Abstract
Differential gene expression in sugarcane during sugarcane-Ustilago scitaminea interaction was conducted in a smut-resistant genotype. Using cDNA-AFLP along with silver staining, a total of 136 transcript-derived fragments (TDFs) were found to be differentially expressed in response to challenge by U. scitaminea. Forty TDFs, 34 newly induced plus six with obvious upregulated expression after infection, were sequenced and validated by RT-PCR analysis. These results demonstrated that the expression of 37 out of these TDFs in RT-PCR analysis was consistent with that in cDNA-AFLP analysis. Based on BlastX in NCBI, 28 TDFs were assumed to function in sugarcane under U. scitaminea stress. Analysis of expression profile of three TDFs revealed that they responded differently after infection with U. scitaminea, and the transcription was significantly enhanced. The response of two TDFs, SUC06 and SUC09, occurred before that of SUC10. This study enriches our knowledge of the molecular basis for sugarcane response to U. scitaminea infection.
1. Introduction
Sugarcane is the principle sugar crop in the world. Cane sugar accounts for over 75% of the sugar production in the world and 92% of that in China. Sugarcane can also be used for producing many industrial products including ethanol, fructo-oligosaccharide, furfural, and glucan. Ethanol produced from sugarcane accounts for about 60% of the world's biomass-based fuel ethanol.
During sugarcane production, its yield is lagging behind due to biotic and abiotic stresses [1–5]. Diseases like mosaic, smut, ratoon stunting disease, and rust are some of the major biotic stresses which can substantially reduce yield. Among these, sugarcane smut caused by the fungus Ustilago scitaminea, is one of the most prevalent diseases and it has been responsible for the demise of several leading varieties [1–3]. Yield losses from 12%–75% have been reported [2, 3]. Total crop failure is possible if susceptible varieties are used and conditions are favorable for infection [4]. Smut has spread to all countries with sugarcane production except Papua New Guinea. Area-wide epidemics have occurred in some sugarcane planting areas. Presently, sugarcane breeding for smut resistance attracts worldwide attention and cultivating smut-resistant varieties is assumed to be the most economic and effective measurement to control this disease [5].
Plant disease resistance is the result of coevolution between the plant and pathogen [6]. During U. scitaminea infection, the fungus grows within the meristematic tissue and induces formation of flowering structures, which it colonises to produce its teliopores. The flowering structures, usually typical grass panicles, are transformed into a whip-like sorus that grows rapidly and protrudes out between the leaf sheaths. The development of sugarcane smut depends on the interaction among environment, the sugarcane variety and the pathogen itself [4, 5]. If the interaction between smut-resistant varieties and the pathogen is nonaffinity, disease resistance occurs; however, if the interaction between smut-susceptible varieties and the pathogen is affinity, disease susceptibility occurs. A series of physiological and biochemical changes, together with the molecular response, occur during the period between the appearance of the stress on plant from the invasion of the pathogen and the subsequent plant-pathogen interaction. The smut-resistant mechanism of sugarcane is reflected at the morphological, physiological and molecular levels [7, 8]. Progress has been made in studies of the molecular basis of sugarcane smut resistance [9–12]. Thokoane and Rutherford (2001) used cDNA-AFLP technique to detect differential gene action in smut resistant and susceptible sugarcane genotypes. Sequence homology analysis showed that a putative chitin receptor kinase, a Pto ser/thr protein kinase interactor, and an active gypsy-type LTR retrotranspon expressed in the resistant variety in response to challenge by the smut organism. Orlando et al. (2005) applied cDNA-AFLP technique to identify sugarcane genes differentially expressed in disease-resistant and susceptible sugarcane somaclones in response to inoculation with U. scitaminea. Eleven differentially expressed transcript-derived fragments (TDFs) isolated from the smut-resistant somaclone showed significant homology with genes involved in defense and/or signal transduction, including a gene encoding an NBS-LRR-like protein. Using cDNA-AFLP analysis, Lao et al. (2008) conducted a differential expression study on the sugarcane-U. scitaminea interaction. A total of 64 TDFs was found to be differentially expressed, among which 67.2% were upregulated in the resistant cultivar. It also suggested a key role for genes involved in the oxidative burst and the lignin pathways in sugarcane defense against the U. scitaminea infection. Previously, our laboratory used DDRT-PCR technique to screen differentially expressed genes associated with smut resistance. Homology analysis revealed that seven TDFs obtained in this study shared high homology with genes encoding cytochrome C oxidase, ribosomal protein, NAD-dependent malic enzyme, aminotransferase, binding protein, RNA polymerase specific transcription initiation factor and retrotransposon [12]. Despite what we have already learned, more studies on the molecular interaction in this pathosystem are needed to discover the mechanisms of smut resistance.
Identification of differentially expressed genes under various stresses can give clues as to what defense mechanisms and biochemical pathways are regulated during different types of stress [13]. To date, differentially expressed genes can be studied using a multitude of methods but mainly involving representational difference analysis of cDNA (RDA) [14], serial analysis of gene expression (SAGE) [15], suppression subtractive hybridization (SSH) [16], cDNA microarray analysis [17], and cDNA-amplified fragment length polymorphism (AFLP) [18]. Among them, cDNA-AFLP has proven to be an excellent tool to identify novel genes related to plant resistance to pathogens [19]. The cDNA-AFLP technique incorporates AFLP for the analysis of the differential expression of mRNA. Combining the advantages of both RT-PCR and AFLP, the cDNA-AFLP technique is reliable and efficient [18, 19]. By using cDNA-AFLP analysis, we were able to survey transcriptional changes with no prior assumptions about which genes might be induced or repressed [19]. In addition, this technique only requires a small amount of starting materials (around 100–200 mg), which is especially important when experimental material is difficult to acquire, thus expanding its application. By means of cDNA-AFLP technique, gene expression can be analyzed comprehensively, and then, the studies on the features of the gene expression during some physiological and biochemical processes can be conducted through analyzing and comparing the obtained TDFs [18]. cDNA-AFLP has been extensively applied in studies of plant gene expression under various exogenous stresses, such as droughts, cold temperatures, and fungal challenge [9–11, 20, 21]. Additionally, the silver staining technique, which can be conducted in a modestly equipped laboratory, can overcome shortcomings associated with the isotope labeling method by providing a faster, safer, and less expensive method that requirs with no special equipment.
