Abstract
Human immunodeficiency virus Rev facilitates the cytoplasmic accumulation of viral RNAs that contain a Rev binding site. A human Rev-interacting protein (hRIP) was originally identified based on its ability to interact with the Rev nuclear export signal (NES) in yeast two-hybrid assays. To date, however, the function of hRIP and a role for hRIP in Rev-directed RNA export have remained elusive. Here we ablate hRIP activity with a dominant-negative mutant or RNA interference and analyze Rev function by RNA in situ hybridization. We find, unexpectedly, that in the absence of functional hRIP, Rev-directed RNAs mislocalize and aberrantly accumulate at the nuclear periphery, where hRIP is localized. In contrast, in the absence of Rev or the Rev cofactor CRM1, Rev-directed RNAs remain nuclear. We further show that the RNA mislocalization pattern resulting from loss of hRIP activity is highly specific to Rev function: the intracellular distribution of cellular poly(A)+ mRNA, nuclear proteins, and, most important, NES-containing proteins, are unaffected. Thus, hRIP is an essential cellular Rev cofactor, which acts at a previously unanticipated step in HIV-1 RNA export: movement of RNAs from the nuclear periphery to the cytoplasm.
Keywords: Rev, HIV-1, RNA export, hRIP
Animal viruses often encode regulatory proteins, which facilitate expression of their viral genes in the host cell. One such example is the human immunodeficiency virus type 1 (HIV-1) Rev protein, which uses a novel mechanism to regulate viral messenger RNA (mRNA) export from the nucleus. Rev is a sequence-specific RNA binding protein that facilitates the cytoplasmic accumulation of the intron-containing HIV-1 gag-pol and env mRNAs (Cullen 2002). Rev interacts with a cis-acting viral RNA element in these mRNAs designated the Rev responsive element (RRE; Pollard and Malim 1998; Hope 1999). Rev contains two distinct functional domains: an arginine-rich RNA binding motif (ARM) and a leucine-rich “effector” domain. Rev's ARM domain confers sequence-specific RNA binding, protein oligomerization, and nuclear targeting (Frankel and Young 1998). Rev's effector domain contains a leucine-rich nuclear export signal (NES), which binds the nuclear export receptor CRM1 (Weis 2002). Other cellular proteins have been shown to interact directly or indirectly with the Rev NES, including the kinesin-like Rev/Rex effector binding protein (REBP) and the eukaryotic initiation factor-5 alpha (eIF-5α; Bevec et al. 1996; Dayton 1996; Kjems and Askjaer 2000; Venkatesh et al. 2003).
The human Rev-interacting protein (hRIP), also known as hRab or Hrb, was identified using a yeast two-hybrid screen with Rev as the bait (Bogerd et al. 1995; Fritz et al. 1995). Additional studies demonstrated a strong correlation between the ability of Rev NES mutants to support Rev function in yeast and their ability to interact with hRIP in the two-hybrid assay (Stutz et al. 1996). However, subsequent genetic analyses indicate the Rev–hRIP interaction is indirect, and is likely bridged by CRM1 (Fritz and Green 1996; Henderson and Percipalle 1997; Neville et al. 1997). hRIP contains multiple phenylalanine-glycine (XXFG) repeat sequences reminiscent of the GLFG and XFXFG repeats found in most yeast and mammalian nuclear pore proteins also known as nucleoporins or “Nups” (Doye and Hurt 1997; Strambio-de-Castillia et al. 1999). The FG repeats in nucleoporins are believed to mediate protein–protein interactions, and in the case of hRIP, are required for its interaction with Rev in vivo. At its N terminus, hRIP contains a zinc finger motif (Krishna et al. 2003), which shares a high degree of sequence identity with a subclass of C4H2-type zinc-finger domains found in proteins involved in vesicular trafficking, intracellular protein localization, cytoskeletal rearrangement, RNA metabolism, or signal transduction (Cukierman et al. 1995; Kirchhausen 2000; Premont et al. 2000; Turner et al. 2001; Nie et al. 2002; Dubois et al. 2003). Within the central body of the protein are stretches of serine/threonine-rich sequences (S/TRR), several of which are substrates for a subset of cellular kinases, and thus, possible targets for regulation (Maraldi et al. 1999; Parker and Parkinson 2001; Biondi and Nebreda 2003). Lastly, there are several asparagine-proline-phenylalanine (SS/TN-PFXX) sequence motifs interspersed among the FG repeats within the most terminal third of the protein. Such NPF motifs are thought to mediate interactions with distinct members of the Eps 15 homology (EH)-domain protein family, which also interact with CRM1 and other nuclear components (Santolini et al. 1999; Hyman et al. 2000; Poupon et al. 2002). Recent microscopic and biochemical experiments indicate that hRIP is localized predominantly at the nuclear periphery, perhaps in a novel perinuclear compartment (Doria et al. 1999).
Based on its putative structural domains and intracellular distribution, hRIP has properties consistent with a role in RNA trafficking or localization. To date, however, the cellular function of hRIP and a role for hRIP in Rev function have remained elusive. Here we describe complementary genetic approaches to inactivate hRIP and analyze the effect of hRIP ablation on Rev function. We find that hRIP is, in fact, an essential cellular cofactor for HIV-1 Rev activity that acts at an unanticipated step in Rev-directed RNA export. Our results have direct relevance to Rev function and general implications for cellular RNA trafficking.
Results
A dominant-negative hRIP mutant interferes with Rev function
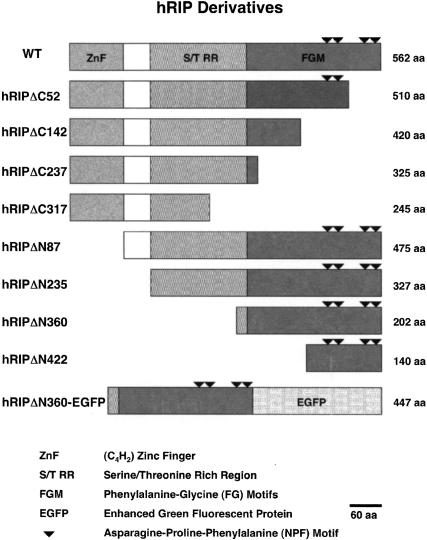
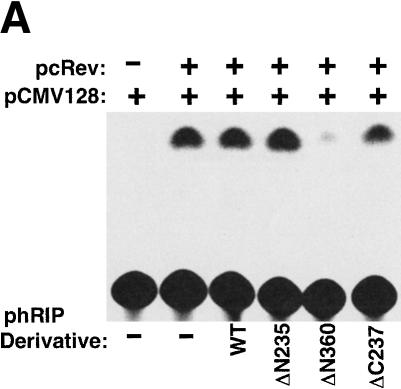
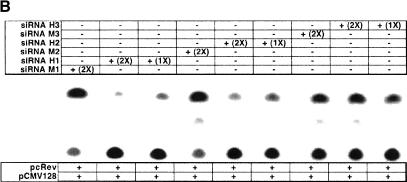
To analyze the function of hRIP, we devised a strategy similar to those traditionally used to study proteins of unknown function, namely, the identification and characterization of a dominant-negative form of hRIP. To accomplish this, we generated a series of N- and C-terminally truncated hRIP derivatives by the systematic removal of the putative hRIP functional domains (Fig. 1). Because hRIP has properties that are consistent with a cellular Rev cofactor, we tested the activity of these derivatives using a standard assay for Rev function (Hope et al. 1990). This mammalian cell transfection assay uses a translation-based reporter gene (pCMV128) that has a single intron containing both the HIV-1 RRE and the bacterial chloramphenicol acetyltransferase (CAT) cDNA as an indicator of Rev function. This intron is efficiently spliced, and therefore cells transfected with pCMV128 express only trace amounts of CAT enzyme activity (Fig. 2A, lane 1). Cotransfection with a Rev expression plasmid (pcRev; Malim et al. 1989) causes unspliced transcripts to enter the cytoplasm, and thus CAT expression is increased (Fig. 2A, lane 2). Most of these hRIP derivatives had no effect; we did, however, identify one, hRIPΔN360, which appeared to interfere with Rev function in this assay (Fig. 2A, lanes 3–5). This hRIP derivative, which retains the Nup-homology region of hRIP required for interaction with Rev in vivo, inhibited Rev function in a dose-dependent manner (Fig. 2B). Because hRIPΔN360 exerted a negative effect on Rev function in the presence of endogenous hRIP, we conclude that it acts as a trans-dominant inhibitor of hRIP function.
Figure 1.
