SUMMARY
The Toll/interleukin-1 receptor (TIR) domain occurs in animal and plant immune receptors. In the animal Toll-like receptors, homodimerization of the intracellular TIR domain is required for initiation of signaling cascades leading to innate immunity. By contrast, the role of the TIR domain in cytoplasmic nucleotide-binding/leucine-rich repeat (NB-LRR) plant immune resistance proteins is poorly understood. L6 is a TIR-NB-LRR resistance protein from flax (Linum usitatissimum) that confers resistance to the flax rust phytopathogenic fungus (Melampsora lini). We determine the crystal structure of the L6 TIR domain and show that, although dispensable for pathogenic effector protein recognition, the TIR domain alone is both necessary and sufficient for L6 immune signaling. We demonstrate that the L6 TIR domain self-associates, most likely forming a homodimer. Analysis of the structure combined with site-directed mutagenesis suggests that self-association is a requirement for immune signaling and reveals distinct surface regions involved in self-association, signaling, and autoregulation.
INTRODUCTION
The Toll/interleukin-1 receptor (TIR) domain occurs in both animal and plant immune receptors. In animal Toll-like receptors (TLRs), this is an intracellular signaling domain that triggers immunity in response to extracellular perception of pathogen-associated molecular patterns (PAMPs) (Tapping 2009). In plants, this domain occurs at the N terminus of a major subclass of cytoplasmic nucleotide-binding leucine-rich repeat (NB-LRR) family resistance receptors (R proteins), which trigger defense responses after perception of pathogen effectors (Chisholm et al., 2006; Jones and Dangl, 2006; Rafiqi et al., 2009; Dodds and Rathjen, 2010). These responses include localized cell death known as the hypersensitive response (HR). The LRR domain of plant R proteins appears to be the major determinant of recognition specificity (Ellis et al., 1999; Dodds et al., 2001; Shen et al., 2003; Rairdan and Moffett, 2006; Padmanabhan et al., 2009). The NB domain is shared with mammalian nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), which also function as regulators of innate immune responses and apoptosis (Inohara et al., 2005). This domain is often referred to as the NB-ARC region (nucleotide-binding adaptor shared by APAF-1, resistance proteins and CED-4; (Van der Biezen and Jones, 1998). Evidence suggests the NB domain can bind and hydrolyse nucleotides, and the presence of bound ATP or ADP may determine whether the R protein is in an active or inactive signaling state (Lukasik and Takken, 2009).
In animal TLRs, PAMP perception by the extracellular LRR region induces homodimerization of the cytoplasmic TIR domain. This provides a new scaffold that binds to adaptor proteins to initiate downstream signaling (Tapping, 2009). However, in plant R proteins, the mechanism by which effector recognition is linked to the activation of defense signaling is poorly understood. Some evidence suggests that the TIR domain mediates R protein signaling. Overexpression of the TIR region plus 40–80 additional residues from different TIR-NB-LRR proteins, from Arabidopsis, tobacco, and flax, triggers an effector-independent cell death response in planta (Frost et al., 2004; Weaver et al., 2006; Swiderski et al., 2009; Krasileva et al., 2010). However, the specific roles of the TIR domain and the additional amino acid sequences required for this function are not clear.
In flax (Linum usitatissimum), the polymorphic L locus encodes TIR-NB-LRR proteins that recognize effector proteins from the flax rust fungus (Melampsora lini) (Ellis et al., 1999; Dodds et al., 2004). For instance, the L5 and L6 proteins interact directly with variants of the AvrL567 effector and trigger resistance to rust strains expressing these effectors (Dodds et al., 2006; Wang et al., 2007). Frost et al. (2004) observed that overexpression of a 248 amino acid N-terminal fragment of L10, containing the predicted TIR region plus an additional 39 amino acids, resulted in an autoactive phenotype including constitutive defense gene expression and cell death induction in tobacco.
In this study, we used the L6 protein as a model to characterize plant TIR domain signaling activity and mode of action. Mutational analysis showed that the TIR domain is dispensable for AvrL567 recognition but is required for induction of defense responses. Expression of truncated fragments defined the minimum functional region required for defense signaling, and crystal structure determination confirmed that this corresponded to the TIR domain alone and redefined the boundaries of this domain. Three different approaches showed that the L6 TIR domain self-associates, and structural and mutational analysis led us to identify distinct surface regions involved in self-association, signaling, and activity regulation.
RESULTS
L6 TIR Domain Mutations Affect Signaling but Not Effector Recognition
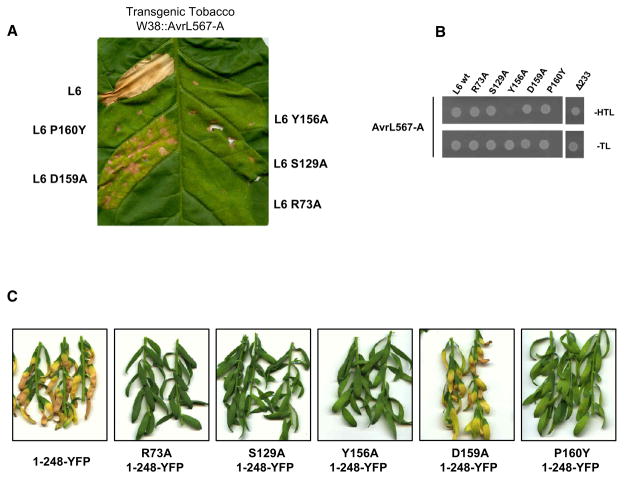
We generated five independent mutations in the TIR region of L6 at positions that are either highly conserved in other plant TIR domains or were shown to be critical for function of the tobacco resistance protein N (Mestre and Baulcombe 2006). L6 TIR domain mutants were tested for their ability to trigger an effector-dependent cell death in planta (Figure 1A). Agrobacterium-mediated transient expression of L6 induces a strong necrotic reaction in transgenic tobacco expressing the corresponding effector protein AvrL567. Four of the TIR domain mutants prevented this cell death reaction, while the fifth (D159A) significantly compromised cell death induction but retained some signaling activity. We also tested the ability of these mutant L6 proteins to interact with AvrL567 in a yeast two-hybrid assay (Figure 1B). Four of the mutants retained the interaction with AvrL567, similar to the wild-type protein, while one (Y156A) disrupted this interaction, possibly as a result of altering the overall protein conformation. A deletion mutant lacking the entire TIR domain (L6 Δ233) also did not affect L6/AvrL567 recognition (Figure 1B). Thus the L6 TIR domain is not required for interaction with AvrL567 but acts downstream of this effector-recognition event in resistance signaling.
Figure 1. Mutations in L6 TIR Domain Affect HR Induction and Signaling Activation but Not Effector Recognition.
(A) Transgenic tobacco W38 expressing AvrL567-A, 4 days after infiltration with A. tumefaciens strains carrying full-length L6 wild-type or TIR domain mutants.
(B) Growth of yeast cells coexpressing GAL4-BD::AvrL567-A with GAL4-AD::full-length L6 wild-type, TIR domain mutants or L6 lacking the TIR domain (Δ233). Growth on media lacking tryptophan and leucine (−TL) confirms yeast viability, while growth on media lacking histidine (−HTL) indicates expression of the HIS3 reporter gene due to interaction between the fusion proteins.
(C) Flax Hosh plants 12 days after infiltration with A. tumefaciens strains carrying L6 TIR1-248 mutants fused to YFP.
