Abstract
We review the application of C. elegans as a model system to understand key aspects of stem cell biology. The only bona fide stem cells in C. elegans are those of the germline, which serves as a valuable paradigm for understanding how stem cell niches influence maintenance and differentiation of stem cells and how somatic differentiation is repressed during germline development. Somatic cells that share stem cell-like characteristics also provide insights into principles in stem cell biology. The epidermalseam cell lineages lend clues to conserved mechanisms of self-renewal and expansion divisions. Principles of developmental plasticity and reprogramming relevant to stem cell biology arise from studies of natural transdifferentiation and from analysis of early embryonic progenitors, which undergo a dramatic transition from a pluripotent, reprogrammable condition to a state of committed differentiation. The relevance of these developmental processes to our understanding of stem cell biology in other organisms is discussed.
Keywords: stem cells, germline, reprogramming, transdifferentiation, microRNA, translational suppressors
I. Introduction
Developmental regulatory cues must be tightly coordinated with cell division programs to ensure that a sufficient number of descendant cells are available to populate tissues and organs prior to their exit from mitosis and differentiation. A conspicuous example of the tight coordination between cell proliferation control and differentiation is seen with the maintenance and differentiation of stem cells (Molofsky et al., 2004; Orford and Scadden, 2008). Stem cells are maintained in a proliferative, undifferentiated state within a “niche” that provides continuous pro-mitotic, anti-differentiation cues (Ohlstein et al., 2004); exit from the niche is often associated with a switch from self-renewal to programs of differentiation. An understanding of how stem cells renew themselves, the regulatory circuitry that maintains their pluripotency, and the events that commit progenitor cells to particular differentiation states is of paramount importance as stem cells are intimately associated with development, maintaining homeostasis, tissue repair and regeneration, aging, and cancer. In addition, it is of great value to learn how fully differentiated cells can be reprogrammed into progenitors of different cell types.
Diverse paradigms and signaling pathways for regulating stem cells have been described (Croce and Calin, 2005; Nystul and Spradling, 2006; Kimble and Crittenden, 2007; Morrison and Spradling, 2008; Spradling et al., 2008; Shenghui et al., 2009). However, the complex architecture of tissues has made it difficult to identify stem cell niches and investigate niche-stem cell interactions in mammalian systems. The relative simplicity of tissue architecture in invertebrate model systems, ease of identification of individual stem cell niches, and highly accessible tools for genetic, cellular and biochemical analysis of regulatory networks, have made invertebrate model organisms powerful systems for studying stem cell biology and deriving general principles for mechanisms of stem cell renewal, maintenance of pluripotency, and cellular reprogramming. In this review, we highlight the strengths of C. elegans as a model system for investigating stem cell properties of self-renewal, maintenance of pluripotency, and reprogramming of differentiation. Given the broad scope of the article and space constraints we direct the reader to more focused topical reviews for a more thorough treatment of specific subjects. In this review, we first focus on the development of the only true stem cells in C. elegans, those of the germline, and discuss the germline as a model system to investigate stem cell-niche interactions and the importance of post-transcriptional regulation and non-coding RNAs in regulating the switch from proliferation to differentiation. We then focus on the specialized epidermal seam cells, the first experimental system to demonstrate a role for microRNAs (miRNAs) in regulating stem cell-like divisions. Finally, we turn to the mechanisms that restrict the pluripotency of progenitor cells and the processes that result in reprogramming of cellular identity, and we discuss how investigating these processes makes it possible to move beyond the transcriptional regulatory cascades that have dominated the field of pluripotency research. We have made an effort to relate the findings made in C. elegans with results from other systems, notably mammalian stem cells.
The C. elegans germline as a model for stem cell biology
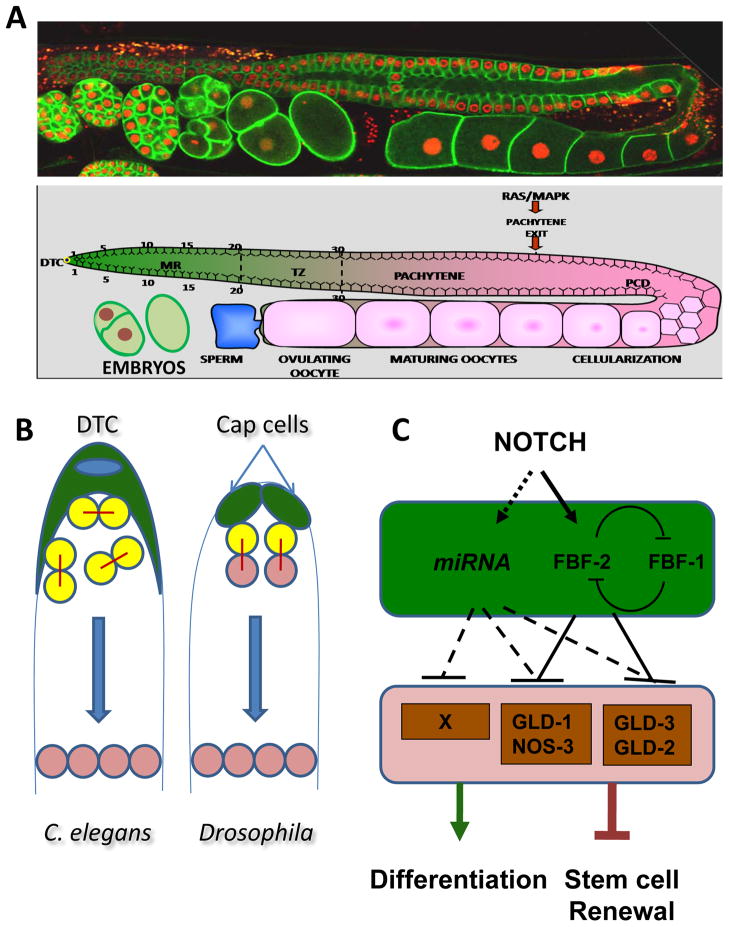
The C. elegans hermaphrodite gonad is a tube with two symmetric U-shaped arms, each with a proximo-distal polarity (Figure 1). Contained within an outer covering of somatic cells, the germline is a syncytium, wherein individual germ cell nuclei that are partially enclosed by cell membranes are arranged circumferentially around a central cytoplasmic core, the rachis. Unlike nuclei in other syncytia, such as those in the pre-cellularization Drosophila embryo which divide synchronously, adjacent germ cell nuclei in the C. elegans syncytium share limited cytoplasm owing to their partial enclosure by a cell membrane, and behave largely as if they were contained in individual cells. They are therefore, by convention, referred to as “germ cells.” The distal-most region of the gonad contains proliferating mitotic germline stem cells (GSCs). As germ cells move proximally, they cease proliferation and progress through successive stages of meiosis and undergo cellularization past the bend in the gonad tube to form mature oocytes that are arrested in diakinesis of prophase I (Figure 1A).
Figure 1. The C. elegans hermaphrodite germline and the network regulating the choice between self-renewal and differentiation.
A) Shown here is a single arm of the C. elegans hermaphrodite gonad which shows a distal to proximal polarity. A pool of germline stem cells is maintained in a proliferative state in response to the GLP-1/Notch signal secreted by the DTC, which serves as a stem cell niche. The mitotic region (green in the cartoon) contains the proliferating germline stem cells, transit amplifying cells as well as cells that are in pre-meiotic S-phase. As cells move away from the DTC they exit mitosis and resume meiosis (Transition Zone). As cells migrate further proximally they progress through successive stages of meiosis –pachytene, diplotene, and diakinesis (Pink). Exit from Pachytene is dependent on MPK signaling pathway. The distal half of the gonad is syncytial; past the bend “germ cells” undergo cellularization and form mature oocytes. B) GSCs in C. elegans gonad undergo symmetric divisions resulting in an expansion in the population of stem cells. As this population of cells is pushed away from the influence of the DTC, they switch from a self renewal to a differentiation program. Further, there is no bias in the orientation of cell divisions. In contrast, germline stem cells in Drosophila gonads undergo oriented asymmetric cell divisions such that one of the daughter cells that remains in contact with the stem cell niche retains its stem cell identity, while the cell that is further away undergoes differentiation. C) Downstream of Notch signaling, a network of translational regulators including RNA-binding proteins and miRNAs function in the switch between self-renewal and differentiation.
II. Germline stem cells and the stem cell - niche interaction
Studies of the C. elegans germline led to what is likely the first stem cell niche to be clearly identified at the cellular level in any metazoan. The germline is the only tissue in C. elegans with a continuously proliferating pool of cells proceeding throughout its life (Hirsh et al., 1976). The germline stem cells (GSCs) are maintained in a proliferative state in the niche as defined by a single somatic cell, the distal tip cell (DTC) that caps the distal end of the gonad and extends fine cytoplasmic processes to about 10 germ cell diameters from the main body of the cell (Kimble, 1981). Experimental removal of the DTC by laser ablation causes the GSCs to cease proliferation and differentiate into gametes. Genetic manipulations using mutations that alter the position or number of DTCs result in ectopic proliferation of germ cells in close proximity to the DTC, thereby establishing the DTC as a bona fide activator of stem cell niche formation. Several excellent reviews detail the germline stem cell niche (Byrd and Kimble, 2009) and the regulation of GSC proliferation and differentiation (Hubbard and Greenstein, 2000; Kimble and Crittenden, 2005; Hansen and Schedl, 2006; Hubbard, 2007; Kimble and Crittenden, 2007). Here, we provide a brief overview of germline stem cell proliferation and differentiation, followed by a discussion of a role for microRNAs and the macroenvironment in regulating GSC maintenance.
The DTC serves as a stem cell niche and maintains a proliferative germline stem cell population via Notch-type signaling. The proliferative zone extends ~20 cells from the DTC. As cells divide, they move away from the influence of the DTC and switch from mitosis into meiosis, as evident by a morphological change in which their nuclei become crescent-shaped, characteristic of germ cells in leptotene and zygotene; this event defines the start of the “transition zone” (Figure 1A). Thus, the “mitotic region (MR)”, defined by the stem cell niche and containing all germline stem cells, refers to the portion of the germline that is distal to the nuclei exhibiting morphological characteristics of early meiotic nuclei.
Recent analysis of cell division kinetics has led to further subdivision of the proliferative region with identification of at least four distinct population of proliferating cells (Crittenden et al., 2006; Maciejowski et al., 2006; Hubbard, 2007; Jaramillo-Lambert et al., 2007): 1. The distal-most population of cells that are in close apposition to the DTC (1–2 cell diameters from the DTC) are associated with the lowest cell division kinetics, 2) immediately following is a population of germ cells extending from 3–10 cell diameters that exhibit the highest average mitotic index, 3) a population of cells with decreasing mitotic index (10 –16 cell diameters) and 4) a population of cells with the lowest average mitotic index extending from 17 –22 cell diameters from the DTC precedes the early meiotic nuclei (reviewed in (Hubbard, 2007). The last two populations of germ cell also include cells that are in meiotic S-phase. This profile of cell division kinetics is similar to the behavior of adult stem cells in mammalian intestinal crypts and hair follicles - bona fide stem cells found in close physical contact with the stem cell niche have significantly longer cell cycle times than an intermediate population of rapidly cycling transit-amplifying stem cells.