Construction of hRIP derivatives. Schematic representation of N- and C-terminally truncated and fluorescent hRIP derivatives. (ZnF) C4H2-type zinc finger domain; (S/TRR) serine/threonine-rich region; (FGM) phenylalanine-glycine motifs; (arrowhead) asparagine-proline-phenylalanine (NPF) motifs; (EGFP) enhanced green fluorescent protein; (WT) hRIP; (aa) amino acids.
Figure 2.
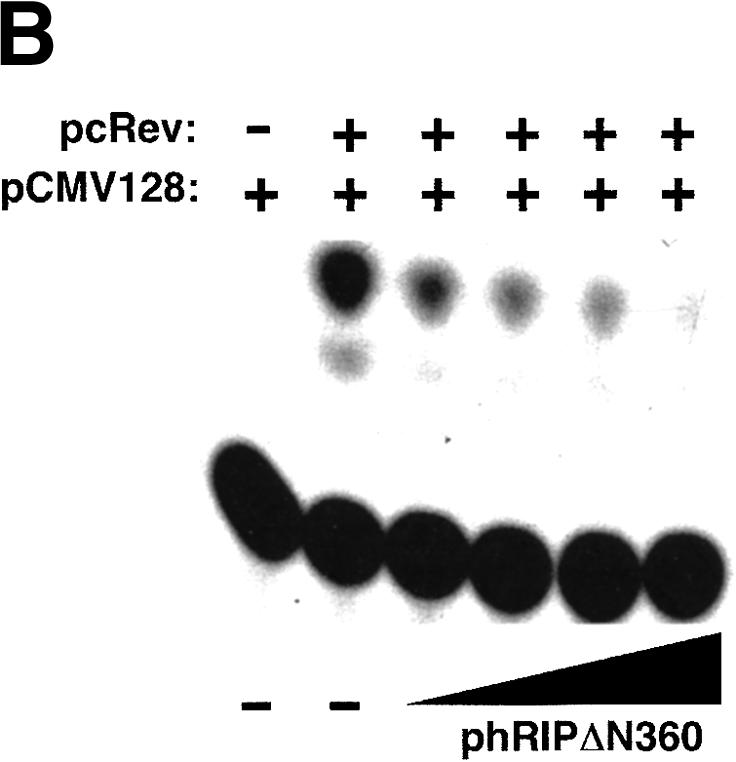
A dominant-negative hRIP mutant interferes with Rev function. (A) A transient transfection assay for Rev function. The CAT reporter plasmid pCMV128 and pCMV β-gal were cotransfected with the Rev expression plasmid pcRev or pcRev and the indicated hRIP expression plasmid into Cos-1 cells. CAT and β-galactosidase enzyme activities were assayed as described in Zapp et al. (1993). (–) Absence; (+) presence; (WT) hRIP. A dominant-negative hRIP mutant interferes with Rev function. (B) The hRIPΔN360 mutant inhibits Rev function in a dose-dependent manner. Cos-1 cells were cotransfected with pCMV128, pcRev, pCMV β-gal, and increasing amounts of phRIPΔN360. Enzyme activities were assayed as in (A). (–) Absence; (+) presence.
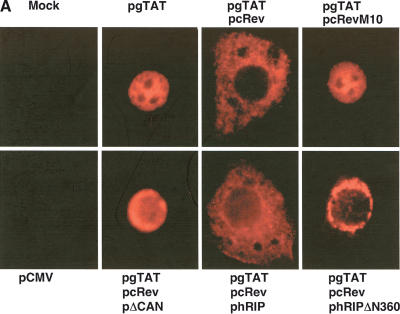
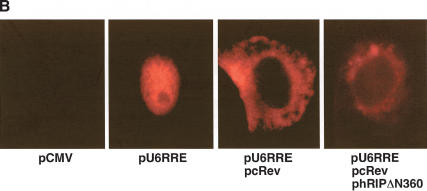
Figure 3.
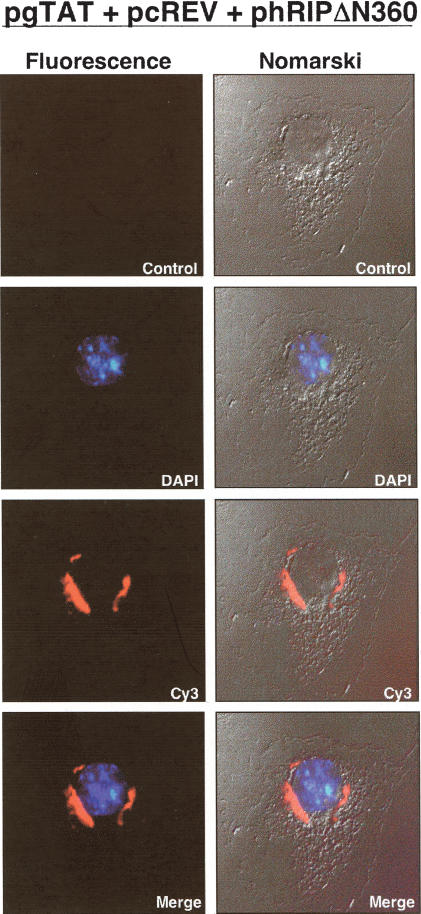
The hRIPΔN360 mutant causes Rev-directed RNAs to mislocalize and aberrantly accumulate at the nuclear periphery. (A) The subgenomic HIV-1 tat expression plasmid (pgTAT) was cotransfected with pcRev or pcRev and the indicated hRIP expression plasmid into Cos-1 cells. Control transfections contained pgTAT alone, pgTAT and pCMV, pgTAT and pRevM10, or pgTAT and pΔCAN. The intracellular distribution of tat RNA was analyzed by in situ hybridization using a Cy3-labeled RRE-specific probe. (B) The U6-RRE expression plasmid (pU6RRE) was cotransfected with pcRev, or pcRev and phRIPΔN360 into Cos-1 cells. Control transfections contained pCMV alone or pU6RRE and pCMV. The intracellular distribution of U6RRE RNA was analyzed as described in A.
To investigate the nature of the block exerted by hRIPΔN360 on Rev function, we analyzed the intracellular distribution of Rev-directed RNAs using in situ hybridization. Cos-1 cells were cotransfected with the subgenomic HIV-1 tat expression plasmid (pgTAT; Malim et al. 1989) and pcRev, in the presence of phRIP or phRIPΔN360. To confirm the specificity of Rev function in this assay, we cotransfected cells with pgTAT and a plasmid that expresses the RevM10 trans-dominant inhibitor (pcRevM10; Malim et al. 1989). Alternatively, cells were cotransfected with pgTAT and pCMVΔCAN, a plasmid that expresses a truncated form of the nucleoporin CAN/Nup214 (ΔCAN). ΔCAN blocks the ability of CRM1 to interact with the nuclear pore complex (NPC) and inhibits the nuclear export of Rev (Fornerod et al. 1997; Bogerd et al. 1998; Yi et al. 2002). The intracellular distribution of tat RNA was visualized using a fluorochrome-labeled oligonucleotide probe complementary to stem loop IIB of the HIV-1 RRE. In the absence of Rev, tat RNAs were nuclear localized (Fig. 3A, first row, second panel). Rev promoted the cytoplasmic accumulation of tat mRNAs in an NES- and CRM1-dependent manner (Fig 3A, first row, third and fourth panels; bottom row, second panel). Cells transfected with reagent alone or an empty DNA vector (pCMV) contained no fluorescent signals after hybridization with the probe, confirming the detection was specific for tat RNAs (Fig. 3A, first and second rows, first panel). Overexpression of hRIP had no discernible effect on the cytoplasmic accumulation of tat RNAs (bottom row, third panel). In contrast, the tat mRNAs were mislocalized and aberrantly accumulated at the nuclear periphery in the presence of hRIPΔN360 (Fig. 3A, second row, fourth panel).
Using the same approach, we tested whether hRIPΔN360 could interfere with the cytoplasmic accumulation of any Rev-directed RNA. We inserted a high-affinity Rev binding site into the U6 small nuclear ribonucleoprotein RNA (U6snRNA) to generate a sequence-minimized RNA polymerase III (pol III)-derived transcript (U6RRE), which is exported from the nucleus in a Rev-directed manner. When expressed from its cognate promoter, the U6RRE transcript is present at high levels in the nuclei of mammalian cells (Fig. 2B, second panel). Cos-1 cells were cotransfected with a U6RRE RNA expression plasmid (pU6RRE) and pcRev in the absence or presence of phRIPΔN360, and the intracellular distribution of the U6RRE RNA analyzed as in the previous experiment. Figure 2B shows that Rev efficiently promoted the cytoplasmic accumulation of U6RRE transcripts in the absence of hRIPΔN360 (third panel). In the presence of hRIPΔN360, however, these RNAs were mislocalized and aberrantly accumulated at the nuclear periphery. Thus, the intracellular distribution of U6RRE RNAs was strikingly similar to that of tat mRNAs in the presence of hRIPΔN360.