In tobacco, overexpression of a 248 amino acid N-terminal fragment of L10 triggers effector-independent cell death (Frost et al., 2004). Transient expression of this L10 fragment, and equivalent regions of L6, L2, and L7, also induced spontaneous cell death in flax leaves (see Figure S1A available online), as did the L61-248 fragment fused to the yellow fluorescent protein (YFP) (Figure 1C). However, incorporation of the R73A, S129A, Y156A, or P160Y mutations abolished L61-248-YFP autoactivity, while the D159A mutation caused a partial reduction of cell death induction (Figure 1C). Immunoblot analysis showed that all constructs were stably expressed in planta and in yeast (Figures S1B–S1D). The correspondence between the phenotypes of these mutations in full-length L6 and in L61-248 suggests that L61-248 autoactivity represents a genuine signaling event, with similar functional requirements as in effector-triggered signaling.
Defining the Functional Boundaries of the Plant Resistance Protein TIR Domain
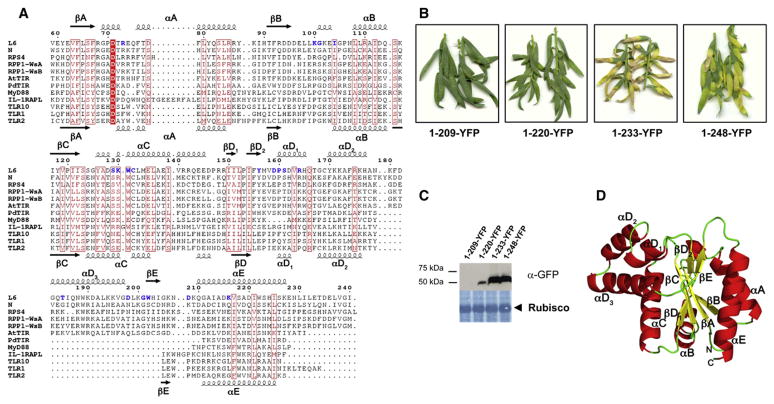
The C-terminal boundary of R protein TIR domains was previously defined to occur at the site of the first intron, based on alignments with animal TIR domain sequences (Whitham et al., 1994). However, we observed that a conserved set of hydrophobic residues in the first 16 amino acids translated from exon 2 aligned with a similar set of residues that form part of the amphipathic α helix E (αE) of the animal TIR domains (Figure 2A). This is consistent with the recently determined crystal structure of NP_177436/At1g72930 (AtTIR) (Chan et al., 2010), an Arabidopsis protein of unknown function comprised of only a TIR domain, as well as with the L6 TIR domain crystal structure described below. To determine the minimum functional region of L6 required for autoactivity, we tested a series of N-terminal fragments fused to YFP for cell death induction in flax (Figure 2B). The smallest fragment that induced cell death was L61-233, which is consistent with the redefined TIR domain sequence boundaries. However, immunoblot analysis (Figure 2C) showed that L61-220 accumulated at lower levels than the autoactive constructs, while L61-209 was not detected, possibly because truncation of the αE helix (see Figure 2A) affects protein folding and stability.
Figure 2. L6 TIR Domain Is Sufficient and Necessary to Trigger Cell Death Signaling.
(A) Multiple sequence alignment of TIR domains. Amino acid sequences from the TIR domains of L6 (residues 59–240), N (10–191), RPS4 (15–191), RPP1-WsA (50–229), and RPP1-WsB (83–262) were aligned with the sequences of the TIR domains with known 3D structures: AtTIR (PDB ID 3JRN), PdTIR (3H16), MyD88 (2Z5V), IL-1RAPL (1T3G), TLR1 (1FYV), TLR2 (1FYW), and TLR10 (2J67) using MUSCLE (Edgar 2004). The positions of the secondary structure elements in L6 and TLR2 are shown at the top and bottom, respectively. The alignment was formatted using ESPript (Gouet et al., 2003). Strictly conserved residues are indicated in white letters with a red box and similar residues are indicated in red letters with a red frame, while mutated residues are indicated in bold blue letters. The intron site is indicated by an arrow head.
(B) Flax Hosh plants 12 days after infiltration with A. tumefaciens strains carrying L6 TIR domain deletion constructs fused to YFP.
(C) Immunoblot detection of L6 TIR-YFP fusions with anti-GFP antibodies 3 days after agroinfiltration in flax leaves. Lower panel shows membrane stained with amido black indicating equal loading of Rubisco.
(D) Crystal structure of L6TIR. Ribbon drawing of one of the two molecules in the asymmetric unit. The secondary structure elements and loops are named according to the nomenclature used for TLR1 TIR domain (Xu et al., 2000) and AtTIR (Chan et al., 2010).
Crystal Structure of the L6 TIR Domain
Several protein fragments including the L6 TIR domain were expressed, purified, and subjected to crystallization trials and one fragment (residues 29–229, designated as L6TIR) yielded plate-like crystals that diffracted to 2.3 Å resolution (Ve et al., 2011). The structure was solved by molecular replacement with the AtTIR structure as template (Chan et al., 2010) and was refined to final Rwork/Rfree values of 17.4%/23.0% (Table S1). The protein crystals have the symmetry of the space-group P21212 and contain two monomers per asymmetric unit. The final model contains residues 59–228, because no electron density was observed for residues 29–58, suggesting that this region has a disordered or flexible conformation in the crystal. Each of the molecules (Figure 2D) consists of a five-stranded parallel β sheet (βA–βE) surrounded by five α-helical regions (αA–αE).
The global fold is similar to the structures of AtTIR; the TIR domains of TLR1, TLR2, TLR10, MyD88, and IL-1RAPL; and the TIR-like protein from the bacterium Paracoccus denitrificans (PdTIR) (Xu et al., 2000; Khan et al., 2004; Nyman et al., 2008; Chan et al., 2009; Ohnishi et al., 2009; Chan et al., 2010). L6TIR exhibits highest structural similarity to AtTIR (40% sequence identity) with an overall Cα rmsd value of 1.3 Å (Figure S2A). However, the L6TIR structure includes the BB loop region and the αB helix, which were absent from the AtTIR structure model, because no electron density was observed for this region (Figure S2B). The L6 TIR domain also has a β bulge in the βD strand due to the presence of two adjacent proline residues (P120 and P153) in the β sheet (Figure 2D). The mammalian and the bacterial TIR domains share less than 20% sequence identity to L6TIR, with the TLR2 TIR domain the most similar among these (by DALI search [Holm and Rosenstrom 2010], Z and rmsd value of 10.9 and 3.2 Å, respectively). Similar to AtTIR, a third αD helix is observed in the L6TIR structure when compared to the mammalian and bacterial TIR domains (Figure 2A, Figure S2C) and is apparently a unique feature of plant TIR domains. The structural definition of the L6 TIR domain corresponds closely to the minimal autoactive region, indicating that this domain alone is sufficient to induce effector-independent cell death.
