Sequencing of multiple mammalian genomes, together with the development of whole-transcriptome profiling technologies, have opened the door to an unprecedented ability to study gene expression in the brain. Transcriptomics refers to a class of high-throughput methods, such as microarray (gene chip), serial analysis of gene expression, and, more recently, whole-transcriptome sequencing (RNA-Seq), which enable measurement of the abundance of tens of thousands of transcribed RNAs in a given sample. Before the development of these technologies, studies of gene expression and function in the brain were restricted to targeted assay of a relatively small number of genes for any given study. Now it is possible to obtain a more panoramic view of gene expression, and potentially to understand the molecular underpinnings of brain function from the viewpoint of gene networks rather than from a viewpoint dominated by the effects of single genes.
Complicating this enterprise is the fact that the brain is comprised of a famously diverse menagerie of cell types, limiting the utility of data obtained from tissue homogenates. For a given experimental condition, gene expression changes occurring in rare cell types may go undetected, as they contribute to only a small fraction of the total tissue RNA. Additionally, genes may be regulated in opposing directions in different cell types, thereby appearing static in composite data. Attempts at deconvolving tissue expression data into independent contributions from constituent cell types are computationally challenging and the results are uncertain. Hence, directly measuring the transcriptomes of specific cell types is crucial to understanding the intracellular gene networks that underlie cellular phenotypes.
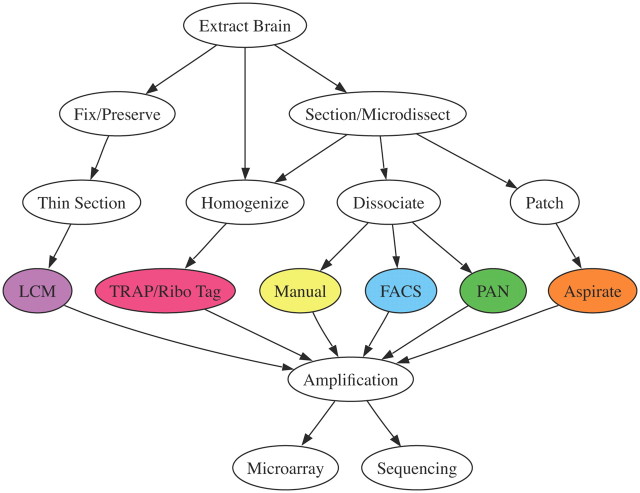
Cell type-specific transcriptomics requires completion of four separable tasks. First, the cell type of interest must be identified and (typically) labeled; second, RNA from the targeted cells must be extricated from that in surrounding cell types; third, because the resulting RNA is typically low in abundance, it must be amplified; and fourth, the isolated sequences must be identified through sequencing or hybridization (i.e., microarray) methods. Recent reviews have focused on the problems of cell identification (Miyoshi and Fishell, 2006; Kuhlman and Huang, 2008; Madisen et al., 2010), and we direct the readers' attention to a number of online resources that provide detailed information about available cell type-specific reporter and cre-driver mouse lines (Table 1). Briefly, cell types are commonly identified by electrophysiological properties using whole-cell patch-clamp recordings, by projection target using retrograde or anterograde tracers, by cell type-specific markers using immunostaining, or through transgenic labeling approaches. The choice of amplification strategies and final readout are interrelated and subject to a number of technical concerns that are outside the scope of this short review. Although the current majority of cell type-specific transcriptomic data comes from microarray studies, RNA-Seq is clearly the next frontier. In particular, RNA-Seq affords a more straightforward ability to study genetic variation, uncover previously uncharacterized transcriptional start and stop sites and splice variants, and discover novel noncoding RNAs (Guttman et al., 2010). However, before performing either microarray or RNA-Seq experiments, the considerable obstacle of tissue heterogeneity must be overcome. Thus, we focus here on methods for extracting cell type-specific RNAs from mammalian brain tissue (although similar approaches have also been applied to nonmammalian species). The typical workflow for each of the reviewed methods is depicted in Figure 1, though this representation is by no means exhaustive of all conceivable experimental designs, and in some cases multiple purification methods may be sequentially applied (Cahoy et al., 2008).
Table 1.
