Abstract
Despite advances in diagnosis and treatment, bacterial sepsis remains a major cause of pediatric morbidity and mortality, particularly among neonates, the critically ill, and the growing immunocompromised patient population. Sepsis is the endpoint of a complex and dynamic series of events in which both host and microbial factors drive the high morbidity and potentially lethal physiological alterations. This article provides a succinct overview of the events that lead to pediatric bloodstream infections (BSI) and sepsis, with a focus on the molecular mechanisms employed by bacteria to subvert host barriers and local immunity to gain access to and persist within the systemic circulation. In the events preceding and during BSI and sepsis, gram-positive and gram-negative pathogens employ a battery of factors for translocation, inhibition of immunity, molecular mimicry, intracellular survival, and nutrient scavenging. Gaps in understanding the molecular pathogenesis of bacterial BSI and sepsis are highlighted as opportunities to identify and develop new therapeutics.
Keywords: bacteremia, bloodstream infections, bacterial sepsis
Introduction
Sepsis is one of the most devastating problems in medicine, stemming from infections by bacteria, viruses, fungi, and parasites, or their toxic products. At the foundation of sepsis is microbial invasion of the bloodstream (bloodstream infection, BSI) or the release of microbial products into the bloodstream, which results in a spectrum of disease from mild systemic symptoms and circulatory compromise including fever, tachycardia, tachypnea, and peripheral vasodilatation to massive circulatory collapse through which multi-organ failure arises and often results in death. Bacterial sepsis is often, but not exclusively, defined by clinical manifestations accompanied by positive isolation of an organism in the circulatory system; however, sepsis can also occur in the absence of organism isolation, in which case, the host is most likely responding to bacterial components (lipoproteins, endotoxins, etc.) in the circulatory system. On a molecular level, a massive deregulated cytokine storm and incoordination of endocrine and hemostatic processes also accompany the progression to sepsis, resulting in one form of severe inflammatory response syndrome (SIRS).1 In addition to multiorgan failure and death, sepsis can lead to severe complications including endocarditis, osteomyelitis, and meningitis. Despite advances in life support and treatment, the associated mortality remains high, as exemplified by neonatal sepsis where the mortality rate is approximately 10–14%.2, 3
As noted earlier, bacterial sepsis is the endpoint of a series of events, most often precipitated by invasion of bacteria into the bloodstream. Thus, understanding the events that lead to bacterial invasion underpins the development of BSI and sepsis, and this will be the focus of this article. The risk for developing BSI and sepsis is dependent on biological age, immunological maturity, the physiological state of the child, and the location of the child (hospital ward, intensive care, community). BSI may arise through multiple routes, including translocation across mucosal-epithelial borders as in the oro-naso-pharynx, gut, lung, or kidney; physical breaks in barriers such as the skin; ascending infection through the urogenital tract of females resulting in dissemination from the fallopian tubes; and through direct access to the bloodstream via indwelling catheters or devices. Clinically, BSI is often categorized in 2 primary ways: 1) by origin of the infection (examples include gut or venous catheter) and 2) by pathogen. In fact, these classifications are highly interrelated, such that origin of infection often portends the etiology. The leading etiologies of BSI also change with the biological age of the child in the context of anatomic and immune alterations. The epidemiology of pediatric BSI and sepsis will now be reviewed to highlight the major etiologies of pediatric BSI.4, 5 which will serve as examples to discuss the molecular mechanisms bacteria employ to gain access to and subsequently persist within the systemic circulation. In many cases, these mechanisms then facilitate the development of sepsis.
Epidemiology
The first several months of life are a high-risk period for BSI and sepsis. Neonates are at particularly high risk of developing BSI as the biochemical and mechanical properties of their mucosal surfaces and immunity (both cellular and humoral immunity, including macrophages, neutrophils, immunoglobulin, and complement) are not fully mature at birth in either function or quantity.6, 7 Sepsis in the neonatal period and shortly beyond is classically divided into three windows of time, reflecting epidemiological shifts in the causative infectious agents. The classifications are: 1) early onset sepsis (EOS), occurring from birth through 6 days of life; 2) late onset sepsis (LOS) from 7–30 days of life; and, as sometimes distinguished from LOS, 3) late, late onset sepsis (LLOS) that occurs at >30 days of life but prior to 3 months. Although these classifications relate to sepsis, most published epidemiological data sets detail infant BSI that may or may not have associated sepsis.
The most common etiologies of EOS are Escherichia coli and group B streptococcus (GBS) at approximately 23% and 47% of cases, respectively.4 Staphylococcus aureus, coagulase negative staphylococci, and Haemophilus influenzae (non-typable, principally Biotypes II–IV), and Enterococcus sp. make up most of the remaining cases in term and preterm infants, representing 13%, 5%, 4.5%, and 4.5% of cases, respectively.4 The overall rates of EOS have declined over the past several decades, most likely related to a decline in EOS attributable to GBS after implementation of widespread maternal screening and prophylactic treatment.8 However, the incidence of EOS among premature and very low birth weight (VLBW) infants has increased (94.9 per 1000 cases vs. 265.94 per 1000 cases, respectively), with a parallel increase in EOS due to gram-negative bacteria, principally E. coli.9 EOS pathogens are primarily acquired from vaginal passage during birth and; thus, the risk of EOS reflects the prevalence of the organism among the mother’s urogenital tract flora as well as specialized virulence of the organism for children in the neonatal period.
After the first week of life, the epidemiology of bacteremia and late-onset sepsis shifts toward organisms now acquired from the environment or caregiver. In LOS, coagulase-negative staphylococci have emerged as major pathogens, composing 39% of cases.4 E. coli, S. aureus, Enterococcus sp. and GBS remain important pathogens during the neonatal period. However during this period, Klebsiella sp., Enterobacter sp., Pseudomonas aeruginosa, and Serratia marcescens emerge as etiologies of LOS that are rare pathogens prior to 7 days of life. This group of pathogens is particularly important among premature infants. In their recent review of neonatal sepsis cases at Yale New Haven Hospital, Bizzarro et al. reported an increased incidence of LOS in both term and premature infants, which doubled during the period from 1997–2006 when compared to 1979–1992.9 Of particular note is that, during the same time period, rates of LOS caused by E. coli increased significantly among neonates in general (4.07 per 100 cases vs. 8.23 per 1000 cases, respectively) and with respect to VLBW infants (10.39 per 1000 vs. 21.66 per 1000 cases, respectively).9 The distribution of pathogens causing LLOS remains similar to that of LOS, and prematurity is highly associated with the risk of LLOS.
The rate of BSI drops dramatically after the first several months of life in otherwise healthy children. However, when previously healthy children develop BSI, the leading etiologies are S. pneumoniae, Neisseria meningitidis, and S. aureus. Medical advances in oncology, stem cell and solid organ transplantation, along with improved survival of children with complex diseases, has led to new cohorts of children at risk of BSI. These children may be at significant risk of BSI due to compromises in mucosal barriers, innate or acquired immunity associated with underlying anatomic defects, primary immunodeficiency, acquired immunodeficiency, or nosocomial immunodeficiency. Among children with these significant underlying defects, a diverse group of virulent but opportunistic pathogens becomes relevant, including P. aeruginosa and Staphylococcus epidermidis, along with an extensive list beyond the scope of this review.
The epidemiology of BSI among children is continually changing based on alterations in medical practices such as antibiotic use and vaccination, regional variations, and emerging organisms. The application of molecular diagnostics has and will continue to significantly alter the epidemiology of BSI and sepsis, especially culture-negative sepsis. For instance, recent molecular-based epidemiological surveys suggest that Kingella kingae is an under recognized pathogen involved in BSI of infants and toddlers and, ultimately, BSI-sequelae including septic arthritis.10, 11 Similarly, Ureaplasma species urealyticum and parvum may emerge as an important and previously unappreciated agent in culture-negative BSI and meningitis of neonates.12 Recent studies comparing PCR as a molecular diagnostic for neonatal sepsis with conventional blood culture revealed bacteria in anywhere from 5 to 35 % of newborns diagnosed with clinical sepsis, indicating that sepsis arises from a variety of causative insults, with a large proportion arising from infection.13, 14 The pathophysiology of culture-negative sepsis is indistinguishable from culture-proven sepsis, but, in many cases, occult infection is still suspected as the primary initiator; although, additional etiologies, like neoplasm, multi-organ trauma, and pancreatitis, can contribute to the same systemic inflammatory response that characterizes sepsis. Many of the same innate immune pathways (eg., pattern recognition receptors, PRR) that are activated upon recognition of bacterial products (explained in detail later) can also be activated by endogenous mediators of inflammation (eg., High Mobility Group Box 1 protein, HMGB1)15 due to trauma, injury, or cellular idiopathy. Epidemiological studies, including the use of cutting-edge molecular diagnostics, remain central to understanding trends in pediatric BSI and sepsis and should improve outcomes through earlier diagnosis and initiation of therapy.
The remainder of this review will focus on how prevalent agents of pediatric bacteremia and sepsis undergo key steps of pathogenesis, including the transition from colonization to mucosal translocation, subversion of the host innate immune system, and dysregulation of host inflammation. The pathophysiology of sepsis is complex, but ultimately results from the host response, mediated by cytokine and chemokine release, to circulating bacteria or their antigenic products. While this review will focus on the bacterial factors that drive successful systemic infection, a companion article reviews current knowledge about the host response to bacteremia and the development of sepsis.
