Abstract
Although Manduca sexta has significantly contributed to our knowledge on a variety of insect physiological processes, the lack of its genome sequence hampers the large-scale gene discovery, transcript profiling, and proteomic analysis in this biochemical model species. Here we report our implementation of the RNA-Seq cDNA sequencing approach based on massively parallel pyrosequencing, which allows us to categorize transcripts based on their relative abundances and to discover process- or tissue-specifically regulated genes simultaneously. We obtained 1,821,652 reads with an average length of 289 bp per read from fat body and hemocytes of naïve and microbe-injected M. sexta larvae. After almost all (92.1%) of these reads were assembled into 19,020 contigs, we identified 528 contigs whose relative abundances increased at least 5- and 8-fold in fat body and hemocytes, respectively, after the microbial challenge. Polypeptides encoded by these contigs include pathogen recognition receptors, extracellular and intracellular signal mediators and regulators, antimicrobial peptides, and proteins with no known sequence but likely participating in defense in novel ways. We also found 250 and 161 contigs that were preferentially expressed in fat body and hemocytes, respectively. Furthermore, we integrated data from our previous study and generated a sequence database to support future gene annotation and proteomic analysis in M. sexta. In summary, we have successfully established a combined approach for gene discovery and expression profiling in organisms lacking known genome sequences.
Keywords: insect immunity, hemolymph proteins, gene discovery, transcript profiling, 454 sequencing, RNA-Seq, functional genomics
1. Introduction
Insects possess an effective defense system to control pathogen invasion, which includes a physical barrier intertwined with biochemical and cellular mechanisms to block penetration and proliferation of infectious agents (Gillespie et al., 1997; Strand, 2008; Lemaitre and Hoffmann, 2007). These mechanisms are mediated by molecules that recognize pathogens, relay or modulate immune signals, and kill the invading pathogens. Most of the molecules are proteins in body fluids (e.g. plasma) and tissues/cells (e.g. fat body and hemocytes), either constitutively produced for responses occurring within minutes or induced within hours to days after the initial encounter of pathogens. Fat body is a major source of plasma proteins, some of which participate in humoral immunity, whereas hemocytes are mainly involved in cellular responses such as phagocytosis and encapsulation.
The tobacco hornworm, Manduca sexta, has been used extensively as a model species to study the biochemical basis of insect immunity (Jiang, 2008; Regan et al., 2009), as well as other physiological processes. Pathogen recognition proteins, hemolymph proteinases (HPs), serpins, phenoloxidases (POs), and antimicrobial peptides (AMPs) have been isolated from larval hemolymph of this insect for functional analysis. A differential expression study uncovered 120 expressed sequence tags (ESTs) identical or similar to immunity-related genes (Zhu et al., 2003). Pyrosequencing of cDNA fragments using the RNAseq approach (Morin, et al. 2008; Mortazavi et al., 2008) from a mixture of eight total RNA samples revealed 218 new EST contigs coding for defense proteins (Zou et al., 2008). Additional immunity-related genes were identified in a gut EST project that combined Sanger and 454 sequencing technologies (Pauchet et al., 2010). Sequences provided by these studies, albeit useful, are limited by the methods used to obtain them, such as low throughput (Zhu et al., 2003), high rate of indels (Zou et al., 2008), gene discovery solely based on homology (Pauchet et al., 2010), and lack of information on relative gene expression levels in all the cases. While these problems can be overcome by the genome sequence and microarray analysis yet to come, is it possible to efficiently discover genes along with their expression profiles using next-generation RNA-Seq technologies without resorting to the reference genome and thereby directly uncover process-related gene expression in non-model organisms whose genome sequences are not yet determined?
Here, we report the results of our ongoing studies aimed at discovering alterations in gene expression in M. sexta larvae before and after a bacterial injection and characterize genes based on their tissue-preferential expression patterns in fat body and hemocytes using the RNAseq approach (Morin, et al. 2008; Mortazavi et al., 2008). Therefore, instead of relying on a priori knowledge of the genome, our approach contributes to future genome annotation, cDNA cloning, and protein identification in this insect and, through extremely deep RNA-Seq studies, reveals novel genes that likely play a role in insect defense, and provides useful leads for functional elucidation of unknown defense proteins in this biochemical model insect. More importantly, this method is applicable to gene discovery and study of process/tissue-related transcriptome changes in all non-model species with no known genome sequences.
2. Methods and materials
2.1. Insect rearing, bacterial injection, RNA isolation, and library construction
M. sexta eggs, purchased from Carolina Biological Supply, were hatched and reared on an artificial diet as described by Dunn and Drake (1983). Each of day 2, 5th instar larvae (60) was injected with a mixture of Escherichia coli (2×107 cells), Micrococcus luteus (20 μg) (Sigma-Aldrich), and curdlan (20 μg, insoluble β-1,3-glucan from Alcaligenes faecalis) (Sigma-Aldrich) in 30 μl H2O. Total RNA samples were extracted from induced hemocytes (IH) and fat body (IF) 24 h later using TRIZOL Reagent (Life Technologies Inc.). Control hemocyte (CH) and fat body (CF) RNA was prepared from day 3, 5th instar naïve larvae (60). PolyA+ RNA was separately purified from the total RNA samples (1.0 mg each) by binding to oligo(dT) cellulose twice in the Poly(A) Purist™ Kit (Ambion). First strand cDNA was synthesized using mRNA (5.0 μg), random dodecanucleotides (100 pmol), and SuperScript™ III reverse transcriptase (1000 U, Life Technologies Inc.). RNase H treatment, second strand synthesis, and gap joining were performed according to the published protocol (Zou et al., 2008). After shearing via nebulization, the four samples were end-repaired (Roe, 2004) and ligated to double-stranded adaptor A and biotinylated adaptor B (Margulies et al., 2005).
2.2. PCR amplification, pyrosequencing, and sequence assembly
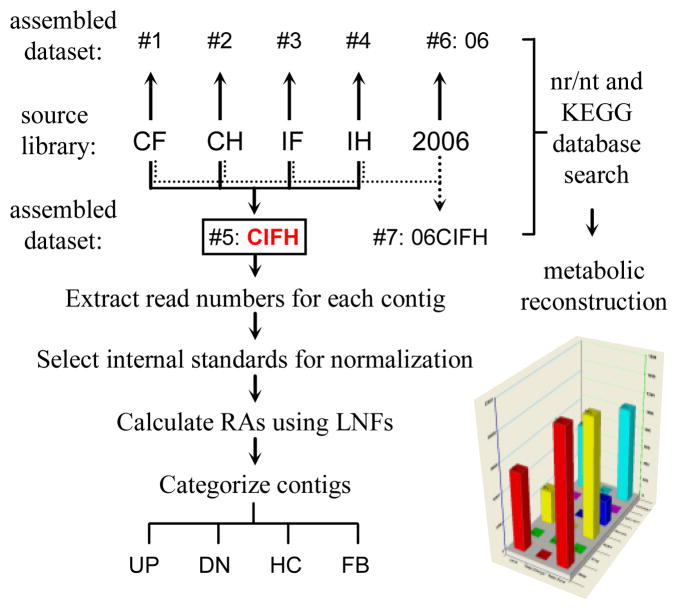
The cDNA with adaptor B attached on one or both ends was isolated using streptavidin-coated magnetic beads, end repaired, and quantified on an Agilent 2100 Bioanalyzer (Agilent Technologies). Diluted DNA molecules, individually captured by beads, were amplified using emulsion PCR with the two primers complementary parts of A and B adaptors (Margulies et al., 2005). After removal of the second strand and empty beads, the sequencing primer identical to another part of A adaptor was used for sequencing. Two full plates were run with one-half plate for each library on a 454 GS-FLX pyrosequencer (Roche Applied Science) using long-read GS-FLX Titanium chemistry. Reads were assembled separately for each library (CF, CH, IF, IH) and collectively (CIFH) using Newbler Assembler (Roche Applied Science) into five datasets: CF, CH, IF, IH, and CIFH (Fig. 1). To improve coverage and quality of the sequence sets, data from our previous run on a 454 GS20 (Zou et al., 2008) were assembled into two datasets (06 for the 2006 data and 06CIFH for the 2006 and 2009 data) using the updated Newbler software. The resulting contigs and singletons from the seven datasets were compared against the NCBI nr/nt and KEGG databases using BLASTN, BLASTP, and BLASTX with a maximum E-value of 1×10−5. For the combined library CIHF, numbers of CH, CF, IH, and IF reads assembled into each contig were extracted from the standard Newbler Assembler output and tabulated using Microsoft Excel.
Fig 1. Scheme of library sequencing, dataset assembling, read normalization, contig categorization, and function prediction.
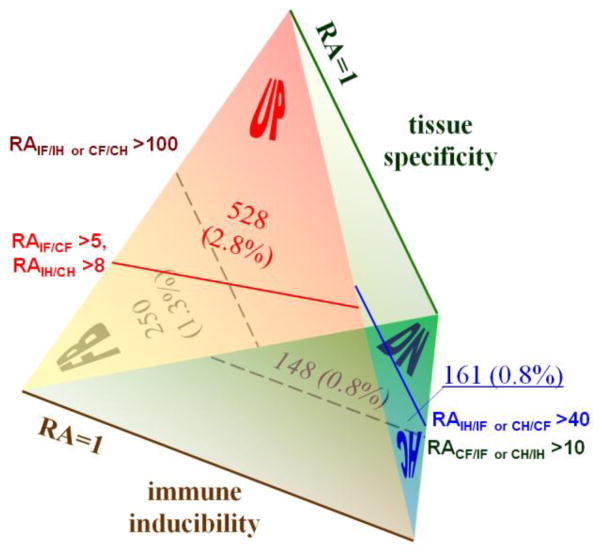
Five cDNA libraries (CF, CH, IF, IH, and 2006) were assembled into seven datasets, one of which (#5: CIFH) was further analyzed by extracting numbers of CF, CH, IF and IH reads assembled into each contig. As described in Section 2.3, read numbers were calibrated using library normalization factors (LNFs) for the calculation of relative abundances (RAs) or adjusted read numbers (ARNs). Based on thresholds set arbitrarily, contigs were categorized into four groups: UP and DN for up- and down-regulated; HC and FB for hemocyte- or fat body-specific.
2.3. Read normalization and ratio calculation
Based on frequencies of several commonly used standards in each of the four libraries (e.g. number of rpS3 reads in CH ÷ number of total reads in CH), a set of six ribosomal protein genes were selected as internal standards, which had high total read numbers and low coefficients of variation (i.e. SD/mean) in their frequencies. The sums of their read numbers for specific libraries, or library normalization factors (LNFs), which already reflected the differences in library sizes, were directly used to calibrate other read numbers in the corresponding libraries. For a specific contig in CIFH, its relative abundance (RA) in libraries X and Y is defined as: RAx/y = (actual read # in library X ÷ LNFx)/(actual read # in library Y ÷ LNFy). In case read # in library Y is zero, adjusted read number (ARN), instead of RA, is calculated as: ARNx = actual read # in library X × LNFy/LNFx. Some of the contigs in CIFH, whose RAs or ARNs are above certain thresholds, are categorized into UP, DN, HC, and FB: UP for up-regulated genes (RAIF/CF >5, RAIH/CH >8, ARNIF >10 when RNCF =0, or ARNIH >10 when RNCH =0), DN for down-regulated genes (RACF/IF >10, RACH/IH >10, ARNCF >20 when RNIF =0, or ARNCH >20 when RNIH =0), HC and FB for genes preferentially expressed in hemocytes (RAIH/IF >40, RACH/CF >40, ARNIH >80 when RNIF =0, or ARNCH >80 when RNCF =0) and fat body (RAIF/IH >100, RACF/CH >100, ARNIF >200 when RNIH =0, or ARNCF >200 when RNCH =0), respectively.
2.4. Sequence extension, database search, and domain prediction
CIFH contigs in UP, DN, HC, and FB categories were used as queries to search local databases of 06CIFH_contigs/singletons, UK_gut_contigs by BLASTN (http://darwin.biochem.okstate.edu/blast/blast.html). The M. sexta midgut ESTs (i.e. UK_gut_contigs) (Pauchet et al., 2009) were kindly provided by Dr. Yannick Pauchet at University of Exeter, UK. The search results were used to extend the CIFH contigs or, in some cases, fill a gap between two contig sequences. The extended sequences were searched against NCBI using BLASTX as described above. For UP CIFH contigs lacking BLAST hits, a set of more stringent conditions was applied to select sequences for further analysis: a) RAIF/CF >15, RAIH/CH >15, ARNIF >30 when RNCF =0, or ARNIH >30 when RNCH =0, b) total read number >70, and c) GC content ≥35% (i.e. coding region-including). Open reading frames in a chosen contig were examined for leader peptide using SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/), which is commonly found in proteins highly induced upon immune challenge (Jiang, 2008; Ragan et al., 2009). The polypeptide sequences were then analyzed to detect conserved domain structures by SMART (http://smart.embl-heidelberg.de/smart/set_mode.cgi).
3. Results
3.1. Identification of differentially regulated genes in M. sexta by pyrosequencing
In order to find immunity-related genes expressed in fat body or hemocytes based on their expression profiles, we isolated mRNA of these two tissues from naïve and bacteria-injected larvae of M. sexta, a lepidopteran insect whose genome sequence has not yet been determined. Using random dodecanucleotide primers that annealed to different regions of mRNA molecules, we generated four cDNA libraries: CF, CH, IF and IH. To facilitate assembly and ORF identification, we adopted long-read Titanium chemistry to sequence these libraries on a 454 GS-FLX pyrosequencer and obtained a total of 227,302 reads from CF, 647,587 reads from CH, 405,739 reads from IF, and 541,024 reads from IH (Table 1). The total number of reads from two plates (0.5 plate per library) was 1,821,652, which was 19.1-fold higher than that from one plate (95,358 reads) sequenced on a 454 GS20 in 2006 (Zou et al., 2008). There also was a substantial increase in average read length from 185 bp to 289 bp, but that was still much lower than what the manufacturer claimed (>400 bp) (http://454.com/about-454/index.asp).
Table 1.
Summary statistics for pyrosequencing analysis of M. sexta ESTs
| 06 a | CF | CH | IF | IH | CIFH b | 06CIFH c | |
|---|---|---|---|---|---|---|---|
| Total number of reads | 95,458 (95,358) | 227,302 | 647,587 | 405,739 | 541,024 | 1,821,652 | 1,917,110 |
| Average reads length (bp) | 185 (185) | 296 | 287 | 293 | 287 | 289 | 284 |
|
| |||||||
| Total number of contigs | 1,471 (7,231) | 2,118 | 11,540 | 4,063 | 10,600 | 19,020 | 19,504 |
| Contigs size (avg./longest in bp) | 391/3,552 (300/3,909) | 770/12,740 | 827/11,667 | 764/8,482 | 832/10,591 | 923/23,095 | 911/23,097 |
| Total assembled reads | 64,874 (69,429) | 191,156 | 561,054 | 349,028 | 465,561 | 1,677,738 | 1,757,333 |
|
| |||||||
| Singlet reads | 28,518 (25,929) | 32,518 | 68,861 | 49,444 | 61,108 | 108,587 | 120,670 |
| Singlet length (avg. in bp) | 179 | 244 | 245 | 235 | 254 | 209 | 200 |
|
| |||||||
| Total BLASTable sequences | 29,989 | 34,636 | 80,401 | 53,507 | 71,708 | 127,607 | 140,174 |
| Orphan sequences (no BLAST match, #/%) | 19,963/67 | 17,982/52 | 51,968/65 | 28,649/54 | 46,521/65 | 73,915/58 | 89,948/64 |
|
| |||||||
| Contigs and reads with functional assignment | 10,026 | 16,654 | 28,433 | 24,858 | 25,187 | 53,692 | 50,226 |
Results from reanalysis of the 2006 sequence data. The numbers in parentheses (adopted from Zou et al., 2008) are listed for comparison with the new results.
Analysis of the 2009 EST sequences of control fat body (CF), control hemocytes (CH), induced fat body (IF), and induced hemocytes (IH) from M. sexta larvae.
Analysis of the combined reads of 2006 (raw flow signals interpreted with the up-graded software) and 2009 (CF, CH, IF, and IH).
We assembled the reads into five datasets: CF, CH, IF, IH, and CIFH (Fig. 1). The first four each came from its respective library, whereas the 5th dataset was assembled from the 1,821,652 reads in the four libraries sequenced in 2009. In CF, CH, IF and IH, 84.1~86.6% of the total reads were incorporated into contigs at average sizes of 764~832 bp; In CIFH, 1,677,738 (92.1%) of the 1,821,652 reads were assembled to 19,020 contigs at 923 bp per contig (Table 1). These assemblies were better than the previous one, that integrated 69,429 (72.8%) of the 95,358 reads into 7,231 contigs at an average length of 300 bp (Zou et al., 2008). To improve the transcriptome coverage, we used the latest version of Newbler to re-analyze the previously generated flowgrams, assembling 64,874 of the reads into 1,471 contigs with an average of 391 bp per contig in the 6th dataset (“06”) (Table 1). Finally we assembled all the source libraries (2006, CF, CH, IF, and IH) into “06CIFH”, which contained 19,504 contigs (average size: 911 bp) and 120,670 singletons.
We used numbers of CH, CF, IH, and IF reads for each CIFH contig to identify differentially regulated genes. Since read numbers depended on library sizes and needed to be normalized against control genes, we compared frequencies of commonly used internal standards in each of the four libraries and found that six ribosomal protein genes (rpS2–rpS5, rpL4 and rpL8) showed low coefficients of variation (<30%) and high total read numbers (>1,000). So, we used the sums of their read numbers 825 (CF), 3,980 (CH), 1,618 (IF), and 3,352 (IH) as library normalization factors (LNFs) to calibrate read numbers and calculate relative abundances (RAs) (Fig. 1). Based on the RA values, 920 or 4.84% of the 19,020 contigs in CIFH were categorized into four groups: UP (Table S1) and DN (Table S2) for up- and down-regulated genes upon immunization; HC (Tables S3) and FB (Table S4) for genes preferentially expressed in hemocytes and fat body, respectively.
