Abstract
Calorie restriction (CR) extends life span in a wide variety of species. Previously, we showed that calorie restriction increases the replicative life span in yeast by activating Sir2, a highly conserved NAD-dependent deacetylase. Here we test whether CR activates Sir2 by increasing the NAD/NADH ratio or by regulating the level of nicotinamide, a known inhibitor of Sir2. We show that CR decreases NADH levels, and that NADH is a competitive inhibitor of Sir2. A genetic intervention that specifically decreases NADH levels increases life span, validating the model that NADH regulates yeast longevity in response to CR.
Keywords: Calorie restriction, longevity regulation, metabolic signaling
Calorie restriction (CR) extends life span in a wide spectrum of organisms and for decades was the only regimen known to promote longevity in mammals (Weindruch and Walford 1998; Roth et al. 2001). CR has also been shown to delay the onset or reduce the incidence of many age-related diseases including cancer and diabetes (Weindruch and Walford 1998; Roth et al. 2001). Although it has been suggested that CR may work by reducing the levels of reactive oxygen species through a slowing in metabolism (Weindruch and Walford 1998), the mechanism by which CR extends life span is still uncertain. To study the mechanism by which CR extends life span, we established a model of CR in the budding yeast Saccharomyces cerevisiae. In this system, life span can be extended by limiting glucose content in the media from 2% to 0.5% or by reducing the activity of the glucose-sensing cAMP-dependent kinase (PKA; Lin et al. 2000).
The benefit of CR requires NAD (nicotinamide adenine dinucleotide, oxidized form) and Sir2 (Lin et al. 2000, 2002), a key regulator of life span in both yeast and animals (Kaeberlein et al. 1999; Tissenbaum and Guarente 2001). Sir2 is an NAD-dependent histone deacetylase and is required for chromatin silencing and life-span extension (Imai et al. 2000; Landry et al. 2000; Smith et al. 2000). The requirement of NAD for Sir2 deacetylase activity suggested CR may activate Sir2 by increasing the available pool of NAD for Sir2 (Guarente 2000).
Our previous studies suggested that there was a second mechanism to activate Sir2 and extend the life span in yeast—osmotic stress (Kaeberlein et al. 2002). This stress mechanism was genetically distinguishable from CR. Mutations knocking out electron transport prevented CR from extending life span, but had no effect on the longevity conferred by osmotic stress (Kaeberlein et al. 2002; Lin et al. 2002). This requirement for electron transport during CR is because of a shunting of carbon metabolism from fermentation to the mitochondrial TCA cycle. The concomitant increase in respiration is necessary and sufficient for the activation of Sir2-mediated silencing and extension in life span (Lin et al. 2002). The fact that respiration produces NAD from NADH (Bakker et al. 2001), reinforces the idea that changes in the NAD/NADH ratio regulate Sir2 during CR.
A recent report, however, challenged this model by claiming that both stress and CR activated Sir2 by a different mechanism, namely, by decreasing intracellular levels of nicotinamide (Anderson et al. 2003), a noncompetitive inhibitor of Sir2 (Bitterman et al. 2002). This change in nicotinamide levels was triggered by activation of the PNC1 gene, encoding a nicotinamidase in the NAD salvage pathway (Ghislain et al. 2002). This gene was strongly activated by osmotic stress, but to a lesser degree by CR. Also consistent with this model, PNC1 was shown to be required for the extension in life span by CR (Anderson et al. 2003). The authors conclude that CR reduces nicotinamide levels by up-regulating PNC1, and this reduction activates Sir2 and extends the life span.
In this report we attempt to determine whether changes in nicotinamide, the NAD/NADH ratio, or both up-regulate Sir2 during CR. We show that NADH is a competitive inhibitor of Sir2, and that CR reduces the level of NADH in cells. These findings provide a simple model for activation of Sir2 and extension of the life span by CR, which we further support by genetic experiments.