Online resources cataloguing available cell type-specific reporter and driver mouse lines
| Resource URL | Available lines |
|---|---|
| http://transgenicmouse.alleninstitute.org | BAC Cre lines and reporters. ISH/IHC atlas available for expression patterns. |
| http://www.bactrap.org | Resources for TRAP. Site in preparation. |
| http://www.credrivermice.org | Knock-in Cre, enhancer trap Cre, enhancer trap Tet and various XFP transgenic lines. Expression pattern viewable in atlas images. |
| http://www.gensat.org | BAC GFP transgenics and BAC Cre lines. Expression atlas available. |
| http://cre.jax.org/index.html | Cre/loxP lines repository. |
| http://jaxmice.jax.org/research/flp_frt.html | Flp/Frt lines repository. |
| http://www.mshri.on.ca/nagy | Nagy Lab Cre driver lines database. Expression pattern annotated by anatomical structure/cell types. |
BAC, Bacterial artificial chromosome; ISH, in situ hybridization; IHC, immunohistochemistry.
Figure 1.
Typical workflow of a cell type-specific transcriptional profiling experiment. Each node represents a key step and the arrows indicate the sequence of steps. The reviewed purification methods are indicated by colored nodes. Notice that there are multiple possible paths for some methods.
Often the preference (if not necessity) for using a given cell type-specific mRNA isolation technique is closely wedded to the means of cellular identification being used. For example, a neuron's electrophysiological properties, characterized in whole-cell patch-clamp recordings from acute slice preparations, are frequently used to classify neural cell types. Historically, there has been considerable interest in reducing a neuron's electrophysiological properties to the specific complement of ion channels it expresses, and in finding genetic signatures that correlate with firing phenotype in general. This led to the development of postrecording, single-cell profiling techniques, in which the cytosol of recorded cells is aspirated through the patch pipette, collected in a buffer, and the mRNA is subsequently isolated (patch/aspirate) (Surmeier et al., 1996; Martina et al., 1998). Given the limited amount of RNA contained in a single cell (5–10 pg), early attempts were only able to profile a small number of candidate genes using reverse-transcription followed by PCR. Over the years, improvements in mRNA amplification and PCR techniques have expanded the scope of the patch/aspirate technique to profiling tens to hundreds of genes using multiplex PCR (Toledo-Rodriguez et al., 2004) and tens of thousands of genes using microarrays and sequencing (Kurimoto and Saitou, 2010; Ozsolak et al., 2010a,b; Subkhankulova et al., 2010). However, due to the small amounts of collected RNA, single-cell methods are generally more prone to producing false negatives (described in more detail below), particularly for low-abundance transcripts, and the profiling results are often less reproducible than transcriptional profiles obtained using pools of cells. The extent to which reduced reproducibility also reflects biological and not just technical variability remains unclear (Raj and van Oudenaarden, 2009; Janes et al., 2010). Given these difficulties, much of our current understanding of the genetic diversity of neural cell types has come from studies predominately using mRNA collected from pooled cells (described below). Moreover, advances in transgenic engineering have made it possible to consistently target electrophysiologically defined cell types (Feng et al., 2000; Oliva et al., 2000; Chattopadhyaya et al., 2004), which offers an alternative strategy for discovering genes underlying electrophysiological phenotypes (Okaty et al., 2009).
Laser-capture microdissection (LCM) and closely related microdissection techniques such as laser microbeam microdissection or laser-directed microdissection (LDM) (Rossner et al., 2006) use a laser to excise cells of interest (identified under a microscope) from mounted thin-tissue sections that have been either fixed or frozen. The methods differ in the type of laser being used and the specific manner in which targeted cells are extracted. In LCM, a low-power infrared laser beam melts selected small (∼7.5 μm) regions of a plastic membrane on the surface of a tissue section that then adhere to the target cell(s) upon cooling. The sheet is then peeled away from the surface of the tissue section, taking with it the adherent cells. An obvious drawback of this method is that it does not allow the user to tailor the laser beam to the particular morphology of any given cell, and thus some cellular domains may be excluded. Additionally, closely apposed off-target cells, such as glia, may become attached to the plastic film, resulting in contamination.
Other laser-based microdissection systems, such as the AS LDM system (Leica) overcome the first limitation by using a much narrower ultraviolet laser (∼0.5 μm), allowing the user to directly trace and cut the outline of the target cell; however, closely apposed glia are often difficult to detect visually, particularly if they are beneath the target cell. Despite these drawbacks, LCM and LDM have greatly facilitated cell type-specific assays of mRNA from human postmortem tissue, as they are ideally suited for obtaining cells from fixed or frozen tissue preparations.