3.2. Sequence analysis and function prediction of UP genes
We discovered 528 CIFH contigs whose RAIF/CF or RAIH/CH was greater than 5 and 8, respectively, or whose adjusted number of IF (or IH) reads (ARN) was >10 when the CF (or CH) read was zero – the adjustment for IF was read # × 825/1618 and that for IH was read # × 3980/3352 (Table S1). As we anticipated, these contigs encoded polypeptides either identical to immunity-related proteins previously isolated from M. sexta (e.g. hemolin), or similar in sequence or domain structure to defense factors found in other insects (e.g. Spodoptera frugiperda X-tox), or related to proteins previously not known to play a role in immune responses (e.g. carboxylesterase), or having no significant sequence similarity to known proteins. In the following, we describe these contigs in the order of their putative immune functions.
A. Recognition of molecular patterns associated with microbes
To reinforce detection of invading organisms, certain pattern recognition receptors (PRRs) are synthesized in insects at higher levels after the initial encounter of foreign entities or abnormal host components. For instance, we found an Ig-domain protein (contig 03442) had an RAIF/CF of 748.5 (Table 2). This protein, M. sexta hemolin, was reported previously as a highly inducible PRR that recognizes LPS of Gram-negative bacteria (Ladendorff and Kanost, 1991). Other PRRs included M. sexta immulectin-2 (contig 04775, RAIF/CF: 45.4), immulectin-4 (contig 04808, ARNIF: 217.2), peptidoglycan recognition protein-1 (PGRP1) (contig 13190, ARNIH: 10.7; contig 14104, RAIF/CF: 6.3; ARNIH: 15.4), PGRP2 (contig 14700, residues 1–96, ARNIF: 93.3; contig 14752, residues 98–196, ARNIF: 60.2), β-1,3-glucan recognition protein-2 (βGRP2) (contig 01326, RAIF/CF: 9.7; RAIH/CH: 9.2). These data not only confirmed the published PRR sequences but also provided information on fold increases in their transcript abundances. Contig 06630 (RAIF/CF: 11.2), 58% identical to M. sexta immulectin-3 (Yu et al., 2005) in residues 1–276, represented a previously unknown immulectin discovered based on its induced expression as well as sequence similarity. Newly identified PRRs also included PGRP3 (contig 00575, RAIF/CF: 44.0), homologs of Bombyx mori PGRP5 (contig 11845, RAIH/CH: 10.1) and PGRP-S6 (contig 08467, ARNIF: 57.6), homologs of B. mori CTL10 (contig 14515, residues 54–182, RAIF/CF: 8.7; contig 15639, residues 233–308, RAIF/CF: 5.6; contig 11458, residues 54–306, ARNIF: 28.0), homolog of B. mori Gram-negative binding protein (contig 08247, RAIH/CH: 10.7) (Tanaka et al., 2008), LPS-binding leureptin (contig 15857, RAIH/CH: 10.7) (Zhu et al., 2010), Ig domain-containing hemicentin-1 (contig 00131, RAIF/CF: 6.4) and -2 (contig 14278, RAIF/CF: 8.7) (Vogel and Hedgecock, 2001). Therefore, expression profiling and sequence similarity together provided a powerful tool to discover process-related genes without a priori genome sequence.
Table 2.
A list of 19 UP CIFH contigs with similarity to pattern recognition receptors*
| CIFH contig # | Original read # | RA or ARN | BLAST results | |||||
|---|---|---|---|---|---|---|---|---|
| CF | CH | IF | IH | Total | IF/CF | IH/CH | ||
| 00131 | 11 | 41 | 137 | 61 | 250 | 6.4 | 1.8 | gi|198430641|ref|XP_002123478.1| hemicentin 1, Ig domains [Ciona intestinalis] |
| 00575 | 3 | 0 | 259 | 5 | 267 | 44.0 | 5.9 | gi|154240658|dbj|BAF74637.1| peptidoglycan recognition protein-D [Samia cynthiaricini] |
| 01326 | 1 | 9 | 19 | 70 | 99 | 9.7 | 9.2 | gi|52782739|sp|Q8ISB6.1|BGBP2_MANSE β-1,3-glucan recognition protein 2 |
| 03442 | 1 | 12 | 1468 | 40 | 1521 | 748.5 | 4.0 | gi|511297|gb|AAC46916.1| hemolin [Manduca sexta] |
| 04775 | 1 | 0 | 89 | 0 | 90 | 45.4 | 0.0 | gi|237869126|gb|AAF91316.3|AF242202_1 immulectin-2 [Manduca sexta] |
| 04808 | 0 | 0 | 426 | 2 | 428 | 217.2 | 2.4 | gi|237861314|gb|AAV41237.2| immulectin-4 [Manduca sexta] |
| 06630 | 2 | 40 | 44 | 77 | 163 | 11.2 | 2.3 | gi|55139125|gb|AAV41236.1| immulectin-3 [Manduca sexta] |
| 08247 | 27 | 2 | 122 | 18 | 169 | 2.3 | 10.7 | gi|208972535|gb|ACI32828.1| β-1,3-glucan recognition protein 3 [Helicoverpa armigera] |
| 08467 | 0 | 0 | 113 | 0 | 113 | 57.6 | 0.0 | gi|112983866|ref|NP_001036858.1| peptidoglycan recognition protein-6 [Bombyx mori] |
| 11458 | 0 | 0 | 55 | 0 | 55 | 28.0 | 0.0 | gi|148298818|ref|NP_001091784.1| multi-binding protein [Bombyx mori] |
| 11845 | 0 | 2 | 9 | 17 | 28 | 4.6 | 10.1 | gi|18202160|sp|O76537.1|PGRP_TRINI peptidoglycan recognition protein |
| 13190 | 15 | 0 | 117 | 9 | 141 | 4.0 | 10.7 | gi|27733423|gb|AAO21509.1|AF413068_1 peptidoglycan recognition protein 1A [Manduca sexta] |
| 14104 | 14 | 0 | 173 | 13 | 200 | 6.3 | 15.4 | gi|27733423|gb|AAO21509.1|AF413068_1 peptidoglycan recognition protein 1A [Manduca sexta] |
| 14278 | 1 | 34 | 17 | 179 | 231 | 8.7 | 6.3 | gi|83583693|gb|ABC24706.1| hemicentin-like protein 1, Ig domains [Spodoptera frugiperda] |
| 14515 | 2 | 0 | 34 | 0 | 36 | 8.7 | 0.0 | gi|148298818|ref|NP_001091784.1| multi-binding protein [Bombyx mori] |
| 14700 | 0 | 0 | 183 | 2 | 185 | 93.3 | 2.4 | gi|260765453|gb|ACX49764.1| peptidoglycan recognition protein 2 [Manduca sexta] |
| 14752 | 0 | 0 | 118 | 2 | 120 | 60.2 | 2.4 | gi|260765453|gb|ACX49764.1| peptidoglycan recognition protein 2 [Manduca sexta] |
| 15639 | 10 | 0 | 109 | 0 | 119 | 5.6 | 0.0 | gi|148298818|ref|NP_001091784.1| multi-binding protein [Bombyx mori] |
| 15857 | 0 | 1 | 0 | 9 | 10 | 0.0 | 10.7 | gi|27733411|gb|AAO21503.1|AF413062_1 leureptin, LPS-binding [Manduca sexta] |
RA and ARN are calculated using original read numbers as described in Section 2.3. Listed here are contigs with RAIF/CF >5, RAIH/CH >8, ARNIF >10 when RNCF =0, or ARNIH >10 when RNCH =0. RAIF/CF and RAIH/CH values are shown in red if they are greater than 5 and 8, respectively. ARNIF and ARNIH values are shown in blue if they are higher than 10. In the columns of RA or ARN, cells shaded yellow and blue represent fat body- and hemocyte-specific gene expression, respectively. The complete list of 528 UP CIFH contigs is in Table S1.
B. Extracellular signal transduction and modulation
Hemolymph proteinases (HPs) in insect plasma form enzyme cascades to detect pathogen-PRR complexes and activate precursors of defense proteins (e.g. PO, spätzle, serine proteinase homolog (SPH), and plasmatocyte-spreading peptide (PSP) by limited proteolysis (Jiang and Kanost, 2000). We found eight HPs in the UP list: M. sexta HP7 (ARNIF: 11.2), HP9 (RAIH/CH: 28.5), HP17 (ARNIH: 15.4), HP18 (RAIH/CH: 40.4), HP19 (RAIF/CF: 7.1), HP22 (RAIF/CF: 5.1), proPO-activating proteinase-2 (PAP2) (ARNIF: 50.0), and PAP3 (ARNIF: 22.9) (Table 3). Expression profiles associated with the immune inducibility agreed well with the RT-PCR and northern blot results published earlier (Jiang et al., 2003a, 2003b, 2005). We also found six contigs encoding isoforms of a strongly inducible protein (scolexin) that contained all three catalytic residues of S1A proteinases but did not display any amidase activity (Finnerty et al., 1999). The high ratios and read numbers of these contigs (RAIF/CF: 338.6 and 551.2; ARNIF: 70.9, 129.5, 145.3, 169.8) suggested that primer binding and reverse transcriptase pausing were biased at certain sites of the template because, otherwise, there should not have been any gap for such a short ORF of ~1.36 kb. The exact role of scolexin in defense is still unclear.
Table 3.
A list of 40 UP CIFH contigs with similarity to extracellular signal modulators*
| CIFH contig # | Original read # | RA or ARN | BLAST results | |||||
|---|---|---|---|---|---|---|---|---|
| CF | CH | IF | IH | Total | IF/CF | IH/CH | ||
| 00915 | 0 | 21 | 21 | 26 | 68 | 10.7 | 1.5 | gi|91084647|ref|XP_966816.1| AGAP002414-PA, Zn protease [Tribolium castaneum] |
| 00940 | 0 | 0 | 209 | 7 | 216 | 106.6 | 8.3 | gi|1352212|sp|P48861.1|DDC_MANSE dopa decarboxylase DDC |
| 02023 | 1 | 0 | 33 | 7 | 41 | 16.8 | 8.3 | gi|148611442|gb|ABQ95973.1| tyrosine hydroxylase isoform A [Manduca sexta] |
| 01667 | 0 | 7 | 98 | 33 | 138 | 50.0 | 5.6 | gi|26006435|gb|AAL76085.1| proPO-activating proteinase-2 [Manduca sexta] |
| 01818 | 0 | 26 | 45 | 66 | 137 | 22.9 | 3.0 | gi|60299972|gb|AAX18637.1| proPO-activating proteinase-3 [Manduca sexta] |
| 02361 | 7 | 4 | 70 | 1 | 82 | 5.1 | 0.3 | gi|56418425|gb|AAV91020.1| hemolymph proteinase 22 [Manduca sexta] |
| 02382 | 0 | 2 | 109 | 69 | 180 | 55.6 | 41.0 | gi|4090964|gb|AAD09279.1| immune-related Hdd1 [Hyphantria cunea] |
| 02693 | 21 | 7 | 310 | 19 | 357 | 7.5 | 3.2 | gi|27733415|gb|AAO21505.1|AF413064_1 serpin 3a [Manduca sexta] |
| 02813 | 108 | 9 | 313 | 72 | 502 | 1.5 | 9.5 | gi|242351233|gb|ACS92763.1| serine proteinase-like protein 1b [Manduca sexta] |
| 02985 | 3 | 0 | 158 | 0 | 161 | 26.9 | 0.0 | gi|56418466|gb|AAV91027.1| serine proteinase-like protein 4 [Manduca sexta] |
| 03018 | 0 | 54 | 22 | 79 | 155 | 11.2 | 1.7 | gi|56418395|gb|AAV91005.1| hemolymph proteinase 7 [Manduca sexta] |
| 03778 | 0 | 11 | 192 | 28 | 231 | 97.9 | 3.0 | gi|74813957|sp|Q86RS3.1|DFP_MANSE immune-related Hdd11, precursor |
| 03989 | 0 | 1 | 8 | 24 | 33 | 4.1 | 28.5 | gi|56418399|gb|AAV91007.1| hemolymph proteinase 9 [Manduca sexta] |
| 05186 | 0 | 0 | 8 | 13 | 21 | 4.1 | 15.4 | gi|56418413|gb|AAV91014.1| hemolymph proteinase 17 [Manduca sexta] |
| 05606 | 1 | 0 | 19 | 4 | 24 | 9.7 | 4.7 | gi|4090968|gb|AAD09281.1| immune-related Hdd13 [Hyphantria cunea] |
| 05831 | 3 | 8 | 97 | 25 | 133 | 16.5 | 3.7 | gi|45594232|gb|AAS68507.1| serpin-5A [Manduca sexta] |
| 06149 | 21 | 22 | 686 | 32 | 761 | 16.7 | 1.7 | gi|27733421|gb|AAO21508.1|AF413067_1 serine protease-like protein [Manduca sexta] |
| 06215 | 29 | 1 | 108 | 8 | 146 | 1.9 | 9.5 | gi|112983872|ref|NP_001036857.1| serpin-like protein (SEP-LP) or serpin-12 [Bombyx mori] |
| 06581 | 0 | 0 | 13 | 10 | 23 | 6.6 | 11.9 | gi|4090970|gb|AAD09282.1| immune-related Hdd23 [Hyphantria cunea] |
| 07639 | 651 | 0 | 1237 | 14 | 1902 | 1.0 | 16.6 | gi|134436|sp|P14754.1|SERA_MANSE serpin-1 |
| 08231 | 0 | 1 | 0 | 34 | 35 | 0.0 | 40.4 | gi|56418417|gb|AAV91016.1| hemolymph proteinase 18 [Manduca sexta] |
| 10791 | 1 | 0 | 1081 | 1 | 1083 | 551.2 | 1.2 | gi|4262357|gb|AAD14591.1| scolexin A [Manduca sexta] |
| 10792 | 0 | 0 | 333 | 0 | 333 | 169.8 | 0.0 | gi|4262357|gb|AAD14591.1| scolexin A [Manduca sexta] |
| 13453 | 5 | 4 | 58 | 7 | 74 | 5.9 | 2.1 | gi|45594232|gb|AAS68507.1| serpin-5 [Manduca sexta] |
| 13454 | 0 | 1 | 17 | 10 | 28 | 8.7 | 11.9 | gi|45594232|gb|AAS68507.1| serpin-5 [Manduca sexta] |
| 14093 | 1 | 0 | 14 | 0 | 15 | 7.1 | 0.0 | gi|56418419|gb|AAV91017.1| hemolymph proteinase 19 [Manduca sexta] |
| 14248 | 0 | 6 | 0 | 196 | 202 | 0.0 | 38.8 | gi|2149091|gb|AAB58491.1| serpin-2 [Manduca sexta] |
| 14393 | 2 | 4 | 132 | 11 | 149 | 33.7 | 3.3 | gi|27733421|gb|AAO21508.1|AF413067_1 serine protease-like protein [Manduca sexta] |
| 14456 | 0 | 0 | 1 | 52 | 53 | 0.5 | 61.7 | gi|2149091|gb|AAB58491.1| serpin-2 [Manduca sexta] |
| 15055 | 1 | 1 | 16 | 0 | 18 | 8.2 | 0.0 | gi|112983896|ref|NP_001037394.1| paralytic peptide binding protein 1 [Bombyx mori] |
| 15111 | 1 | 48 | 8 | 800 | 857 | 4.1 | 19.8 | gi|2149091|gb|AAB58491.1| serpin-2 [Manduca sexta] |
| 16520 | 1 | 0 | 664 | 1 | 666 | 338.6 | 1.2 | gi|4262357|gb|AAD14591.1| scolexin A [Manduca sexta] |
| 16917 | 0 | 40 | 2 | 519 | 561 | 1.0 | 15.4 | gi|2149091|gb|AAB58491.1| serpin-2 [Manduca sexta] |
| 17048 | 0 | 1 | 0 | 95 | 96 | 0.0 | 112.8 | gi|2149091|gb|AAB58491.1| serpin-2 [Manduca sexta] |
| 17058 | 0 | 32 | 4 | 545 | 581 | 2.0 | 20.2 | gi|2149091|gb|AAB58491.1| serpin-2 [Manduca sexta] |
| 17751 | 0 | 24 | 1 | 269 | 294 | 0.5 | 13.3 | gi|2149091|gb|AAB58491.1| serpin-2 [Manduca sexta] |
| 18441 | 0 | 1 | 0 | 65 | 66 | 0.0 | 77.2 | gi|2149091|gb|AAB58491.1| serpin-2 [Manduca sexta] |
| 18669 | 0 | 0 | 285 | 0 | 285 | 145.3 | 0.0 | gi|4262357|gb|AAD14591.1| scolexin A [Manduca sexta] |
| 18670 | 0 | 0 | 139 | 0 | 139 | 70.9 | 0.0 | gi|4262357|gb|AAD14591.1| scolexin A [Manduca sexta] |
| 18963 | 0 | 0 | 254 | 0 | 254 | 129.5 | 0.0 | gi|4262357|gb|AAD14591.1| scolexin A [Manduca sexta] |
RA and ARN are calculated using original read numbers as described in Section 2.3. Listed here are contigs with RAIF/CF >5, RAIH/CH >8, ARNIF >10 when RNCF =0, or ARNIH >10 when RNCH =0. RAIF/CF and RAIH/CH values are shown in red if they are greater than 5 and 8, respectively. ARNIF and ARNIH values are shown in blue if they are higher than 10. In the columns of RA or ARN, cells shaded yellow and blue represent fat body- and hemocyte-specific gene expression, respectively. The complete list of 528 UP CIFH contigs is in Table S1.
In the reaction of proPO activation, a high molecular weight complex of SPH1 and SPH2 has to be present along with PAP and proPO to generate active PO (Gupta et al., 2005). In this study, we identified SPH1 (contig 02813, RAIH/CH: 9.5) and SPH2 (contig 6149, RAIF/CF: 16.7; contig 14393, RAIF/CF: 33.7) and confirmed their induced expression (Yu et al., 2003). Contig 02985 (RAIF/CF: 27) contained a complete ORF coding for a regulatory clip domain followed by a serine proteinase-like domain. The protein, designated M. sexta SPH4, is 49% and 92% identical to SPH1 in the amino- and carboxyl-terminal domains, respectively. Such a disparity in sequence alterations suggests that the selection pressures or structural constraints for these two regions differ dramatically.
