Abstract
BACKGROUND:
Mitochondrial DNA (mtDNA) and Y-DNA analysis have been widely used to predict ancestral origin. Genetic anthropologists predict that human civilizations may have originated in central Africa one to two million years previously. Primary iron overload is not a common diagnosis among indigenous people of northern Africa, but hereditary hemochromatosis is present in approximately one in 200 people in northern Europe. MtDNA analysis has the potential to determine whether contemporary hemochromatosis patients have an ancient ancestral linkage.
METHODS:
DNA was obtained from buccal smears for mtDNA and Y-DNA analysis. Y-DNA analysis included examination of 20 short tandem repeat markers on the Y chromosome. Analysis of mtDNA involved sequencing of the HVR-1 genetic sequence (nucleotides 16001 to 16520) and was compared with the Cambridge Reference Sequence. MtDNA ancestral haplotypes were predicted from the analysis of the HVR-1 sequence.
RESULTS:
Twenty-six male C282Y homozygotes were studied. There were 28 polymorphisms present in the HVR-1 sequence of these participants. The most common polymorphism was present at position 16519 in 15 participants and at position 16311 in eight participants. There were 12 different ancestral haplotypes predicted by mtDNA analysis, with the K haplotype being present in five participants. Y-DNA analysis revealed eight different haplotypes, with R1b being found in 11 of the 26 participants.
CONCLUSION:
Analysis of mtDNA and Y-DNA in 26 hemochromatosis patients suggested that they did not all originate from the same ancestral tribe in Africa. These findings were consistent with the theory that the original hemochromatosis mutation occurred after migration of these ancestral people to central Europe, possibly 4000 years previously.
Keywords: Ancestry, Hemochromatosis, Mitochondrial DNA
Abstract
HISTORIQUE :
L’analyse de l’ADN mitochondrial (ADNmt) et de l’ADN-Y est largement utilisée pour prédire l’origine ancestrale. Selon les anthropologues génétiques, les civilisations humaines auraient vu le jour en Afrique centrale il y a de un à deux millions d’années. La surcharge en fer primaire n’est pas un diagnostic courant chez les peuples indigènes d’Afrique du Nord, mais on constate une hémochromatose héréditaire chez environ une personne sur 200 en Europe du Nord. L’analyse de l’ADNmt a le potentiel de déterminer si les patients actuels présentant une hémochromatose partagent la même origine ancestrale.
MÉTHODOLOGIE :
Les chercheurs ont fait un prélèvement buccal de l’ADN afin d’analyser l’ADNmt et l’ADN-Y des sujets. L’analyse de l’ADN-Y incluait l’examen de 20 marqueurs répétés courts sur le chromosome Y. L’analyse de l’ADNmt incluait le séquençage génétique HVR-1 (nucléotides 16001 à 16520) et a été comparée à la séquence de référence de Cambridge. Les chercheurs ont prédit les haplotypes d’ADNmt ancestral à partir de l’analyse de la séquence HVR-1.
RÉSULTATS :
Les chercheurs ont étudié 26 homozygotes C282Y mâles. Ils ont observé 28 polymorphismes dans la séquence HVR-1 de ces participants. Le principal polymorphisme était présent à la position 16519 chez 15 participants et à la position 16311 chez huit participants. Ils ont repéré 12 haplotypes ancestraux différents selon l’analyse de l’ADNmt, l’haplotype K étant présent chez cinq participants. L’analyse de l’ADN-Y a révélé huit haplotypes différents, le R1b étant présent chez 11 des 26 participants.
CONCLUSION :
L’analyse de l’ADNmt et de l’ADN-Y chez 26 patients ayant une hémochromatose indiquait qu’ils ne provenaient pas tous de la même tribu ancestrale d’Afrique. Ces observations correspondent à la théorie selon laquelle une mutation de l’hémochromatose originale s’est produite après la migration de ces peuples ancestraux en Europe centrale, il y a peut-être 4 000 ans.
Hereditary hemochromatosis is an autosomal recessive disorder of iron overload resulting in significant morbidity including liver cirrhosis, diabetes, arthropathy and cardiomyopathy (1). The most common genetic variant of hemochromatosis – C282Y – is the result of a cysteine to tyrosine substitution at position 282 of the HFE gene. Populations of northern European ancestry have a high prevalence of hemochromatosis, with one in 10 individuals being heterozygote carriers, and 0.3% to 0.5% having a homozygote mutation for C282Y (2).
According to linkage disequilibrium analysis (3), the C282Y mutation dates back approximately 60 to 70 generations. Mapping allele frequencies across Europe has ignited a scholarly debate regarding a Celtic versus Viking origin of the mutation (4–8). Mitochondrial DNA (mtDNA) and Y-DNA analysis have emerged as widely used tools to predict ancestral origin (Genebase, Vancouver, British Columbia). Genetic anthropologists have predicted that human civilizations may have originated in central Africa one to two million years ago. Primary iron overload is not a common diagnosis among indigenous people of northern Africa. MtDNA and Y-DNA analysis have the potential to determine whether current hemochromatosis patients have a common ancient ancestral linkage.
