Abstract
In plant immunity, recognition of pathogen effectors by plant resistance proteins leads to the activation of plant defenses and a localized cell death response. The AvrM effector from flax rust is a small secreted protein that is recognized by the M resistance protein in flax. Here, we investigate the mechanism of M–AvrM recognition and show that these two proteins directly interact in a yeast two-hybrid assay, and that this interaction correlates with the recognition specificity observed for each of the different AvrM variants. We further characterize this interaction by demonstrating that the C-terminal domain of AvrM is required for M-dependent cell death, and show that this domain also interacts with the M protein in yeast. We investigate the role of C-terminal differences among the different AvrM proteins for their involvement in this interaction and establish that M recognition is hindered by an additional 34 amino acids present at the C terminus of several AvrM variants. Structural characterization of recombinant AvrM-A protein revealed a globular C-terminal domain that dimerizes.
Plants have a variety of highly regulated defense strategies to evade pathogen invasion. One widely studied strategy is effector-triggered immunity, in which plant resistance (R) proteins recognize specific pathogen effector proteins, termed avirulence (Avr) proteins, produced during infection. This recognition triggers a rapid defense response, including a localized necrosis called a hypersensitive response (HR), which restricts pathogen growth. Most R proteins are large cytoplasmic proteins containing nucleotide-binding (NB) and leucine-rich-repeat (LRR) domains (Martin et al. 2003). This NB-LRR class can then be subdivided based on the presence of either an additional N-terminal coiled-coil (CC) or toll-interleukin 1 receptor (TIR) domain, which has homology to the Toll and interleukin-1 receptors. Conversely, pathogen Avr proteins share very few common features and appear to perform diverse pathogenicity functions during infection (Chisholm et al. 2006; Bent and Mackey 2007; Kamoun 2007). The most extensively characterized Avr elicitors are the type III effectors from bacteria, which are delivered directly into host cells, where they act to facilitate disease. Avr proteins from fungi and oomycetes also enter plant cells; however, their translocation mechanisms are not yet understood (Catanzariti et al. 2007; Tyler 2009).
Despite the identification of many R and Avr genes, in most cases, the recognition event that occurs between these cognate proteins is not understood. However, it is now evident that R proteins are able to recognize effectors either directly or indirectly. The latter, know as the “guard model”, has been demonstrated for several resistance proteins identified from Arabidopsis thaliana, including RPM1, RPS2, and RPS5, which provide resistance to the bacterium Pseudomonas syringae (Axtell et al. 2003; Mackey et al. 2003; Shao et al. 2003). These CC-NB-LRR proteins directly associate with a host protein that is targeted by the Avr effector for modification during infection, and are activated by perturbations to that host protein. Direct physical association between cognate R and Avr proteins has also been demonstrated in four cases involving NB-LRR proteins: Pi-ta from rice with Avr-Pita from Magnaporthe grisea (Jia et al. 2000); the A. thaliana protein RRS1 with PopP2 from Ralstonia solanacearum (Deslandes et al. 2003); the flax L5, L6, and L7 proteins with AvrL567 proteins of flax rust (Dodds et al. 2006); and N from tobacco with the p50 elicitor from Tobacco mosaic virus (Ueda et al. 2006), although here the association appears to occur after an indirect recognition event and may be involved in N activation (Caplan et al. 2008).
Effector-triggered immunity, also known as gene-for-gene resistance, has been extensively studied in the interaction between flax (Linum usitatissimum) and the obligate biotrophic fungal rust pathogen Melampsora lini. Several flax rust Avr genes (AvrL567, AvrM, AvrP123, and AvrP4) have been identified and encode small secreted proteins, which are recognized by TIR-NB-LRR proteins inside the plant cell (Dodds et al. 2004; Catanzariti et al. 2006; Wang et al. 2007). AvrM is a small gene family, which in the heterozygous flax rust strain CH5 consists of AvrM-A, AvrM-B, AvrM-C, AvrM-D, and AvrM-E at the avirulence allele and a single member, avrM, at the virulence allele (Catanzariti et al. 2006). Among these proteins, avrM differs most with a 69-residue internal deletion in the N-terminal region and 10 unique polymorphic residues concentrated in the C-terminal region, which appear to be the result of diversifying selection (Catanzariti et al. 2006). All the AvrM variants, except avrM and AvrM-E, which is a truncated protein lacking the C-terminal region, are recognized by the flax M resistance protein to activate a resistance response. However, the recognized AvrM proteins differ in the degree of the response they elicit. When transiently expressed in planta, AvrM-A triggers the strongest cell death response, followed by AvrM-D, whereas AvrM-B and AvrM-C are significantly weaker (Catanzariti et al. 2006).
In this study, we investigate the mechanism of M–AvrM recognition and further characterize the structure and function of this fungal effector. We show that AvrM directly interacts with the M protein in yeast and that this interaction correlates with in planta recognition specificities. We also demonstrate that the C–terminal domain of AvrM is required for M-dependent cell death and also physically associates with M in yeast.
RESULTS
The AvrM and M proteins interact in yeast
To investigate whether AvrM recognition involves a direct interaction with the M protein, we employed a yeast two-hybrid assay. Bait and prey constructs were made expressing each of the five AvrM variants, avrM, and M proteins fused C-terminal to either the GAL4 DNA-binding domain (BD) or the transcriptional activation domain (AD). These constructs were co-transformed into yeast to test for activation of expression from the GAL4 upstream activation sequence. Co-expression of the M fusion proteins with either the AvrM-A or –D fusion proteins activated the HIS3 reporter gene (Fig. 1A and B), indicating that these R and Avr proteins can physically interact in yeast. However, this interaction appears to be weak because the proteins were unable to activate the ADE2 or LacZ reporter genes, which are more stringent assays (data not shown). No protein interaction was detected between M and avrM (Fig. 1), which is unable to induce an M-dependant HR when expressed in planta, and supports a direct interaction for the basis of recognition specificity. Co-expression of the M fusion with AvrM-B, AvrM-C, or AvrM-E fusion proteins were also unable to activate reporter gene expression (Fig. 1B). However, these three AvrM proteins could not be detected by immunoblot analysis (Fig. 1C); therefore, insufficient protein accumulation in yeast may explain why no interaction with the M protein was detected for these AvrM variants.
Fig. 1.