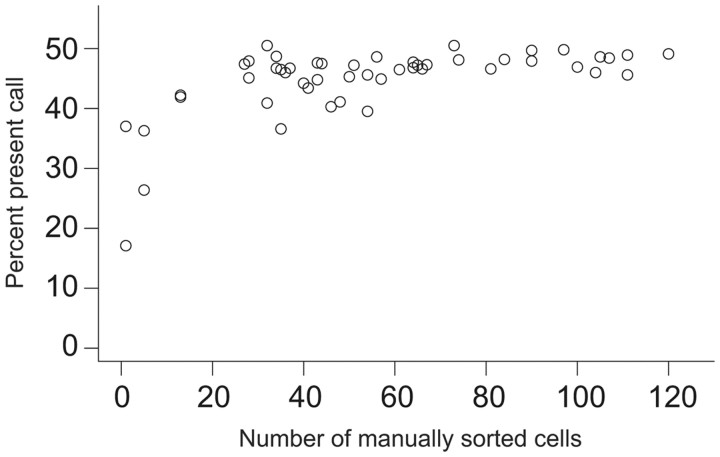
Given that tissue fixation can degrade nucleic acids, and the heightened risk of contamination when extracting cells from intact tissue, cellular dissociation-based methods may be better suited for studies in which live tissue is available. As a first step to performing fluorescence-activated cell sorting (FACS), immunopanning (PAN), and manual cell sorting (Manual), acutely dissected brain tissue is digested in a protease solution containing artificial CSF (ACSF) and in some cases ion channel and receptor blockers that promote the health of dissociated cells (Hempel et al., 2007). In the Manual technique, dissociated cells are deposited in a small Petri dish and viewed under a fluorescence microscope. Fluorescent cells are aspirated using a small glass pipette, washed in ACSF in a series of clean dishes to ensure purity, and finally collected in a cell lysis buffer. RNA collected from even a single manually sorted cell can be amplified using a two-step in vitro transcription amplification method and yield sufficient quantities of labeled RNA for hybridizing to microarrays (≥10 μg). However, comparison between the microarray present call, or the percentage of probes on the Affymetrix chip that register a signal level above an algorithmically determined threshold for being expressed in a given experiment, and the number of manually sorted cells used as input demonstrates that a minimum of 30 cells is generally required to achieve the highest reproducible sensitivity, indicating that this level of input minimizes false negatives (Fig. 2; note the saturating present call between 30 and 60 cells, though the precise number of cells and the resulting amount of collected RNA and present call depend on the particular cell type). Furthermore, detecting a single nonfluorescent cell out of a population of 30–100 dissociated cells under a microscope is relatively straight forward, ensuring that false positives resulting from contaminating cell types are kept to a minimum. Thus, Manual sorting is particularly well suited to profiling rare and low-abundance cell types, as well as more plentiful cell populations. However, a high level of proficiency is generally required on the part of the sorting technician.
Figure 2.
Percentage microarray present call as a function of the number of manually sorted cells used as input.
FACS requires a flow cytometry platform and can typically sort thousands of cells in a relatively short amount of time, provided the proportion of the labeled cell is reasonably high (∼1 h with ∼0.1% labeled cells) (Lobo et al., 2006; Cahoy et al., 2008; Marsh et al., 2008). Streams of dissociated cells are ushered into a fluidic channel where a fluorescence detector sorts cells based on user-specified criteria. This is often achieved by forming a droplet that only contains a single cell just after it has passed through the beam of the fluorometer. A charge is then applied to the droplet, depending on whether or not the target spectrum was detected, and the cell-containing droplet is then diverted into the appropriate receptacle. Multiple excitation lasers and fluorescence detectors can be incorporated into a single flow cytometry platform, allowing for simultaneous detection of multiple spectra, and therefore multiple labeled cell markers. This is especially useful given that many cell types can only be identified by combinatorial expression of multiple genes, rather than expression of a single, unique marker gene. Additionally, properties of the light scatter may be used to infer cell size, morphology, and intracellular complexity (granularity); thus, FACS allows for multiparametric analysis across an array of cellular properties. A drawback of FACS sorting is that it is often too stressful for mature neurons (Arlotta et al., 2005; Lobo et al., 2006; Heiman et al., 2008) and special care must be taken to ensure that neurons remain healthy (Lobo et al., 2006). Given that the Manual method has been successfully implemented for acquiring microarray data from numerous cell types from mature mice (Sugino et al., 2006), the high-throughput nature of FACS may be more of an advantage in applications that require a greater number of starting cells than required for microarrays, such as assays of genomic DNA, like chromatin immunoprecipitation.