Colonization
While some episodes of BSI and sepsis occur after direct introduction of bacteria into the bloodstream, most arise from bacteria colonizing biotic (living tissues) or abiotic (hardware, artificial materials) surfaces. Many bacteria producing BSI are commensals of the mucosal surfaces of children, and BSI is the consequence of bacteria breaching the mucosal-epithelial barrier. Mucous membranes cover the respiratory, gastrointestinal, and urogenital tracts, maintaining complex microbial communities while providing effective mechanical and chemical barriers to bacterial translocation. Thus, pathogens must persist at the mucosal surface despite host inhibition and competition with other microbes. One of the most important steps in colonization is adherence of bacteria to mucosal surfaces, which is usually accomplished through expression of receptor-specific and non-specific adhesive factors. Gram-positive and gram-negative bacteria decorate their surfaces with a wide array of adhesive factors, ranging from filamentous, multimeric surface fibers such as the prototypical type 1 pili of E. coli16 to the high molecular weight autotransporter proteins exemplified in H. influenzae (HMW1/HMW2, Hia/Hsf, Hap),17 and the “anchorless” adhesive factors of gram-positive organisms such as PavA of S. pneumoniae.18 Adherence is a unifying theme for successful colonization and persistence. For example, without the RlrA pili of S. pneumoniae, bacteria are effectively outcompeted by other endogenous flora and displaced at the mucosal surface of the nasopharynx.19 Colonization is a complex process, and some organisms require multiple adherence factors for successful colonization. For instance, S. aureus requires at least four surface factors to adhere to and colonize the desquamated epithelial barrier of the nasal passage,20 illustrating that S. aureus nasal colonization is a multifactorial process involving multiple ligands that differentially affect colonization and persistence.21
Commensal-pathogens must also compete with endogenous flora and amongst themselves. For example, S. pneumoniae produces NanA, which strips sialic acid from the LPS of H. influenzae and N. meningitides, thus decreasing their biofilm-forming capacity, exposing these bacteria to host recognition, and ultimately targeting them for elimination.22, 23 Conversely, H. influenzae may promote localized inflammation, recruiting neutrophils that more selectively eliminate S. pneumoniae.24 Antibiotics may shift the balance in this competition25 and eliminate organisms that are normally responsible for keeping potential pathogens in check. Among many exciting opportunities, the new roadmap initiative of the NIH to define the human microbiome may reveal patterns of microbial communities that preclude or limit pathogen colonization. This ongoing research has the potential to reveal novel probiotic approaches to therapy that effectively eliminate potential pathogens prior to the onset of invasive disease.
Not only must pathogens adhere to the mucosal surface and compete with endogenous microbial communities, but they must also survive the potent mucosal immune system. The mucosal membranes have specific mucosal-associated lymphoid tissue (MALT) that protects mucous membranes from assault by pathogens while distinguishing between colonization by commensal organisms. Thus, bacteria employ an array of mechanisms to avoid being eradicated by the mucosal-associated local innate immune system. For example, diverse BSI-associated organisms, including nasopharyngeal N. meningitidis, non-typeable H. influenzae, S. pneumoniae, and urogenital U. urealyticum, produce IgA proteases to avoid neutralizing antibodies (detailed in the excellent review by Mistry and Stockley, 2006).26–28 Similarly, complement inhibition is critical for the persistence of pneumococci in the nasopharynx.24, 29 A recently recognized family of multi-functional histidine triad (Pht) proteins may support initial mucosal colonization, cleave complement C3, and promote translocation from sites of colonization.30 Capsules, such as the K1 capsule of neonatal meningitis isolates of E. coli, exclude complement from the bacterial surface.31, 32 Some of these mechanisms are not exclusive to colonization, but are also instituted by invading bacteria at later steps of BSI and can be important for the pathogenesis of sepsis.
Translocation of Epithelial-Mucosal Barriers
In the absence of catheters and medical devices that serve as conduits for bacteria to directly access the bloodstream, the mucosa serves as the major source from which BSI/sepsis arise. Because the gastrointestinal tract collectively harbors the greatest number and density of bacteria in the human body (part of the enteric microbiome), it is a major reservoir for BSI organisms and serves to illustrate numerous mechanisms of bacterial translocation. To some extent, bacterial translocation from the gut may occur regularly even in healthy individuals, but bacteremia is generally limited by an intact immune system.33–35 However, immune immaturity, anatomic insufficiency of the mucosal barrier, or alterations of the microbial ecology may increase the frequency of translocation events.
Several bacterial mechanisms facilitate translocation across the gut mucosa. First, bacteria may bypass the enterocyte border either by invading through a cell or past the junctional complexes between cells. The autoaggregative protein Hek from BSI- and neonatal meningitis-causing E. coli is an example of a factor promoting adherence to and invasion into cultured colonic epithelial cells by binding to glycosaminoglycans such as heparin, a key constituent of mucous; however, a definitive role for Hek in the transcytosis of E. coli across the intestine epithelial barrier has yet to be demonstrated.36, 37 Bacteria may also produce a localized breech in the mucosa. For instance, lipopolysaccharide (LPS; also called endotoxin) of gram-negative bacteria produces localized dysfunction of the gut barrier that, in turn, facilitates bacterial translocation. In recent work, LPS challenge was shown to reduce bile flow rate, increase mucosal permeability, and subsequently increase bacterial translocation, each of which was dependent on the expression of the inflammatory cytokine high-mobility group box 1 (HMGB1)38 and the subsequent release of mast cell proteases into the colonic lumen.39
Further knowledge about bacterial translocation comes from molecular studies of respiratory organisms at the nasopulmonary-mucosal interface. Organisms such as S. pneumoniae frequently colonize the nasopharynx or pharynx, but only infrequently subvert the mucosal barrier and produce invasive disease, highlighting that it is a combination of stochastic events, bacterial virulence, and host immunity that ultimately leads to translocation and the establishment of invasive disease. However, specialized virulence and co-infections increase the likelihood of translocation. Many of the known bacterial virulence determinants among respiratory pathogens produce localized epithelial damage. For instance, a common mechanism is secretion of toxins that produce local epithelial damage and lead to local tissue and bloodstream invasions. For example, K. kingeae has recently been shown to produce an RTX-like toxin that damages the respiratory epithelium and may provide increased access to the local capillaries.40 In another example, P. aeruginosa employs the needle-like injection system of type III secretion (TTSS) to directly instill potent effector molecules into target host cells and cause acute epithelial damage and cell death.41 A similar outcome may arise due to co-infection with a respiratory virus such as influenza, resulting in cytopathic effects on the respiratory epithelium that facilitates invasion and dissemination of nasopharyngeal bacteria such as S. pneumoniae or H. influenzae.42, 43
In addition to the enteric and respiratory tracts, the urinary tract in children serves as a common portal for bacterial translocation into the circulatory system, leading to sepsis (urosepsis). In general, uropathogens are presumed to enter the bloodstream by translocating across the renal tubular epithelium and local capillary endothelium, which are in close proximity. The molecular mechanisms employed by bacteria that produce urosepsis remain poorly understood; however, recent studies using real-time in vivo microscopy have suggested that E. coli, by far the leading uropathogen, produce a hemolysin toxin that cause local renal epithelial damage and may facilitate the translocation process.44 Rapid, local renal ischemia was shown to be an essential host response necessary to block translocation and prevent lethal urosepsis.45
Bacterial Pathogen Interactions with Innate Immunity: A Balancing Act
Four general mechanisms with varying degrees of overlap are frequently employed by bacteria to avoid detection by the innate immune system: 1) subversion of detection and modification of inflammation, 2) inhibition of phagocytosis, 3) resistance to intracellular killing, and 4) resistance to or escape from innate effectors. Avoiding detection by the host immune system may provide a window of opportunity for the pathogen that makes the difference between successful infection and clearance.
Subverting detection and modifying inflammation
The development of BSI and the transition from bacteremia to sepsis involves a combination of bacterial and host factors. Recognition of pathogen-associated molecular patterns, or PAMPs, by the Toll-like receptors (TLRs) leads to induction of inflammation, phagocytosis, and bactericidal action (Figure 1, panel B). The host recognition events and inflammatory responses are only touched on briefly in this review as the topics are covered in much greater detail in a companion article. Certain bacterial factors are known to trigger the physiology of sepsis, such as gram-negative LPS, which stimulates a robust and complex inflammatory response through host Toll-like receptor 4 (TLR4) and other independent targets.46, 47 Despite its ubiquity, LPS has subtle structural distinctions among different gram-negative species, notably in the lipid A moiety, that produce different effects on TLR 4 signaling, providing one explanation for the disproportionate host response to certain pathogens.47 The peptidoglycan and lipoproteins of gram-positive organisms also promote inflammation through Toll-like receptor pattern recognition (TLR2, TLR1/6, TLR2/6).48 The major molecules of bacterial pathogens recognized by the innate immune system include LPS, peptidoglycan, lipoproteins, flagellin, and CpG DNA (Figure 1, panel A). Recognition of bacteria through innate immune receptors is critical to early detection of potential pathogens; however, these pathways, when left unchecked, may result in a cascade of physiological effectors that produce an overwhelming inflammatory response.