Next, we used differential interference contrast (DIC) microscopy to define the perinuclear localization of Rev-directed RNAs more precisely. Cos-1 cells were cotransfected with pgTAT, pcRev, and phRIPΔN360 and in situ RNA hybridization analysis was performed as in the previous experiments. The results in Figure 4 clearly show that the accumulation of Rev-directed RNAs is on the outside of the nucleus (Fig. 4, third and fourth rows). Collectively, our results indicate that hRIPΔN360 exerts its inhibitory activity on RRE-containing RNAs at the cytoplasmic side of the NPC. This previously unanticipated step further implies an extended role for Rev in the movement of Rev-directed RNAs from the nuclear periphery to the cytoplasm.
Figure 4.
Rev-directed RNAs accumulate on the outside of the nucleus in the presence of the hRIPΔN360 mutant. Cos-1 cells were transfected with the indicated plasmids and RNA localization analyzed by fluorescent in situ hybridization as in the previous experiment (Fluorescence, left column) and differential interference contrast (DIC) microscopy (Nomarski, right panel). (Control panels) No DAPI, no probe; (DAPI) (+) DAPI; (Cy3) (+) probe; (merge) (+) DAPI and probe.
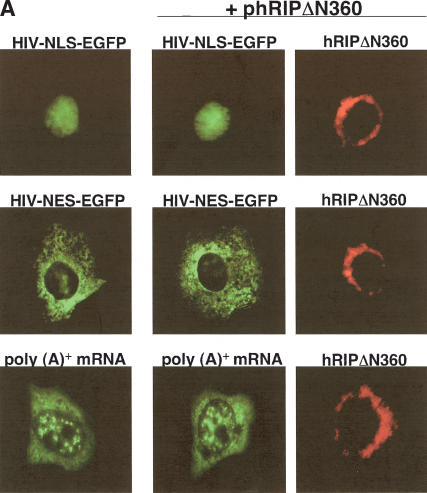
We next performed a series of experiments to confirm that the inhibition exerted by hRIPΔN360 on Rev-directed RNA export was specific. The presence of both a nuclear localization signal (NLS) and an NES enables Rev to shuttle continuously between the nucleus and cytoplasm, an activity required for its function (Kalland et al. 1994; Meyer and Malim 1994; Richard et al. 1994). In view of this requirement, we examined whether hRIPΔN360 could interfere with the general NLS-dependent protein import or NES-dependent protein export pathways. Cos-1 cells were cotransfected with phRIPΔN360 and an HIV-NLS- or HIV-NES-containing enhanced green fluorescent protein (EGFP) expression plasmid (pHIV-NLS-EGFP-NLS, pHIV-NES-EGFP; Dupont et al. 1999). The intracellular distributions of these HIV-EGFP fusion proteins and hRIPΔN360 were analyzed by fluorescence microscopy. Consistent with previous findings, HIV-NLS-EGFP was localized in the nucleus, and HIV-NES-EGFP in the cytoplasm of transfected cells (Fig. 5A). Importantly, the intracellular distributions of these compartment-restricted reporter proteins were unaffected by hRIPΔN360, which was localized at the nuclear periphery (Fig. 5A, first and second rows, center and right panels). Similarly, hRIPΔN360 had no effect on the intracellular distribution of ectopically expressed Rev, endogenous CRM1, and other NES-containing cellular proteins in subsequent localization studies (data not shown). These results provide evidence that hRIPΔN360's ability to interfere with Rev function is not simply due to a general block in the transport of NLS- or NES-containing proteins.
Figure 5.
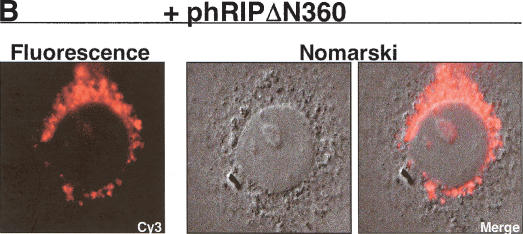
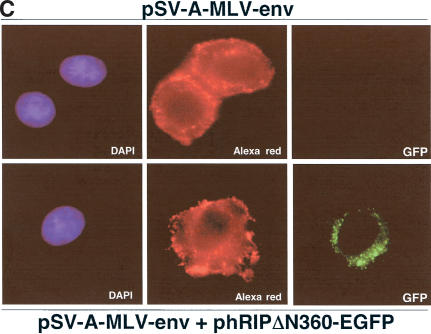
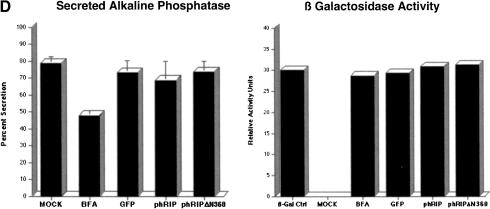
The hRIPΔN360 mutant specifically inhibits Rev-directed RNA export. (A) The hRIPΔN360 mutant does not affect the intracellular distribution of NLS- or NES-containing proteins or total poly(A)+ mRNA in mammalian cells. Cos-1 cells were transfected with the indicated EGFP-fusion reporter plasmid in the absence or presence of phRIPΔN360 (first and second rows, left and center panels). HeLa cells were transfected with phRIPΔN360 and the intracellular distribution of poly(A)+ mRNA was analyzed by in situ hybridization using an Alexa Green-labeled oligo dT52 probe (third row, left and center panels). The intracellular distribution of hRIPΔN360 was analyzed by indirect immunofluorescence microscopy using anti-HA (primary) and Cy3-labeled anti-mouse (secondary) antibodies (right panels). (B) The hRIPΔN360 mutant is localized at the nuclear periphery in mammalian cells. Cos-1 cells were transfected with phRIPΔN360 and analyzed by indirect immunofluorescence as in A (Fluorescence, left panel) and DIC microscopy (Nomarski, center and right panels). (C) The hRIPΔN360 mutant does not affect the intracellular distribution of MLV envelope glycoproteins in mammalian cells. HeLa cells were transfected with an MLV-envelope expression plasmid (pSV-A-MLV-env) alone (first row) or cotransfected with phRIPΔN360-EGFP (second row). The intracellular distribution of nascent MLV envelope glycoproteins was analyzed by indirect immunofluorescence microscopy using anti-MLV (primary) and anti-rat Alexa Red (secondary) antibodies. (D) The hRIPΔN360 mutant does not affect constitutive secretion of SEAP in mammalian cells. 293-T cells were cotransfected with the indicated plasmid in the presence of pCMV β-gal. Untransfected cells were incubated with Brefeldin A (BFA) as a positive control for SEAP inhibition. Endogenous SEAP activity (left panel) and β-galactosidase expression (right panel) were monitored using standard enzyme assays. SEAP secretion levels were quantified as the ratio of SEAP activity in culture supernatants to the sum of SEAP activity in both the culture supernatants and cell lysates.
We also examined whether hRIPΔN360 could interfere with the general mRNA export pathway. HeLa cells were transfected with pCMV or phRIPΔN360 in the presence of β-galactosidase expression plasmid (pCMV β-gal). The intracellular distribution of total cellular poly(A)+ mRNA was analyzed by in situ hybridization with a fluorochrome-labeled oligonucleotide dT(52) probe. In contrast to our findings for Rev-directed RNAs, there was no significant change in the intracellular distribution of poly(A)+ mRNA in the presence of hRIPΔN360 (Fig. 5A, third row, left and center panels). Similarly, the cytoplasmic accumulation of heat shock protein 70 (hsp70) mRNA or RNA containing the Mason–Pfizer monkey virus (MPMV) constitutive transport element (CTE) were also unaffected by hRIPΔN360 (data not shown). These results provide additional evidence that the nature of the inhibition exerted by hRIPΔN360 on Rev function is specific.
