Abstract
Argentine population genetic structure was examined using a set of 78 ancestry informative markers (AIMs) to assess the contributions of European, Amerindian, and African ancestry in 94 individuals members of this population. Using the Bayesian clustering algorithm STRUCTURE, the mean European contribution was 78%, the Amerindian contribution was 19.4%, and the African contribution was 2.5%. Similar results were found using weighted least mean square method: European, 80.2%; Amerindian, 18.1%; and African, 1.7%. Consistent with previous studies the current results showed very few individuals (four of 94) with greater than 10% African admixture. Notably, when individual admixture was examined, the Amerindian and European admixture showed a very large variance and individual Amerindian contribution ranged from 1.5 to 84.5% in the 94 individual Argentine subjects. These results indicate that admixture must be considered when clinical epidemiology or case control genetic analyses are studied in this population. Moreover, the current study provides a set of informative SNPs that can be used to ascertain or control for this potentially hidden stratification. In addition, the large variance in admixture proportions in individual Argentine subjects shown by this study suggests that this population is appropriate for future admixture mapping studies.
Keywords: ancestry informative markers, admixture, population stratification
Determining the composition of different populations using DNA polymorphisms is receiving considerable attention for the value in elucidating the history of populations, identifying ethnicity, and the potential application in assessing phenotypic relationships (Rosenberg et al., 2002; Burchard et al., 2003; Shriver et al., 2003; Bonilla et al., 2004; Yang et al., 2005). In particular, the examination of complex genetic diseases is potentially greatly enhanced by understanding population genetic structure and substructure. This is perhaps particularly true of population groups that reflect admixtures between peoples of different continents where population stratification in case-control studies can lead to erroneous conclusions if admixture is not examined (Pritchard et al., 2000b; Satten et al., 2001; Kittles et al., 2002; Hoggart et al., 2003; Hinds et al., 2004; Tang et al., 2005). Newly developed methods of admixture mapping also suggest that recently admixed populations can also be useful for defining the chromosomal location of ancestry associated traits (Hoggart et al., 2004; Montana and Pritchard, 2004; Patterson et al., 2004; Seldin et al., 2004; Zhang et al., 2004; Zhu et al., 2004; McKeigue, 2005). These admixture mapping methods that rely on identifying linkage of a trait to ancestry at specific chromosomal locations has shown potential value in recent studies of both hypertension and multiple sclerosis in the African American admixed population (Reich et al., 2005; Zhu et al., 2005).
The current study was undertaken to assess the population genetic structure and potential value of complex disease studies in one such admixed population, the Argentine population that has not been fully characterized. The study has used a set of particularly informative DNA polymorphisms that have been termed Ancestry Informative Markers (AIMs) because of their information content for distinguishing particular ancestral groups that correspond to continental populations. Previous studies by our group and others have assessed population genetic structure in several “Hispanic” population groups using these and similar genomic nuclear DNA polymorphisms (Bonilla et al., 2004; Collins-Schramm et al., 2004; Yang et al., 2005). These studies have shown rather disparate characteristics of these different groups as might be anticipated by the differencesin the history of European invasionand migration, as well as the transatlantic slave trade. For Mexican American and Mexican populations the Amerindian ancestry contribution is around 50% (modestly higher in Mexican) and the African contribution is less than 5%, in contrast for Puerto Ricans the Amerindian component is about 12% and the African component is over 20% (Yang et al., 2005).
Previous studies have examined the overall admixture in the Argentine population using a very limited set of markers that included only nine blood group antigens and the Km/Gm haplotypes (Avena et al., 2001). Recently, population genetic structure in an Argentine population from Buenos Aires was examined to assess the African contribution to individual subjects using 12 markers that discriminated between African and European ancestry, but this study did not examine the individual Amerindian contribution (Fejerman et al., 2005). The current results complement these studies by confirming the limited African contribution to the nonindigenous Argentine population and providing an accurate assessment of Amerindian contribution as well as the individual variation in admixture proportions using a much larger and informative panel of SNP AIMs.