NCo376 is the highly resistant standard control variety currently adopted in China in sugarcane smut resistance identification field trials using artificial inoculation techniques. To gain more insight into the genetic background of the enhanced resistance in a certain variety, one approach would be to perform analysis of differential gene expression in sugarcane challenged by U. scitaminea. We report herein results investigating the differential gene expression in NCo376 with and without U. scitaminea inoculation employing the combined method of cDNA-AFLP technique and silver staining. The differential expression of obtained TDFs was validated with RT-PCR analysis. Finally, the expression profiling of several differentially expressed TDFs was conducted by RT-PCR. The objective of this study is to determine those molecular events associated with the sugarcane resistance after infection with U. scitaminea by isolating TDFs which are differentially expressed in a highly smut-resistant genotype of NCo376.
2. Materials and Methods
2.1. Sugarcane Plant and Smut Inoculation
Plant material utilized for this study was from the smut-resistant sugarcane variety NCo376. The source of U. scitaminea Race 2 was inoculum collected from sugarcane variety F134.
Healthy, vigorous sugarcane plants with uniform growth were selected, and the stems were cut into double bud stem segments. Following 50°C hot water treatment for 2 h, buds were manually pinpricked with three punctures per bud using a pin dipped in the teliospore suspension at the concentration of 5 × 106 spores mL−1. The inoculated sugarcane was cultured in pots containing sand and placed in a illumination incubator with constant temperature at 27°C and light intensity at 2000 Lx, and a photoperiod of 13 : 11 (L : D). Previous studies showed that the sampling times sufficient to detect gene expression in sugarcane in bud tissue was at 0 h, 6 h, 12 h, 24 h, 48 h, 60 h, and 72 h after infection [22, 23]. A mixture of an equal amount from the seven samples was used for cDNA-AFLP analysis and RT-PCR validation, while expression profiling analysis was performed on an individual samples. Sugarcane buds inoculated with sterile ddH2O were used as control. The detection protocol developed by Albert and Schenck was used to confirm that the hot water disinfection was complete [24]. Collected samples were frozen in liquid nitrogen immediately and stored at −80°C in a freezer until RNA extraction.
2.2. RNA Extraction and cDNA Synthesis
Trizol (Invitrogen) was used to extract the total RNA of those samples following procedures as described by Que et al. [25]. The quantity and quality of extracted RNA was then determined by UV spectrophotometer. Finally, the first strand cDNA from 10 μg DNase-I-treated RNA and the second strand of cDNA was prepared using SMART cDNA synthesis kit (Clontech, USA).
2.3. cDNA-AFLP Analysis
Template preparation for cDNA-AFLP reaction and silver staining: The template for cDNA-AFLP analysis was prepared according to Bachem et al. [18]. cDNA was digested by Eco RI and Mse I end ligated by Eco RI and Mse I adaptors, and then, the PCR reactions were performed using combinations of the Eco RI-Mse I primers. The primers used in the AFLP reaction included the following (N = any nucleotide): Eco RI preamplifying primer, 5-GACTGCGTACCAATTC-3; Mse I preamplifying primer, 5-GACG ATGAGTCCTGAGTAA-3; Eco RI selectively amplifying primer, 5-GACTGCG TACCAATTCNN(N)-3 and Mse I selectively amplifying primer, 5-GACGATGAGTCCTGAGTAANN (N)-3. PCR profiles were obtained as follows: 11 cycles at 94°C, 30 s; 65°C [−0.7°C/cycle], 30 s; 72°C, 1 min and 30 cycles at 94°C, 30 s; 56°C, 30 s; 72°C, 1 min [18]. cDNA-AFLP products from inoculated and uninoculated bud tissues were separated by electrophoresis on a 6% denaturing gel run at 100 W until the bromophenol blue reached the bottom of the gel. The separated DNA fragments were then detected by silver staining and imaging. Since we have greater interest in genes enhancing smut resistance, only those TDFs with obvious upregulated expression or newly induced after infection were selected for further study.
Isolation and sequencing of TDFs: The bands of interest were marked, cut out from the gel and incubated in 100 μL TE (10 mmol/L Tris, pH 7.5, and 1 mmol/LM EDTA, pH 8.0) at 37°C overnight. DNA fragments were reamplified, subcloned into pMD18-T and sent for sequencing.
2.4. RT-PCR Validation and Bioinformatics Analysis of TDFs
For all of the 40 TDFs screened by cDNA-AFLP, differential expression was validated by RT-PCR technique. Sugarcane 25S rRNA was used for the normalization of reactions [26]. RT-PCR primers were designed according to the sequences of TDFs and the primer sequences are listed in Table 1. The 20 μL RT-PCR reaction system contained 2 μL 10xPCR Buffer, 0.5 μL dNTP Mixtures (2.5 μmol/L each), 1.0 μL Primer F (10 μmol/L), 1.0 μL Primer R (10 μmol/L), 0.3 μL Taq DNA polymerase (5 U/μL), 2.0 μL cDNA template, and 13.2 μL ddH2O. The following thermal cycling protocol was used: 94°C 5 min, 30 cycles of 94°C for 30 s, X°C for 30 s, 72°C 1 min, and then final extension at 72°C for 5 min (X stands for the annealing temperature of each primer listed in Table 1).
Table 1.