Unlike FACS and Manual, PAN does not rely on a fluorescent signal to detect specific cell populations, and therefore may be used to purify unlabeled cell types (Barres et al., 1988, 1992). PAN uses antibodies raised against cell type-specific surface proteins to select various subsets of cells from a heterogeneous cell suspension. Panning plates are first coated with antibodies and the dissociated cells are deposited on the plate for 30 min to 1 h to allow sufficient time for antibodies to bind. The plate is then washed and either the adherent or nonadherent cells (depending on the target population) can then be used for downstream assay. Often, multiple plates with multiple antibodies are necessary to sequentially enrich the target population of cells (Barres et al., 1988, 1992; Cahoy et al., 2008). Thus, PAN can be more time consuming than the other techniques and potentially exposes dissociated cells to more reagents, both of which could in theory induce aberrant transcriptional responses. Another important consideration is that a given cell type is only amenable to immunopanning purification if it can be identified by a unique cell surface antigen, and if a suitable antibody exists for that antigen. Ultimately, the purity of the resulting cell population depends on the specificity of the antibody. Alternatively, an exogenously derived cell surface antigen for which a highly specific antibody exists may be introduced to a particular population of cells through retrograde transport of the injected antigen adsorbed to fluorescent beads (Dugas et al., 2008), or conceivably by introduction of an exogenous epitope through viral transfection or transgenic methods.
Whereas LCM, FACS, Manual, and PAN harvest total RNA from pooled sorted cells, translating ribosome affinity purification (TRAP) immunoprecipitates labeled polysomes directly from tissue homogenates obtained from special transgenic mice. These mice are engineered using bacterial artificial chromosomes (Shizuya et al., 1992; Gong et al., 2003) to target an EGFP-L10a ribosomal transgene to restricted cell populations in the CNS (Doyle et al., 2008; Heiman et al., 2008). Thus, TRAP detects only ribosome-associated mRNA, and therefore only transcripts that are actively being translated, rather than the full population of transcribed RNA. Data obtained from some of these lines appear to be somewhat prone to background contamination and post hoc cleanup of the data must be applied (Dougherty et al., 2010). However, TRAP and another closely related method called RiboTag (Sanz et al., 2009) significantly improve on pre-existing translational profiling techniques, such as polysome fractionation (Darnell et al., 2009), by adding cell type-specificity and offering considerably greater throughput. Detecting the presence of polysomal mRNA for a given gene is a stronger indicator of protein expression than the detection of the mRNA itself, and thus TRAP offers an advantage over other methods in this regard. Conversely, the inability to detect noncoding RNAs using the TRAP method may also be construed as a limitation insofar as noncoding RNAs are critically important for gene regulation. Lastly, TRAP and RiboTag require the use of special mouse lines, whereas the other methods are currently applicable to a wider array of cell types; however, several new TRAP- and RiboTag-compatible mouse lines are in development. Importantly, RiboTag mice and the newer generation of TRAP mice incorporate cre-responsive ribosomal transgenes, which may be crossed with cell type-specific cre-driver mice, providing a modular experimental design strategy that will greatly expand the applicability of ribosomal pull-down methods.