The architecture of the C. elegans germline stem cell niche with germline stem cells at one end of a blind tube and progressively differentiating cells near the open end shows similarities with germline stem cell niche in other model systems including male and female germline stem cell niches in Drosophila - the hub (Tulina and Matunis, 2001) and the germarial tip (Xie and Spradling, 2000) respectively (Figure 1B). Adhesion between the GSC and the niche, mediated by E cadherin, β-catenin and integrins, retains GSCs within the niche (Song and Xie, 2002; Song et al., 2002; Wang et al., 2006; Issigonis et al., 2009). Adhesion also orients the axis of division resulting in asymmetric cell division. This division gives rise to a cell that remains in contact with the niche and consequently persists as a stem cell (Yamashita et al., 2007) (Figure 1B). Analysis of the cell division plane of GSCs in the distal-most region of the hermaphrodite gonad did not reveal any bias in orientation of cell division plane along the distal-proximal axis of the gonad (Crittenden et al., 2006). By pulse-chase BrdU labeling of proliferating cells, the same study reported the lack of any quiescent cells or retention of labeled cells within the stem cell niche region (Crittenden et al., 2006). While all cells do eventually leave the niche, it has yet to be determined whether there are differences in retention of one of the daughter cells as a consequence of its close apposition to the DTC. Lack of GSC-specific markers, or an easy way to label individual cells in the germline specifically, has hindered the ability to trace the lineage of a given cell and correlate it with its ultimate fate; however, the recent adaptation in C. elegans of the widely used FLP/FRT based system for permanently marking individual cells (Davis et al., 2008; Voutev and Hubbard, 2008) provides a promising tool for analyzing germline development at the resolution of individual cells.
Extrinsic regulators of GSC proliferation
The distal tip cell (DTC) maintains the mitotically proliferating population of cells in the MR through a Notch-type signaling pathway mediated by Delta/Serrate-like ligands, LAG-2 (Henderson et al., 1994) and APX-1 (Nadarajan et al., 2009) produced by the DTC and the GLP-1 (germline proliferation-1)/Notch receptor present on the surface of the germ cells (Austin and Kimble, 1987; Nadarajan et al., 2009). Mutations that compromise GLP-1/Notch signaling result in too few germ cells as a consequence of premature exit from mitosis and entry into meiosis. A constitutively active form of GLP-1 leads to a germline tumor phenotype (tum), owing to the continued proliferation of germline stem cells and their failure to enter meiosis (Berry et al., 1997). How, then, does GLP-1/Notch signaling regulate the proliferation of GSCs? To date a direct link between cell cycle control machinery and transcriptional regulation by the LAG-1/SEL-8/Notch-Intracellular domain complex has not been established. The entire purpose of Notch/GLP-1 signaling seems to be to repress the function of gld-1 and gld-2, two genes that promote entry into meiosis (see below). Indeed, the requirement for GLP-1 in germ cell proliferation is eliminated in animals that are doubly mutant for gld-1 and gld-2, in which the entire germline becomes tumorous owing to excessive proliferation (Kadyk and Kimble, 1998). Interestingly, the GSCs that are in the closest proximity to the DTC do not divide more frequently in a glp-1(gf) mutant. However, no other signaling pathway has been identified that, when mutated, affects the GSC renewal/differentiation switch as severely as the Notch signaling pathway. Identification of other signaling factors or cell cycle regulatory components that result in a reduced rate of proliferation would be of considerable interest with potential clinical implications for aging and cancer.
In addition to close-range signaling of the type just described, proliferation of stem cells can also be regulated by long-range-acting factors such as hormones, cytokines and neurotransmitters (Shenghui et al., 2009). Neural insulin-like peptides signal nutrient availability in Drosophila. They also directly regulate the rate of GSC division and ovulation, thereby controlling egg production in response to nutritional status (Drummond-Barbosa and Spradling, 2001; LaFever and Drummond-Barbosa, 2005; Hsu et al., 2008; Hsu and Drummond-Barbosa, 2009). This arrest is dependent on the CDK inhibitor p21/Dacapo and is regulated by miRNAs (Yu et al., 2009). Under conditions of nutritional deprivation, C. elegans can enter diapause at several stages in its life cycle; the L1 larval stage, the L2 stage to form dauers, and as adults. During L1 diapause the germ cells are arrested in G2 phase; while in mutants lacking the worm PTEN ortholog, DAF-18, germ cells continue to proliferate by a process dependent on AKT-1 and AGE-1, two kinases involved in regulating insulin signaling in C. elegans (Fukuyama et al., 2006). A similar arrest of GSC proliferation was observed during dauer diapause (Narbonne and Roy, 2006b; Narbonne and Roy, 2006a). A role for translational regulation in mediating germ cell proliferation arrest during the dauer stage is suggested by the low protein expression profile of germline proteins despite elevated transcript levels, as determined by microarray profiling of dauer larvae (Jeong et al., 2009)(unpublished data).
During adult reproductive diapause, all germ cells except the GSCs undergo apoptosis and on restoration to food these surviving GSCs repopulate the entire gonad in a manner that may be Notch-dependent (Angelo and Van Gilst, 2009). While entry into adult reproductive diapause is mediated by the nuclear receptor HNF4α homolog NHR-49, GSC survival and proliferation are regulated by a mechanism independent of NHR-49 (Angelo and Van Gilst, 2009). What are the mechanisms by which GSCs are kept in a quiescent state during the period of starvation? Do all germ cells proliferate or do only the distal-most GSCs contribute to repopulating the germline? How is normal development achieved even in the presence of multiple LAG-2-expressing cells (see “latent niches” below)? Analysis of germline “regeneration” in animals exiting adult reproductive diapause holds the promise of shedding light on these and other aspects of stem cell biology.
Intrinsic regulators of GSC proliferation – a preponderance of translational regulators
A prominent role for RNA binding proteins in specification and maintenance of germ cell fate is observed in all sexually reproducing metazoans (Seydoux and Braun, 2006; Ewen-Campen et al., 2009); indeed, the RNA binding proteins Vasa and Nanos are evolutionarily conserved markers of germ cell identity (Extavour and Akam, 2003). Post-transcriptional control by binding of factors to the 3′ UTR of target mRNAs is a predominant mechanism by which germline gene expression is regulated in C. elegans (Merritt et al., 2008) and in Drosophila (Rangan et al., 2008; Rangan et al., 2009). Downstream of GLP-1-mediated transcriptional control, the mitosis/meiosis decision is regulated by a network of RNA binding proteins that bifurcates into two partially redundant branches - GLD-1 and NOS-3 in one pathway and GLD-2 and GLD-3 in the other (Fig. 1C) (Kimble and Crittenden, 2005; Hansen and Schedl, 2006; Kimble and Crittenden, 2007). Germ cells in null mutants of any of these genes are still able to initiate germline meiosis; however, they prematurely exit meiosis and resume mitosis resulting in a germline tumor. In double mutant combinations, in which both branches are abolished, a majority of germ cells apparently never enter meiosis and remain mitotic. Elimination of both pathways does not, however, completely eliminate all indications of meiotic initiation, implying that yet another pathway may act in parallel to these two (Hansen et al., 2004). GLD-1, a KH domain-containing RNA binding protein functions as a translational repressor of mitosis-promoting genes. One of the targets of GLD-1 is glp-1 itself, constituting a feedback system that causes GLP-1 to be expressed only in the proliferative cells despite the presence of glp-1 transcript throughout the germline. NOS-3, a member of the Nanos family of translational repressors, activates GLD-1 possibly through the inhibition of an unidentified inhibitor. GLD-2, a poly(A) polymerase, and GLD-3 an ortholog of Drosophila Bicaudal-C RNA binding protein function in a complex to activate meiosis-promoting genes.
Negative regulation of the meiosis-promoting, mitosis-inhibiting factors by GLP-1/NOTCH is mediated by two nearly identical Pumilio family of RNA binding proteins that are critical for maintaining adult stem cells, FBF-1 and FBF-2. Only FBF-2 is the direct target of GLP-1 signaling and is the only direct target of GLP-1/LAG-1 signaling involved in the mitosis/meiosis decision. FBF proteins bind to the 3′ UTRs of gld-1 and gld-3 transcripts, thereby preventing their translation. Thus, the balance between GSC renewal and differentiation is an outcome of opposing networks of translational repressors. FBF associates with >1000 mRNAs, including those encoding members of the EGF/MPK pathway (Byrd and Kimble, 2009). Thus, FBF constitutes a prominent hub in the mitosis/meiosis regulatory network, which is characterized by multiple redundancies and feedback loops. This network architecture presumably ensures that the maintenance of adult GSCs is a very robust process, resulting in only modest changes when any one component is perturbed. At the same time these changes are readily quantifiable, making GSC proliferation an extremely sensitive reporter of signal flux through the network. An experimental system that is at once both robust and sensitive is ideally suited for a systems level analysis of how developmental programs ensure precision while being responsive to a large dynamic range in signal intensity.
While FBF proteins are critical for the maintenance of adult GSCs, they do not play a significant role in GSC proliferation during early larval stages. Even when all FBF activity is eliminated, 100s of germ cells are made, in contrast to <10 GSCs in glp-1 null mutants, implying that there are additional Notch targets that regulate germline proliferation. Factors that mediate Notch signaling to promote GSC proliferation during the L2 and L3 larval stage have not been identified. In addition to FBFs, another Pumilio family protein, PUF-8, acts in conjunction with MEX-3, a KH domain-containing RNA binding protein, to promote GSC proliferation (Ariz et al., 2009).
As in C. elegans, translational repression may also be a critical mode of GSC fate regulation in Drosophila. The balance between GSC renewal and differentiation is an outcome of opposing interactions between the translational repressor complex of Pumilio and Nanos proteins, which function to promote GSC renewal by suppressing the translation of differentiation-promoting factors, and that of Bam and BGCN, which promote differentiation through an unknown mechanism (Lin and Spradling, 1997; Szakmary et al., 2005). NOS and BAM/BGCN show a reciprocal expression pattern. A recent study suggests that Bam/Bgcn act as a complex to suppress Nos expression in a manner dependent on the Nos 3′ UTR (Li et al., 2009). Bam and Bgcn directly repress translation initiation of E-cadherin, which in turn is critical for maintaining contact of GSCs with the niche, by binding to its 3′ UTR (Shen et al., 2009).
