Abstract
Recent studies indicated that recombination is strongly mutagenic. In particular, data from the mouse pseudoautosomal boundary (PAB) suggested that locally intensive recombination increased the nucleotide substitution rate by more than 100-fold and greatly increased the GC content. Here we study the rates of nucleotide substitution in eight introns of the human and great ape XG gene, which spans the boundary between the pseudoautosomal region 1 (PAR1) and the X-specific region. Contrary to what is expected under the above hypothesis, our sequence data from humans and great apes reveal that the PAR1 introns of XG have actually evolved slightly slower than X-specific introns. Only when a New World monkey was compared with hominoids were the rates slightly increased in the PAR1 introns. In terms of base composition, although the intergenic regions of the human PAR1 show a significant increase of G and C nucleotides, the base composition of the surveyed PAR1 introns is similar to that of the X-specific introns. Direct and indirect evidence indicates that the recombination rate is, indeed, much higher in PAR1 introns than in X-specific introns, and that the present PAB has persisted since the common ancestor of hominoids. Therefore, the mutagenic effect of recombination is far weaker than previously proposed, at least in hominoid PABs.
The role of recombination in nucleotide sequence evolution is a fundamental issue in molecular evolution. During the last decade, experimental and theoretical work has led to the proposal that recombination facilitates adaptive evolution, by enhancing the efficacy of natural selection on molecular variants (e.g., Marais and Charlesworth 2003). Recently, the role of recombination in neutral evolution has also drawn great interest (e.g., Perry and Ashworth 1999; Fullerton et al. 2001; Marais et al. 2001; Lercher and Hurst 2002; Filatov and Gerrard 2003; Hellmann et al. 2003; Montoya-Burgos et al. 2003).
The mammalian pseudoautosomal regions (PARs) provide an ideal system to study the role of recombination in molecular evolution. PARs are regions of homology between otherwise nonhomologous sex chromosomes and undergo meiotic recombination. Pairing and crossover in the mammalian PARs may ensure proper segregation of sex chromosomes during male meiosis (Gabriel-Robez et al. 1990; Mohandas et al. 1992). Because obligatory meiotic crossover between the mammalian X- and Y-chromosomes is confined to the short PARs, these regions undergo more intensive recombination than do adjacent X-chromosome/Y-chromosome-specific regions (Harbers et al. 1986; Soriano et al. 1987; Lien et al. 2000). Therefore, any mutational effect of recombination may manifest as evolutionary rate disparity between PARs and adjacent sex-chromosome-specific regions.
Dramatic effects of recombination on evolutionary rate were seen in the recent partitioning of the house mouse (Mus musculus domesticus) FXY locus into X-specific and pseudoautosomal regions. In the wild mouse (Mus spretus), FXY is strictly X-linked, but spans the PAB in M. m. domesticus (Perry and Ashworth 1999). In M. m. domesticus, the first three exons of FXY are X-specific, whereas the others lie within the PAR. Perry and Ashworth (1999) reported that the nucleotide substitution rate of the PAR-borne portion of house mouse FXY had increased 170-fold since the divergence of M. m. domesticus from M. spretus, between 1 to 3 million years ago (Mya; Ferris et al. 1983). Furthermore, Montoya-Burgos et al. (2003) demonstrated that the PAR-borne portion of FXY has undergone substantial molecular evolutionary changes and exhibits several characteristics typical of GC rich isochores. In particular, the PAR-borne FXY shows a highly elevated GC content, rapid nucleotide substitution, and shorter introns than the X-specific regions.
Human sex chromosomes have two PARs. The ∼2.6-Mb long PAR1 comprises the tip of the X/Y short arm (Xp22.3 and Yp11.32), and contains at least 12 genes. Meiotic recombination in the human PAR1 has been well studied (see Discussion), and is reportedly ∼20-fold more frequent than the genome average during male meiosis. Human PAR1 is homologous to the lone PAR of other great apes and Old World monkeys (Ellis et al. 1990; Graves et al. 1998); the ∼330-kb-long PAR2 at the tip of the X/Y long arm is human-specific (and contains four genes; Charchar et al. 2003). Recombination in PAR2 is reportedly much rarer than in PAR1 (Li and Hamer 1995; Lien et al. 2000).
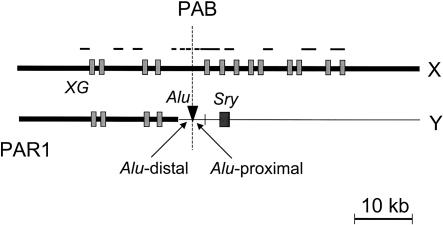
In humans and great apes, the PAB (PAB1 in human) divides the XG blood group gene, also called PBDX (pseudoautosomal boundary divided on the X-chromosome; Ellis et al. 1994). The first four exons of this gene lie within PAR1, whereas the remaining nine exons are X-specific. Salient homology between Xp and Yp sequences vanishes centromeric of PAB1 (Ellis et al. 1994; see Fig. 1; also, see Discussion). As noted above, the recombination rate in PAB1 is ≥20 times the genome average in males. Therefore, comparison of evolutionary rates in PAR1-borne versus X-specific portions of XG should reveal any local effect of recombination on neutral substitution rate.
Figure 1.
