Abstract
The study of Hox clusters and genes provides insights into the evolution of genomic regulation of development. Derived ray-finned fishes (Actinopterygii, Teleostei) such as zebrafish and pufferfish possess duplicated Hox clusters that have undergone considerable sequence evolution. Whether these changes are associated with the duplication(s) that produced extra Hox clusters is unresolved because comparison with basal lineages is unavailable. We sequenced and analyzed the HoxA cluster of the bichir (Polypterus senegalus), a phylogenetically basal actinopterygian. Independent lines of evidence indicate that bichir has one HoxA cluster that is mosaic in its patterns of noncoding sequence conservation and gene retention relative to the HoxA clusters of human and shark, and the HoxAα and HoxAβ clusters of zebrafish, pufferfish, and striped bass. HoxA cluster noncoding sequences conserved between bichir and euteleosts indicate that novel cis-sequences were acquired in the stem actinopterygians and maintained after cluster duplication. Hence, in the earliest actinopterygians, evolution of the single HoxA cluster was already more dynamic than in human and shark. This tendency peaked among teleosts after HoxA cluster duplication.
Hox genes, which share sequence homology with the Hom-C genes of Drosophila and are clustered in the genome, form a distinct class of transcription factors that play an essential role in embryonic patterning (McGinnis and Krumlauf 1992). Hox clusters display the phenomenon of colinearity, in which the position of a gene in the cluster is related to its spatiotemporal pattern of expression along the anterior-posterior (A-P) axis (Lufkin 1996). Hox genes have a dynamic evolutionary history hallmarked by tandem (Kappen et al. 1989) and whole-cluster duplications (Holland and Garcia-Fernandez 1996; Ruddle et al. 1999). Whereas protostome taxa possess at most a single Hox cluster, the number of Hox clusters in different vertebrate lineages is varied (Ruddle et al. 1999). Vertebrate Hox clusters and their genes, therefore, are good models for identifying putative correlations between genomic and phenotypic evolution (Hughes and Kaufman 2002).
The ray-finned fishes (Actinopterygii; Fig. 1) are well suited to such studies. Teleost fishes (Nelson 1994) are the most diverse extant vertebrates, with >25,000 species. Moreover, the genomes of derived teleost fishes such as zebrafish, “euteleosts” sensu (Nelson 1994), contain duplicated genes of several different gene families (Postlethwait et al. 1998; Robinson-Rechavi et al. 2001; Taylor et al. 2001) and extra Hox clusters (Amores et al. 1998), as compared with their single orthologs in the genome of humans. The zebrafish (Danio rerio) possesses at least seven unlinked Hox clusters (Aα, Aβ, Bα, Bβ, Cα, Cβ, and D; Amores et al. 1998). Evidence for the presence of more than four Hox clusters also has been reported for additional euteleosts, including pufferfish (Takifugu rubripes; Aparicio et al. 2002; Amores et al. 2003), medaka (Oryzias latipes; Naruse et al. 2000), striped bass (Morone saxatilis; Snell et al. 1999), killifish (Fundulus heteroclitus; Misof and Wagner 1996), and tilapia (Oreochromis niloticus; Malaga-Trillo and Meyer 2001; Santini et al. 2003).
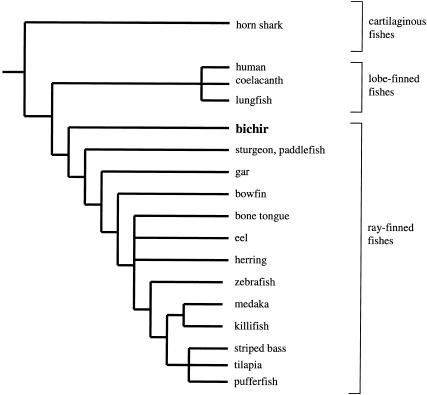
Figure 1.
Overview of jawed-vertebrate phylogenetic relationships with focus on the ray-finned fishes (Actinopterygii; Patterson 1982; Nelson 1994; Bartsch and Britz 1997; Bemis et al. 1997). The other two living sister groups to the ray-finnedfishes among gnathostomes are the cartilaginous (Chondrichthyes) andlobe-finned(Sarcopterygii) fishes, respectively. Note that only some euteleost clades are well represented to date, but remaining higher actinopterygian groups, Chondrostei (sturgeon and paddlefish), Neopterygii (gar, bowfin, and teleosts), andbasal teleost clades are virtually unresolved. This general outline of actinopterygian phylogeny is supportedin three of the most recent hypotheses basedon molecular data (Le et al. 1993; Venkatesh et al. 2001; Inoue et al. 2003). But, as with some conflicting morphologically basedhypotheses in the past, these also still indicate changing positions or lack of resolution among other basal actinopterygian fauna: sturgeon and paddlefish, gar, and bowfin.