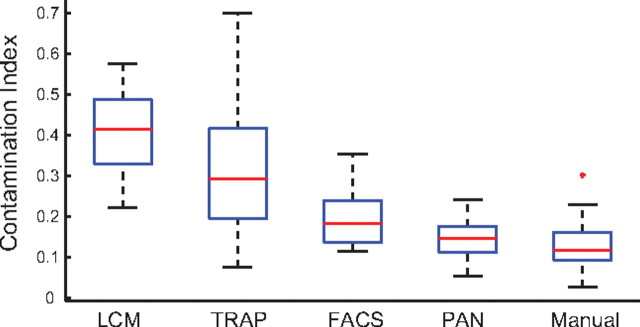
Recently, we reanalyzed all of the publically available mouse brain, cell type-specific microarray data (Affymetrix platforms only) obtained by each of the described methods (with the exception of patch/aspirate) to quantify potential differences in repeatability, contamination, and stress effects (Okaty et al., 2011). We found that all methods demonstrated a comparably high degree of repeatability as measured by the correlation between biological replicate samples (>0.94), but we detected significant differences in the levels of contamination. Using the expression levels of well established cell-type-specific marker genes for GABAergic cells, astrocytes, and oligodendrocytes to calculate contamination indices for non-GABAergic, nonastrocyte, and nonoligodendrocyte cell types profiled by each method, we found that LCM and TRAP samples showed significantly higher levels of contamination than FACS, PAN, and Manual. On average, the effects were higher in LCM samples and contamination of TRAP samples was highly variable, indicating that the level of contamination may vary depending on the cell type and/or transgenic mouse line being used. A summary of these results can be found in Figure 3 (note that using an expanded list of genes selected by unsupervised clustering to calculate contamination indices did not alter the key finding, namely that LCM and TRAP samples showed significantly higher levels of contamination). Although comparisons were largely made between different cell types, in a handful of cases common cell types were profiled by different methods, and in each case LCM or TRAP data showed evidence of higher contamination than data acquired by other methods, suggesting that differential contamination stems from the purification method, rather than the cell type. Also, differences in the translational activities of different transcripts may be reflected in TRAP versus non-TRAP data. Expression of noncoding RNAs, for example, was detected in LCM-, Manual-, FACS-, and PAN-purified samples but not in TRAP samples, as expected. We also detected heightened expression of immediate early genes, stress-related genes, and apoptosis genes in some PAN samples; however, the effect was modest. Additional sources of differences observed in the data may be differential sensitivities between methods or more idiosyncratic differences deriving from other experimental conditions rather than the purification methods per se.
Figure 3.
Box-and-whisker plots depicting contamination indices for LCM, TRAP, FACS, PAN, and Manual methods. Briefly, contamination indices were computed using the expression levels of well defined marker genes for GABAergic cells (Slc32a1, Gad1, Gad2), astrocytes (Aqp4, Gfap, Fgfr3, Slc1a2, Gjb6), and oligodendrocytes (Mbp, Sox10, Mag, Mog) in non-GABAergic, nonastrocyte, and nonoligodendrocyte samples. A contamination index of 1 indicates expression of marker genes at the same level in the target population (e.g., non-GABAergic) as in the nontarget population (e.g., GABAergic), whereas 0 indicates no expression. The bottom and top edges of each box indicate the 25th and 75th percentiles, respectively; the red line indicates the median value; and the whiskers indicate the minimum and maximum values not considered outliers. The red cross indicates an outlier (a data point >1.5 times the interquartile range outside the box).
Ultimately, the suitability of each method for use in a given study is a function of the particular goals of the study, the cell type(s) of interest, and the availability of resources, such as equipment and/or mouse lines (Tables 1, 2). For instance, all of the discussed methods can be used to detect enrichment of strong marker genes for a given cell type. However, comparison between different conditions for the same cell type, such as a disease state, activity deprivation, or other perturbations in which expression differences are generally smaller than between-cell type differences requires a method achieving high purity to ensure that contamination will not skew the results. Additionally, not all methods are equally compatible with all cell populations. Cells identified by retrograde or anterograde tracer injection or by viral transfection of a fluorescent reporter construct are more readily collected by LCM, FACS, PAN, or Manual, and only LCM and Manual are well suited to small, sparsely labeled populations. However, for assays requiring greater amounts of genetic material, FACS, PAN, and TRAP may be more appropriate. Regardless of the sorting method used, cell type-specific transcriptomic data are a valuable resource to the neuroscience community.
Table 2.