Role of small ncRNA in regulating GSC proliferation
Another prominent paradigm of translational control is regulation by microRNAs (miRNAs) (Ambros, 2004; Bartel, 2004; Bushati and Cohen, 2007). MicroRNAs are ~21–22 nucleotide long RNA molecules that primarily inhibit gene expression by binding to 3′ UTRs, thereby inhibiting translation or destabilizing target mRNA. miRNAs were first found in C. elegans by classical developmental genetics when it was discovered that the lin-4 gene, a regulator of stage-specific switches during larval development (Lee et al., 1993), encodes not a protein, but a miRNA. The lin-14 gene, which encodes a protein that regulates the same developmental switches in the opposite direction, was subsequently identified as a target of lin-4 action: the 3′ untranslated region (UTR) of the lin-14 mRNA was found to contain sequences that are complementary to the lin-4 miRNA, through which it binds and represses translation of thismessage (Wightman et al., 1993). These seminal findings established a new paradigm for gene regulation in metazoans: inhibition of translation by the binding of endogenous RNA molecules to the 3′ UTR of mRNAs (Lee et al., 1993; Wightman et al., 1993). A number of studies have revealed that miRNAs regulate cell proliferation, particularly in the context of stem cell populations and tumorigenesis (Nimmo and Slack, 2009). A role for miRNAs in maintaining stem cell populations in mice is implicated by the loss of Oct4-expressing pluripotent stem cells in dicer1 knockout embryos (Bernstein et al., 2003). Similarly, a reduction in germ cyst production was observed in GSCs of dicer(−) mutants in Drosophila as the result of mitotic arrest mediated by Dacapo/p27, an inhibitor of cyclin-dependent kinase activity, implicating miRNAs in the control of GSC proliferation in Drosophila (Hatfield et al., 2005; Yu et al., 2009). Genetic screens in C. elegans have identified additional components required for GLP-1-mediated regulation of germline proliferation including EGO-1 (enhancer of glp), an RNA-dependent RNA polymerase that is a critical component of RNA interference (Qiao et al., 1995; Smardon et al., 2000; Vought et al., 2005). The observation that EGO-1 is required for efficient GLP-1 signaling suggests a role for endogenous RNAi and/or miRNAs in regulating germ line stem cell proliferation. In addition to EGO-1, CSR-1, a Piwi/PAZ/Argonaute protein, EKL-1, a Tudor domain-containing RBP, and DRH-3, a DEAH box helicase have also been implicated in regulating germ cell proliferation (She et al., 2009). CSR-1 has a Slicer function and is involved in generating 20 siRNAs, while DRH-3 is known to be involved in generating multiple classes of sncRNAs and interacts with EGO-1. Thus, miRNAs or endogenous siRNAs might be involved in regulating GSC renewal and differentiation. Several genes involved in the mitosis/meiosis decision have predicted binding sites for multiple miRNAs in their 3′ UTRs. Consistent with a role for miRNAs in regulating GSCs, work from our lab has identified at least 13 miRNAs that function to maintain adult GSC population in part by targeting GLD-1 mRNA for degradation (P.J. and J.R., unpublished data). A similar stage-specific requirement for Dicer and bantam miRNA in maintaining GSCs was observed in Drosophila (Shcherbata et al., 2007). As mentioned previously, the Pumilio-Nos translational repressor complex promotes Drosophila GSC proliferation by repressing pro-differentiation factors. It was found that ectopic expression of Ago1, one of five Argonaute proteins that are critical for RNAi, but not Nos leads to extra GSCs, suggesting that in Drosophila, miRNAs might also function prominently with FBF proteins to maintain GSCs (Yang et al., 2007). Computational analysis of 3′ UTRs of human FBF/Pumilio targets also show an enrichment for miRNA binding sites surrounding the FBF binding sites, also suggesting extensive interactions between FBF and miRNA translational repressors. Thus, it appears that interactions between two tiers of translational regulators, the FBF family of RNA binding proteins and microRNAs, is an evolutionarily conserved mechanism regulating stem cell proliferation. The mechanistic details of this interaction await elucidation.
Latent niches and effect of macroenvironment on GSC biology
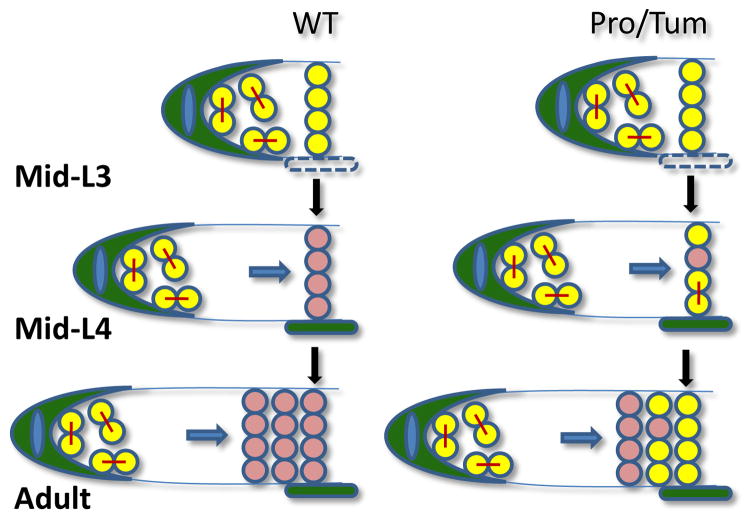
Cancer cells from malignant tumors metastasize to different parts of the body (Hess et al., 2006). Certain tumors show preference for the regions to which they metastasize. For example, lung cancers show increased propensity to metastasize to the brain, while metastases of colon cancers are frequently found in the liver. Tumors in breast cancer show increased propensity to metastasize to the lung, liver and brain (Lu and Kang, 2007). Elegant work by McGovern et al. on proximal tumors in the C. elegans germline, wherein there arise two centers of proliferative germ cells separated by differentiating cells instead of one, suggests a “latent niche” as a possible mechanism for the initiation of these neoplasias following metastasis (McGovern et al., 2009). During early stages of gonadal development (i.e., early larval stages-L2 and L3), the gonad is small and all germ cells are still under the influence of the pro-mitotic signals LAG-2 and APX-1 expressed by the DTC and remain proliferative (Figure 2). As development proceeds, germ cells continue to proliferate and the gonad continues to elongate. Consequently, some of the germ cells are now no longer under the influence of the pro-mitotic signals from the DTC and begin to differentiate. During the L4 stage, additional somatic gonadal cells that express LAG-2 and APX-1, the proximal sheath cells, are formed. Unlike the DTC, these cells do not behave like stem cell niches in normal animals as the cells that are in proximity to the proximal sheath cells are differentiating, non-responsive germ cells. However, in certain mutant backgrounds, the coordination between somatic and germline development is disrupted such that LAG-2/APX-1-responsive germ cells are now found in proximity to the proximal sheath cells, thereby establishing a second niche or proliferative center in addition to that created by the DTC. These cells continue to proliferate and form the “seed” of the proximal tumor (Figure 2). Thus, a latent niche is a cell that normally by itself does not act as a niche, but under certain circumstances can provide the necessary microenvironment to support cells capable of self-renewal. It was proposed that inappropriate contact of a proliferation-competent cell, such as cancer stem cell or a metastasizing tumor cell capable of self-renewal within a latent niche, would serve as seeds for new tumors. In this model, it was stressed that neither the tumor cell nor the niche cell need be genetically compromised to sustain tumorous growth and that aberrant development itself may result in tumors.
Figure 2. Latent niche model of ectopic germline proliferation.
In normally developing L1 to L3 larvae, the gonad is small and all germ cells are in the proximity of the pro-mitotic signals LAG-2 and APX-1 which cause them to remain proliferative. As development proceeds, the germ cells continue to proliferate and are pushed away from the influence of the proliferative signals expressed by the DTC causing the germ cells to differentiate. During the L4 stage, additional somatic gonadal cells express pro-mitotic signals, but do not normally become centers of new proliferation as the neighboring cells are differentiating, non-responsive germ cells. However in certain mutant backgrounds that lead to proximal tumors (Pro), the coordination between somatic and germline development is disrupted such that “proliferation competent” cells are now found in proximity to the proximal sheath cells, thereby establishing a second “latent niche” or proliferative center in addition to that created by the DTC.
III. Analysis of stem cell-like lineages: the seam cells as a paradigm
The epidermal seam cells as models for stem cell-like lineages
Proliferation and differentiation are modulated in stem cell lineages to ensure adequate production of specialized differentiated cells while maintaining a constant supply of undifferentiated stem cells. Stem cells can ensure a balance of proliferating and differentiating cells through asymmetric division patterns, in which one daughter differentiates into a specialized type and the other continues to function as a proliferating stem cell (self-renewal maintenance). A stem cell can also divide symmetrically to increase the stem cell pool (self-renewal expansion). The molecular mechanisms used by stem cells to regulate these patterns of division remain poorly understood and research into these mechanisms may allow us to control the choice between self-renewal expansion and differentiation for the purpose of regenerative medicine. The self-renewal maintenance and expansion patterns through which stem cells divide is mimicked by the C. elegans lateral epidermal cells, called seam cells, which both proliferate to self-renew and divide asymmetrically to produce differentiated neural and epidermal cells through defined cell lineages. Thus, the seam cell lineages are attractive simplified model for studying such stem cell-like division patterns.
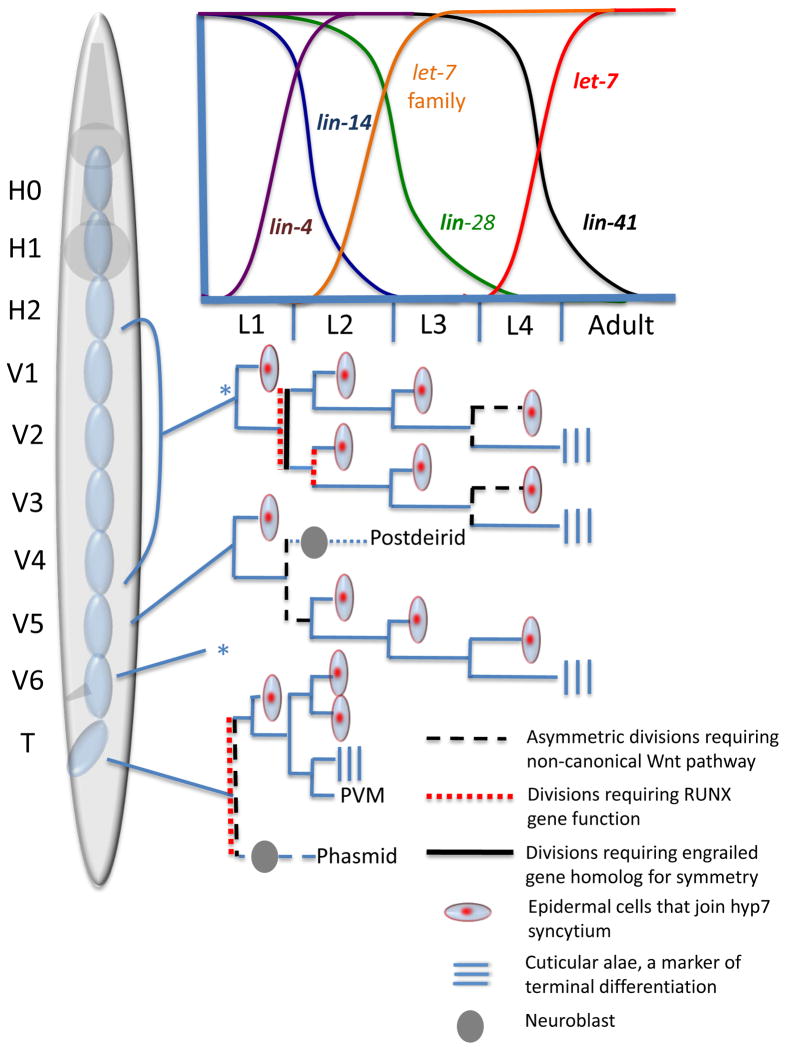
The ten bilateral pairs of seam cells (H0–H2, V1–V6, and T) born during embryogenesis are blast cells that undergo further post-embryonic divisions in a manner reminiscent of stem cell lineages. During the first larval stage, the V1–V6 cells divide and their anterior daughters fuse to the surrounding hyp 7 syncytium, exit the cell cycle, and differentiate while the posterior daughter continues to function as a seam cell (Figure 3). At the second larval stage V1–V4, V6, and T divide in a symmetric self-renewal expansion pattern, thereby doubling the seam cell number in those lineages. An exception to this pattern is the V5 cell, whose anterior daughter forms a neuroblast, which subsequently divides to produce the postdeirid (a neuronal sensory structure) in the hermaphrodite. In the male, V5 and V6 instead form tail sensory rays needed for mating. During the third and fourth larval stages, the V1–V6 descendant cells divide again by a self-renewal maintenance pattern, each time producing an anterior daughter that fuses to the hyp7 syncytium and a posterior daughter that remains in a proliferative state. At the end of the fourth larval stage, both daughters terminally differentiate when the anterior daughters fuse to hyp7 and the posterior seam cells fuse into a single seam cell syncytium and form specialized cuticular structures, the alae (Sulston and Horvitz, 1977). Mechanisms controlling the choice between seam cell maintenance division and expansion division have been uncovered and this knowledge can be used to gain insights into how stem cells make similar choices.