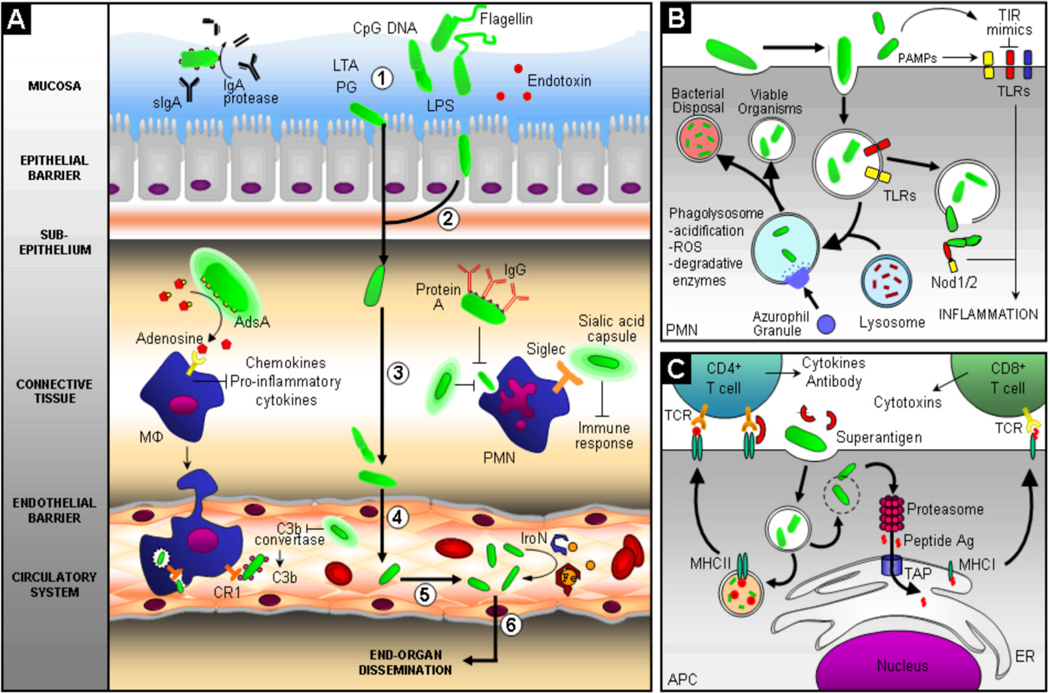
Figure 1. Bacterial-host interactions during the progression of BSI and sepsis.
Panel (A) depicts the pathogenic steps that bacteria must undergo to gain access to and persist within the circulatory system, with pathogen interaction with the host innate and adaptive immune system highlighted. Bacteria often initiate the infection process through colonization of the mucosal surface of the intestine, respiratory tract, or urogenital tract (1), after which, bacteria can traverse the epithelial barrier transcellularly, paracellularly, or as intracellular passengers within granulocytes and mononuclear cells (2). Bacteria must then survive in connective tissues (3), cross the endothelial-blood barrier (4), and survive and persist in the circulatory system (5). Bacterial pathogens have evolved multiple mechanisms to avoid clearance by professional phagocytes, like neutrophils and macrophages, by synthesizing the immunosuppressant nucleoside adenosine, assembling sialic acid or haluronic acid capsules to inhibit phagocytosis and dampen the immune response, limiting deposition of complement factor C3b, and coating themselves with incorrectly-oriented IgG antibody to limit opsonophagocytosis. Bacteria have evolved mechanisms to survive within the systemic circulation by scavaging metals, eg. iron, with high-affinity molecules called siderophores. Finally, bacterial pathogens can exit the circulatory system, causing end-organ dissemination and/or failure (6). Panel (B) illustrates host mechanisms employed to contain infection, including phagocytosis and innate pathogen-associated molecular pattern (PAMP) recognition through the Toll-like receptors (TLR) and intracellular nucleotide oligomerization domain/caspase recruitment domain (NOD/CARD) proteins, such as Nod1/2. Phagocytosis and lysosomal fusion are typically part of a unified pathway to clear pathogens, but bacteria have evolved countermeasures to avoid being killed. Panel (C) demonstrates antigen-dependent (MHC presentation) and –independent (superantigen) activation of the adaptive immune system by phagosome-contained and cytoplasmic bacteria. Successful activation of the adaptive immune branch is a key step in bacterial clearance; however, non-antigen-dependent activation of T helper cells via bacterial superantigens elaborates a destructive cascade of cytokine release and non-specific T cell proliferation. Antigens from intracellular pathogens are processed through the proteasome into peptides, transported into the endoplamic reticulum (ER), captured by MHCI molecules, and presented to CD8+ T cells that elicit a barrage of cytotoxic effectors. Alternatively, antigens from extracellular pathogens are captured by MHCII molecules after digestion in the phagolysosome and presented to CD4+ that elicit a burst of cytokines that activate B cells to produce an antibody response. Abbreviations and notations; bold arrow ( ) indicates steps during the progression of BSI and sepsis; unbolded arrow (
) indicates steps during the progression of BSI and sepsis; unbolded arrow ( ) indicates activation of pathway; (
) indicates activation of pathway; ( ) indicates inhibition of pathway; curved unbolded arrow (
) indicates inhibition of pathway; curved unbolded arrow ( ) indicates production; MΦ, macrophage; PMN, polymorphonuclear neutrophil; LTA, lipotechoic acid; PG, petidoglycan; LPS, lipopolysaccharide; IgG, Immunoglobulin G; IgA, Immunoglobulin A; sIGA, secretory IgA; CR1, Complement Receptor 1; Fe, iron; IroN, siderophore; TIR, TLR/Interleukin-1 receptor; Nod1/2, cytoplasmic PAMP receptors; ROS, reactive oxygen species; TCR, T cell receptor; TAP, transporter associated with antigen processing; ER, endoplasmic reticulum; APC, antigen presenting cell; MHCI/II, major histocompatability complex I/II; Ag, antigen; CD4+, T cell co-receptor distinguishing T helper cells; CD8+ T cell co-receptor distinguishing cytotoxic T cells.
) indicates production; MΦ, macrophage; PMN, polymorphonuclear neutrophil; LTA, lipotechoic acid; PG, petidoglycan; LPS, lipopolysaccharide; IgG, Immunoglobulin G; IgA, Immunoglobulin A; sIGA, secretory IgA; CR1, Complement Receptor 1; Fe, iron; IroN, siderophore; TIR, TLR/Interleukin-1 receptor; Nod1/2, cytoplasmic PAMP receptors; ROS, reactive oxygen species; TCR, T cell receptor; TAP, transporter associated with antigen processing; ER, endoplasmic reticulum; APC, antigen presenting cell; MHCI/II, major histocompatability complex I/II; Ag, antigen; CD4+, T cell co-receptor distinguishing T helper cells; CD8+ T cell co-receptor distinguishing cytotoxic T cells.
In response to the host sentinel system, bacteria have acquired strategies for subversion of host innate immunity. For example, recent work has shown that urinary tract isolates of E. coli secrete inhibitory mimics of the TLR/Interleukin-1 receptor (TIR) domain, called Tcps, which directly interfere with the recognition of pathogens and downstream TLR signaling, and, in the case of E. coli, lead to more severe pyelonephritis and renal abscess formation, which may portend increased risk for urosepsis (Figure 1, panel B).49 TIR mimicry is not exclusive to E. coli. Salmonella enterica also employs this countermeasure by secreting a TIR-like protein called TlpA that impairs TLR signaling and induces host cell apoptosis.50 Although non-typhoidal salmonellae (NTS) are a relatively uncommon cause of bacteremia and sepsis in developed countries, NTS (particularly S. enterica serovar Typhimurium and serovar Enteritidis) is emerging as a common cause of bacteremia in tropical Africa, particularly among children aged 6 months to 3 years.51, 52 Genomic analyses have revealed a variety of bacterial pathogens may produce TIR mimics, suggesting this may be a broadly conserved mechanism of immune evasion. However, as sepsis is a syndrome primarily characterized by an overwhelming host immune response directed at invading bacteria or bacterial products in the systemic circulation, clearly, bacteria are not always successful in evading the sentinel innate immune system.
Recently, a novel mechanism for evading the immune response in the bloodstream was identified for S. aureus. In particular, S. aureus is able to escape phagocytic clearance in the blood by expressing the cell wall-anchored enzyme AdsA, which converts adenosine monophosphate to adenosine (Figure 1, panel A).53 Adenosine is a potent immunosuppressant that engages receptors on the surface of leukocytes that inhibit proinflammatory responses.54–56 The ability of S. aureus to escape phagocytic clearance and form organ abscesses were dependent on synthesis of adenosine by AdsA and a genetic deletion could be rescued by an exogenous supply of adenosine.53 Other gram-positive pathogenic bacteria that cause pediatric sepsis like E. faecalis, S. epidermidis, and S. pyogenes also have homologues of AdsA; however, an adenosine-dependent regulatory system with an AdsA homologue has not yet been identified in gram-negative pathogens, suggesting that alteration of adenosine levels may be an exclusive gram-positive pathogenic paradigm. Interestingly, recent work has focused on taking advantage of the immune suppressive effects of the adenosine pathway as a therapeutic for sepsis. Treatment with an adenosine receptor 2A agonist was able to improve the outcome of gram-positive, gram-negative, and LPS-induced septic shock experimentally in mice.57 Adenosine production may be a conserved, yet previously unrecognized, immunomodulatory factor for gram-positive invasive bacterial disease, but may actually reduce the induction of host inflammatory-mediated sepsis syndrome. Further investigation of the adenosine pathway including a focus on receptor distribution, activation, and signaling in the pathophysiology and treatment of gram-positive and gram-negative sepsis is warranted.