L6 TIR Domain Self-Associates
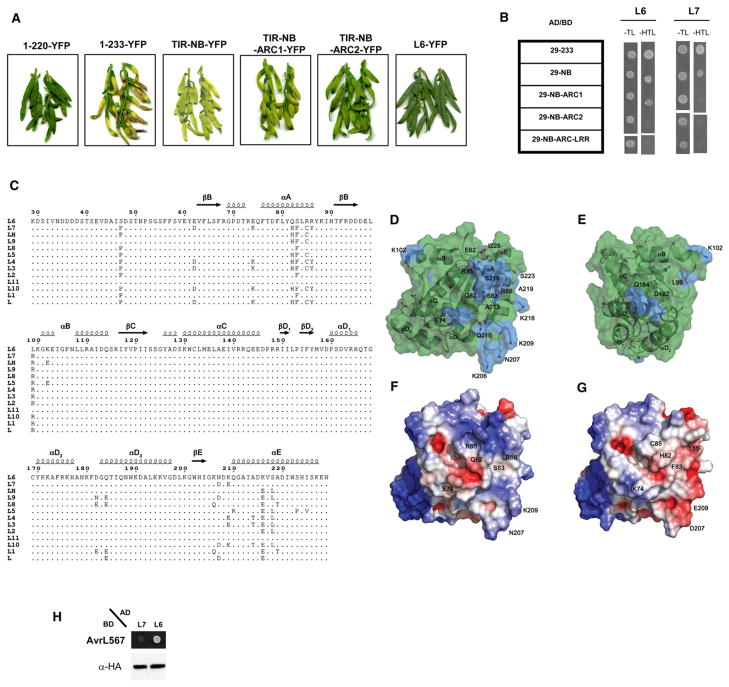
Upon PAMP perception, animal TLR activation leads to homodimerization of the intracellular TIR domain, which initiates down-stream signaling pathways (Brikos and O’Neill, 2008; Monie et al., 2009; Tapping, 2009). Yeast two-hybrid assays showed L629-233 or L629-248 (corresponding to the autoactive TIR domain fragments but lacking a 29 residue membrane anchor; D. Takemoto, M. Rafiqi, U. Hurley, G.J. Lawrence, M.B., A.R. Hardham, J.G.E., P.N.D., and D.A. Jones, unpublished data) could self-associate, while the nonautoactive fragments, L629-209, L629-220, and full-length L6 (L629-end) did not (Figure 3A). Furthermore, a construct lacking the N-terminal half of L6 TIR domain (L6114-248) also failed to interact with itself. All fusion proteins were stable in yeast (Figure 3B). These results indicate that the L6 TIR domain is required and sufficient for both effector-independent cell death signaling and self-association.
Figure 3. L6 TIR Domain Self-Associates.
(A) Growth of yeast cells coexpressing GAL4-BD and GAL4-AD L6 TIR domain fusions on synthetic media lacking tryptophan and leucine (−TL) or selective media additionally lacking histidine (−HTL).
(B) Immunoblot detection of GAL4-AD and GAL4-BD fusion proteins in yeast. Proteins were detected with anti-HA and anti-Myc antibodies, respectively.
(C–F) Solution properties of L6TIR. Red lines indicate the trace from the refractive index detector (arbitrary units) during size exclusion chromatography, and the blue lines are the weight-average molecular weight (Mw; y axis) distribution across the peak determined by MALLS. In (C), the initial concentration of L6TIR is 2 mg/ml and the buffer consists of 10 mM HEPES (pH 7.4) and 150 mM NaCl. (D) Same buffer condition as (C), but the initial protein concentration was 1 mg/ml. (E) Initial protein concentration is 2 mg/ml; buffer contains 0 mM NaCl. (F) Initial protein concentration is 2 mg/ml; buffer contains 500 mM NaCl.
We also examined the self-association of the purified L6 TIR protein in solution by gel filtration chromatography coupled to multiangle laser light scattering (MALLS). At an initial protein concentration of 2 mg/ml and in 150 mM NaCl, the elution peak showed a molecular mass of 35.5 kDa (Figure 3C), between the expected mass of the dimeric (46.8 kDa) and monomeric (23.4 kDa) states. At a lower concentration (1 mg/ml) L6 TIR elutes slightly later, and the estimated molecular mass is 29.5 kDa (Figure 3D). In both cases the elution peak is asymmetric, indicating a polydisperse population of molecules. Furthermore, the molecular mass distribution is lower on each side of the peak maximum where the local protein concentration is lower. These data suggest that monomeric and oligomeric species are not partitioned on the size exclusion column and are in a rapid reversible equilibrium under these conditions. The maximum peak concentrations in the two experiments were 11.0 and 4.9 μM, respectively, indicating that the dissociation constant for oligomerization is in the micromolar range. Reducing the ionic strength by removal of NaCl from the mobile phase resulted in a faster elution time and a molecular mass of 49.5 kDa, close to the expected dimer mass (Figure 3E), while higher ionic strength (500 mM NaCl) resulted in slower elution, a more symmetric elution peak, and a molecular mass of 25.7 kDa, close to the expected monomer size (Figure 3F). The dependence of oligomerization on ionic strength suggests that self-association is predominantly governed by electrostatic interactions. L6TIR self-association was also analyzed by sedimentation equilibrium analytical ultracentrifugation (AUC), the data from which fit best with a monomer-dimer equilibrium model with an estimated Kd of 20.3 μM (Figure S3).
Identification of TIR/TIR Interfaces in the Crystal Structure
Two different TIR/TIR domain interfaces are observed in the crystal (Figure 4A). The two chains in the asymmetric unit form a two-fold symmetrical dimer with a buried surface area of approximately 890 Å2 (interface 1). Both of these chains also interact with an additional TIR domain on the opposite side of the molecule with two-fold symmetry and a buried surface area of 780 Å2 (interface 2).
Figure 4. TIR/TIR Domain Interfaces in the L6 TIR Crystal.
(A) Ribbon representation of the asymmetric unit interfaces (green and yellow) and the interface related by crystallographic symmetry (yellow and cyan).
(B) As in (A), with the residues involved in close contacts (at distances <4 Å ) shown in wireframe. Hydrogen bonds/salt bridges are shown as dotted red lines.
An extensive network of 15 hydrogen bonds and electrostatic interactions are observed in interface 1, involving residues from the αD1, αD3, and αE helices; the βE strand; and the DE and EE loops (Figure 4B). At the core of the interface, the βE strands are connected by hydrogen bonds between the two G201 residues. The carbon chains of the K200 residues are in close proximity to each other and also stack against the side chain of W202 from the second molecule, creating a hydrophobic sandwich with the two lysine side chains in the middle. W202 also forms a hydrogen bond with the main-chain carbonyl of D198. On either side of the interface, electrostatic interactions are observed between D198 at the end of the αD3 helix and K216 in the αE helix, and between R164 in the αD1 helix and D208 in the EE loop. R164 also forms two hydrogen bonds with the carbonyl oxygen of H203 in the βE strand, while D208 forms a hydrogen bond with S161 in the αD1 helix.
Interface 2 has a hydrophobic core consisting of I104, L108, L109, and W131 from both molecules (Figure 4B), stabilized on both sides by hydrophobic contacts between F181 and L98, and hydrogen bonds between the side chains of K100 and T185, and between the side chain of K130 and the main-chain carbonyl oxygens of E97 and L99.