An overview of cell type-specific mRNA purification methods reviewed from the literature
| Method | Preamplification yield of total RNA | Pros | Cons | References |
|---|---|---|---|---|
| Patch/aspirate | 5–10 pg (1 cell) | Allows direct comparison between electrophysiology and gene expression at the single-cell level | Lowest preamplification yield, potentially leading to false negatives and low reproducibility | (Subkhankulova et al., 2010; Toledo-Rodriguez et al., 2004) |
| LCM | 1–10 ng (100–700 cells) | Readily applied to preserved human postmortem tissue | Tissue fixation degrades mRNA | (Chung et al., 2005; Rossner et al., 2006; Pietersen et al., 2009) |
| Manual | 0.25–1 ng (30–100 cells) | Compatible with dim, sparsely labeled cell populations | Low preamplification yield | (Sugino et al., 2006; Hempel et al., 2007; Okaty et al., 2009) |
| FACS | 10–500 ng (103-105 cells) | Highest preamplification yield | Processing samples from mature animals requires optimization to overcome stress of procedure | (Arlotta et al., 2005; Lobo et al., 2006; Cahoy et al., 2008; Marsh et al., 2008; Molyneaux et al., 2009) |
| Immunopanning | 30–50 ng (103-105 cells) | Compatible with unlabeled cells | Cell type must be distinguishable by a surface protein | (Barres et al., 1988, 1992; Cahoy et al., 2008) |
| Procedure often requires multiple iterations of plating and antibody reaction steps which may result in stress effects | ||||
| TRAP/RiboTag | >15 ng (>103 cells) | Targets actively translated transcripts | Requires specific transgenic mouse lines | (Doyle et al., 2008; Heiman et al., 2008; Sanz et al., 2009; Dougherty et al., 2010) |
| Incompatible with detection of noncoding RNAs |
Preamplification yield of total RNA indicates the reported quantity of RNA that served as input to the two round in vitro transcription amplifications used by each method. Cell numbers were either given explicitly in the references or were estimated from the amount of RNA, using 10 pg per cell as an approximation (however, in general, the quantity of total RNA in a given cell varies by cell type). Pros and Cons are by no means complete, but are provided to illustrate some of the key differences between methods, particularly as they may relate to deciding which method is best suited to a given scientific question.
Footnotes
Editor's Note: Toolboxes are intended to describe and evaluate methods that are becoming widely relevant to the neuroscience community or to provide a critical analysis of established techniques. For more information, see http://www.jneurosci.org/misc/ifa_minireviews.dtl.
This work was supported by funding from the National Institute of Mental Health.
The authors declare no competing financial interests.
References
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Barres BA, Silverstein BE, Corey DP, Chun LL. Immunological, morphological, and electrophysiological variation among retinal ganglion cells purified by panning. Neuron. 1988;1:791–803. doi: 10.1016/0896-6273(88)90127-4. [DOI] [PubMed] [Google Scholar]
- Barres BA, Hart IK, Coles HS, Burne JF, Voyvodic JT, Richardson WD, Raff MC. Cell death and control of cell survival in the oligodendrocyte lineage. Cell. 1992;70:31–46. doi: 10.1016/0092-8674(92)90531-g. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyaya B, Di Cristo G, Higashiyama H, Knott GW, Kuhlman SJ, Welker E, Huang ZJ. Experience and activity-dependent maturation of perisomatic GABAergic innervation in primary visual cortex during a postnatal critical period. J Neurosci. 2004;24:9598–9611. doi: 10.1523/JNEUROSCI.1851-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Fraser CE, Mostovetsky O, Darnell RB. Discrimination of common and unique RNA-binding activities among Fragile X mental retardation protein paralogs. Human Molecular Genetics. 2009;18:3164–3177. doi: 10.1093/hmg/ddp255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty JD, Schmidt EF, Nakajima M, Heintz N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res. 2010;38:4218–4230. doi: 10.1093/nar/gkq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, Gong S, Greengard P, Heintz N. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Mandemakers W, Rogers M, Ibrahim A, Daneman R, Barres BA. A novel purification method for CNS projection neurons leads to the identification of brain vascular cells as a source of trophic support for corticospinal motor neurons. J Neurosci. 2008;28:8294–8305. doi: 10.1523/JNEUROSCI.2010-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, Rinn JL, Lander ES, Regev A. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suárez-Fariñas M, Schwarz C, Stephan DA, Surmeier DJ, Greengard P, Heintz N. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel CM, Sugino K, Nelson SB. A manual method for the purification of fluorescently labeled neurons from the mammalian brain. Nat Protoc. 2007;2:2924–2929. doi: 10.1038/nprot.2007.416. [DOI] [PubMed] [Google Scholar]