Figure 3. The C. elegans hermaphrodite epidermal stem cell lineage.
The seam cells divide asymmetrically at each stage in a self-renewal pattern, producing an undifferentiated seam cell that continues to divide and a epidermal cell that will fuse to the hyp7 syncytium and terminally differentiate. A symmetric seam cell expansion division occurs at the end of the L1 larval stage to produce two undifferentiated cells that remain in a proliferative state. In the V5 and T lineages, seam cells give rise to neuronal cell types that produce the postdeirid and phasmid sensory structures. Reciprocal temporal expression of miRNAs and their heterochronic targets are required for the proper timing of seam cell divisions; the relative expression levels of these genes is indicated by the graph. The V6 lineage pattern (*) is the same as V1–V4. (Adapted from Nimmo and Slack, 2009)
The timing of seam cell proliferation and differentiation is regulated by miRNAs
The discovery of miRNA-mediated regulation of gene expression has had a broad impact on biology, including stem cell biology (reviewed in Nimmo and Slack, 2009). miRNAs were first discovered in screens for genes that control developmental timing in the C. elegans seam cell lineages. The founding miRNAs, lin-4 and let-7, control timing and terminal divisions of seam cells by inhibiting translation of several heterochronic target genes (lin-14, lin-28, lin-41, hbl-1, daf-12 and lin-29) at specific developmental stages (Slack and Ruvkun, 1997). These miRNAs, and several of their target genes, are conserved across metazoa. Mutations in let-7 or lin-4 in C. elegans result in seam cell overproliferation (Figure 3). This phenotype may reflect a conserved function in cell proliferation, as downregulation of the let-7 homologue in humans is associated with cancer, and expression of let-7 suppresses tumor formation in lung and breast cancers (Nimmo and Slack, 2009). The cancerous state is often considered to be an atavistic cellular behavior in which an earlier developmental program is inappropriately expressed late in development. It is, therefore, not surprising that heterochronic regulatory genes play important roles in cancer progression.
In the seam cells, the let-7 and lin-4 miRNAs promote differentiation and inhibit seam cell self-renewal. Similarly, the vertebrate equivalents of these miRNAs, let-7 and lin-4/mir-125, are largely absent in embryonic stem (ES) cells and become expressed at later embryonic developmental stages (Nimmo and Slack, 2009). Expression of let-7 and lin-4 in both C. elegans and mammals is reciprocal to that of their target genes lin-28 and lin-41, respectively, and depletion of these miRNAs abrogates the downregulation of target genes that occurs during normal development. LIN-28 is one of the four factors that promotes reprogramming of differentiated human somatic cells into induced pluripotent stem (iPS) cells and is implicated in maintaining pluripotent states based on many studies. LIN-41, another target of let-7, co-operates with LIN-28 to repress let-7 activity in both worms and vertebrates. LIN-41 is an E3-ubiquitin ligase that targets Ago-2/let-7-containing complexes for degradation (Rybak et al., 2009). Based on these studies, it appears that differentiation induced by the downregulation of lin-28 by let-7 is common to worms and humans.
Regulation of seam cell expansion by RUNX and Engrailed-type transcription factors
Runx transcription factors are conserved in metazoa and function as the alpha subunit of an enhancer-binding complex with CBFβ/Bro to promote or repress transcription, depending on cellular context (Nimmo and Woollard, 2008). The C. elegans Runx transcription factor, rnt-1, is required for proliferation of precursor cells in the V and T seam cell lineages (Nimmo and Woollard, 2008)(Figure 3). Loss of bro-1 (the C. elegans CBFβ/Bro homologue) results in the same phenotype and RNT-1 and BRO-1 interact in vitro (Kagoshima et al., 2007). Moreover, overexpression of either rnt-1 or bro-1 results in seam cell hyperplasia. The KIP/CIP family cyclin-dependent kinase inhibitor, CKI-1, which is required for G1 cell-cycle arrest during normal development (Hong et al., 1998), is upregulated in rnt-1 mutants and its depletion rescues the seam cell proliferation defect of rnt-1 mutants. Thus, rnt-1 and bro-1 may function to downregulate cki-1, thereby stimulating cell division.
RUNX1 and RUNX2, two of the three mammalian RUNX homologs, have also been shown to regulate expression of cyclin-dependent kinase inhibitors; however, the outcome of this regulation varies in mammals (Nimmo and Woollard, 2008). In osteoprogenitor cells, Runx2 expression is downregulated during proliferative stages of the cell cycle and Runx2 acts to suppress proliferation. In contrast, in both endothelial cells and hematopoietic cells, Runx levels are high during the G1/S transition and Runx promotes proliferation similar to its action in C. elegans seam cells. Thus, Runx genes can act as key regulatory switches of the proliferation vs. differentiation decision. Studies of the simplified C. elegans seam cell lineage, involving only one Runx gene and a small number of cells may help to uncover the cell context-specific function of these transcription factors in stem cell development.
The seam cell under-proliferation phenotype observed in Runx mutants is also seen in mutants for the C. elegans engrailed homolog ceh-16 (Huang et al., 2009). In ceh-16 mutants, seam cells do not undergo the self-renewal expansion division at the second larval stage but instead divide asymmetrically, ultimately resulting in decreased seam cell number (Figure 3). Overexpression of rnt-1 is able to rescue this ceh-16 mutant phenotype. It is also interesting to note that the Drosophila Runx gene has been shown to repress engrailed in flies (Durst and Hiebert, 2004). However, the reduction in seam cell number in ceh-16(−) mutants does not appear to be related to the defects seen in rnt-1(−) or bro-1(−) mutants, as double mutations of ceh-16(−) with either rnt-1(−) or bro-1(−) further reduce the number of seam cells, implying that these pathways act in parallel. Moreover, the proliferation-promoting pathways involving rnt-1, bro-1, and ceh-16 differ in that rnt-1(−) and bro-1(−) mutants can be rescued by inhibiting negative regulators of the cell cycle (lin-35 or fzr-1), whereas loss of these factors in a ceh-16(−) mutant does not restore seam cell number. In addition the RNT-1/BRO-1 complex does not interact with CEH-16 in vitro. These findings suggest that the function of ceh-16 in promoting proliferation may not reflect its direct action on regulation of the cell cycle.
The human engrailed homolog En2 rescues the ceh-16 underproliferation phenotype in C. elegans demonstrating functional conservation between these proteins. In mammals, En2 normally promotes survival of neural cell types during development (Morgan, 2006). However, En2 expression is also associated with many cancers and ectopic expression of En2 promotes proliferation and inhibits differentiation in epithelial cell lines (Martin et al., 2005). Overexpression of either ceh-16 or En2 results in extra symmetric seam cell divisions in C. elegans (Huang et al., 2009) Intriguingly, this effect was observed only if ceh-16 was expressed during a short time window between late L1 and early L2 larval stages; however, overexpression of En2 at any stage causes seam cell hyperplasia. Engrailed or its targets could potentially be used as a means of increasing the pool of stem cells, a possibility that has not been explored to our knowledge.
Seam cell expansion and proliferation are regulated by Wnt signaling
Analysis of the seam cell lineages has also provided insights into the role of Wnt-type signaling in stem cell lineage patterns. Wnt signaling has been intensively investigated in human stem cells and appears to be critical for both maintenance and differentiation (Kleber and Sommer, 2004) Loss of the Tcf transcription factor, which mediates Wnt signaling, leads to depletion of stem cells in the human colon, and hyperactivation of Wnt signaling through loss of the inhibitory component, APC, or gain of β-catenin activity, is associated with excessive stem cells and cancer (van de Wetering et al., 2002). Wnts are also implicated in the normal expansion of neural stem cells, as reduced Wnt function results in loss of midbrain structures and their overexpression leads to expansion of brain owing to an increase in the neural stem cell pool (Ikeya et al., 1997; Chenn and Walsh, 2002). It is also clear, however, that Wnts have a role in promoting fate decisions in the same cells. For example Wnts function in a stage-specific manner in cortical neuron development, promoting precursor cell expansion at early stages and at later stages inhibiting pluripotency and instructing neural precursor cells to commit to the neural lineage (Hirabayashi et al., 2004).
Wnt signaling appears to control seam cell expansion in C. elegans, as hyperactivation of Wnt signaling by inactivation of apr-1, the C. elegans APC gene, causes excessive division in the seam cell lineages. In apr-1(RNAi) animals, the normal asymmetric seam cell division at the early L4 stage is transformed into a self-renewal expansion division, resulting in seam cell hyperplasia (Huang et al., 2009)(Figure 3). An extra symmetric division of V5.p is also frequently observed in these mutants. This results in formation of two postdeirids with both of the daughters developing into neuronal structures. Thus, in seam cell lineages, Wnt signaling appears to select between symmetric, expansion divisions and asymmetric, differentiation-promoting divisions, consistent with its role in asymmetric cell division throughout much of C. elegans development (Eisenmann, 2005) and also similar to some of its functions in vertebrate development.
It has been suggested that formation of the postdeirid in the hermaphrodite and the sensory rays in the male is dependent on contacts between seam cells, and that ablation of contacting cells activates the canonical Wnt signaling pathway, leading to an increase in mab-5 expression and transformation to symmetric cell division. The phenotype of apr-1-disrupted mutants supports these observations; however, the absence of self-renewal expansion division in ceh-16 mutants is not rescued in canonical Wnt ligand or in β-catenin mutants. This apparent discrepancy was resolved by the finding that the non-canonical Wnt-signaling (Wnt/MAPK) pathway functions in asymmetric cell division in this lineage. The non-canonical pathway in C. elegans, which uses wrm-1/β-catenin, pop-1/Tcf and the kinase lit-1/Nlk, controls most asymmetric divisions in C. elegans and is distinct from non-canonical Wnt signaling in other species (Eisenmann, 2005). In contrast to the canonical Wnt pathway, in which a Tcf transcription factor is activated by Wnt signal, in the non-canonical pathway, POP-1 acts normally as a transcriptional repressor and Wnt signaling leads to its displacement from the nucleus of one daughter cell. Mutations in wrm-1, lit-1, or pop-1 partially rescue the absence of the postdeirid in ceh-16 mutants by restoring asymmetric cell divisions. In addition, asymmetric nuclear localization of POP-1 is disrupted in a ceh-16 mutant. These findings made by Huang et al. (2009) suggested that non-canonical Wnt signaling is necessary for the asymmetric division resulting in postdeirid formation. The Wnt/MAPK pathway is also required at the L4 stage seam cell asymmetric division. Kanamori et al. (2008) found that this asymmetry is regulated by the phospholipase IPLA-1, which may function in membrane trafficking and polarization of β-catenin/WRM-1. This possibility is supported by the observation that ipla-1 mutants are suppressed by mutations in mon-2 (which encodes an Arf GEF-like protein) and tbc-3 (which encodes Rab GSP), both of which function in endosome-to-Golgi traffic.