Functions of serine proteinases are modulated not only by SPHs but also by their inhibitors. Particularly, some members of the serpin superfamily regulate serine proteinase activities by forming covalent complexes with their cognate enzymes (Kanost, 1999). We have identified six serpins in the UP list (Table 3), five of which are known as M. sexta serpin-1 (contig 7639: ARNIH: 16.6), serpin-2 (four contigs, ARNIH: 61.7, RAIH/CH: 13.3, 15.4, 20.2), serpin-2 homolog (four contigs, RAIH/CH: 19.8, 38.8, 77.2, 112.8), serpin-3 (contig 2693, RAIF/CF: 7.5), serpin-5 (three contigs, RA: 5.9, 11.9, 16.5). We have found a new serpin (contig 6215, RAIH/CH: 9.5) and its ortholog in B. mori, SLP or serpin-12. The silkworm serpin was expressed in fat body of bacteria-injected larvae but not in fat body of naïve ones (Zou et al., 2009). Its transcription in hemocytes also was similar to that of the M. sexta serpin: the mRNA was low in naïve larvae and became higher in induced ones.
Besides serine proteinases, SPHs and serpins, we also have found other proteins that either mediate or regulate immune responses in M. sexta or other moths (Table 3). These include: tyrosine hydroxylase (contig 2023, RAIF/CF: 16.8) (Gorman et al., 2007), dopa decarboxylase (contig 00940, ARNIF: 106.6) (Noguchi et al., 2003), PSP-binding protein (contig 15055, RAIF/CF: 8.2) (Matsumoto et al., 2003), and Zn proteinase (contig 0915, ARNIF: 11) (Altincicek and Vilcinskas, 2008). Four immunity-related proteins, Hdd1, Hdd11, Hdd13, and Hdd23 (Shin et al., 1998), are included here even though their functions remain unknown.
C. Intracellular signaling pathways and their components
Pathogen recognition and signal transduction can either go through a PRR-SP system in insect plasma (e.g. spätzle processing for Toll activation) or directly binds to PRRs on the surface of immune tissues/cells (e.g. PGRP-LC binding for Imd activation in Drosophila). After that, intracellular proteins are mobilized to relay signals into the cell nucleus where transcriptional regulation occurs. As shown in Table 4, we have detected increase in transcript levels of the putative pathway members: Toll-like receptors (contigs 06893 and 18001, 68% and 94% similar in amino acid sequence to ABO21763) (Ao et al., 2008), cactus (contig 01044) (Furukawa et al., 2009), relish (contigs 04802 and 15532) (Tanaka et al., 2007), and eiger (contig 01020, a membrane-bound TNF homolog) (Kauppila et al., 2003). Other intracellular proteins possibly involved in signal transduction or modulation include a Ser/Thr protein kinase, GTP/GDP exchange factors, a receptor Tyr phosphatase, a protein phosphatase 2c, ankyrin repeat proteins, and vrille transcription factor.
Table 4.
A list of 18 UP CIFH contigs with similarity to intracellular signal transducers*
| CIFH contig # | Original read # | RA or ARN | BLAST results | |||||
|---|---|---|---|---|---|---|---|---|
| CF | CH | IF | IH | Total | IF/CF | IH/CH | ||
| 00461 | 1 | 48 | 14 | 108 | 171 | 7.1 | 2.7 | gi|47217104|emb|CAG02605.1| unnamed protein product, integrin β6 precursor [Tetraodon nigroviridis] |
| 00537 | 1 | 32 | 10 | 32 | 75 | 5.1 | 1.2 | gi|270009406|gb|EFA05854.1| TcasGA2_TC008649 Tyr protein kinase [Tribolium castaneum] |
| 00671 | 1 | 12 | 10 | 46 | 69 | 5.1 | 4.6 | gi|189235637|ref|XP_967498.2| ral guanine nucleotide exchange factor [Tribolium castaneum] |
| 01020 | 42 | 4 | 63 | 27 | 136 | 0.8 | 8.0 | gi|91082721|ref|XP_972476.1| ~ eiger CG12919-PA, JNK [Tribolium castaneum] |
| 01044 | 9 | 70 | 163 | 105 | 347 | 9.2 | 1.8 | gi|289629214|ref|NP_001166191.1| cactus [Bombyx mori] |
| 01313 | 2 | 52 | 33 | 35 | 122 | 8.4 | 0.8 | gi|242009174|ref|XP_002425367.1| Ser-Thr protein kinase, plant-type [P. humanus corporis] |
| 01390 | 1 | 31 | 14 | 19 | 65 | 7.1 | 0.7 | gi|46403173|gb|AAS92609.1| vrille transcription factor [Antheraea pernyi] |
| 01970 | 1 | 16 | 12 | 29 | 58 | 6.1 | 2.2 | gi|157118595|ref|XP_001659169.1| guanine nucleotide exchange factor [Aedes aegypti] |
| 04802 | 2 | 42 | 25 | 66 | 135 | 6.4 | 1.9 | gi|157412326|ref|NP_001098704.1| Relish2 [Bombyx mori] |
| 05836 | 2 | 1 | 0 | 7 | 10 | 0.0 | 8.3 | gi|189235110|ref|XP_971078.2| receptor tyrosine phosphatase type r2a [Tribolium castaneum] |
| 06304 | 1 | 0 | 11 | 1 | 13 | 5.6 | 1.2 | gi|170038257|ref|XP_001846968.1| dipeptidyl peptidase 4, apoptosis, immunity [Culex quinquefasciatus] |
| 06868 | 0 | 1 | 1 | 11 | 13 | 0.5 | 13.1 | gi|193713771|ref|XP_001946690.1| ankyrin repeat domain 54 [Acyrthosiphon pisum] |
| 06893 | 0 | 1 | 1 | 20 | 22 | 0.5 | 23.7 | gi|126635756|gb|ABO21763.1| Toll receptor [Manduca sexta] |
| 11311 | 0 | 1 | 3 | 9 | 13 | 1.5 | 10.7 | gi|189237512|ref|XP_972880.2| protein phosphatase type 2c [Tribolium castaneum] |
| 11356 | 0 | 1 | 4 | 7 | 12 | 2.0 | 8.3 | gi|156551808|ref|XP_001603899.1| arf6 guanine nucleotide exchange factor [Nasonia vitripennis] |
| 13966 | 0 | 1 | 0 | 9 | 10 | 0.0 | 10.7 | gi|190570736|ref|YP_001975094.1| ankyrin repeat protein [Wolbachia of C. quinquefasciatus Pel] |
| 15532 | 1 | 19 | 12 | 9 | 41 | 6.1 | 0.6 | gi|157412326|ref|NP_001098704.1| Relish2 [Bombyx mori] |
| 18001 | 0 | 1 | 0 | 7 | 8 | 0.0 | 8.3 | gi|126635756|gb|ABO21763.1| Toll receptor [Manduca sexta] |
RA and ARN are calculated using original read numbers as described in Section 2.3. Listed here are contigs with RAIF/CF >5, RAIH/CH >8, ARNIF >10 when RNCF =0, or ARNIH >10 when RNCH =0. RAIF/CF and RAIH/CH values are shown in red if they are greater than 5 and 8, respectively. ARNIF and ARNIH values are shown in blue if they are higher than 10. In the columns of RA or ARN, cells shaded yellow and blue represent fat body- and hemocyte-specific gene expression, respectively. The complete list of 528 UP CIFH contigs is in Table S1.
D. Antimicrobial peptides/proteins
Overproduction of effector proteins that immobilize pathogens, block their proliferation, or directly kill them is a hallmark of insect immunity (Bulet et al., 2004). Consistent with this notion, we have detected 65 UP contigs encoding: A) antimicrobial peptides, B) low molecular weight proteinase inhibitors, C) lysozymes, and D) transferrins (Table 5). In group A, twenty-five contigs (06782, 07203, 08902, 11040, 11711, 13563, 14343, 14380, 14641, 15159, 15732, 15744, 15953, 15997, 16129, 16150, 16576, 17135, 17304, 17350, 17632, 17705, 18324, 18814, 18977) code for at least six attacins, eight (03746, 14568, 15998, 16292, 17184, 18150, 18699, 18819) for at least three X-tox (Girard et al., 2008), six (04903, 07116, 10853, 13916, 17301, 17434) for four lebocin-related proteins (Rayaprolu et al., 2010), four (12151, 13894, 14997, 15041) for three cecropins (Zhu et al., 2003), two (09484, 17439) for two moricins (Dai et al., 2008), and one (02067) for gloverin (Zhu et al., 2003). Group B consists of eight contigs (03142, 03674, 04175, 05197, 08286, 10722, 13936, 16018) encoding proteinase inhibitor-like proteins which may block proteinases released by bacteria, fungi, or parasites (Armstrong, 2006; Zang and Maizels, 2001). Group C has three contigs (08421, 15931, 16133) coding for two lysozymes (Mulnix and Dunn, 1994) that hydrolyze bacterial peptidoglycans. Group D includes seven contigs (02145, 11027, 14937, 16606, 17206, 18239, 18308) encoding at least two transferrins that may sequester iron and, by doing so, prevent bacteria from proliferation (Nichol et al., 2002).
Table 5.
A list of 65 UP CIFH contigs with similarity to antimicrobial proteins*
| CIFH contig # | Original read # | RA or ARN | BLAST results | |||||
|---|---|---|---|---|---|---|---|---|
| CF | CH | IF | IH | Total | IF/CF | IH/CH | ||
| 02067 | 1 | 0 | 280 | 82 | 363 | 142.8 | 97.4 | gi|110649240|emb|CAL25129.1| gloverin [Manduca sexta] |
| 02145 | 0 | 15 | 20 | 95 | 130 | 10.2 | 7.5 | gi|157134051|ref|XP_001663123.1| transferrin [Aedes aegypti] |
| 03142 | 1 | 7 | 420 | 121 | 549 | 214.2 | 20.5 | gi|33860163|sp|P82176.2|IMPI_GALME inducible metalloproteinase inhibitor IMPIα precursor |
| 03674 | 1 | 0 | 5 | 21 | 27 | 2.5 | 24.9 | gi|110347837|gb|ABG72720.1| protease inhibitor-like protein [Antherae amylitta] |
| 03746 | 0 | 7 | 55 | 389 | 451 | 28.0 | 66.0 | gi|148298709|ref|NP_001091749.1| possible antimicrobial peptide [Bombyx mori] |
| 04175 | 0 | 7 | 40 | 45 | 92 | 20.4 | 7.6 | gi|114052803|ref|NP_001040277.1| salivary Cys-rich peptide [Bombyx mori] |
| 04903 | 0 | 0 | 279 | 6 | 285 | 142.3 | 7.1 | gi|187281722|ref|NP_001119732.1| lebocin 3 precursor [Bombyx mori] |
| 05197 | 0 | 0 | 20 | 1 | 21 | 10.2 | 1.2 | gi|115392217|gb|ABI96910.1| brasiliensin precursor, thrombin inhibitor [Triatoma brasiliensis] |
| 06782 | 0 | 0 | 102 | 17 | 119 | 52.0 | 20.2 | gi|67906420|gb|AAY82587.1| attacin-1 [Manduca sexta] |
| 07116 | 1 | 4 | 902 | 3 | 910 | 459.9 | 0.9 | gi|171262319|gb|ACB45566.1| lebocin-like protein [Antheraea pernyi] |
| 07203 | 2 | 3 | 312 | 22 | 339 | 79.5 | 8.7 | gi|67906420|gb|AAY82587.1| attacin-1 [Manduca sexta] |
| 08286 | 0 | 0 | 139 | 23 | 162 | 70.9 | 27.3 | gi|56462340|gb|AAV91453.1| protease inhibitor 6 [Lonomia obliqua] |
| 08421 | 4 | 2 | 28 | 99 | 133 | 3.6 | 58.8 | gi|7327646|gb|AAB31190.2| lysozyme [Manduca sexta] |
| 08902 | 0 | 0 | 164 | 14 | 178 | 83.6 | 16.6 | gi|67906420|gb|AAY82587.1| attacin-1 [Manduca sexta] |
| 09484 | 1 | 0 | 134 | 56 | 191 | 68.3 | 66.5 | gi|29469961|gb|AAO74637.1| antimicrobial peptide moricin [Manduca sexta] |
| 10234 | 0 | 1 | 249 | 7 | 257 | 127.0 | 8.3 | gi|169264911|dbj|BAG12297.1| gallerimycin [Samia cynthia ricini] |
| 10722 | 9 | 3 | 102 | 3 | 117 | 5.8 | 1.2 | gi|110347833|gb|ABG72718.1| protease inhibitor-like protein [Antherae amylitta] |
| 10853 | 0 | 0 | 113 | 1 | 114 | 57.6 | 1.2 | gi|171262319|gb|ACB45566.1| lebocin-like protein [Antheraea pernyi] |
| 11027 | 59 | 0 | 694 | 0 | 753 | 6.0 | 0.0 | gi|136206|sp|P22297.1|TRF_MANSE transferrin precursor |
| 11040 | 0 | 4 | 51 | 249 | 304 | 26.0 | 73.9 | gi|29469969|gb|AAO74640.1| antimicrobial protein attacin 2 [Manduca sexta] |
| 11711 | 0 | 7 | 85 | 1317 | 1409 | 43.3 | 223.4 | gi|29469969|gb|AAO74640.1| antimicrobial protein attacin 2 [Manduca sexta] |
| 12151 | 0 | 0 | 153 | 0 | 153 | 78.0 | 0.0 | gi|116084|sp|P14665.1|CEC5_MANSE bactericidin B-5P, cecropin-like |
| 13563 | 0 | 0 | 657 | 0 | 657 | 335.0 | 0.0 | gi|110347786|gb|ABG72695.1| attacin-like protein [Antheraea mylitta] |
| 13894 | 0 | 0 | 48 | 29 | 77 | 24.5 | 34.4 | gi|112984238|ref|NP_001037460.1| cecropin B precursor [Bombyx mori] |
| 13916 | 1 | 0 | 741 | 0 | 742 | 377.8 | 0.0 | gi|219958086|gb|ACL68097.1| lebocin-related protein precursor [Manduca sexta] |
| 13936 | 0 | 0 | 25 | 0 | 25 | 12.7 | 0.0 | gi|123725|sp|P26227.1|HTIB_MANSE trypsin inhibitor B, BPTI-type |
| 14343 | 0 | 0 | 186 | 7 | 193 | 94.8 | 8.3 | gi|67906420|gb|AAY82587.1| attacin-1 [Manduca sexta] |
| 14380 | 0 | 0 | 106 | 0 | 106 | 54.0 | 0.0 | gi|67906420|gb|AAY82587.1| attacin-1 [Manduca sexta] |
| 14568 | 0 | 0 | 2 | 68 | 70 | 1.0 | 80.7 | gi|148298709|ref|NP_001091749.1| possible antimicrobial peptide [Bombyx mori] |
| 14641 | 0 | 0 | 157 | 0 | 157 | 80.1 | 0.0 | gi|67906420|gb|AAY82587.1| attacin-1 [Manduca sexta] |
| 14937 | 13 | 0 | 164 | 0 | 177 | 6.4 | 0.0 | gi|136206|sp|P22297.1|TRF_MANSE: transferrin precursor |
| 14997 | 0 | 0 | 34 | 10 | 44 | 17.3 | 11.9 | gi|29469965|gb|AAO74638.1| antimicrobial peptide cecropin 6 [Manduca sexta] |
| 15041 | 0 | 0 | 36 | 0 | 36 | 18.4 | 0.0 | gi|116084|sp|P14665.1|CEC5_MANSE bactericidin B-5P, cecropin-like |
| 15159 | 0 | 0 | 0 | 15 | 15 | 0.0 | 17.8 | gi|15963410|dbj|BAB69462.1| attacin [Samia cynthia ricini] |
| 15732 | 0 | 1 | 253 | 43 | 297 | 129.0 | 51.1 | gi|67906420|gb|AAY82587.1| attacin-1 [Manduca sexta] |
| 15744 | 0 | 0 | 0 | 35 | 35 | 0.0 | 41.6 | gi|29469969|gb|AAO74640.1| antimicrobial protein attacin 2 [Manduca sexta] |
| 15931 | 40 | 37 | 1504 | 364 | 1945 | 19.2 | 11.7 | gi|7327646|gb|AAB31190.2| lysozyme [Manduca sexta] |
| 15953 | 1 | 0 | 43 | 6 | 50 | 21.9 | 7.1 | gi|67906420|gb|AAY82587.1| attacin-1 [Manduca sexta] |
| 15997 | 0 | 0 | 142 | 4 | 146 | 72.4 | 4.7 | gi|67906420|gb|AAY82587.1| attacin-1 [Manduca sexta] |
| 15998 | 0 | 0 | 1 | 10 | 11 | 0.5 | 11.9 | gi|73921456|gb|AAZ94260.1| immune related protein X-tox [Spodoptera frugiperda] |
| 16018 | 0 | 0 | 40 | 12 | 52 | 20.4 | 14.2 | gi|116833115|gb|ABK29470.1| immune reactive putative protease inhibitor [Helicoverpa armigera] |
| 16129 | 1 | 0 | 212 | 35 | 248 | 108.1 | 41.6 | gi|67906420|gb|AAY82587.1| attacin-1 [Manduca sexta] |
| 16133 | 47 | 57 | 1719 | 440 | 2263 | 18.6 | 9.2 | gi|233964|gb|AAB19535.1| lysozyme |
| 16150 | 0 | 1 | 145 | 3 | 149 | 73.9 | 3.6 | gi|67906420|gb|AAY82587.1| attacin-1 [Manduca sexta] |
| 16292 | 0 | 0 | 1 | 34 | 35 | 0.5 | 40.4 | gi|148298709|ref|NP_001091749.1| possible antimicrobial peptide [Bombyx mori] |
| 16576 | 0 | 0 | 0 | 18 | 18 | 0.0 | 21.4 | gi|74767320|sp|Q5MGE6.1|DFP3_LONON defense protein 3 precursor, attacin E |
| 16606 | 8 | 0 | 164 | 0 | 172 | 10.5 | 0.0 | gi|136206|sp|P22297.1|TRF_MANSE transferrin precursor |
| 17135 | 0 | 9 | 103 | 1157 | 1269 | 52.5 | 152.6 | gi|110649242|emb|CAL25130.1| attacin II [Manduca sexta] |
| 17184 | 0 | 11 | 76 | 449 | 536 | 38.8 | 48.5 | gi|73921456|gb|AAZ94260.1| immune related protein, X-tox [Spodoptera frugiperda] |
| 17206 | 3 | 0 | 136 | 0 | 139 | 23.1 | 0.0 | gi|136206|sp|P22297.1|TRF_MANSE transferrin precursor |
| 17301 | 1 | 0 | 272 | 0 | 273 | 138.7 | 0.0 | gi|219958086|gb|ACL68097.1| lebocin-related protein precursor [Manduca sexta] |
| 17304 | 0 | 1 | 412 | 13 | 426 | 210.1 | 15.4 | gi|67906420|gb|AAY82587.1| attacin-1 [Manduca sexta] |
| 17350 | 0 | 0 | 205 | 0 | 205 | 104.5 | 0.0 | gi|29469969|gb|AAO74640.1| antimicrobial protein attacin 2 [Manduca sexta] |
| 17434 | 1 | 0 | 314 | 0 | 315 | 160.1 | 0.0 | gi|219958086|gb|ACL68097.1| lebocin-related protein precursor [Manduca sexta] |
| 17439 | 0 | 0 | 98 | 31 | 129 | 50.0 | 36.8 | gi|110649236|emb|CAL25127.1| moricin [Manduca sexta] |
| 17632 | 0 | 0 | 83 | 6 | 89 | 42.3 | 7.1 | gi|67906420|gb|AAY82587.1| attacin-1 [Manduca sexta] |
| 17705 | 0 | 0 | 36 | 0 | 36 | 18.4 | 0.0 | gi|67906420|gb|AAY82587.1| attacin-1 [Manduca sexta] |
| 18150 | 0 | 0 | 0 | 18 | 18 | 0.0 | 21.4 | gi|148298709|ref|NP_001091749.1| possible antimicrobial peptide [Bombyx mori] |
| 18239 | 3 | 0 | 67 | 0 | 70 | 11.4 | 0.0 | gi|136206|sp|P22297.1|TRF_MANSE transferrin precursor |
| 18308 | 15 | 0 | 169 | 0 | 184 | 5.7 | 0.0 | gi|136206|sp|P22297.1|TRF_MANSE transferrin precursor |
| 18324 | 0 | 0 | 25 | 0 | 25 | 12.7 | 0.0 | gi|67906420|gb|AAY82587.1| attacin-1 [Manduca sexta] |
| 18699 | 0 | 1 | 26 | 114 | 141 | 13.3 | 135.4 | gi|148298709|ref|NP_001091749.1| possible antimicrobial peptide [Bombyx mori] |
| 18814 | 0 | 0 | 235 | 29 | 264 | 119.8 | 34.4 | gi|67906420|gb|AAY82587.1| attacin-1 [Manduca sexta] |
| 18819 | 0 | 5 | 59 | 405 | 469 | 30.1 | 96.2 | gi|73921456|gb|AAZ94260.1| immunity-related protein X-tox [Spodoptera frugiperda] |
| 18977 | 0 | 1 | 20 | 2 | 23 | 10.2 | 2.4 | gi|67906420|gb|AAY82587.1| attacin-1 [Manduca sexta] |
RA and ARN are calculated using original read numbers as described in Section 2.3. Listed here are contigs with RAIF/CF >5, RAIH/CH >8, ARNIF >10 when RNCF =0, or ARNIH >10 when RNCH =0. RAIF/CF and RAIH/CH values are shown in red if they are greater than 5 and 8, respectively. ARNIF and ARNIH values are shown in blue if they are higher than 10. In the columns of RA or ARN, cells shaded yellow and blue represent fat body- and hemocyte-specific gene expression, respectively. The complete list of 528 UP CIFH contigs is in Table S1.