In our recent analysis of patterns of noncoding sequence evolution in the duplicated HoxAα and HoxAβ clusters of zebrafish and in the single HoxA clusters of horn shark (Heterodontus francisci) and mammals, we proposed that following Hox cluster duplication, the noncoding control elements of zebrafish undergo extensive remodeling (Chiu et al. 2002). But, as indicated in Figure 1, there are several basal and intermediate actinopterygian lineages for which we do not, at present, have definitive knowledge on Hox cluster number. Hence, major questions remain. When, during ray-finned fish phylogeny, did the duplications that produced extra Hox clusters occur? Is the loss of Hox cluster noncoding sequence conservation observed in euteleosts also present in primitive actinopterygians? That is, is this phenomenon a characteristic of all ray-finned fishes, independent of duplication? How have Hox clusters evolved after duplication? To address these questions, we sequenced and analyzed the complete HoxA cluster of the bichir (Polypterus senegalus), a representative of the most basal extant ray-finned fish lineage as inferred from independent molecular data sets (Le et al. 1993; Venkatesh et al. 2001; Inoue et al. 2003) as well as its possession of ancestral morphological characters (Patterson 1982; Nelson 1994; Bartsch and Britz 1997; Bemis et al. 1997).
RESULTS AND DISCUSSION
Bichir HoxA Cluster Architecture
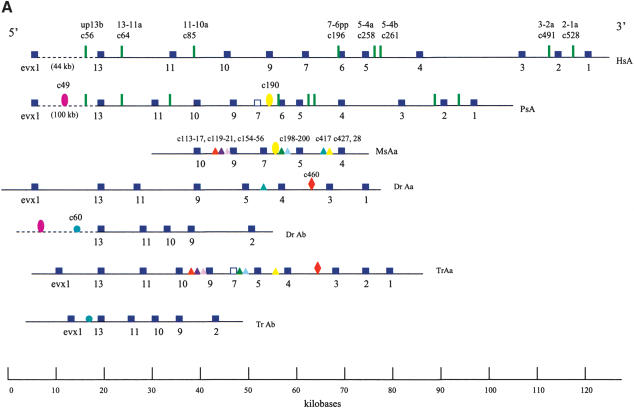
We screened a BAC genomic library from P. senegalus and isolated a 180-kb HoxA contig spanning Evx1 to HoxA1 (Fig. 2). An additional HoxA cluster was not identified in our screenings. The architecture and gene complement of the HoxA cluster closely resemble that of the single HoxA clusters of human and horn shark (Kim et al. 2000; Chiu et al. 2002) and of the hypothetical gnathostome ancestor (Stellwag 1999). Disparate patterns of retention were observed for two medial group genes, HoxA6 and HoxA7. The bichir HoxA cluster encodes an intact HoxA6 gene, which has not been found in any euteleost examined. Remnants of the HoxA7 gene were observed in the bichir (Fig. 2A), similar to the situation observed in pufferfish (Amores et al. 2003). No obvious traces of an HoxA7 ortholog have been found in either the zebrafish HoxAα and HoxAβ clusters (Amores et al. 1998) or the pufferfish HoxAβ cluster (Fig. 2A; Aparicio et al. 2002; Amores et al. 2003). The HoxAα clusters of striped bass (Snell et al. 1999) and tilapia (Santini et al. 2003), on the other hand, each house an intact HoxA7 gene. Thus the HoxA7 gene has been independently lost in at least three distinct actinopterygian lineages, at least once in a basal group (bichir) and at least twice in teleosts (zebrafish, pufferfish). These two genes, HoxA6 and HoxA7, underscore that parallel events of gene losses within actinopterygians cannot be inferred solely on the basis of character reconstructions using parsimony.
Figure 2.