Inhibition of phagocytosis
Despite mechanisms to dampen and evade immune recognition, bacteria often come to encounter professional phagocytes such as neutrophils and macrophages. Thus, bacterial pathogens have evolved mechanisms to inhibit and avoid phagocytosis. The polysaccharide capsules of S. pneumoniae, H. influenzae, N. meningitidis, and E. coli, among many, are known inhibitors of phagocytosis.58 Another clever strategy involves Protein A of S. aureus, which binds to the Fc portion of IgG, coating the bacteria in incorrectly-oriented immunoglobulins that cannot be recognized and bound by Fc receptors on phagocytes (both examples illustrated in Figure 1, panel A).59, 60
Recent research has focused on a family of sialic-acid-binding Ig-like lectins (Siglecs) differentially expressed on hematopoetic and immune cells that regulate the function of cells in the innate and adaptive immune systems. Siglecs bind host cell sialic acid and relay a signal that represses proinflammatory activation signals. It has been shown that the sialic acid capsule of different GBS serotypes can bind multiple human Siglecs, which then suppresses phagocytic cell function.61, 62 More than 20 pathogens have been shown to decorate their surfaces with sialic acid, including N. meningidis, H. influenzae, and E. coli. Future studies will reveal if these sialic acid glycosylations act as ligands for Siglecs, thus demonstrating a conserved mechanism employed by both gram-negative and gram-positive bacterial pathogens to evade the host immune system.63
Intracellular survival
Once bacteria are taken up through the process of phagocytosis, the host cell begins a cascade of events that ultimately leads to the fusion of the bacteria-containing phagosome with a lysosome, resulting in what is called a phagolysosome. In the phagolysosome, the invading bacteria are exposed to a battery of antimicrobial factors, including acidification of the compartment, oxidative stress, and antimicrobial peptides (Figure 1, panel B). Some bacterial pathogens have developed countermeasures to ensure intracellular survival, including 1) inhibition of phagolysosome fusion or maturation, 2) resistance to acidification and oxidative burst, and 3) escape from the compartment. Although, there are several classical examples of pathogenic bacteria that exploit these mechanisms for intracellular survival, several representative examples specifically relevant to pediatric BSI are highlighted in the text that follows.
S. pyogenes is one of many organisms producing BSI in children that has adapted to survival and replication in the phagolysosome of granulocytes. Within the neutrophil phagolysosome, S. pyogenes modulates membrane traffic and inhibits fusion of microbicidal granules with the phagosome.64, 65 Recent work has shown that S. pyogenes M or M-like proteins are capable of selectively inhibiting azurophilic granules, which contain reactive oxygen species, with the phagosome. Another mechanism of intracellular survival common to many gram-positive and gram-negative pathogens is resistance to intracellular killing by antimicrobial peptides in the phagolysosome. For instance, S. aureus has several families of proteins (Dlt proteins and MprF) that modify surface lipoproteins with the positively charged amino acids D-alanine or L-lysine, respectively, which partially neutralize the negative charge of the cell surface that attracts cationic antimicrobial peptides.66–68
Some pathogens can also escape from the phagolysosome into the cytosol. Listeria monocytogenes, a rare, but serious, gram-positive pathogen causing BSI and meningitis in infants and the immunocompromised, exerts well defined steps to disrupt the endocytic vesicle, including the expression of Lyseriolysin O that perforates the vesicle membrane. K1 encapsulated E. coli, a leading agent of invasive disease in neonates, is able to actively replicate in the phagolysosome of macrophages and ultimately escape into the cytosol.69 The consequences of intracellular survival are several-fold. Survival of bacteria in phagocytes not only limits clearance of localized infections, in the case of E. coli, it provides a specialized “taxi service” for trafficking through the bloodstream and lymphatics, resulting in rapid, wide-spread dissemination.
Inhibition of innate effectors
Bacterial pathogens must evade non-cellular, antimicrobial small molecules (referred to here generically as innate effectors), such as complement components and antimicrobial peptides. In some cases, evasion of innate effectors entails inactivating complement components. For instance, as part of the response to being in the bloodstream, S. pyogenes expresses the complement inhibiting protein, C5a peptidase.70 The C5a peptidase of S. pyogenes cleaves and thereby inactivates C5a, a potent proinflammatory molecule that recruits phagocytic cells. S. aureus also interferes with C5a activity by producing a chemotaxis inhibitory protein (CHIPS) that binds the C5a receptor, blocking C5a binding to neutrophils, resulting in inhibition of cellular migration towards S. aureus.71 Consistent with the importance of C5a in vivo was the recent demonstration that C5a-deficient animals rapidly succumb to S. aureus bacteremia.72
Bacteria also restrict access of complement and antimicrobial peptides by way of polysaccharide capsules. The complement component C3b is deposited on the surface of the bacteria, which leads to Complement Receptor 1 (CR1) recognition of C3b-opsonized bacteria, initiating engulfment by macrophages (Figure 1, panel A). However, the GBS capsule restricts deposition of the complement factor C3b on the surface of the bacterium, and this limited opsonization, in turn, attenuates recognition by macrophages.73 In another example, the negatively charged capsules of K. pneumoniae and P. aeruginosa have been demonstrated to act as a biochemical sponge or “sink” to absorb potentially toxic cationic antibacterial peptides produced by the host.74 These bacterial “shields” against innate immune effectors allow invasive bacteria to disseminate and replicate with limited impediments.
Adaptive Immunity
Adaptive immunity is generally separated into antibody-mediated (ABI) and cell-mediated immunity (CMI), which directs host immune responses primarily against extracellular versus intracellular pathogens, respectively. Many pathogens that cause BSI and sepsis have both extracellular and intracellular lifecycles. Thus, the coordination of these two branches of the adaptive immune system is necessary for successful clearance of these pathogens. Several classical mechanisms of avoiding the adaptive branch of the immune response, such as antigenic disguise, antigenic variation (eg., phase variation), and superantigen T-cell proliferation are discussed below, as are new insights into bacterial inhibition of the transition between the innate and adaptive responses.
Antigenic Disguise: A Game of Hide and Seek
Some pathogens have evolved clever ways to hide surface antigens from antibody-mediated immunity using a shroud of host proteins such as fibrin, fibronectin, or immunoglobulin molecules. Surface-localized Proteins A & G produced by S. aureus and S. pyogenes, respectively, bind the Fc portion of circulating IgG, thereby, coating the bacteria in incorrectly-oriented antibody and blocking their opsonizing capacity.59, 60 S. aureus also releases soluble forms of Protein A that can agglutinate circulating IgG, thereby, neutralizing the activity of immunoglobulins.
S. aureus is also able to produce cell-bound coagulase and clumping factors (ClfA and ClfB) that cause host fibrin to clot and precipitate on the bacterial surface, effectively interfering with neutrophil and macrophage phagocytosis.75–77 This is an example of bacteria co-opting host systems to disguise themselves from immune recognition. Some pathogens, however, endogenously produce antigens that resemble host antigens, referred to as molecular mimicry. For instance, the sialic capsules of E. coli and GBS and the hyaluronic acid capsule of group A Streptococcus (GAS) provide an antigenic disguise the “mimics” naturally occurring host molecules. The antigenic determinants of the bacteria and host are essentially indistinguishable; thus, immune tolerance to “self” extinguishes the potential immunological responses to these bacterial antigens.
Antigenic Variation: Why Phase Matters
One way bacteria can escape detection and avoid potent neutralizing antibodies is to periodically change major antigens (antigenic variation) or turn off expression of a highly antigenic factor (phase variation). Antigenic variation may occur in the host during the course of infection, in which case a particular antigen actually “changes” over the course of infection, or the organism can exist in nature as multiple, fixed antigenic types (eg., serovars). Some types of antigenic variation result from site-specific inversions or gene rearrangements in the DNA of the microorganism. One of the classic and well-studied examples of antigenic variation is the outer membrane proteins (OMPs) of N. meningiditis. The antigenic variation of serogroup A meningococcal OMPs arise due to horizontal genetic exchange between species (reviewed in Achtman, 1995).78 For instance, opacity (Opa) family proteins are antigenically-variable Neisseria OMPs that can undergo genetic recombination, changing the inventory of Opa proteins that can be expressed. Even among clonally-related, epidemic meningococcal isolates, an ongoing process of recombination and genetic exchange contributes to significant variability in surface antigens, which can hide the pathogenic bacteria from the host immune system.79
An alternative approach for pathogenic bacteria is to exist in nature as multiple different serotypes as to avoid a previously-activated adaptive response to a different serovar. Many pathogenic bacteria, including E. coli, S. aureus, and S. pyogenes exist in multiple serotypes based on the complement of surface proteins. For instance, S. pyogenes has 80 different antigenic types based on the surface complement of M proteins, while there are over one hundred serotypes of S. pneumoniae based on variants of the capsular polysaccharide antigens.