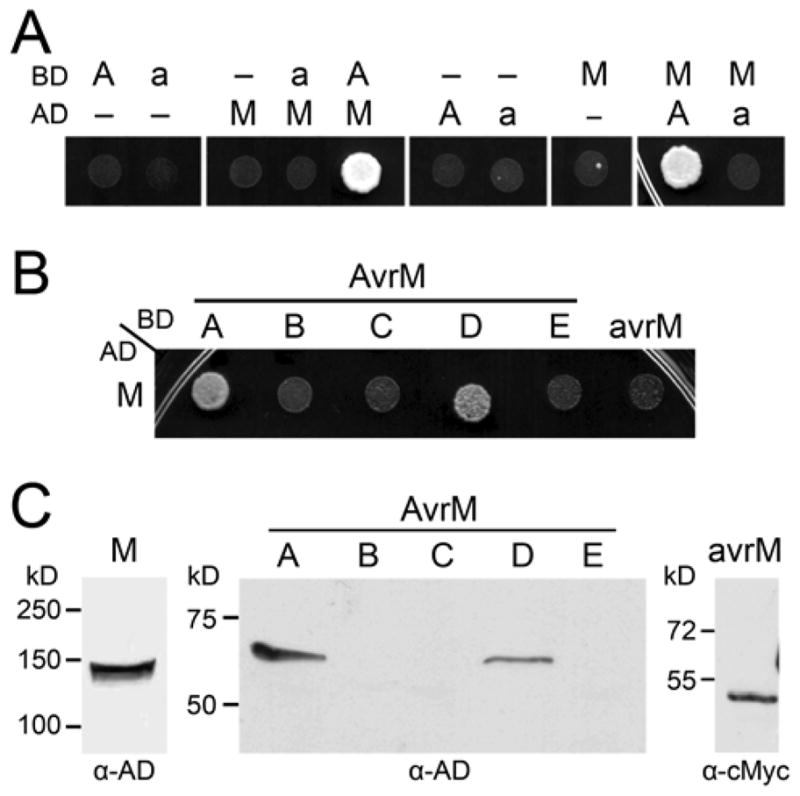
AvrM proteins interact with the M protein in yeast. A and B, Growth of yeast strain AH109 expressing GAL4-activation domain (AD) and -binding domain (BD) fusion proteins on minimal media lacking histidine. Growth indicates expression of the HIS3 reporter gene as a result of interaction between the GAL4-AD and GAL4-BD fusion proteins. A, AD and BD proteins were expressed alone (–) or as fusions to AvrM-A (A), avrM (a), or M. B, Co-expression of the AD–M fusion protein and the BD–AvrM (A to E) or BD–avrM fusion proteins. C, Protein extracts from yeast strain AH109 expressing AD–M, AD–AvrM (A to E), or BD–avrM were analyzed by immunoblotting with anti-GAL4-AD (α-AD) or anti-cMyc (α-cMyc). Positions and sizes of protein molecular mass standards are indicated.
The C-terminal domain of AvrM-A is required for M-mediated cell death
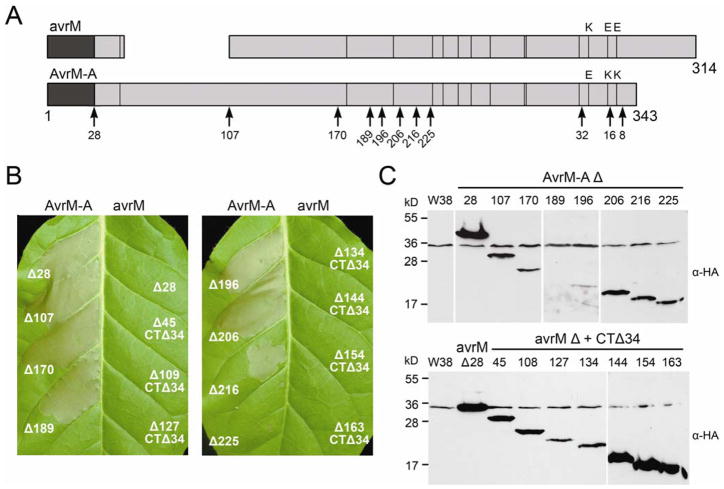
Agrobacterium-mediated transient expression of AvrM-A in Nicotiana tabacum carrying the M gene triggers a very strong HR (Catanzariti et al. 2006). Therefore, we utilized this robust response as a convenient experimental system to further characterize the structure and function of this family of flax rust effectors. To determine the region recognized by the M protein, seven AvrM-A N-terminally truncated proteins were assayed for their ability to induce an M-dependent HR in Agro-bacterium-infiltrated N. tabacum. The first, AvrM-A Δ107, corresponds to the end of an N-terminal inframe deletion within the virulent avrM gene that occurs just after the signal peptide sequence (Fig. 2A). To assist in the demarcation of additional truncations, the AvrM-A sequence was analyzed by GlobPlot 2.3 (Linding et al. 2003b), which predicts regions of globularity and disorder. Based on this output, we constructed AvrM-A Δ170 and AvrM-A Δ189 as potential globular domains. Subsequent truncations were made at regular intervals. T-DNA plasmids were constructed for each of the truncations with the Cauliflower mosaic virus (CaMV) 35S promoter and an N-terminal hemagglutinin (HA) tag. Agrobacterium tumefaciens strains containing the AvrM-A constructs were infiltrated into the leaves of W38 tobacco and a transgenic W38 line containing the M gene (W38::M) and compared with the full-length protein lacking the signal peptide (Δ28).
Fig. 2.
C-terminal domain of AvrM elicits M-dependent cell death. A, Schematic diagram showing variation between AvrM-A and avrM proteins with the size of each protein as amino acid length shown at the end. The signal peptide is shaded dark and lines indicate positions of polymorphic residues; the last three variable residues are given. The position of the N-terminal 69-amino-acid (aa) deletion and the 34-aa extension of avrM are shown. N- and C-terminal truncation sites are indicated by arrows along with the amino acid number, and the size of each full-length protein in amino acids is given. B, Leaves of transgenic W38 tobacco carrying the M gene were infiltrated with Agrobacterium cultures containing T-DNA plasmids encoding AvrM-A (left side of leaf) and avrM (right side of leaf) truncations with an N-terminal hemagglutinin (HA) epitope tag. The size of the N-terminal truncation (Δ) is given and CTΔ34 indicates that the avrM truncations were made with the 34-aa C-terminal extension removed. Leaves were photographed 3 days after infiltration. C, Protein extracts from tobacco leaf tissue (W38) and leaf tissue transiently expressing HA–AvrM-A (upper) or HA–avrM (lower) truncations were analyzed by immunoblotting with anti-HA (α-HA). Positions and sizes of protein molecular mass standards are indicated.
Transient expression of all the AvrM-A constructs in the leaves of W38 gave no phenotype (data not shown). Conversely, when expressed in the leaves of W38::M, each of the N-terminal truncations induced some level of necrotic response, indicating that all seven truncated proteins are recognized by the M protein; however, this detection was significantly weaker for the larger truncations (Fig. 2B). Expression of the first five truncated proteins (Δ107, Δ170, Δ189, Δ196, and Δ206) all induced a necrotic response equal to that of AvrM-A Δ28, although the response observed with Δ206 was slower to develop. The M-dependent HR seen in tobacco with AvrM-A Δ28 can be first observed approximately 20 to 24 h after infiltration whereas, for AvrM-A Δ206, this time was delayed by at least 5 h. The response seen when expressing the last two truncations (Δ216 and Δ225) was extremely variable. AvrM-A Δ216 was able to induce a necrotic response equivalent to that of AvrM-A Δ28 but approximately 60% of the time this response was weaker, with only small patches of cell death observed, or absent. However, AvrM-A Δ225 was unable to cause an HR with the same intensity of AvrM-A Δ28. At most, this protein induced a weak cell death as described for Δ216 but, more frequently, no response was observed. When present, the response of AvrM-A Δ216 and Δ225 was also delayed by at least 8 and 25 h, respectively. Immunoblot analysis indicated that the truncated proteins do not all accumulate to the same level in A. tumefaciens-infiltrated leaves (Fig. 2C). However, these differences do not correlate with the differences observed in the level of HR induction, because the truncated proteins Δ216 and Δ225, which showed weaker cell death activity, are more abundant than Δ189 and Δ196, which induce a strong HR yet are not or barely detected.
The majority of the polymorphic sites between AvrM-A and avrM occur within the C-terminal region; therefore, to further investigate the basis of recognition specificity, the corresponding N-terminal truncations were also made with avrM. Because this protein has an additional 34 amino acids (aa) at the C-terminus which are absent from AvrM-A, each of the truncations were made with the C-terminal extension removed (CTΔ34), allowing inferences to be made about the specific amino acid differences within the minimal recognized region of AvrM-A. Expression of the corresponding avrM truncations did not induce necrosis in either W38 or W38::M (data not shown and Fig. 2B), although immunoblot analysis showed that all were expressed at high levels in infiltrated leaves (Fig. 2C).