Mutational Analysis of L6 TIR Self-Association and Signaling
The dependence of L6 TIR self-association on ionic strength suggests that it depends largely on electrostatic interactions, which is more consistent with interface 1 representing the self-association interface. To test this directly, we analyzed the effects of amino acid substitutions in both putative interfaces on L6 TIR domain self-association and autoactivity (Table 1, Figure S4). Out of eight sites mutated in interface 1, mutations of residues R164, K200, G201, W202, D208, and K216 disrupted self-association in yeast. Most of these residues occupy a central location in interface 1, and R164, W202, and D208 are predicted as binding hot spots by the KFC and HotPoint web servers (Darnell et al., 2008; Tuncbag et al., 2010). K216 however is more peripheral to the interface, and while a glutamate substitution at this position disrupted self-association, alanine substitution did not. Likewise, alanine substitution of residues S161 and D198, which are also involved in hydrogen bonds on the periphery of interface 1, did not affect self-association (Table 1). The P160Y and Y156A mutations also disrupted both autoactivity and self-association (Table 1, Figure 1C). These residues are not directly involved in interactions between molecules, but are important for maintaining the DD loop, the αD1 helix, and the surrounding surface in the correct conformation. None of the mutations introduced in residues involved in interface 2 affected self-association, suggesting that this interface does not represent a true interaction. However, mutations of several conserved residues in the BB loop and αC helix completely disrupt the autoactive phenotype in flax (Figure S4A), suggesting that this region may be important for signaling downstream of the self-association event.
Table 1.
Mutational Analysis of L6 TIR Domain
| Mutation Position | Location in the Crystal Structure | Interface | Autoactivity | Self-Association |
|---|---|---|---|---|
| WT | +++ | + | ||
| R73A | αA helix | − | + | |
| K100A | BB loop | 2 | ++ | + |
| G101C | BB loop | − | + | |
| I104A | BB loop | 2 | − | + |
| S129A | αC helix | − | + | |
| K130A | αC helix | 2 | +/− | + |
| W131A | αC helix | 2 | − | + |
| C132S | αC helix | − | + | |
| Y156A | DD loop | − | − | |
| D159A | DD loop | + | + | |
| P160Y | DD loop | − | − | |
| S161A | αD1 helix | 1 | + | + |
| R164A | αD1 helix | 1 | + | − |
| R164E | αD1 helix | 1 | + | − |
| T185A | αD3 helix | 2 | + | + |
| D198A | αD3 helix | 1 | ++ | + |
| K200E | DE loop | 1 | +/− | − |
| G201Y | DE loop | 1 | +/− | +/− |
| G201R | DE loop | 1 | − | − |
| G201C | DE loop | 1 | + | + |
| W202A | DE loop | 1 | +/− | − |
| D208A | EE loop | 1 | + | − |
| K216A | αE helix | 1 | ++ | + |
| K216E | αE helix | 1 | +++ | − |
List of single mutations introduced in the L6 TIR domain. Residues showing polar contacts in dimer interface 1 or 2 are indicated in the interface column. Autoactivity column shows strength of cell death symptoms as compared to wild-type (WT) protein; comparable to wild-type (+++), weaker than wild-type (++), chlorosis symptoms (+), very weak chlorosis symptoms (+/−), no symptoms (−). Self-association column shows ability of the TIR domain mutants to self-associate in yeast.
MALLS and AUC studies conducted on the P160Y, R164A, K200E, D208A, and K216E mutants (Table S2 and Figures S4E and S4F) showed that these interface 1 mutants had reduced average molecular mass at the gel filtration peak and significantly higher monomer-dimer dissociation constants than the wild-type. By contrast, the W131A mutation in interface 2 had no effect on average molecular mass and only a slight increase in the monomer-dimer dissociation constant. With the exception of K216E, all of the mutations that disrupted self-association also compromised autoactivity in flax (Table 1 and Figure S4), suggesting that self-association is critical for signaling activity. The K216E mutation had a smaller effect on the dimer binding affinity measured by AUC than other mutations in this region (Table S2), so it is possible that the binding affinity is reduced just below the threshold required for yeast two-hybrid detection, but remains sufficient to induce cell death. Interestingly, the L10 TIR domain, which contains a glutamate at this position, did not self-associate in the Y2H assay, but an E216K mutation allowed detection of this interaction (Figure S5A). However, this substitution also reduced the cell death induced by the L10 TIR domain (Figure S5B). This residue is located at the periphery of the dimerization interface and remains largely solvent exposed even in the dimer. Thus it is possible that it has an additional role in affecting the interaction with a signaling partner, and that glutamic acid at this position is more favorable to this signaling interaction than lysine.
Mapping of sequence conservation among plant TIR domains onto the structure of L6TIR using ConSurf (Ashkenazy et al., 2010) revealed that P160, S161, R164, and G201 constitute a conserved surface patch in interface 1, while I104, L108, and W131 are part of a relatively conserved patch in interface 2 (Figures S6A–S6D). Several other surface-exposed residues are also conserved in this area, including R68, R73, G101, S124, and D135. Homology modeling of the RPS4 and N protein TIR domains based on the L6TIR structure, and mapping of existing mutational data, suggest that these regions may be important for self-association and signaling in other TIR domains as well (Figures S6E–S6J).
The NB-ARC Domain Inhibits L6 TIR Domain Autoactivity and Self-Association
L6 signaling activity normally requires effector recognition, suggesting that other parts of the protein may inhibit TIR domain signaling. We therefore tested autoactivity and self-association of larger L6 N-terminal fragments (Figure 5A). Homology modeling of plant NB-ARC domains with the crystal structures of APAF-1 and CED-4 predicts three subdomains (NB, ARC1, and ARC2) that together form a nucleotide-binding pocket (van Ooijen et al., 2008). In flax leaves, transient expression of L6 fragments containing the TIR domain plus the NB (TIR-NB) or NB and ARC1 (TIR-NB-ARC1) regions induced similar chlorotic responses, weaker than the cell death triggered by the autoactive L61-233 fragment. The presence of the ARC2 subdomain significantly reduced chlorosis but still allowed a weak phenotype relative to the completely inactive L61-220 truncation or full-length L6. Similarly, in yeast two-hybrid assays, the L6 TIR-NB and L6 TIR-NB-ARC1 fragments showed partial inhibition of self-association, while the presence of the complete NB-ARC domain (L6 TIR-NB-ARC2) completely prevented self-association (Figure 5B). The larger L6 constructs were less stable than L61-233 in planta, but all protein fusions were expressed at similar levels in yeast (Figure S7). Inhibition of TIR domain self-association by the NB-ARC domain may contribute to the negative autoregulation of nonactivated L6.
Figure 5. The NB-ARC Domain Regulates TIR Autoactivity and Self-Association.
(A) Flax Hosh plants 12 days after infiltration with A. tumefaciens strains carrying truncated L6 constructs fused to YFP.
(B) Growth of yeast cells coexpressing GAL4-BD and GAL4-AD fused to truncated L6 and L7 constructs on synthetic media lacking tryptophan and leucine (−TL) or selective media additionally lacking histidine (−HTL).
(C) Comparison of allelic variants of the L6 TIR domain. Sequence alignment of the TIR domain region of different L alleles. The positions of polymorphic residues are highlighted.
(D and E) Transparent surface representation of L6 TIR, polymorphic regions present in allelic variants are shown in blue. The molecule in E is oriented 135° around the vertical axis compared to (D).
(F and G) Surface representations of L6TIR (F) and a homology model of the L7 TIR domain (G), with electrostatic potential (calculated using APBS; Baker et al., 2001) mapped to the surface. Coloring is continuous going from blue (potential +5 kt/e) through white to red (potential −5 kt/e). The molecules are oriented as in (D).