Schematic diagram of the XG regions. Filled boxes indicate the exons of the XG gene as identified in Ellis et al. (1994). The regions of the XG gene sequenced in this study are shown as lines above the diagram. Thick bars represent the sequences on or homologous to the X-chromosome. On the Y-chromosome, the Alu insertion site that separates the pseudoautosomal boundary is indicated. Alu-distal and Alu-proximal regions are not drawn to scale; their actual sizes are 234 bp and 221 bp, respectively.
We estimated the effect of recombination on neutral substitution in PAR1 and adjacent regions of the human sex chromosomes by several methods. First, we sequenced 12 intronic segments of XG from diverse catarrhines, and compared patterns of nucleotide substitution within PAR1-linked and X-linked regions. We found recombination to have little or no effect on nucleotide substitution rate in great ape XG introns. Second, we took a genomic approach to assess the effects of recombination on base composition across the human X-chromosome; in contrast to the results of the small-scale analysis of XG, recombination appeared to influence sequence evolution at a chromosomal scale. Finally, we inferred evolutionary distances between New World monkey and ape XG sequences and found a weak mutagenic effect of recombination. Within the anthropoid clade, therefore, the mutagenic effect of recombination near the PAB may be less pervasive than has been posited.
RESULTS
Evolutionary Rates in XG Introns in Great Apes
Overall averages of XG intron Tamura-Nei distances from human to bonobo (Pan paniscus), western lowland gorilla (Gorilla gorilla gorilla), and Bornean orangutan (Pongo pygmaeus pygmaeus) were 0.0157 (±0.007), 0.0189 (±0.007), and 0.0350 (±0.010), respectively. These differences slightly exceed published estimates from autosomal intergenic regions in the same taxa (Chen and Li 2001). When repetitive sequences and similar motifs were excluded via the RepeatMasker program, average Tamura-Nei divergences between human and bonobo, gorilla, and orangutan were 0.0129, 0.0159, and 0.0315, respectively, which are in good agreements with earlier estimates (Chen and Li 2001). Excluding repeats did not affect any of the conclusions reported below (data not shown).
We sought to discern any evolutionary rate difference between the great ape PAR1-linked and X-specific introns of XG. Tamura-Nei distances for taxon pairs, estimated from concatenated sequences from these two regions, are shown in Table 1. No tendency toward increased evolutionary rates in PAR1-linked sequences is evident. Unlike results from mouse FXY (Perry and Ashworth 1999; Montoya-Burgos et al. 2003), our findings do not support the hypothesis that recombination is globally mutagenic (i.e., Lercher and Hurst 2002; Hellmann et al. 2003). Filatov and Gerrard (2003) also reported no substantial difference in evolutionary rates between PAR-borne and X-specific sequences within short segments of human and orangutan XG.
Table 1.
The Average Tamura-Nei Evolutionary Distances Between Species and the GC Contents for the PAR1 (PAR)- and X-Linked XG Sequences
| Species pair | PAR | X-specific | ||
|---|---|---|---|---|
| Number of base pairs | 4216 | 7546 | ||
| T—N distance | Human—bonobo | 0.014 (0.010) | 0.015 (0.011) | |
| Human—gorilla | 0.019 (0.013) | 0.018 (0.012) | ||
| Human—orangutan | 0.032 (0.023) | 0.041 (0.028) | ||
| Bonobo—gorilla | 0.016 (0.011) | 0.016 (0.011) | ||
| Bonobo—orangutan | 0.031 (0.023) | 0.039 (0.027) | ||
| Gorilla—orangutan | 0.034 (0.025) | 0.036 (0.026) | ||
| % GC | Human | 45.6 (43.9) | 48.4 (46.5) | |
| Bonobo | 45.3 (43.9) | 48.0 (46.5) | ||
| Gorillas | 45.1 (43.5) | 48.5 (46.3) | ||
| Orangutan | 44.2 (43.5) | 47.5 (45.9) | ||
Exons are excluded from the analyses. Within parentheses are the evolutionary distances estimated after excluding the CpG dinucleotides.
In fact, between human and bonobo or orangutan, X-specific XG introns appear to have evolved faster than their pseudoautosomal counterparts, although between human and gorilla, the substitution rate appears slightly slower in X-specific sequences (Table 1). These results show no statistically significant trend that distinguishes great ape PAR1 from X-specific regions.
In addition, we estimated genetic distances after excluding all sites of a CpG dinucleotide in any of the sequences examined. Evolutionary distances estimated from such non-CpG sequences are shown in parentheses in Table 1. As expected, sitewise evolutionary distances for non-CpG sequences are significantly lower than for CpG-containing sequences. Overall, we found a 32% reduction in estimated evolutionary distance after removal of CpG sites, which occupy on average only ∼4% of all sites in our data. The proportion of CpG sites in XG introns closely approximates that in other introns of several primates, as reported in Subramanian and Kumar (2003).
Montoya-Burgos et al. (2003) showed that the GC content of sequences in mouse FXY increased as a direct consequence of increased recombination. However, our great ape XG data showed no such pattern. In fact, the GCcontent of the sampled XG regions was consistently lower in PAR-linked sequences than in X-linked sequences (Table 1).