MATERIALS AND METHODS
Populations samples
European American (EURA) (88 subjects), Mexican American (MAM) (89 subjects), Mexican (MXN) (94 subsjects), Amerindian (AMI) (70 subjects), West African (AFR) (95 subjects), were included in this study. These populations were based on self-identified ethnic affiliation. The MAM were recruited from California; the AMI subjects were self identified as Mayan (Kachiquel language group) and were recruited in Chimaltenango, Guatemala; the West African subjects were collected in Nigeria and were from the Edo (Bini) ethnic group; and the Mexican subjects were recruited from Mexico City, all as previously reported (Collins-Schramm et al., 2004; Yang et al., 2005). Blood- or buccal-cell samples were obtained from all individuals, according to protocols and informed-consent procedures approved by institutional review boards, and were labeled with an anonymous code number. The Argentine subjects were recruited in a multicenter collaborative network aimed at collecting individuals with systemic lupus erythematosus (SLE) and matched controls. The subjects studied here were all healthy unrelated controls matched with the SLE individuals for age, sex, and ethnicity. The ascertainment criteria was the same in each region and resulted in a similar age (mean age 63 years) and gender (95% female) distribution at each recruitment center. The individuals originate from Buenos Aires (15 subjects), Córdoba (33 subjects), Santa Fé (33 subjects), Mar del Plata (11 subjects), and La Plata (2 subjects) and not only Buenos Aires as in the work of Avena et al. (2001) or Fejerman et al. (2005). Dr. Bernardo Pons-Estel coordinated the collection of samples in Argentina.
Laboratory analysis
Ancestry informative markers
Single nucleotide polymorphisms (SNPs) with large frequency differences between continental populations were selected based on previous studies showing Fst values >0.4 for either European/African or European/Amerindian analyses (Collins-Schramm et al., 2004; Yang et al., 2005, and Seldin unpublished). Fst values for European/African, European/Amerindian, and African/Amerindian and allele frequencies for all ancestry informative markers (AIMs) in the current study are provided in Table 1. These studies included 29 SNPs not reported in our previous studies (see notation in Table 1). Additional marker information including primers is provided in supplemental Table 1.
TABLE 1.
Summary of ancestry informative marker genotyping results
| Allele frequencya |
Fstb |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs number | Chr | Mbc | EURA | AMI | WAFR | ARG | MAM | MXN | EURA/ WAFR |
EURA/ AMI |
WAFR/ AMI |
Newd | P1e | P2f |