E. Other up-regulated genes
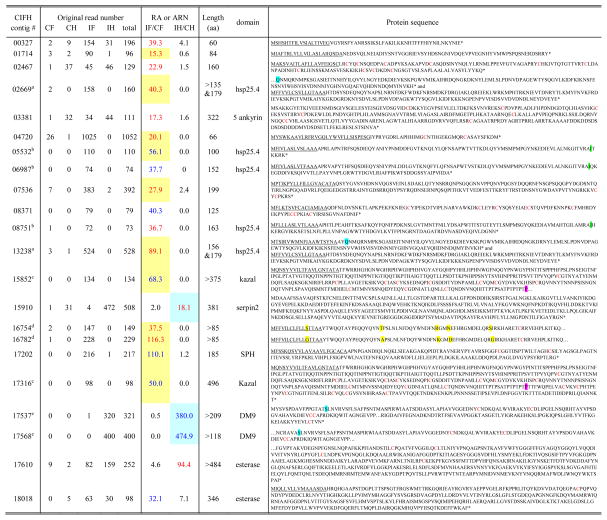
Among the 528 UP contigs, 177 did not have any BLAST hits (Table S1), indicating that some of them may encode polypeptides previously not known to be involved in immunity. To ensure these sequences are indeed up-regulated, we selected contigs with RA >15 (or ARN >30) and total read numbers >70. We then extended these contigs, if possible, with sequences in dataset “06” (Table 1) and in the M. sexta gut EST dataset (Pauchet et al., 2010). After eliminating the contigs with GC-contents <35% (hence, likely representing 5′ or 3′ AT-rich untranslated regions of up-regulated genes), we examined the remaining ones in greater detail (Table 6). Contigs 00327, 01714, 04720, 05532, and 07536 contain ORFs with a secretion signal peptide. The putative mature proteins (41, 61, 37, 86, 179 residues long) could be novel AMPs or in other ways involved in immunity. Contig 02467 encodes a secreted protein containing ten Cys that may tether the 139-residue polypeptide into a stable domain functioning as a proteinase inhibitor or an antifungal protein (Kanost, 1999). Contigs 15852 and 17316 encode proteins with 2 and 3 Kazal-type proteinase inhibitor domains, respectively. Contigs 17537 and 17568 encode proteins with a DM9 domain. Contigs 03381 and 15910, after extension, are found to be a part of cactus and serpin-2 transcripts. The other contigs encode sequences similar to B. mori heat shock protein 25.4, SPH, and esterases.
Table 6.
A list of 22 UP CIFH contigs without BLAST hit*
|
RA and ARN are calculated using original read numbers as described in Section 2.3. Listed here are contigs with RAIF/CF >15, RAIH/CH >15, ARNIF >30 when RNCF =0, or ARNIH >30 when RNCH =0. Contigs with total read numbers lower than 70 or GC content lower than 35% are not listed. Some of the contig sequences have been extended using sequences in dataset “06” (Table 1, Zou et al., 2008) and in the M. sexta gut EST dataset (Pauchet et al., 2009). RAIF/CF and RAIH/CH values are shown in red if they are greater than 15, while ARNIF and ARNIH values are shown in blue if they are higher than 30. In the two columns of RA or ARN, cells shaded yellow and blue represent fat body- and hemocyte-specific gene expression, respectively. The complete list of 528 UP CIFH contigs is in Table S1. The contigs labeled with the same letter (a to e) in superscript indicate high sequence similarity between them, as highlighted with different colors at certain key sites of the protein sequences. Underlined sequences represent putative signal peptides. The * indicates the end of protein sequence (stop codon).
3.3. Sequence analysis and function prediction of DN genes
The analysis of down-regulated genes yielded results that surprised us at first: among the 53 DN CIFH contig groups with BLAST hits, ten were closely related to immune responses (Table 7). A contig group represents a single contig in most cases but, in other times, has multiple contigs with the same BLAST hit, which may come from different genes. They include lectins (06497, 07642, 11280, 13813, 14570, 14760), lacunin (00015), HP1 (16288), and proPOs (17085 and 17958). A closer inspection of the data indicated that the decreases in mRNA levels seem to always occur in fat body instead of hemocytes. Since these genes were all expressed at much higher levels in hemocytes than fat body (RACH/CF or IH/IF >40), we suggest the apparent down regulation in fat body were caused by unequal contamination of fat body tissue by hemocytes: somehow there was much less contamination in induced fat body of these hemocyte-specific transcripts. In hemocytes, their average RACH/IH was only 2.1 – no major down-regulation was observed for these immunity-related genes in cells mainly expressing them. It is likely that similar contamination of fat body tissue by hemocytes also resulted in the observation of genes not known to be directly related to immunity, which includes 11 contig groups (00010, 00248, 00379, 00623, 00628, 03286, 03654, 07139, 08686, 10124, 13842) with RACH/CF or IH/IF >40 (hemocyte-specific) and RACF/IF >10 (fat body DN) but RACH/IH <3.
Table 7.
A list of DN CIFH contigs with BLAST hits*
| CIFH contig # | Original read # | RA or ARN | BLAST results | |||||
|---|---|---|---|---|---|---|---|---|
| CF | CH | IF | IH | Total | CF/IF | CH/IH | ||
| 00010 | 29 | 464 | 3 | 286 | 782 | 19.0 | 1.4 | gi|242005387|ref|XP_002423550.1| cAMP-dependent protein kinase subunit [Pediculus humanus corporis] |
| 00015 & | 112 | 4705 | 17 | 3918 | 8752 | 12.9 | 1.0 | gi|6164595|gb|AAF04457.1|AF078161_1 lacunin [Manduca sexta] [00015, 02717] |
| 00248 | 7 | 200 | 1 | 155 | 363 | 13.7 | 1.1 | gi|157113908|ref|XP_001657920.1| n-acetyllactosaminide β-1,3-NAG transferase [Aedes aegypti] |
| 00379 | 10 | 308 | 1 | 184 | 503 | 19.6 | 1.4 | gi|170037242|ref|XP_001846468.1| Leu-rich repeat-containing protein 1 [Culex quinquefasciatus] |
| 00623 | 12 | 527 | 1 | 443 | 983 | 23.5 | 1.0 | gi|157132531|ref|XP_001656056.1| odd Oz protein [Aedes aegypti] |
| 00628 | 7 | 38 | 1 | 39 | 85 | 13.7 | 0.8 | gi|170030982|ref|XP_001843366.1| rho/rac/cdc GTPase-activating protein [Culex quinquefasciatus] |
| 00773 | 49 | 12 | 93 | 1 | 155 | 1.0 | 10.1 | gi|157103945|ref|XP_001648193.1| dihydropyrimidine dehydrogenase [Aedes aegypti] |
| 00851 | 6 | 42 | 1 | 26 | 75 | 11.8 | 1.4 | gi|158300087|ref|XP_320080.3| AGAP009284-PA [Anopheles gambiae] |
| 01289 | 7 | 45 | 1 | 31 | 84 | 13.7 | 1.2 | gi|187281809|ref|NP_001119723.1| kinesin-like protein Ncd [Bombyx mori] |
| 02637 | 5 | 12 | 9 | 1 | 27 | 1.1 | 10.1 | gi|116789445|gb|ABK25249.1| unknown [Picea sitchensis] |
| 02730 | 8 | 15 | 8 | 1 | 32 | 2.0 | 12.6 | gi|2970687|gb|AAC06038.1| β-glucosidase precursor [Spodoptera frugiperda] |
| 03286 etc. | 62 | 1976 | 7 | 586 | 2631 | 17.4 | 2.8 | gi|254746344|emb|CAX16637.1| C1A Cys protease [Manduca sexta] [03286, 05560, 15201, 17978] |
| 03654 | 21 | 686 | 2 | 647 | 1356 | 20.6 | 0.9 | gi|157134123|ref|XP_001663157.1| atlastin [Aedes aegypti] |
| 03792 | 7 | 20 | 1 | 5 | 33 | 13.7 | 3.4 | gi|91090218|ref|XP_968156.1| E1a binding protein P400 [Tribolium castaneum] |
| 03996 | 6 | 6 | 1 | 6 | 19 | 11.8 | 0.8 | gi|170052039|ref|XP_001862040.1| small GTP-binding protein [Culex quinquefasciatus] |
| 05824 | 8 | 0 | 1 | 4 | 13 | 15.7 | 0.0 | gi|116326818|ref|YP_803355.1| hypothetical TNAV2c gp132 [Trichoplusia ni ascovirus 2c] |
| 06497 etc. | 225 | 10451 | 12 | 4266 | 14954 | 36.8 | 2.1 | gi|217262|dbj|BAA03124.1| lectin [Bombyx mori] [06497, 15047, 15764, 16677, 16801, 16877, 16886, 17700] |
| 06713 | 0 | 12 | 0 | 1 | 13 | 0.0 | 10.1 | gi|193613364|ref|XP_001943860.1| limkain b1 [Acyrthosiphon pisum] |
| 06902 | 12 | 3 | 2 | 0 | 17 | 11.8 | 2.5 | gi|114050917|ref|NP_001040414.1| 3-hydroxyacyl-CoA dehydrogenase [Bombyx mori] |
| 07139 | 21 | 767 | 2 | 262 | 1052 | 20.6 | 2.5 | gi|110649216|emb|CAL25117.1| dVA-AP3 [Manduca sexta] |
| 07515 | 7 | 1 | 1 | 0 | 9 | 13.7 | 0.8 | gi|158295141|ref|XP_316035.4| AGAP005993-PA [Anopheles gambiae] |
| 07642 | 9 | 601 | 1 | 153 | 764 | 17.7 | 3.3 | gi|55139125|gb|AAV41236.1| immulectin-3 [Manduca sexta] |
| 07754 | 0 | 12 | 1 | 1 | 14 | 0.0 | 10.1 | gi|71895231|ref|NP_001026433.1| coiled-coil domain containing 93 [Gallus gallus] |
| 08686 & | 21 | 854 | 3 | 680 | 1558 | 13.7 | 1.1 | gi|82880638|gb|ABB92836.1| scavenger receptor C-like protein [Spodoptera frugiperda] [08686, 15116] |
| 08705 | 8 | 10 | 1 | 5 | 24 | 15.7 | 1.7 | gi|224084416|ref|XP_002192181.1| selenium binding protein 1 [Taeniopygia guttata] |
| 08707 | 6 | 9 | 1 | 13 | 29 | 11.8 | 0.6 | gi|24585081|ref|NP_609923.2| CG10639 [Drosophila melanogaster] |
| 08801 | 1 | 14 | 1 | 1 | 17 | 2.0 | 11.8 | gi|91081401|ref|XP_972667.1| exosome component 8 [Tribolium castaneum] |
| 09847 | 0 | 13 | 0 | 1 | 14 | 0.0 | 10.9 | gi|194745608|ref|XP_001955279.1| GF16313 [Drosophila ananassae] |
| 10124 etc. | 115 | 4638 | 8 | 2848 | 7609 | 28.2 | 1.4 | gi|114050871|ref|NP_001040411.1| carboxylesterase [Bombyx mori] [10124, 16922, 17330, 18860] |
| 10316 | 0 | 13 | 1 | 1 | 15 | 0.0 | 10.9 | gi|157106599|ref|XP_001649397.1| hypothetical protein AaeL_AAEL004554 [Aedes aegypti] |
| 10439 | 12 | 0 | 1 | 0 | 13 | 23.5 | 0.0 | gi|183979241|dbj|BAG30782.1| cuticular protein CPR41B [Papilio xuthus] |
| 11030 | 13 | 0 | 2 | 0 | 15 | 12.7 | 0.0 | gi|3121953|sp|Q25504.1|CU16_MANSE larval cuticle protein 16/17 precursor |
| 11098 | 40 | 0 | 3 | 0 | 43 | 26.1 | 0.0 | gi|159526|gb|AAA29320.1| Met-rich storage protein 1 [Manduca sexta] |
| 11161 | 0 | 12 | 1 | 1 | 14 | 0.0 | 10.1 | gi|125808686|ref|XP_001360831.1| GA18253 [Drosophila pseudoobscura] |
| 11280 etc. | 143 | 7866 | 11 | 2589 | 10609 | 25.5 | 2.6 | gi|91090548|ref|XP_971239.1| hemolectin CG7002-PA [Tribolium castaneum] [11280, 15506, 15594, 18551] |
| 12095 | 10 | 0 | 1 | 0 | 11 | 19.6 | 0.0 | gi|194741936|ref|XP_001953465.1| GF17208 [Drosophila ananassae] |
| 12848 | 0 | 16 | 0 | 1 | 17 | 0.0 | 13.5 | gi|2822109|sp|P14730.2|EXPI_RAT extracellular peptidase inhibitor, WDNM1 precursor |
| 13013 | 7 | 1 | 1 | 0 | 9 | 13.7 | 0.8 | gi|189031278|gb|ACD74812.1| cuticle protein 1 [Helicoverpa armigera] |
| 13094 | 15 | 10 | 1 | 5 | 31 | 29.4 | 1.7 | gi|183979298|dbj|BAG30762.1| similar to CG5304-PA [Papilio xuthus] |
| 13813 | 31 | 2398 | 4 | 848 | 3281 | 15.2 | 2.4 | gi|110758905|ref|XP_395067.3| hemolectin CG7002-PA [Apis mellifera] |
| 13842 | 14 | 677 | 2 | 228 | 921 | 13.7 | 2.5 | gi|138601|sp|P19616.1|VITM_MANSE microvitellogenin precursor |
| 14129 | 7 | 0 | 1 | 0 | 8 | 13.7 | 0.0 | gi|91078692|ref|XP_971204.1| phospholipase A2, grp VI (cytosolic, Ca-independent) [Tribolium castaneum] |
| 14570 etc. | 559 | 28386 | 29 | 10677 | 39651 | 37.8 | 2.2 | gi|162462371|ref|NP_001104817.1| lectin [Bombyx mori] [14570, 15250, 15380, 15792, 16289, 16291, 16594, 16842, 17159, 17421, 17471, 17732, 17769, 18032, 18067, 18097, 18286, 18326, 18719, 18721, 18794, 18997] |
| 14760 etc. | 57 | 3372 | 3 | 1184 | 4616 | 37.3 | 2.4 | gi|156545430|ref|XP_001606650.1| CG7002-PA [Nasonia vitripennis] [14760, 18045] |
| 14781 | 28 | 0 | 3 | 1 | 32 | 18.3 | 0.0 | gi|114052677|ref|NP_001040269.1| phosphoserine aminotransferase 1 [Bombyx mori] |
| 15132 | 9 | 0 | 1 | 0 | 10 | 17.7 | 0.0 | gi|112984526|ref|NP_001037199.1| promoting protein [Bombyx mori] |
| 15465 | 6 | 0 | 1 | 1 | 8 | 11.8 | 0.0 | gi|170574840|ref|XP_001892989.1| hypothetical protein Bm1_07595 [Brugia malayi] |
| 16105 | 10 | 23 | 1 | 42 | 76 | 19.6 | 0.5 | gi|91087179|ref|XP_975411.1| CG9471-PB [Tribolium castaneum] |
| 16288 etc. | 63 | 3044 | 4 | 1126 | 4237 | 30.9 | 2.3 | gi|2738863|gb|AAB94557.1| hemocyte protease-1 [Manduca sexta] [16288, 16719, 17102] |
| 17085 etc. | 236 | 11035 | 27 | 7455 | 18753 | 17.1 | 1.2 | gi|74763772|sp|O44249.3|MANSE proPO-p1 [17085, 17315, 17420, 17612, 17629, 18065, 18463, 18887] |
| 17958 etc. | 130 | 5309 | 19 | 3669 | 9127 | 16.3 | 1.2 | gi|75038472|sp|Q25519.3|MANSE proPO-p2 [17958, 18004, 18516] |
| 18482 | 11 | 0 | 0 | 0 | 11 | 21.6 | 0.0 | gi|114240|sp|P14296.1|ARYA_MANSE arylphorin α subunit precursor |
| 18611 | 0 | 12 | 4 | 1 | 17 | 0.0 | 10.1 | gi|12585261|sp|Q9U639.1|HSP7D_MANSE heat shock 70 kDa protein cognate 4 (Hsp70-4) |
RA and ARN are calculated using original read numbers as described in Section 2.3. Listed here are contigs with RACF/IF >10, RACH/IH >10, ARNCF >20 when RNIF =0, or ARNCH >20 when RNIH =0. RACF/IF and RACH/IH values are shown in red if they are greater than 10, whereas ARNCF and ARNCH values are shown in blue if they are higher than 20. In the two columns of RA or ARN, cells shaded yellow and blue represent fat body- and hemocyte-specific gene expression, respectively. Contigs with identical BLAST results are combined, with their average RAs or ARNs calculated based on the sums of original reads in CF, CH, IF, and IH for each group. Contigs with no BLAST hit can be found in Table S2, a complete list of 148 DN CIFH contigs.