METHODS
Male patients with hemochromatosis recruited from London Health Sciences Centre (London, Ontario), were invited to participate in the present study. All participants were homozygous for the C282Y mutation of the HFE gene. The study was approved by The University of Western Ontario Health Sciences Research Ethics Board (London, Ontario).
Genetic testing
MtDNA and Y-DNA analysis was performed with DNA obtained from buccal smears. MtDNA and Y-DNA follow maternal and paternal lineage, respectively. Genealogical DNA testing involves comparing the nucleotide sequence of mtDNA and Y-DNA of a sample population with a known historic reference – the Cambridge Reference Sequence (CRS). The CRS was the first-ever published mtDNA sequence (9). Comparing more genetic markers improves the resolution of the predictive capability of the origin of the sample. The combination kit used in the present study (Genebase Combo Ancestry) can only be applied to men. Buccal smears taken from patients using the combination kit were sent to Genebase Laboratories in British Columbia for analysis. Results from the genetic analysis were then interpreted descriptively.
MtDNA is considered to be extremely stable, with little genetic recombination over many centuries. MtDNA analysis included genetic sequencing of HVR-1 (nucleotides 16001 to 16520) and comparing the results with the CRS (Genebase) (9). MtDNA ancestral haplogroups were then predicted from the HVR-1 analysis. A haplo-group is a group of similar haplotypes and genetic polymorphisms that share a common ancestor. The HVR-1 sequence is part of the D-loop or hypervariable region of mtDNA (Genebase). The D-loop does not carry any functional genes and, thus, mutations in this region are preserved over many generations, providing an ancestral footprint of each individual. In addition, mutation rates in HVR-1 are higher than in other parts of mtDNA and thus, are a good starting point in ancestral investigations.
Y-DNA analysis included 20 short tandem repeat (STR) markers on the Y chromosome. Based on these data, Y-DNA ancestral haplotypes were subsequently predicted.
RESULTS
Twenty-six male C282Y homozygotes were studied. This included participants with self-reported ancestors from the United Kingdom, Portugal, Poland, Netherlands and Belgium.
MtDNA
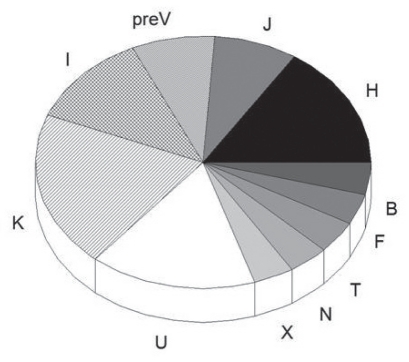
There were 28 polymorphisms present in the HVR-1 sequence of the 26 participants. The most common polymorphism was present at position 16519 in 15 participants and at position 16311 in eight participants. There were 12 different ancestral haplogroups predicted by mtDNA analysis, with the K haplogroup present in five participants (Figure 1).
Figure 1).
Mitochondrial DNA haplogroups
Y-DNA
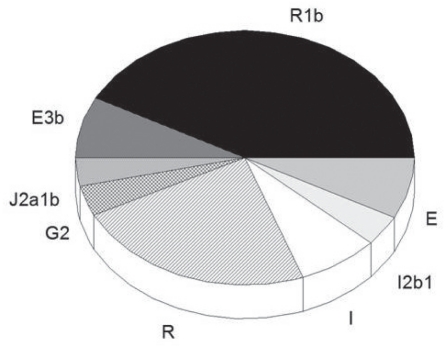
Y-DNA analysis revealed eight different haplotypes, with R1b (Cro-Magnons, 35,000 years ago) being found in 11 of 26 participants (Figure 2).
Figure 2).
Y-DNA haplotypes
DISCUSSION
Mapping the ancestral haplogroups and haplotypes predicted from the mtDNA and Y-DNA analysis, respectively, of 26 patients with hemochromatosis demonstrated a broad range of lineages. All of the participants belonged to an ancestral branch that emerged following the migration from Africa approximately 4000 years ago. This finding supports the theory that the hemochromatosis mutation occurred following settlement in Europe (8). Because the C282Y mutation did not interfere with reproductive capacity and, potentially, conferred advantages to the host such as resistance to dietary iron deficiency and infection, hemochromatosis provides the opportunity for an interesting anthropological study (10).