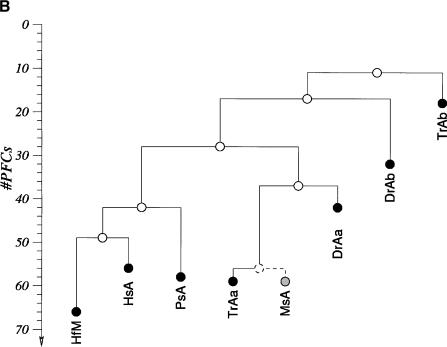
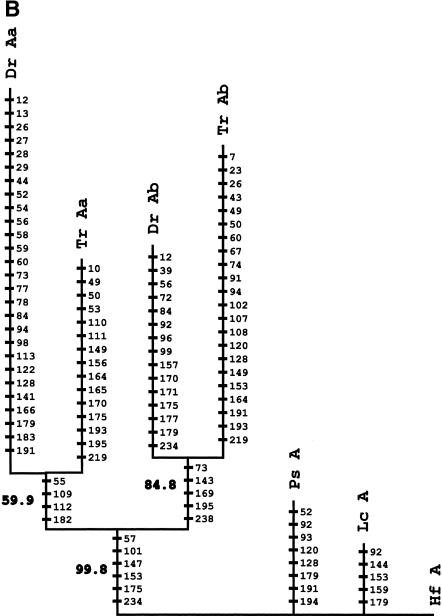
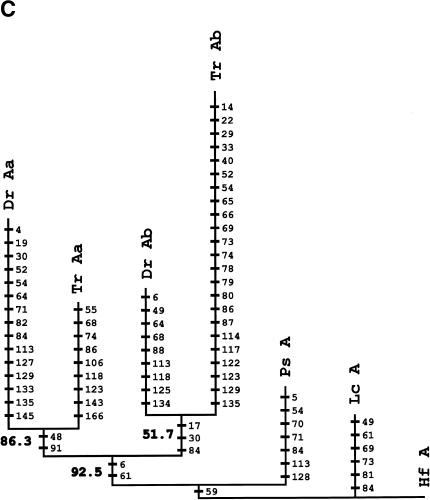
HoxA clusters and PFCs in human, shark, bichir, andeuteleosts. (Hs) Homo sapiens; (Hf) Heterodontus francisci; (Ps) Polypterus senegalus; (MsAa) Morone saxatilis HoxAα; (DrAa) Danio rerio HoxAα; (DrAb) D. rerio HoxAβ; (TrAa) Takifugu rubripes HoxAα; (TrAb) T. rubripes HoxAβ. (A) Hox genes are indicatedby blue rectangles. PFCs sharedexclusively between human andbichir are indicatedby green bars. PFCs sharedexclusively between bichir andeuteleosts are indicatedby coloredellipses. PFCs sharedbetween the HoxAα clusters of striped bass, zebrafish, and/or pufferfish are indicated by coloredtriangles. The PFC sharedbetween only zebrafish andpufferfish HoxAα clusters is indicated by a red diamond. The PFC shared between zebrafish andpufferfish HoxAβ clusters is indicated by an aqua blue circle. See text for description. (B) The co-occurrences of the PFCs in different clusters (Supplemental Table 1) can be representedas a tree. The height of an internal node is the average number of PFCs shared by two clusters in the two different subtrees. The position of the tips gives the total number of PFCs in the segment of the HoxA cluster that spans from Evx1 to HoxA1. The nonduplicated HoxA regions of human, shark, andbichir form one group. The secondsignificant group consists of the HoxAα sequences of the euteleosts. The position of the incomplete stripedbass (Morone saxatilis) sequence is estimatedby assuming that in a complete sequence we wouldhave foundroughly the same number of PFCs as in pufferfish (T. rubripes). The HoxAβ clusters are much further divergedanddo not appear to group together because euteleost-specific PFCs in the HoxAβ cluster are very rare (Supplemental Table 1). The smaller rate of PFC loss in the zebrafish HoxAβ cluster is the dominating effect.