Primers used in RT-PCR analysis.
| Clone no. | Primer name | Sequence | Anneal temperature (%) |
|---|---|---|---|
| SUC01 | C418-F | 5′-TGAAAGCCTCCACATCCT-3′ | 55°C |
| C418-R | 5′-CAACGAGCAGAAGACACG-3′ | 55°C | |
| SUC02 | C165-F | 5′-TATGGCCGTGTGTGTTATGG-3′ | 60°C |
| C165-R | 5′-AACGGCTCTTTTTGCAGCTA-3′ | 60°C | |
| SUC03 | C220-F | 5′-CAGGAACCTTCTGGCATCAT-3′ | 60°C |
| C220-R | 5′-ATGGGCATCAACAGACTTCC-3′ | 60°C | |
| SUC04 | C203-F | 5′-TGTTTTCACCGTTCAGTTCCT-3′ | 59°C |
| C203-R | 5′-ATCAGTCGCACCCTGAAGAT-3′ | 59°C | |
| SUC05 | C149-F | 5′-CGATTACGGATTACTAGGTATTAGGA-3′ | 58°C |
| C149-R | 5′-CGATTCATTTCAATATGAGGACA-3′ | 58°C | |
| SUC06 | C312-F | 5′-TGGCTTAGGGCTGATACTGG-3′ | 58°C |
| C312-R | 5′-TCAAACGCTGACCCGAGA-3′ | 58°C | |
| SUC07 | C362-F | 5′-TGGGATGACGATACACCG-3′ | 56°C |
| C362-R | 5′-AGGCTCCTTCAAATGTAGATAG-3′ | 56°C | |
| SUC08 | C217-F | 5′-TCCTTTGGTTCCATTTGCTC-3′ | 58°C |
| C217-R | 5′-CGTGCAAATTAGGACTGTCG-3′ | 58°C | |
| SUC09 | C308-F | 5′-TGTCTCCAAACCACGAAC-3′ | 55°C |
| C308-R | 5′-TACAGAAGCCACAAGGGA-3′ | 55°C | |
| SUC10 | C243-F | 5′-TGGTTATTGGGCAGTCAA-3′ | 55°C |
| C243-R | 5′-ACGGCAGTAAGCAGAGGT-3′ | 55°C | |
| SUC11 | C266-F | 5′-AAAGCAACCCGAGATTTTCC-3′ | 60°C |
| C266-R | 5′-TAGGCGGAGAGGATGTAGGA-3′ | 60°C | |
| SUC12 | C363-F | 5′-AGTTTGCACCCGGTAGTGAC-3′ | 59°C |
| C363-R | 5′-ATGATGCCACCCTACCACTC-3′ | 59°C | |
| SUC13 | C141-F | 5′-GACCCAAGGGTCAATGTCTG-3′ | 57°C |
| C141-R | 5′-GCCCATCATACAATCCCTTC-3′ | 57°C | |
| SUC14 | C275-F | 5′-ACCAGTGCCACTTCATCCTC-3′ | 60°C |
| C275-R | 5′-TGCTGGCTATTGTTGGACAG-3′ | 60°C | |
| SUC15 | C217-F | 5′-TATGGCCATGGTGTTTATGG-3′ | 56°C |
| C217-R | 5′-AACGGTCCTTTTTGCACCTA-3′ | 56°C | |
| SUC16 | C214-F | 5′-GGCCATTGAGCACCTAACAT-3′ | 56°C |
| C214-R | 5′-CATGGGTCAGCATCATCAAC-3′ | 56°C | |
| SUC17 | C404-F | 5′-TAGTGGTGCGACCACACAAT-3′ | 60°C |
| C404-R | 5′-CGAACAGGGCCACTATGTCT-3′ | 60°C | |
| SUC18 | C190-F | 5′-CCGTGACCCTTCAATGTTTT-3′ | 58°C |
| C190-R | 5′-CCAACTTCCAAAGGGCTACA-3′ | 58°C | |
| SUC19 | C211-F | 5′-TTAGACCTTGCCCCCTTTTT-3′ | 57°C |
| C211-R | 5′-CCTGAGTAACATGATGGGTCCT-3′ | 57°C | |
| SUC20 | C288-F | 5′-GCATAGCCTTTCAGGTGG-3′ | 56°C |
| C288-R | 5′-TCGGGTGAACGATGAGGT-3′ | 56°C | |
| SUC21 | C274-F | 5′-AAACACTGGCTTAGGGCTGA-3′ | 58°C |
| C274-R | 5′-CAGCTCAGATCAAACGCTGA-3′ | 58°C | |
| SUC22 | C256-F | 5′-CTGATTCTTACTATTGGTGGTG-3′ | 55°C |
| C256-R | 5′-TGGATGCTCTGCTGCTAT-3′ | 55°C | |
| SUC23 | C363-F | 5′-AGTTTGCACCCGGTAGTGAC-3′ | 58°C |
| C363-R | 5′-ATGATGCCACCCTACCACTC-3′ | 58°C | |
| SUC24 | C244-F | 5′-GACCTCTCCACCCCTAGACC-3′ | 57°C |
| C244-R | 5′-GGAGAAGCATGGTTCCAAAA-3′ | 57°C | |
| SUC25 | C169-F | 5′-GTTGGTGATCCTGAGGGAAC-3′ | 56°C |
| C169-R | 5′-AGTCGAGTGCAGCCCTACAT-3′ | 56°C | |
| SUC26 | C436-F | 5′-TCCAAAGGCATGAAGGAAAC-3′ | 58°C |
| C436-R | 5′-CGTCGTCATCTACCTGCTCA-3′ | 58°C | |
| SUC27 | C436-F | 5′-TCCAAAGGCATGAAGGAAAC-3′ | 57°C |
| C436-R | 5′-CGTCGTCTACCTGCTCATCA-3′ | 57°C | |
| SUC28 | C315-F | 5′-CTCCACCAGCTGATGTTCCT-3′ | 60°C |
| C315-R | 5′-ACCACAACATCACGGAATGA-3′ | 60°C | |
| SUC29 | C364-F | 5′-AGTTTGCACCCGGTAGTGAC-3′ | 60°C |
| C364-R | 5′-GACGATGAGCCTCGTTTAGG-3′ | 60°C | |
| SUC30 | C217-F | 5′-TATCCGGGGTGTTGTTATGG-3′ | 58°C |
| C217-R | 5′-AACGTCGCTTTTCGTAGCTA-3′ | 58°C | |
| SUC31 | C427-F | 5′-CCCGTCCTAGAGAGGAGGTC-3′ | 58°C |
| C427-R | 5′-AGGTCTTTCAGGCAACATCG-3′ | 58°C | |
| SUC32 | C255-F | 5′-TAAAGGCGACACCTCTAGGC-3′ | 57°C |
| C255-R | 5′-AACTTCAGGCTTTGGTGCTC-3′ | 57°C | |
| SUC33 | C291-F | 5′-CACACAATGACCATGCAACA-3′ | 58°C |
| C291-R | 5′-AATGGCGAAGCAATGTTTTT-3′ | 58°C | |
| SUC34 | C312-F | 5′-GGGTCACCGTCAGCCTACT-3′ | 56°C |
| C312-R | 5′-GGAGCCAGCACTCCATTT-3′ | 56°C | |
| SUC35 | C312-F | 5′-TCCTTATGCCAGAGGGTCAC-3′ | 59°C |
| C312-R | 5′-GGAGCCAGCACTCCATTTTA-3′ | 59°C | |
| SUC36 | C270-F | 5′-AAGGATCTTTGCAAGCATGG-3′ | 57°C |
| C270-R | 5′-AGGTCTTTCAGGCAACATCG-3′ | 57°C | |
| SUC37 | C206-F | 5′-TCCTATGATGACTGCCAAGC-3′ | 58°C |
| C206-R | 5′-CAAACTTCCATGTTTCCTCCA-3′ | 58°C | |
| SUC38 | C217-F | 5′-TATGGCCTGTGTGGTTATGG-3′ | 60°C |
| C217-R | 5′-AACGTCTCGTTTTGCAGCTA-3′ | 60°C | |
| SUC39 | C121-F | 5′-TGAGGGAACCTGTTCGAG-3′ | 55°C |
| C121-R | 5′-GAGTGCAGCCCTACATTG-3′ | 55°C | |
| SUC40 | C142-F | 5′-TTAGGAGATGTTGCTCCAAGC-3′ | 57°C |
| C142-R | 5′-GCCTCTTTCTTTCTTCCATGC-3′ | 57°C | |
| 25S rRNA | 25S rRNA-F | 5′-GCAGCCAAGCGTTCATAG-3′ | 56°C |
| 25S rRNA-R | 5′-CGGCACGGTCATCAGTAG-3′ | 56°C |