Wnt signaling may provide an attractive means of coaxing cells into adopting different fates or maintaining pluripotency because it involves signaling molecules that can be administered externally to cells, thereby circumventing the need for genetic manipulation. For example, it has been shown that treatment of ESCs with an inhibitor of GSK3β is sufficient to sustain pluripotency and self-renewal by activating the canonical Wnt pathway (Sato et al., 2004). However Wnt signaling in mammalian cells can have various outcomes that likely reflect temporal or spatial differences in cells (for review see Kleber and Sommer, 2004). Determining how these differences regulate a cells response to Wnt signal will be assisted by in vivo, temporally and spatially modulated developmental programs, such as that seen with the C. elegans seam cells.
Summary: relationship between seam cells and stem cell lineages
Seam cell division patterns resemble the division patterns of stem cells and data continue to accumulate on the similarity of genes that control the timing and outcome of these divisions in both cases. In both seam cells and stem cells, the transition from pluripotency and proliferation to differentiation appears to be tuned by miRNAs and is regulated by RUNX transcription factors. It is also clear that Wnt signaling is important for determining the outcome of a division and creating asymmetry in both seam and stem cells. A major challenge will be to understand how these pathways are linked and coordinated, providing a broad picture of how self-renewal and maintenance division are regulated. The principles gleaned from studying the seam cell lineages, in which single cell resolution, simplified genetics, and methods for manipulation are available, may help to guide studies in mammalian cells and lead to a better understanding of stem cell development and creation of methods for stem cell manipulation.
IV. Pluripotency, Transdifferentiation, and Developmental Reprogramming
In the foregoing sections, we described mechanisms that specify bona fide C. elegans stem cells, the germline stem cells, and considered regulation of self-renewing stem cell-like lineages that arise during post-embryonic development of the seam cells. A key problem in stem cell biology is to understand how stem cells maintain multipotentiality and avoid committing to a unique differentiated fate. With the knowledge of the molecular regulatory circuitry that allows stem cells to remain pluripotent, and how this circuitry becomes modified when cells switch from a multipotential state to a committed pathway of differentiation, it may be possible to produce stem cells from virtually any differentiated cell type. A major technological advance in this field was achieved with the discovery of methods for generating induced pluripotent (iPS) cells from fully differentiated cells of adult animals (Takahashi and Yamanaka, 2006; Huangfu et al., 2008; Nakagawa et al., 2008) by expressing just a single factor, Oct4 (Kim et al., 2009a; Kim et al., 2009b), in neural stem cells. While production of iPS cells is a powerful method that promises to lead to the creation of a great variety of new stem cell types for clinical applications, there is much to be learned about the molecular processes that distinguish multipotential stem cells and their committed, differentiated descendants and how such processes may be reversed or altered, resulting in cellular transdifferentiation. This information may make it possible to reprogram fully differentiated cells into new cell types which can then populate functioning tissues. Using C. elegans, it has been possible to analyze the steps that occur during natural transdifferentiation, regulatory events that repress somatic differentiation and maintain pluripotency during germline development, and molecular processes that convert pluripotent progenitors to cells of restricted differentiation potential during embryogenesis.
Natural transdifferentiation during C. elegans development: an epithelial-to-neural transformation
While the phenomenon of transdifferentiation has been known for many years (Pritchard et al., 1978; Okada, 1980; Slack and Tosh, 2001; Slack, 2007), very few examples of bona fide transdifferentiation have been observed during normal animal development. For example, the process of larval metamorphosis, in which entire new tissues or organs are born in a differentiated animal, does not necessitate that fully differentiated cells of one type become transformed into cells of an altogether different cell type; rather, the newly differentiated tissues often arises from uncommitted progenitor or stem cells that had been set aside at earlier developmental stages (Cohen, 1993). Genuine transdifferentiation is most convincingly demonstrated only when a cell is observed continuously and found to convert from one fully differentiated cell type to another. Such observations require cell lineage analysis, a technique that has been accomplished par excellence with C. elegans, in which the history and fate of every somatic cell observed continuously through development have been documented. The conversion of the “Y” rectal epithelial cell into a neuron, called PDA, during C. elegans larval development (Jarriault et al., 2008), is among the clearest examples of a true transdifferentiation event in any animal.
The Y cell, born during embryogenesis, is an essential structural cell of the rectal epithelium in the newly hatched larva (Sulston and Horvitz, 1977; Sulston et al., 1980; White, 1988), and displays all the characteristic morphological features of a fully differentiated epithelial cell (Jarriault et al., 2008). As post-embryonic development proceeds through the L2 stage, the Y cell withdraws from the rectum, migrates away from the rectal region, and apparently transdifferentiates into the PDA motor neuron, which projects processes that synapse with other neurons by the L3 larval stage. Significantly, this transdifferentiation process occurs in the absence of division of the Y cell; rather, it results from the complete remodeling of an extant post-mitotic epithelial cell into a neuron. Another lineally unrelated cell, P12.pa, which is born shortly before this event is initiated, replaces the departing Y cell in the rectal epithelium. Concomitant with its morphological transdifferentiation, this cell loses expression of all tested epithelial-specific markers, including proteins involved in epithelial polarity and transcription factors that specify epithelial fates, by the time it has become a neuron. Moreover, the trans-fated PDA neuron expresses a number of neuron-specific genes that are not detectable in the Y cell before this event has occurred. The transformation of a Y epithelial cell into a PDA motor neuron has been divided into five phases: establishment of Y cell identity, establishment of competence to undergo transdifferentiation, retraction from the rectum, migration of Y from the rectum, and establishment of PDA identity. Dissecting the molecular and cellular events that direct the transformation of Y into PDA will help to unveil the mechanisms underlying natural transdifferentiation and cellular plasticity.
Several experiments in which local cellular interactions were interrupted, including ablation of cells surrounding the Y cell, failed to prevent the Y-to-PDA transdifferentiation event; moreover, blocking the normal anteriorward migration of the erstwhile Y cell does not abrogate its transdifferentiation into a PDA neuron. Thus, region-specific cues do not appear to be essential for this event to proceed, which may be directed exclusively by cell-intrinsic programs. Indeed, two transcription factors, the homebox protein EGL-5/Abd-B, and the zinc finger transcription factor SEM-4/spalt are required for transdifferentiation of Y into PDA: loss of either in a Y cell causes it to remain epithelial throughout development and prevents formation of the PDA neuron. However, while essential, these factors are not sufficient for transdifferentiation and are therefore not specific to the process, as they are also expressed in other epithelial cells that do not transdifferentiate. Thus, EGL-5 and SEM-4 appear to function early by making Y competent for transformation into PDA. Though no evidence for cell-cell signaling in the transdifferentiation event has been obtained, the identity of the Y cell is specified in part by a LIN-12/Notch-dependent lateral interaction between the progenitors of Y and a neuron called DA9. Removal of lin-12 function causes what would normally have become the Y cell instead to adopt the fate of the DA9 neuron (Greenwald et al., 1983). This signaling through LIN-12/Notch also appears to impart to the Y cell the ability to transdifferentiate during later development, as constitutively active LIN-12 results in production of an extra Y cell, from what would normally be the DA9 neuron. Along with the normal Y cell, this ectopic Y transdifferentiates into PDA later in development. It is not yet known whether LIN-12 endows the Y cell with the general susceptibility for transdifferentiation or instead specifically allows it to become only PDA as development ensues. However, it is not the case that all conditions in which an extra Y-like cell is generated allow Y to transdifferentiate into PDA: in egl-38(−) and mab-9(−) mutants, in which the rectal cells U and B, respectively, are converted to Y-like cells based on morphology and marker expression, extra PDAs are not generated. Thus, other conditions must be met to predispose the Y cell to undergo later transdifferentiation that are distinct from those that dictate at least some Y-specific characteristics. Finally, it is unclear whether Y dedifferentiates before it becomes PDA or instead undergoes direct transdifferentiation without a dedifferentiation intermediate. If the former is correct, Y may be subject to reprogramming into other cell types when presented with another lineage-specific fate directing factor, a possibility that has not yet been tested.
Control of totipotency and exclusion of somatic development in the germline
Germ cells are capable of giving rise to an entire organism and hence may be viewed as the ultimate totipotent stem cells. While the mechanisms that specify the embryonic germline progenitors, the primordial germ cells (PGCs), vary between species, they share several common characteristics. The most striking of these commonalities is the transcriptional quiescence of early germ cells mediated by direct repression of RNA Pol II, as seen in C. elegans and Drosophila, or by silencing mediated in part by the transcriptional repressor Blimp1(Nakamura and Seydoux, 2008). These regulatory mechanisms result in total repression of all somatic programs of differentiation and also contribute to the maintenance of totipotency of the PGCs.
In addition to regulation at the transcriptional level, post-transcriptional regulatory processes, mediated in part by miRNAs, are of crucial importance to the development of germline stem cells (Gangaraju and Lin, 2009), as has also shown to be the case with embryonic stem cells (Gangaraju and Lin, 2009; Qi et al., 2009). Repression of let-7 miRNA function by the LIN-28 RNA binding protein, as first observed in the stem cell-like divisions of the C. elegans seam cells, was also found to be critical for germ cell specification and maintenance (West et al., 2009); moreover, LIN-28 overexpression is associated with increased proliferation, leading to germ cell tumors (Viswanathan et al., 2009; West et al., 2009). This system is also relevant to reactivation of pluripotency in differentiated cells: LIN-28, when coexpressed with the transcription factors OCT4, SOX2, and NANOG is sufficient to reprogram human fibroblasts into ES–like induced pluripotent stem cells (iPS). Likewise, totipotent PGCs continue to express the pluripotency-determining genes Nanog and Oct4 (Seydoux and Braun, 2006). This close relationship between germline and embryonic stem cells is underscored by expression profile studies of mRNAs and miRNAs (Zovoilis et al., 2008), which suggest that ES cells are most closely related to GSCs (Zwaka and Thomson, 2005). Further, ES cells, germ cells and PGCs all can give rise to teratomas, rare germline tumors containing differentiated somatic cells representative of all three germ layers, suggesting a conserved pluripotency module.
The discovery of teratomas in the C. elegans germline offers new perspectives into the molecular circuitry that regulates totipotency. Ciosk et al. discovered that GLD-1 and MEX-3, two RNA-binding translational repressors involved in regulating GSC proliferation, also function together critically in maintaining germ cell totipotency by repressing somatic differentiation programs. Simultaneous removal of both factors results in the appearance of germline teratomas, containing differentiated cells characteristic of all three germ layers. Thus, translational repression of presumably many target transcripts by MEX-3 and GLD-1 is a key mechanism that prevents somatic differentiation, and maintains pluripotency, in the developing germline.
MEX-3 and GLD-1 function in part by repressing translation of somatically active transcription factors, such as the mesectoderm-promoting caudal-type homedomain protein PAL-1 (Ciosk et al., 2006). In addition, elimination of GLD-1 function results in inappropriate translation of its direct target, the message encoding CYC-E (cyclin E), resulting in mitotic reentry of normally meiotic cells and premature activation of somatic gene expression (Biedermann et al., 2009). Activation of somatic gene expression is not a consequence of inappropriate cell proliferation per se, but is a consequence of activation of the CYC-E/CDK2 complex, as somatic gene transcripts continue to be expressed even when germ cell proliferation is blocked. A similar requirement for a cyclin/cdk complex, specifically cyclinA2/CDK2, has been reported for transcriptional activation of embryonic gene expression in the one-cell mouse embryo (Hara et al., 2005).