To further define the region of AvrM-A required for recognition by M, three C-terminal truncations (CTΔ) were also made. Removal of the last 8 aa from the C terminus of AvrM-A did not affect the intensity or the timing of the HR response when expressed in the leaves of W38::M (Fig. 3A). However, expression of AvrM-A CTΔ16 resulted in a variable response, giving either weak or no necrosis, whereas AvrM-A CTΔ32 consistently gave no response (Fig. 3A). No responses were observed in W38, and immunoblot analysis showed the proteins to be equally abundant (Fig. 3B).
Fig. 3.
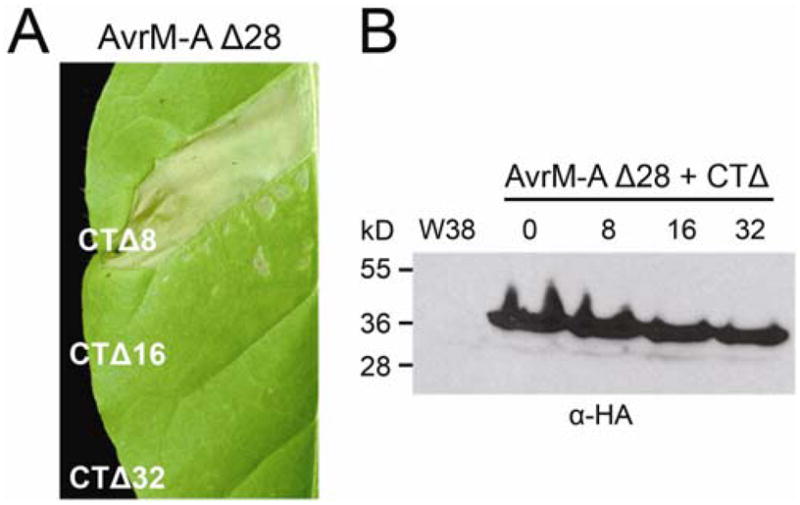
C-terminal deletions of AvrM-A prevent M-recognition. A, Leaves of transgenic W38 tobacco carrying the M gene were infiltrated with Agrobacterium cultures containing T-DNA plasmids encoding AvrM-A C-terminal truncations (CTΔ) without the signal peptide (Δ28) and with an N-terminal hemagglutinin (HA) epitope tag. Leaves were photographed 5 days after infiltration. B, Protein extracts from tobacco leaf tissue (W38) and leaf tissue transiently expressing HA–AvrM-A C-terminal truncations were analyzed by immunoblotting with anti-HA (αHA). Positions and sizes of protein molecular mass standards are indicated.
The C-terminal domain of AvrM-A directly interacts with M in yeast
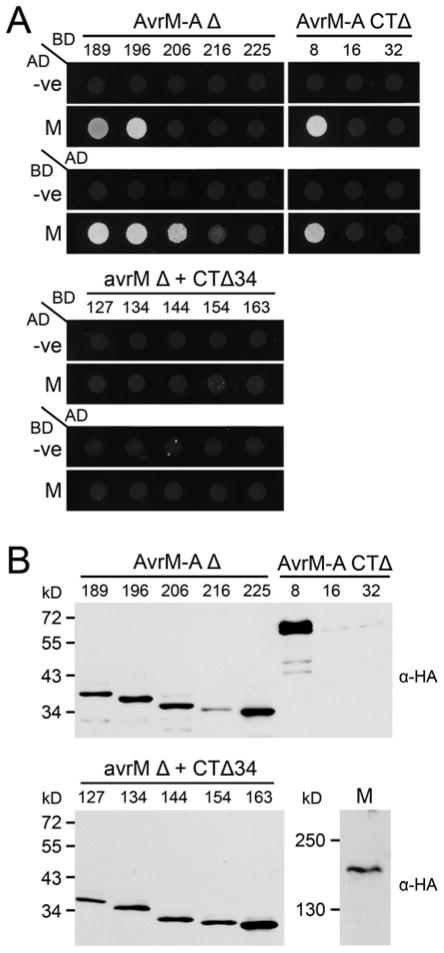
The AvrM-A Δ189 to Δ225 and CTΔ8 to Δ32 truncated proteins were also tested for interaction with M in yeast. Of the eight truncations assayed, AvrM-A Δ189, Δ196, Δ206, and the CTΔ8 fusion proteins retained their ability to physically interact with the M protein and activate the HIS3 reporter gene (Fig. 4A). AvrM-A Δ216 also showed a small amount of growth on –His selection, although it was considerably less than those mentioned above. As for AvrM-A Δ28, none of the truncated proteins were able to activate expression of the ADE2 or LacZ reporter genes (data not shown). These yeast two-hybrid results correlate well with the induction of HR during transient expression in planta. All the M-interacting truncations also caused a strong M-dependent cell death. AvrM-A Δ216 showed a weaker interaction with the M protein and a weaker cell-death response, while AvrM-A Δ225, CTΔ16, and CTΔ32 did not interact with the M protein and gave either weak or no necrotic response. In contrast, none of the corresponding avrM truncated proteins were able to interact with M in yeast (Fig. 4A). Immunoblot analysis showed that most of the GAL4-BD and GAL4-AD fusion proteins were expressed to detectable levels, although AvrM-A CTΔ16 and CTΔ32 did show reduced protein accumulation, which may be responsible for the lack of interaction with the M protein (Fig. 4B).
Fig. 4.
Interactions of AvrM-A truncations with M protein in yeast. A, Growth of yeast strain AH109 expressing GAL4-activation domain (AD) and -binding domain (BD) fusion proteins on minimal media lacking histidine. Upper panel: strains expressing BD–AvrM-A N-terminal (Δ; left) and C-terminal (CTΔ; right) truncations with AD alone (-ve) or AD–M fusion protein. Strains expressing the reversed fusions are shown below; AD–AvrM-A truncations with BD–green fluorescent protein (-ve) or BD–M. Lower panel: strains expressing the corresponding BD–avrM truncated proteins (N-terminal [Δ] plus C-terminal [CTΔ34] to remove an additional 34-amino-acid extension) with AD alone (-ve) or AD–M. Strains expressing the reversed fusions are shown below; AD–avrM truncations with BD alone (-ve) or BD–M. B, Protein extracts from yeast strain AH109 expressing AD–AvrM-A (upper), AD–avrM, or AD–M (lower) fusion proteins were analyzed by immunoblotting with anti-hemagglutinin (α-HA). Positions and sizes of protein molecular mass standards are indicated.