With the basic local alignment search tool (BLAST) program, sequences of TDFs confirmed by RT-PCR were subjected to BlastX similarity search against the NCBI protein database and then annotated according to their homologies with known function protein sequences. A search for sequence homologies in the sugarcane EST database of TIGR gene indices (SoGI) was also carried out for those TDFs. The functional classification of the identified TDFs was performed using the gene ontology (GO) database.
2.5. Expression Profiling of Three Differentially Expressed TDFs
Samples were collected at the seven time points (0 h, 12 h, 24 h, 36 h, 48 h, 60 h, and 72 h). RT-PCR expression profiling was conducted for three differentially expressed TDFs (SUC09-encoding bZIP transcription factor, SUC06-encoding ribosomal protein, and SUC10-encoding cytochrome). The protocol of expression profiling analysis of these TDFs was the same as that in RT-PCR validation. The quantity of RT-PCR product (expression value) was then calculated by Quantity One (Bio-Rad).
3. Results and Analysis
3.1. cDNA-AFLP Analysis
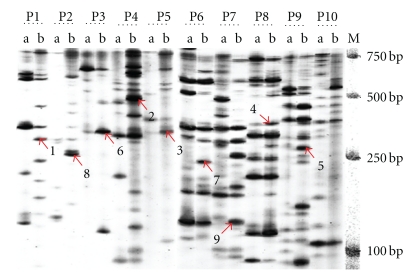
In the present study, eight pairs of selective amplification primers (64 primer combinations) were used for cDNA-AFLP analysis between U. scitaminea inoculated and uninoculated sugarcane. There were 1258 amplified bands ranging from 11 to 28 bands per primer combination with an average of 20. Figure 1 shows the partial results of cDNA-AFLP analysis. It should be noted that two types of differential TDFs were obtained in the study, polymorphic bands (newly induced or totally inhibited after infection) and bands with different expression value (upregulated or downregulated). A total of 136 differential TDFs and 1122 common bands were screened, in which the differential TDFs accounted for 10.8% of the total. Among these differential TDFs, there were 91 polymorphic (69.9%) and 45 upregulated (33.1%).
Figure 1.
PAGE electrophoresis patterns of some cDNA-AFLP amplification products. Notes: primers of P1–P10 represent E-AAC/M-CAG, E-ACA/M-CAG, E-ACA/M-CTC, E-AAC/M-CTT, E-AGG/M-CTC, E-AAG/M-CAT, E-AAG/M-CTA, E-AGC/M-CAG, E-AGG/M-CAT, and E-AGG/M-CTG, respectively; a, b represent control group and treat group, respectively; 1–9 represent SUC09, SUC01, SUC34, SUC07, SUC20, SUC06, SUC10, SUC22, and SUC39, respectively; M represents molecular weight marker.
3.2. Reamplification of the Differentially Expressed TDFs and RT-PCR Validation
A total of 40 TDFs from cDNA-AFLP analysis were selected for further study, including 34 newly induced and 6 upregulated after infection (all of them were from the inoculation treatment group). Re-amplification was performed on the TDFs with the same primer and reaction system used in 2.3. The result revealed that a clear band was amplified and the length of the band was consistent with that of the corresponding band in cDNA-AFLP analysis, indicating that the TDFs tested were correct. Recovery, purification, and subcloning were then performed for the TDFs which were sent for sequencing following identification by PCR and enzyme digestion.
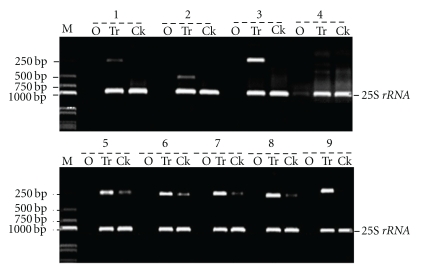
In order to ensure the reliability of these TDFs, specific primers were designed according to the sequence of corresponding TDFs. The differential expression of these 40 TDFs was validated by RT-PCR, and the electrophoresis results of partial RT-PCR products are shown in Figure 2. In RT-PCR validation, eight TDFs coded SUC02, SUC06, SUC07, SUC10, SUC18, SUC20, SUC33, and SUC39 were upregulated in the pathogen inoculation treatment group, while the remaining 29 TDFs were only expressed in the treatment group, specifically polymorphic TDFs. The results demonstrated that among these TDFs, the expression of 37 out of 40 TDFs in RT-PCR analysis was consistent with that of the corresponding TDFs in cDNA-AFLP analysis. The remaining three TDFs (No. SUC15, SUC30, and SUC38) having no amplified bands and were presumed to be false positives at the rate of 7.5%. This rate, when compared with a false positive rate of almost 70% in DDRT-PCR, is distinct advantage [12]. As illustrated in Figure 2, the expression of five TDFs (SUC01, SUC09, SUC22, SUC34, and SUC39) was newly induced after infection, but that of the other four TDFs (SUC06, SUC07, SUC10, and SUC20) was upregulated after infection. The above results suggested that 37 truly differentially expressed TDFs were obtained in this study and the application of cDNA-AFLP in the study of differential gene expression in sugarcane during sugarcane-U. scitaminea interaction was feasible.