Phylogenetic Analysis Supports a Single HoxA Cluster in the Bichir
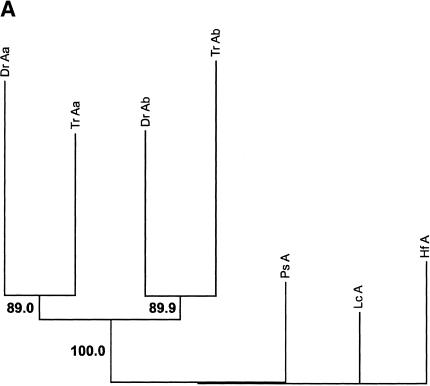
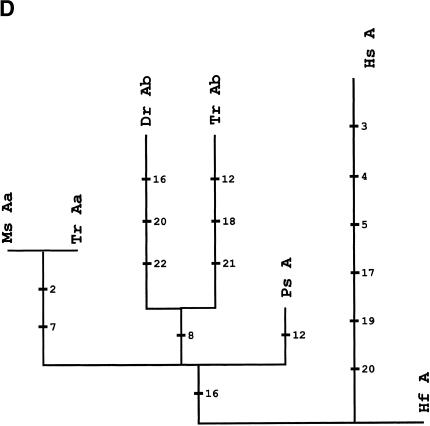
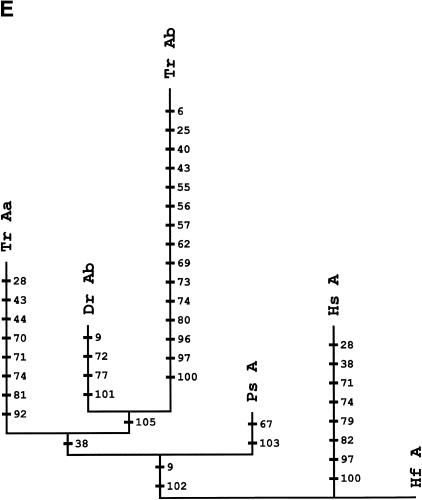
Phylogenetic analyses of HoxA cluster coding sequences of human, shark, bichir, and euteleosts are consistent with the inference that the bichir has a single HoxA cluster (Figs. 3A-E). We examined trees for HoxA13, HoxA11, HoxA10, and HoxA2 amino acid sequences of shark (Kim et al. 2000), human (Venter et al. 2001), coelacanth (Koh et al. 2003; C. T. Amemiya and T. Powers, unpubl.), bichir (this study), zebrafish (Amores et al. 1998), and pufferfish (Fig. 3; Aparicio et al. 2002). The topology of a neighbor-joining tree of concatenated HoxA11 and HoxA13 exon 1 coding sequences indicates that the bichir and teleosts last shared a common ancestor prior to the duplication event that gave rise to the duplicated HoxA clusters in zebrafish and pufferfish (Fig. 3A). This leaves open the possibility that bichir has independently acquired an HoxA cluster duplication. To address this possibility, we examined the rate of nonsynonymous substitutions (Figs. 3B-E) because our earlier findings for the duplicated HoxA11α and HoxA11β paralogs of zebrafish (Chiu et al. 2000a) showed that gene duplication is associated with an increased rate of replacement substitutions. Amino acid character reconstructions of HoxA13 (Fig. 3B), HoxA11 (Fig. 3C), and HoxA2 (Fig. 3E) confirm a higher rate of nonsynonymous substitutions for the duplicated paralogs of zebrafish and pufferfish. The orthologous bichir sequences accumulated significantly fewer amino acid changes (Figs. 3A-E). This finding is consistent with our library screening results and gene tree reconstructions indicating only a single HoxA cluster in bichir.
Figure 3.
Phylogenetic analysis andcharacter reconstructions of Hox coding sequences. (A) Neighbor-joining tree of concatenated HoxA13 and HoxA11 exon 1 coding sequences. Bootstrap support (1000 replications) for the euteleost nodes are shown. (Hf) Heterodontus francisci; (Lc) Latimeria chalumnae; (Ps) Polypterus senegalus; (TrAb) Takifugu rubripes HoxAβ; (DrAb) Danio rerio HoxAβ; (TrAa) T. rubripes HoxAα; (DrAa) D. rerio HoxAα. (B-E) Character reconstructions of exon 1 coding sequences under constraint trees for HoxA13, HoxA11, HoxA10, and HoxA2, respectively. Taxa abbreviations are as in A above, including (Hs) Homo sapiens. The indicated substitutions are only those that map unambiguously to the branches of the trees. The numbers of analyzedamino acidresidues andassumedunambiguous changes for each gene are (B) HoxA13, 244 amino acids and 196 steps; (C) HoxA11, 192 amino acids and 151 steps; (D) HoxA10, 24 amino acids and 35 steps; (E) HoxA2, 105 amino acids and 59 steps.