Superantigens: Why sometimes it is better to overstimulate
The superantigens (SAgs) of staphylococci and streptococci (particularly group A Streptococcus) have been implicated in the severe systemic manifestations and pathogenesis of toxic shock and are likely also involved in sepsis syndrome initiated by bloodstream infections (bacterial superantigens and superantigen-like proteins are reviewed in Fraser and Proft, 2008).80 SAgs stimulate a robust and nonproductive host immune response by complexing major histocompatability complex II (MHCII) to a limited number of Vβ regions of CD4+ T helper cells, resulting in excessive inflammatory cytokine production (predominantly tumor necrosis factor alpha, TNFα, and interferon gamma, IFNγ) and robust proliferation of non-specific T cells (Figure 1, panel C). In some patients with toxic shock syndrome, research has shown a selective depletion of certain Vβ subgroups of T cells, consistent with a role for SAgs in an unproductive adaptive immune response.81
Immunosuppression
Bacterial metabolites are also important in regulating adaptive immunity. In addition to supporting enterocyte growth and integrity in the colon, recent work has shown that the short chain fatty acid n-butyrate (a histone deacetylase (HDAC) inhibitor) produced by multiple members of the microbiota in the human gastrointestinal tract alters the maturation of monocyte-derived dendritic cells (DC) by modulating the epigenetic acetylation state of histones, thereby affecting host gene expression.82 HDAC inhibitors have also been shown to decrease the expression of various cytokines in both macrophages and DCs in response to LPS stimulation, which could lead to defects in the initiation of an adaptive immune response.83 For instance, recent work has shown that n-butyrate is able to induce proliferative unresponsiveness, or anergy, in antigen-stimulated murine CD4+ T cells, which demonstrated reduced cytokine secretion upon reactivation.84 The immunosuppressive effect of n-butyrate on both the maturation and cytokine production of DCs may have profound consequences for the activation of the Th1 versus Th2 adaptive immune response through DC-derived co-stimulatory signals and antigen presentation. n-butyrate provides key nutritional support for the mucosal barrier and suppresses inappropriate inflammation; however, when out of balance, the immunoregulatory property of butyrate may allow bacteria to escape the potent mucosal immune system in the gastrointestinal tract and disseminate.
Nutritional Requirements
Once bacteria transit to the circulatory system and/or intracellular environment, certain key nutrients are required for bacterial survival and proliferation. Divalent metals are sequestered by the host and, thus, are generally in limited in supply to invasive bacteria. One mechanism that the host has evolved to combat bacterial proliferation in the blood is to tightly regulate and store iron (Fe), but this homeostasis can be perturbed through dietary overload, poor iron regulation in the premature infant, or hemoglobinopathies (sickle cell disease), leading to an increased risk of bacterial infection (review of neonatal iron homeostasis in Collard, 2009).85
The pathogenesis of human infections caused by S. aureus and E. coli has been previously shown to rely on the acquisition of iron from host hemoproteins. In S. aureus, the iron-regulated surface determinant system Lsd, a heme transport apparatus anchored to the cell wall, is able to mediate utilization of the host hemoproteins hemoglobin and myoglobin as iron sources for bacterial growth.86 In E. coli, the siderophore IroN, which scavenges host iron, has been shown in the neonatal rat model of sepsis to be important for full virulence.87 Iron scavenging from host hemoglobin represents an important virulence strategy for S. aureus, E. coli, Salmonella sp., as well as other bacterial pathogens (Figure 1, panel A).
In addition to iron, the host and bacteria battle to sequester other essential metals. Recent work has shown that several other transitional metals like manganese (Mn2+) and zinc (Zn2+) are also important for bacterial replication and virulence. Work in Salmonella has shown that Mn2+ transport is particularly important for intracellular replication in macrophages88 and that bacteria lacking a functional Zn2+ transporter were attenuated after oral or intraperitoneal infection of mice.89 The role of metal homeostasis in bacterial pathogenesis seems logical given that many bacteria have evolved elegant gene regulatory networks that coordinate metal acquisition with virulence factor regulation. For example, in a zinc-dependent manner, S. pneumoniae regulates the expression of virulence genes to escape complement action during colonization and initial disease.30 S. pneumoniae is also able to regulate virulence genes through a Mn-responsive regulator.90 This is an example where niche-specific nutritional requirements may also serve as signals for virulence networks.
A recently identified manganese efflux system in S. pneumoniae plays a role in both nasal colonization and bloodstream invasion as a strain defective in manganese export was more resistant to oxidative stress in vitro and, yet, was attenuated in a mouse intranasal challenge. Further investigation revealed that S. pneumoniae can differentiate between extracellular and intracellular manganese, activating efflux-dependent regulation of several known and putative virulence factors.91, 92 In addition to their role as essential co-factors in biochemical processes, transitional metals, and their homeostasis, is of potential importance in pathogenesis.
Prospectus
Sepsis is the result of a complex and dynamic series of events in which a combination of host responses, bacterial virulence factors, and stochastic events ultimately drive forward the progression of disseminated infection. Bloodstream infections (BSI) and sepsis represent a continuum of disease in which the host response plays an essential role in determining the series of events leading to sepsis. Through multiple convergent pathways, a wide variety of gram-negative and gram-positive organisms may instigate exuberant physiological responses that typify the host state during sepsis. Many would-be pathogens associated with BSI and subsequent sepsis are commensal organisms that typically respect niche boundaries in the host; however, the balance between commensalism and pathogenesis can be tipped by a combination of factors: generalized loss of protective immunity, physical or immune breaches to host barriers, specialized bacterial virulence, or breakout phenotypically-invasive variants in the bacterial population. The pathogen must walk a fine line between successful infection and host mortality by employing a range of survival tactics that include mimicry of the host, subversion of immunity, and defection to new niches (a representative summary of the mechanisms and exemplary virulence factors are provided in Table 1).
Table 1.
Examples of bacterial virulence factors important during the molecular pathogenesis of BSI and sepsis
| FACTOR | ORGANISM | FUNCTION | REF. |
|---|---|---|---|
| Colonization/Adherence | |||
| RlrA, type IV pili, type I pili | S. pneumoniae, K. kingae, E. coli | Adherence to epithelial cells like nasal, respiratory, and bladder cells, respectively | 19, 93, 16 |
| NanA | S. pneumoniae | Desialidation of LPS of H.influenzae and N. meningitidis, exposing them to host recognition and decreasing their biofilm-forming capacity | 22, 23 |
| Epithelial Damage/Transcytosis | |||
| LPS | gram-negative bacteria | Produces gut-barrier dysfunction mediated by HMGB1 and the release of mast cell proteases causing increased bacterial translocation | 38, 39 |
| ExoU/ExoT | P. aeruginosa | TTSS effectors that cause acute epithelial damage through apoptosis, producing access to deeper tissue | 41, 94 |
| Hek | E. coli | Adherence and invasion into colonic epithelial cells in vitro | 36, 37 |
| Intracellular Survival | |||
| MntH | Salmonella sp. | Manganese transport; Inhibition of oxidative killing resulting in intracellular replication | 88 |
| Protein M | S. pyogenes | Selective inhibition of azurophilic granules with the phagosome | 64, 65 |
| Complement Inhibitors | |||
| Pht/Psp proteins | S. pneumoniae | Mucosal colonization, cleavage of complement protein C3, and promotion of translocation from sites of colonization | 29, 30 |
| C5a peptidase | S. pyogenes | Cleavage of complement protein C5a, a potent chemoattractant molecule | 70 |
| Polysaccharide capsule | GBS, E. coli | Inhibition of C3b deposition, limiting opsonization through the macrophage receptor CR1 | 73 |
| Innate Immunity Effectors | |||
| Protein A | S. aureus | Binding to Fc portion of IgG, masking itself in incorrectly oriented antibody | 59, 60 |
| AdsA | S. aureus, E. faecalis, S. pyogenes, S. epidermiditis | Synthesis of adenosine, which engages receptors on the surface of leukocytes that inhibit proinflammatory responses | 53–56 |
| IgA protease | N. meningitides, U. urealyticum | Cleavage of IgA, avoiding a neutralizing antibody response | 26–28 |
| Tcps, TlpA | UPEC, S. enterica | Inhibitory mimics of the TLR/Interleukin-1 (TIR) receptor domain to impair TLR signaling, inducing host cell apoptosis and downregulate proinflammatory signals | 49, 50 |
| LPS | gram-negative bacteria | Heighten inflammatory response through TLR4, leading to increased BT | 38, 46, 47, 39 |
| Peptidoglycan, lipoproteins | gram-positive bacteria | Proinflammatory signaling through TLR2, inducing host cell apoptosis | 48 |
| Polysaccharide capsule | S. pneumoniae, N. meningitides, E. coli, GBS | Direct inhibition of phagocytosis; block Siglecs (sialic acid capsule), dampen proinflammatory responses and suppress phagocytosis; act as an antimicrobial peptide ’sink’ | 58, 61–63, 74 |
| Adaptive Immunity Effectors | |||
| n-butyrate synthesis | E. coli, | Inhibit the maturation of monocyte-derived DCs and expression of cytokines by macrophages and DCs in response to LPS | 82, 83 |
| CflA/CflB | S. aureus | Antigenic disguise by precipitate and clot host fibrin on the surface of the bacterium | 75, 76, 77 |
| Omp proteins | N. meningiditis | Antigenic variation of outer membrane proteins disguises host antibody targets | 78, 79 |
| SAgs | S. aureus, S. pyogenes | Non-specific proliferation of T helper cells due to cross-linking of MHCII molecule and T cell receptor | 95, 59 |
| Nutrient Acquisition | |||
| IroN | E. coli, Salmonella sp., S. aureus | Scavenge and transport iron from host hemoproteins; necessary for bacterial growth | 86, 87 |
| ZnuABC | Salmonella sp., S. pneumoniae | High affinity zinc transport system; necessary for full virulence in mice; proteins regulates complement inhibiting | 30, 89 |
NOTE.