Correlation Between GC Content and Evolutionary Rate
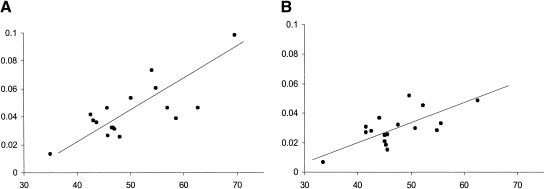
We assessed the relation between GCcontent and evolutionary distance between human and orangutan XG sequences. We chose this species pair because they are the most divergent among the orthologous sequences compared and therefore may yield the most statistical power for this type of analysis. In addition to the 12 aforementioned segments newly sequenced in this work, we added five segments from PAR1 sequenced by Filatov and Gerrard (2003). They explored the pattern of molecular evolution of nine DNA segments between the human and orangutan. From these we excluded three segments from the XG region that overlap with our data, and one from the SYBL1 locus, which occupies PAR2. We found a strong correlation between GC content and the Tamura-Nei evolutionary distance (p < 0.01; Fig. 2A); this correlation remained significant when we excluded all CpG sites in either lineage (Fig. 2B).
Figure 2.
Relationship between the rate of nucleotide substitution (estimated by the Tamura-Nei method, Y-axis) and the GC content (%, X-axis) of intron sequences. (A) The entire sequence of each segment was used. Pearson's correlation coefficient is 0.76 (p < 0.01). (B) CpG dinucleotides were excluded. Pearson's correlation coefficient is 0.67 (p < 0.01).
GC Content of the Human Pseudoautosomal Regions
From the entire human X-chromosome, we extracted 45,153,479 bp of intergenic DNA (see Methods), of which 467,756 bp, 166,183 bp, and 44,519,539 bp are in PAR1, PAR2, and the X-specific region, respectively. The GCcontents of the three regions are 46.5%, 38.9%, and 38.9%, respectively. Thus, intergenic sequences from PAR1 have a much higher GCcontent than do those from PAR2 or the rest of the X-chromosome (see Table 2; the data when repetitive sequences are excluded are also shown).
Table 2.
The GC Content (%) Estimated From the Intergenic Regions Extracted From the Human X-Chromosome
|
Entire sequence
|
CpG, TpG, GpA dinucleotides excluded
|
|||
|---|---|---|---|---|
| Repeats included | Repeats excluded | Repeats included | Repeats excluded | |
| Entire X | 38.94 | 37.68 | 33.04 | 31.68 |
| PAR1 | 46.51 | 46.64 | 41.86 | 42.23 |
| PAR2 | 38.91 | 37.83 | 33.13 | 32.00 |
| Non-PAR | 38.86 | 37.57 | 32.95 | 31.55 |
Both known and annotated genes (from GenScan) are excluded, and the proportions of G and C nucleotides are counted.
To assess the significance of the observed high GCcontent in PAR1, we first conducted a Monte Carlo analysis. Briefly, we randomly sampled PAR1-sized DNA segments from the X-specific sequences. We computed the GCcontent of each sampled segment and scored whether it was greater or less than the GCcontent of PAR1. From several replicates of 1000 randomly chosen segments, only 30 cases per average replicate showed a higher GC content than that of PAR1. That is, the probability of obtaining a DNA segment with a GCcontent at least as high as that of PAR1, given the sequence context of the X-specific region, was ∼0.03.
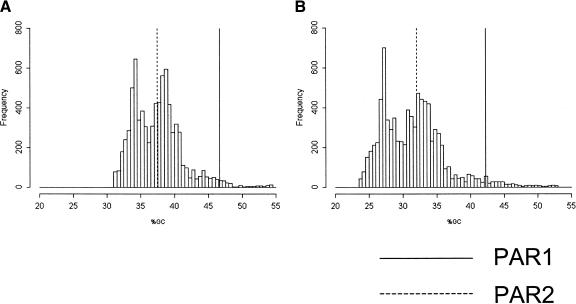
Next, we derived a sliding-window frequency distribution of X-specific GCcontent. A roughly PAR1-sized window was advanced across the X-specific published human genome sequence by 1% of the window size per step. Figure 3A summarizes this analysis. The GCcontent of PAR1 lies within the upper 0.7% of this distribution.
Figure 3.
Frequency distribution of the %GC of sequence windows of the same length as PAR1 in the human X-chromosome. (A) Distribution of %GC of intergenic nonrepetitive sequences. Each window is 190 kb, the number of intergenic nonrepetitive nucleotides from PAR1. We slid each window by 1% of the window size. The %GC of PAR1 is >99.33% of the total distribution, whereas that of PAR2 is only >55.64%. (B) Distribution of %GC of intergenic nonrepetitive sequences after excluding CpG, TpG, and GpA dinucleotides. Each window is 125 kb, the number of intergenic nonrepetitive nucleotides after excluding those three types of dinucleotides from PAR1. The %GC of human PAR1 lies above 99.05% of the distribution, and that of PAR2 is only above 57.09%.
From these analyses, we conclude that the GCcontent of PAR1 significantly exceeds the expected value for a random segment from the X-specific region. This conclusion holds whether we consider the intergenic or the intronic regions, and regardless of whether repetitive sequences are included (Table 2). In contrast, the GCcontent of PAR2 lies near the middle of the X-derived distribution in all such analyses.
We also checked for any significant difference in the GC content between PAR1 and the adjacent X-specific segment of the same size. Whereas the GCcontent in human PAR1 is 46.5%, that of the adjacent X-specific segment is 41.3%. These values differ significantly (p < 0.01). In contrast, the GCcontent of PAR2 is statistically equivalent to that of the adjacent same-sized X-linked region.