Noncoding DNA Sequence Comparisons Suggest the Single HoxA Cluster in Bichir Is Mosaic Between Human and Derived Ray-Finned Fishes
We next examined the evolution of noncoding sequences to determine whether the single HoxA cluster of bichir exhibits the dramatic loss of noncoding sequence conservation observed in the duplicated HoxA clusters of euteleosts (Chiu et al. 2002). We used a new software package, Tracker (Prohaska et al. 2003), to identify conserved Hox noncoding sequence tracts (potential cis-regulatory elements) in all the taxa shown in Figure 2A. Here, we report phylogenetic footprint clusters (PFCs; Chiu et al. 2002) of each HoxA cluster spanning from Evx1 to HoxA1 (Fig. 2A) counted by Tracker in pairwise sequence alignments of intergenic sequences between orthologous HoxA gene pairs (Supplemental Table 1). Figure 2B diagrammatically illustrates the number of PFCs that each pair of sequences (e.g., human, bichir) has in common. Several notable patterns of conservation of PFCs are evident. First, the HoxA clusters of human and horn shark share the most PFCs, consistent with our earlier findings (Chiu et al. 2002). Second, the bichir HoxA cluster shares 44 PFCs with horn shark and 40 PFCs with human; these numbers are much higher than the number of PFCs shared between any of the euteleosts and human and/or shark. A subset of these PFCs conserved between bichir and human are completely absent in euteleosts (Fig. 2A). Two extensive PFCs, of 11 total, shared exclusively between bichir and euteleosts (c49, c190 of Supplemental Table 1 available online at www.genome.org) are illustrated in Figure 2A. These observations indicate that the bichir exhibits a mosaic pattern of conservation of HoxA noncoding sequence tracts with human and derived actinopterygians. Third, the common ancestor of bichir and teleosts possessed cis-sequence elements that were largely acquired before the duplication and were retained in the duplicated HoxAβ clusters only. Most of the changes are located between Evx1 and HoxA13 (Fig. 2A; c36,37,38,47,49,50,54 of Supplemental Table 1), indicating that this modification was a single event. Fourth, 24 PFCs are shared only among the different euteleosts examined (Fig. 2A; Supplemental Table 1). Based on parsimony, these PFCs were uniquely derived in the stem lineage of euteleosts prior to the HoxA cluster duplication. We conclude that the loss of conservation in zebrafish, bass, and pufferfish is a derived state for teleost fishes and that the bichir is already more derived in its noncoding sequence than human and shark.
Conclusions
In this study we found independent lines of evidence indicating that the bichir has one HoxA cluster that is mosaic in its patterns of noncoding sequence conservation and gene retention relative to the single HoxA cluster of human and shark, and the duplicated HoxAα and HoxAβ clusters of zebrafish, pufferfish, and striped bass. Our findings show that the duplication that produced additional HoxA clusters in derived actinopterygians occurred after the bichir diverged from the rest of the ray-finned fishes (Fig. 1). Hence, it is important to investigate the genome situation in additional basal ray-finned fishes such as the paddlefish, bowfin, sturgeon, and gar, as well as basal teleosts such as the eel (Fig. 1). Some of the changes that distinguish zebrafish and fugu from human, however, were acquired prior to the teleost HoxA cluster duplication, as shown in the pattern of cis-sequence conservation between teleosts and bichir. Hence the divergence of actinopterygian Hox clusters from the gnathostome archetype already began before the duplication that produced duplicate Hox clusters in teleost fishes.
In this genome-enabled era, two major problems remain: how evolutionary forces such as mutation, duplication, and selection shape genomes, and how genomic variation relates to phenotypic alterations in different lineages. This study reinforces the view that the ray-finned fishes provide good opportunities for addressing these challenges. Comparisons on a genome-wide basis are urgently needed to understand the complex interactions between evolution of genomes and body plans in this speciose assemblage.
METHODS
Construction and Screening of the Bichir BAC Genomic Library
A 5× coverage BAC genomic library, with an average insert size of 130 kb, was constructed for the bichir (P. senegalus) as described (Strong et al. 1997; Osoegawa et al. 1998). High-density 5 × 5 arrayed filters were made by the RZPD (http://www.rzpd.de). Hybridization using nonradioactive DIG-labeled probes was done following methods described in Chiu et al. (2000b). The first screen of this library was carried out using a pool of bichir (gift of C. Ledje and F. Ruddle; Ledje and Ruddle 2002) and coelacanth (gift of T. Powers [Benaroya Research Institute at Virginia Mason, Seattle, WA] and C. Amemiya) homeobox sequences isolated in genome-wide PCR surveys. This screen identified nine BAC clones of P. senegalus. A PCR survey of each BAC clone was done using a degenerate homeobox primer pair (334, 5′-GARYTIGARAARGARTTY-3′; 335, 5′-ICKICKRTTYT GRAACAA-3′). One clone containing the posterior part of the HoxA (Evx1, HoxA13, HoxA11, HoxA10) cluster was identified. Bichir HoxB, HoxC, and HoxD cluster sequences are presently being analyzed (C.-H. Chiu, K. Dewar, and P.F. Stadler, unpubl.).