Figure 2.
Results of RT-PCR validation of partial differential TDFs. Notes: M represents molecular weight marker; O, Tr, and Ck represent negative control, fungus infected, and control group, respectively; 1–9 represent SUC09, SUC01, SUC34, SUC07, SUC20, SUC06, SUC10, SUC22, and SUC39, respectively.
3.3. Homology Analysis and Functional Classification of Differentially Expressed TDFs
In order to acquire the function information of these 37 differentially expressed TDFs, their sequences were submitted into NCBI database for Blastx homology analysis. The result showed that these TDFs, with different functions annotated, might play a role in the interaction response of sugarcane-U. scitaminea (Table 2) [9–12]. Among these TDFs, nine of them were assumed to be new genes with unknown functions and the homologous genes of all the other 28 TDFs could be retrieved in GenBank, accounting for 24.3% and 75.7% of the 37 TDFs, respectively. Based on the functions of homologous genes or the similarity of translated proteins, and referring to the functional classification of GO database, these genes could be classified into eight groups with the results shown in Table 2. They were as follows: (1) cell rescue and defense related genes, such as aldehyde oxidase gene and aldose reductase gene, (2) energy metabolism related gene, such as cytochrome b6, (3) intracellular transport related gene, such as intracellular protein translocator gene and sucrose translocator gene, (4) signal transduction related gene, such as calmodulin-binding protein gene, (5) nucleic acid metabolism-related gene, such as SAM-dependent methyltransferase gene, (6) transcription related gene, such as RNA recognition protein gene; (7) protein synthesis and modification related gene, such as ribosomal protein gene, and (8) nine protein genes whose function unknown. Among these 28 TDFs with their function known, only seven (SUC06, SUC07, SUC09, SUC10, SUC13, SUC25, and SUC36) had been reported in previous studies on sugarcane-U. scitaminea interaction [10–12]. It should also be noted that several TDFs showed different base composition but encoded identical proteins, including two TRP proteins (SUC26 and SUC27), two serine/threonine protein kinase (SUC13 and SUC36), two phytoene dehydrogenase (SUC20 and SUC24), and three reverse transcriptase (SUC12, SUC23, and SUC19).
Table 2.
Homology of 37 differentially expressed TDFs identified in cDNA-AFLP analysis.
| Clone ID (GenBank Accn) | Size (bp) | Function description | Species | Homology (%) |
|---|---|---|---|---|
| Cell rescue and defense (Two) | ||||
| SUC01 (HS977224) | 418 bp | putative aldehyde oxidase | O. sativa | 78% |
| SUC28 (HS977227) | 315 bp | putative aldose reductase-related protein | Z. mays | 80% |
| Energy metabolism (Six) | ||||
| SUC10 (HS977228) | 243 bp | cytochrome | M. indica | 98% |
| SUC20 (HS977229) | 288 bp | phytoene desaturase | Z. mays | 96% |
| SUC21 (HS977230) | 274 bp | polyphosphate kinase | S.typhimurium | 29% |
| SUC22 (HS977231) | 256 bp | 3-dehydroquinate synthase | C. botulinum | 36% |
| SUC24 (HS977232) | 244 bp | phytoene desaturase | Z. mays | 100% |
| SUC31 (HS977233) | 427 bp | putative acid phosphatase | E. nidulans | 28% |
| Intracellular transport (Two) | ||||
| SUC02 (HS977225) | 165 bp | intracellular protein transport protein | O. sativa | 32% |
| SUC39 (HS977234) | 121 bp | sugar transporter | Y. pestis | 57% |
| Signal transduction (Six) | ||||
| SUC07 (HS977235) | 362 bp | putative calmodulin-binding protein | O. sativa | 92% |
| SUC13 (HS977236) | 141 bp | serine/threonine kinase protein | Z. mays | 64% |
| SUC14 (HS977237) | 275 bp | F-box domain containing protein | O. sativa | 59% |
| SUC26 (HS977238) | 436 bp | tetratricopeptide repeat domain protein | M. marina | 23% |
| SUC27 (HS977239) | 437 bp | tetratricopeptide repeat domain protein | M. marina | 24% |
| SUC36 (HS977240) | 270 bp | serine/threonine kinase protein | Z. mays | 49% |
| Nucleic acid metabolization (Four) | ||||
| SUC12 (HS977241) | 363 bp | putative reverse transcriptase | O. sativa | 70% |
| SUC23 (HS977242) | 365 bp | putative reverse transcriptase | O. sativa | 69% |
| SUC25 (HS977243) | 169 bp | putative SAM-dependent methyltransferase | W. pipientis | 35% |
| SUC29 (HS977244) | 364 bp | putative reverse transcriptase | O. sativa | 70% |
| Transcription (Five) | ||||
| SUC09 (HS977245) | 308 bp | bZIP transcription factor | G. max | 32% |
| SUC11 (HS977246) | 266 bp | promoter-binding-like protein | O. sativa | 66% |
| SUC35 (HS977247) | 312 bp | RNA recognition motif family protein | O. sativa | 45% |
| SUC37 (HS977248) | 206 bp | maturase K | A. scandens | 31% |
| SUC40 (HS977249) | 142 bp | putative ternary complex factor | A. majus | 63% |
| Protein synthesis and modulation (Three) | ||||
| SUC06 (HS977250) | 317 bp | ribosomal protein | Z. mays | 73% |
| SUC16 (HS977251) | 214 bp | pentatricopeptide repeat-containing protein | A. thaliana | 26% |
| SUC34 (HS977252) | 312 bp | translation elongation factor | N. paludosum | 40% |
| Unknown (Nine) | ||||
| SUC03 (HS977226) | 220 bp | unknown protein (AT2G32240) | A. thaliana | 53% |
| SUC04 (HS977258) | 203 bp | hypothetical protein PCO123869 | Z. mays | 80% |
| SUC05 (HS977259) | 149 bp | unknown protein | S. officinarum | 96% |
| SUC08 (HS977253) | 217 bp | hypothetical protein | O. sativa | 81% |
| SUC17 (HS977254) | 404 bp | hypothetical protein | C. immitis | 37% |
| SUC18 (HS977255) | 190 bp | hypothetical protein | O. sativa | 83% |
| SUC19 (HS977256) | 211 bp | hypothetical protein | D. vulgaris | 39% |
| SUC32 (HS977260) | 255 bp | hypothetical protein | Z. mays | 86% |
| SUC33 (HS977257) | 291 bp | hypothetical protein | O. sativa | 45% |
3.4. Expression Profiling of Three Differentially Expressed TDFs
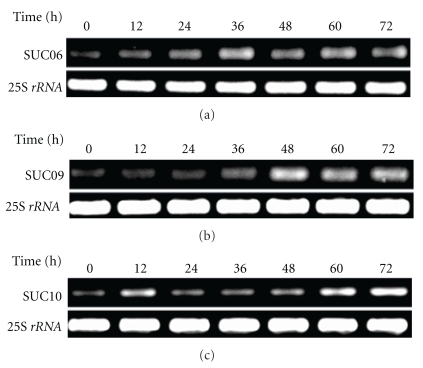
In order to elucidate the expression profiling of the different TDFs in the interaction between sugarcane and U. scitaminea, three TDFs of which the differential expression had been confirmed by RT-PCR analysis (SUC09 newly induced and SUC06 and SUC10 with upregulation after infection) were selected for the analysis of expression profiling through RT-PCR technique.