(H) Growth of yeast cells coexpressing GAL4-BD::AvrL567-A with GAL4-AD::full-length L7 or L6 on selective media lacking histidine (−HTL). GAL4 AD fusion proteins were detected with anti-HA antibodies.
TIR Domain Allelic Variants Identify a Potential Interaction Site with the NB-LRR Region
Twenty-one residues in the TIR domain are polymorphic among the 12 L alleles. The majority of these are located in the adjacent αA and αE helices, and the EE loop region (Figure 5C). They cluster to a surface region that is distinct from the TIR/TIR domain interfaces observed in the crystal (Figures 5D and 5E). The L6 and L7 proteins differ only in the TIR region, and show similar recognition specificity, but with L7 mediating a weaker resistance response (Ellis et al., 1999; Luck et al., 2000). Seven of the ten amino acid changes between L6 and L7 domain are in the αA and αE helices and EE loop, and lead to significant differences in both hydrophobicity and electrostatic potential (Figures 5F and 5G). The L7 TIR domain was just as autoactive and able to dimerize in yeast as the L6 TIR domain (Figure S1A, Figure 5B), but the full-length L7 protein showed much weaker interaction with AvrL567 than L6 (Figure 5H). This shows that the phenotypic difference is due to a weaker recognition of the effector by L7 and not weaker signaling, even though the TIR domain is not required for recognition. This suggests that the L7 TIR domain interferes with AvrL567 recognition through interaction with other parts of the protein. Indeed, we found that inclusion of the L6 NB and NB-ARC regions inhibited L7 TIR dimerization more strongly than that of L6 TIR (Figure 5B). Thus this polymorphic region may be involved in autoregulation through intramolecular interaction with the NB-ARC and/or the LRR domains.
DISCUSSION
Plant disease resistance (R) proteins mediate strong immune responses to pathogens, but their activity is associated with cell death, so tight regulation of their signaling activity is essential. However, the molecular mechanisms involved in the repression of R protein signaling in the native state, and its activation by effector recognition, remain largely unknown. Here we show that the L6 TIR domain is necessary and sufficient for cell death signaling. TIR domain mutations had similar effects on effector-dependent cell-death signaling by the full-length L6 protein and on effector-independent signaling by the TIR domain alone, suggesting that TIR autoactivity represents a genuine signaling event, with similar functional requirements as in effector-triggered signaling. An alternative possibility is that the TIR domain may interact with endogenous TIR-NB-LRR proteins and cause their activation. However, the L6 TIR did not interact with full-length L6 or with other known autoactive TIR domains (from RPS4 and N) in yeast (our unpublished data), suggesting that this explanation is unlikely.
Previously, the sequence boundaries of plant TIR domains were predicted to correspond to the exon 1 product of TIR-NB-LRR genes (Whitham et al., 1994), but autoactive N-terminal fragments of these proteins contained about 40 additional amino acids, and shorter fragments were not autoactive and often not stable (Mestre and Baulcombe, 2006; Weaver et al., 2006; Swiderski et al., 2009; Krasileva et al., 2010). The L6TIR crystal structure now reveals that this domain extends to amino acid 228 of L6 (exon 1 product plus 20 amino acids), and this corresponds closely to the minimal domain required for autoactivity (Figure 2B). Thus the TIR domain alone is sufficient for signaling.
Yeast two-hybrid, MALLS, and AUC analysis showed that the L6 TIR domain is able to self-associate in yeast and in vitro (Figure 3, Figure S3). The crystal structure revealed a TIR/TIR domain interface in the crystal asymmetric unit (interface 1), involving residues from the αD1 and αE helices, the βE strand, and the DE and EE loops. Mutation of key residues in this interface, as well as deletion of parts of αE, abolished TIR domain self-association, supporting the role of this interface in this interaction. There was a strong correlation between self-association of L6 TIR domain fragments in yeast and in vitro and their autoactivity in planta (Table 1), suggesting that this is a key event in TIR domain signaling. Some of the mutations that result in loss of function of the RPS4 and N resistance proteins (Dinesh-Kumar et al., 2000; Mestre and Baulcombe, 2006; Swiderski et al., 2009) are also located in the L6 TIR domain dimer interface (Figures S6H–S6J), suggesting that this interface and its role in activation may be conserved among other TIR-NB-LRR proteins.
Signaling by animal TLRs is mediated by homodimerization of the TIR domain. Our yeast two-hybrid and MALLS data do not clearly distinguish between dimerization or higher-order oligomerization, although the molecular weight of the complex identified by MALLS at low ionic strength is close to the expected size of the dimer and the sedimentation equilibrium AUC data are most consistent with a monomer-dimer equilibium model. Likewise, the experimentally verified crystal interface involves only two molecules, suggesting that dimer formation is involved in L6 TIR domain self-association. However, it remains possible that higher-order TIR domain complexes form after this initial dimerization event. Whether other autoactive plant TIR domains also self-associate remains an open question. Mestre and Baulcombe (2006) demonstrated that the tobacco N protein oligomerizes in the presence of the tobacco mosaic virus p50 protein and that the N TIR domain self-associates in planta. However, by comparison with the L6 TIR domain structure, the N TIR domain construct lacked the last α helix and furthermore was not autoactive, so the relevance of this result to TIR domain signaling is not clear. Given that the N protein TIR domain and the TMV p50 protein independently associate with the chloroplast NRIP protein (Caplan et al., 2008), these observations may be related to the formation of TIR-NRIP-p50 recognition complexes rather than specifically related to a signaling event.
Despite many efforts, we could not detect L6 TIR domain oligomerization in planta by coimmunoprecipitation (data not shown). Other attempts to coimmunoprecipitate autoactive plant TIR domains were similarly unsuccessful (Swiderski et al., 2009; Krasileva et al., 2010). This could be explained by the observation that the L6 TIR domain self-association in vitro is of relatively low affinity and readily reversible (Figure 3), suggesting that any in vivo oligomer may not persist long enough to be detected by coimmunoprecipitation. The low affinity of self-association may be explained by the small size (890 Å2) of the dimerization interface, because more than 1000 Å2 is normally required to form stable protein-protein interactions (Kobe et al., 2008). Such transient low-affinity oligomerization may be a key part of the regulation of the R protein function, as it would prevent inappropriate cell death signaling.
Mutations in the αC helix and the BB loop do not affect L6 TIR domain self-association, but do interfere with effector-independent cell death induction in planta (Table 1). Many loss-of-function mutations in RPS4 and N also map to this region (Figures S6E–S6G). In both N and L6, mutation of the conserved and surface-exposed tryptophan in the αC helix to an alanine results in loss of function, while increased autoactivity is observed for the corresponding mutation in RPS4. Mutation of the neighboring cysteine and serine residues also results in loss of activity, suggesting that the αC helix plays an important role in plant TIR domain-dependent signaling. Several of the loss- or gain-of-function mutations in RPS4 are located in the unique αD3 helical region, indicating that this region also may be important for signaling. Upon ligand-induced activation in animal TLRs, TIR domain dimerization is thought to provide a new scaffold that is able to bind downstream signaling proteins, which in turn initiate downstream cascades leading to immune responses. Thus, the αC helix and the BB loop region of the L6 TIR domain may represent an interacting surface for downstream signaling components.