To find overlapping BAC clone(s) that contain the rest of the HoxA cluster, PCR primers specific to bichir HoxA10 exon 1 were designed (PseA10F, 5′-ATGTCATGCTCAGATAGCCCGG-3′; PseA10R, 5′-TGATGTTTTGTATAAGGACATCG-3′). Using these primers, exon 1 of HoxA10 was amplified from whole-genomic DNA of the bichir (P. senegalus), cloned, and five independent colonies were sequenced on both strands. All five clones from genomic DNA were identical in sequence to each other and to the HoxA10 sequence on the BAC clones isolated using hybridization (described above). The PseA10F/R PCR primer pair was then used to PCR-screen DNA pools of the bichir BAC library (the library consists of 216 pools, with each pool equivalent to one 384-well plate). The screen with HoxA10 primers yielded two clones, the original BAC clone described above (containing Evx1 to HoxA10) and one overlapping clone, spanning HoxA10 to HoxA1. These two clones overlap ∼4.0 kb encompassing the entire HoxA10 locus with 5′- and 3′-flanking sequences. Concurrent with PCR screening of the library pools, the 5 × 5 high-density filters of the bichir BAC library were also screened by hybridization, using bichir-specific HoxA10 exon 1 (from genomic DNA) as probe. Hybridization yielded the identical two HoxA cluster clones identified above that overlap over HoxA10. Finally, bichir HoxA2 exon 1 was amplified from genomic DNA (P. senegalus) using universal primers we designed (PseA2UF, 5′-AATAG TCAGCCR TCGCTYGCTGAG-3′; PseA2UR, 5′-CTTGGAHGCYTT TTTCTCKTTC-3′). The bichir HoxA2 exon 1 PCR product was cloned, sequenced, and used to screen the bichir BAC library filters by hybridization. Only one strong hybridization signal was detected, corresponding to the same BAC clone (spanning HoxA10 to HoxA1) identified above.
Phylogenetic Analyses
Phylogenetic reconstructions were conducted using the neighbor-joining method (Saitou and Nei 1987). Character reconstructions were done using MacClade Version 4.03 (Maddison and Maddison 2001) under constraint trees that retain bony fishes (ray-finned fishes and lobe-finned fishes) as monophyletic to the exclusion of horn shark. The indicated substitutions are only those that map unambiguously to the branches of the trees. Supplemental Table 2 contains the alignments used for phylogenetic tree reconstructions.
Noncoding Sequence Analyses
The program Tracker is based on BLAST (Altschul et al. 1990) searches of all possible sequence pairs. The resulting list of pairwise sequence alignments is then assembled into groups of partially overlapping regions that are subsequently passed through several filtering steps. There are three levels of resolution of conserved noncoding sequences that are analyzed. (1) Individual phylogenetic footprints (PFs) are blocks of 6 bp or more of DNA sequence 100% conserved in taxa that have an additive evolutionary time of 250 million years (Tagle et al. 1988). PFs are considered to be putative transcription-factor-binding sites. (2) Cliques (CCs) are groups of contiguous footprints (Prohaska et al. 2003). (3) Phylogenetic footprint clusters (PFCs; Chiu et al. 2002) are composed of at least two PFs that are separated by <100 nt. CCs and PFCs are considered to be putative enhancer/promoter regions.