Results and Discussion
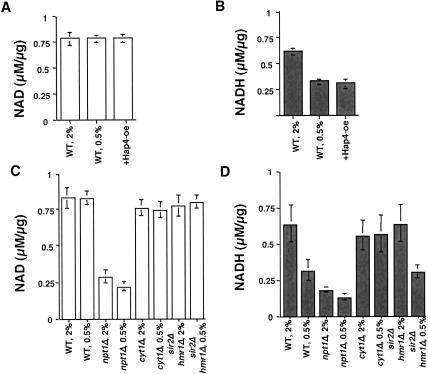
To study whether CR activates Sir2 by increasing the NAD/NADH ratio, we first measured the NAD and NADH levels in cells grown in 2% and 0.5% glucose (CR). Surprisingly, the NAD levels in cells under CR were not changed (Fig. 1A), whereas the NADH levels were decreased to 50% of non-CR cells (Fig. 1B). To validate the efficacy and sensitivity of our assay, we measured the NAD levels of the npt1Δ mutant. It has been shown that cells lacking the NPT1 gene (encoding nicotinic acid phosphoribosyl transferase in the NAD salvage pathway) exhibit a 40% to 60% decrease in NAD levels (Smith et al. 2000; Anderson et al. 2002). Consistent with previous reports (Smith et al. 2000; Anderson et al. 2002), we detected a similar decrease in the NAD levels in npt1Δ mutants. As a further test, we measured NAD and NADH levels in cells that overexpress the transcription factor Hap4, which activates nuclear genes encoding mitochondrial proteins (Forsburg and Guarente 1989; de Winde and Grivell 1993). This manipulation was shown to extend life span in a Sir2-dependent manner (Lin et al. 2002). Similar to CR, Hap4 overexpression causes a switch of metabolism from fermentation toward respiration (Blom et al. 2000) and, as shown in Figure 1B, triggered a 50% decrease in NADH levels without altering NAD concentration (Fig. 1A). Because respiration is required for life-span extension in CR, we tested whether a functional electron transport chain is required for the decrease in NADH levels. As shown in Figure 1D, deleting the CYT1 gene, which encodes the cytochrome c1, abolished the decrease in NADH levels induced by CR. All these studies suggest that CR increases the intracellular NAD/NADH ratio by up-regulating respiration, thereby decreasing NADH levels. We also measured NAD and NADH levels in a sir2Δ mutant grown in 2% and 0.5% glucose. The sir2Δ mutant exhibited the normal reduction in NADH levels in response to CR (shown in Fig. 1C,D), indicating that it functions downstream of the metabolic changes.
Figure 1.
Calorie restriction decreases intracellular NADH levels. Measurements of intracellular levels of NAD (A,C) and NADH (B,D) in various yeast strains grown in 2% or 0.5% glucose. Results show average of three independent experiments, each conducted in duplicate. Error bars denote standard deviations. NAD and NADH levels are shown as levels of NAD or NADH (in micromolar) normalized to the concentrations of proteins from the extracts (in micrograms) present in the reaction. Reactions contain cell extracts from 105 cells. (WT) Wild-type yeast strain PSY316; (+Hap4-oe) wild-type cells overexpressing Hap4; (2%) cells grown in 2% glucose; (0.5%) cells grown in 0.5% glucose. npt1Δ, cyt1Δ, and sir2Δ hmr1Δ are isogenic derivatives of PSY316.
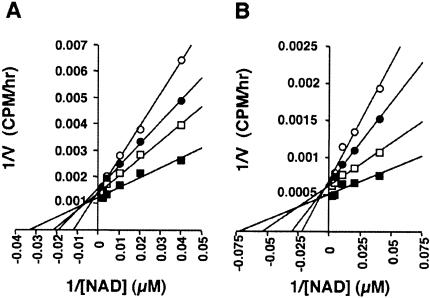
Because NADH levels responded to CR but NAD levels did not, it seemed possible that NADH, and not NAD, regulated Sir2, perhaps by inhibiting its activity. To test this hypothesis, we measured the activity of Sir2 in the presence of various concentrations of NADH using purified recombinant GST-tagged Sir2 proteins. Sir2 activity was determined by quantitating the NAD-dependent deacetylation of histone H4 peptides labeled with [3H]-acetyl coenzyme A (Armstrong et al. 2002). If NADH is, indeed, a competitive inhibitor of Sir2 activity, the presence of NADH should increase the apparent KM (the Michaelis-Menten binding constant) of Sir2 for NAD without affecting the Vmax (maximum velocity; Stryer 1995). Kinetic analysis with a wide range of substrate (NAD) concentrations is shown as a Lineweaver-Burk double reciprocal plot (Fig. 2A). This analysis estimates the KM (Fig. 2A, X intercept) of Sir2 to be 30 μM, consistent with previous reports (Imai et al. 2000; Tanner et al. 2000). As NADH concentrations were stepped up, the apparent KM for NAD was increased, reaching an increase of threefold at 250 μM NADH. The Vmax (Fig. 2A, Y intercept) of Sir2 activity was not significantly changed in the presence of NADH. These data suggest that NADH functions as a competitive inhibitor of Sir2. As shown in Figure 2B, NADH also competitively inhibited the human SIRT1. In the presence of 300 μM NADH, the KM of SIRT1 increased threefold.