The three C-terminal amino acid polymorphisms are not involved in recognition specificity
The truncated AvrM-A proteins revealed the C-terminal domain as being required for M recognition. Of those tested, AvrM-A CTΔ32 was the only truncation to completely abolish HR activity. Within this deleted region are three polymorphic sites, E316, K326, and K333, which, in avrM, correspond to amino acids K253, E263, and E270. Interestingly, the glutamic acid at position 316 distinguishes both AvrM-A and AvrM-D from the other active AvrM variants (AvrM-B and AvrM-C) which, like avrM, have a lysine at this position, and both AvrM-A and AvrM-D were found to have the strongest necrotic response when expressed in flax leaves containing the M gene (Catanzariti et al. 2006). Therefore, we carried out site-directed mutagenesis in AvrM-A Δ107 and avrM Δ45/CTΔ34 to evaluate the involvement of these three polymorphic sites in the M–AvrM interaction. T-DNA plasmids were constructed for each mutant with the 35S promoter and an N-terminal HA tag, and expressed in W38 and W38::M tobacco leaves. Expression of avrMEEE, avrMKKE, and avrMKEK, as well as AvrM-AKKK, AvrM-AEEK and AvrM-AEKE caused no change in recognition specificity or the intensity of the HR (data not shown). To further assess this result, double and triple mutants of AvrM-A Δ107 were generated to cover all possible combinations of these three residues. Expression of each of the mutated proteins (AvrM-AKEK, AvrM-AEEE, AvrM-AKKE, and AvrM-AKEE) by Agrobacterium infiltration caused an M-dependent HR equivalent to that of the wild-type protein (data not shown), suggesting these three most distal amino acid polymorphisms do not directly affect recognition by M.
The C-terminal extension interferes with recognition
The avrM protein contains a 34-aa C-terminal extension that is absent in AvrM-A and AvrM-D but is present in AvrM-B and AvrM-C, which induce a weaker necrotic response. To test whether this extension affects recognition, we generated a chimeric gene encoding AvrM-A Δ28 protein with the avrM C-terminal extension (AvrM-A+34aa). We also generated single, double, and triple mutations at the three C-terminal polymorphic sites mentioned above in this protein context. In particular, this included AvrM-A+34aaKEE (to represent the residues in avrM) and AvrM-A+34aaKKK (to represent the residues in AvrM-B and AvrM-C). These were expressed with an N-terminal HA tag in W38 and W38::M tobacco. Compared with the wild-type AvrM-A Δ28 protein, all eight constructs triggered a significantly delayed, often weaker and occasionally absent M-dependent HR (data not shown). Cell death from the transient expression of these chimeric genes did not begin until 4 days after infiltration regardless of the intensity of the response which, within each experiment, was the same for all constructs. Immunoblot analysis showed all AvrM-A+34aa proteins to be equally abundant (Supplementary Fig. 1). These results show that recognition by M is affected by the presence of the C-terminal extension and further supports a negligible role for the three polymorphic residues residing immediately upstream of this extension. Furthermore, the AvrM-A+34aa proteins were unable to interact with M in yeast when tested in a yeast two-hybrid assay (data not shown).
The C-terminal region of AvrM-A is a structured domain that forms a dimer
AvrM-A Δ28 protein expressed and purified from Escherichia coli was found to degrade over time into a stable product of approximately 25 kDa (as judged by sodium dodecyl sulfate polyacrylamide gel electrophoresis [SDS-PAGE]) when stored at 4°C. Characterization of the degradation product by N-terminal sequencing and electrospray ionization mass spectrometry (ESI-MS) revealed that the first 74 N-terminal amino acids of AvrM-A had been cleaved off (Fig. 5A), whereas the C-terminal end of the protein was still intact. Treatment of purified AvrM-A with the proteases Glu-C, Lys-C, and chymotrypsin also resulted in only N-terminal degradation (Fig. 5A), indicating that the C-terminal part of the protein (residues 104 to 343) forms a domain protected from proteolysis while the N-terminal part of the protein (residues 29 to 103) may be unstructured and flexible. This is also supported by disordered region predictors such as DisEMBL and GlobPlot (Linding et al. 2003a and b).
Fig. 5.
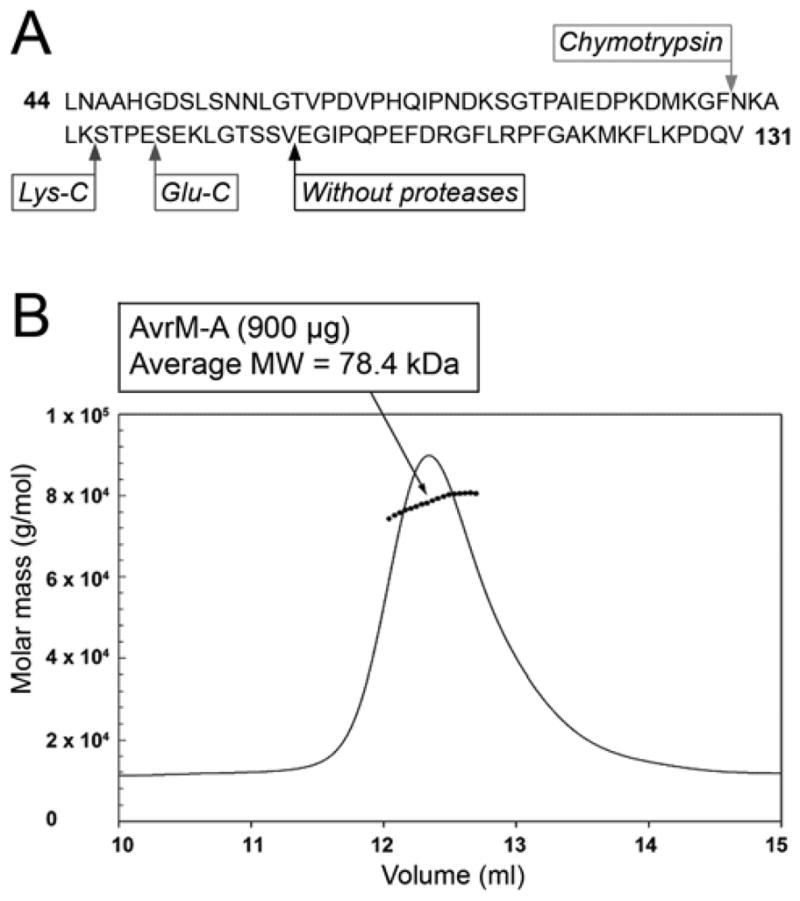
Structural characterization of recombinant AvrM-A protein. A, N-terminal cleavage sites in the AvrM-A protein after limited proteolysis or storage at 4°C are indicated by arrows (only residues 29 to 148 are shown): chymotrypsin (cleaves C terminal to bulky hydrophobic residues), Lys-C (cleaves C terminal to lysine), Glu-C (cleaves C terminal to glutamate), without proteases corresponds to the cleavage site when AvrM-A was stored at 4°C without proteases. B, Determination of the molecular weight of AvrM-A: molar mass distribution plot. The solid lines indicate the trace from the refractive index detector, and the dots are the weight-average molecular weights for each slice (measured every 0.5 s). AvrM-A was analyzed on a Superdex 200 10/300 column in a buffer consisting of 20 mM Tris (pH 8.0) and 300 mM NaCl.
Purified AvrM-A was analyzed for secondary structure content by circular dichroism (CD). The CD spectrum profile (Supplementary Fig. 2) is typical for a folded protein dominated by an α-helical secondary structure, with negative maxima at 222 and 208 nm and a positive maximum near 190 nm. The α-helical content of the protein was estimated to be approximately 50% by the programs ContinII, Selcon3, and CDSSTR (Sreerama and Woody 2000).
Comparison of the AvrM-A gel-filtration profile to a set of standard proteins (catalase, aldolase, bovine serum albumin, ovalbumin, and chymotrypsin) suggests that it may exist as an oligomer in solution (data not shown). To determine the oligomeric state of AvrM-A, the protein was analyzed on a Superdex 200 10/300 column coupled with inline multi-angle laser light scattering and refractive index detection (Fig. 5B). Molecular weights determined by this method depend only on the readings from the laser light and the refractive index detectors and, thus, are independent of the shape of the molecules (Folta-Stogniew and Williams 1999). The molecular weight calculated was 78.4 kDa, close to the expected molecular weight (77.1 kDa) for a dimer of AvrM-A.