Acknowledgments
We are grateful to April Cook and the Whitehead Institute Center for Genome Research sequencing teams for the complete sequencing of the bichir HoxA cluster; and Tom Powers and Allan Force for discussion of portions of this manuscript. C.-H.C. and G.P.W. thank Austin Hughes for providing the SCR3 software. Funding for this research is gratefully acknowledged from the Busch Biomedical Research Fund to C.-H.C.; NSF IBN-9905403 to F.R. and G.P.W.; IBN-9905403 and NIH R24-RR14085 to C.T.A.; and DFG Bioinformatics Initiative BIZ 6/1-2 and FWF P#15893 to P.F.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1712904.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- Amores, A., Force, A., Yan, Y.-L., Jolly, L., Amemiya, C.T., Fritz, A., Ho, R.K., Langeland, J., Prince, V., Wang, Y.L., et al. 1998. Zebrafish hox clusters and vertebrate genome evolution. Science 282: 1711-1714. [DOI] [PubMed] [Google Scholar]
- Amores, A., Suzuki, T., Yan, Y-L., Pomeroy, J., Singer, A., Amemiya, C.T., and Postlethwait, J.H. 2003. Developmental roles of pufferfish Hox clusters and genome evolution in ray-fin fish. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301-1310. [DOI] [PubMed] [Google Scholar]
- Bartsch, P. and Britz, R. 1997. A single micropyle in the eggs of the most basal living actinopterygian fish, Polypterus (Actinopterygii, Polypteriformes). J. Zool. London 241: 589-592. [Google Scholar]
- Bemis, W.E., Findeis, E.K., and Grande, L. 1997. An overview of Acipenseriformes. Env. Biol. Fish 48: 25-71. [Google Scholar]
- Chiu, C.-H., Nonaka, D., Xue, L., Amemiya, C.T., and Wagner, G.P. 2000a. Evolution of Hoxa-11 in lineages phylogenetically positioned along the fin-limb transition. Mol. Phyl. Evol. 17: 305-316. [DOI] [PubMed] [Google Scholar]
- Chiu, C.-H., Amemiya, C.T., Carr, J.L., Bhargava, J., Hwang, J.K., Shashikant, C.S., Ruddle, F.H., and Wagner, G.P. 2000b. A recombinogenic targeting method to modify large-inserts for cis-regulatory analysis in transgenic mice: Construction and expression of a 100-kb, zebrafish Hoxa-11b-lacZ reporter gene. Dev. Genes Evol. 210: 105-109. [DOI] [PubMed] [Google Scholar]
- Chiu, C.-H., Amemiya, C.T., Dewar, K., Kim, C.-B., Ruddle, F.H., and Wagner, G.P. 2002. Molecular evolution of the HoxA cluster in the three major gnathostome lineages. Proc. Natl. Acad. Sci. 99: 5492-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland, P.W. and Garcia-Fernandez, J. 1996. Hox genes and chordate evolution. Dev. Biol. 173: 382-395. [DOI] [PubMed] [Google Scholar]
- Hughes, C.L. and Kaufman, T.C. 2002. Hox genes and the evolution of the arthropod body plan. Evol. Dev. 4: 459-499. [DOI] [PubMed] [Google Scholar]
- Inoue, J.G., Masaki, M., Tsukamoto, K., and Nishida, M. 2003. Basal actinopterygian relationships: A mitogenomic perspective on the phylogeny of the “ancient fish.” Mol. Phyl. Evol. 26: 110-120. [DOI] [PubMed] [Google Scholar]
- Kappen, C., Schughart, K., and Ruddle, F.H. 1989. Two steps in the evolution of Antennapedia-class vertebrate homeobox genes. Proc. Natl. Acad. Sci. 86: 5459-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C.B., Amemiya, C.T., Bailey, W., Kawasaki, K., Mezey, J., Miller, W., Minoshima, S., Shimizu, N., Wagner, G.P., and Ruddle, F.H. 2000. Hox cluster genomics in the horn shark, Heterodontus francisci. Proc. Natl. Acad. Sci. 97: 1655-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, E.G., Lam, K., Christoffels, A., Erdmann, M.V., Brenner, S., and Venkatesh, B. 2003. Hox gene clusters in the Indonesian coelacanth, Latimeria menadoensis. Proc. Natl. Acad. Sci. 100: 1084-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, H.L.V., Lecointre, G., and Perasso, R. 1993. A 28S rRNA-based phylogeny of the gnathostomes: First steps in the analysis of conflict and congruence with morphologically based cladograms. Mol. Phyl. Evol. 2: 31-51. [DOI] [PubMed] [Google Scholar]
- Ledje, C. and Ruddle, F.H. 2002. Characterization of Hox genes in the bichir, Polypterus palmas. J. Exp. Zool. 294: 107-111. [DOI] [PubMed] [Google Scholar]
- Lufkin, T. 1996. Transcriptional control of Hox genes in the vertebrate nervous system. Curr. Opin. Genet. Dev. 6: 575-580. [DOI] [PubMed] [Google Scholar]
- Maddison, D.R. and Maddison, W.P. 2001. MacClade 4: Analysis of phylogeny and character evolution. Version 4.03. Sinauer Associates, Sunderland, MA.