Abbreviations. HMGB1, inflammatory cytokine high-mobility group box 1; TTSS, type III secretion system; IgG, immunoglobulin G; IgA, immunoglobulin A; UPEC, uropathogenic E. coli; TLR, Toll-like receptor; DC, dendritic cell; CflA/B, clumping factor A/B of S. aureus; SAgs, superantigens; MHCII, major histocompatability complex II; IroN, bacterial siderophore. This table summarizes bacterial virulence factors described during this review, but is not intended to be a comprehensive list of all bacterial factors involved in the pathogenesis of sepsis.
Medical advances in neonatology, transplantation, oncology, and critical care medicine, among other fields, continue to broaden the scope of patients at risk for sepsis; thus, new therapeutics for the prevention and treatment of culture-proven sepsis must accompany these innovations. Infection control, prudent use of medical devices such as catheters, and the responsible administration of antimicrobials, which modify the resident microbiota and alter mucosal barriers, will continue to be mainstays in the prevention and attenuation of sepsis. However, dissecting the regulatory networks and functional components involved in bacterial transitions from colonization to infection, subversion of innate immunity, and persistence, may reveal novel strategies to limit early events leading to the initiation of BSI and, ultimately, sepsis. Advanced genomics, proteomics, metabolomics, and molecular epidemiology may point toward commonalities amongst pathogens, paving the way toward designing precision anti-infectives and broadly neutralizing vaccines. Similarly, knowledge of bacterial community dynamics in competitive niches, such as the colon, and understanding the molecular basis for probiotic effects on community structure will make significant contributions to reducing the risk of BSI and sepsis. Therapeutic strategies arising from these discoveries, merged with novel approaches to abrogate, modulate, or activate specific host pathways engaged early during infection, will address the contemporary and future threat of BSI and sepsis.
Acknowledgements
We would like to thank Tiffany Prest, Carlos Goller, Joseph St. Geme, and Ravi Jhaveri for their helpful suggestions and editing of earlier versions of this manuscript.
Abbreviations
- BSI
bloodstream infection
- SIRS
severe inflammatory response syndrome
Footnotes
Financial Disclosures: None declared
Conflicts of Interest: None declared
References
- 1.Jean-Baptiste E. Cellular mechanisms in sepsis. J Intensive Care Med. 2007;22(2):63–72. doi: 10.1177/0885066606297123. [DOI] [PubMed] [Google Scholar]
- 2.Watson RS, Carcillo JA, Linde-Zwirble WT, Clermont G, Lidicker J, Angus DC. The epidemiology of severe sepsis in children in the United States. Am J Respir Crit Care Med. 2003;167(5):695–701. doi: 10.1164/rccm.200207-682OC. [DOI] [PubMed] [Google Scholar]
- 3.Wisplinghoff H, Seifert H, Tallent SM, Bischoff T, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in pediatric patients in United States hospitals: epidemiology, clinical features and susceptibilities. Pediatr Infect Dis J. 2003;22(8):686–691. doi: 10.1097/01.inf.0000078159.53132.40. [DOI] [PubMed] [Google Scholar]
- 4.Bizzarro MJ, Raskind C, Baltimore RS, Gallagher PG. Seventy-five years of neonatal sepsis at Yale: 1928–2003. Pediatrics. 2005;116(3):595–602. doi: 10.1542/peds.2005-0552. [DOI] [PubMed] [Google Scholar]
- 5.Lee CY, Chen PY, Huang FL, Lin CF. Microbiologic spectrum and susceptibility pattern of clinical isolates from the pediatric intensive care unit in a single medical center - 6 years' experience. J Microbiol Immunol Infect. 2009;42(2):160–165. [PubMed] [Google Scholar]
- 6.Clapp DW. Developmental regulation of the immune system. Semin Perinatol. 2006;30(2):69–72. doi: 10.1053/j.semperi.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Wynn J, Seed P, Cotten C. Does IVIg administration yield improved immune function in very premature neonates? J Perinatol. 2010 doi: 10.1038/jp.2009.197. [DOI] [PubMed] [Google Scholar]
- 8.Klinger G, Levy I, Sirota L, Boyko V, Reichman B, Lerner-Geva L. Epidemiology and risk factors for early onset sepsis among very-low-birthweight infants. Am J Obstet Gynecol. 2009;201(1):38 e31–38 e36. doi: 10.1016/j.ajog.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 9.Bizzarro MJ, Dembry LM, Baltimore RS, Gallagher PG. Changing patterns in neonatal Escherichia coli sepsis and ampicillin resistance in the era of intrapartum antibiotic prophylaxis. Pediatrics. 2008;121(4):689–696. doi: 10.1542/peds.2007-2171. [DOI] [PubMed] [Google Scholar]
- 10.Yagupsky P, Dagan R, Prajgrod F, Merires M. Respiratory carriage of Kingella kingae among healthy children. Pediatr Infect Dis J. 1995;14(8):673–678. doi: 10.1097/00006454-199508000-00005. [DOI] [PubMed] [Google Scholar]
- 11.Yagupsky P, Peled N, Katz O. Epidemiological features of invasive Kingella kingae infections and respiratory carriage of the organism. J Clin Microbiol. 2002;40(11):4180–4184. doi: 10.1128/JCM.40.11.4180-4184.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viscardi RM, Hashmi N, Gross GW, Sun CC, Rodriguez A, Fairchild KD. Incidence of invasive ureaplasma in VLBW infants: relationship to severe intraventricular hemorrhage. J Perinatol. 2008;28(11):759–765. doi: 10.1038/jp.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jordan JA, Durso MB. Comparison of 16S rRNA gene PCR and BACTEC 9240 for detection of neonatal bacteremia. J Clin Microbiol. 2000;38(7):2574–2578. doi: 10.1128/jcm.38.7.2574-2578.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reier-Nilsen T, Farstad T, Nakstad B, Lauvrak V, Steinbakk M. Comparison of broad range 16S rDNA PCR and conventional blood culture for diagnosis of sepsis in the newborn: a case control study. BMC Pediatr. 2009;9:5. doi: 10.1186/1471-2431-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park JS, Svetkauskaite D, He Q, et al. Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J Biol Chem. 2004;279(9):7370–7377. doi: 10.1074/jbc.M306793200. [DOI] [PubMed] [Google Scholar]
- 16.Schilling JD, Mulvey MA, Hultgren SJ. Structure and function of Escherichia coli type 1 pili: new insight into the pathogenesis of urinary tract infections. J Infect Dis. 2001;183 Suppl 1:S36–S40. doi: 10.1086/318855. [DOI] [PubMed] [Google Scholar]
- 17.Dautin N, Bernstein HD. Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu Rev Microbiol. 2007;61:89–112. doi: 10.1146/annurev.micro.61.080706.093233. [DOI] [PubMed] [Google Scholar]
- 18.Chhatwal GS. Anchorless adhesins and invasins of Gram-positive bacteria: a new class of virulence factors. Trends Microbiol. 2002;10(5):205–208. doi: 10.1016/s0966-842x(02)02351-x. [DOI] [PubMed] [Google Scholar]
- 19.Barocchi MA, Ries J, Zogaj X, et al. A pneumococcal pilus influences virulence and host inflammatory responses. Proc Natl Acad Sci U S A. 2006;103(8):2857–2862. doi: 10.1073/pnas.0511017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corrigan RM, Miajlovic H, Foster TJ. Surface proteins that promote adherence of Staphylococcus aureus to human desquamated nasal epithelial cells. BMC Microbiol. 2009;9:22. doi: 10.1186/1471-2180-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weidenmaier C, Kokai-Kun JF, Kulauzovic E, et al. Differential roles of sortase-anchored surface proteins and wall teichoic acid in Staphylococcus aureus nasal colonization. Int J Med Microbiol. 2008;298(5–6):505–513. doi: 10.1016/j.ijmm.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 22.Shakhnovich EA, King SJ, Weiser JN. Neuraminidase expressed by Streptococcus pneumoniae desialylates the lipopolysaccharide of Neisseria meningitidis and Haemophilus influenzae: a paradigm for interbacterial competition among pathogens of the human respiratory tract. Infect Immun. 2002;70(12):7161–7164. doi: 10.1128/IAI.70.12.7161-7164.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swords WE, Moore ML, Godzicki L, Bukofzer G, Mitten MJ, VonCannon J. Sialylation of lipooligosaccharides promotes biofilm formation by nontypeable Haemophilus influenzae. Infect Immun. 2004;72(1):106–113. doi: 10.1128/IAI.72.1.106-113.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lysenko ES, Ratner AJ, Nelson AL, Weiser JN. The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PLoS Pathog. 2005;1(1):e1. doi: 10.1371/journal.ppat.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettigrew MM, Gent JF, Revai K, Patel JA, Chonmaitree T. Microbial interactions during upper respiratory tract infections. Emerg Infect Dis. 2008;14(10):1584–1591. doi: 10.3201/eid1410.080119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mistry D, Stockley RA. IgA1 protease. Int J Biochem Cell Biol. 2006;38(8):1244–1248. doi: 10.1016/j.biocel.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulks MH, Plaut AG. IgA protease production as a characteristic distinguishing pathogenic from harmless Neisseriaceae. N Engl J Med. 1978;299(18):973–976. doi: 10.1056/NEJM197811022991802. [DOI] [PubMed] [Google Scholar]