Figure 2.
NADH inhibits Sir2 NAD-dependent histone deacetylase activity. (A) Kinetic analysis of the Sir2 NAD-dependent histone deacetylase activities in the presence of 0 μM (filled squares), 50 μM (open squares); 100 μM (filled circles), or 250 μM (open circles) NADH. Here 150 ng of recombinant GST-tagged yeast Sir2 protein was assayed with various concentrations of NAD and NADH for 2.5 h at 30°C. Data are shown as a Lineweaver-Burk double reciprocal plot of 1/V (CPM/h) versus 1/[NAD] (μM). The results show the average of three independent experiments, each measured in duplicate. (B) Kinetic analysis of the human SIRT1 NAD-dependent histone deacetylase activities in the presence of 0 μM (filled squares), 50 μM (open squares), 150 μM (filled circles), or 300 μM (open circles) NADH. Experiments were carried out as in A, except that 50 ng of recombinant GST-tagged human SIRT1 protein (Takata and Ishikawa 2003) was used, and the reaction was carried out for 2 h at 37°C.
Intracellular concentrations of total NAD plus NADH of ∼1 mM have been reported (de Koning and van Dam 1992; Richard et al. 1993). We have calculated values for NAD and NADH from the data in Figure 1, assuming a yeast cell size of 70 μm3 (Sherman 2002). Table 1 shows our values and those of other recent reports. The data are in reasonable agreement with the previous reports and with each other, and small differences may be partly due to different estimates of cell size. The concentration of free NADH in cells must be less than the values in Table 1, and appears to be in a sensitive range to regulate Sir2 activity.
Table 1.
Intracellular concentrations of NAD and NADH
| Normal condition (mM)
|
Calorie restriction (mM)
|
|||||
|---|---|---|---|---|---|---|
| NAD | NADH | NAD/NADH ratio | NAD | NADH | NAD/NADH ratio | References |
| 2.14a,b | — | — | — | — | — | Smith et al. 2000 |
| 1.14a,c | — | — | — | — | — | Ashrafi et al. 2000 |
| 2-3 | — | — | 2-3 | — | — | Lin et al. 2001 |
| 2a,d | 0.78a,d | 2.56 | — | — | — | Anderson et al. 2002 |
| 1.26 ± 0.06e | 0.85 ± 0.13 | 1.48 | 1.19 ± 0.08 | 0.39 ± 0.11 | 3.05 | This study |
We converted reported units into millimolar (per cell) assuming a yeast cell size of 70 fL (70 μm3) and the total protein of 6 pg/cell (Sherman 2002).
Reported unit was 1.5 × 10-4 pmole/cell.
Reported unit was ∼80 amole/cell.
Reported units were 23.7 amole/pg protein and 9.3 amole/pg protein for NAD and NADH, respectively.
Numbers show mean ± standard deviations derived from data shown in Fig. 1.