- Malaga-Trillo, E. and Meyer, A. 2001. Genome duplications and accelerated evolution of Hox genes and cluster architecture in teleost fishes. Amer. Zool. 41: 676-686. [Google Scholar]
- McGinnis, W. and Krumlauf, R. 1992. Homeobox genes and axial patterning. Cell 68: 283-302. [DOI] [PubMed] [Google Scholar]
- Misof, B.Y. and Wagner, G.P. 1996. Evidence for four Hox clusters in the killifish, Fundulus heteroclitus (Teleostei). Mol. Phyl. Evol. 5: 309-322. [DOI] [PubMed] [Google Scholar]
- Naruse, K., Fukamachi, S., Mitani, H., Kondo, M., Matsuoka, T., Kondo, S., Hanamura, N., Morita, Y., Hasegawa, K., Nishigaki, R., et al. 2000. A detailed linkage map of medaka, Oryzias latipes: Comparative genomics and genome evolution. Genetics 154: 1773-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, J.S. 1994. Fishes of the world, 3rd ed. John Wiley & Sons, New York.
- Osoegawa, K., Woon, P.Y., Zhao, B., Frengen, E., Tateno, M., Catanese, J.J., and de Jong, P. 1998. An improved approach for construction of bacterial artificial chromosome libraries. Genomics 52: 1-8. [DOI] [PubMed] [Google Scholar]
- Patterson, C. 1982. Morphology and interrelationships of primitive actinopterygian fishes. Am. Zool. 22: 241-259. [Google Scholar]
- Postlethwait, J.H., Yan, Y.L., Gates, M.A., Horne, S., Amores, A., Brownlie, A., Donovan, A., Egan, E.S., Force, A., Gong, Z., et al. 1998. Vertebrate genome evolution and the zebrafish gene map. Nat. Genet. 18: 345-349. [DOI] [PubMed] [Google Scholar]
- Prohaska, S., Fried, C., Flamm, C., Wagner, G.P., and Stadler, P.F. 2003. Surveying phylogenetic footprints in large gene clusters: Applications to Hox cluster duplications. Santa Fe Institute (SFI) working paper 03-02-011 (in press). [DOI] [PubMed]
- Robinson-Rechavi, M., Marchand, O., Escriva, H., Bradet, P.-L., Zelus, D., Hughes, S., and Laudet, V. 2001. Euteleost fish genomes are characterized by expansion of gene families. Genome Res. 11: 781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruddle, F.H., Amemiya, C.T., Carr, J.L., Kim, C.B., Ledje, C., Shashikant, C.S., and Wagner, G.P. 1999. Evolution of chordate hox gene clusters. Ann. NY Acad. Sci. 870: 238-248. [DOI] [PubMed] [Google Scholar]
- Saitou, N. and Nei, M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 407-425. [DOI] [PubMed] [Google Scholar]
- Santini, S., Boore, J.L., and Meyer, A. 2003. Evolutionary conservation of regulatory elements in vertebrate hox gene clusters. Genome Res. 13: 1111-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell, E.A., Scemama, J.L, and Stellwag, E.J. 1999. Genomic organization of the Hoxa4-Hoxa10 region from Morone saxatilis: Implications for Hox gene evolution among vertebrates. J. Exp. Zool. 285: 41-49. [PubMed] [Google Scholar]
- Stellwag, E. 1999. Hox gene duplication in fish. Semin. Cell Dev. Biol. 10: 531-540. [DOI] [PubMed] [Google Scholar]
- Strong, S.J., Ohta, Y., Litman, G.W., and Amemiya, C.T. 1997. Marked improvement of PAC and BAC cloning is achieved using electroelution of pulsed-field gel-separated partial digests of genomic DNA. Nucleic Acids Res. 25: 3959-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagle, D.A., Koop, B.F., Goodman, M., Slightom, J.L., Hess, D.L., and Jones, R.T. 1988. Embryonic ε and γ globin genes of a prosimian primate (Galago crassicaudatus). Nucleotide and amino acid sequences, developmental regulation and phylogenetic footprints. J. Mol. Biol. 203: 439-455. [DOI] [PubMed] [Google Scholar]
- Taylor, J.S., van der Peer, Y., Braasch, I., and Meyer, A. 2001. Comparative genomics provides evidence for an ancient genome duplication event in fish. Phil. Trans. R. Soc. Lond. B 356: 1661-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh, B., Erdmann, M.V., and Brenner, S. 2001. Molecular synapomorphies resolve evolutionary relationships of extant jawed vertebrates. Proc. Natl. Acad. Sci. 98: 11382-11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter, J., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.rzpd.de; RZPD.