As shown in Figure 3, three TDFs showed weak expression at the beginning of interaction (0 h), but at later time points during the 72 h, their expression increased, which suggested the differential expression resulted from the challenge of U. scitaminea. Just like in the cDNA-AFLP analysis, these three TDFs were expression-induced or upregulated. It was also found that different TDFs had different levels of induced expression (Figure 3). Without pathogen attack, these TDFs only had a minor expression, while after pathogen infection, various TDFs had different inducement levels. SUC06 had the highest expression peak at 36 h after infection, dropped at 48 h, and then gradually upregulated (Figure 3(a)). SUC09 had a constant increase in expression after pathogen infection, reached at the peak value at 48 h, and then during 60–72 h period the expression was inhibited with the expression value decreased during 60–72 h (Figure 3(b)). The expression of SUC10 was initially inhibited and then upregulated; that is, it was upregulated at 12 h, inhibited at 24 h, and then the expression level kept increasing (Figure 3(c)). The expression of this TDF was induced 12 h after infection, and the inducible expression lasted for at least 72 h. These genes responded in the early period of inducement (12 h) when challenged by U. scitaminea, and the expression was significantly enhanced, but with different modes. The response of SUC06 and SUC09 to U. scitaminea was earlier than that of SUC10. From above, these genes probably took part in the sugarcane defense response against U. scitaminea, with different regulation modes.
Figure 3.
Expression profiling of three TDFs during sugarcane-U. scitaminea interaction based on RT-PCR analysis. Notes: (a) expression profiling of SUC06; (b) expression profiling of SUC09; (c) expression profiling of SUC10.
4. Discussion and Conclusions
Despite what we have already learned [9–12], little is known about the molecular background of the interaction between sugarcane and U. scitaminea. In the present study, cDNA-AFLP coupled with silver staining analysis was performed to identify genes that possibly contribute to the near-complete and stable smut resistance in NCo376, a variety that is highly resistant to sugarcane smut. Theoretically, the identification of differentially expressed TDFs has great potential to helping to understand the molecular mechanism of smut resistance. Furthermore, the full-length genes of those TDFs that correlate with smut resistance can even be cloned and transferred to target sugarcane variety to improve smut resistance [27].
Signal transduction, such as protein phosphorylation and dephosphorylation and Ca2+-dependent reaction, is important for the plant to perceive exogenous stress [28, 29]. Thokoane and Rutherford (2001) identified several TDFs related to signal transduction during smut reaction in a resistant variety N52/219. In the study by Orlando et al. (2005), most of the TDFs differentially expressed in smut-resistant somaclones were related to defense or signaling. In this study, six signal transduction-related TDFs (SUC07, SUC 13, SUC14, SUC26, SUC27, and SUC 36) were found to be upregulated during sugarcane-U. scitaminea interaction. Serine/threonine (Ser/Thr) protein kinase encoded by SUC 13 and SUC 36 can function in the midstream and downstream of signal transduction [28]. In two previous reports [9, 10], it is interesting to note that one TDF corresponding to Ser/Thr protein was found in both. Calmodulin binding protein (CaMBP) encoded by SUC07 is the target protein that directly bound and affected by Ca2+-CaM during signal transduction [30, 31]. TPR (tetratricopeptide repeat domain) protein encoded by SUC26 and SUC27 can play a role in physiological processes [32]. Besides, F-box protein coded by SUC14 is involved in the processes of plant stress and defensive reactions [33]. The above suggests that the differential expression of signal transduction-related TDFs may trigger a series of signal transductions to enable the resistant sugarcane variety to quickly respond to the challenge of U. scitaminea, thus improve its smut resistance [9, 10].
Previous research showed that transcription elements were a key factor in the signal transduction of the plant under stress [34]. One TDF encoding a transcription factor was found to function in sugarcane-U. scitaminea interaction [11]. In this study, three TDFs, one newly induced (SUC09) and two upregulated (SUC11 and SUC40) TDFs after infection related to transcription elements, were identified during sugarcane-U. scitaminea interaction, which also suggests the role of signal transduction in this pathosystem.
Metabolism would be accelerated under exogenous stress [35, 36]. In this study, we obtained five upregulated metabolism-related TDFs, SUC20, SUC24, SUC21, SUC22, and SUC31. Phytoene dehydrogenase encoded by SUC20 and SUC24 is a key enzyme involving lycopene synthesis. Phytoene can absorb light to protect plants from photo-oxidation [35]. Polyphosphate kinase (PPK) encoded by SUC21 can substitute for ATP and act as the energy source and regulator for exogenous stress. Dehydroquinic acid synthetase encoded by SUC22 is a key enzyme in the pathway of shikimic acid synthesis. Acid phosphatase encoded by SUC31 is associated with phosphate decomposition in plant, which has been demonstrated to function in carbohydrate transforming and protein synthesis [36]. These differentially expressed TDFs indicated that sugarcane-U. scitaminea interaction did affect the metabolism in sugarcane.