The NB-ARC domain may negatively regulate L6 TIR domain autoactivity by preventing the dimerization event (Figure 5). Comparison of L6 and L7 TIR domains suggests that a third distinct surface region on this domain, involving the αA and αE helices, is involved in intramolecular interactions with the NB and/or LRR domains, which may be important for autoregulation of the resistance protein. The animal NB-ARC proteins APAF-1 and CED-4 respond to apoptotic stimuli by oligomerizing through their NB-ARC domains (Kim et al., 2005; Yu et al., 2005; Bao and Shi, 2007; Petrilli et al., 2007; Qi et al., 2010; Yuan et al., 2010), which brings their N-terminal caspase recruitment domains (CARDs) into close proximity to form an active signaling platform. By analogy, it is possible that plant TIR-NB-LRRs form an NB-ARC-dependent oligomeric complex once activated, leading to proximity-induced dimerization of the TIR domain and activation of defense pathways. This hypothesis is consistent with the finding that RPP1-WsB autoactive TIR domain phenotype depends on the presence of a dimeric form of GFP fused to its C-terminal extremity (Krasileva et al., 2010).
Some NB-LRR plant resistance proteins contain a coiled-coil (CC) or an uncharacterized domain instead of the TIR domain. However, characterization of the potato virus resistance protein Rx provided some evidence that the NB domain and not the CC domain functions as the signaling domain (Rairdan et al., 2008). However, Maekawa et al. (2011) recently solved the 3D structure of the CC domain of the barley MLA protein conferring resistance to powdery mildew. This revealed a homodimeric association of the CC domain which was required for cell death activity of an autoactive MLA mutant. Thus dimerization of N-terminal domains may be a critical event in signaling by both CC and TIR-NB-LRR resistance proteins.
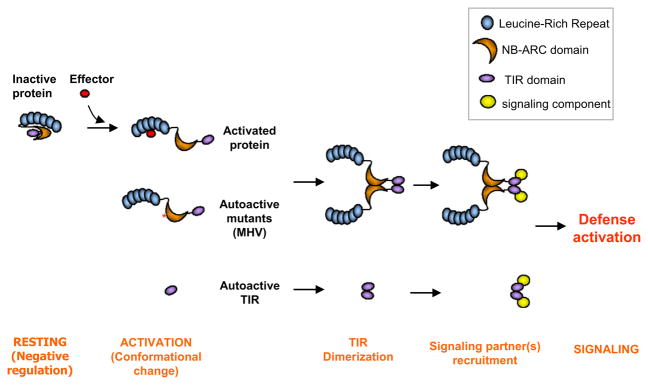
We have identified important regions of the L6 TIR domain that are involved in self-association, signaling, and autoregulation, and have provided clues to how resistance protein activation is linked to induction of signaling. These data are consistent with a model (Figure 6) in which resistance protein activation is driven by a conformational change of the NB-ARC domain upon pathogen perception, reversing the intramolecular negative regulation and leading to exposure of the TIR signaling domain. The latter becomes available for self-association and recruitment of other signaling partners. The identity of potential downstream signaling partners in plants remains unknown, and their identification will greatly enhance our knowledge of plant immunity.
Figure 6. Proposed Model for Activation of TIR-NB-LRRs Proteins.
In the absence of a recognized effector protein, intramolecular interactions keep the protein in a resting conformation where the TIR domain dimerization interface is not exposed. Upon activation, negative regulation is released, probably through nucleotide exchange in the NB domain, and conformational change exposes the TIR domain for homodimerization and the TIR domain signaling interface for interaction with signaling proteins. This induces a signaling cascade leading to innate immunity. Autoactive variants, such as the MHV mutant (contains a D-to-V mutation in the MHD motif in the ARC2 subdomain) or the TIR domain alone, circumvent the need for effector recognition.
EXPERIMENTAL PROCEDURES
Vectors and Constructs
L6 TIR domain site-directed mutants were generated with the Gene-Tailor kit (Invitrogen) according to the manufacturer’s instructions. The R73A, S129A, Y156A, D159A, and P160Y mutations were generated separately in L6 cDNA and genomic fragments and inserted by restriction cloning into the pGBT9-L6 and pGADT7-L6 vectors (Dodds et al., 2006) for yeast two-hybrid assays or the pTNL6ΔEB binary vector containing a 9 kbp L6 genomic fragment (Ellis et al., 1999) with a C-terminal triple hemagglutinin (HA) tag for in planta assays. Other yeast two-hybrid and transient expression plasmids were constructed by Gateway cloning (GWY; Invitrogen). PCR products flanked by the attB sites were recombined into pDONR207 (Invitrogen) and then into corresponding destination vectors. Gateway-compatible yeast two-hybrid vectors based on pGADT7 and pGBKT7 (Clontech) were kindly provided by Laurent Deslandes (INRA Toulouse, France). L6-YFP fusions were generated by insertion into the Gateway binary vector pAM-PAT-35 s-GWY-YFPv (Bernoux et al., 2008). Primer sequences used to generate L6 constructs are shown in the Supplemental Experimental Procedures. Mutations were introduced directly in the plant expression plasmids pAM-PAT-35 s-L61-233-YFPv or in the pENTR-L629-233 and then recombined in the yeast two-hybrid Gateway destination vectors.
Transient Expression and Yeast Two-Hybrid Assays
Agrobacterium tumefaciens cells were grown for 36 hr at 28°C in LB media containing appropriate antibiotic selections. Cells were pelleted, resuspended in infiltration medium (10 mM MgCl2, 200 μM acetosyringone), adjusted to OD600 = 1, and incubated for 2 hr at room temperature. Resuspended cells were infiltrated with a 1 ml needleless syringe into leaves of 3-week-old tobacco or 4-week-old flax (Hoshangabad) plants. Transgenic W38 tobacco expressing AvrL567-A was described by Dodds et al. (2004). Yeast transformation and growth assays were performed as described in the Yeast Protocols Handbook (Clontech).
Immunoblot Analysis
Yeast proteins were extracted by the trichloroacetic acid method (Yeast Protocols Handbook). Plant proteins were extracted by grinding two flax leaves collected 3 days after agroinfiltration in loading buffer. Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes (Pall). Membranes were blocked in 5% skim milk and probed with anti-GFP, anti-HA, or anti-Myc mouse monoclonal antibodies (Roche), followed by goat anti-mouse antibodies conjugated with horseradish peroxidase (Pierce). Labeling was detected with the SuperSignal West Pico chemiluminescence kit (Pierce). Membranes were stained with amido black for protein loading.
Production and Purification of L6TIR
The L6 TIR domain (residues 29–229) and mutant derivatives were expressed in E. coli BL21(DE3) cells and purified by Ni affinity chromatography followed by TEV protease cleavage and gel filtration as described (Ve et al., 2011). The resulting proteins contain three additional residues (SNA) at the N terminus and were stored at 6 mg/ml at −80°C.