How might inappropriate activation of cyclin/CDK complexes result in somatic gene expression in the germline? While cyclins and cdks were initially identified as cell cycle regulators, the known repertoire of their action has greatly expanded. Cyclin/cdk complexes have been shown to influence transcription by directly regulating specific transcription factors and generally by phosphorylating Ser2 and Ser5 in the carboxy-terminal domain (CTD) of RNA Pol II. These complexes have also been implicated in regulating splicing through the phosphorylation of splicing machinery components. These observations indicate that coordination of the regulatory machinery for the cell cycle, translation, and transcription is critical for regulating germ cell totipotency and for repression of somatic differentiation in the germline.
The multipotency → commitment transition in the early C. elegans embryo
Given the close relationship between pluripotent stem cells and early embryonic cells, much can be learned about the mechanisms controlling stem cell pluripotency by studying the plasticity of cells in early embryos. In the early C. elegans embryo, progenitor cells with distinct lineage identities are born at each round of cell division, beginning at the first cleavage (Laufer et al., 1980; Sulston et al., 1983); there are no fields of equivalent self-renewing cells that are subsequently induced to adopt more specialized fates. Moreover, the stereotypic pattern of cell divisions and fates reveal a deterministic program of development (Sulston et al., 1983). Combined with the lack of a system for cell culture, C. elegans embryonic development might therefore seem to be poorly suited for the study of stem cell pluripotency and self-renewal. However, a number of studies have demonstrated that, while specification of distinct differentiation pathways apparently occurs very early in embryogenesis, cells nonetheless maintain pluripotency throughout much of the first half of embryogenesis as evidenced by their ability to be reprogrammed into alternative pathways of development when forced to express cell fate regulators that normally function in different lineages. Later in embryonic development, cells become restricted in their capacity to become redirected down alternative developmental pathways and firmly commit to their appropriate differentiation programs. This transition from multipotency to a state of restricted differentiation in the C. elegans embryo provides a useful system for probing mechanisms that control pluripotency.
The evidence for a dramatic switch from a developmentally plastic to a committed state during embryogenesis has been obtained in a variety of cell fate reprogramming experiments. The five somatic and one germline “founder cells,” each which transmits a distinct cell cycle clock to its descendants and gives rise to a unique set of cell types and lineages, are born during the first several embryonic cell divisions. Ectopic expression of the END-1 GATA-type transcription factor, which is normally expressed only in the E founder cell lineage shortly after it is established (Zhu et al., 1997), causes virtually all cells in the early embryo to become reprogrammed and to differentiate into intestine (endoderm) (Zhu et al., 1998). In extreme examples, virtually every cell in terminal embryos differentiates into an intestinal cell, at the expense of all mesodermal and ectodermal differentiation. Thus, although only the E lineage normally engenders endoderm, every somatic cell in the early embryo has the capacity to do so. Similarly, ectopic expression of the PHA-4/FoxA transcription factor, which is essential to establish organ-specific identify of the various cells of the pharynx (Horner et al., 1998a; Kalb et al., 1998), leads to ectopic production of pharyngeal tissues and repression of non-pharyngeal differentiation; however, in contrast to the experiments with END-1, only a subset of embryonic cells are responsive to PHA-4-directed reprogramming (Horner et al., 1998b). Subsequent experiments have revealed that early embryonic cells can also be reprogrammed into epidermal and other epithelial cells (Gilleard and McGhee, 2001; Quintin et al., 2001), as well as body wall muscle cells (Fukushige and Krause, 2005), when challenged with the appropriate cell-fate-promoting transcription factors. The competence of cells of the early C. elegans embryo to be redirected from their normal developmental fates into cell types representing all three germ layers demonstrates that they are genuinely pluripotent.
Studies in which the ectopic expression of specification factors is temporally varied demonstrated that embryonic cells are competent to be reprogrammed only during a restricted window of time, beyond which they become refractory to reprogramming (Horner et al., 1998a; Zhu et al., 1998; Gilleard and McGhee, 2001; Quintin et al., 2001; Fukushige and Krause, 2005). This window of competency lasts until approximately 3 hours after the first cell division, during which dramatic changes in gene expression are occurring as a result of the widespread mobilization of differentiation programs throughout the embryo (Baugh et al., 2003; Yuzyuk et al., 2009). This period of developmental plasticity ends shortly after the founder cell identities are established and lineages become restricted to undergo differentiation into particular cell types (e.g., nervous system, epidermis, muscle, pharynx, and intestine). It is not known whether this developmental plasticity ends at precisely the same time in each lineage or, as seems likely based on the differences in their times of birth and lineage specification, the boundaries of the competency window vary somewhat between founder cell lineages. Further, it is not known whether the ability to become reprogrammed to a variety of cell types is equally distributed among the founder cell lineages. Nevertheless, the observation that the window of susceptibility to reprogramming is similar regardless of the cell fate specification factor used points to existence of a major transition from a pluripotent, developmentally plastic state to a committed state during embryogenesis.
The observation that the period of developmental plasticity correlates with the time during which restricted differentiation patterns are being specified in the embryo raises the possibility that the complex transcriptional regulatory networks activated by cell fate specification factors per se result in the pluripotency → commitment switch. Such gene regulatory networks are known to include positive transcriptional feedback regulatory loops that “lock down” differentiation pathways during specification (e.g. Davidson and Erwin, 2006; McGhee, 2007) and the lockdown of one gene regulatory state might be sufficient to prevent the activation of others. If this is the case, then eliminating the function of genes essential for the specification of a cell type might be expected to cause the descendant cells to remain pluripotent. A recent study suggests that this may not be the case at least for pharyngeal cell fates: for example, elimination of the pha-4/FoxA, critical for pharynx specification, did not result in an extension of the window during which the affected embryonic cells are capable of being reprogrammed (Yuzyuk et al., 2009). Thus, there may exist global mechanisms controlling pluripotency that are independent of the known cell fate regulatory programs.
Such a global mechanism controlling pluripotency might be expected to reside at the level of changes in chromatin organization. Indeed, Yuzyuk et al. found that, concomitant with the embryonic pluripotent → commitment transition, nuclear chromatin appears to become more condensed, based both on alterations in the morphological appearance of extrachromosomal transgenic elements and on the propinquity of endogenous chromosomal genes detected by DNA in situ hybridization of chromosomes. Thus, chromatin appears to undergo dramatic reorganization as cells lose pluripotency during this transition.
One component that might be expected to direct changes in chromatin organization during the transition from pluripotency to commitment is the polycomb repressor complex, which was first identified in D. melanogaster based on its role in maintaining differentiation (Lewis, 1978; Struhl, 1981), and which has subsequently been shown to be important for pluripotency. In both mouse and human stem cell models (Pasini et al., 2008), the polycomb complex prevents differentiation of ES cells by repressing genes involved in differentiation and also functions in the stem cell niche in plants (Xu and Shen, 2008). Members of the polycomb complex have been shown to be indispensable for the self-renewal of neural progenitor cells (Roman-Trufero et al., 2009) and restrict differentiation potential in neural cell lineages (Hirabayashi et al., 2009). Studies by Yuzyuk et al. in C. elegans showed that components of the PRC2 polycomb complex, which methylates histone H3, is not required to maintain developmental plasticity or specification per se, but is necessary for the switch from pluripotency to commitment.
Transcriptional profiling experiments revealed a number of genes expressed in early C. elegans embryos that are downregulated during the pluripotency → commitment transition. Mutants lacking MES-2, the PRC2 repressor complex protein E(z), which has also been implicated in repression of HOX gene expression (Ross and Zarkower, 2003), show prolonged expression of these normally early-specific genes, demonstrating that PRC2 is required for their downregulation. Further, genes associated with ongoing differentiation that are normally expressed late in the transition fail to reach normal expression levels at this time. These findings suggest that mes-2(−) embryos retain characteristics of early embryos that have not yet committed to particular differentiation pathways. Indeed, the transition from a pluripotent condition into a committed state fails to occur at the normal time in these mutants: cells remain competent to become reprogrammed by heterologous cell fate regulators of muscle and intestinal differentiation beyond the normal window of plasticity (Yuzyuk et al., 2009). In addition, the mutants do not undergo the same extent of chromatin condensation during the pluripotency → commitment transition, and this apparently less condensed chromatin is associated with higher transcriptional activity based on the presence of phosphoserine2 on the RNA pol II CTD. These findings argue that PC2-directed remodeling of chromatin is responsible for the transition from a plastic, pluripotent state to a committed state of differentiation.
Future prospects
C. elegans is likely to continue to be a useful tool for illuminating the biology of stem cells, not only by providing a system for analyzing bona fide stem cells generated by the germline stem cell niche, but also by making it possible to dissect the parameters that control stem cell-like lineage patterns, as we have described with the larval seam cells, and the processes that convert precursor cells in the early embryo from a multipotential state into a committed pathway of differentiation. Several of the attributes that have made C. elegans a popular model system for the reductionist piece-meal deconstruction of signaling pathways and developmental mechanisms are also conducive for a “systems” level analysis of regulatory circuits in conjunction with macroenvironmental influences that result in strongly canalized developmental programs regulating pluripotency and totipotency of the soma and germline respectively.