The present results also permit us to hypothesize about the key role of protein synthesis and modulation on sugarcane defense against U. scitaminea infection. Three TDFs, SUC06, SUC16, and SUC34, were also identified in this study. Ribosomal protein encoded by SUC06 is an important component of ribosome, and it binds with rRNA via noncovalent bond to enhance the function of rRNA, and thus it functions in transcription, replication, translation regulation, and DNA repair [37]. Previously, we have identified a TDF of which the corresponding gene shows homology to a ribosomal protein gene during sugarcane-U. scitaminea interaction [12]. Pentatricopeptide repeat-containing protein (PPRs) encoded by SUC16 is mainly located on chloroplast and mitochondria. It participates in the translation and posttranslational modification of the gene transcript, reversely influencing its expression [38]. Besides, the translation elongation factor encoded by SUC34 employs energy from GTP hydrolyzation to transfer nascent peptide chain to site P to elongate the protein in protein synthesis.
During the expression profiling analysis of three TDFs (SUC09, SUC06, and SUC10), all had a background level of trace expression at 0 h in both the treatment and control groups. At the other time points after the challenge of U. scitaminea, their expression was induced in the treatment but inhibited in the control group. Previously, we have also identified a TDF of which the corresponding genes show homologies to an Oryza sativa cytochrome C oxidase in sugarcane-U. scitaminea interaction [12]. Caldo et al. (2004) demonstrated that the genes related to the defense response would have different expression after powdery mildew inoculation in barley [39]. Jantasuriyarat et al. (2005) found that a multitude of genes with various expression modes changed noticeably during rice-blast interaction [40]. For sugarcane, the expression pattern of SUC09, SUC06, and SUC10 show differential responses to interactions with U. scitaminea, which suggests that they may participate in specific cascade(s) that can identify or resist a pathogenic infection [9–12].
In conclusion, we concur with previous studies that the gene regulatory network during the interaction of sugarcane-U. scitaminea is complicated [9–12]. This study contributes new insight into the molecular basis of sugarcane's response to the infection of U. scitaminea. Nevertheless, more detailed studies are needed to be conducted so that more differential genes can be identified and the corresponding regulatory network be investigated over the duration of the interaction—from initiation to termination.
Acknowledgments
This research was funded by the earmarked fund for Modern Agro-industry Technology Research System (CARS-20), National Natural Science Fund (no. 30170639), Research Fund for the Doctoral Program of Higher Education (no. 20103515120006) and National High Technology Research and Development Program of China (863 Program) Project (no. 2007AA100701). The authors appreciate all ideas and constructive criticism from the reviewers, especially Dr. William H. White, Michael P. Grisham and Thomas L. Tew, three research scientists in USDA-ARS, Sugarcane Research Unit, Louisiana, USA. Que You-Xiong and Lin Jian-Wei contributed equally to this paper.
References
- 1.J. W. Hoy, Hollier CA, Fontenot DB, Grelen LB. Incidence of sugarcane smut in Louisiana and its effects on yield. Plant Disease. 1986;70(1):59–60. [Google Scholar]
- 2.Sandhu SA, Bhatti DS, Rattan BK. Extent of losses caused by red (Physalosporatucumane NSis Speg.) and smut (Ustilago scitaminea Syd.) Journal of Research (Punjab Agricultural University) 1969;6:341–344. [Google Scholar]
- 3.Whittle AM. Yield loss in sugar-cane due to culmicolous smut infection. Tropical Agriculture. 1982;59(3):239–242. [Google Scholar]
- 4.Lee-Lovick G. Smut of sugarcane-Ustilago scitaminea. Review of Plant Pathology. 1978;147:181–188. [Google Scholar]
- 5.Chao CP, Hoy JW, Saxton AM, Martin FA. Heritability of resistance and repeatability of clone reactions to sugarcane smut in Louisiana. Phytopathology. 1990;80(7):622–627. [Google Scholar]
- 6.Graham TL, Graham MY. Cellular coordination of molecular responses in plant defense. Molecular Plant-Microbe Interactions. 1991;4(5):415–421. [Google Scholar]
- 7.Piñon D, de Armas R, Vicente C, Legaz ME. Role of polyamines in the infection of sugarcane buds by Ustilago scitaminea spores. Plant Physiology and Biochemistry. 1999;37(1):57–64. [Google Scholar]
- 8.de Armas R, Santiago R, Legaz ME, Vicente C. Levels of phenolic compounds and enzyme activity can be used to screen for resistance of sugarcane to smut (Ustilago scitaminea) Australasian Plant Pathology. 2007;36(1):32–38. [Google Scholar]
- 9.Thokoane LN, Rutherford RS. cDNA-AFLP differential display of sugarcane (Saccharum spp, hybrids) genes induced by challenge with the fungal pathogen Ustilago scitaminea (sugarcane smut) Proceedings of the South African Sugar Technologists Association. 2001;75:104–107. [Google Scholar]
- 10.Orlando BH, Thomma BP, Carmona E, et al. Identification of sugarcane genes induced in disease-resistant somaclones upon inoculation with Ustilago scitaminea or Bipolaris sacchari. Plant Physiology and Biochemistry. 2005;43(12):1115–1121. doi: 10.1016/j.plaphy.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 11.LaO M, Arencibia AD, Carmona ER, et al. Differential expression analysis by cDNA-AFLP of Saccharum spp. after inoculation with the host pathogen Sporisorium scitamineum. Plant Cell Reports. 2008;27(6):1103–1111. doi: 10.1007/s00299-008-0524-y. [DOI] [PubMed] [Google Scholar]
- 12.Que YX, Yang ZX, Xu LP, Chen RK. Isolation and identification of differentially expressed genes in sugarcane infected by Ustilago scitaminea. Acta Agronomica Sinica. 2009;35(3):452–458. [Google Scholar]