Crystallization and Structure Solution
Crystals were obtained by hanging-drop vapor diffusion after 4–5 days by mixing 1 μl of protein solution with 1 μl of reservoir containing 36% PEG 200, 0.1 M sodium acetate (pH 5.2), and 10 mM hexammine cobalt(III) chloride. X-ray diffraction data were measured at the MX2 beamline of the Australian synchrotron using the Blu-Ice software (McPhillips et al., 2002), and were processed and scaled using XDS (Kabsch, 2010) and Scala (CPP4, 2003), respectively. The structure was solved by molecular replacement using Phaser (McCoy et al., 2007) and the AtTIR structure (PDB ID 3JRN; Chan et al., 2010) as a template. Automatic model building was performed with ARP/WARP (Langer et al., 2008) within the CCP4 package. The resulting model was refined with data between 19.3 and 2.3 Å using Phenix (Adams et al., 2010), and iterative model building between refinement rounds was carried out in Coot (Emsley and Cowtan 2004). Structure validation was performed using MolProbity (Chen et al., 2010). Coordinates and structure factors have been deposited in the Protein Data Bank with the ID 3OZI. Structure analysis was done with Coot and PyMOL (http://www.pymol.org/; DeLano Scientific LLC). Molecular models of the TIR domains from L7, RPS4, and N were built using the L6TIR structure as the template with the program Modeler (Eswar et al., 2006).
Multiangle Laser Light Scattering and Size-Exclusion Chromatography
MALLS coupled with size-exclusion chromatography was performed using a Dawn Heleos II 18-angle light-scattering detector coupled with an Optilab rEX refractive index detector (Wyatt Technology, Santa Barbara, CA, USA) and combined inline with a Superdex 200 10/300 size exclusion column (GE Healthcare). The L6TIR experiments were performed at room temperature at a flow rate of 0.5 ml/min in prefiltered (0.2 μm) 10 mM HEPES (pH 7.4) buffer with 1 mM DTT and various concentrations of NaCl (0–500 mM). The sample volume was 500 μl. Molecular mass calculations were performed using the Astra 5.3 software. Input of the refractive increment (dn/dc values) was set at 0.186 in the molecular mass calculations, based on the premise that dn/dc is constant for unmodified proteins (Wen et al., 1996).
Analytical Ultracentrifugation
Sedimentation equilibrium AUC was performed with the protein at 0.5 mg/mL in 10 mM HEPES (pH 7.4), 150 mM NaCl in two-channel quartz-window sedimentation velocity cells in an XLA analytical ultracentrifuge (Beckman/Coulter). Two scans were performed at equilibrium (24 hr) at 12,000 and 18,000 rpm and 20°C, as well as scanned at 42,000 rpm to determine the baseline. Protein and buffer properties were calculated using the software Sednterp (John Philo, Alliance Protein Laboratories), and the data were analyzed using the software Sedphat v6.5 (Peter Shuck, NIH). The 12,000 rpm and 18,000 rpm data were fit globally together to a monomer-dimer equilibrium model. The baseline and concentration were floated for each experiment, with the best-fit baselines 0.0648 and 0.066 for the low- and high-speed experiments, respectively.
Supplementary Material
Acknowledgments
This work was supported by the Australian Research Council (ARC). B.K. is an ARC Federation Fellow and National Health and Medical Research Council (NHMRC) Honorary Research Fellow. We thank Laurent Deslandes (CNRS, Toulouse, France) for providing the Gateway vectors for yeast and in planta expression. The X-ray diffraction data collection was undertaken on the MX2 beamline at the Australian Synchrotron, Victoria, Australia. We also acknowledge the use of the UQ ROCX Diffraction Facility. Kim Newell provided excellent technical assistance. Brit Winnen and Robert Simpson (University of Queensland) helped with multiangle laser light scattering and analytical ultracentrifugation, respectively.
Footnotes
Supplemental Information includes six figures, two tables, and Supplemental Experimental Procedures and can be found with this article at doi:10.1016/j.chom.2011.02.009.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res Suppl. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Q, Shi Y. Apoptosome: a platform for the activation of initiator caspases. Cell Death Differ. 2007;14:56–65. doi: 10.1038/sj.cdd.4402028. [DOI] [PubMed] [Google Scholar]
- Bernoux M, Timmers T, Jauneau A, Briere C, de Wit PJ, Marco Y, Deslandes L. RD19, an Arabidopsis cysteine protease required for RRS1-R-mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell. 2008;20:2252–2264. doi: 10.1105/tpc.108.058685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brikos C, O’Neill LA. Signalling of toll-like receptors. Handb Exp Pharmacol. 2008;2008:21–50. doi: 10.1007/978-3-540-72167-3_2. [DOI] [PubMed] [Google Scholar]
- Caplan JL, Mamillapalli P, Burch-Smith TM, Czymmek K, Dinesh-Kumar SP. Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell. 2008;132:449–462. doi: 10.1016/j.cell.2007.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Low LY, Hsu S, Li S, Liu T, Santelli E, Le Negrate G, Reed JC, Woods VL, Jr, Pascual J. Molecular mimicry in innate immunity: crystal structure of a bacterial TIR domain. J Biol Chem. 2009;284:21386–21392. doi: 10.1074/jbc.C109.007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Mukasa T, Santelli E, Low LY, Pascual J. The crystal structure of a TIR domain from Arabidopsis thaliana reveals a conserved helical region unique to plants. Protein Sci. 2010;19:155–161. doi: 10.1002/pro.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- CPP4 (Collaborative Computational Project Number 4) An overview of the CCP4 project in protein crystallography: an example of a collaborative project. J Synchrotron Radiat. 2003;10:23–25. doi: 10.1107/s0909049502017235. [DOI] [PubMed] [Google Scholar]
- Darnell SJ, LeGault L, Mitchell JC. KFC Server: interactive forecasting of protein interaction hot spots. Nucleic Acids Res. 2008;36:W265–W269. doi: 10.1093/nar/gkn346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar SP, Tham WH, Baker BJ. Structure-function analysis of the tobacco mosaic virus resistance gene N. Proc Natl Acad Sci USA. 2000;97:14789–14794. doi: 10.1073/pnas.97.26.14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Ellis JG. Six amino acid changes confined to the leucine-rich repeat beta-strand/beta-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell. 2001;13:163–178. doi: 10.1105/tpc.13.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Catanzariti AM, Ayliffe MA, Ellis JG. The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell. 2004;16:755–768. doi: 10.1105/tpc.020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CI, Ayliffe MA, Kobe B, Ellis JG. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci USA. 2006;103:8888–8893. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JG, Lawrence GJ, Luck JE, Dodds PN. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell. 1999;11:495–506. doi: 10.1105/tpc.11.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006;chapter 5(unit 5.6) doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost D, Way H, Howles P, Luck J, Manners J, Hardham A, Finnegan J, Ellis J. Tobacco transgenic for the flax rust resistance gene L expresses allele-specific activation of defense responses. Mol Plant Microbe Interact. 2004;17:224–232. doi: 10.1094/MPMI.2004.17.2.224. [DOI] [PubMed] [Google Scholar]
- Gouet P, Robert X, Courcelle E. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res Suppl. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara Chamaillard, McDonald C, Nunez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan JA, Brint EK, O’Neill LA, Tong L. Crystal structure of the Toll/interleukin-1 receptor domain of human IL-1RAPL. J Biol Chem. 2004;279:31664–31670. doi: 10.1074/jbc.M403434200. [DOI] [PubMed] [Google Scholar]