To investigate whether AvrM-A can dimerize in planta, we co-expressed two different epitope-tagged AvrM-A Δ28 proteins in tobacco for co-immunoprecipitation (co-IP) analysis. The HA–AvrM-A protein was found to associate with immunoprecipitated yellow fluorescent protein (YFP)-AvrM-A (Fig. 6A), indicating that AvrM-A can form dimers or possibly oligomers in planta and validating the structural characterization of the recombinant protein. Furthermore, this interaction is specific to AvrM-A because HA–avrM did not co-IP with YFP-AvrM-A. In the equivalent co-IP experiment with the avrM protein, only a very small amount of HA–avrM was found to associate with the immunoprecipitated YFP-avrM, as evident by a very faint band (Fig. 6A). Thus, avrM may only weakly dimerize.
Fig. 6.
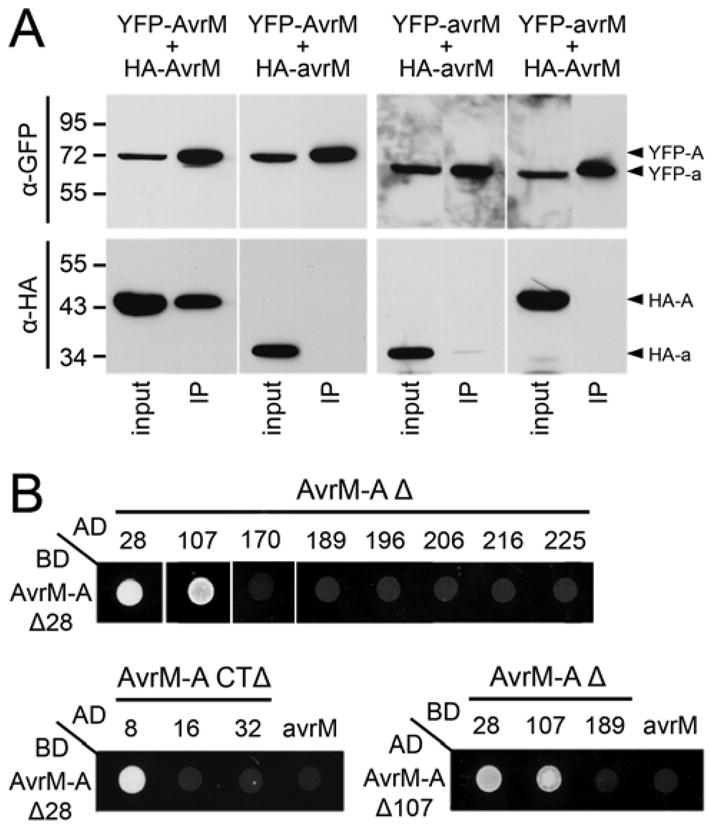
AvrM-A self-associates in planta and in yeast. A, Immunoprecipitation of AvrM and avrM from total plant protein extracts. Proteins extracted from W38 tobacco leaves expressing pairwise combinations of N-terminal tagged (yellow fluorescent protein [YFP] or hemagglutinin [HA]) AvrM-A and avrM fusion proteins were immunoprecipitated using anti-green fluorescent protein (GFP) beads and magnetic capture. Total extracted protein (input) and immunoprecipitated proteins (IPs) were analyzed by immunoblotting with anti-GFP (α-GFP; upper) and anti-HA (αHA; lower). Proteins corresponding to YFP–AvrM-A (YFP-A), HA–AvrM-A (HA–A), YFP–avrM (YFP-a), and HA–avrM (HA–a) are indicated by arrowheads on the right. Positions and sizes of protein molecular mass standards are shown. B, Growth of yeast strain AH109 expressing GAL4-activation domain (AD) and -binding domain (BD) fusion proteins on minimal media lacking histidine. Strains expressing BD–AvrM-A Δ28 with AD–AvrM-A N-terminal (Δ; upper) or C-terminal (CTΔ; lower) truncations, or AD–avrM Δ28. Strains expressing the AD–AvrM-A Δ107 with BD–AvrM-A N-terminal truncations or corresponding BD–avrM truncated protein (Δ45/CTΔ34).
To identify the region involved in AvrM-A dimerization, the AvrM-A Δ28, Δ107–225, and CTΔ8–32 truncated proteins as well as avrM Δ28 were expressed as GAL4-AD fusions in yeast cells expressing the GAL4-BD–AvrM-A Δ28 fusion protein. Of the 12 proteins assayed, only AvrM-A Δ28, Δ107, and CTΔ8 were able to interact with the BD–AvrM-A Δ28 protein (Fig. 6B). We also tested AvrM-A Δ107 for self-interaction in yeast (Fig. 6B), which was also able to associate with itself but not AvrM-A Δ189 or the equivalently truncated avrM (Δ45/CTΔ34). Furthermore, no interaction was detected between AD– and BD–AvrM-A+34aaEKK fusion proteins, suggesting that the C-terminal extension interferes with dimerization; however, the avrM protein lacking this 34-aa extension was also unable to self-associate in yeast (data not shown).
DISCUSSION
A critical component of plant resistance in effector-triggered immunity is the ability for R proteins to recognize pathogen effectors, yet how this recognition occurs is understood in only a few cases. Here, we provide evidence for a direct recognition mechanism between M, a TIR-NB-LRR protein from flax, and AvrM from the flax rust pathogen. Using a yeast-two hybrid system, we show that the M and AvrM proteins physically interact and that this interaction correlates with recognition specificity. Taking advantage of the ability to reconstruct the M–AvrM interaction in N. tabacum, we further characterized the elicitor activity of AvrM. By expressing truncated AvrM-A proteins in transgenic tobacco containing the M gene, we confirm that the C-terminal domain is necessary and sufficient for triggering M-dependent HR and show that this region also retains its ability to interact with M in yeast. Together, these results support a physical association as the mechanism of AvrM detection by the M protein.
The M and AvrM proteins directly interact in a recognition-specific manner
Of the AvrM variants, four induce an M-dependent HR when expressed in planta; AvrM-A and AvrM-D give a very fast and strong necrotic response, whereas the response of AvrM-B and AvrM-C is considerably less robust. The two variants that escape M recognition include the truncated AvrM-E protein and avrM from the virulent allele. Co-expression of the M and AvrM proteins in yeast two-hybrid assays detected a protein–protein interaction for M with AvrM-A and AvrM-D but not with any other variant (Fig. 1B). These interactions could not be detected in the more stringent Ade and lacZ assays and, therefore, appear to involve a relatively weak association. The lack of M interaction with AvrM-B and AvrM-C may be due to insufficient protein expression in yeast, because these proteins were undetectable by immunoblot analysis (Fig. 1C). It is also possible that the weaker HR observed for AvrM-B and AvrM-C is due to a weaker physical interaction with the M protein, which is below the threshold level of detection by the yeast two-hybrid system. Nonetheless, there is a correlation between the interaction with M in yeast and the induction of an HR when expressed in planta, suggesting that M activation is governed by its binding affinity to the AvrM protein.