To estimate the effect of CpG methylation on the heterogeneity of the base composition of the X-chromosome, we performed the same analyses after excluding all CpG, TpG, and CpA dinucleotides. Directional hypermutability of CpG dinucleotides to TpG and CpA is well-known (Bird 1980; Hendrich et al. 1999). By excluding such dinucleotides, we may remove the effect of CpG methylation during the recent evolution of the human X-chromosome. We excluded ∼30% of our DNA sequence in this procedure. Figure 3B (see also Table 2) shows the resulting frequency distribution of GCproportion. The GCcontent of human PAR1 still lies within the upper 1% of the distribution. Therefore, the GC richness of PAR1 is not entirely due to the accumulation of CpG or CpG-derived dinucleotides.
Evolutionary Rates From a New World Monkey Outgroup
We obtained XG sequences from a New World monkey, (the spider monkey, Ateles geoffroyi), for 1200 bp from PAR1, as well as two X-specific DNA segments totaling 2400 bp. After excluding exon sequences, we estimated taxon-pairwise Tamura-Nei distances (Table 3). The genetic distances among hominoids showed the same pattern as in the overall data set: Evolutionary rates were slightly higher in X-specific regions. But when the distance from the New World monkey sequence was estimated, the hominoid PAR1 sequence appeared to have evolved on average ∼19% faster than the X-specific sequence. This disparity remained after we excluded dinucleotides in CpG contexts as above.
Table 3.
The Average Tamura–Nei Evolutionary Distances Between Species and the GC Contents for the Subset of the Data Set for the PAR- and X-Linked XG Sequences Where the Spider Monkey Sequence Is Available
| PAR | X specific | |
|---|---|---|
| Number of base pairs | 1050 (1016) | 2314 (2150) |
| T—N distance | ||
| Human—bonobo | 0.012 (0.005) | 0.012 (0.008) |
| Human—gorillas | 0.013 (0.005) | 0.018 (0.009) |
| Human—orangutan | 0.027 (0.016) | 0.041 (0.026) |
| Human—spider monkey | 0.138 (0.112) | 0.116 (0.088) |
| Bonobo—gorilla | 0.013 (0.006) | 0.015 (0.008) |
| Bonobo—orangutan | 0.029 (0.017) | 0.038 (0.025) |
| Bonobo—spider monkey | 0.138 (0.114) | 0.116 (0.089) |
| Gorilla—orangutan | 0.026 (0.013) | 0.034 (0.022) |
| Gorilla—spider monkey | 0.132 (0.109) | 0.115 (0.087) |
| Orangutan—spider monkey | 0.135 (0.108) | 0.113 (0.088) |
| % GC | ||
| Human | 47.3 (45.1) | 48.7 (46.3) |
| Bonobo | 47.9 (45.5) | 48.3 (46.2) |
| Gorillas | 47.8 (45.3) | 48.1 (46.0) |
| Orangutan | 47.2 (45.4) | 47.6 (45.7) |
| Spider monkey | 49.7 (47.1) | 48.3 (46.2) |
Exons are excluded from the analyses.
DISCUSSION
Is Recombination Mutagenic in Hominoid PAB?
The most striking finding of this study, unexpected under the widely accepted hypothesis that recombination is mutagenic (Perry and Ashworth 1999; Fullerton et al. 2001; Lercher and Hurst 2002; Montoya-Burgos et al. 2003), is the lack of acceleration of sequence evolution in PAR1-linked segments of the human XG gene. When evolutionary rates of human and great ape sequences were compared, PAR1-linked and X-specific regions showed no clear-cut rate difference, contrary to what has been observed surrounding the house mouse PAB. Nor did the PAR1 and X-specific regions differ significantly in GCcontent, unlike the case of the PAR-linked portions of the FXY locus in the mouse (Montoya-Burgos et al. 2003).
What might explain these discrepancies? Under the view that recombination is mutagenic and that XG intronic sequence is evolving neutrally, there are at least two alternative hypotheses. First, recombination rates in human PAR1 versus X-specific regions may not differ significantly. We show below that this is unlikely, in view of existing literature and our unpublished data. Second, human PAB1 may be too young to show a significant effect of differential recombination. This too is highly unlikely; we show below that the catarrhine PAB appears to have a long history.
Recombination Rate in Human PAR1
Lien et al. (2000) estimated subregional male meiotic recombination rates in human PAR1 by genotyping more than 1900 single sperm for 25 PAR1 markers. They inferred overall recombination frequencies in PAR1 to be 13- to 38-fold higher than the genome average. Similar estimates were reported in studies using long-range restriction mapping (Brown 1988; Petit et al. 1988). More importantly, according to Lien et al. (2000), the PAB1 region is one of the most recombinant regions in PAR1. They suggested that the male meiotic recombination frequency in this region is 26- to 38-fold higher than the genome average.
Other data on recombination rate come from within-population haplotype distributions. We are in the process of collecting nucleotide polymorphism data from >100 chromosomes from the regions surveyed in this paper (N.M. Pearson, P.R. Obara, S.Yi, B. Nikbin, P.A. Underhill, L.L. Cavelli-Sforza, M. Kreitman, and B.T. Lahn, unpubl.). The SNPs from the X-specific regions exhibit linkage disequilibria over significantly longer physical distances than those on PAR1, indicating that the PAR1 region has a significantly higher level of recombination than the X-specific region. Therefore, the first hypothesis above is not supported.