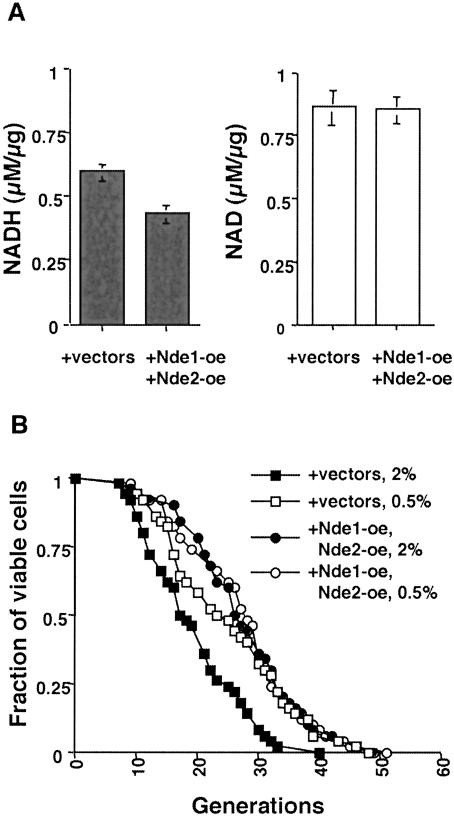
We sought genetic data to support the idea that the increase in the NAD/NADH ratio is what extends life span in CR. During respiratory growth, both cytosolic and mitochondrial NADH are reoxidized, in part by the NADH dehydrogenases in the respiratory chain (Bakker et al. 2001). To determine whether yeast life span could be extended by simply activating NADH dehydrogenase, we overexpressed two related mitochondrial NADH dehydrogenases, Nde1 and Nde2 (Luttik et al. 1998). Similarly to CR, overexpressing Nde1 and Nde2 significantly decreased the NADH levels without changing the NAD levels (Fig. 3A). Moreover, cells overexpressing Nde1 and Nde2 exhibited a longer life span on 2% glucose to a degree similar to cells grown on 0.5% glucose (Fig. 3B). Furthermore, 0.5% glucose did not extend the life span of cells overexpressing the NADH dehydrogenases (Fig. 3B), suggesting that CR and the NADH dehydrogenases function in the same pathway, that is, to decrease NADH levels and extend the life span.
Figure 3.
Overexpressing NADH dehydrogenase increases the intracellular NAD/NADH ratio and life span. (A) Measurements of NADH (closed bars, left panel) and NAD (open bars, right panel) levels in cells overexpressing Nde1 and Nde2. The results show the average of four independent experiments, each conducted in duplicate. Error bars denote standard deviations. (B) Life-span analysis of cells overexpressing Nde1 and Nde2 grown in 2% and 0.5% glucose. Average life spans on 2% glucose: +vectors, 19.4; +Nde1-oe, +Nde2-oe: 27.2. Average life spans on 0.5% glucose: +vectors, 25.8; +Nde1-oe, +Nde2-oe: 27.1. (+ vectors) Cells carrying control vectors ppp35 and ppp81; (+Nde1-oe, +Nde2-oe) cells overexpressing Nde1 and Nde2.
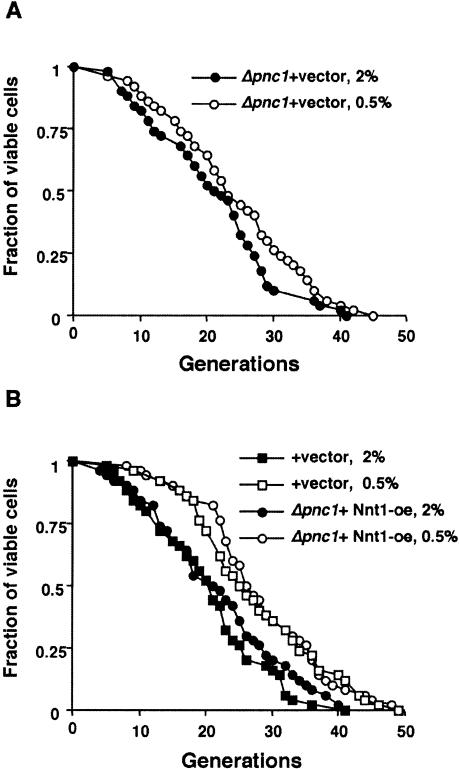
Our findings appear to challenge the claim that the Pnc1 nicotinamidase triggers the life-span extension in CR (Anderson et al. 2003). To address this paradox, we first repeated life-span analysis of the pnc1Δ mutants on 2% and 0.5% glucose. Consistent with the previous report, the pnc1Δ mutation largely prevented the life-span extension by 0.5% glucose (at best a 10%–15% increase; Fig. 4A) when compared with wild-type cells (∼30% increase; Fig. 4B). Because the above data indicated that CR functions by decreasing NADH, we surmised that hyperaccumulation of nicotinamide in the pnc1Δ mutants might mask regulation by NADH. Nnt1, a putative nicotinamide methyl transferase, appears to modify nicotinamide in yeast (Anderson et al. 2003). Thus, we overexpressed Nnt1 to reduce nicotinamide levels in the pnc1Δ mutant. Strikingly, overexpressing Nnt1 restored the ability of CR to extend life span (∼30%) in pnc1Δ mutants (Fig. 4B). These data show that the reduction in NADH can activate Sir2 and give a full extension of the life span in a pnc1Δ mutant, as long as the excess nicotinamide is depleted. This result shows that PNC1 and glucose-dependent changes in nicotinamide levels are not required for the extension of life span by CR.