| 424436 | 1 | 8.0 | 0.00 | 0.72 | 0.06 | 0.08 | 0.28 | 0.46 | 0.05 | 0.73 | 0.64 | 1 | 1 | |
| 7504 | 1 | 26.9 | 0.22 | 0.95 | 0.36 | 0.45 | 0.63 | 0.78 | 0.04 | 0.70 | 0.54 | 1 | 1 | |
| 1931059 | 1 | 35.0 | 0.81 | 0.23 | 0.92 | 0.65 | 0.56 | 0.47 | 0.05 | 0.50 | 0.67 | 1 | 1 | |
| 596985 | 1 | 64.0 | 0.97 | 0.99 | 0.13 | 0.99 | 0.93 | 0.94 | 0.83 | 0.00 | 0.84 | 1 | ||
| 5025718 | 1 | 120.2 | 0.87 | 0.88 | 0.03 | 0.89 | 0.77 | 0.79 | 0.84 | 0.00 | 0.85 | 1 | ||
| 2274533 | 1 | 148.2 | 0.85 | 0.07 | 0.49 | 0.71 | 0.37 | 0.30 | 0.24 | 0.75 | 0.34 | 1 | 1 | |
| 2065160 | 1 | 201.5 | 0.91 | 0.08 | 0.60 | 0.81 | 0.49 | 0.39 | 0.22 | 0.81 | 0.43 | 1 | 1 | |
| 883399 | 2 | 9.6 | 0.59 | 0.01 | ND | 0.51 | 0.29 | ND | ND | 0.57 | ND | 1 | 1 | |
| 300152 | 2 | 17.9 | 0.85 | 0.08 | 0.77 | 0.62 | 0.41 | 0.34 | 0.01 | 0.73 | 0.64 | 1 | 1 | |
| 2384319 | 2 | 26.1 | 0.07 | 0.83 | 0.06 | 0.23 | 0.46 | 0.48 | 0.00 | 0.73 | 0.76 | 1 | 1 | |
| 3768641 | 2 | 72.3 | 0.08 | 0.00 | 0.99 | 0.11 | 0.08 | 0.06 | 0.91 | 0.07 | 0.99 | 1 | ||
| 260714 | 2 | 109.0 | 0.88 | 0.02 | ND | 0.69 | 0.39 | ND | ND | 0.85 | ND | 1 | ||
| 2305260 | 2 | 128.8 | 0.77 | 0.14 | ND | 0.69 | 0.44 | ND | ND | 0.57 | ND | 1 | 1 | |
| 901304 | 2 | 163.2 | 0.86 | 0.26 | ND | 0.73 | 0.49 | ND | ND | 0.54 | ND | 1 | 1 | |
| 9847748 | 3 | 69.0 | 0.63 | 0.04 | ND | 0.51 | 0.45 | ND | ND | 0.55 | ND | 1 | 1 | |
| 13069719 | 3 | 71.6 | 0.85 | 0.09 | 0.84 | 0.65 | 0.47 | 0.48 | 0.00 | 0.73 | 0.72 | 1 | 1 | |
| 1352158 | 3 | 98.8 | 0.81 | 0.07 | 0.30 | 0.77 | 0.45 | 0.30 | 0.42 | 0.71 | 0.14 | 1 | 1 | |
| 6437783 | 3 | 109.7 | 0.13 | 0.86 | 0.31 | 0.29 | 0.44 | 0.56 | 0.08 | 0.69 | 0.46 | 1 | 1 | |
| 2165139 | 3 | 140.7 | 0.88 | 0.04 | 0.98 | 0.84 | 0.52 | 0.32 | 0.07 | 0.82 | 0.94 | 1 | 1 | |
| 9290363 | 3 | 170.5 | 0.09 | 0.10 | 0.88 | 0.10 | 0.10 | 0.05 | 0.78 | 0.00 | 0.76 | 1 | ||
| 11723316 | 4 | 184.8 | 0.40 | 0.97 | ND | 0.57 | 0.75 | ND | ND | 0.54 | ND | 1 | 1 | |
| 814597 | 5 | 10.5 | 0.88 | 0.30 | 0.76 | 0.80 | 0.55 | 0.47 | 0.05 | 0.52 | 0.34 | 1 | 1 | |
| 35395 | 5 | 34.0 | 0.06 | 0.97 | 0.83 | 0.26 | 0.56 | 0.67 | 0.74 | 0.91 | 0.10 | 1 | 1 | |
| 1551765 | 5 | 153.2 | 0.23 | 0.84 | ND | 0.24 | 0.55 | ND | ND | 0.54 | ND | 1 | 1 | |
| 262838 | 5 | 169.1 | 0.87 | 0.21 | ND | 0.80 | 0.48 | ND | ND | 0.61 | ND | 1 | 1 | |
| 9356944 | 6 | 24.8 | 1.00 | 0.26 | ND | 0.90 | 0.68 | ND | ND | 0.74 | ND | 1 | ||
| 1266874 | 6 | 51.9 | 0.67 | 0.03 | ND | 0.55 | 0.28 | ND | ND | 0.62 | ND | 1 | 1 | |
| 218867 | 6 | 121.4 | 0.13 | 0.91 | ND | 0.29 | 0.52 | ND | ND | 0.75 | ND | 1 | ||
| 1744173 | 6 | 158.5 | 0.12 | 0.81 | ND | 0.25 | 0.49 | ND | ND | 0.64 | ND | 1 | 1 | |