After eliminating contigs whose RACH/CF or IH/IF calculated from low read numbers, we have found four DN contigs: 02730 encodes a β-glucosidase, 11098 a Met-rich storage protein, 12848 a proteinase inhibitor, and 14781 a phosphoserine amino transferase. Follow-up studies are needed to confirm their down-regulation and explore physiological relevance of the decrease in transcript levels.
3.4. Tissue-specifically regulated genes in larval hemocytes
Using the same set of read numbers in CIFH, we found 45 contig groups representing genes preferentially expressed in hemocytes. Interestingly, this tissue-specific pattern (RA >40 or ARNIH >80) was only found in the induced samples but not in the control ones (Table 8). A closer examination of the data uncovered the possible reason for this bias: although fat body was collected under the same conditions, more hemocytes attached to the control fat body tissue than the induced one. Consequently, higher read numbers from contaminating hemocytes in control fat body led to much lower RACH/CF values than their corresponding RAIH/IF’s. While the same reason caused wrong identification of some contigs as down-regulated ones (Table 7), the skewing of RAs against the control samples (i.e. lower RACH/CF) did not seem to affect the correct calling of hemocyte-specificity in a qualitative term. For the entire contig groups, the sums of CF and CH reads were 2173 and 105143, respectively. The average RACH/CF of 10.0 was much lower than the cutoff value of 40 but still substantially higher than 2–5, thresholds commonly used in microarray or qPCR studies to assess differential expression. In comparison, the sum of IF and IH reads were 302 and 62907, respectively, and their average RAIH/IF was 100.5.
Table 8.
A list of HC CIFH contigs with BLAST hits*
| CIFH contig # | Original read # | RA or ARN | BLAST results | |||||
|---|---|---|---|---|---|---|---|---|
| CF | CH | IF | IH | Total | CH/CF | IH/IF | ||
| 00010 | 29 | 464 | 3 | 286 | 782 | 3.3 | 46.0 | gi|242005387|ref|XP_002423550.1| cAMP-dependent protein kinase catalytic subunit [Pediculus humanus corporis] |
| 00015 etc. | 119 | 5073 | 20 | 4227 | 9439 | 8.8 | 102.0 | gi|6164595|gb|AAF04457.1|AF078161_1 lacunin [Manduca sexta] (00015, 02717, 15269) |
| 00028 | 13 | 958 | 4 | 754 | 1729 | 15.3 | 91.0 | gi|91081003|ref|XP_975140.1| odd Oz protein [Tribolium castaneum] |
| 00248 | 7 | 200 | 1 | 155 | 363 | 5.9 | 74.8 | gi|157113908|ref|XP_001657920.1| n-acetyllactosaminide β-1,3-NAG transferase [Aedes aegypti] |
| 00379 | 10 | 308 | 1 | 184 | 503 | 6.4 | 88.8 | gi|170037242|ref|XP_001846468.1| Leu-rich repeat-containing protein 1 [Culex quinquefasciatus] |
| 00541 | 14 | 567 | 7 | 760 | 1348 | 8.4 | 52.4 | gi|170029717|ref|XP_001842738.1| Leu-rich repeat-containing G-protein coupled receptor 4 [Culex quinquefasciatus] |
| 00569 | 4 | 182 | 1 | 176 | 363 | 9.4 | 85.0 | gi|283135216|ref|NP_001164363.1| homeobox protein prospero [Nasonia vitripennis] |
| 00623 | 12 | 527 | 1 | 443 | 983 | 9.1 | 213.8 | gi|157132531|ref|XP_001656056.1| odd Oz protein [Aedes aegypti] |
| 00752 | 0 | 38 | 1 | 164 | 203 | 7.9 | 79.2 | gi|194859640|ref|XP_001969420.1| GG23966 [Drosophila erecta] |
| 00802 | 3 | 203 | 3 | 253 | 462 | 14.0 | 40.7 | gi|260840271|ref|XP_002613791.1| hypothetical BRAFLDRAFT_85332 [Branchiostoma floridae] |
| 00839 | 3 | 340 | 1 | 226 | 570 | 23.5 | 109.1 | gi|242021897|ref|XP_002431379.1| conserved hypothetical protein [Pediculus humanus corporis] |
| 00882 | 7 | 268 | 0 | 261 | 536 | 7.9 | 126.0 | gi|112983326|ref|NP_001037620.1| ras-related GTP-binding protein Rab3 [Bombyx mori] |
| 01064 | 5 | 134 | 1 | 116 | 256 | 5.6 | 56.0 | gi|48095930|ref|XP_394560.1| Jagged-1 precursor (Jagged1, hJ1, CD339 antigen) [Apis mellifera] |
| 01429 & | 27 | 924 | 4 | 827 | 1782 | 7.1 | 99.8 | gi|157134123|ref|XP_001663157.1| atlastin [Aedes aegypti] (01429, 03654) |
| 01609 | 1 | 71 | 1 | 144 | 217 | 14.7 | 69.5 | gi|134001247|gb|ABO45233.1| reverse transcriptase [Ostrinia nubilalis] |
| 02159 | 3 | 101 | 1 | 144 | 249 | 7.0 | 69.5 | gi|114052056|ref|NP_001040346.1| septin [Bombyx mori] |
| 02473 | 10 | 255 | 2 | 382 | 649 | 5.3 | 92.2 | gi|281362668|ref|NP_651533.2| eater [Drosophila melanogaster] |
| 02852 | 23 | 1128 | 7 | 885 | 2043 | 10.2 | 61.0 | gi|66391199|ref|YP_239364.1| hypothetical protein [Microplitis demolitorbracovirus] |
| 03225 | 1 | 25 | 1 | 143 | 170 | 5.2 | 69.0 | gi|195445668|ref|XP_002070431.1| GK11035 [Drosophila willistoni] |
| 03246 & | 4 | 182 | 2 | 245 | 433 | 9.4 | 59.1 | gi|83583697|gb|ABC24708.1| G protein-coupled receptor [Spodoptera frugiperda] (03246, 06319) |
| 03287 | 7 | 493 | 0 | 237 | 737 | 14.6 | 114.4 | gi|114052174|ref|NP_001040228.1| aminoacylase [Bombyx mori] |
| 04085 | 0 | 34 | 3 | 268 | 305 | 7.0 | 43.1 | gi|206725499|ref|NP_001128673.1| cathepsin L like protein [Bombyx mori] |
| 04278 | 3 | 141 | 1 | 154 | 299 | 9.7 | 74.3 | gi|270001550|gb|EEZ97997.1| hypothetical TcasGA2_TC000395 [Tribolium castaneum] |
| 04746 etc. | 0 | 0 | 16 | 1939 | 1955 | 0.0 | 58.5 | gi|195486646|ref|XP_002091593.1| GE13745 [Drosophila yakuba] (04746, 13353, 14100) |
| 05560 | 24 | 965 | 4 | 440 | 1433 | 8.3 | 53.1 | gi|254746344|emb|CAX16637.1| putative C1A Cys protease precursor [Manduca sexta] |
| 05577 | 4 | 157 | 22 | 1895 | 2078 | 8.1 | 41.6 | gi|254746342|emb|CAX16636.1| putative C1A Cys protease precursor [Manduca sexta] |
| 05933 etc. | 39 | 1862 | 8 | 1395 | 3304 | 9.9 | 84.2 | gi|82880638|gb|ABB92836.1| SRC-like protein [Spodoptera frugiperda] (05933, 08686, 13271, 15116, 15350, 15564) |
| 06497 etc. | 237 | 11297 | 15 | 4531 | 16080 | 9.9 | 145.8 | gi|217262|dbj|BAA03124.1| lectin [Bombyx mori] (06497, 15047, 15764, 15986, 16677, 16801, 16877, 16886, 17700) |
| 07139 | 21 | 767 | 2 | 262 | 1052 | 7.6 | 63.2 | gi|110649216|emb|CAL25117.1| dVA-AP3 [Manduca sexta] |
| 07199 | 2 | 73 | 1 | 102 | 178 | 7.6 | 49.2 | gi|110649250|emb|CAL25134.1| immulectin III [Manduca sexta] |
| 07480 | 3 | 248 | 2 | 193 | 446 | 17.1 | 46.6 | gi|91086517|ref|XP_971701.1| NtR CG6698-PA [Tribolium castaneum] |
| 07642 etc. | 17 | 1246 | 3 | 562 | 1828 | 15.2 | 90.4 | gi|55139125|gb|AAV41236.1| immulectin-3 [Manduca sexta] (07642, 13452, 14991) |
| 07883 | 0 | 0 | 3 | 792 | 795 | 0.0 | 127.4 | gi|157128533|ref|XP_001661472.1| hypothetical protein AaeL_AAEL011180 [Aedes aegypti] |
| 08524 etc. | 74 | 3481 | 7 | 1984 | 5546 | 9.8 | 136.8 | gi|2738863|gb|AAB94557.1| hemocyte protease-1 [Manduca sexta] (08524, 12527, 16288, 16719, 17102) |
| 10124 etc. | 162 | 6970 | 18 | 4204 | 11354 | 8.9 | 112.7 | gi|114050871|ref|NP_001040411.1| carboxylesterase [Bombyx mori] (10124, 15112, 16627, 16922, 17330, 18860) |
| 11280 etc. | 143 | 7866 | 11 | 2589 | 10609 | 11.4 | 113.6 | gi|91090548|ref|XP_971239.1| hemolectin CG7002-PA [Tribolium castaneum] (11280, 15506, 15594, 18551) |
| 13813 | 31 | 2398 | 4 | 848 | 3281 | 16.0 | 102.3 | gi|110758905|ref|XP_395067.3| ~ hemolectin CG7002-PA [Apis mellifera] |
| 13842 | 14 | 677 | 2 | 228 | 921 | 10.0 | 55.0 | gi|138601|sp|P19616.1|VITM_MANSE microvitellogenin precursor |
| 14248 etc. | 1 | 150 | 15 | 2329 | 2495 | 31.1 | 74.9 | gi|2149091|gb|AAB58491.1| serpin-2 [Manduca sexta] (14248, 15111, 16917, 17058, 17751) |
| 14570 etc. | 562 | 29402 | 26 | 11144 | 41134 | 10.8 | 206.9 | gi|162462371|ref|NP_001104817.1| lectin [B. mori] (14570, 15250, 15380, 15792, 16278, 16289, 16291, 16594, 16842, 17159, 17421, 17471, 17732, 17769, 18032, 18067, 18073, 18089, 18097, 18286, 18326, 18719, 18721, 18794) |
| 14760 & | 57 | 3372 | 3 | 1184 | 4616 | 12.3 | 190.5 | gi|156545430|ref|XP_001606650.1| ~ CG7002-PA [Nasonia vitripennis] (14760, 18045) |
| 14811 | 5 | 136 | 1 | 121 | 263 | 5.6 | 58.4 | gi|221055473|ref|XP_002258875.1| hypothetical protein, conserved in Plasmodium [Plasmodium knowlesi] |
| 15584 | 3 | 241 | 1 | 202 | 447 | 16.7 | 97.5 | gi|66535330|ref|XP_623280.1| atlastin CG6668-PA, isoformA [Apis mellifera] |
| 16815 etc. | 208 | 9161 | 39 | 6243 | 15651 | 9.1 | 77.3 | gi|75038472|sp|Q25519.3|PRP2_MANSE proPO-2 (16815, 17417, 17958, 18004, 18516, 18811) |
| 17085 etc. | 261 | 12058 | 33 | 8286 | 20638 | 9.6 | 121.2 | gi|74763772|sp|O44249.3|PRP1_MANSE proPO-1 (17085, 17315, 17420, 17612, 17629, 17562, 18065, 18463, 18887) |
RA and ARN are calculated using original read numbers as described in Section 2.3. Listed here are contigs with RAIH/IF >40, RACH/CF >40, ARNIH >80 when RNIF =0, or ARNCH >80 when RNCF =0. RAIH/IF and RACH/CF values are shown in red if they are greater than 40, whereas ARNIH and ARNCH values are shown in blue if they are higher than 80. In the columns of RA or ARN, cells shaded green and orange represent down- and up-regulated gene expression, respectively. Contigs with identical BLAST results are combined, with their average RAs or ARNs calculated based on the sums of original reads in CF, CH, IF, and IH for each group. Contigs with no BLAST hit can be found in Table S3, a complete list of 161 HC CIFH contigs.
The hemocyte-specific gene expression is, in several cases, supported by previous studies on M. sexta defense proteins such as lacunin (Nardi et al., 1999), HP1 (Jiang et al., 1999), serpin-2 (Gan et al., 2001), and proPO (Jiang et al., 1997). Lacunin is an extracellular matrix protein responsible for transforming circulating non-adhesive hemocytes to adhesive ones that aggregate on foreign surfaces (Nardi et al., 2005). Contigs 16288, 16719 and 17102 encodes clip-domain HP1; contigs 08524 and 12527 encode an HP1 homolog ~97% identical in sequence to the published one (Jiang et al., 1999). HP1 may be involved in a serine proteinase cascade that proteolytically activates proPO in plasma. Hemolymph proPO is synthesized in oenocytoids only (Jiang et al., 1997): 6 contigs encode proPO subunit-1 and 9 encode proPO subunit-2.
Based on sequence homology, we also discovered 51 contigs that were not known to be related to hemocyte-mediated immunity in M. sexta (Table 8). Contigs 11280, 13813, 15506, 15594, and 18551 probably encode parts of hemolectin or hemocytin, a >300 kDa protein participating in hemolymph coagulation (Lesch et al., 2007; Kanost and Nardi, 2010). As many as 37 contigs encode multiple lectins that bind to carbohydrates. Contigs 05933, 08686, 13271, 15116, 15350, and 15564 encode scavenger receptor C-like proteins that could also recognize carbohydrates. Apparently, hemocytes play critical roles in the recognition of pathogens that are covered with polysaccharides on the surface. Contig 02473 encodes a protein homologous to Drosophila eater that mediates bacteria phagocytosis by hemocytes (Kocks et al., 2005). Contigs 03287 and 07139 may be related to antiviral and antiparasitoid responses, respectively (Abdel-latief and Hilker, 2008; Liu et al., 2010).
Inside hemocytes, proteins may relay signals in a cell-specific manner. These include contigs 00541, 00752, 03246, 06319 (G-protein coupled receptors), 00882 (GTP-binding protein) 00010 (cAMP-dependent kinase), 00839 (receptor-type Tyr-protein phosphatase), 02159 (septin for ubiquitination), 15584 (GTPase atlastin), 14248, 15111, 16917, 17058, and 17751 (serpin-2 and 2′). It is unclear how these two highly inducible, intracellular serpins may inhibit a proteinase during apoptosis (Bird, 1998). Nor is it known how the other proteins may transduce signals dependent on the immune status of hemocytes.
3.5. Specific gene expression in fat body from feeding larvae
Because hemocyte samples collected through cut prolegs of feeding larvae were unlikely contaminated with fat body tissue, the 132 fat body-specific (i.e. FB) contig groups had high RACF/CH or IF/IH values (Table 9). Moreover, since chances for such contamination were equal for hemocytes from naïve and challenged M. sexta larvae, there was no globally uneven distribution of RAs or ARNs between the CF/CH and IF/IH groups. In other words, the data on fat body-specific gene expression were unbiased and reliable.
Table 9.