- Kim HE, Du F, Fang M, Wang X. Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc Natl Acad Sci USA. 2005;102:17545–17550. doi: 10.1073/pnas.0507900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B, Guncar G, Buchholz R, Huber T, Maco B, Cowieson N, Martin JL, Marfori M, Forwood JK. Crystallography and protein-protein interactions: biological interfaces and crystal contacts. Biochem Soc Trans. 2008;36:1438–1441. doi: 10.1042/BST0361438. [DOI] [PubMed] [Google Scholar]
- Krasileva KV, Dahlbeck D, Staskawicz BJ. Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell. 2010;22:1–15. doi: 10.1105/tpc.110.075358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck JE, Lawrence GJ, Dodds PN, Shepherd KW, Ellis JG. Regions outside of the leucine-rich repeats of flax rust resistance proteins play a role in specificity determination. Plant Cell. 2000;12:1367–1377. doi: 10.1105/tpc.12.8.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasik E, Takken FL. STANDing strong, resistance proteins instigators of plant defence. Curr Opin Plant Biol. 2009;12:427–436. doi: 10.1016/j.pbi.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Maekawa T, Cheng W, Spiridon LN, Töller A, Lukasik E, Saijo Y, Liu P, Shen QH, Micluta MA, Somssich IE, et al. Coiled-coil domain-dependent homodimerization of intracellular MLA immune receptors defines a minimal functional module for triggering. Cell Host Microbe. 2011 doi: 10.1016/j.chom.2011.02.008. in press. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhillips TM, McPhillips SE, Chiu HJ, Cohen AE, Deacon AM, Ellis PJ, Garman E, Gonzalez A, Sauter NK, Phizackerley RP, et al. Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. J Synchrotron Radiat. 2002;9:401–406. doi: 10.1107/s0909049502015170. [DOI] [PubMed] [Google Scholar]
- Mestre P, Baulcombe DC. Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell. 2006;18:491–501. doi: 10.1105/tpc.105.037234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monie TP, Bryant CE, Gay NJ. Activating immunity: lessons from the TLRs and NLRs. Trends Biochem Sci. 2009;34:553–561. doi: 10.1016/j.tibs.2009.06.011. [DOI] [PubMed] [Google Scholar]
- Nyman T, Stenmark P, Flodin S, Johansson I, Hammarstrom M, Nordlund P. The crystal structure of the human toll-like receptor 10 cytoplasmic domain reveals a putative signaling dimer. J Biol Chem. 2008;283:11861–11865. doi: 10.1074/jbc.C800001200. [DOI] [PubMed] [Google Scholar]
- Ohnishi H, Tochio H, Kato Z, Orii KE, Li A, Kimura T, Hiroaki H, Kondo N, Shirakawa M. Structural basis for the multiple interactions of the MyD88 TIR domain in TLR4 signaling. Proc Natl Acad Sci USA. 2009;106:10260–10265. doi: 10.1073/pnas.0812956106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan M, Cournoyer P, Dinesh-Kumar SP. The leucine-rich repeat domain in plant innate immunity: a wealth of possibilities. Cell Microbiol. 2009;11:191–198. doi: 10.1111/j.1462-5822.2008.01260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrilli V, Dostert C, Muruve DA, Tschopp J. The inflammasome: a danger sensing complex triggering innate immunity. Curr Opin Immunol. 2007;19:615–622. doi: 10.1016/j.coi.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Qi S, Pang Y, Hu Q, Liu Q, Li H, Zhou Y, He T, Liang Q, Liu Y, Yuan X, et al. Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell. 2010;141:446–457. doi: 10.1016/j.cell.2010.03.017. [DOI] [PubMed] [Google Scholar]
- Rafiqi M, Bernoux M, Ellis JG, Dodds PN. In the trenches of plant pathogen recognition: role of NB-LRR proteins. Semin Cell Dev Biol. 2009;20:1017–1024. doi: 10.1016/j.semcdb.2009.04.010. [DOI] [PubMed] [Google Scholar]
- Rairdan GJ, Moffett P. Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell. 2006;18:2082–2093. doi: 10.1105/tpc.106.042747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan GJ, Collier SM, Sacco MA, Baldwin TT, Boettrich T, Moffett P. The coiled-coil and nucleotide binding domains of the Potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell. 2008;20:739–751. doi: 10.1105/tpc.107.056036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QH, Zhou F, Bieri S, Haizel T, Shirasu K, Schulze-Lefert P. Recognition specificity and RAR1/SGT1 dependence in barley Mla disease resistance genes to the powdery mildew fungus. Plant Cell. 2003;15:732–744. doi: 10.1105/tpc.009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiderski MR, Birker D, Jones JD. The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol Plant Microbe Interact. 2009;22:157–165. doi: 10.1094/MPMI-22-2-0157. [DOI] [PubMed] [Google Scholar]
- Tapping RI. Innate immune sensing and activation of cell surface Toll-like receptors. Semin Immunol. 2009;21:175–184. doi: 10.1016/j.smim.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Tuncbag N, Keskin O, Gursoy A. HotPoint: hot spot prediction server for protein interfaces. Nucleic Acids Res Suppl. 2010;38:W402–W406. doi: 10.1093/nar/gkq323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Biezen EA, Jones JD. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci. 1998;23:454–456. doi: 10.1016/s0968-0004(98)01311-5. [DOI] [PubMed] [Google Scholar]
- van Ooijen G, Mayr G, Kasiem MM, Albrecht M, Cornelissen BJ, Takken FL. Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J Exp Bot. 2008;59:1383–1397. doi: 10.1093/jxb/ern045. [DOI] [PubMed] [Google Scholar]
- Ve T, Williams S, Valkov E, Ellis JG, Dodds PN, Kobe B. Crystallization, X-ray diffraction analysis, and preliminary structure determination of the TIR domain from the flax resistance protein L6. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:237–240. doi: 10.1107/S1744309110051006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CI, Guncar G, Forwood JK, Teh T, Catanzariti AM, Lawrence GJ, Loughlin FE, Mackay JP, Schirra HJ, Anderson PA, et al. Crystal structures of flax rust avirulence proteins AvrL567-A and -D reveal details of the structural basis for flax disease resistance specificity. Plant Cell. 2007;19:2898–2912. doi: 10.1105/tpc.107.053611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LM, Swiderski MR, Li Y, Jones JD. The Arabidopsis thaliana TIR-NB-LRR R-protein, RPP1A; protein localization and constitutive activation of defence by truncated alleles in tobacco and Arabidopsis. Plant J. 2006;47:829–840. doi: 10.1111/j.1365-313X.2006.02834.x. [DOI] [PubMed] [Google Scholar]
- Wen J, Arakawa T, Philo JS. Size-exclusion chromatography with on-line light-scattering, absorbance, and refractive index detectors for studying proteins and their interactions. Anal Biochem. 1996;240:155–166. doi: 10.1006/abio.1996.0345. [DOI] [PubMed] [Google Scholar]
- Whitham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr C, Baker B. The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell. 1994;78:1101–1115. doi: 10.1016/0092-8674(94)90283-6. [DOI] [PubMed] [Google Scholar]
- Xu Y, Tao X, Shen B, Horng T, Medzhitov R, Manley JL, Tong L. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 2000;408:111–115. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- Yu X, Acehan D, Menetret JF, Booth CR, Ludtke SJ, Riedl SJ, Shi Y, Wang X, Akey CW. A structure of the human apoptosome at 12.8 A resolution provides insights into this cell death platform. Structure. 2005;13:1725–1735. doi: 10.1016/j.str.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Yuan S, Yu X, Topf M, Ludtke SJ, Wang X, Akey CW. Structure of an apoptosome-procaspase-9 CARD complex. Structure. 2010;18:571–583. doi: 10.1016/j.str.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.