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Angelo G, Van Gilst MR. Starvation Protects Germline Stem Cells and Extends Reproductive Longevity in C. elegans. Science. 2009 doi: 10.1126/science.1178343. [DOI] [PubMed] [Google Scholar]
- Ariz M, Mainpal R, Subramaniam K. C. elegans RNA-binding proteins PUF-8 and MEX-3 function redundantly to promote germline stem cell mitosis. Dev Biol. 2009;326:295–304. doi: 10.1016/j.ydbio.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin J, Kimble J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell. 1987;51:589–599. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Slonim DK, Brown EL, Hunter CP. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development. 2003;130:889–900. doi: 10.1242/dev.00302. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Berry LW, Westlund B, Schedl T. Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development. 1997;124:925–936. doi: 10.1242/dev.124.4.925. [DOI] [PubMed] [Google Scholar]
- Biedermann B, Wright J, Senften M, Kalchhauser I, Sarathy G, Lee MH, Ciosk R. Translational repression of cyclin E prevents precocious mitosis and embryonic gene activation during C. elegans meiosis. Dev Cell. 2009;17:355–364. doi: 10.1016/j.devcel.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Byrd DT, Kimble J. Scratching the niche that controls Caenorhabditis elegans germline stem cells. Semin Cell Dev Biol. 2009 doi: 10.1016/j.semcdb.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- Ciosk R, DePalma M, Priess JR. Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science. 2006;311:851–853. doi: 10.1126/science.1122491. [DOI] [PubMed] [Google Scholar]
- Cohen SM, editor. Imaginal disc development. Cold Spring Harbor Press; 1993. [Google Scholar]
- Crittenden SL, Leonhard KA, Byrd DT, Kimble J. Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol Biol Cell. 2006;17:3051–3061. doi: 10.1091/mbc.E06-03-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- Davis MW, Morton JJ, Carroll D, Jorgensen EM. Gene activation using FLP recombinase in C. elegans. PLoS Genet. 2008;4:e1000028. doi: 10.1371/journal.pgen.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond-Barbosa D, Spradling AC. Stem cells and their progeny respond to nutritional changes during Drosophila oogenesis. Dev Biol. 2001;231:265–278. doi: 10.1006/dbio.2000.0135. [DOI] [PubMed] [Google Scholar]
- Durst KL, Hiebert SW. Role of RUNX family members in transcriptional repression and gene silencing. Oncogene. 2004;23:4220–4224. doi: 10.1038/sj.onc.1207122. [DOI] [PubMed] [Google Scholar]
- Eisenmann DM. Wnt signaling. WormBook. 2005:1–17. doi: 10.1895/wormbook.1.7.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen-Campen B, Schwager EE, Extavour CG. The molecular machinery of germ line specification. Mol Reprod Dev. 2009;77:3–18. doi: 10.1002/mrd.21091. [DOI] [PubMed] [Google Scholar]
- Extavour CG, Akam M. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- Fukushige T, Krause M. The myogenic potency of HLH-1 reveals wide-spread developmental plasticity in early C. elegans embryos. Development. 2005;132:1795–1805. doi: 10.1242/dev.01774. [DOI] [PubMed] [Google Scholar]
- Fukuyama M, Rougvie AE, Rothman JH. C. elegans DAF-18/PTEN mediates nutrient-dependent arrest of cell cycle and growth in the germline. Curr Biol. 2006;16:773–779. doi: 10.1016/j.cub.2006.02.073. [DOI] [PubMed] [Google Scholar]
- Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleard JS, McGhee JD. Activation of hypodermal differentiation in the Caenorhabditis elegans embryo by GATA transcription factors ELT-1 and ELT-3. Mol Cell Biol. 2001;21:2533–2544. doi: 10.1128/MCB.21.7.2533-2544.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald IS, Sternberg PW, Horvitz HR. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell. 1983;34:435–444. doi: 10.1016/0092-8674(83)90377-x. [DOI] [PubMed] [Google Scholar]
- Hansen D, Hubbard EJ, Schedl T. Multi-pathway control of the proliferation versus meiotic development decision in the Caenorhabditis elegans germline. Dev Biol. 2004;268:342–357. doi: 10.1016/j.ydbio.2003.12.023. [DOI] [PubMed] [Google Scholar]
- Hansen D, Schedl T. The regulatory network controlling the proliferation-meiotic entry decision in the Caenorhabditis elegans germ line. Curr Top Dev Biol. 2006;76:185–215. doi: 10.1016/S0070-2153(06)76006-9. [DOI] [PubMed] [Google Scholar]
- Hara KT, Oda S, Naito K, Nagata M, Schultz RM, Aoki F. Cyclin A2-CDK2 regulates embryonic gene activation in 1-cell mouse embryos. Dev Biol. 2005;286:102–113. doi: 10.1016/j.ydbio.2005.07.012. [DOI] [PubMed] [Google Scholar]
- Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Gao D, Lambie EJ, Kimble J. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development. 1994;120:2913–2924. doi: 10.1242/dev.120.10.2913. [DOI] [PubMed] [Google Scholar]
- Hess KR, Varadhachary GR, Taylor SH, Wei W, Raber MN, Lenzi R, Abbruzzese JL. Metastatic patterns in adenocarcinoma. Cancer. 2006;106:1624–1633. doi: 10.1002/cncr.21778. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Itoh Y, Tabata H, Nakajima K, Akiyama T, Masuyama N, Gotoh Y. The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development. 2004;131:2791–2801. doi: 10.1242/dev.01165. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Suzki N, Tsuboi M, Endo TA, Toyoda T, Shinga J, Koseki H, Vidal M, Gotoh Y. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63:600–613. doi: 10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- Hirsh D, Oppenheim D, Klass M. Development of the reproductive system of Caenorhabditis elegans. Dev Biol. 1976;49:200–219. doi: 10.1016/0012-1606(76)90267-0. [DOI] [PubMed] [Google Scholar]
- Hong Y, Roy R, Ambros V. Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development. 1998;125:3585–3597. doi: 10.1242/dev.125.18.3585. [DOI] [PubMed] [Google Scholar]
- Horner MA, Quintin S, Domeier ME, Kimble J, Labouesse M, Mango SE. pha-4, an HNF-3 homolog, specifies pharyngeal organ identity in Caenorhabditis elegans. Genes Dev. 1998a;12:1947–1952. doi: 10.1101/gad.12.13.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner MA, Quintin S, Domeier ME, Kimble J, Labouesse M, Mango SE. pha-4, an HNF-3 homolog, specifies pharyngeal organ identity in Caenorhabditis elegans. Genes Dev. 1998b;12:1947–1952. doi: 10.1101/gad.12.13.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HJ, Drummond-Barbosa D. Insulin levels control female germline stem cell maintenance via the niche in Drosophila. Proc Natl Acad Sci U S A. 2009;106:1117–1121. doi: 10.1073/pnas.0809144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HJ, LaFever L, Drummond-Barbosa D. Diet controls normal and tumorous germline stem cells via insulin-dependent and -independent mechanisms in Drosophila. Dev Biol. 2008;313:700–712. doi: 10.1016/j.ydbio.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Tian E, Xu Y, Zhang H. The C. elegans engrailed homolog ceh-16 regulates the self-renewal expansion division of stem cell-like seam cells. Dev Biol. 2009 doi: 10.1016/j.ydbio.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- Hubbard EJ. Caenorhabditis elegans germ line: a model for stem cell biology. Dev Dyn. 2007;236:3343–3357. doi: 10.1002/dvdy.21335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard EJ, Greenstein D. The Caenorhabditis elegans gonad: a test tube for cell and developmental biology. Dev Dyn. 2000;218:2–22. doi: 10.1002/(SICI)1097-0177(200005)218:1<2::AID-DVDY2>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Ikeya M, Lee SM, Johnson JE, McMahon AP, Takada S. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- Issigonis M, Tulina N, de Cuevas M, Brawley C, Sandler L, Matunis E. JAK-STAT signal inhibition regulates competition in the Drosophila testis stem cell niche. Science. 2009;326:153–156. doi: 10.1126/science.1176817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo-Lambert A, Ellefson M, Villeneuve AM, Engebrecht J. Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev Biol. 2007;308:206–221. doi: 10.1016/j.ydbio.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Schwab Y, Greenwald I. A Caenorhabditis elegans model for epithelial-neuronal transdifferentiation. Proc Natl Acad Sci U S A. 2008;105:3790–3795. doi: 10.1073/pnas.0712159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong PY, Kwon MS, Joo HJ, Paik YK. Molecular time-course and the metabolic basis of entry into dauer in Caenorhabditis elegans. PLoS ONE. 2009;4:e4162. doi: 10.1371/journal.pone.0004162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk LC, Kimble J. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development. 1998;125:1803–1813. doi: 10.1242/dev.125.10.1803. [DOI] [PubMed] [Google Scholar]
- Kagoshima H, Nimmo R, Saad N, Tanaka J, Miwa Y, Mitani S, Kohara Y, Woollard A. The C. elegans CBFbeta homologue BRO-1 interacts with the Runx factor, RNT-1, to promote stem cell proliferation and self-renewal. Development. 2007;134:3905–3915. doi: 10.1242/dev.008276. [DOI] [PubMed] [Google Scholar]
- Kalb JM, Lau KK, Goszczynski B, Fukushige T, Moons D, Okkema PG, McGhee JD. pha-4 is Ce-fkh-1, a fork head/HNF-3α, β, γ homolog that functions in organogenesis of the C. elegans pharynx. Development. 1998;125:2171–2180. doi: 10.1242/dev.125.12.2171. [DOI] [PubMed] [Google Scholar]
- Kim JB, Greber B, Arauzo-Bravo MJ, Meyer J, Park KI, Zaehres H, Scholer HR. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009a;461:649–643. doi: 10.1038/nature08436. [DOI] [PubMed] [Google Scholar]
- Kim JB, Sebastiano V, Wu G, Arauzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D, Meyer J, Hubner K, Bernemann C, Ortmeier C, Zenke M, Fleischmann BK, Zaehres H, Scholer HR. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009b;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Kimble J. Alterations in cell lineage following laser ablation of cells in the somatic gonad of Caenorhabditis elegans. Dev Biol. 1981;87:286–300. doi: 10.1016/0012-1606(81)90152-4. [DOI] [PubMed] [Google Scholar]
- Kimble J, Crittenden SL. Germline proliferation and its control. WormBook. 2005:1–14. doi: 10.1895/wormbook.1.13.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J, Crittenden SL. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu Rev Cell Dev Biol. 2007;23:405–433. doi: 10.1146/annurev.cellbio.23.090506.123326. [DOI] [PubMed] [Google Scholar]
- Kleber M, Sommer L. Wnt signaling and the regulation of stem cell function. Curr Opin Cell Biol. 2004;16:681–687. doi: 10.1016/j.ceb.2004.08.006. [DOI] [PubMed] [Google Scholar]
- LaFever L, Drummond-Barbosa D. Direct control of germline stem cell division and cyst growth by neural insulin in Drosophila. Science. 2005;309:1071–1073. doi: 10.1126/science.1111410. [DOI] [PubMed] [Google Scholar]
- Laufer JS, Bazzicalupo P, Wood WB. Segregation of developmental potential in early embryos of Caenorhabditis elegans. Cell. 1980;19:569–577. doi: 10.1016/s0092-8674(80)80033-x. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Li Y, Minor NT, Park JK, McKearin DM, Maines JZ. Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc Natl Acad Sci U S A. 2009;106:9304–9309. doi: 10.1073/pnas.0901452106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Lu X, Kang Y. Organotropism of breast cancer metastasis. J Mammary Gland Biol Neoplasia. 2007;12:153–162. doi: 10.1007/s10911-007-9047-3. [DOI] [PubMed] [Google Scholar]
- Maciejowski J, Ugel N, Mishra B, Isopi M, Hubbard EJ. Quantitative analysis of germline mitosis in adult C. elegans. Dev Biol. 2006;292:142–151. doi: 10.1016/j.ydbio.2005.12.046. [DOI] [PubMed] [Google Scholar]
- Martin NL, Saba-El-Leil MK, Sadekova S, Meloche S, Sauvageau G. EN2 is a candidate oncogene in human breast cancer. Oncogene. 2005;24:6890–6901. doi: 10.1038/sj.onc.1208840. [DOI] [PubMed] [Google Scholar]