Our in situ hybridization assays revealed that the mislocalized Rev-directed RNAs have a similar intracellular distribution to that of hRIPΔN360 (Figs. 4 and 5B). Given hRIPΔN360's proximity to the endoplasmic reticulum (ER), Golgi apparatus, and vesicular structures of the endosome-lysosome system, we considered whether its intracellular location could affect cellular processes mediating vesicular transport and protein secretion. Disruption of such trafficking pathways might prevent movement of Rev-directed RNAs from the nuclear periphery to the cytoplasm. To explore this possibility, we tested whether hRIPΔN360 could affect the intracellular distribution of the murine leukemia virus envelope glycoprotein (MLV-Env), an integral membrane protein that uses cellular vesicular pathways for transport from the trans Golgi network (TGN) to the plasma membrane (Miller et al. 1994). HeLa cells were transfected with an MLV-Env expression plasmid (pSA-A-MLV-env; Landau et al. 1991) in the absence or presence of an hRIPΔN360-EGFP expression plasmid (phRIPΔN360-EGFP). Similar to the untagged derivative, hRIPΔN360-EGFP causes Rev-directed RNAs to mislocalize and aberrantly accumulate at the nuclear periphery of transfected cells (data not shown). The intracellular distribution of nascent MLV-Env proteins was analyzed using indirect immunofluorescence microscopy with anti-MLV-Env antibodies (Evans et al. 1990). Consistent with previous reports, the MLV-Env proteins were localized predominantly at the plasma membrane (Fig. 5C, first row). hRIPΔN360-EGFP has no discernible effect on the intracellular distribution of the MLV-Env proteins (Fig. 5C, second row). Importantly, the intracellular localization of hRIPΔN360-EGFP was spatially distinct from that of MLV-Env. These results further indicate that hRIPΔN360-GFP failed to disrupt the cytoplasmic accumulation and expression of MLV-env mRNAs.
As a complementary approach, we examined whether hRIPΔN360 could block an early step in the constitutive secretory pathway using a secreted alkaline phosphatase assay (SEAP). In brief, 293T cells, which secrete appreciable levels of SEAP (Pertea et al. 2003), were transfected with pCMV, phRIP, phRIPΔN360, or pEGFP in the presence of pCMV β-gal, and endogenous SEAP activity was monitored using a standard chromogenic enzyme assay (Liu et al. 2001). To control the specificity of inhibition, we treated mock transfected cells with Brefeldin A (BFA), a fungal toxin that selectively disrupts trafficking through the Golgi (Chardin and McCormick 1999). Figure 5D (left panel) shows the constitutive SEAP secretion in 293T was reduced following a mild BFA treatment. In contrast, SEAP levels were unchanged in cells expressing hRIP or the hRIPΔN360 mutant. The cytoplasmic accumulation and expression of the internal control β-galactosidase mRNA was also unaffected in the presence of hRIPΔN360 (Fig. 5D, right panel). Collectively, these results indicate that the perinuclear presence of hRIPΔN360 does not interfere with vesicular trafficking pathways used for intracellular distribution of integral membrane proteins or constitutive protein secretion. Thus, we conclude that the inhibition exerted by hRIPΔN360 on Rev-directed RNA export is specific.
Ablation of hRIP activity by RNA interference results in the loss of Rev function
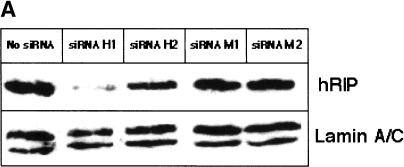
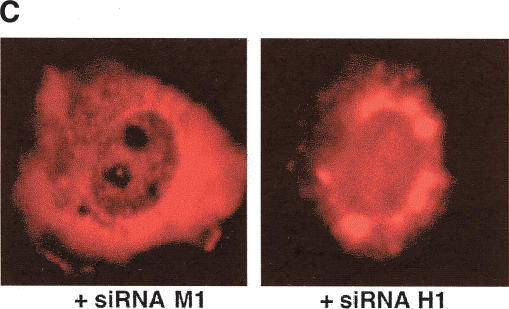
To further substantiate the requirement for hRIP in Rev function, we examined the intracellular distribution of Rev-directed RNAs in cells depleted of endogenous hRIP by RNA interference (Elbashir et al. 2001, 2002). Mammalian cells were transfected with synthetic small interfering RNA duplexes (siRNAs) homologous to nucleotides 1593–1613, 1106–1126, or 463–483 of the hRIP coding sequence (hRIP-specific siRNA HI, H2, and H3, respectively). Alternatively, cells were transfected with siRNAs harboring a single base pair mismatch within the same hRIP sequences (mutant siRNA M1, M2, and M3) using Oligofectamine. Cellular hRIP levels were analyzed by Western blotting of cell extracts from the bulk culture using anti-hRIP polyclonal antibodies. The results in Figure 6A show that hRIP was efficiently depleted following treatment with H1 siRNAs, but unaffected by Oligofectamine alone (first row, lanes 1 and 2). Depletion of cellular hRIP was also achieved, albeit to a lesser extent, using H2 siRNAs (Fig. 6A, lane 3), whereas the H3 or M3 siRNA had no effect on cellular hRIP levels (data not shown). Importantly, transfection of the M1 or M2 siRNAs failed to reduce the level of cellular hRIP (Fig. 6A, lanes 4 and 5). Additionally, the level of endogenous control protein Lamin A/C was unchanged by any siRNA treatment, further demonstrating the RNA interference is specific for hRIP expression (Fig. 6A, second row). Using these optimized conditions, we tested whether ablation of hRIP activity could interfere with Rev function. Cells were treated with the hRIP-specific or mutant siRNAs, then cotransfected with pCMV128 and pcRev or pgTAT and pcRev. CAT activity and intracellular RNA localization were analyzed as in previous experiments. Figure 6B shows that hRIP-depleted cells failed to support Rev function in this assay. Moreover, we observed a dose-dependent correlation between the level of hRIP depletion and the loss of Rev function (Fig. 6B, lanes 2, 3, 5, and 6). Most important, in situ hybridization analysis revealed that Rev-directed RNAs were mislocalized and aberrantly accumulated at the nuclear periphery in hRIP-depleted cells (Fig. 6C). Thus, the intracellular distribution of Rev-directed RNAs in hRIP-depleted cells was strikingly similar to that observed in the presence of the hRIP dominant-negative mutant. It is also worth noting that ablation of hRIP activity by RNA interference had no effect on the intracellular distribution of the HIV-based and cellular NLS- or NES-containing proteins or the internal control β-galactosidase mRNA (data not shown). Collectively, our results indicate that hRIP is an essential cellular cofactor for Rev-directed RNA export, which acts at a previously unanticipated step: movement of RNAs from the nuclear periphery to the cytoplasm.
Figure 6.
Ablation of hRIP activity inhibits HIV Rev function by mislocalizing Rev-directed RNAs to the nuclear periphery. (A) Depletion of cellular hRIP using RNA interference. HeLa cells were transfected with hRIP-specific (H1 or H2) or mutant siRNA duplexes (M1 or M2) using Oligofectamine. Control cells were transfected with Oligofectamine alone (no siRNA). hRIP and Lamin A/C levels in cell lysates prepared from the bulk culture were analyzed by Western blotting using goat anti-hRIP or anti-Lamin A/C polyclonal antibodies, respectively, and visualized by ECL. (B) Ablation of hRIP activity inhibits Rev function in mammalian cells. HeLa cells treated with the indicated siRNA duplexes were cotransfected with pCMV128, pcREV, and pCMV β-gal. CAT and β-galactosidase enzyme activities were assayed as in Figure 2,Figure 2. (1X) 50 nM; (2X) 100 nM; (–) absence; (+) presence. (C) Rev-directed RNAs mislocalize and aberrantly accumulate at the nuclear periphery of hRIP-depleted cells. HeLa cells treated with siRNA M1 or H1 were cotransfected with pgTAT and pcRev and RNA localization analyzed as in Figure 3,Figure 3.
Discussion
We have determined a function for the cellular HIV-1 Rev interacting protein, hRIP, during HIV-1 Rev-directed RNA export. Ablation of hRIP activity by either a trans-dominant-negative mutant or siRNAs inhibits HIV-1 Rev function by mislocalizing RRE-containing RNAs to the nuclear periphery. Comparable ablation of hRIP activity did not affect intracellular localization or trafficking of a variety of proteins or cellular poly(A)+ mRNA. Thus, hRIP is an essential cellular cofactor for HIV-1 Rev function that acts by promoting movement of RRE-containing RNAs from the nuclear periphery to the cytoplasm.