Figure 4.
Calorie restriction in pnc1Δ mutants. (A) Calorie restriction slightly extends life span in a pnc1Δ mutant. (B) Overexpressing Nnt1 restores the full life-span extension by calorie restriction in a pnc1Δ mutant. Average life spans on 2% glucose: pnc1Δ + vector, 20.9; + vector, 20.16; Δpnc1 +Nnt1-oe, 21.6. Average life spans on 0.5% glucose: pnc1Δ + vector, 23.9; + vector, 27.3; pnc1Δ +Nnt1-oe, 28. (+ vector) cells carrying a control vector ppp35; (+Nnt1-oe) cells overexpressing Nnt1. pnc1Δ was derived from PSY316.
Our studies show that a switch to oxidative metabolism during CR increases the NAD/NADH ratio by decreasing NADH levels. NADH is a competitive inhibitor of Sir2, implying that a reduction in this dinucleotide activates Sir2 to extend the life span in CR. Indeed, overexpression of the NADH dehydrogenase specifically lowers NADH levels and extends the life span, providing strong support for this hypothesis. Regulation of the life span by NADH is also consistent with the earlier finding that electron transport is required for longevity during CR (Lin et al. 2002). The NAD/NADH ratio reflects the intracellular redox state and is a readout of metabolic activity. Our findings suggest that this ratio can serve a critical regulatory function, namely, the determination of the life span of yeast mother cells. It remains to be seen whether this ratio will serve related regulatory functions in higher organisms.
Materials and methods
Yeast strain PSY316 MATα ura3-52 leu2-3, 112 his3-Δ200 ade2-101 lys2-801 RDN1::ADE2 has been previously described (Park et al. 1999). Rich media YPD and synthetic media were made as described (Sherman et al. 1978). The ADH1-driven integrating ppp35 (URA3) vector is a derivative of the ppp81 (LEU2) vector (Lin et al. 2002) made by Peter Park. Overexpression constructs of Nde1, Nde2, and Nnt1 were made as described previously (Lin et al. 2002) using ppp81 (Nde1) or ppp35 (Nde2 and Nnt1). All constructs made for this study were verified by sequencing. The yeast Sir2 expression construct pGEX-SIR2 was a gift from J. Tanny and D. Moazed at Harvard Medical School (Tanny et al. 1999). The human SIRT1 expression construct pGEX-GST-hSIRT1 was a gift from F. Ishikawa at Tokyo Institute of Technology, Japan (Takata and Ishikawa 2003). All gene deletions in this study were done by replacing the wild-type genes with the Kanr marker as described in Guldener et al. (1996) and were verified by polymerase chain reaction (PCR) using oligonucleotides flanking the genes of interest.
Life-span analyses were carried out as previously described (Lin et al. 2000). All life-span analyses in this study were carried out on YPD plates at least twice independently with >45 cells per strain per experiment. The results from a single experiment are shown.
Yeast GST-Sir2 and human GST-SIRT1 fusion proteins were made using the T7 expression system as previously described (Ford and Hernandez 1997) with the following modifications: pGEX-SIR2 and pGEX-GST-hSIRT1 were transformed into the Escherichia coli strains BL21(DE3) Codon-Plus-RIL (Stratagene) and BL21(DE3), respectively. In addition, the procedure for yeast GST-Sir2 was scaled up to a 10-L culture with 5 mL of glutathione-agarose resin. The purified protein was concentrated on a Centriprep-30 (Amacon) column.
The deacetylation assay was carried out as described previously (Armstrong et al. 2002). In brief, histone deacetylase activity was measured using a peptide corresponding to the N-terminal tail of the histone H4 (Upstate Biotechnology) labeled with tritiated Acetyl Coenzyme A (PerkinElmer). Reactions were carried out in 50 μL of buffer containing 50 mM Tris-HCl (pH 8), 4 mM MgCl2, 0.2 mM DTT, and a variable concentration of NAD and NADH for 2.5 h at 30°C (yeast Sir2) or for 2 h at 37°C (human SIRT1).
