictyostelium discoideum . Mol Cell Biol. 1989;9:1965–1968. doi: 10.1128/mcb.9.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Troll H, Malchow D, Müller-Taubenberger A, Humbel B, Lottspeich F, Ecke M, Gerisch G, Schmid A, Benz R. Purification, functional characterization, and cDNA sequencing of mitochondrial porin from Dictyostelium discoideum . J Biol Chem. 1992;267:21072–21079. [PubMed] [Google Scholar]
- 38.Pagh K, Gerisch G. Monoclonal antibodies binding to the tail of Dictyostelium discoideum myosin: their effects on antiparallel and parallel assembly and actin-activated ATPase activity. J Cell Biol. 1986;103:1527–1538. doi: 10.1083/jcb.103.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schleicher M, Noegel A, Schwarz T, Wallraff E, Brink M, Faix J, Gerisch G, Isenberg G. A Dictyostelium mutant with severe defects in alpha-actinin: its characterization using cDNA probes and monoclonal antibodies. J Cell Sci. 1988;90:59–71. doi: 10.1242/jcs.90.1.59. [DOI] [PubMed] [Google Scholar]
- 40.Neuhaus EM, Horstmann H, Almers W, Maniak M, Soldati T. Ethane-freezing/methanol-fixation of cell monolayers: a procedure for improved preservation of structure and antigenicity for light and electron microscopies. J Struct Biol. 1998;121:326–342. doi: 10.1006/jsbi.1998.3971. [DOI] [PubMed] [Google Scholar]
- 41.Griffith GR, Consigli RA. Isolation and characterization of monopinocytotic vesicles containing polyomavirus from the cytoplasm of infected mouse kidney cells. J Virol. 1984;50:77–85. doi: 10.1128/jvi.50.1.77-85.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber I, Wallraff E, Albrecht R, Gerisch G. Motility and substratum adhesion of Dictyostelium wild-type and cytoskeletal mutant cells: a study by RICM/bright-field double-view image analysis. J Cell Sci. 1995;108:1519–1530. doi: 10.1242/jcs.108.4.1519. [DOI] [PubMed] [Google Scholar]
- 43.Rasband WS (1997–2009) ImageJ, U.S. National Institutes of Health, Bethesda, Maryland, USA. http://rsb.info.nih.gov/ij/index.html. Accessed 4 Oct 2010
- 44.Troll H, Winckler T, Lascu I, Müller N, Saurin W, Véron M, Mutzel R. Separate nuclear genes encode cytosolic and mitochondrial nucleoside diphosphate kinase in Dictyostelium discoideum . J Biol Chem. 1993;268:25469–25475. [PubMed] [Google Scholar]
- 45.Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- 46.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 47.Gavel Y, von Heijne G. Cleavage-site motifs in mitochondrial targeting peptides. Protein Eng. 1990;4:33–37. doi: 10.1093/protein/4.1.33. [DOI] [PubMed] [Google Scholar]
- 48.Deleage G, Roux B. An algorithm for protein secondary structure prediction based on class prediction. Protein Eng. 1987;1:289–294. doi: 10.1093/protein/1.4.289. [DOI] [PubMed] [Google Scholar]
- 49.Cerveny KL, Tamura Y, Zhang Z, Jensen RE, Sesaki H. Regulation of mitochondrial fusion and division. Trends Cell Biol. 2007;17:563–569. doi: 10.1016/j.tcb.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 50.Delille HK, Alves R, Schrader M. Biogenesis of peroxisomes and mitochondria: linked by division. Histochem Cell Biol. 2009;131:441–446. doi: 10.1007/s00418-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 51.Hoepfner D, van den Berg M, Philippsen P, Tabak HF, Hettema EH. A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae . J Cell Biol. 2001;155:979–990. doi: 10.1083/jcb.200107028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lazarow PB. Rat liver peroxisomes catalyze the beta oxidation of fatty acids. J Biol Chem. 1978;253:1522–1528. [PubMed] [Google Scholar]
- 53.Singh H, Derwas N, Poulos A. Very long chain fatty acid beta-oxidation by rat liver mitochondria and peroxisomes. Arch Biochem Biophys. 1987;259:382–390. doi: 10.1016/0003-9861(87)90504-2. [DOI] [PubMed] [Google Scholar]