Movement from the perinuclear region as a step in nuclear RNA export
Our results identify release from the nuclear periphery as an essential step in HIV-1 Rev-directed RNA nuclear export. Moreover, this perinuclear step may link nuclear export with cytoplasmic RNA localization and function. The RNA mislocalization phenotype resulting from hRIP inhibition is strikingly different from the strictly nuclear mislocalization previously observed due to inhibition of Rev mutants or CRM1. This mislocalization pattern was particularly unexpected because CRM1 is thought to bridge the interaction between Rev and hRIP, and therefore one would predict the pattern to resemble that of CRM1 inhibition. Furthermore, it is significant that this mislocalization phenotype requires both Rev and an RRE-containing RNA. This high degree of specificity verifies that this RNA mislocalization represents a block in an intermediate step in Rev-directed RNA export.
The results from our localization studies with poly(A)+ mRNA suggest that hRIP is not a general mRNA export factor. Nevertheless, cellular RNAs would, presumably, have to undergo a similar perinuclear release step. We therefore predict there are proteins with analogous functions to hRIP which act on cellular mRNAs to promote their movement from the nuclear periphery to the cytoplasm.
An extended role for Rev
A role for the perinuclear region in Rev-directed RNA export is consistent with several previous findings. First, Rev is required for efficient translation of the intron-containing gag-pol and env RNAs (Arrigo and Chen 1991; Lawrence et al. 1991; D'Agostino et al. 1992; Favaro et al. 1999) and has been proposed to direct viral RNAs to the translation machinery (Kimura et al. 1996; Coyle et al. 2003). Second, the cellular protein SAM68 (src-associated in mitosis) can partially substitute for Rev in transient transfection assays (Reddy et al. 1999; Soros et al. 2001). Interestingly, a dominant-negative SAM68 mutant also mislocalizes Rev-directed RNAs to the nuclear periphery (Soros et al. 2001). Third, hRIP has been reported to interact with Eps15 (Doria et al. 1999), an EH-domain containing protein that is concentrated at the perinuclear region and can bind to CRM1 (Santolini et al. 1999; Vecchi et al. 2001).
Mechanism of hRIP action
Compared with our understanding of protein and RNA transport across the NPC, relatively little is known about the factors and mechanisms that facilitate release of RNAs from the cytoplasmic side of the NPC and their subsequent movement through the cytoplasm. Models for hRIP function must account for the finding that the mislocalization phenotype requires both Rev and an RRE-containing RNA. In addition, standard NES-containing protein substrates were not mislocalized in the absence of functional hRIP. Thus, hRIP is specifically involved in Rev-directed RNA localization.
One can invoke at least two models for how hRIP promotes movement of Rev-directed RNAs from the nuclear periphery into the cytoplasm. In the first model, hRIP could promote the disassembly of Rev-directed RNA from the export complex at the cytoplasmic side of the NPC. Several factors are known to be involved in disassembly of transport complexes following movement through the NPC, including RanBP1, RanBP2, and NXT1 (Kehlenbach et al. 1999; Black et al. 2001; Bednenko et al. 2003). If the function of hRIP were to remove Rev from its cognate RNA, failure to completely disassemble the export complex would result in retention of these RNAs at the nuclear periphery. Consistent with such a model, we note that hRIP contains a C4H2-type zinc finger motif, which has been previously implicated in several activities, including the disassembly of macromolecular complexes (Spang 2002).
Alternatively, hRIP might function after disassembly of the export complex to facilitate the movement of Rev-directed RNA from the nuclear periphery into the cytoplasm. In this model, hRIP could act as a positive regulator for site-specific RNA localization within the cytoplasm. One pertinent example is Staufen, an RNA binding protein that has been implicated in the cytoplasmic trafficking of HIV-1 genomic RNA. Interestingly, like hRIP, Staufen is concentrated at the perinuclear region (Wickham et al. 1999), where HIV-1 RNAs are thought to be assembled into large Staufen-containing granules for subsequent trafficking out to the cell periphery (Mouland et al. 2000). Another example of a positive regulator is the zipcode binding protein 1 (ZBP1), which mediates actin-dependent localization of β-actin mRNA in chicken embryo fibroblasts (CEFs; for review, see Jansen 2001). Predominantly a cytoplasmic protein, ZBP1 binds to a 54-nt cis-acting element or “zipcode” located in the 3′ UTR of the β-actin mRNA and colocalizes with the RNA to the leading lamellae (Farina et al. 2003). ZBP1 appears in large cytoplasmic granules, which codistribute with β-actin mRNA granules thought to contain components necessary for transport, localization, and translation of specific mRNAs. Based on these observations, it has been suggested that interaction of ZBP1 with the β-actin zipcode facilitates granule formation and cytoskeletal attachment, two processes that, ultimately, lead to the localization of the RNA.
Possible cellular function of hRIP
An important future objective will be to determine the cellular function of hRIP. Loss of hRIP activity does not affect the distribution of bulk poly(A)+ mRNA, indicating it does not play a general role in nuclear mRNA export. However, hRIP could act on a distinct subset of cellular mRNAs. In this regard, in several experiments, we observed small, discrete patches of RNA at the nuclear periphery of cells expressing the hRIP dominant-negative mutant. Perhaps these represent the subset of mRNAs whose cytoplasmic localization is mediated by a CRM1-dependent pathway (Brennan et al. 2000; Gallouzi and Steitz 2001).
hRIP can interact via CRM1 with nuclear export factor 3 (NXF3), a protein implicated as a factor involved in mRNA export (Herold et al. 2001; Yang et al. 2001). NXF3 is predominantly expressed in human testis and has been suggested to play a role in male germ cell development, perhaps as a tissue-specific nuclear RNA export factor. hRIP is also expressed at high levels in testis (Pertea et al. 2003) and hRIP-deficient male mice are infertile, suggesting a role for hRIP during mouse spermatogenesis (Kang-Decker et al. 2001).
Regardless of the cellular role of hRIP, our results clearly show that it is required for the RNA transport function of HIV-1 Rev. It is well established that Rev is essential for HIV-1 replication and that inhibition of Rev function interferes with virus production. These considerations raise the possibility that hRIP is a cellular cofactor required for HIV-1 replication and a potential target for new antiviral strategies.
Materials and methods
Plasmid construction
N- and C-terminal truncated hRIP cDNAs were generated by PCR amplification [hRIP nts (Accession No. X89478): 1–1530(ΔC52), 1–1260(ΔC142), 1–975(ΔC237), 1–735(ΔC317), 262–1686(ΔN87), 706–1686(ΔN235), 1081–1686(ΔN360), and 1267–1686(ΔN422)] and inserted in the HindIII/XbaI restriction sites of pCMV-HA-Flag (Roberts et al. 1995). phRIPΔN360-EGFP contains nucleotides 1081–1686 of hRIP inserted into HindIII/BamHI restriction sites of pEGFP-N1 (Clontech). All recombinant expression plasmids were analyzed by DNA sequencing prior to individual transfection into Cos-1, HeLa, or 293-T cells. Protein levels in cell lysates (50–75 μg total protein) were analyzed by Western blotting using anti-HA (12CA5 at 1:1200; BABCO) or anti-Flag (M5 at 1:1000; Sigma) monoclonal antibodies and visualized by enhanced chemiluminescence (ECL; Amersham). The mutant hRIP protein levels were comparable to full-length hRIP in these mammalian cell lines.
Cell lines and media
Monkey Cos-1, human HeLa, and 293-T fibroblasts were grown in DMEM (Life Technologies) supplemented with 25 mM Hepes (pH 7.2), 44 mM sodium bicarbonate, 11% fetal bovine serum (Clontech), 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C, 10% CO2.
Transient transfection assays
Transient transfection assays of Rev function in Cos-1 cells were performed as described (Zapp et al. 1993). Transfection mixtures contained 0.5 μg pCMV128 (Hope et al. 1990), 0.5 μg pCMV128, and 1 μg pcRev (Malim et al. 1989); 0.5 μg pCMV128, 1 μg pcRev, and 2.5 μg phRIP; or 0.5 μg pCMV128, 1 μg pcRev, and 2.5 μg phRIPΔN235, phRIPΔN360, or phRIPΔC237. Additional transfection mixtures contained 0.5 μg pCMV128, 1 μg pcRev, and 0 to 8 μg of phRIPΔN360. All transfection mixtures contained 0.2 μg pCMV β-gal (Clontech), a β-galactosidase expression plasmid as an internal control for transfection efficiency, protein normalization, and nonspecific effects of the CMV promoter. Total DNA was adjusted to 10 μg using pBC-IL2 (Cullen 1986), pCMV-HA-Flag, and pUC118. All CAT assays were normalized to the expression of β-galactosidase activity as described in Hope et al. (1990). Transient transfection assays were performed in duplicate, with the data shown in Figure 2,Figure 2 representing six independent experiments.