This finding is further strengthened by results from additional yeast two-hybrid assays using truncated and chimeric AvrM-A proteins, which also found a correlation between M recognition in planta and in yeast. AvrM-A truncations that had lost their necrotic function, or gave a variable and overall weaker cell death response, also lost their ability to interact with M in the yeast assay (Fig. 4A). Similarly, a chimeric AvrM-A protein containing the 34-aa C-terminal extension from avrM also gave a weaker HR in planta and, again, did not interact with the M protein in yeast. We have also investigated the M–AvrM-A interaction in N. tabacum using reciprocal co-IP analysis and a split luciferase complementation assay (Chen et al. 2008). Despite confirming the protein fusions were active (by HR) and detectable by immunoblot analysis, both approaches failed to show a direct interaction. Although these methods did not provide experimental evidence for an in planta interaction, this does not rule out that a direct association of these proteins occurs during infection. As mentioned, the M–AvrM interaction in yeast is weak and, therefore, it is possible that the interaction in planta is below the threshold of detection. It is also possible that these proteins interact only transiently to initiate a resistance response, which may also explain why we were unable to detect an in planta interaction.
The association of M and AvrM proteins is the second example of a flax rust effector directly interacting with the cognate R protein in a yeast two-hybrid assay. A direct interaction was previously shown for the variable flax rust AvrL567 proteins and the flax L5, L6, and L7 resistance proteins (Dodds et al. 2006). For the 12 AvrL567 variants tested, the protein–protein interactions that were detected in yeast were the same combinations of AvrL567 and L proteins that induced HR in transient expression assays in planta. In this case, both the Avr and R loci are highly polymorphic, which has resulted from diversifying selection and led to differences in recognition specificity. The sequence variation seen at the AvrM locus also appears to be the result of diversifying selection, as indicated by a significant excess of nucleotide substitutions in the coding regions relative to flanking DNA. There is also a significant excess of nonsynonymous over synonymous changes between avrM and the other AvrM genes. Given these two examples, it will be interesting to uncover whether a direct R–Avr recognition is a common feature in the flax rust disease system, and if it extends to other obligate plant pathogens.
AvrM has a structured C-terminal effector domain
A sequence alignment of the AvrM proteins shows that most of the polymorphic residues occur within the C-terminal region, suggesting that this region is involved in recognition specificity. To test this hypothesis, we made a series of N-terminal truncations of AvrM-A and used Agrobacterium transient expression in tobacco to look for an M-dependent cell death response. The removal of over half of the predicted mature protein from the N-terminus did not alter its ability to trigger HR, confirming the C-terminal domain as the region responsible for HR-triggering activity (Fig. 2). However, the three largest truncations (Δ206, Δ216, and Δ225) displayed incremental reductions in their response, showing both limited and delayed necrotic symptoms compared with the full-length mature protein (Δ28). To evaluate the role of specific amino acid differences between AvrM-A and avrM, equivalent avrM truncations were made that included the removal of the 34-aa C-terminal extension present in this protein. None of the corresponding avrM truncations induced HR in tobacco carrying the M gene and all the proteins were detectable by immunoblot analysis. This result suggests that the polymorphic residues that exist within the minimal HR-eliciting region play an important role in determining recognition specificity. Defining the C-terminal domain as the region involved in M-activated HR is further supported by three C-terminal truncations, which showed that the removal of as few as 16 aa weakened the necrotic response, whereas the deletion of 32 aa completely abolished its elicitor activity.
The above finding agrees with computational analysis of the AvrM-A sequence for regions of globularity within a protein (Linding et al. 2003b) and also with structural characterization of recombinantly expressed AvrM-A protein. AvrM-A produced in E. coli was found to degrade, through the loss of the N-terminal region, to yield a stable Δ103 protein. Furthermore, results from limited proteolysis indicated that the C-terminal region of this protein corresponds to a proteolytically resistant structure, suggesting that this region of AvrM forms a globular structured domain and that the N-terminal region of approximately 90 residues is flexible and less ordered, making it more accessible to protease cleavage. Several Avr proteins identified from other eukaryotic plant pathogens have also been shown to consist of different functional domains. For example, two Avr proteins from extracellular fungal pathogens undergo post-translational processing to remove either the N-terminal (Avr9 from Cladosporium fulvum) (Van den Ackerveken et al. 1993), or both N- and C-terminal regions (Avr3 from Fusarium oxysporum) (Rep et al. 2004). In another example, the 147-aa Avr3a protein from the oomycete pathogen Phytophthora infestans only requires the last 75 aa of the C-terminal domain for effector activity (Bos et al. 2006). Avr elicitors from oomycetes appear to contain two functional domains, with the N-terminal domain involved in secretion and host cell targeting and the C-terminal domain containing effector activity, although it is not known if they are processed (Whisson et al. 2007; Allen et al. 2008; Grouffaud et al. 2008). Flax rust effectors are also recognized within the host cell (Catanzariti et al. 2006; Dodds et al. 2006), and the N-terminal region of AvrM is sufficient to direct uptake of a secreted fusion to a green fluorescent protein (GFP) into plant cells (P. N. Dodds, unpublished data), suggesting that this protein also has distinct N-terminal uptake and C-terminal recognition domains.
The role of sequence variation in AvrM recognition
The AvrM-A truncated proteins revealed that an intact C-terminal domain is required for both elicitor function and binding to the M protein in yeast, and the removal of 32 aa from the C-terminus was sufficient to abolish both activities. Within this, region there are three polymorphic residues, one of which (E316) differentiates AvrM-A and AvrM-D from the other variants and is also the only amino acid difference between AvrM-A and AvrM-C. However, mutation of this amino acid, as well as the other two polymorphic residues within the last 32-aa region, did not alter the recognition specificity of AvrM-A or avrM. The elimination of the AvrM-A E316 polymorphism as influencing recognition specificity suggested that the additional 34 aa at the C terminus of AvrM-B, AvrM-C, and avrM must play a role in recognition, particularly because this is the only other sequence difference between AvrM-A and AvrM-C. Indeed, addition of this 34-aa extension to AvrM-A impaired its recognition, causing a delayed and often weaker necrotic response in W38::M tobacco and abolishing interaction with M in yeast. This provides convincing evidence that the weaker elicitor activity observed for both AvrM-B and AvrM-C is due to their C-terminal extension, which may sterically impede the interaction with M.
Even though the last three polymorphic residues do not appear to contribute directly to recognition specificity, evidence suggests that amino acid variation does govern detection by the M protein. The polymorphic residues present within this family of effectors are concentrated within the C-terminal domain, and nearly all the variation is seen between avrM and the other AvrM variants. The smallest truncated AvrM-A protein that still retained elicitor activity was AvrM-A Δ225. The equivalent avrM protein (Δ163/CTΔ34) is unable to elicit any degree of cell death; therefore, polymorphic residues in this region appear to be responsible for preventing recognition by the M protein. Excluding the last three polymorphic amino acids examined here, this corresponding avrM region contains six unique residues and a single residue deletion. Several pieces of evidence (Figs. 5 and 6) suggest that the AvrM-A protein forms a dimer, mediated largely through the C-terminal domain. By contrast, avrM appears to dimerize only very weakly. Although the M protein may interact with a dimeric AvrM-A, this oligomeric state does not appear to be important for recognition because several of the N-terminally truncated AvrM-A proteins are recognized by M in planta (Fig. 2) yet do not self-associate in yeast (Fig. 6).
Our results indicate that a direct recognition mechanism underlies M-mediated flax rust resistance, which occurs through binding of M protein to the C-terminal domain of the AvrM protein. Work to solve the structure of this pathogen protein is currently underway and may help determine the involvement of polymorphic residues within this region, and provide insight into the function of this pathogen effector.