The Age of the Present-Day PAB in Hominoids
We now consider the hypothesis that the effect of recombination on XG neutral substitution rates is not yet conspicuous because human PAB1 has arisen only recently. The structure of the PAB in nonhuman anthropoids (homologous to human PAB1) has been well studied (see Fig. 1; Ellis et al. 1989, 1990). On the Y-chromosome, the present-day PAB is conventionally marked by an Alu element, flanked centromerically by ∼220 bp of X-Y-homologous sequence (Ellis et al. 1990). Farther toward the centromere, the sex-chromosome-specific sequence, which shows no clear X-Y homology, begins. In particular, the sex-determining locus, SRY, follows on the Y-chromosome.
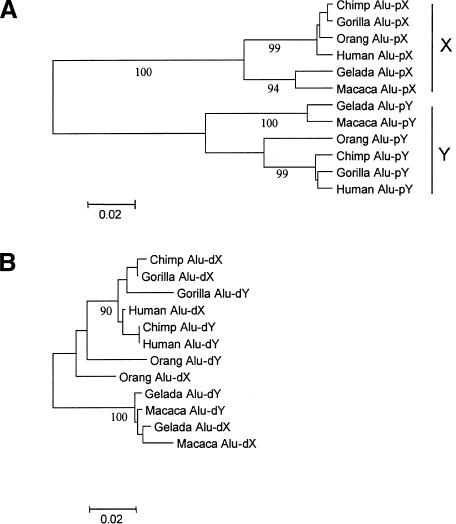
Although the age of the present-day PAB is unknown, available sequence data from the PAB of the X- and the Y-chromosomes of diverse catarrhines (Ellis et al. 1990) provide much insight when we consider the phylogenies of the Alu-proximal and the Alu-distal regions separately. The Alu-proximal region shows, throughout catarrhines, ∼10% X-Y sequence divergence, roughly the same as that observed in genes comprising “stratum 4” of the human sex chromosomes (Lahn and Page 1999; Iwase et al. 2003). The phylogenetic tree of catarrhine Alu-proximal regions shows distinct clades of X-versus Y-derived sequences, well supported by high bootstrap values (Fig. 4A), even though only 234 bp were aligned. In sharp contrast, the phylogenetic tree of the Alu-distal region, although comparable in length to that of the Alu-proximal region, shows a mixture of the X- and Y-derived sequences (Fig. 4B), implying local recombination events between the two chromosomes.
Figure 4.
Neighbor-joining trees based on Kimura's two-parameter distances. (A) Alu-proximal regions, 234 bp. (B) Alu-distal regions, 221 bp. Bootstrap values >90% after 1000 replications are shown below the branches.
Moreover, the total branch length of the Alu-proximal regions greatly exceeds that of the Alu-distal region (Fig. 4). In the former, the branch leading to the ancestor of the X- and Y-chromosomes is notably extended, and substitution rates are clearly higher among Y-derived than X-derived branches. These observations are in excellent accord with the hypothesis of male-driven evolution (Makova and Li 2002). Clearly, Figure 4 shows that the Y-sequences have been evolving much faster, implying that the Alu-proximal sequence has had male-limited transmission for a substantially long period of time.
Thus, a boundary between Alu-proximal and the Alu-distal regions appears to have arisen before the divergence of hominoids from Old World monkeys. For this reason, the lack of rate difference between the PAR-linked and X-specific introns of the XG gene cannot be explained by the hypothesis of late emergence of the PAB1 in the human and ape lineages.
As noted, the rate of recombination in PAB1 exceeds that of the X-linked region in our data set, and the emergence of PAB1 appears to predate the last common ancestor of hominoids. The lack of a noticeable mutagenic effect of recombination on the sequence evolution of PAB1 is therefore puzzling. One possible explanation is that recombination and mutation do correlate in the regions surveyed, but are localized to a scale undetected in our survey. We note that May et al. (2002) posited highly patchy recombination in the SHOX gene region of human PAR1. However, it is rather unlikely that we have sampled more than 30% of XG intron sequences without hitting such a hotspot.
Substitution Rates in Mouse PAB and the New World Monkey
The glaring disparity between the house mouse PAB and the human PAB1 in the inferred mutagenic effect of recombination is quite difficult to explain, especially as house mouse FXY likely moved to a PAB less than 3 million years ago, according to molecular divergence. Even if we take into account the differences in generation times between primates and rodents, the contrast between the analogous human and the mouse PABs is hard to reconcile.
However, the mouse PAR is shorter than human PAR1 (720 kb vs. 2.6 Mb; Perry et al. 2001). Therefore, the effect of condensed obligatory male meiotic recombination may be stronger, and perhaps more homogeneous, in the mouse PAR than in human PAR1. It is also possible that recombination-induced mutagenesis may depend on other biological factors such as a certain chromatin structure, which may differ between human and mouse PABs.