In situ hybridization assays
Cos-1 cells were incubated with the transfection mixture for 8 to 12 h, washed with PBS (1 mM KH2PO4, 10 mM Na2HPO4, 130 mM NaCl, 2.7 mM KCl at pH 7.0), then re-fed with 3 mL DMEM. Transfection mixtures contained 1 μg pgTAT; 1 μg pg-TAT and 2.5 μg pcRev; or 1 μg pgTAT, 2.5 μg pcRev, and 4 μg phRIP or phRIPΔN360. Control mixtures contained 1 μg pgTAT and 2.5 μg pcRev M10, or 1 μg pgTAT and 3 μg pCMVΔCAN. Total DNA was adjusted to 10 μg using pBC-IL2, pCMV, and pUC118. At 18 to 22 h posttransfection, cells were treated with 0.5% trypsin/0.1 mM EDTA, and resuspended in 5 mL media, plated onto three to five sterile glass coverslips in 60-mm dishes containing 2.5 mL of media, and incubated further for 48–72 h. Cells were then washed in PBS, incubated in fixation solution (4% EM-grade paraformaldehyde [EMS], 1× PBS, 5 mM MgCl2) for 15 min, rinsed, and stored in 70% ethanol at 4°C until use. Hybridization reactions were performed in triplicate as described previously (Zhang et al. 1996) and contained 60–120 ng of labeled probe. Following a posthybridization step, coverslips were mounted on glass slides using 10 μL Vectashield containing 10 μg/mL DAPI (Vector Laboratories). Hybridization signals were visualized using a Zeiss Axioplan 2 fluorescence microscope equipped with a 63× plan Apochromat objective. Images were acquired and analyzed using Open Lab 2.2.5 software.
Oligonucleotide probes
In situ oligonucleotide probes were labeled with Cy3 (Amersham) or Alexa Green 488 (Molecular Probes) according to the manufacturer's instructions. The RRE-specific probe is complementary to nucleotides 7353 through 7420 of the HIV-1LAI RNA (Accession No. K03455). The probe used to detect cellular poly(A)+ mRNA consisted of 52 thymidine residues (dT52).
Immunofluorescence analysis
HIV-NLS-and NES-EGFP reporter proteins Cos-1 cells were transfected with pHIV-NLS-EGFP or pHIV-NES-EGFP (1–2 μg) alone or in the presence of phRIPΔN360 (4 μg) as described in Bartz et al. (1996). After 12 h, the transfection mixture was removed, and the cells washed twice with PBS. Following treatment with 0.5% trypsin/0.1 mM EDTA, cells were suspended in 1 mL media and plated onto three to five sterile coverslips in 60-mm dishes. After 30 min at 37°C, 2 mL media was added to each dish, and the cells were incubated further for 24–48 h. For analysis, cells were incubated in ice-cold methanol for 7 min, then treated with cytoskeletal (CSK) buffer (10 mM PIPES at pH 6.9, 100 mM KCl, 300 mM sucrose, 5 mM MgCl2, 2 mM EGTA, 0.5% Triton-X-100) for 10 min at 25°C. Cells were blocked in Buffer B (0.2% bovine serum albumin, PBS, 0.1% Triton-X-100) for 30 min, then incubated in Buffer B containing an anti-HA antibody (at 1:500) for 1 h. Following washing in PBS, cells were incubated in Buffer B containing affinity-purified donkey anti-mouse Cy3-labeled antibodies (at 1:750; Kirkeguard and Perry Laboratories). After 30 min, the cells were washed in PBS and mounted as described for in situ hybridization.
MLV envelope proteins HeLa cells grown to 50% confluence were cotransfected with pSV-A-MLV-env (5 μg) and pCMV (5 μg), pSV-A-MLV-env (5 μg) and phRIPΔN360-EGFP (5 μg), or pCMV (10 μg) using the Calcium Phosphate Transfection System (Life Technologies). At 12 h posttransfection, cells were processed as described in the previous section using an anti-MLV envelope rat monoclonal antibody (83A25, Evans et al. 1990, primary at 1:2000) and an anti-rat Alexa Red 594 (Molecular Probes, secondary at 1:5000).
SEAP assays 293T cells were transfected with pCMV, phRIP, phRIPΔN360, or pEGFP expression plasmids (5 μg) as described by Bartz et al. (1996). Duplicate transfection reactions also contained pCMV β-gal (0.2 μg) as an internal control for transfection efficiency and protein normalization. Control untransfected cells were treated with BFA (2.5 μg/mL, Sigma) as described previously (Ding et al. 2001). Cell viability (>99.4%) was monitored by trypan blue staining following DNA transfection or BFA treatment. Cells and culture supernatants were harvested at 48 and 72 h posttransfection and SEAP activity assays were performed as described in Liu et al. (2001). The level of SEAP secretion was quantified as the ratio of SEAP activity in culture supernatants to the sum of SEAP activity in both the culture supernatants and cell lysates. Data shown in Figure 5D are representative of four independent experiments.
Small interfering RNA (siRNA)
Synthetic 21-nt RNA duplexes with symmetric 2-nt 3′(2′deoxy) thymidine overhangs correspond to nts 1593–1613, 1106–1126, or 463–483 of the hRIP mRNA, which are unique in the human genome database. hRIP siRNA duplex sequences written in the 5′ to 3′ direction are as follows: (H1) CAGCCCAAUGGUGCA GGUUTT, AACCUGCACCAUUGGGCUGTT; (M1) GAGCC CAAUCGUGCAGGUUTTGTT, AACCUGCACGAUUGGGC UGTT; (H2) CUGGCUUUGGGACCACAGGTT, CCUGUGG UCCCAAAGCCACTT; (M2) CUGGCUUCGGGACCACAG GTT, CCUGUGGUCCCGAAGCCACTT; (H3) GCCAAAGU CCUGGCAUCAGTT, CUGAUGCCAGGACUUUGGCTT; (M3) GCCAAAGUCCUCGCAUCAGTT, CUGAUGCGAGGACUUU GGCTT. Oligonucleotides were annealed as described in Elbashir et al. (2001) and duplex formation was monitored using nondenaturing gel electrophoresis and ethidium bromide staining.
siRNAs transfections
HeLa cells grown to 50% confluence in 60-mm dishes were transfected with hRIP-specific siRNA duplexes (50–100 nM) using Oligofectamine as described previously (Elbashir et al. 2002). At 16–20 h posttransfection, cells were retransfected with 1.4 μg pCMV128, 3 μg pcRev, and 0.2 μg pCMV β or 1 μg pgTAT and 2.5 μg pcRev using Gene-PORTER (GTS). Cell viability (>98.7%) was monitored by trypan blue staining following siRNA treatment and DNA transfection. Transfected cells were harvested after 48–72 h at 37°C, suspended in 450 μL RIPA buffer (10 mM Tris-HCl at pH 7.4, 1 mM EDTA, 0.1% SDS, 1% TX-100, 1% sodium deoxycholate, 150 mM NaCl, and 0.2 mM PMSF), incubated for 30 min on ice, then centrifuged at 13,000g for 15 min at 4°C. Supernatants (lysates) were quantified using Total Protein Reagent (Sigma), and equal amounts of protein (25–50 μg) resolved on a 10% SDS denaturing polyacrylamide gel (National Diagnostics). Protein compositions were analyzed by Western blotting using goat anti-hRIP (Santa Cruz Biochemical, C-19 at 1:750; goat anti-Lamin A/C, N-18 at 1:500) and visualized by ECL/Amersham. CAT activity assays and RNA in situ hybridization analysis using hRIP-depleted cells were performed as described in a previous section.
Acknowledgments
We are grateful to Drs. Britt, Green, and Noonburg for providing antibodies and plasmids. pSV-A-MLV-env DNA was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pSV-A-MLV-env from Drs. Landau and Littman. We acknowledge the UMASS Center for AIDS Research (CFAR) Molecular Biology Core for oligonucleotide synthesis and DNA sequence analysis. We are grateful to K. Long and E. Kittler for critical reading and preparation of the manuscript. This work was supported by an NIAID/NIH grant (5R01 AI43208) and CFAR Developmental Core (UMass CFAR 5P30 AI42845) to M.L.Z.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1149704.