MATERIALS AND METHODS
Transient in planta expression assays
Gene expression constructs of AvrM-A and avrM truncations were generated by polymerase chain reaction (PCR) from cDNA clones. Forward primers for N-terminal truncations incorporated a Met codon at the beginning of the sequence while reverse primers for C-terminal truncations incorporated a stop codon at the end of the sequence (Supplementary Table 1). PCR products were cloned into pENTR/D-TOPO using the Directional TOPO cloning kit (Invitrogen, Carlsbad, CA, U.S.A.). The AvrM-A/avrM chimeric gene was generated by replacing the HindIII-PvuI fragment from pENTR-AvrM-A Δ28 with the same fragment from pENTR-avrM Δ28. Site-directed mutations were generated using the Quick Change PCR mutagenesis kit (Stratagene, La Jolla, CA, U.S.A.) on the pENTR-AvrM-A Δ107, AvrM-A/avrM, and avrM Δ45/CTΔ45 constructs. LR Clonase recombination (Invitrogen) was used to transfer pENTR-AvrM-A and avrM constructs to pEarley-Gate binary vectors (Earley et al. 2006) containing the CaMV 35S promoter with an N-terminal HA (pEarleyGate 201) or YFP (pEarleyGate 104) tag. Agrobacterium cultures of strain GV3101-pMP90 containing the binary vector expression constructs were prepared at an optical density at 600 nm (OD600) of 1.0 in 10 mM MES (pH 5.6) buffer with 10 mM MgCl2 and 150 μM acetosyringone for infiltration into W38 and W38::M tobacco leaves (Catanzariti et al. 2006).
Yeast two-hybrid assay
The M cDNA was amplified by PCR from the pPICz-cM7 plasmid (Schmidt et al. 2007) using primers TTTGTGCGA ATTCAGACCAAATCTCAACAATGACAAC (For) and CA TCCTGCAGTCACTTATATTTCTCGTCCGGCCATC (Rev), which omitted the first 21 aa of the protein due to a potential signal anchor function that may interfere with localization of the fusion protein. The PCR product was cloned into the pGBT9 and pGADT7 vectors (Clontech, Mountain View, CA, U.S.A.) as an EcoRI-PstI fragment to generate the GAL4 DNA BD and transcriptional AD fusions, respectively. For AvrM and avrM constructs, fusions were prepared in the pGBKT7 and pGADT7 vectors (Clontech), which were modified to be Gateway compatible by inserting the ccdB cassette reading frame C (Invitrogen) into digested restriction sites (NdeI-BamHI and NdeI-XhoI, respectively) filled in with T4 DNA polymerase. The pENTR-AvrM-A and avrM constructs were transferred to pGBKT7 and pGADT7 destination vectors by LR recombination. Transformations of AH109 yeast cells were performed using the Frozen-EZ Yeast Transformation II kit (Zymo Research, Orange, CA 92867 U.S.A.). All yeast transformants were grown on minimal media lacking tryptophan and leucine to select for the presence of both plasmids, and to confirm equal growth of transformed cells. His/Ade growth and lacZ assays were performed as described in the Yeast Protocols Handbook (Clontech).
Immunoblot analysis
Tobacco leaf tissue and yeast cells were frozen in liquid nitrogen and ground in 3× Laemmli buffer with β-mercapto-ethanol (16%) and urea (10 M) to extract proteins. Samples were boiled for 5 min and then spun to pellet leaf or cell debris. Proteins were separated by SDS-PAGE and transferred by electroblotting to nitrocellulose membranes. Protein blots were probed with anti-HA horseradish peroxidase (Roche, Branchburg, NJ, U.S.A.), anti-cMyc (Covance, Princeton, NJ, U.S.A.), anti-GFP (Covance), or anti-GAL4-AD. Goat anti-mouse immunoglobulin G antibody conjugated to horseradish peroxidase was used to detect cMyc, GFP, and GAL4-AD primary antibodies. Immunoblots were visualized with SuperSignal West Pico chemiluminescence as described by the manufacturer (Pierce, Rockford, IL, U.S.A.).
Co-IP
Protein extracts were prepared from tobacco leaf tissue by grinding 0.5 g of tissue in 1 ml of IP buffer (100 mM Tris, pH 8; 150 mM NaCl; 10 mM EDTA; 5 mM dithiothreitol; 0.5% Triton X-100; 1× protease inhibitor cocktail [Roche]; leupeptin at 2 μg/ml; and aprotinin at 2 μg/ml). Extracts were centrifuged at 10,000 × g for 10 min at 4°C. The supernatant was incubated on ice for 30 min with μMACS anti-GFP micro-beads (Miltenyi Biotec, Cologne, Germany) and bead-proteins complexes were captured by magnetic separation according to the manufacturer. IP buffer was used to equilibrate the column and for all washes. Samples were analyzed by SDS-PAGE and immunoblotting as described above.
Expression and purification of recombinant AvrM-A
AvrM-A with an N-terminal hexahistidine tag was expressed in E. coli BL21(DE3) pLysS strain from the vector pET15b. Luria-Bertani broth cultures were grown at 37°C to mid-exponential phase (OD600 of approximately 0.6 to 0.8) and induced with 1 mM isopropylthio-β-D-galactoside for 16 h at 18°C. Cells were harvested by centrifugation and resuspended in 1/20 of the original culture volume with 20 mM Tris (pH 8.0), 300 mM NaCl, 10 mM imidazole, and 1 mM phenylmethylsulfonyl fluoride. The cells were lysed and the soluble fraction was incubated with Talon Co2+ resin for 1 h (BD Biosciences, San Jose, CA, U.S.A.). The resin was then washed with 20 mM Tris (pH 8.0), 300 mM NaCl, and 20 mM imidazole. AvrM-A was eluted with 20 mM Tris (pH 8.0), 300 mM NaCl, and 250 mM imidazole, and further purified by size-exclusion chromatography by using a Superdex 200 26/60 column (GE Healthcare, Piscataway, NJ, U.S.A.) pre-equilibrated with 20 mM Tris (pH 8.0) and 300 mM NaCl. Purified protein was concentrated to 20 mg/ml using Amicon Ultra centrifugal filter devices (Millipore, Bedford, MA, U.S.A.), snap frozen in liquid nitrogen, and stored at –80°C.
Limited proteolysis
Purified AvrM-A (1 mg/ml) was treated with chymotrypsin, GluC, or LysC (Sigma [St. Louis], sequencing grade) at room temperature. The protease to substrate ratio was 1:100 for GluC and 1:1000 for chymotrypsin and LysC. Aliquots of the reactions were removed at regular time intervals (30 s to 24 h) and analyzed by SDS-PAGE. Protected fragments were detected by SDS-PAGE and further characterized using N-terminal sequencing and mass spectrometry. Protein samples for N-terminal sequencing were transferred to Hybond-P polyvinylidene difluoride membranes (GE Healthcare) (Tous et al. 1989) and stained with Ponceau-S (Sigma), and excised bands were sequenced with an Applied Biosystems (Foster City, CA, U.S.A.) Procise 492 cLC protein sequencer. For mass spectrometry, protein samples were desalted by reverse-phase high-performance liquid chromatography using a C18 capillary column (Agilent Technologies, Santa Clara, CA, U.S.A.) with a linear gradient of 0 to 60% (vol/vol) acetonitrile in 0.1% aqueous acetic acid, then analyzed with an Applied Biosystems API QSTARPulsar1 ESI mass spectrometer.