A list of FB CIFH contigs with BLAST hits*
| CIFH contig # | Original read # | RA or ARN | BLAST results | |||||
|---|---|---|---|---|---|---|---|---|
| CF | CH | IF | IH | Total | CF/CH | IF/IH | ||
| 00051 | 291 | 1 | 329 | 0 | 621 | 1403.9 | 681.6 | gi|183979376|dbj|BAG30740.1| muscle myosin heavy chain [Papilio xuthus] |
| 00153 etc. | 2069 | 4 | 2563 | 1 | 4637 | 2495.3 | 5309.8 | gi|2498144|sp|Q25490.1 apoLp (00153 02405 02406 03748 04510 06831 06834 07770 14087 14589) |
| 00194 | 37 | 0 | 81 | 1 | 119 | 178.5 | 167.8 | gi|48476133|gb|AAT44358.1| calcium-activated potassium channel α subunit [Manduca sexta] |
| 00285 & | 298 | 23 | 921 | 5 | 1247 | 62.5 | 381.6 | gi|73921301|gb|AAG42021.2|AF327882_1 JHE precursor [Manduca sexta] (00285, 00859) |
| 00409 | 168 | 0 | 216 | 0 | 384 | 810.5 | 447.5 | gi|110750043|ref|XP_394261.3| plexin A CG11081-PA, isoform A [Apis mellifera] |
| 00414 | 58 | 1 | 50 | 0 | 109 | 279.8 | 103.6 | gi|195382713|ref|XP_002050074.1| GJ21937 [Drosophila virilis] |
| 00423 | 149 | 0 | 220 | 0 | 369 | 718.8 | 455.8 | gi|158295580|ref|XP_316291.4| AGAP006225-PA [Anopheles gambiae] |
| 00465 | 134 | 1 | 230 | 0 | 365 | 646.4 | 476.5 | gi|149755131|ref|XP_001491560.1| hemicentin 1 [Equus caballus] |
| 00535 | 67 | 1 | 100 | 0 | 168 | 323.2 | 207.2 | gi|242015135|ref|XP_002428229.1| thrombospondin-3 precursor [Pediculus humanus corporis] |
| 00575 | 3 | 0 | 259 | 5 | 267 | 14.5 | 107.3 | gi|154240658|dbj|BAF74637.1| peptidoglycan recognition protein-D [Samia cynthiaricini] |
| 00609 | 324 | 0 | 762 | 0 | 1086 | 1563.1 | 1578.6 | gi|225542786|gb|ACN91276.1| dentin sialophosphoprotein precursor [Bos taurus] |
| 00737 | 2 | 4 | 131 | 2 | 139 | 2.4 | 135.7 | gi|198466442|ref|XP_002135189.1| GA23919 [Drosophila pseudoobscura] |
| 00748 | 131 | 4 | 118 | 2 | 255 | 158.0 | 122.2 | gi|29346557|ref|NP_810060.1| glycine dehydrogenase [Bacteroides thetaiotaomicron] |
| 00766 | 45 | 0 | 74 | 1 | 120 | 217.1 | 153.3 | gi|158293377|ref|XP_314728.3| AGAP008632-PA [Anopheles gambiae] |
| 00773 | 49 | 12 | 93 | 1 | 155 | 19.7 | 192.7 | gi|157103945|ref|XP_001648193.1| dihydropyrimidine dehydrogenase [Aedes aegypti] |
| 00785 | 120 | 2 | 139 | 2 | 263 | 289.5 | 144.0 | gi|193795848|gb|ACF21977.1| paramyosin [Bombyx mandarina] |
| 00884 | 39 | 1 | 23 | 0 | 63 | 188.1 | 47.6 | gi|156553304|ref|XP_001599652.1| GA21752-PA [Nasonia vitripennis] |
| 00960 | 52 | 2 | 99 | 1 | 154 | 125.4 | 205.1 | gi|157107996|ref|XP_001650030.1| sarcosine dehydrogenase [Aedes aegypti] |
| 01095 | 64 | 0 | 99 | 1 | 164 | 308.8 | 205.1 | gi|169639235|gb|ACA60733.1| venom acid phosphatase [Pteromalus puparum] |
| 01097 | 134 | 2 | 436 | 5 | 577 | 323.2 | 180.7 | gi|55139125|gb|AAV41236.1| immulectin-3 [Manduca sexta] |
| 01127 | 41 | 1 | 52 | 1 | 95 | 197.8 | 107.7 | gi|189491898|gb|ACE00761.1| adipokinetic hormone receptor [Manduca sexta] |
| 01454 | 599 | 3 | 1337 | 3 | 1942 | 963.2 | 923.3 | gi|91082539|ref|XP_973726.1| inter-α (globulin) inhibitor H4 (Kallikrein-sensitive) [T. castaneum] |
| 01480 | 211 | 0 | 729 | 0 | 940 | 1017.9 | 1510.3 | gi|183979392|dbj|BAG30748.1| hypothetical protein [Papilio xuthus] |
| 01601 | 60 | 1 | 79 | 0 | 140 | 289.5 | 163.7 | gi|270005801|gb|EFA02249.1| hypothetical protein TcasGA2 TC007912 [Tribolium castaneum] |
| 01742 | 65 | 0 | 75 | 0 | 140 | 313.6 | 155.4 | gi|283100192|gb|ADB08386.1| sugar transporter 4 [Bombyx mori] |
| 01743 | 27 | 0 | 112 | 0 | 139 | 130.3 | 232.0 | gi|134252572|gb|ABO65045.1| β-hexosaminidase [Ostrinia furnacalis] |
| 01870 | 184 | 0 | 323 | 0 | 507 | 887.7 | 669.2 | gi|242010783|ref|XP_002426138.1| conserved hypothetical protein [Pediculus humanus corporis] |
| 01892 | 82 | 0 | 108 | 0 | 190 | 395.6 | 223.7 | gi|158289807|ref|XP_311448.4| AGAP010734-PA [Anopheles gambiae] |
| 01915 | 85 | 2 | 275 | 0 | 362 | 205.0 | 569.7 | gi|110757936|ref|XP_623940.2| peroxidase precursor (DmPO) [Apis mellifera] |
| 01956 | 127 | 0 | 99 | 0 | 226 | 612.7 | 205.1 | gi|156551746|ref|XP_001602035.1| ENSANGP00000015052 [Nasonia vitripennis] |
| 01972 etc. | 383 | 0 | 3327 | 0 | 3710 | 1847.7 | 6892.5 | gi|136206|sp|P22297.1|transferrin (01972 10382 11027 14937 17193 17206 17395 16606 18234 18308) |
| 02101 | 51 | 0 | 75 | 0 | 126 | 246.0 | 155.4 | gi|186909546|gb|ACC94296.1| glucose oxidase-like enzyme [Helicoverpa armigera] |
| 02104 | 59 | 1 | 67 | 1 | 128 | 284.6 | 138.8 | gi|91079628|ref|XP_967731.1| AGAP002355-PA [Tribolium castaneum] |
| 02137 | 101 | 0 | 24 | 0 | 125 | 487.2 | 49.7 | gi|91084191|ref|XP_967340.1| AGAP002557-PA [Tribolium castaneum] |
| 02144 | 82 | 0 | 132 | 3 | 217 | 395.6 | 91.2 | gi|62002223|gb|AAX58711.1| pheromone-degrading enzyme 1 [Antheraea polyphemus] |
| 02166 | 60 | 0 | 57 | 0 | 117 | 289.5 | 118.1 | gi|193876254|gb|ACF24761.1| lipid storage droplet protein 1 [Manduca sexta] |
| 02184 | 53 | 2 | 111 | 1 | 167 | 127.8 | 230.0 | gi|226342886|ref|NP_001139705.1| serpin 13 [Bombyx mori] |
| 02219 | 454 | 3 | 971 | 3 | 1431 | 730.1 | 670.5 | gi|219815604|gb|ACL36977.1| putative ecdysone oxidase [Helicoverpa zea] |
| 02329 | 143 | 0 | 411 | 0 | 554 | 689.9 | 851.5 | gi|112984054|ref|NP_001037422.1| yellow1 [Bombyx mori] |
| 02337 & | 107 | 2 | 170 | 7 | 286 | 258.1 | 50.3 | gi|91079867|ref|XP_967070.1| AGAP005945-PB [Tribolium castaneum] (02337, 15796) |
| 02361 | 7 | 4 | 70 | 1 | 82 | 8.4 | 145.0 | gi|56418425|gb|AAV91020.1| hemolymph proteinase 22 [Manduca sexta] |
| 02393 | 45 | 0 | 77 | 5 | 127 | 217.1 | 31.9 | gi|156545523|ref|XP_001607196.1| dihydroxyacetone kinase-2 homolog (yeast) [Nasonia vitripennis] |
| 02394 | 28 | 1 | 23 | 0 | 52 | 135.1 | 47.6 | gi|91077746|ref|XP_966706.1| conserved hypothetical protein [Tribolium castaneum] |
| 02409 | 113 | 0 | 187 | 0 | 300 | 545.1 | 387.4 | gi|109502352|gb|ABE01157.2| carboxylesterase [Spodoptera litura] |
| 02482 | 63 | 0 | 85 | 1 | 149 | 303.9 | 176.1 | gi|66519258|ref|XP_625210.1| ~ CG6188-PA [Apis mellifera] |
| 02609 | 97 | 0 | 146 | 2 | 245 | 468.0 | 151.2 | gi|156968285|gb|ABU98614.1| α-amylase [Helicoverpa armigera] |
| 02638 & | 241 | 0 | 206 | 0 | 447 | 1162.6 | 426.8 | gi|41016826|sp|Q27772.3|C1TC_SPOFR C-1-THF synthase, cytoplasmic (02638, 07658) |
| 02651 | 24 | 0 | 124 | 0 | 148 | 115.8 | 256.9 | gi|5326830|gb|AAD42058.1|AF122899_1 plasmatocyte-spreading peptide precursor [Manduca sexta] |
| 02800 | 28 | 0 | 97 | 0 | 125 | 135.1 | 201.0 | gi|260765449|gb|ACX49762.1| β-fructofuranosidase 1 [Manduca sexta] |
| 02847 | 33 | 0 | 103 | 0 | 136 | 159.2 | 213.4 | gi|114051702|ref|NP_001040423.1| zinc-containing alcohol dehydrogenase [Bombyx mori] |
| 02931 & | 187 | 0 | 429 | 0 | 616 | 902.1 | 888.8 | gi|1658003|gb|AAB18243.1| microsomal epoxide hydrolase [Trichoplusia ni] (02931, 04388) |
| 02947 | 518 | 21 | 981 | 56 | 1576 | 119.0 | 36.3 | gi|259493819|gb|ACW82749.1| hemocyte aggregation inhibitor protein precursor [Manduca sexta] |
| 02979 | 49 | 0 | 92 | 4 | 145 | 236.4 | 47.6 | gi|52782757|sp|Q9NJ98.1|BGRP1_MANSE β-1,3-glucan recognitionprotein 1 βGRP-1 |
| 02985 | 3 | 0 | 158 | 0 | 161 | 14.5 | 327.3 | gi|56418466|gb|AAV91027.1| serine proteinase-like protein 4 [Manduca sexta] |
| 03185 | 106 | 0 | 234 | 10 | 350 | 511.4 | 48.5 | gi|157117489|ref|XP_001658792.1| 3-hydroxyacyl-CoA dehyrogenase [Aedes aegypti] |
| 03224 | 98 | 0 | 477 | 0 | 575 | 472.8 | 988.2 | gi|226342906|ref|NP_001139715.1| serpin 22 [Bombyx mori] |
| 03226 | 222 | 0 | 663 | 0 | 885 | 1071.0 | 1373.5 | gi|153791757|ref|NP_001093275.1| myo-inositol oxygenase [Bombyx mori] |
| 03395 | 22 | 1 | 24 | 0 | 47 | 106.1 | 49.7 | gi|157908523|dbj|BAF81491.1| juvenile hormone epoxide hydrolase [Bombyx mori] |
| 03415 | 190 | 0 | 216 | 1 | 407 | 916.6 | 447.5 | gi|2708688|gb|AAB92583.1| acyl-CoA 9 desaturase [Trichoplusia ni] |
| 03434 | 1 | 0 | 387 | 0 | 388 | 4.8 | 801.7 | gi|189234566|ref|XP_001815977.1| Kaz1-ORFB CG1220-PE [Tribolium castaneum] |
| 03454 | 28 | 0 | 102 | 0 | 130 | 135.1 | 211.3 | gi|6560669|gb|AAF16712.1|AF117590_1 unknown [Manduca sexta] |
| 03483 | 280 | 0 | 374 | 0 | 654 | 1350.8 | 774.8 | gi|283558277|gb|ADB27116.1| aliphatic nitrilase [Bombyx mori] |
| 03712 | 49 | 2 | 157 | 5 | 213 | 118.2 | 65.1 | gi|170779021|gb|ACB36909.1| glutathione S-transferase θ [Antheraea pernyi] |
| 03737 | 167 | 1 | 197 | 0 | 365 | 805.6 | 408.1 | gi|56462300|gb|AAV91433.1| putative serine protease-like protein 2 [Lonomia obliqua] |
| 03776 etc. | 204 | 8 | 960 | 51 | 1223 | 123.0 | 39.0 | gi|112983872|ref|NP_001036857.1| serpin-like protein [Bombyx mori] (03776, 06215, 06531, 17814) |
| 04012 & | 167 | 3 | 727 | 11 | 908 | 268.5 | 136.9 | gi|27733411|gb|AAO21503.1|AF413062_1 leureptin, LPS binding [Manduca sexta] (04012, 08453) |
| 04413 | 69 | 1 | 133 | 1 | 204 | 332.9 | 275.5 | gi|194743582|ref|XP_001954279.1| GF18195 [Drosophila ananassae] |
| 04424 | 72 | 0 | 64 | 0 | 136 | 347.3 | 132.6 | gi|114052020|ref|NP_001040445.1| tropomyosin 1 [Bombyx mori] |
| 04430 | 74 | 0 | 68 | 0 | 142 | 357.0 | 140.9 | gi|114052573|ref|NP_001040481.1| phosphoribosyl pyrophosphate synthetase [Bombyx mori] |
| 04498 | 46 | 0 | 115 | 0 | 161 | 221.9 | 238.2 | gi|90025232|gb|ABD85119.1| JH epoxide hydrolase [Spodopteraexigua] |
| 04504 | 53 | 0 | 135 | 0 | 188 | 255.7 | 279.7 | gi|7239259|gb|AAF43151.1|AF226857_1 hemolymph JHBP precursor [Manduca sexta] |
| 04722 & | 578 | 0 | 861 | 0 | 1439 | 2788.4 | 1783.7 | gi|116791778|gb|ABK26104.1| unknown [Picea sitchensis] (04722, 04994) |
| 04781 | 56 | 0 | 237 | 0 | 293 | 270.2 | 491.0 | gi|118359591|ref|XP_001013035.1| PHD-finger family protein [Tetrahymenathermophila] |
| 04786 | 61 | 0 | 62 | 0 | 123 | 294.3 | 128.4 | gi|219686082|emb|CAW30924.1| putative aldo-ketose reductase 1 [Papilio dardanus] |
| 04791 | 144 | 0 | 200 | 0 | 344 | 694.7 | 414.3 | gi|116788175|gb|ABK24783.1| unknown [Picea sitchensis] |
| 04806 | 518 | 1 | 372 | 0 | 891 | 2499.0 | 770.7 | gi|157122933|ref|XP_001659963.1| actin [Aedes aegypti] |
| 04808 | 0 | 0 | 426 | 2 | 428 | 0.0 | 441.3 | gi|237861314|gb|AAV41237.2| immulectin-4 [Manduca sexta] |
| 04830 etc. | 59 | 2 | 755 | 6 | 822 | 142.3 | 260.7 | gi|169646838|ref|NP_001112375.1| heat shock protein 25.4 [Bombyx mori] (04830, 04887, 05717) |
| 05038 & | 101 | 0 | 175 | 1 | 277 | 487.2 | 362.5 | gi|110759694|ref|XP_394781.3| rTS β protein [Apis mellifera] (05038, 05832) |
| 05136 | 1074 | 11 | 1041 | 37 | 2163 | 471.0 | 58.3 | gi|114051966|ref|NP_001040198.1| mitochondrial aldehyde dehydrogenase [Bombyx mori] |
| 05324 | 68 | 0 | 88 | 0 | 156 | 328.0 | 182.3 | gi|225346695|gb|ACN86370.1| troponin I transcript variant C [Bombyx mandarina] |
| 05348 | 50 | 0 | 67 | 0 | 117 | 241.2 | 138.8 | gi|189234391|ref|XP_974849.2| GA16498-PA [Tribolium castaneum] |
| 05417 etc. | 273 | 0 | 917 | 0 | 1190 | 1317.0 | 1899.7 | gi|260907784|gb|ACX53694.1| alcohol DH [Heliothis virescens] (05417, 05461, 07389, 07432) |
| 05984 | 89 | 0 | 97 | 0 | 186 | 429.4 | 201.0 | gi|56462260|gb|AAV91413.1| myosin 3 light chain [Lonomia obliqua] |
| 06175 | 11 | 0 | 52 | 1 | 64 | 53.1 | 107.7 | gi|170070451|ref|XP_001869584.1| conserved hypothetical protein [Culex quinquefasciatus] |
| 06227 | 251 | 1 | 715 | 0 | 967 | 1210.9 | 1481.3 | gi|124527|sp|Q00630.1|ICYB_MANSE insecticyanin-B, blue biliprotein |
| 06251 | 66 | 2 | 57 | 7 | 132 | 159.2 | 16.9 | gi|158289206|ref|XP_310956.4| AGAP000179-PA [Anopheles gambiae] |
| 06394 | 51 | 0 | 228 | 0 | 279 | 246.0 | 472.3 | gi|110611262|gb|ABG77980.1| alanine-glyoxylate transaminase 1 [Glossinamorsitans morsitans] |
| 06588 | 60 | 0 | 75 | 0 | 135 | 289.5 | 155.4 | gi|56462256|gb|AAV91411.1| myosin 1 light chain [Lonomia obliqua] |
| 06597 | 60 | 0 | 200 | 0 | 260 | 289.5 | 414.3 | gi|56462320|gb|AAV91443.1| secreted peptide 30 [Lonomia obliqua] |
| 06732 | 115 | 1 | 244 | 0 | 360 | 554.8 | 505.5 | gi|25090512|sp|Q25513.1|HGLY_MANSE 27 kDa hemolymph glycoprotein |
| 06789 & | 159 | 0 | 460 | 0 | 619 | 767.1 | 953.0 | gi|156968291|gb|ABU98617.1| unknown [Helicoverpa armigera] (06789, 06876) |
| 06975 & | 106 | 2 | 212 | 2 | 322 | 255.7 | 219.6 | gi|189237651|ref|XP_001813448.1| N-acetyl neuraminatelyase [Tribolium castaneum] (06975, 14637) |
| 07116 & | 1 | 4 | 1015 | 4 | 1024 | 1.2 | 525.7 | gi|171262319|gb|ACB45566.1| lebocin-like protein [Antheraea pernyi] (07116, 10853) |
| 07565 | 24 | 1 | 14 | 0 | 39 | 115.8 | 29.0 | gi|7862150|gb|AAF70499.1|AF255341_1 3-dehydroecdysone 3α-reductase [Spodoptera littoralis] |
| 07608 etc. | 353 | 3 | 3931 | 0 | 4287 | 567.7 | 8143.9 | gi|159526|gb|AAA29320.1| Met-rich storage protein 1 (07608, 07975, 08141, 14688) |
| 07629 | 65 | 0 | 82 | 0 | 147 | 313.6 | 169.9 | gi|77415676|emb|CAJ01507.1| hypothetical protein [Manduca sexta] |
| 07639 & | 811 | 0 | 1616 | 18 | 2445 | 3912.5 | 186.0 | gi|134436|sp|P14754.1| alaserpin or serpin-1 (07639, 15891) |
| 07671 | 227 | 3 | 450 | 3 | 683 | 365.0 | 310.8 | gi|195164814|ref|XP_002023241.1| GL21066 [Drosophila persimilis] |
| 08076 & | 47 | 3 | 115 | 2 | 167 | 75.6 | 119.1 | gi|226342878|ref|NP_001139701.1| serpin 7 [Bombyx mori] (08076, 14528) |
| 08224 etc. | 7528 | 8 | 10093 | 0 | 17629 | 4539.6 | 20909.2 | gi|1168527|sp|P14297.2| arylphorin β subunit (08224, 16474, 16501, 16664, 16715, 16764, 18695) |
| 08467 | 0 | 0 | 113 | 0 | 113 | 0.0 | 234.1 | gi|112983866|ref|NP_001036858.1| T7 lysozyme-like protein 1 (BTL-LP1) [Bombyx mori] |
| 08500 | 138 | 0 | 407 | 0 | 545 | 665.7 | 843.2 | gi|156406857|ref|XP_001641261.1| predicted protein [Nematostella vectensis] |
| 08821 | 246 | 0 | 436 | 2 | 684 | 1186.8 | 451.6 | gi|112983550|ref|NP_001036879.1| fibrillin-like protein [Bombyx mori] |
| 08845 | 27 | 0 | 130 | 0 | 157 | 130.3 | 269.3 | gi|195029763|ref|XP_001987741.1| GH19797 [Drosophila grimshawi] |
| 08854 & | 302 | 5 | 5234 | 0 | 5541 | 291.4 | 10843.3 | gi|5869985|emb|CAB55603.1| moderately Met-rich storage protein [Spodoptera litura] (08854, 15324) |
| 09928 | 30 | 0 | 106 | 0 | 136 | 144.7 | 219.6 | gi|242090851|ref|XP_002441258.1| hypothetical SORBIDRAFT_09g023310 [Sorghum bicolor] |