References
- Arrigo S.J. and Chen, I.S. 1991. Rev is necessary for translation but not cytoplasmic accumulation of HIV-1 vif, vpu, and env/vpu 2 RNAs. Genes & Dev. 5: 808–819. [DOI] [PubMed] [Google Scholar]
- Bartz S.R., Rogel, M.E., and Emerman, M. 1996. Human immunodeficiency virus type 1 cell cycle control: Vpr is cytostatic and mediates G2 accumulation by a mechanism which differs from DNA damage checkpoint control. J. Virol. 70: 2324–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednenko J., Cingolani, G., and Gerace, L. 2003. Nucleocytoplasmic transport: Navigating the channel. Traffic 4: 127–135. [DOI] [PubMed] [Google Scholar]
- Bevec D., Jaksche, H., Oft, M., Wohl, T., Himmelspach, M., Pacher, A., Schebesta, M., Koettnitz, K., Dobrovnik, M., Csonga, R., et al. 1996. Inhibition of HIV-1 replication in lymphocytes by mutants of the Rev cofactor eIF-5A. Science 271: 1858–1860. [DOI] [PubMed] [Google Scholar]
- Biondi R.M. and Nebreda, A.R. 2003. Signalling specificity of Ser/Thr protein kinases through docking-site-mediated interactions. Biochem. J. 372: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B.E., Holaska, J.M., Levesque, L., Ossareh-Nazari, B., Gwizdek, C., Dargemont, C., and Paschal, B.M. 2001. NXT1 is necessary for the terminal step of Crm1-mediated nuclear export. J. Cell Biol. 152: 141–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd H.P., Fridell, R.A., Madore, S., and Cullen, B.R. 1995. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell 82: 485–494. [DOI] [PubMed] [Google Scholar]
- Bogerd H.P., Echarri, A., Ross, T.M., and Cullen, B.R. 1998. Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J. Virol. 72: 8627–8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C.M., Gallouzi, I.E., and Steitz, J.A. 2000. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 151: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardin P. and McCormick, F. 1999. Brefeldin A: The advantage of being uncompetitive. Cell 97: 153–155. [DOI] [PubMed] [Google Scholar]
- Coyle J.H., Guzik, B.W., Bor, Y.C., Jin, L., Eisner-Smerage, L., Taylor, S.J., Rekosh, D., and Hammarskjold, M.L. 2003. Sam68 enhances the cytoplasmic utilization of intron-containing RNA and is functionally regulated by the nuclear kinase Sik/BRK. Mol. Cell Biol. 23: 92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukierman E., Huber, I., Rotman, M., and Cassel, D. 1995. The ARF1 GTPase activating protein: Zinc finger motif and Golgi complex localization. Science 270: 1999–2002. [DOI] [PubMed] [Google Scholar]
- Cullen B.R. 1986. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell 46: 973–982. [DOI] [PubMed] [Google Scholar]
- ____. 2002. Using retroviruses to study the nuclear export of mRNA. Results Probl. Cell Differ. 35: 151–168. [DOI] [PubMed] [Google Scholar]
- D'Agostino D.M., Felber, B.K., Harrison, J.E., and Pavlakis, G.N. 1992. The Rev protein of human immunodeficiency virus type 1 promotes polysomal association and translation of gag/pol and vpu/env mRNAs. Mol. Cell Biol. 12: 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayton A.I. 1996. The Rev axis of HIV-1 and its associated host cofactors: A viral window onto the workings of eukaryotic posttranscriptional RNA processing. J. Biomed. Sci. 3: 69–77. [DOI] [PubMed] [Google Scholar]
- Ding W., Albrecht, B., Luo, R., Zhang, W., Stanley, J.R., New-bound, G.C., and Lairmore, M.D. 2001. Endoplasmic reticulum and cis-Golgi localization of the human T-lymphotropic virus type 1 p12(I): Association with calreticulin and calnexin. J. Virol. 75: 7672–7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria M., Salcini, A.E., Columbo, E., Parslow, T.G., Pelicci, P.G., and Di Fiore, P.P. 1999. The eps15 homology (EH) domain-based interaction between Eps15 and Hrb connects the molecular machinery of endocytosis to that of nucleocytosolic transport. J. Cell Biol. 147: 1379–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye V. and Hurt, E. 1997. From nucleoporins to nuclear pore complexes. Curr. Opin. Cell Biol. 9: 401–411. [DOI] [PubMed] [Google Scholar]
- Dubois T., Zemlickova, E., Howell, S., and Aiken, A. 2003. Centaurin-α1 associates in vitro and in vivo with nucleolin. Biochem. Biophys. Res. Commun. 301: 502–508. [DOI] [PubMed] [Google Scholar]
- Dupont S., Sharova, N., DeHoratius, C., Virbasius, C.M., Zhu, X., Bukrinskaya, A.G., Stevenson, M., and Green, M.R. 1999. A novel nuclear export activity in HIV-1 matrix protein required for viral replication. Nature 402: 681–685. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth, J., Weber, K., and Tuschl, T. 2002. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26: 199–213. [DOI] [PubMed] [Google Scholar]
- Evans L.H., Morrison, R.P., Malik, F.G., Portis, J., and Britt, W.J. 1990. A neutralizable epitope common to the envelope glycoproteins of ecotropic, polytropic, xenotropic, and amphotropic murine leukemia viruses. J. Virol. 64: 6176–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina K.L., Hüttelmaier, S., Musunuru, K., Darnell, R., and Singer, R.H. 2003. Two ZBP1 KH domains facilitate β-actin mRNA localization, granule formation and cytoskeletal attachment. J. Cell Biol. 160: 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro J.P., Maldarelli, F., Arrigo, S.J., and Schmidt, M.G. 1999. Effect of rev on the cytoplasmic localization of intron-containing human immunodeficiency virus type 1 RNA. Virology 255: 237–249. [DOI] [PubMed] [Google Scholar]
- Fornerod M., Ohno, M., Yoshida, M., and Mattaj, I.W. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90: 1051–1060. [DOI] [PubMed] [Google Scholar]
- Frankel A.D. and Young, J.A. 1998. HIV-1: Fifteen proteins and an RNA. Annu. Rev. Biochem. 67: 1–25. [DOI] [PubMed] [Google Scholar]
- Fritz C.C. and Green, M.R. 1996. HIV Rev uses a conserved cellular protein export pathway for the nucleocytoplasmic transport of viral RNAs. Curr. Biol. 6: 848–854. [DOI] [PubMed] [Google Scholar]
- Fritz C.C., Zapp, M.L., and Green, M.R. 1995. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature 376: 530–533. [DOI] [PubMed] [Google Scholar]
- Gallouzi I.E. and Steitz, J.A. 2001. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science 294: 1895–1901. [DOI] [PubMed] [Google Scholar]
- Henderson B.R. and Percipalle, P. 1997. Interactions between HIV Rev and nuclear import and export receptors: The Rev nuclear localization signal mediates specific binding to human importin-beta. J. Mol. Biol. 274: 693–707. [DOI] [PubMed] [Google Scholar]
- Herold A., Klymenko, T., and Izaurralde, E. 2001. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA 7: 1768–1780. [PMC free article] [PubMed] [Google Scholar]
- Hope T.J. 1999. The ins and outs of HIV Rev. Arch. Biochem. Biophys. 365: 186–191. [DOI] [PubMed] [Google Scholar]
- Hope T.J., Huang, X.J., McDonald, D., and Parslow, T.G. 1990. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: Mapping cryptic functions of the arginine-rich motif. Proc. Natl. Acad. Sci. 87: 7787–7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman J., Chen, H., Di Fiore, P.P., De Camilli, P., and Brunger, A.T. 2000. Epsin 1 undergoes nucleocytosolic shuttling and its eps15 interactor NH (2)-terminal homology (ENTH) domain, structurally similar to Armadillo and HEAT repeats, interacts with the transcription factor promyelocytic leukemia Zn (2)+ finger protein (PLZF). J. Cell Biol. 149: 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R.P. 2001. mRNA localization: Message on the move. Nat. Rev. Mol. Cell Biol. 2: 247–256. [DOI] [PubMed] [Google Scholar]
- Kalland K.H., Szilvay, A.M., Brokstad, K.A., Saetrevik, W., and Haukenes, G. 1994. The human immunodeficiency virus type 1 Rev protein shuttles between the cytoplasm and nuclear compartments. Mol. Cell Biol. 14: 7436–7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang-Decker N., Mantchev, G.T., Juneja, S.C., McNiven, M.A., and van Deursen, J.M. 2001. Lack of acrosome formation in Hrb-deficient mice. Science 294: 1531–1533. [DOI] [PubMed] [Google Scholar]