CD
AvrM-A protein was dialyzed into 10 mM potassium phosphate buffer, pH 8.0, and diluted to 4 μM. CD spectra were recorded on a Jasco J710 spectropolarimeter at 25°C at 0.5-nm wavelength increments from 260 to 190 nm at 50 nm/min, using a 0.1-mm path length cell, 0.5-nm band width, 1-s response time, and five accumulations. The spectra were corrected for buffer base line contribution, and analyzed by ContinII, Selcon 3 and CDSSTR (Sreerama and Woody 2000).
Multi-angle laser light scattering coupled with size-exclusion chromatography
Multi-angle laser light scattering coupled with size-exclusion chromatography was accomplished using a DAWN HELEOS II 18-angle light-scattering detector coupled with an Optilab rEX refractive index detector (Wyatt Technology, Santa Barbara, CA, U.S.A.) and combined inline with a Superdex 200 10/300 gel filtration column (GE Healthcare). The chromatographic separations were performed at room temperature at a flow-rate of 0.5 ml/min in prefiltered (0.2 μm) 20 mM Tris (pH 8.0) and 300 mM NaCl buffer. The initial AvrM-A concentration was set to 9 mg/ml. Molecular mass calculations were performed using the Astra 5.3 software. Input of the refractive increment (dn/dc values) was set at 0.186 in the molecular mass calculations, based on the premise that dn/dc is constant for unmodified proteins (Wen et al. 1996).
Supplementary Material
Acknowledgments
We thank G. King (University of Queensland) for help with multi-angle laser light scattering. B. Kobe is an Australian Research Council Federation Fellow and a National Health and Medical Research Council Honorary Research Fellow.
Footnotes
The e-Xtra logo stands for “electronic extra” and indicates that supplementary material comprising two figures and one table is published online.
LITERATURE CITED
- Allen RL, Meitz JC, Baumber RE, Hall SA, Lee SC, Rose LE, Beynon JL. Natural variation reveals key amino acids in a downy mildew effector that alters recognition specificity by an Arabidopsis resistance gene. Mol Plant Pathol. 2008;9:511–523. doi: 10.1111/j.1364-3703.2008.00481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Chisholm ST, Dahlbeck D, Staskawicz BJ. Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol Microbiol. 2003;49:1537–1546. doi: 10.1046/j.1365-2958.2003.03666.x. [DOI] [PubMed] [Google Scholar]
- Bent AF, Mackey D. Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol. 2007;45:399–436. doi: 10.1146/annurev.phyto.45.062806.094427. [DOI] [PubMed] [Google Scholar]
- Bos JI, Kanneganti TD, Young C, Cakir C, Huitema E, Win J, Armstrong MR, Birch PR, Kamoun S. The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 2006;48:165–176. doi: 10.1111/j.1365-313X.2006.02866.x. [DOI] [PubMed] [Google Scholar]
- Caplan JL, Mamillapalli P, Burch-Smith TM, Czymmek K, Dinesh-Kumar SP. Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell. 2008;132:449–462. doi: 10.1016/j.cell.2007.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti A, Dodds PN, Lawrence GJ, Ayliffe MA, Ellis JG. Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell. 2006;18:243–256. doi: 10.1105/tpc.105.035980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti A, Dodds PN, Ellis JG. Avirulence proteins from haustoria-forming pathogens. FEMS (Fed Eur Microbiol Soc) Microbiol Lett. 2007;269:181–188. doi: 10.1111/j.1574-6968.2007.00684.x. [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008;146:368–376. doi: 10.1104/pp.107.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host–microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA. 2003;100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Catanzariti A, Ayliffe MA, Ellis JG. The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell. 2004;16:755–768. doi: 10.1105/tpc.020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Catanzariti A, Teh T, Wang CI, Ayliffe MA, Kobe B, Ellis JG. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci USA. 2006;103:8888–8893. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Folta-Stogniew E, Williams K. Determination of molecular masses of protein in solution: implementation of an HPLC size exclusion chromatography and laser light scattering in a core laboratory. J Biomol Technol. 1999;10:51–63. [PMC free article] [PubMed] [Google Scholar]
- Grouffaud S, van West P, Avrova AO, Birch PR, Whisson SC. Plasmodium falciparum and Hyaloperonospora parasitica effector translocation motifs are functional in Phytophthora infestans. Microbiology. 2008;154:3743–3751. doi: 10.1099/mic.0.2008/021964-0. [DOI] [PubMed] [Google Scholar]
- Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO (Eur Mol Biol Organ) J. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun S. Groovy times: filamentous pathogen effectors revealed. Curr Opin Plant Biol. 2007;10:358–365. doi: 10.1016/j.pbi.2007.04.017. [DOI] [PubMed] [Google Scholar]
- Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003a;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003b;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- Martin GB, Bogdanove AJ, Sessa G. Understanding the functions of plant disease resistance proteins. Annu Rev Plant Biol. 2003;54:23–61. doi: 10.1146/annurev.arplant.54.031902.135035. [DOI] [PubMed] [Google Scholar]
- Rep M, van der Does HC, Meijer M, van Wijk R, Houterman PM, Dekker HL, de Koster CG, Cornelissen BJ. A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol Microbiol. 2004;53:1373–1383. doi: 10.1111/j.1365-2958.2004.04177.x. [DOI] [PubMed] [Google Scholar]
- Schmidt SA, Williams SJ, Wang CI, Sornaraj P, James B, Kobe B, Dodds PN, Ellis JG, Anderson PA. Purification of the M flax-rust resistance protein expressed in Pichia pastoris. Plant J. 2007;50:1107–1117. doi: 10.1111/j.1365-313X.2007.03104.x. [DOI] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science. 2003;301:1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- Tous GI, Fausnaugh JL, Akinyosoye O, Lackland H, Winter-Cash P, Vitorica FJ, Stein S. Amino acid analysis on polyvinylidene difluoride membranes. Anal Biochem. 1989;179:50–55. doi: 10.1016/0003-2697(89)90198-x. [DOI] [PubMed] [Google Scholar]
- Tyler BM. Entering and breaking: virulence effector proteins of oomycete plant pathogens. Cell Microbiol. 2009;11:13–20. doi: 10.1111/j.1462-5822.2008.01240.x. [DOI] [PubMed] [Google Scholar]
- Ueda H, Yamaguchi Y, Sano H. Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein, N factor during the hypersensitive response in tobacco plants. Plant Mol Biol. 2006;61:31–45. doi: 10.1007/s11103-005-5817-8. [DOI] [PubMed] [Google Scholar]
- Van den Ackerveken GF, Vossen P, De Wit PJ. The AVR9 race-specific elicitor of Cladosporium fulvum is processed by endogenous and plant proteases. Plant Physiol. 1993;103:91–96. doi: 10.1104/pp.103.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CI, Guncar G, Forwood JK, Teh T, Catanzariti A, Lawrence GJ, Loughlin FE, Mackay JP, Schirra HJ, Anderson PA, Ellis JG, Dodds PN, Kobe B. Crystal structures of flax rust avirulence proteins AvrL567-A and -D reveal details of the structural basis for flax disease resistance specificity. Plant Cell. 2007;19:2898–2912. doi: 10.1105/tpc.107.053611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Arakawa T, Philo JS. Size-exclusion chromatography with on-line light-scattering, absorbance, and refractive index detectors for studying proteins and their interactions. Anal Biochem. 1996;240:155–166. doi: 10.1006/abio.1996.0345. [DOI] [PubMed] [Google Scholar]
- Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, Gilroy EM, Armstrong MR, Grouffaud S, van West P, Chapman S, Hein I, Toth IK, Pritchard L, Birch PR. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.