Measurement of the NAD and NADH nucleotides was performed as described previously (Lin et al. 2001) with a few modifications. In brief, cells were grown to an OD600 of 0.5, then 107 cells were harvested in duplicates by centrifugation in 2 × 1.5-mL tubes. Acid extraction was performed in one tube to obtain NAD and alkali extraction was performed in the other to obtain NADH, with 2 μL of neutralized cell extract (∼105 cells) used for enzymatic cycling reaction as previously described (Lin et al. 2001). The concentration of nucleotides was measured fluorometrically with excitation at 365 nm and emission monitored at 460 nm. Standard curves for determining NAD and NADH concentrations were obtained as follows: NAD and NADH were added into the acid and alkali buffer to a final concentration of 0 μM, 2.5 μM, 5 μM, and 7.5 μM, which were then treated with the same procedure along with other samples. The fluorometer was calibrated each time before use with 0 μM, 5 μM, 10 μM, 20 μM, 30 μM, and 40 μM NADH to ensure the detection was within a linear range.
Acknowledgments
We thank members of the Guarente laboratory for discussions; J. Gordon for advice with the NAD nucleotide measurements; J. Tanny and D. Moazed for the yeast Sir2 expression construct and discussions; and F. Ishikawa for the human SIRT1 expression construct. This work was supported by the NIH, the Ellison Medical Foundation, the Seaver Institute, and the Howard and Linda Stern Fund (L.G.); and the Individual National Research Service Award (S.-J.L., E.F., M.H.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1164804.
Corresponding authors.
References
- Anderson R.M., Bitterman, K.J., Wood, J.G., Medvedik, O., Cohen, H., Lin, S.S., Manchester, J.K., Gordon, J.I., and Sinclair, D.A. 2002. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 277: 18881–18890. [DOI] [PubMed] [Google Scholar]
- Anderson R.M., Bitterman, K.J., Wood, J.G., Medvedik, O., and Sinclair, D.A. 2003. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 423: 181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong C.M., Kaeberlein, M., Imai, S.I., and Guarente, L. 2002. Mutations in Saccharomyces cerevisiae gene SIR2 can have differential effects on in vivo silencing phenotypes and in vitro histone deacetylation activity. Mol. Biol. Cell 13: 1427–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K., Lin, S.S., Manchester, J.K., and Gordon, J.I. 2000. Sip2p and its partner snf1p kinase affect aging in S. cerevisiae. Genes & Dev. 14: 1872–1885. [PMC free article] [PubMed] [Google Scholar]
- Bakker B.M., Overkamp, K.M., van Maris, A.J., Kotter, P., Luttik, M.A., van Dijken, J.P., and Pronk, J.T. 2001. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25: 15–37. [DOI] [PubMed] [Google Scholar]
- Bitterman K.J., Anderson, R.M., Cohen, H.Y., Latorre-Esteves, M., and Sinclair, D.A. 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast Sir2 and human SIRT1. J. Biol. Chem. 277: 45099–45107. [DOI] [PubMed] [Google Scholar]
- Blom J., De Mattos, M.J., and Grivell, L.A. 2000. Redirection of the respiro-fermentative flux distribution in Saccharomyces cerevisiae by overexpression of the transcription factor Hap4p. Appl. Environ. Microbiol. 66: 1970–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning W. and van Dam, K. 1992. A method for the determination of changes of glycolytic metabolites in yeast on a subsecond time scale using extaction at neutral pH. Anal. Biochem. 204: 118–123. [DOI] [PubMed] [Google Scholar]
- de Winde J.H. and Grivell, L.A. 1993. Global regulation of mitochondrial biogenesis in Saccharomyces cerevisiae. Prog. Nucleic Acid Res. Mol. Biol. 46: 51–91. [DOI] [PubMed] [Google Scholar]
- Ford E. and Hernandez, N. 1997. Characterization of a trimeric complex containing Oct-1, SNAPc, and DNA. J. Biol. Chem. 272: 16048–16055. [DOI] [PubMed] [Google Scholar]
- Forsburg S.L. and Guarente, L. 1989. Identification and characterization of HAP4: A third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes & Dev. 3: 1166–1178. [DOI] [PubMed] [Google Scholar]