In addition, in the mouse PAR, the GCcontent at the third codon positions of the FXY was on average 30.1% higher than in the X-specific orthologs (Montoya-Burgos et al. 2003), whereas in XG, no such difference was found. Unlike the case of mouse FXY, we do not know the GClandscape of the ancestral XG locus, which presumably resided in a relatively uniform recombination environment. Therefore, we cannot quantify the rate of GC change of XG compared with its ancestral situation. However, although we do not know the GCcontent beyond orangutan, we can see clearly the GCcontent in the common ancestor of orangutan, chimpanzee, bonobos, and humans, which has little changed. Thus, there is little evidence that the GCcontent of the XG has substantially changed in hominoids. As exon sequences tend to accumulate more CpG dinucleotides than do noncoding sequences (Subramanian and Kumar 2003), assessing the evolutionary rates from intron sequences of FXY may offer an interesting and informative contrast in this respect.
We are further intrigued by the increased evolutionary rate in PAR1 among the larger anthropoid clade, at least for the smaller portion of XG sequenced in New World monkeys. This may indicate that in some primates recombination is indeed mutagenic in PAB1; however, such an increased rate cannot be ascribed with certainty to recombination rate, because XG may have come to straddle the pseudoautosomal boundary after the divergence of catarrhines from the New World monkeys. As noted above, the GCcontent of human PAR1 significantly exceeds that of the rest of the X-chromosome, consistent with the mutagenic effect of recombination. Moreover, Filatov and Gerrard (2003) reported faster evolutionary rates in PAR1-linked sequences than in X-specific regions, as expected if recombination is mutagenic; however, we note, again, that in their analyses the partial XG region stood out as an outlier as having no difference in evolutionary rates between PAR-borne and the X-specific segments. In view of these observations, it is a great puzzle why human PAB1 shows no obvious mutagenic effect of recombination on evolutionary rate.
Another issue that might complicate the analyses of evolutionary rates in the XG has to do with the fact that the XG gene was duplicated in some cattarhines (Weller et al. 1995; Gläser et al. 1997). Briefly, Weller et al. (1995) described several male-specific transcripts of XG-related sequences. They concluded that those sequences were the results of alternative splicing of a gene mapped to Yq11.21. All of the transcripts encoded premature stop codons; hence, the duplicated copy is named XGPY (XG pseudogene on the Y-chromosome). The XGPY resides within a larger segmental duplication, which encompasses at least the entire XG region (S. Yi, unpubl.). This could have originated from the X-chromosome copy or from the ancestral copy before the differentiation between the X-chromosome and Y-chromosome XG loci. If gene conversion occurs between the XG and XGPY similar to what has been reported to occur between paired palindromes on the human Y-chromosome (Rozen et al. 2003), then only the PAR-borne XG portion is affected, because the second half of the XG locus is truncated on the Y-chromosome (Fig. 1). However, if such gene conversion between the PAR-borne XG and the Y-specific copy does occur, it is most likely to increase divergence between species in this region, which is the opposite of what we observed. Also, whether such gene conversion events occur in non-palindromic sequences such as XG and XGPY remains an open question at this point.
Recombination and GC Content in Human PAR1
It is informative to note the differences in GCcontents of human PAR1 and PAR2. Although PAR1 has a significantly higher GC content than the rest of the X-chromosome, PAR2 is on the whole indistinguishable from the rest of the X-chromosome in this respect (Fig. 3; Table 3). Recombination frequencies in PAR2 have been estimated to be about an order of magnitude lower than that of PAR1 (Li and Hamer 1995; Lien et al. 2000). Also, PAR2 is not implicated in mediating meiosis (Charchar et al. 2003). Furthermore, PAR2 has a short history, as it is human-specific. Therefore, the elevated GCcontent of PAR1 appears correlated with substantially intensified recombination in PAR1, whereas the GCcontent of PAR2 is not affected.
In addition, we found significant correlation between the GCcontent and the evolutionary rate for the human and orangutan XG comparison (Fig. 2). GCcontent explained ∼58% of the relationship observed (that figure decreased to 45% when we excluded all CpG sites from our data set). In fact, for historically CpG sites only (sequences that code CpG dinucleotides in any species), Tamura-Nei distances between human and bonobo or between human and orangutan were greater in PAR1 XG introns than in X-specific XG introns. In other words, at least over short divergence times, evolutionary rates in highly recombining regions are inflated, partially reflecting fast-evolving CpG contexts.
METHODS
PCR, Cloning, and Sequencing
Genomic DNA samples of male and female bonobo (Pan paniscus), western lowland gorilla (Gorilla gorilla gorilla), and Bornean orangutan (Pongo pygmaeus pygmaeus) were used separately as templates in polymerase chain reaction (PCR). We used the Expand High Fidelity PCR system (Roche) to minimize errors in amplification. We designed PCR primer pairs to amplify ∼1 kb of DNA sequences from each of 13 roughly evenly spaced regions spanning the ∼64-kb XG region. Several of the original primer pairs did not amplify in some taxa (data not shown). Additionally, we obtained larger sequence segments spanning the proposed PAB1. As a result, we amplified and sequenced a total of ∼14 kb throughout the XG region, including >5 kb flanking the PAB1, from male and female hominoids. The most disparate nucleotides sequenced are 57,930 bp apart (Fig. 1). Additionally, we sampled 4 kb from the spider monkey (Ateles geoffroyi), a New World monkey.
The primers used in this work are shown in the Supplemental material, available online at www.genome.org. The regions covered in our collecting scheme are depicted in Figure 1. Some segments were cloned using the TA cloning kit (Invitrogen). We used the BigDye v3 sequencing kit (ABI), run on an ABI 377 for both strands.