- 28.Kapatais-Zoumbos K, Chandler DK, Barile MF. Survey of immunoglobulin A protease activity among selected species of Ureaplasma and Mycoplasma: specificity for host immunoglobulin A. Infect Immun. 1985;47(3):704–709. doi: 10.1128/iai.47.3.704-709.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Glover DT, Szalai AJ, Hollingshead SK, Briles DE. PspA and PspC minimize immune adherence and transfer of pneumococci from erythrocytes to macrophages through their effects on complement activation. Infect Immun. 2007;75(12):5877–5885. doi: 10.1128/IAI.00839-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogunniyi AD, Grabowicz M, Mahdi LK, et al. Pneumococcal histidine triad proteins are regulated by the Zn2+-dependent repressor AdcR and inhibit complement deposition through the recruitment of complement factor H. FASEB J. 2009;23(3):731–738. doi: 10.1096/fj.08-119537. [DOI] [PubMed] [Google Scholar]
- 31.Van Dijk WC, Verbrugh HA, van der Tol ME, Peters R, Verhoef J. Role of Escherichia coli K capsular antigens during complement activation, C3 fixation, and opsonization. Infect Immun. 1979;25(2):603–609. doi: 10.1128/iai.25.2.603-609.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verweij-Van Vught AM, Namavar F, Peerbooms PG, Sparrius M, Maclaren DM. The role of different K antigens of Escherichia coli in phagocytosis by polymorphonuclear leukocytes. J Med Microbiol. 1984;17(2):141–150. doi: 10.1099/00222615-17-2-141. [DOI] [PubMed] [Google Scholar]
- 33.Nikkari S, McLaughlin IJ, Bi W, Dodge DE, Relman DA. Does blood of healthy subjects contain bacterial ribosomal DNA? J Clin Microbiol. 2001;39(5):1956–1959. doi: 10.1128/JCM.39.5.1956-1959.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sedman PC, Macfie J, Sagar P, et al. The prevalence of gut translocation in humans. Gastroenterology. 1994;107(3):643–649. doi: 10.1016/0016-5085(94)90110-4. [DOI] [PubMed] [Google Scholar]
- 35.O'Boyle CJ, MacFie J, Mitchell CJ, Johnstone D, Sagar PM, Sedman PC. Microbiology of bacterial translocation in humans. Gut. 1998;42(1):29–35. doi: 10.1136/gut.42.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fagan RP, Smith SG. The Hek outer membrane protein of Escherichia coli is an auto-aggregating adhesin and invasin. FEMS Microbiol Lett. 2007;269(2):248–255. doi: 10.1111/j.1574-6968.2006.00628.x. [DOI] [PubMed] [Google Scholar]
- 37.Fagan RP, Lambert MA, Smith SG. The hek outer membrane protein of Escherichia coli strain RS218 binds to proteoglycan and utilizes a single extracellular loop for adherence, invasion, and autoaggregation. Infect Immun. 2008;76(3):1135–1142. doi: 10.1128/IAI.01327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang R, Miki K, Oksala N, et al. Bile high-mobility group box 1 contributes to gut barrier dysfunction in experimental endotoxemia. Am J Physiol Regul Integr Comp Physiol. 2009;297(2):R362–R369. doi: 10.1152/ajpregu.00184.2009. [DOI] [PubMed] [Google Scholar]
- 39.Moriez R, Leveque M, Salvador-Cartier C, et al. Mucosal mast cell proteases are involved in colonic permeability alterations and subsequent bacterial translocation in endotoxemic rats. Shock. 2007;28(1):118–124. doi: 10.1097/SHK.0b013e3180315ba9. [DOI] [PubMed] [Google Scholar]
- 40.Kehl-Fie TE, St Geme JW., 3rd Identification and characterization of an RTX toxin in the emerging pathogen Kingella kingae. J Bacteriol. 2007;189(2):430–436. doi: 10.1128/JB.01319-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faure K, Sawa T, Ajayi T, et al. TLR4 signaling is essential for survival in acute lung injury induced by virulent Pseudomonas aeruginosa secreting type III secretory toxins. Respir Res. 2004;5:1. doi: 10.1186/1465-9921-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee LN, Dias P, Han D, et al. A mouse model of lethal synergism between influenza virus and Haemophilus influenzae. Am J Pathol. 2010;176(2):800–811. doi: 10.2353/ajpath.2010.090596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LeVine AM, Koeningsknecht V, Stark JM. Decreased pulmonary clearance of S. pneumoniae following influenza A infection in mice. J Virol Methods. 2001;94(1–2):173–186. doi: 10.1016/s0166-0934(01)00287-7. [DOI] [PubMed] [Google Scholar]
- 44.Mansson LE, Melican K, Boekel J, et al. Real-time studies of the progression of bacterial infections and immediate tissue responses in live animals. Cell Microbiol. 2007;9(2):413–424. doi: 10.1111/j.1462-5822.2006.00799.x. [DOI] [PubMed] [Google Scholar]
- 45.Melican K, Boekel J, Mansson LE, et al. Bacterial infection-mediated mucosal signalling induces local renal ischaemia as a defence against sepsis. Cell Microbiol. 2008;10(10):1987–1998. doi: 10.1111/j.1462-5822.2008.01182.x. [DOI] [PubMed] [Google Scholar]
- 46.Van Amersfoort ES, Van Berkel TJ, Kuiper J. Receptors, mediators, and mechanisms involved in bacterial sepsis and septic shock. Clin Microbiol Rev. 2003;16(3):379–414. doi: 10.1128/CMR.16.3.379-414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dixon DR, Darveau RP. Lipopolysaccharide heterogeneity: innate host responses to bacterial modification of lipid a structure. J Dent Res. 2005;84(7):584–595. doi: 10.1177/154405910508400702. [DOI] [PubMed] [Google Scholar]
- 48.Yoshimura A, Lien E, Ingalls RR, Tuomanen E, Dziarski R, Golenbock D. Cutting edge: recognition of Gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2. J Immunol. 1999;163(1):1–5. [PubMed] [Google Scholar]
- 49.Cirl C, Wieser A, Yadav M, et al. Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat Med. 2008;14(4):399–406. doi: 10.1038/nm1734. [DOI] [PubMed] [Google Scholar]
- 50.Newman RM, Salunkhe P, Godzik A, Reed JC. Identification and characterization of a novel bacterial virulence factor that shares homology with mammalian Toll/interleukin-1 receptor family proteins. Infect Immun. 2006;74(1):594–601. doi: 10.1128/IAI.74.1.594-601.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gordon MA, Graham SM, Walsh AL, et al. Epidemics of invasive Salmonella enterica serovar Enteritidis and S. enterica serovar Typhimurium infection associated with multidrug resistance among adults and children in Malawi. Clin Infect Dis. 2008;46(7):963–969. doi: 10.1086/529146. [DOI] [PubMed] [Google Scholar]
- 52.Graham SM, English M. Non-typhoidal salmonellae: a management challenge for children with community-acquired invasive disease in tropical African countries. Lancet. 2009;373(9659):267–269. doi: 10.1016/S0140-6736(09)60073-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thammavongsa V, Kern JW, Missiakas DM, Schneewind O. Staphylococcus aureus synthesizes adenosine to escape host immune responses. J Exp Med. 2009;206(11):2417–2427. doi: 10.1084/jem.20090097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khoa ND, Montesinos MC, Reiss AB, Delano D, Awadallah N, Cronstein BN. Inflammatory cytokines regulate function and expression of adenosine A(2A) receptors in human monocytic THP-1 cells. J Immunol. 2001;167(7):4026–4032. doi: 10.4049/jimmunol.167.7.4026. [DOI] [PubMed] [Google Scholar]
- 55.Panther E, Idzko M, Herouy Y, et al. Expression and function of adenosine receptors in human dendritic cells. FASEB J. 2001;15(11):1963–1970. doi: 10.1096/fj.01-0169com. [DOI] [PubMed] [Google Scholar]
- 56.McColl SR, St-Onge M, Dussault AA, et al. Immunomodulatory impact of the A2A adenosine receptor on the profile of chemokines produced by neutrophils. FASEB J. 2006;20(1):187–189. doi: 10.1096/fj.05-4804fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moore CC, Martin EN, Lee GH, Obrig T, Linden J, Scheld WM. An A2A adenosine receptor agonist, ATL313, reduces inflammation and improves survival in murine sepsis models. BMC Infect Dis. 2008;8:141. doi: 10.1186/1471-2334-8-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lindberg AA. Polyosides (encapsulated bacteria) C R Acad Sci III. 1999;322(11):925–932. doi: 10.1016/s0764-4469(00)87188-7. [DOI] [PubMed] [Google Scholar]
- 59.Foster TJ. Immune evasion by staphylococci. Nat Rev Microbiol. 2005;3(12):948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 60.Palmqvist N, Foster T, Tarkowski A, Josefsson E. Protein A is a virulence factor in Staphylococcus aureus arthritis and septic death. Microb Pathog. 2002;33(5):239–249. doi: 10.1006/mpat.2002.0533. [DOI] [PubMed] [Google Scholar]