- 54.van den Bosch H, Schutgens RB, Wanders RJ, Tager JM. Biochemistry of peroxisomes. Annu Rev Biochem. 1992;61:157–197. doi: 10.1146/annurev.bi.61.070192.001105. [DOI] [PubMed] [Google Scholar]
- 55.Ponte E, Bracco E, Faix J, Bozzaro S. Detection of subtle phenotypes: the case of the cell adhesion molecule csA in Dictyostelium . Proc Natl Acad Sci USA. 1998;95:9360–9365. doi: 10.1073/pnas.95.16.9360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heuser J, Zhu Q, Clarke M. Proton pumps populate the contractile vacuoles of Dictyostelium amoebae. J Cell Biol. 1993;121:1311–1327. doi: 10.1083/jcb.121.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heuser J. Evidence for recycling of contractile vacuole membrane during osmoregulation in Dictyostelium amoebae––a tribute to Günther Gerisch. Eur J Cell Biol. 2006;85:859–871. doi: 10.1016/j.ejcb.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 58.Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci USA. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barth C, Le P, Fisher PR. Mitochondrial biology and disease in Dictyostelium . Int Rev Cytol. 2007;263:207–252. doi: 10.1016/S0074-7696(07)63005-8. [DOI] [PubMed] [Google Scholar]
- 60.Gilson PR, Yu XC, Hereld D, Barth C, Savage A, Kiefel BR, Lay S, Fisher PR, Margolin W, Beech PL. Two Dictyostelium orthologs of the prokaryotic cell division protein FtsZ localize to mitochondria and are required for the maintenance of normal mitochondrial morphology. Eukaryot Cell. 2003;2:1315–1326. doi: 10.1128/EC.2.6.1315-1326.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nishida K, Takahara M, Miyagishima SY, Kuroiwa H, Matsuzaki M, Kuroiwa T. Dynamic recruitment of dynamin for final mitochondrial severance in a primitive red alga. Proc Natl Acad Sci USA. 2003;100:2146–2151. doi: 10.1073/pnas.0436886100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neuspiel M, Schauss AC, Braschi E, Zunino R, Rippstein P, Rachubinski RA, Andrade-Navarro MA, McBride HM. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr Biol. 2008;18:102–108. doi: 10.1016/j.cub.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 63.Schrader M. Shared components of mitochondrial and peroxisomal division. Biochim Biophys Acta. 2006;1763:531–541. doi: 10.1016/j.bbamcr.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 64.Wanders RJ, Waterham HR. Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem. 2006;75:295–332. doi: 10.1146/annurev.biochem.74.082803.133329. [DOI] [PubMed] [Google Scholar]
- 65.Kessen U, Schaloske R, Aichem A, Mutzel R. Ca(2+)/calmodulin-independent activation of calcineurin from Dictyostelium by unsaturated long chain fatty acids. J Biol Chem. 1999;274:37821–37826. doi: 10.1074/jbc.274.53.37821. [DOI] [PubMed] [Google Scholar]
- 66.Ezratty EJ, Partridge MA, Gundersen GG. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat Cell Biol. 2005;7:581–590. doi: 10.1038/ncb1262. [DOI] [PubMed] [Google Scholar]
- 67.Damke H, Baba T, Warnock DE, Schmid SL. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mooren OL, Kotova TI, Moore AJ, Schafer DA. Dynamin2 GTPase and cortactin remodel actin filaments. J Biol Chem. 2009;284:23995–24005. doi: 10.1074/jbc.M109.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bretschneider T, Diez S, Anderson K, Heuser J, Clarke M, Müller-Taubenberger A, Kohler J, Gerisch G. Dynamic actin patterns and Arp2/3 assembly at the substrate-attached surface of motile cells. Curr Biol. 2004;14:1–10. doi: 10.1016/j.cub.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 70.Uchida KS, Yumura S. Dynamics of novel feet of Dictyostelium cells during migration. J Cell Sci. 2004;117:1443–1455. doi: 10.1242/jcs.01015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 5 (MOV 10,014 kb)