Sequence Analyses
Sequence data were curated using the Sequencher 4.0 program (GeneCode) to make and visually confirm or correct base calls. Sequences were then exported and aligned using CLUSTAL W (Thompson et al. 1994) with default parameters. All exons were excluded from the alignments before further analyses. Repeat elements in these sequences were identified using the Repeat-Masker program (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker) and removed for some analyses. To infer the CpG context of sequences, we first identified CpG dinucleotides in the multiple alignments; if any sequence within the alignment showed a CpG dinucleotide, all orthologous dinucleotides were designated as CpG sites for removal from further analysis. We used the Tamura-Nei method (Tamura and Nei 1993) to correct for multiple hits. This method takes into account GCbias. We concatenated sequences from the PAR1 versus X-specific regions to estimate average genetic distances.
X-Chromosome Data Analyses
We downloaded the entire X-chromosome from the UCSC Genome Browser (http://genome.ucsc.edu) using the April 2003 release of the human genome. In this release, repetitive sequences are identified as lower-case letters, using the RepeatMasker program (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker). We used the available data tables to set up a relational database of the coordinates and sequences of all known and predicted regulatory regions, exons, and introns. For most of our analysis, we excluded all annotated/predicted gene regions, including regulatory regions, exons, and introns from the KnownGenes and GeneScan annotations.
Phylogenetic Analyses of the PAB in Higher Primates
The sequences for the PAB boundary from several higher primate species were downloaded from GenBank (http://ncbi.nih.gov/Genbank, accession numbers M54450–M54462). They were aligned using CLUSTAL W (Thompson et al. 1994). We estimated sequence divergences using Kimura's two-parameter method (Kimura 1980). The neighbor-joining (NJ) method implemented in MEGA2 (http://www.megasoftware.net; Kumar et al. 2000) was used to draw phylogenetic trees.
Acknowledgments
Genomic DNA samples of bonobos, gorillas, and orangutans were purchased from the San Diego Zoological Society. We thank Melissa McMahill for her rotation work in the Li lab, during which she generated some sequence data; Dmitry Filatov for sharing unpublished results; an anonymous reviewer for an interesting suggestion; and Peter Bouman for discussions. This study was supported by NIH grants GM65499 and GM30998.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC-section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1777204. Article published online before print in December 2003.
Footnotes
[Supplemental material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: D. Filatov.]
References
- Bird, A.P. 1980. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 8: 1499-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, W.R. 1988. A physical map of the human pseudoautosomal region. EMBO J. 7: 2377-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charchar, F.J., Svartman, M., El-Mogharbel, N., Ventura, M., Kirby, P., Matarazzo, M.R., Ciccodicola, A., Rocchi, M., D'Esposito, M., and Graves, J.A. 2003. Complex events in the evolution of the human PAR2. Genome Res. 13: 281-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F.-C. and Li, W.-H. 2001. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 68: 444-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, N.A., Goodfellow, P.J., Pym, B., Smith, M., Palmer, M., Frischauf, A.M., and Goodfellow, P.N. 1989. The pseudoautosomal boundary in man is defined by an Alu repeat sequence inserted on the Y chromosome. Nature 337: 81-84. [DOI] [PubMed] [Google Scholar]
- Ellis, N., Yen, P., Neiswanger, K., Shapiro, L.J., and Goodfellow, P.N. 1990. Evolution of the pseudoautosomal boundary in Old World monkeys and great apes. Cell 63: 977-986. [DOI] [PubMed] [Google Scholar]
- Ellis, N.A., Ye, T.Z., Patton, S., German, J., Goodfellow, P.N., and Weller, P. 1994. Cloning of PBDX, an MIC2-related gene that spans the pseudoautosomal boundary on chromosome Xp. Nat. Genet. 6: 394-400. [DOI] [PubMed] [Google Scholar]
- Filatov, D.A. and Gerrard, D.T. 2003. High mutation rates in humans and ape pseudoautosomal genes. Gene 317: 67-77. [DOI] [PubMed] [Google Scholar]
- Ferris, S.D., Sage, R.D., Prager, E.M., Ritte, U., and Wilson, A.C. 1983. Mitochondrial DNA evolution in mice. Genetics 105: 681-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton, S.M., Bernardo Carvalho, A., and Clark, A.G. 2001. Local rates of recombination are positively correlated with GCcontent in the human genome. Mol. Biol. Evol. 18: 1139-1142. [DOI] [PubMed] [Google Scholar]
- Gabriel-Robez, O., Rumpler, Y., Ratomponirina, C., Levilliers, J., Croquette, M.F., and Couturier, J. 1990. Deletion of the pseudoautosomal region and lack of sex-chromosome pairing at pachytene in two infertile men carrying an X;Y translocation. Cytogenet. Cell Genet. 54: 38-42. [DOI] [PubMed] [Google Scholar]
- Gläser, B., Grützner, F., Taylor, K., Schiebel, K., Meroni, G., Tsioupra, K., Pasantes, J., Rietschel, W., Toder, R., Willmann, U., et al. 1997. Comparative mapping of Xp22 genes in hominoids—Evolutionary linear instability of their Y homologues. Chromo. Res. 5: 167-176. [DOI] [PubMed] [Google Scholar]
- Graves, J.A.M., Wakefield, M.J., and Toder, R. 1998. The origin and evolution of the pseudoautosomal regions of human sex chromosomes. Hum. Mol. Genet. 7: 1991-1996. [DOI] [PubMed] [Google Scholar]
- Harbers, K., Soriano, P., Muller, U., and Jaenisch, R. 1986. High frequency of unequal recombination in pseudoautosomal region shown by proviral insertion in transgene mouse. Nature 324: 682-685. [DOI] [PubMed] [Google Scholar]
- Hellmann, I., Ebersberger, I., Ptak, S.E., Pääbo, S., and Przeworski, M. 2003. A neutral explanation for the correlation of diversity with recombination rates in humans. Am. J. Hum. Genet. 72: 1527-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich, B., Hardeland, Y., Ng, H.H., Jiricny, J., and Bird, A. 1999. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature 401: 301-304. [DOI] [PubMed] [Google Scholar]
- Iwase, M., Satta, Y., Hirai, Y., Hirai, H., Imai, H., and Takahata, N. 2003. The amlenogenin loci span an ancient pseudoautosomal boundary in diverse mammalian species. Proc. Natl. Acad. Sci. 100: 5258-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16: 111-120. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., Jakobsen, I., and Nei, M. 2000. MEGA: Molecular evolutionary genetics analysis. Pennsylvania State University, University Park, PA.