- 61.Carlin AF, Lewis AL, Varki A, Nizet V. Group B streptococcal capsular sialic acids interact with siglecs (immunoglobulin-like lectins) on human leukocytes. J Bacteriol. 2007;189(4):1231–1237. doi: 10.1128/JB.01155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carlin AF, Uchiyama S, Chang YC, Lewis AL, Nizet V, Varki A. Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood. 2009;113(14):3333–3336. doi: 10.1182/blood-2008-11-187302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7(4):255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 64.Staali L, Bauer S, Morgelin M, Bjorck L, Tapper H. Streptococcus pyogenes bacteria modulate membrane traffic in human neutrophils and selectively inhibit azurophilic granule fusion with phagosomes. Cell Microbiol. 2006;8(4):690–703. doi: 10.1111/j.1462-5822.2005.00662.x. [DOI] [PubMed] [Google Scholar]
- 65.Karlsson A, Dahlgren C. Assembly and activation of the neutrophil NADPH oxidase in granule membranes. Antioxid Redox Signal. 2002;4(1):49–60. doi: 10.1089/152308602753625852. [DOI] [PubMed] [Google Scholar]
- 66.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Gotz F. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem. 1999;274(13):8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 67.Peschel A, Jack RW, Otto M, et al. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J Exp Med. 2001;193(9):1067–1076. doi: 10.1084/jem.193.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Collins LV, Kristian SA, Weidenmaier C, et al. Staphylococcus aureus strains lacking D-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J Infect Dis. 2002;186(2):214–219. doi: 10.1086/341454. [DOI] [PubMed] [Google Scholar]
- 69.Sukumaran SK, Shimada H, Prasadarao NV. Entry and intracellular replication of Escherichia coli K1 in macrophages require expression of outer membrane protein A. Infect Immun. 2003;71(10):5951–5961. doi: 10.1128/IAI.71.10.5951-5961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gleich-Theurer U, Aymanns S, Haas G, Mauerer S, Vogt J, Spellerberg B. Human serum induces streptococcal c5a peptidase expression. Infect Immun. 2009;77(9):3817–3825. doi: 10.1128/IAI.00826-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Postma B, Poppelier MJ, van Galen JC, et al. Chemotaxis inhibitory protein of Staphylococcus aureus binds specifically to the C5a and formylated peptide receptor. J Immunol. 2004;172(11):6994–7001. doi: 10.4049/jimmunol.172.11.6994. [DOI] [PubMed] [Google Scholar]
- 72.Maren von Kockritz-Blickwede SK, Gessner Engelbert, Medina Eva. Experimental Model of Staphylococcus aureus Bacteremia. Journal of Innate Immunology. 2010;2:87–92. doi: 10.1159/000247157. [DOI] [PubMed] [Google Scholar]
- 73.Marques MB, Kasper DL, Pangburn MK, Wessels MR. Prevention of C3 deposition by capsular polysaccharide is a virulence mechanism of type III group B streptococci. Infect Immun. 1992;60(10):3986–3993. doi: 10.1128/iai.60.10.3986-3993.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Llobet E, Tomas JM, Bengoechea JA. Capsule polysaccharide is a bacterial decoy for antimicrobial peptides. Microbiology. 2008;154(Pt 12):3877–3886. doi: 10.1099/mic.0.2008/022301-0. [DOI] [PubMed] [Google Scholar]
- 75.Palmqvist N, Josefsson E, Tarkowski A. Clumping factor A-mediated virulence during Staphylococcus aureus infection is retained despite fibrinogen depletion. Microbes Infect. 2004;6(2):196–201. doi: 10.1016/j.micinf.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 76.Palmqvist N, Patti JM, Tarkowski A, Josefsson E. Expression of staphylococcal clumping factor A impedes macrophage phagocytosis. Microbes Infect. 2004;6(2):188–195. doi: 10.1016/j.micinf.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 77.Higgins J, Loughman A, van Kessel KP, van Strijp JA, Foster TJ. Clumping factor A of Staphylococcus aureus inhibits phagocytosis by human polymorphonuclear leucocytes. FEMS Microbiol Lett. 2006;258(2):290–296. doi: 10.1111/j.1574-6968.2006.00229.x. [DOI] [PubMed] [Google Scholar]
- 78.Achtman M. Epidemic spread and antigenic variability of Neisseria meningitidis. Trends Microbiol. 1995;3(5):186–192. doi: 10.1016/s0966-842x(00)88918-0. [DOI] [PubMed] [Google Scholar]
- 79.Hobbs MM, Seiler A, Achtman M, Cannon JG. Microevolution within a clonal population of pathogenic bacteria: recombination, gene duplication and horizontal genetic exchange in the opa gene family of Neisseria meningitidis. Mol Microbiol. 1994;12(2):171–180. doi: 10.1111/j.1365-2958.1994.tb01006.x. [DOI] [PubMed] [Google Scholar]
- 80.Fraser JD, Proft T. The bacterial superantigen and superantigen-like proteins. Immunol Rev. 2008;225:226–243. doi: 10.1111/j.1600-065X.2008.00681.x. [DOI] [PubMed] [Google Scholar]
- 81.Watanabe-Ohnishi R, Low DE, McGeer A, et al. Selective depletion of V beta-bearing T cells in patients with severe invasive group A streptococcal infections and streptococcal toxic shock syndrome. Ontario Streptococcal Study Project. J Infect Dis. 1995;171(1):74–84. doi: 10.1093/infdis/171.1.74. [DOI] [PubMed] [Google Scholar]
- 82.Wang B, Morinobu A, Horiuchi M, Liu J, Kumagai S. Butyrate inhibits functional differentiation of human monocyte-derived dendritic cells. Cell Immunol. 2008;253(1–2):54–58. doi: 10.1016/j.cellimm.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 83.Brogdon JL, Xu Y, Szabo SJ, et al. Histone deacetylase activities are required for innate immune cell control of Th1 but not Th2 effector cell function. Blood. 2007;109(3):1123–1130. doi: 10.1182/blood-2006-04-019711. [DOI] [PubMed] [Google Scholar]
- 84.Dagtas AS, Gilbert KM. p21(Cip1) up-regulated during histone deacetylase inhibitor-induced CD4(+) T-cell anergy selectively associates with mitogen-activated protein kinases. Immunology. 2009 doi: 10.1111/j.1365-2567.2009.03161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Collard KJ. Iron homeostasis in the neonate. Pediatrics. 2009;123(4):1208–1216. doi: 10.1542/peds.2008-1047. [DOI] [PubMed] [Google Scholar]
- 86.Torres VJ, Pishchany G, Humayun M, Schneewind O, Skaar EP. Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J Bacteriol. 2006;188(24):8421–8429. doi: 10.1128/JB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Negre VL, Bonacorsi S, Schubert S, Bidet P, Nassif X, Bingen E. The siderophore receptor IroN, but not the high-pathogenicity island or the hemin receptor ChuA, contributes to the bacteremic step of Escherichia coli neonatal meningitis. Infect Immun. 2004;72(2):1216–1220. doi: 10.1128/IAI.72.2.1216-1220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Papp-Wallace KM, Maguire ME. Manganese transport and the role of manganese in virulence. Annu Rev Microbiol. 2006;60:187–209. doi: 10.1146/annurev.micro.60.080805.142149. [DOI] [PubMed] [Google Scholar]
- 89.Ammendola S, Pasquali P, Pistoia C, et al. High-affinity Zn2+ uptake system ZnuABC is required for bacterial zinc homeostasis in intracellular environments and contributes to the virulence of Salmonella enterica. Infect Immun. 2007;75(12):5867–5876. doi: 10.1128/IAI.00559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johnston JW, Briles DE, Myers LE, Hollingshead SK. Mn2+-dependent regulation of multiple genes in Streptococcus pneumoniae through PsaR and the resultant impact on virulence. Infect Immun. 2006;74(2):1171–1180. doi: 10.1128/IAI.74.2.1171-1180.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rosch JW, Gao G, Ridout G, Wang YD, Tuomanen EI. Role of the manganese efflux system mntE for signalling and pathogenesis in Streptococcus pneumoniae. Mol Microbiol. 2009;72(1):12–25. doi: 10.1111/j.1365-2958.2009.06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jakubovics NS, Valentine RA. A new direction for manganese homeostasis in bacteria: identification of a novel efflux system in Streptococcus pneumoniae. Mol Microbiol. 2009;72(1):1–4. doi: 10.1111/j.1365-2958.2009.06637.x. [DOI] [PubMed] [Google Scholar]
- 93.Kehl-Fie TE, Miller SE, St Geme JW., 3rd Kingella kingae expresses type IV pili that mediate adherence to respiratory epithelial and synovial cells. J Bacteriol. 2008;190(21):7157–7163. doi: 10.1128/JB.00884-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sutterwala FS, Mijares LA, Li L, Ogura Y, Kazmierczak BI, Flavell RA. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J Exp Med. 2007;204(13):3235–3245. doi: 10.1084/jem.20071239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alouf JE, Muller-Alouf H. Staphylococcal and streptococcal superantigens: molecular, biological and clinical aspects. Int J Med Microbiol. 2003;292(7–8):429–440. doi: 10.1078/1438-4221-00232. [DOI] [PubMed] [Google Scholar]