| 10071 & | 15 | 1 | 1243 | 0 | 1259 | 72.4 | 2575.1 | gi|228382|prf||1803340A Met-rich storage protein SP1A (10071, 17516) |
| 10326 | 284 | 4 | 299 | 11 | 598 | 342.5 | 56.3 | gi|56462160|gb|AAV91363.1| hypothetical protein 10 [Lonomia obliqua] |
| 10791 etc. | 2 | 0 | 2756 | 2 | 2760 | 9.6 | 2854.8 | gi|4262357|gb|AAD14591.1| scolexin A [Manduca sexta] (10791, 10792, 16520, 18669, 18670, 18963) |
| 11039 etc. | 13962 | 11 | 19836 | 0 | 33809 | 6123.3 | 41094.2 | gi|114240|sp|P14296.1| arylphorin α subunit (11039 16171 16537 16814 17492 18240 18257 18556) |
| 11830 | 26 | 1 | 33 | 0 | 60 | 125.4 | 68.4 | gi|260780799|ref|XP_002585527.1| hypothetical protein BRAFLDRAFT_89257 [B. floridae] |
| 11922 & | 901 | 12 | 1052 | 3 | 1968 | 362.2 | 726.5 | gi|114058|sp|P13276.1| apoLp-III (11922, 13093) |
| 12005 | 154 | 0 | 2177 | 0 | 2331 | 742.9 | 4510.1 | gi|2625150|gb|AAB86646.1| moderately Met-rich hexamerin precursor [Hyalophora cecropia] |
| 12151 | 0 | 0 | 153 | 0 | 153 | 0.0 | 317.0 | gi|116084|sp|P14665.1|bactericidin B-5P, cecropin-like peptide precursor |
| 12749 | 135 | 0 | 1462 | 0 | 1597 | 651.3 | 3028.8 | gi|159530|gb|AAA29322.1| Met-rich storage protein 3 [Manduca sexta] |
| 13563 | 0 | 0 | 657 | 0 | 657 | 0.0 | 1361.1 | gi|110347786|gb|ABG72695.1| attacin-like protein [Antheraea mylitta] |
| 13916 etc. | 3 | 0 | 1327 | 0 | 1330 | 14.5 | 2749.1 | gi|219958086|gb|ACL68097.1| lebocin-related protein precursor [M. sexta] (13916, 17301, 17434) |
| 13994 | 57 | 0 | 62 | 0 | 119 | 275.0 | 128.4 | gi|112983654|ref|NP_001036872.1| bombyrin [Bombyx mori] |
| 14173 | 45 | 0 | 32 | 0 | 77 | 217.1 | 66.3 | gi|153792114|ref|NP_001093267.1| phosphatidylethanolamine binding protein [Bombyx mori] |
| 14375 etc. | 400 | 0 | 681 | 0 | 1081 | 1929.7 | 1410.8 | gi|400673|sp|P31420|OMBP ommochrome-binding protein precursor (14375, 14659, 17494, 17813) |
| 14380 etc. | 0 | 1 | 408 | 3 | 412 | 0.0 | 281.8 | gi|67906420|gb|AAY82587.1| attacin-1 [Manduca sexta] (14380, 14641, 16150) |
| 14700 & | 0 | 0 | 301 | 4 | 305 | 0.0 | 155.9 | gi|260765453|gb|ACX49764.1| peptidoglycan recognition protein 2 [Manduca sexta] (14700, 14752) |
| 15089 | 271 | 0 | 194 | 0 | 465 | 1307.4 | 401.9 | gi|158293921|ref|XP_315269.4| AGAP011516-PA [Anopheles gambiae] |
| 15639 | 10 | 0 | 109 | 0 | 119 | 48.2 | 225.8 | gi|148298818|ref|NP_001091784.1| multi-binding protein [Bombyx mori] |
| 16000 | 61 | 0 | 138 | 0 | 199 | 294.3 | 285.9 | gi|109458629|ref|XP_001073545.1| hypothetical protein [Rattus norvegicus] |
| 16223 | 22 | 1 | 47 | 2 | 72 | 106.1 | 48.7 | gi|242003442|ref|XP_002422733.1| bifunctional purine biosynthesis protein [Pediculus corporis] |
| 16281 & | 358 | 0 | 541 | 0 | 899 | 1727.1 | 1120.8 | gi|134103857|gb|ABO60878.1| cationic peptide CP8 precursor [Manduca sexta] (16281, 17312) |
| 16849 | 134 | 0 | 541 | 0 | 675 | 646.4 | 1120.8 | gi|114051738|ref|NP_001040426.1| alcohol dehydrogenase [Bombyx mori] |
| 17199 | 42 | 2 | 33 | 4 | 81 | 101.3 | 17.1 | gi|3108073|gb|AAC15763.1| putative multifunctional protein ADE2 [Manduca sexta] |
| 17350 | 0 | 0 | 205 | 0 | 205 | 0.0 | 424.7 | gi|29469969|gb|AAO74640.1| antimicrobial protein attacin 2 [Manduca sexta] |
| 18797 | 9 | 0 | 549 | 0 | 558 | 43.4 | 1137.4 | gi|39843367|gb|AAR32136.1| VHDL receptor [Helicoverpa zea] |
RA and ARN are calculated using original read numbers as described in Section 2.3. Listed here are contigs with RAIF/IH >100, RACF/CH >100, ARNIF >200 when RNIH =0, or ARNCF >200 when RNCH =0. RAIF/IH and RACF/CH values are shown in red if they are greater than 100, whereas ARNIF and ARNCF values are shown in blue if they are higher than 200. In the columns of RA or ARN, cells shaded green and orange represent down- and up-regulated gene expression, respectively. Contigs with identical BLAST results are combined, with their average RAs or ARNs calculated based on the sums of original reads in CF, CH, IF, and IH for each group. Contigs with no BLAST hit can be found in Table S4, a complete list of 250 FB CIFH contigs.
Insect fat body, analogous to combined mammalian liver and adipose tissue, is the site where most intermediary metabolism takes place (Arrese and Soulages, 2010). It also is the principal source of plasma proteins, including those participating in innate immune responses (Jiang, 2008; Ragan et al., 2009). These notions are strongly supported by the identification of FB contigs and BLAST search: 61 or 46% of the 132 FB contig groups are metabolism-related, whereas 32 or 24% are immunity-related (Table 9). Since metabolism-related genes and their transcript level changes after the immune challenge will be reported elsewhere, we only discuss fat body-specific gene expression involved in antimicrobial defense responses and the UP contigs covered in Section 3.2 are not repeated here. β-1,3-glucan recognition protein-1 (02979) (Ma and Kanost, 2000), immulectin-3 (01097) (Yu et al., 2005), and leureptin (04012 and 08453) (Zhu et al., 2010) are pattern recognition receptors binds fungi and bacteria (Table 9). HAIP (02947), a chitinase-like protein, inhibits hemocyte aggregation (Kanost et al., 1994). Contig 05348 encodes a protein with at least three Ig domains. Contig 00535 encodes a thrombospodin-like protein with eight EGF-like domains and one coiled coil for protein-protein interaction. Contig 07671, after extension, is found to encode a >60 kDa protein with at least four EGF domains. Hemicentin (00465) is a cell adhesion protein containing a von Willebrand A domain (Vogel and Hedgecock, 2001). Contig 08821 encodes a fibrillin-like nimrod B which may play a role in pathogen recognition and phagocytosis (Kurucz et al., 2007).
We have found six proteinase inhibitor-like proteins, including homologs of B. mori serpin12 (or SLP: 03776, 06215, 06531, 17814), serpin13 (02184) and serpin22 (03224) (Zou et al., 2009), two Cys-rich secreted protein (06175, 06597), and cationic protein-8 (16281, 17312) (Ling et al., 2009). Contig 02651 encodes three cytokines that may regulate cellular immune responses (Kanamori et al., 2010).
4. Discussion
Next-generation sequencing has been increasingly used for profiling gene expression in the past few years (Costa et al., 2010; Marguerat and Bähler, 2010). While its advantages over microarray analysis are obvious in some aspects, this technique has been so far, to the best of our knowledge, only applied to species with known genome sequences for transcript profiling. (It has also been used in other species for general transcriptome analysis but not for systematically studying mRNA level changes.) In this study, we have extended massive sequencing and data analysis to a new dimension in which gene discovery, expression profiling, and function prediction are done at the same time in a lepidopteran insect lacking known genome sequence. This technical improvement, mathematically simple, generated a wealth of information in a cost-effective manner and opens a door for similar studies in non-model organisms of practical importance.
Using numbers of reads assembled into specific contigs to calculate RAs provides a genome- independent way of looking at gene expression in relation to specific physiological processes and tissue/cell types. This perspective becomes more relevant when homology-based search results are also taken into consideration, even though expression pattern by itself is an autonomous parameter for gene discovery (Table 6). Systematic examination of transcript level changes in conjunction with sequence comparison can be extremely powerful in terms of gene discovery and functional prediction. It is our general impression that details of microarray data were, in many cases, overlooked when they were merely considered as spots rather than specific genes associating with functions. This tends to be true, especially when expression of genes does not strongly correlate with a treatment due to biological or technical reasons. For example, real expression changes of certain genes are sometimes too small to be distinguished from background caused by nonspecific binding in hybridization-based methods. In contrast, since read numbers for individual contigs are “digital”, their corresponding RAs or ARNs have a greater dynamic range in measurement of relative transcript abundance and do not carry noise or artifact and can, thus, be calculated and compared with high confidence. In this study, we set cutoff RA or ARN values more or less empirically based on preliminary tests with different thresholds, presence or absence of immunity-related hits in the results, and number of contigs appropriate for manual checking and tabulation. If necessary, however, we can conveniently use lower RA cutoffs to increase detection sensitivity to discover subtle changes in transcript levels. The depth of our datasets surely allows us to explore in greater details the immunity-relatedness and tissue specificity of gene expression. For instance, some metabolic enzymes do not change much in their transcript levels. By mapping their contigs onto metabolic pathways with close-to-one RA values, we may detect trends from multiple members of specific pathways, which reveal impacts of immune response on general metabolism of the insect.
In essence, we have projected RA values of our dataset (“CIFH”) onto four surfaces of a tetrahedron: two for UP and DN, two for HC and FB (Fig. 2). If each contig is represented by a point, the data we examined are located far away from the axis of immune inducibility (RAIF/CF or IH/CH =1) or tissue specificity (RAIF/IH or CF/CH =1). The remaining >95% of the 19,020 data points are densely packed along these two axes. The large number of such contigs reflects the depth of our sequence data, as also revealed by studying a number of selected contigs. For example, we have found in our dataset four lebocin-related sequences: the first one encodes a precursor protein which is processed by an intracellular processing proteinase into 4 peptides and 2 are antibacterial (Rayaprolu et al., 2010). The other three lebocin precursors are anticipated to be processed in the same manner, based on their sequences and conserved cleavage sites (data not shown). Further evidence in support of sequence depth came from the detection of highly similar contigs assembled from large numbers of reads, such as attacins (data not shown).
Fig 2. Analysis of the CIFH dataset in terms of immune inducibility and tissue specificity.
Examination of the RA values is a process of projecting the CIFH dataset onto four surfaces of a tetrahedron. Two of them represent deviations from 1 (brown line) regarding mRNA level changes before and after the immune challenge: UP (red) for RAIF/CF or IH/CH above a set value of greater than 1; DN (green) for RAIF/CF or IH/CH below a set value of smaller than 1. The other two surfaces represent deviations from 1 (green line) regarding abundance changes in the two tissues: FB (yellow) for RAIF/IH or CF/CH above a set value of greater than 1; HC (blue) for RAIF/IH or CF/CH below a set value of smaller than 1. In this study, we used RAIF/CF >5 and RAIH/CH >8 as cutoff values for UP (red line), RACF/IF or CH/IH >10 for DN (dashed green line), RAIH/IF or CH/CF >40 for HC (blue line), and RAIF/IH or CF/CH >100 for FB (dashed yellow line). The regions above these lines contain 528 UP, 148 DN, 161 HC, and 250 FB contigs (accounting for 2.8%, 0.8%, 0.8%, and 1.3% of the 19,020 contigs, respectively), whereas the regions below represent >94.3% of the total contigs, not analyzed in this study.
Our data also indicate that not all immunity-related genes are up-regulated after the injection of microbes – some of them do not increase much and others may even decrease. For instance, HP6 and HP8 mRNA levels did not significantly change (RAs for CIFH 00540, 05370, and 09086: 1.6, 2.3, and 1.3, respectively) after the challenge but they are involved in spätzle and proPO activation (An et al., 2009 and 2010). In order to identify gene products that may be involved in pathogen recognition or signal transduction but not significantly increase in mRNA levels, we have searched the entire dataset (“06CIFH”) using the B. mori sequences (Tanaka et al., 2008; Zou et al., 2009) in conjunction with M. sexta HP1 through HP24 and PAPs. Of the 534 contigs identified, 368 do not belong to the highly induced (UP: RAIF/CF >5 or RAIH/CH >8) or suppressed groups (DN: RACF/IF or CH/IH >20): 119, 193, 62, and 8 contigs have their RA values falling into moderately induced (RAIF/CF: 2~5, RAIH/CH 2~8, or ARNIF or IH: 4~10), no change (RAIF/CF or IH/CH: 2~0.5 or ARNIF, IH, CF, or CH <4), slightly suppressed (RACF/IF or CH/IH: 2~6 or ARNCF, or CH: 4~12), and moderately suppressed (RACF/IF or CH/IH: 6~20 or ARNCF, or CH: 12~40) groups, respectively. While most of the highly suppressed group (“DN”) (RACF/IF or CH/IH: >20 or ARNCF, or CH: >40) turned out to be false positive (Table 7), mRNA levels of 48 and 3 contigs were indeed reduced slightly and moderately (data not shown), respectively.
Beyond method development, this study has established a foundation for genome annotation, especially for genes that are induced or suppressed in response to the immune challenge and for genes preferentially expressed in fat body or hemocytes. As a new version of the transcriptome analysis, it provided two high-quality cDNA datasets (“CIFH” and “06CIFH”) that can be used for identification of plasma proteins from naïve and induced feeding larvae. We detected a lot fewer indels in these contigs than those from our previous work (Zou et al., 2008). The coverage of our datasets, as well as expression profiles from the read comparisons, is anticipated to not only facilitate proteomic analysis but also assist cDNA cloning, recombinant expression, and functional elucidation of immunity-related genes in this biochemical model insect. It is our sincere hope that similar experiments including data processing will be performed in non-model organisms to discover genes of critical importance.