- McGhee JD. The C. elegans intestine. WormBook. 2007:1–36. doi: 10.1895/wormbook.1.133.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern M, Voutev R, Maciejowski J, Corsi AK, Hubbard EJ. A “latent niche” mechanism for tumor initiation. Proc Natl Acad Sci U S A. 2009;106:11617–11622. doi: 10.1073/pnas.0903768106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, Seydoux G. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol. 2008;18:1476–1482. doi: 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Morrison SJ. Diverse mechanisms regulate stem cell self-renewal. Current Opinion in Cell Biology. 2004;16:700–707. doi: 10.1016/j.ceb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Morgan R. Engrailed: complexity and economy of a multi-functional transcription factor. FEBS Lett. 2006;580:2531–2533. doi: 10.1016/j.febslet.2006.04.053. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajan S, Govindan JA, McGovern M, Hubbard EJ, Greenstein D. MSP and GLP-1/Notch signaling coordinately regulate actomyosin-dependent cytoplasmic streaming and oocyte growth in C. elegans. Development. 2009;136:2223–2234. doi: 10.1242/dev.034603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Seydoux G. Less is more: specification of the germline by transcriptional repression. Development. 2008;135:3817–3827. doi: 10.1242/dev.022434. [DOI] [PubMed] [Google Scholar]
- Narbonne P, Roy R. Inhibition of germline proliferation during C. elegans dauer development requires PTEN, LKB1 and AMPK signalling. Development. 2006a;133:611–619. doi: 10.1242/dev.02232. [DOI] [PubMed] [Google Scholar]
- Narbonne P, Roy R. Regulation of germline stem cell proliferation downstreamof nutrient sensing. Cell Div. 2006b;1:29. doi: 10.1186/1747-1028-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo R, Woollard A. Worming out the biology of Runx. Dev Biol. 2008;313:492–500. doi: 10.1016/j.ydbio.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Nimmo RA, Slack FJ. An elegant miRror: microRNAs in stem cells, developmental timing and cancer. Chromosoma. 2009;118:405–418. doi: 10.1007/s00412-009-0210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystul TG, Spradling AC. Breaking out of the mold: diversity within adult stem cells and their niches. Curr Opin Genet Dev. 2006;16:463–468. doi: 10.1016/j.gde.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Ohlstein B, Kai T, Decotto E, Spradling A. The stem cell niche: theme and variations. Current Opinion in Cell Biology. 2004;16:693–699. doi: 10.1016/j.ceb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Okada TS. Cellular metaplasia or transdifferentiation as a model for retinal cell differentiation. Curr Top Dev Biol. 1980;16:349–380. [PubMed] [Google Scholar]
- Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Agger K, Christensen J, Hansen K, Cloos PA, Helin K. Regulation of stem cell differentiation by histone methyltransferases and demethylases. Cold Spring Harb Symp Quant Biol. 2008;73:253–263. doi: 10.1101/sqb.2008.73.009. [DOI] [PubMed] [Google Scholar]
- Pritchard DJ, Clayton RM, de Pomerai DI. ‘Transdifferentiation’ of chicken neural retina into lens and pigment epithelium in culture: controlling influences. J Embryol Exp Morphol. 1978;48:1–21. [PubMed] [Google Scholar]
- Qi J, Yu JY, Shcherbata HR, Mathieu J, Wang AJ, Seal S, Zhou W, Stadler BM, Bourgin D, Wang L, Nelson A, Ware C, Raymond C, Lim LP, Magnus J, Ivanovska I, Diaz R, Ball A, Cleary MA, Ruohola-Baker H. microRNAs regulate human embryonic stem cell division. Cell Cycle. 2009;8:3729–3741. doi: 10.4161/cc.8.22.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao L, Lissemore JL, Shu P, Smardon A, Gelber MB, Maine EM. Enhancers of glp-1, a gene required for cell-signaling in Caenorhabditis elegans, define a set of genes required for germline development. Genetics. 1995;141:551–569. doi: 10.1093/genetics/141.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintin S, Michaux G, McMahon L, Gansmuller A, Labouesse M. The Caenorhabditis elegans gene lin-26 can trigger epithelial differentiation without conferring tissue specificity. Dev Biol. 2001;235:410–421. doi: 10.1006/dbio.2001.0294. [DOI] [PubMed] [Google Scholar]
- Rangan P, DeGennaro M, Jaime-Bustamante K, Coux RX, Martinho RG, Lehmann R. Temporal and spatial control of germ-plasm RNAs. Curr Biol. 2009;19:72–77. doi: 10.1016/j.cub.2008.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P, DeGennaro M, Lehmann R. Regulating gene expression in the Drosophila germ line. Cold Spring Harb Symp Quant Biol. 2008;73:1–8. doi: 10.1101/sqb.2008.73.057. [DOI] [PubMed] [Google Scholar]
- Roman-Trufero M, Mendez-Gomez HR, Perez C, Hijikata A, Fujimura Y, Endo T, Koseki H, Vicario-Abejon C, Vidal M. Maintenance of undifferentiated state and self-renewal of embryonic neural stem cells by Polycomb protein Ring1B. Stem Cells. 2009;27:1559–1570. doi: 10.1002/stem.82. [DOI] [PubMed] [Google Scholar]
- Ross JM, Zarkower D. Polycomb group regulation of Hox gene expression in C. elegans. Dev Cell. 2003;4:891–901. doi: 10.1016/s1534-5807(03)00135-7. [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Hadian K, Smirnova L, Wulczyn EA, Michel G, Nitsch R, Krappmann D, Wulczyn FG. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat Cell Biol. 2009;11:1411–1420. doi: 10.1038/ncb1987. [DOI] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Braun RE. Pathway to totipotency: lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Shcherbata HR, Ward EJ, Fischer KA, Yu JY, Reynolds SH, Chen CH, Xu P, Hay BA, Ruohola-Baker H. Stage-specific differences in the requirements for germline stem cell maintenance in the Drosophila ovary. Cell Stem Cell. 2007;1:698–709. doi: 10.1016/j.stem.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X, Xu X, Fedotov A, Kelly WG, Maine EM. Regulation of heterochromatin assembly on unpaired chromosomes during caenorhabditis elegans meiosis by components of a small RNA-mediated pathway. PLoS Genet. 2009;5:e1000624. doi: 10.1371/journal.pgen.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R, Weng C, Yu J, Xie T. eIF4A controls germline stem cell self-renewal by directly inhibiting BAM function in the Drosophila ovary. Proc Natl Acad Sci U S A. 2009;106:11623–11628. doi: 10.1073/pnas.0903325106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenghui H, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- Slack F, Ruvkun G. Temporal pattern formation by heterochronic genes. Annu Rev Genet. 1997;31:611–634. doi: 10.1146/annurev.genet.31.1.611. [DOI] [PubMed] [Google Scholar]
- Slack JM. Metaplasia and transdifferentiation: from pure biology to the clinic. Nat Rev Mol Cell Biol. 2007;8:369–378. doi: 10.1038/nrm2146. [DOI] [PubMed] [Google Scholar]
- Slack JM, Tosh D. Transdifferentiation and metaplasia--switching cell types. Curr Opin Genet Dev. 2001;11:581–586. doi: 10.1016/s0959-437x(00)00236-7. [DOI] [PubMed] [Google Scholar]
- Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- Song X, Xie T. DE-cadherin-mediated cell adhesion is essential for maintaining somatic stem cells in the Drosophila ovary. Proc Natl Acad Sci U S A. 2002;99:14813–14818. doi: 10.1073/pnas.232389399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Zhu CH, Doan C, Xie T. Germline stem cells anchored by adherens junctions in the Drosophila ovary niches. Science. 2002;296:1855–1857. doi: 10.1126/science.1069871. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Nystul T, Lighthouse D, Morris L, Fox D, Cox R, Tootle T, Frederick R, Skora A. Stem cells and their niches: integrated units that maintain Drosophila tissues. Cold Spring Harb Symp Quant Biol. 2008;73:49–57. doi: 10.1101/sqb.2008.73.023. [DOI] [PubMed] [Google Scholar]
- Struhl G. A gene product required for correct initiation of segmental determination in Drosophila. Nature. 1981;293:36–41. doi: 10.1038/293036a0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Albertson DG, Thomson JN. The Caenorhabditis elegans male: postembryonic development of nongonadal structures. Dev Biol. 1980;78:542–576. doi: 10.1016/0012-1606(80)90352-8. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Szakmary A, Cox DN, Wang Z, Lin H. Regulatory relationship among piwi, pumilio, and bag-of-marbles in Drosophila germline stem cell self-renewal and differentiation. Curr Biol. 2005;15:171–178. doi: 10.1016/j.cub.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tulina N, Matunis E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science. 2001;294:2546–2549. doi: 10.1126/science.1066700. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, Tjon-Pon-Fong M, Moerer P, van den Born M, Soete G, Pals S, Eilers M, Medema R, Clevers H. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O’Sullivan M, Lu J, Phillips LA, Lockhart VL, Shah SP, Tanwar PS, Mermel CH, Beroukhim R, Azam M, Teixeira J, Meyerson M, Hughes TP, Llovet JM, Radich J, Mullighan CG, Golub TR, Sorensen PH, Daley GQ. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vought VE, Ohmachi M, Lee M-H, Maine EM. EGO-1, a putative RNA-directed RNA polymerase, promotes germline proliferation in parallel with GLP-1/Notch signaling and regulates the spatial organization of nuclear pore complexes and germline P granules in Caenorhabditis elegans. Genetics:genetics. 2005;105:042135. doi: 10.1534/genetics.105.042135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voutev R, Hubbard EJ. A “FLP-Out” system for controlled gene expression in Caenorhabditis elegans. Genetics. 2008;180:103–119. doi: 10.1534/genetics.108.090274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Singh SR, Zheng Z, Oh SW, Chen X, Edwards K, Hou SX. Rap-GEF signaling controls stem cell anchoring to their niche through regulating DE-cadherin-mediated cell adhesion in the Drosophila testis. Dev Cell. 2006;10:117–126. doi: 10.1016/j.devcel.2005.11.004. [DOI] [PubMed] [Google Scholar]
- West JA, Viswanathan SR, Yabuuchi A, Cunniff K, Takeuchi A, Park IH, Sero JE, Zhu H, Perez-Atayde A, Frazier AL, Surani MA, Daley GQ. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 2009;460:909–913. doi: 10.1038/nature08210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. The Anatomy. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1988. pp. 81–122. [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Xie T, Spradling AC. A niche maintaining germ line stem cells in the Drosophila ovary. Science. 2000;290:328–330. doi: 10.1126/science.290.5490.328. [DOI] [PubMed] [Google Scholar]
- Xu L, Shen WH. Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr Biol. 2008;18:1966–1971. doi: 10.1016/j.cub.2008.11.019. [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Chen D, Duan R, Xia L, Wang J, Qurashi A, Jin P. Argonaute 1 regulates the fate of germline stem cells in Drosophila. Development. 2007;134:4265–4272. doi: 10.1242/dev.009159. [DOI] [PubMed] [Google Scholar]
- Yu JY, Reynolds SH, Hatfield SD, Shcherbata HR, Fischer KA, Ward EJ, Long D, Ding Y, Ruohola-Baker H. Dicer-1-dependent Dacapo suppression acts downstream of Insulin receptor in regulating cell division of Drosophila germline stem cells. Development. 2009;136:1497–1507. doi: 10.1242/dev.025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzyuk T, Fakhouri TH, Kiefer J, Mango SE. The polycomb complex protein mes-2/E(z) promotes the transition from developmental plasticity to differentiationin C. elegans embryos. Dev Cell. 2009;16:699–710. doi: 10.1016/j.devcel.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Fukushige T, McGhee JD, Rothman JH. Reprogramming of early embryonic blastomeres into endodermal progenitors by a Caenorhabditis elegans GATA factor. Genes Dev. 1998;12:3809–3814. doi: 10.1101/gad.12.24.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Hill RJ, Heid PJ, Fukuyama M, Sugimoto A, Priess JR, Rothman JH. end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes and Development. 1997;11 doi: 10.1101/gad.11.21.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovoilis A, Nolte J, Drusenheimer N, Zechner U, Hada H, Guan K, Hasenfuss G, Nayernia K, Engel W. Multipotent adult germline stem cells and embryonic stem cells have similar microRNA profiles. Mol Hum Reprod. 2008;14:521–529. doi: 10.1093/molehr/gan044. [DOI] [PubMed] [Google Scholar]
- Zwaka TP, Thomson JA. A germ cell origin of embryonic stem cells? Development. 2005;132:227–233. doi: 10.1242/dev.01586. [DOI] [PubMed] [Google Scholar]