- Ghislain M., Talla, E., and Francois, J.M. 2002. Identification and functional analysis of the Saccharomyces cerevisiae nicotinamidase gene, PNC1. Yeast 19: 215–224. [DOI] [PubMed] [Google Scholar]
- Guarente L. 2000. Sir2 links chromatin silencing, metabolism, and aging. Genes & Dev. 14: 1021–1026. [PubMed] [Google Scholar]
- Guldener U., Heck, S., Fielder, T., Beinhauer, J., and Hegemann, J.H. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24: 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Armstrong, C.M., Kaeberlein, M., and Guarente, L. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M., McVey, M., and Guarente, L. 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & Dev. 13: 2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., Andalis, A.A., Fink, G.R., and Guarente, L. 2002. High osmolarity extends life span in Saccharomyces cerevisiae by a mechanism related to calorie restriction. Mol. Cell. Biol. 22: 8056–8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J., Sutton, A., Tafrov, S.T., Heller, R.C., Stebbins, J., Pillus, L., and Sternglanz, R. 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. 97: 5807–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.J., Defossez, P.A., and Guarente, L. 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289: 2126–2128. [DOI] [PubMed] [Google Scholar]
- Lin S.S., Manchester, J.K., and Gordon, J.I. 2001. Enhanced gluconeogenesis and increased energy storage as hallmarks of aging in Saccharomyces cerevisiae. J. Biol. Chem. 276: 36000–36007. [DOI] [PubMed] [Google Scholar]
- Lin S.-J., Kaeberlein, M., Andalis, A.A., Sturtz, L.A., Defossez, P.-A., Culotta, V.C., Fink, G.R., and Guarente, L. 2002. Calorie restriction extends life span by shifting carbon toward respiration. Nature 418: 344–348. [DOI] [PubMed] [Google Scholar]
- Luttik M.A., Overkamp, K.M., Kotter, P., de Vries, S., van Dijken, J.P., and Pronk, J.T. 1998. The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH. J. Biol. Chem. 273: 24529–24534. [DOI] [PubMed] [Google Scholar]
- Park P.U., Defossez, P.A., and Guarente, L. 1999. Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 3848–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P., Teusink, B., Westerhoff, H.V., and van Dam, K. 1993. Around the growth phase transition S. cerevisiae's make-up favours sustained oscillations of intracellular metabolites. FEBS Lett. 318: 80–82. [DOI] [PubMed] [Google Scholar]
- Roth G.S., Ingram, D.K., and Lane, M.A. 2001. Caloric restriction in primates and relevance to humans. Ann. NY Acad. Sci. 928: 305–315. [DOI] [PubMed] [Google Scholar]
- Sherman F. 2002. Guide to yeast genetics and molecular and cellular biology. In Methods in enzymology (eds. C. Guthrie and G.R. Fink), pp. 3–41. Academic Press, San Diego.12073320
- Sherman F., Fink, G.R., and Lawrence, C.W. 1978. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Smith J.S., Brachmann, C.B., Celic, I., Kenna, M.A., Muhammad, S., Starai, V.J., Avalos, J.L., Escalante-Semerena, J.C., Grubmeyer, C., Wolberger, C., et al. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. 97: 6658–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryer L. 1995. Biochemistry. W.H. Freeman and Company, New York.
- Takata T. and Ishikawa, F. 2003. Human Sir2-related protein SIRT1 associates with the bHLH repressors HES1 and HEY2 and is involved in HES1- and HEY2-mediated transcriptional repression. Biochem. Biophys. Res. Commun. 301: 250–257. [DOI] [PubMed] [Google Scholar]
- Tanner K.G., Landry, J., Sternglanz, R., and Denu, J.M. 2000. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. 97: 14178–14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny J.C., Dowd, G.J., Huang, J., Hilz, H., and Moazed, D. 1999. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99: 735–745. [DOI] [PubMed] [Google Scholar]
- Tissenbaum H.A. and Guarente, L. 2001. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410: 227–230. [DOI] [PubMed] [Google Scholar]
- Weindruch W. and Walford, R.L. 1998. The retardation of aging and diseases by dietary restriction. Charles C. Thomas, Springfield, IL.