- Janes KA, Wang CC, Holmberg KJ, Cabral K, Brugge JS. Identifying single-cell molecular programs by stochastic profiling. Nat Methods. 2010;7:311–317. doi: 10.1038/nmeth.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman SJ, Huang ZJ. High-resolution labeling and functional manipulation of specific neuron types in mouse brain by Cre-activated viral gene expression. PLoS One. 2008;3:e2005. doi: 10.1371/journal.pone.0002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Saitou M. Single-cell cDNA microarray profiling of complex biological processes of differentiation. Curr Opin Genet Dev. 2010;20:470–477. doi: 10.1016/j.gde.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh ED, Minarcik J, Campbell K, Brooks-Kayal AR, Golden JA. FACS-array gene expression analysis during early development of mouse telencephalic interneurons. Dev Neurobiol. 2008;68:434–445. doi: 10.1002/dneu.20602. [DOI] [PubMed] [Google Scholar]
- Martina M, Schultz JH, Ehmke H, Monyer H, Jonas P. Functional and molecular differences between voltage-gated K+ channels of fast-spiking interneurons and pyramidal neurons of rat hippocampus. J Neurosci. 1998;18:8111–8125. doi: 10.1523/JNEUROSCI.18-20-08111.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi G, Fishell G. Directing neuron-specific transgene expression in the mouse CNS. Curr Opin Neurobiol. 2006;16:577–584. doi: 10.1016/j.conb.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Okaty BW, Miller MN, Sugino K, Hempel CM, Nelson SB. Transcriptional and electrophysiological maturation of neocortical fast-spiking GABAergic interneurons. J Neurosci. 2009;29:7040–7052. doi: 10.1523/JNEUROSCI.0105-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okaty BW, Sugino K, Nelson SB. A quantitative comparison of cell-type-specific microarray gene expression profiling methods in the mouse brain. PLoS One. 2011;6:e16493. doi: 10.1371/journal.pone.0016493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva AA, Jr, Jiang M, Lam T, Smith KL, Swann JW. Novel hippocampal interneuronal subtypes identified using transgenic mice that express green fluorescent protein in GABAergic interneurons. J Neurosci. 2000;20:3354–3368. doi: 10.1523/JNEUROSCI.20-09-03354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Goren A, Gymrek M, Guttman M, Regev A, Bernstein BE, Milos PM. Digital transcriptome profiling from attomole-level RNA samples. Genome Res. 2010a;20:519–525. doi: 10.1101/gr.102129.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Ting DT, Wittner BS, Brannigan BW, Paul S, Bardeesy N, Ramaswamy S, Milos PM, Haber DA. Amplification-free digital gene expression profiling from minute cell quantities. Nat Methods. 2010b;7:619–621. doi: 10.1038/nmeth.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A. Single-molecule approaches to stochastic gene expression. Annu Rev Biophys. 2009;38:255–270. doi: 10.1146/annurev.biophys.37.032807.125928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossner MJ, Hirrlinger J, Wichert SP, Boehm C, Newrzella D, Hiemisch H, Eisenhardt G, Stuenkel C, von Ahsen O, Nave KA. Global transcriptome analysis of genetically identified neurons in the adult cortex. J Neurosci. 2006;26:9956–9966. doi: 10.1523/JNEUROSCI.0468-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shizuya H, Birren B, Kim UJ, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci U S A. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subkhankulova T, Yano K, Robinson HP, Livesey FJ. Grouping and classifying electrophysiologically-defined classes of neocortical neurons by single cell, whole-genome expression profiling. Front Mol Neurosci. 2010;3:10. doi: 10.3389/fnmol.2010.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- Surmeier DJ, Song WJ, Yan Z. Coordinated expression of dopamine receptors in neostriatal medium spiny neurons. J Neurosci. 1996;16:6579–6591. doi: 10.1523/JNEUROSCI.16-20-06579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Rodriguez M, Blumenfeld B, Wu C, Luo J, Attali B, Goodman P, Markram H. Correlation maps allow neuronal electrical properties to be predicted from single-cell gene expression profiles in rat neocortex. Cereb Cortex. 2004;14:1310–1327. doi: 10.1093/cercor/bhh092. [DOI] [PubMed] [Google Scholar]