- Lahn, B.T. and Page, D.C. 1999. Four evolutionary strata on the human X chromosome. Science 286: 964-967. [DOI] [PubMed] [Google Scholar]
- Lercher, M.J. and Hurst, L.D. 2002. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 18: 337-340. [DOI] [PubMed] [Google Scholar]
- Li, L. and Hamer, D.H. 1995. Recombination and allelic association in the Xq/Yq homology region. Hum. Mol. Genet. 4: 2013-2016. [DOI] [PubMed] [Google Scholar]
- Lien, S., Szyda, J., Schechinger, B., Rappold, G., and Arnheim, N. 2000. Evidence for heterogeneity in recombination in the human pseudoautosomal region: High resolution analysis by sperm typing and radiation-hybrid mapping. Am. J. Hum. Genet. 66: 557-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makova, K.D. and Li, W.-H. 2002. Strong male-driven evolution of DNA sequences in humans and apes. Nature 416: 624-626. [DOI] [PubMed] [Google Scholar]
- Marais, G. and Charlesworth, B. 2003. Genome evolution: Recombination speeds up adaptive evolution. Curr. Biol. 13: R68-R70. [DOI] [PubMed] [Google Scholar]
- Marais, G., Mouchiroud, D., and Duret, L. 2001. Does recombination improve selection on codon usage? Lessons from nematode and fly complete genomes. Proc. Natl. Acad. Sci. 98: 5688-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, C.A., Shone, A.C., Kalaydjieva, L., Sajantila, A., and Jeffreys, A.J. 2002. Crossover clustering and rapid decay of linkage disequilibrium in the Xp/Yp pseudoautosomal gene SHOX. Nat. Genet. 31: 272-275. [DOI] [PubMed] [Google Scholar]
- Mohandas, T.K., Speed, R.M., Passage, M.B., Yen, P.H., Chadley, A.C., and Shapiro, L.J. 1992. Role of the pseudoautosomal region in sex-chromosome pairing during male meiosis: Meiosis studies in a man with a deletion of distal Xp. Am. J. Hum. Genet. 51: 526-533. [PMC free article] [PubMed] [Google Scholar]
- Montoya-Burgos, J.I., Boursot, P., and Galtier, N. 2003. Recombination explains isochores in mammalian genomes. Trends. Genet. 19: 128-130. [DOI] [PubMed] [Google Scholar]
- Perry, J. and Ashworth, A. 1999. Evolutionary rate of a gene affected by chromosomal position. Curr. Biol. 9: 987-989. [DOI] [PubMed] [Google Scholar]
- Perry, J., Palmer, S., Gabriel, A., and Ashworth, A. 2001. A short pseudoautosomal region in laboratory mice. Genome Res. 11: 1826-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit, C., Levilliers, J., and Weissenbach, J. 1988. Physical mapping of the human pseudo-autosomal region; comparison with genetic linkage map. EMBO J. 7: 2369-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S., Skaletsky, H., Marszalek, J.D., Minx, P.J., Cordum, H.S., Waterson, R.H., Wilson, R.K., and Page, D.C. 2003. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 423: 873-876. [DOI] [PubMed] [Google Scholar]
- Soriano, P., Keitgesm E.A., Schorderet, D.F., Harbers, K., Gartler, S.M., and Jaenisch, R. 1987. High rate of recombination and double crossovers in the mouse pseudoautosomal region during male meiosis. Proc. Natl. Acad. Sci. 84: 7218-7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, S. and Kumar, S. 2003. Neutral substitutions occur at a faster rate in exons than in noncoding DNA in primate genomes. Genome Res. 13: 838-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. and Nei, M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10: 512-526. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller, P.A., Critcherm, R., Goodfellow, P.N., German, G., and Ellis, N.A. 1995. The human Y chromosome homologue of XG: Transcription of a naturally truncated gene. Hum. Mol. Genet. 4: 859-868. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://ftp.genome.washington.edu/cgi-bin/RepeatMasker; RepeatMasker program.
- http://genome.ucsc.edu/; UCSC Genome Bioinformatics site.
- http://www.megasoftware.net/; MEGA (Molecular Evolutionary Genetic Analysis).
- http://www.ncbi.nih.gov/Genbank; GenBank.