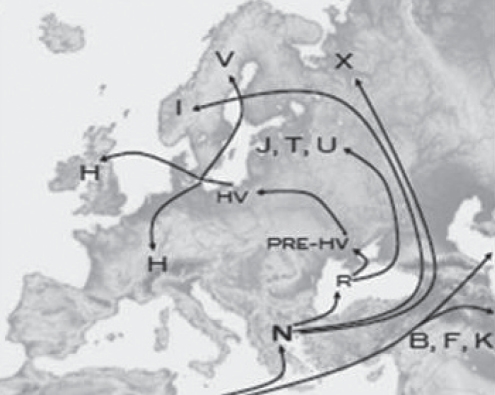
The diverse range of haplogroups predicted in the current study support estimates of the age of hereditary hemochromatosis at approximately 1200 to 2100 years. Allele age is indirectly measured by haplotype diversity (3,8). With younger allele mutations, there have been fewer opportunities for mutations to accumulate within chromosomes not implicated in the disease pathology (3). The ancestry of participants in the current investigation diffusely span the European continent, in comparison with previous studies localizing the origin of hemochromatosis to a northern European geography (7) (Figure 3). This may be explained by the small sample size used in the current study.
Figure 3).
Geographical representation of mitochondrial (mt) DNA haplo-group ancestral origins. Adapted from Genebase mtDNA Haplogroup Migration Map <http://upload.wikimedia.org/wikipedia/commons/b/b7/Europe_topography_map.png>
The K and H haplogroups and R1b haplotype were the most common identifications demonstrated in the present study. Hill et al (11) described the highest frequency of the haplotype R1b3 – a branch of the R1b haplotype – in individuals of western Irish descent. Similar studies have yet to be performed in Scandinavian nations linked with the Viking theory of hemochromatosis origin. In addition, the H haplogroups and R1b haplotype are two of the most common ancestral lineages in Europe (Genebase).
Some limitations to the current investigation include a small sample size and the selection of few markers. Selecting fewer high-yield markers was appropriate for an introductory investigation in this arena. However, we would recommend using additional genetic markers and expanding the size of the population tested to improve the predictive value for the ancestry of hemochromatosis. Another limitation was that our sample was only recruited from Southwestern Ontario. Future studies should aim to recruit their subjects from a wider geographical distribution. Future studies can also use other genetic testing approaches to predict ancestry. For example, the testing of non-mtDNA using single nucleotide polymorphisms markers can be used.
Correlating the C282Y mutation with the ancestral footprint provided by mtDNA and Y-DNA provides strong evidence to support the origin of hereditary hemochromatosis as occurring after the migration of people from a central African tribe.
REFERENCES
- 1.Adams PC, Barton JC. Haemochromatosis. Lancet. 2007;370:1855–60. doi: 10.1016/S0140-6736(07)61782-6. [DOI] [PubMed] [Google Scholar]
- 2.Adams PC, Reboussin DM, Barton JC, et al. Hemochromatosis and iron-overload screening in a racially diverse population. N Engl J Med. 2005;352:1769–78. doi: 10.1056/NEJMoa041534. [DOI] [PubMed] [Google Scholar]
- 3.Whittington C. Was the C282Y mutation and Irish Gaelic mutation that the Vikings help disseminate? Medical Hypotheses. 2006;67:1270–3. doi: 10.1016/j.mehy.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 4.Lucotte G. Celtic origin of the C282Y mutation of hemochromatosis. Blood Cells Mol Dis. 1998;24:433–8. doi: 10.1006/bcmd.1998.0212. [DOI] [PubMed] [Google Scholar]
- 5.Lucotte G, Dieterlen F. A European allele map of the C282Y mutation of hemochromatosis: Celtic versus Viking origin of the mutation? Blood Cells Mol Dis. 2003;31:262–7. doi: 10.1016/s1079-9796(03)00133-5. [DOI] [PubMed] [Google Scholar]
- 6.Rochette J, Pointon JJ, Fisher CA, et al. Multicentric origin of hemochromatosis gene (HFE) mutations. Am J Hum Genet. 1999;64:1056–62. doi: 10.1086/302318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merryweather-Clarke A, Pointon J, Jouanolle A, Rochette J, Robson K. Geography of HFE C282Y and H63D mutations. Genet Test. 2000;4:183–98. doi: 10.1089/10906570050114902. [DOI] [PubMed] [Google Scholar]
- 8.Distante S, Robson K, Graham-Campbell J, Arnaiz-Villena A, Brissot P, Worwood M. The origin and spread of the HFE-C282Y haemochromatosis mutation. Hum Genet. 2004;115:269–79. doi: 10.1007/s00439-004-1152-4. [DOI] [PubMed] [Google Scholar]
- 9.Anderson S, Bankier AT, Barrell BG, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–65. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 10.Pietrangelo A. Hereditary Hemochromatosis: A new look at an old disease. N Eng J Med. 2004;350:2383–97. doi: 10.1056/NEJMra031573. [DOI] [PubMed] [Google Scholar]
- 11.Hill EW, Jobling MA, Bradley DG. Y-chromosome variation and Irish origins. Nature. 2000;404:351–2. doi: 10.1038/35006158. [DOI] [PubMed] [Google Scholar]