Supplementary Material
Acknowledgments
We thank Drs. Ulrich Melcher and Jack Dillwith for their critical comments on the manuscript. This work was supported by National Institutes of Health Grants GM58634 (to H. Jiang). This article was approved for publication by the Director of the Oklahoma Agricultural Experiment Station and supported in part under project OKLO2450.
Abbreviations
- CF, IF, CH and IH
control and induced fat body or hemocytes
- RA
relative abundance
- HP
hemolymph proteinase
- PO and proPO
phenoloxidase and its precursor
- AMP
antimicrobial peptides
- EST
expressed sequence tag
- rpS/Lx
ribosomal small or large subunit protein X
- LNF
library normalization factor
- ARN
adjusted read number
- UP and DN
up- and down- regulated
- FB and HC
fat body- and hemocyte-specific
- ORF
open reading frame
- PRR
pattern recognition receptors
- LPS
lipopolysaccharide
- PGRP
peptidoglycan recognition protein
- βGRP
β-1,3-glucan recognition protein
- CTL
C-type lectin
- SPH
serine proteinase homolog
- PSP
plasmatocyte-spreading peptide
- PAP
proPO-activating proteinase
- HAIP
hemocyte aggregation inhibitor protein
- EGF
epidermal growth factor
- RT-PCR
reverse transcription-polymerase chain reaction
Footnotes
The sequences of raw reads are deposited in the NCBI SRA (SRS167319) and the 19,020 CIFH contig sequences are available at ftp.genome.ou.edu/pub/for_Haobo/manduca/FourLibrariesAssembly/ and http://entoplp.okstate.edu/profiles/jiang.htm.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdel-latief M, Hilker M. Innate immunity: eggs of Manduca sexta are able to respond to parasitism by Trichogramma evanescens. Insect Biochem Mol Biol. 2008;38:136–145. doi: 10.1016/j.ibmb.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Altincicek B, Vilcinskas A. Identification of a lepidopteran matrix metalloproteinase with dual roles in metamorphosis and innate immunity. Dev Comp Immunol. 2008;32:400–9. doi: 10.1016/j.dci.2007.08.001. [DOI] [PubMed] [Google Scholar]
- An C, Ishibashi J, Ragan EJ, Jiang H, Kanost MR. Functions of Manduca sexta hemolymph proteinases HP6 and HP8 in two innate immune pathways. J Biol Chem. 2009;284:19716–19726. doi: 10.1074/jbc.M109.007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An C, Jiang H, Kanost MR. Proteolytic activation and function of the cytokine Spätzle in innate immune response of a lepidopteran insect, Manduca sexta. FEBS J. 2010;277:148–162. doi: 10.1111/j.1742-4658.2009.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao JQ, Ling E, Yu XQ. A Toll receptor from Manduca sexta is in response to Escherichia coli infection. Mol Immunol. 2008;45:543–52. doi: 10.1016/j.molimm.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Armstrong PB. Proteases and protease inhibitors: a balance of activities in host-pathogen interaction. Immunobiol. 2006;211:263–281. doi: 10.1016/j.imbio.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Arrese EL, Soulages JL. Insect fat body: energy, metabolism, and regulation. Ann Rev Entomol. 2010;55:207–225. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird PI. Serpins and regulation of cell death. Results Probl Cell Differ. 1998;24:63–89. doi: 10.1007/978-3-540-69185-3_4. [DOI] [PubMed] [Google Scholar]
- Bulet P, Stöcklin R, Menin L. Anti-microbial peptides: from invertebrates to vertebrates. Immunol Rev. 2004;198:169–184. doi: 10.1111/j.0105-2896.2004.0124.x. [DOI] [PubMed] [Google Scholar]
- Costa V, Angelini C, De Feis I, Ciccodicola A. Uncovering the complexity of transcriptomes with RNA-Seq. J Biomed Biotechnol. 2010:853916. doi: 10.1155/2010/853916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Rayaprolu S, Gong Y, Huang R, Prakash O, Jiang H. Solution structure, antibacterial activity, and expression profile of Manduca sexta moricin. J Pept Sci. 2008;14:855–863. doi: 10.1002/psc.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn PE, Drake D. Fate of bacteria injected into naive and immunized larvae of the tobacco hornworm, Manduca sexta. J Invertebr Pathol. 1983;41:77–85. [Google Scholar]
- Finnerty CM, Karplus PA, Granados RR. The insect immune protein scolexin is a novel serine proteinase homolog. Protein Sci. 1999;8:242–248. doi: 10.1110/ps.8.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa S, Tanaka H, Ishibashi J, Imanishi S, Yamakawa M. Functional characterization of a cactus homolog from the silkworm Bombyx mori. Biosci Biotechnol Biochem. 2009;73:2665–2670. doi: 10.1271/bbb.90511. [DOI] [PubMed] [Google Scholar]
- Gan H, Wang Y, Jiang H, Mita K, Kanost MR. A bacteria-induced, intracellular serpin in granular hemocytes of Manduca sexta. Insect Biochem Mol Biol. 2001;31:887–898. doi: 10.1016/s0965-1748(01)00034-0. [DOI] [PubMed] [Google Scholar]
- Gillespie JP, Kanost MR, Trenczek T. Biological mediators of insect immunity. Ann Rev Entomol. 1997;42:611–643. doi: 10.1146/annurev.ento.42.1.611. [DOI] [PubMed] [Google Scholar]
- Girard P, Boublik Y, Wheat CW, Volkoff AN, Cousserans F, Brehélin M, Escoubas JM. X-tox: an atypical defensin derived family of immune-related proteins specific to Lepidoptera. Dev Comp Immunol. 2008;32:575–584. doi: 10.1016/j.dci.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Gorman MJ, An C, Kanost MR. Characterization of tyrosine hydroxylase from Manduca sexta. Insect Biochem Mol Biol. 2007;37:1327–37. doi: 10.1016/j.ibmb.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Wang Y, Jiang H. Manduca sexta prophenoloxidase (proPO) activation requires proPO-activating proteinase (PAP) and serine proteinase homologs (SPHs) simultaneously. Insect Biochem Mol Biol. 2005b;35:241–248. doi: 10.1016/j.ibmb.2004.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H. The biochemical basis of antimicrobial responses in Manduca sexta. Insect Sci. 2008;15:53–66. [Google Scholar]
- Jiang H, Kanost MR. The clip-domain family of serine proteinases in arthropods. Insect Biochem Mol Biol. 2000;30:95–105. doi: 10.1016/s0965-1748(99)00113-7. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Kanost MR. Four serine proteinases expressed in Manduca sexta hemocytes. Insect Mol Biol. 1999;8:39–53. doi: 10.1046/j.1365-2583.1999.810039.x. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Gu Y, Guo X, Zou Z, Scholz F, Trenczek TE, Kanost MR. Molecular identification of a bevy of serine proteinases in Manduca sexta hemolymph. Insect Biochem Mol Biol. 2005;35:931–943. doi: 10.1016/j.ibmb.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Ma C, Kanost MR. Subunit composition of pro-phenol oxidase from Manduca sexta: molecular cloning of subunit proPO-p1. Insect Biochem Mol Biol. 1997;27:835–850. doi: 10.1016/s0965-1748(97)00066-0. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Kanost MR. Prophenoloxidase-activating proteinase-2 from hemolymph of Manduca sexta: a bacteria-inducible serine proteinase containing two clip domains. J Biol Chem. 2003a;278:3552–3561. doi: 10.1074/jbc.M205743200. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Yu XQ, Zhu Y, Kanost MR. Prophenoloxidase-activating proteinase-3 (PAP-3) from Manduca sexta hemolymph: a clip-domain serine proteinase regulated by serpin-1J and serine proteinase homologs. Insect Biochem Mol Biol. 2003b;33:1049–1060. doi: 10.1016/s0965-1748(03)00123-1. [DOI] [PubMed] [Google Scholar]
- Kanamori Y, Hayakawa Y, Matsumoto H, Yasukochi Y, Shimura S, Nakahara Y, Kiuchi M, Kamimura M. A eukaryotic (insect) tricistronic mRNA encodes three proteins selected by context-dependent scanning. J Biol Chem. 2010;285:36933–36944. doi: 10.1074/jbc.M110.180398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanost MR, Nardi JB. Innate immune responses of Manduca sexta. In: Goldsmith MR, Marec F, editors. Molecular Biology and Genetics of Lepidoptera. CRC Press; New York: 2010. pp. 271–292. [Google Scholar]
- Kanost MR, Zepp MK, Ladendorff NE, Andersson LA. Isolation and characterization of a hemocyte aggregation inhibitor from hemolymph of Manduca sexta larvae. Arch Insect Biochem Physiol. 1994;27:123–136. doi: 10.1002/arch.940270205. [DOI] [PubMed] [Google Scholar]
- Kanost MR. Serine proteinase inhibitors in arthropod immunity. Dev Comp Immunol. 1999;23:291–301. doi: 10.1016/s0145-305x(99)00012-9. [DOI] [PubMed] [Google Scholar]
- Kauppila S, Maaty WS, Chen P, Tomar RS, Eby MT, Chapo J, Chew S, Rathore N, Zachariah S, Sinha SK, Abrams JM, Chaudhary PM. Eiger and its receptor, Wengen, comprise a TNF-like system in Drosophila. Oncogene. 2003;22:4860–4867. doi: 10.1038/sj.onc.1206715. [DOI] [PubMed] [Google Scholar]
- Kocks C, Cho JH, Nehme N, Ulvila J, Pearson AM, Meister M, Strom C, Conto SL, Hetru C, Stuart LM, Stehle T, Hoffmann JA, Reichhart JM, Ferrandon D, Rämet M, Ezekowitz RA. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell. 2005;123:335–346. doi: 10.1016/j.cell.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Kurucz E, Márkus R, Zsámboki J, Folkl-Medzihradszky K, Darula Z, Vilmos P, Udvardy A, Krausz I, Lukacsovich T, Gateff E, Zettervall CJ, Hultmark D, Andó I. Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr Biol. 2007;17:649–654. doi: 10.1016/j.cub.2007.02.041. [DOI] [PubMed] [Google Scholar]
- Ladendorff NE, Kanost MR. Bacteria-induced protein P4 (hemolin) from Manduca sexta: a member of the immunoglobulin superfamily which can inhibit hemocyte aggregation. Arch Insect Biochem Physiol. 1991;18:285–300. doi: 10.1002/arch.940180410. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Ann Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Lesch C, Goto A, Lindgren M, Bidla G, Dushay MS, Theopold U. A role for hemolectin in coagulation and immunity in Drosophila melanogaster. Dev Comp Immunol. 2007;31:1255–1263. doi: 10.1016/j.dci.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Ling E, Rao XJ, Ao JQ, Yu XQ. Purification and characterization of a small cationic protein from the tobacco hornworm Manduca sexta. Insect Biochem Mol Biol. 2009;39:263–271. doi: 10.1016/j.ibmb.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yao Q, Wang Y, Chen K. Proteomic analysis of nucleopolyhedrovirus infection resistance in the silkworm, Bombyx mori. J Invertebr Pathol. 2010;105:84–90. doi: 10.1016/j.jip.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Ma C, Kanost MR. A beta-1,3-glucan recognition protein from an insect, Manduca sexta, agglutinates microorganisms and activates the phenoloxidase cascade. J Biol Chem. 2000;275:7505–7514. doi: 10.1074/jbc.275.11.7505. [DOI] [PubMed] [Google Scholar]
- Marguerat S, Bähler J. RNA-Seq: from technology to biology. Cell Mol Life Sci. 2010;67:569–579. doi: 10.1007/s00018-009-0180-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Bader JS, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Oda Y, Uryu M, Hayakawa Y. Insect cytokine growth-blocking peptide triggers a termination system of cellular immunity by inducing its binding protein. J Biol Chem. 2003;278:38579–38585. doi: 10.1074/jbc.M305986200. [DOI] [PubMed] [Google Scholar]
- Morin RD, Aksay G, Dolgosheina E, Ebhardt HA, Magrini V, Mardis ER, Sahinalp SC, Unrau PJ. Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res. 2008;18:571–584. doi: 10.1101/gr.6897308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Mulnix AB, Dunn PE. Structure and induction of a lysozyme gene from the tobacco hornworm, Manduca sexta. Insect Biochem Mol Biol. 1994;24:271–281. doi: 10.1016/0965-1748(94)90007-8. [DOI] [PubMed] [Google Scholar]
- Nardi JB, Martos R, Walden KK, Lampe DJ, Robertson HM. Expression of lacunin, a large multidomain extracellular matrix protein, accompanies morphogenesis of epithelial monolayers in Manduca sexta. Insect Biochem Mol Biol. 1999;29:883–897. doi: 10.1016/s0965-1748(99)00064-8. [DOI] [PubMed] [Google Scholar]
- Nardi JB, Zhuang S, Pilas B, Bee CM, Kanost MR. Clustering of adhesion receptors following exposure of insect blood cells to foreign surfaces. J Insect Physiol. 2005;51:555–564. doi: 10.1016/j.jinsphys.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Nichol H, Law JH, Winzerling JJ. Iron metabolism in insects. Ann Rev Entomol. 2002;47:535–559. doi: 10.1146/annurev.ento.47.091201.145237. [DOI] [PubMed] [Google Scholar]
- Noguchi H, Tsuzuki S, Tanaka K, Matsumoto H, Hiruma K, Hayakawa Y. Isolation and characterization of a dopa decarboxylase cDNA and the induction of its expression by an insect cytokine, growth-blocking peptide in Pseudaletia separata. Insect Biochem Mol Biol. 2003;33:209–217. doi: 10.1016/s0965-1748(02)00192-3. [DOI] [PubMed] [Google Scholar]
- Pauchet Y, Wilkinson P, Vogel H, Nelson DR, Reynolds SE, Heckel DG, ffrench-Constant RH. Pyrosequencing the Manduca sexta larval midgut transcriptome: messages for digestion, detoxification and defence. Insect Mol Biol. 2010;19:61–75. doi: 10.1111/j.1365-2583.2009.00936.x. [DOI] [PubMed] [Google Scholar]
- Ragan EJ, An C, Jiang H, Kanost MR. Roles of hemolymph proteins in antimicrobial defenses of Manduca sexta. In: Reynolds S, Rolff J, editors. Insect Infection and Immunity. Oxford University Press; 2009. pp. 34–48. [Google Scholar]
- Rayaprolu S, Wang Y, Kanost MR, Hartson S, Jiang H. Functional analysis of four processing products from multiple precursors encoded by a lebocin-related gene from Manduca sexta. Dev Comp Immunol. 2010;34:638–647. doi: 10.1016/j.dci.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe B. Shotgun library construction for DNA sequencing. Methods Mol Biol. 2004;255:171–187. doi: 10.1385/1-59259-752-1:171. [DOI] [PubMed] [Google Scholar]
- Shin SW, Park SS, Park DS, Kim MG, Kim SC, Brey PT, Park HY. Isolation and characterization of immune-related genes from the fall webworm, Hyphantria cunea, using PCR-based differential display and subtractive cloning. Insect Biochem Mol Biol. 1998;28:827–37. doi: 10.1016/s0965-1748(98)00077-0. [DOI] [PubMed] [Google Scholar]
- Strand MR. The insect cellular immune response. Insect Sci. 2008;15:1–15. [Google Scholar]
- Tanaka H, Ishibashi J, Fujita K, Nakajima Y, Sagisaka A, Tomimoto K, Suzuki N, Yoshiyama M, Kaneko Y, Iwasaki T, Sunagawa T, Yamaji K, Asaoka A, Mita K, Yamakawa M. A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect Biochem Mol Biol. 2008;38:1087–1110. doi: 10.1016/j.ibmb.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Matsuki H, Furukawa S, Sagisaka A, Kotani E, Mori H, Yamakawa M. Identification and functional analysis of Relish homologs in the silkworm, Bombyx mori. Biochim Biophys Acta. 2007;1769:559–568. doi: 10.1016/j.bbaexp.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Vogel BE, Hedgecock EM. Hemicentin, a conserved extracellular member of the immunoglobulin superfamily, organizes epithelial and other cell attachments into oriented line-shaped junctions. Development. 2001;128:883–894. doi: 10.1242/dev.128.6.883. [DOI] [PubMed] [Google Scholar]
- Yu XQ, Jiang H, Wang Y, Kanost MR. Nonproteolytic serine proteinase homologs involved in phenoloxidase activation in the tobacco hornworm, Manduca sexta. Insect Biochem Mol Biol. 2003;33:197–208. doi: 10.1016/s0965-1748(02)00191-1. [DOI] [PubMed] [Google Scholar]
- Yu XQ, Tracy ME, Ling E, Scholz FR, Trenczek T. A novel C-type immulectin-3 from Manduca sexta is translocated from hemolymph into the cytoplasm of hemocytes. Insect Biochem Mol Biol. 2005;35:285–295. doi: 10.1016/j.ibmb.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Zang X, Maizels RM. Serine proteinase inhibitors from nematodes and the arms race between host and pathogen. Trends Biochem Sci. 2001;26:191–197. doi: 10.1016/s0968-0004(00)01761-8. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Johnson TJ, Myers AA, Kanost MR. Identification by subtractive suppression hybridization of bacteria-induced genes expressed in Manduca sexta fat body. Insect Biochem Mol Biol. 2003;33:541–559. doi: 10.1016/s0965-1748(03)00028-6. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Ragan EJ, Kanost MR. Leureptin: a soluble, extracellular leucine-rich repeat protein from Manduca sexta that binds lipopolysaccharide. Insect Biochem Mol Biol. 2010;40:713–722. doi: 10.1016/j.ibmb.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Najar F, Wang Y, Roe B, Jiang H. Pyrosequence analysis of expressed sequence tags for Manduca sexta hemolymph proteins involved in immune responses. Insect Biochem Mol Biol. 2008;38:677–682. doi: 10.1016/j.ibmb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Zhao P, Weng H, Mita K, Jiang H. A comparative analysis of serpin genes in the silkworm genome. Genomics. 2009;93:367–375. doi: 10.1016/j.ygeno.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.