- Kehlenbach R.H., Dickmanns, A., Kehlenbach, A., Guan, T., and Gerace, L. 1999. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J. Cell Biol. 145: 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Hashimoto, I., Nishikawa, M., and Fujisawa, J.I. 1996. A role for Rev in the association of HIV-1 gag mRNA with the cytoskeletal beta-actin and viral protein expression. Biochemie 78: 1075–1080. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T. 2000. Three ways to make a vesicle. Nat. Rev. Mol. Cell Biol. 1: 187–198. [DOI] [PubMed] [Google Scholar]
- Kjems J. and Askjaer, P. 2000. Rev protein and its cellular partners. Adv. Pharmacol. 48: 251–298. [DOI] [PubMed] [Google Scholar]
- Krishna S.S., Majumdar, I., and Grishin, N.V. 2003. Structural classification of zinc fingers: Survey and summary. Nucleic Acids Res. 31: 532–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau N.R., Page, K.A., and Littman, D.R. 1991. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J. Virol. 65: 162–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J.B., Cochrane, A.W., Johnston, C.V., Perkins, A., and Rosen, C.A. 1991. The HIV-1 Rev protein: A model system for coupled RNA transport and translation. New Biol. 3: 1220–1232. [PubMed] [Google Scholar]
- Liu X., Zhang, C., Xing, G., Chen, Q., and He, F. 2001. Functional characterization of novel human ARFGAP3. FEBS Lett. 490: 79–83. [DOI] [PubMed] [Google Scholar]
- Malim M.H., Hauber, J., Le, S.-Y., Maizel, J.V., and Cullen, B.R. 1989. The HIV-1 Rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338: 254–257. [DOI] [PubMed] [Google Scholar]
- Maraldi N.M., Zini, N., Santi, S., and Manzoli, F.A. 1999. Topology of inositol lipid signal transduction in the nucleus. J. Cell. Physiol. 181: 203–217. [DOI] [PubMed] [Google Scholar]
- Meyer B.E. and Malim, M.H. 1994. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes & Dev. 8: 1538–1547. [DOI] [PubMed] [Google Scholar]
- Miller D.G., Edwards, R.H., and Miller, A.D. 1994. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc. Natl. Acad. Sci. 91: 78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouland A.J., Mercier, J., Luo, M., Bernier, L., DesGroseillers, L., and Cohen, E.A. 2000. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: Evidence for a role in genomic RNA encapsidation. J. Virol. 74: 5441–5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville M., Stutz, F., Lee, L., Davis, L.I., and Rosbash, M. 1997. The importin-beta family member Crmlp bridges the interaction between Rev and the nuclear protein complex during nuclear export. Curr. Biol. 7: 767–775. [DOI] [PubMed] [Google Scholar]
- Nie Z., Stanley, K.T., Stauffer, S., Jacques, K.M., Hirsch, D.S., Takei, J., and Randazzo, P.A. 2002. AGAP1, an endosome-associated, phosphoinositide-dependent ADP-ribosylation factor GTPase-activating protein that affects actin cytoskeleton. J. Biol. Chem. 277: 48965–48975. [DOI] [PubMed] [Google Scholar]
- Parker P.J. and Parkinson, S.J. 2001. AGC protein kinase phosphorylation and protein kinase C. Biochem. Soc. Trans. 29: 860–863. [DOI] [PubMed] [Google Scholar]
- Pertea G., Huang, X., Liang, F., Antonescu, V., Sultana, R., Karamycheva, S., Lee, Y., White, J., Cheung, F., Parvizi, B., et al. 2003. TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 19: 651–652. [DOI] [PubMed] [Google Scholar]
- Pollard V.W. and Malim, M.H. 1998. The HIV-1 Rev protein. Annu. Rev. Microbiol. 52: 491–532. [DOI] [PubMed] [Google Scholar]
- Poupon V., Polo, S., Vecchi, M., Martin, G., Dautry-Varsat, A., Cerf-Bensussan, N., DiFiore, P.P., and Benmerah, A. 2002. Differential nucleocytoplasmic trafficking between the related endocytic proteins Eps15 and Eps15R. J. Biol. Chem. 277: 8941–8948. [DOI] [PubMed] [Google Scholar]
- Premont R.T., Claing, A., Vitale, N., Perry, S.J., and Lefkowitz, R.J. 2000. The GIT family of ADP-ribosylation factor GTPase-activating proteins. Functional diversity of GIT2 through alternative splicing. J. Biol. Chem. 275: 22373–22380. [DOI] [PubMed] [Google Scholar]
- Reddy T.R., Xu, W., Mau, J.K., Goodwin, C.D., Suhasini, M., Tang, H., Frimpong, K., Rose, D.W., and Wong-Staal, F. 1999. Inhibition of HIV replication by dominant negative mutants of Sam68, a functional homolog of HIV-1 Rev. Nat. Med. 5: 635–642. [DOI] [PubMed] [Google Scholar]
- Richard N., Iacampo, S., and Cochrane, A. 1994. HIV-1 Rev is capable of shuttling between the nucleus and cytoplasm. Virology 204: 123–131. [DOI] [PubMed] [Google Scholar]
- Roberts S.G., Choy, B., Walker, S.S., Lin, Y.S., and Green, M.R. 1995. A role for activator-mediated TFIIB recruitment in diverse aspects of transcriptional regulation. Curr. Biol. 5: 508–516. [DOI] [PubMed] [Google Scholar]
- Santolini E., Salcini, A.E., Kay, B.K., Yamabhai, M., and DiFiore, P.P. 1999. The EH network. Exp. Cell Res. 253: 186–209. [DOI] [PubMed] [Google Scholar]
- Soros V.B., Carvajal, H.V., Richard, S., and Cochrane, A.W. 2001. Inhibition of human immunodeficiency virus type 1 Rev function by a dominant-negative mutant of Sam68 through sequestration of unspliced RNA at perinuclear bundles. J. Virol. 75: 8203–8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A. 2002. ARF1 regulatory factors and COPI vesicle formation. Curr. Opin. Cell Biol. 14: 423–427. [DOI] [PubMed] [Google Scholar]
- Strambio-de-Castillia C., Blobel, G., and Rout, M.P. 1999. Proteins connecting the nuclear pore complex with the nuclear interior. J. Cell Biol. 144: 839–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F., Izaurralde, E., Mattaj, I.W., and Rosbash, M. 1996. A role for nucleoporin FG repeat domains in export of human immunodeficiency virus type 1 Rev protein and RNA from the nucleus. Mol. Cell Biol. 16: 7144–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner C.E., West, K.A., and Brown, M.C. 2001. Paxillin-ARF GAP signaling and the cytoskeleton. Curr. Opin. Cell Biol. 13: 593–599. [DOI] [PubMed] [Google Scholar]
- Vecchi M., Polo, S., Poupon, V., van de Loo, J.W., Benmerah, A., and DiFiore, P.P. 2001. Nucleocytoplasmic shuttling of endocytic proteins. J. Cell Biol. 153: 1511–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh L.K., Gettemeier, T., and Chinnadurai, G. 2003. A nuclear kinesin-like protein interacts with and stimulates the activity of the leucine-rich nuclear export signal of the human immunodeficiency virus type 1 rev protein. J. Virol. 77: 7236–7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K. 2002. Nucleocytoplasmic transport: Cargo trafficking across the border. Curr. Opin. Cell Biol. 14: 328–335. [DOI] [PubMed] [Google Scholar]
- Wickham L., Duchaine, T., Luo, M., Nabi, I.R., and DesGroseillers, L. 1999. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol. Cell Biol. 19: 2220–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Bogerd, H.P., Wang, P.J., Page, D.C., and Cullen, B.R. 2001. Two closely related human nuclear export factors utilize entirely distinct export pathways. Mol. Cell 8: 397–406. [DOI] [PubMed] [Google Scholar]
- Yi R., Bogerd, H.P., and Cullen, B.R. 2002. Recruitment of the Crm1 nuclear export factor is sufficient to induce cytoplasmic expression of incompletely spliced human immunodeficiency virus mRNAs. J. Virol. 76: 2036–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapp M.L., Stern, S., and Green, M.R. 1993. Small molecules that selectively block RNA binding of HIV-1 Rev protein inhibit Rev function and viral production. Cell 74: 969–978. [DOI] [PubMed] [Google Scholar]
- Zhang G., Zapp, M.L., Yan, G., and Green, M.R. 1996. Localization of HIV-1 RNAs in mammalian nuclei. J. Cell Biol. 135: 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]