- 13.Staskawicz BJ. Genetics of plant-pathogen interactions specifying plant disease resistance. Plant Physiology. 2001;125(1):73–76. doi: 10.1104/pp.125.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hubank M, Schatz DG. Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Research. 1994;22(25):5640–5648. doi: 10.1093/nar/22.25.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto M, Wakatsuki T, Hada A, Ryo A. Use of serial analysis of gene expression (SAGE) technology. Journal of Immunological Methods. 2001;250(1-2):45–66. doi: 10.1016/s0022-1759(01)00305-2. [DOI] [PubMed] [Google Scholar]
- 16.Aharoni A, Vorst O. DNA microarrays for functional plant genomics. Plant Molecular Biology. 2002;48(1-2):99–118. doi: 10.1023/a:1013734019946. [DOI] [PubMed] [Google Scholar]
- 17.Diatchenko L, Lau YFC, Campbell AP, et al. Suppression subtractive hybridization:a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(12):6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bachem CWB, Oomen RJFJ, Visser RGF. Transcript imaging with cDNA-AFLP: a step-by-step protocol. Plant Molecular Biology Reporter. 1998;16(2):157–173. [Google Scholar]
- 19.Durrant WE, Rowland O, Piedras P, Hammond-Kosack KE, Jones JDG. cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell. 2000;12(6):963–978. doi: 10.1105/tpc.12.6.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin J, Wang G, Xiao J, et al. Identification of genes involved in stem rust resistance from wheat mutant D51 with the cDNA-AFLP technique. Molecular Biology Reports. 2010;37(2):1111–1117. doi: 10.1007/s11033-009-9870-2. [DOI] [PubMed] [Google Scholar]
- 21.Gao F, Zhang H, Wang H, Gao H, Li Z. Comparative transcriptional profiling under drought stress between upland and lowland rice (Oryza sativa L.) using cDNA-aflp. Chinese Science Bulletin. 2009;54(19):3555–3571. [Google Scholar]
- 22.Que YX, Lin JW, Zhang JS, Ruan MH, Xu LP, Zhang MQ. Molecular cloning and characterisation of a non-TIR-NBS-LRR type disease resistance gene analogue from sugarcane. Sugar Tech. 2008;10(1):71–73. [Google Scholar]
- 23.Que YX, Xu LP, Lin JW, Chen RK. Isolation and characterization of NBS-LRR resistance gene analogs from sugarcane. Acta Agronomica Sinica. 2009;35(4):631–639. [Google Scholar]
- 24.Albert HH, Schenck S. PCR amplification from a homolog of the bE mating-type gene as a sensitive assay for the presence of Ustilago scitaminea DNA. Plant Disease. 1996;80(10):1189–1192. [Google Scholar]
- 25.Que YX, Li W, Xu JS, Xu LP, Zhang MQ, Chen RK. A simple and versatile protocol for isolation of RNA from plant, fungi and animal. Journal of Agriculture Science and Technology. 2008;2(1):p. 63. [Google Scholar]
- 26.Que YX, Xu LP, Xu JS, Zhang JS, Zhang MQ, Chen RK. Selection of control genes in real-time qPCR analysis of gene expression in sugarcane. Chinese Journal of Tropical Crops. 2009;30(3):274–278. [Google Scholar]
- 27.Butterfield MK, Rutherford RS, Carson DL, Huckett BI. Application of gene discovery to varietal improvement in sugarcane. South African Journal of Botany. 2004;70(1):167–172. [Google Scholar]
- 28.Song WY, Wang GL, Chen LL, et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270(5243):1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- 29.Reddy ASN. Calcium: silver bullet in signaling. Plant Science. 2001;160(3):381–404. doi: 10.1016/s0168-9452(00)00386-1. [DOI] [PubMed] [Google Scholar]
- 30.Zielinski RE. Calmodulin and calmodulin-binding proteins in plants. Annual Review of Plant Biology. 1998;49:697–725. doi: 10.1146/annurev.arplant.49.1.697. [DOI] [PubMed] [Google Scholar]
- 31.Gul SA, Reddy VS, Lindgren PB, Jakobek JL, Ŕeddy ASN. Differential expression of genes encoding calmodulin-binding proteins in response to bacterial pathogens and inducers of defense responses. Plant Molecular Biology. 2003;51(6):803–815. doi: 10.1023/a:1023001403794. [DOI] [PubMed] [Google Scholar]
- 32.Scheufler C, Brinker A, Bourenkov G, et al. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101(2):199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 33.del Pozo JC, Estelle M. F-box proteins and protein degradation: an emerging theme in cellular regulation. Plant Molecular Biology. 2000;44(2):123–128. doi: 10.1023/a:1006413007456. [DOI] [PubMed] [Google Scholar]
- 34.Singh KB, Foley RC, Onate-Sanchez L. Transcription factors in plant defense and stress responses. Current Opinion in Plant Biology. 2002;5(5):430–436. doi: 10.1016/s1369-5266(02)00289-3. [DOI] [PubMed] [Google Scholar]
- 35.Velayos A, Blasco JL, Alvarez MI, Iturriaga EA, Eslava AP. Blue-light regulation of phytoene dehydrogenase (carB) gene expression in Mucor circinelloides. Planta. 2000;210(6):938–946. doi: 10.1007/s004250050701. [DOI] [PubMed] [Google Scholar]
- 36.Stephen MGD, Gautam S, William CP. The role of acid phosphatases in plant phosphorus metabolism. Physiology Planta. 2006;90(4):791–800. [Google Scholar]
- 37.Wool IG. Extraribosomal functions of ribosomal proteins. Trends in Biochemical Sciences. 1996;21(5):164–165. [PubMed] [Google Scholar]
- 38.Bentolila S, Alfonso AA, Hanson MR. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(16):10887–10892. doi: 10.1073/pnas.102301599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caldo RA, Nettleton D, Wise RP. Interaction-dependent gene expression in Mla-specified response to barley powdery mildew. Plant Cell. 2004;16(9):2514–2528. doi: 10.1105/tpc.104.023382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jantasuriyarat C, Gowda M, Haller K, et al. Large-scale identification of expressed sequence tags involved in rice and rice blast fungus interaction. Plant Physiology. 2005;138(1):105–115. doi: 10.1104/pp.104.055624. [DOI] [PMC free article] [PubMed] [Google Scholar]