| 9295009 | 6 | 169.8 | 0.38 | 0.96 | ND | 0.46 | 0.69 | ND | ND | 0.54 | ND | 1 | 1 | |
| 1880550 | 7 | 14.5 | 0.18 | 0.84 | ND | 0.26 | 0.46 | ND | ND | 0.59 | ND | 1 | 1 | |
| 6601288 | 8 | 9.0 | 0.33 | 0.94 | ND | 0.37 | 0.60 | ND | ND | 0.56 | ND | 1 | 1 | |
| 11778591 | 8 | 12.8 | 0.85 | 0.02 | ND | 0.76 | 0.48 | ND | ND | 0.82 | ND | 1 | 1 | |
| 2439522 | 8 | 97.6 | 0.85 | 0.26 | ND | 0.77 | 0.66 | ND | ND | 0.52 | ND | 1 | 1 | |
| 1871534 | 8 | 145.6 | 0.01 | 0.03 | 0.95 | 0.03 | 0.07 | 0.02 | 0.94 | 0.02 | 0.91 | 1 | ||
| 4478653 | 9 | 21.8 | 0.41 | 0.97 | ND | 0.50 | 0.74 | ND | ND | 0.52 | ND | 1 | 1 | |
| 1417999 | 9 | 101.2 | 0.32 | 0.91 | ND | 0.40 | 0.63 | ND | ND | 0.54 | ND | 1 | 1 | |
| 587364 | 9 | 122.8 | 0.86 | 0.44 | 0.03 | 0.84 | 0.60 | 0.53 | 0.83 | 0.32 | 0.41 | 1 | 1 | |
| 1951936 | 10 | 28.4 | 0.85 | 0.06 | 0.29 | 0.62 | 0.48 | 0.37 | 0.48 | 0.77 | 0.16 | 1 | 1 | |
| 10748592 | 10 | 94.9 | 0.74 | 0.03 | ND | 0.63 | 0.45 | ND | ND | 0.69 | ND | 1 | 1 | |
| 1572396 | 10 | 117.3 | 0.70 | 0.06 | 0.36 | 0.64 | 0.39 | 0.31 | 0.20 | 0.59 | 0.22 | 1 | 1 | |
| 6485600 | 11 | 12.2 | 0.31 | 0.98 | ND | 0.38 | 0.59 | ND | ND | 0.65 | ND | 1 | 1 | |
| 1638567 | 11 | 66.9 | 0.98 | 0.36 | ND | 0.82 | 0.53 | ND | ND | 0.61 | ND | 1 | ||
| 2458640 | 11 | 77.7 | 0.26 | 0.87 | ND | 0.38 | 0.47 | ND | ND | 0.55 | ND | 1 | 1 | |
| 533571 | 11 | 100.4 | 0.28 | 0.94 | 0.12 | 0.37 | 0.63 | 0.67 | 0.07 | 0.61 | 0.79 | 1 | 1 | |
| 4936512 | 11 | 119.7 | 0.18 | 0.96 | 0.38 | 0.32 | 0.54 | 0.68 | 0.09 | 0.75 | 0.52 | 1 | 1 | |
| 1648180 | 11 | 127.6 | 0.24 | 0.92 | 0.42 | 0.37 | 0.64 | 0.73 | 0.06 | 0.62 | 0.42 | 1 | 1 | |
| 984303 | 12 | 15.6 | 1.00 | 0.98 | 0.21 | 0.98 | 0.97 | 0.95 | 0.79 | 0.02 | 0.74 | 1 | 1 | |
| 2293048 | 12 | 116.1 | 0.92 | 0.39 | 0.48 | 0.83 | 0.53 | 0.48 | 0.37 | 0.50 | 0.01 | 1 | 1 | |
| 7995033 | 13 | 24.7 | 0.82 | 0.19 | ND | 0.68 | 0.49 | ND | ND | 0.56 | ND | 1 | 1 | |
| 2065982 | 13 | 33.8 | 0.06 | 0.81 | 0.09 | 0.20 | 0.47 | 0.57 | 0.00 | 0.74 | 0.70 | 1 | 1 | |
| 1540979 | 13 | 93.9 | 0.82 | 0.21 | 0.97 | 0.73 | 0.49 | 0.46 | 0.11 | 0.55 | 0.77 | 1 | ||
| 8003430 | 14 | 23.9 | 0.15 | 0.80 | ND | 0.25 | 0.41 | ND | ND | 0.59 | ND | 1 | 1 | |
| 730570 | 14 | 100.2 | 0.87 | 0.10 | ND | 0.68 | 0.49 | ND | ND | 0.74 | ND | 1 | ||
| 2714758 | 15 | 23.0 | 0.98 | 0.99 | 0.13 | 0.94 | 0.92 | 0.95 | 0.84 | 0.00 | 0.84 | 1 | ||
| 1129038 | 15 | 26.0 | 0.27 | 1.00 | ND | 0.65 | 0.84 | ND | ND | 0.66 | ND | 1 | 1 | |
| 1426654 | 15 | 46.2 | 1.00 | 0.05 | 0.02 | 0.87 | 0.50 | 0.38 | 0.98 | 0.96 | 0.01 | 1 | 1 | |
| 11073967 | 15 | 89.4 | 0.63 | 0.01 | ND | 0.48 | 0.27 | ND | ND | 0.60 | ND | 1 | 1 | |
| 9937955 | 16 | 10.9 | 0.23 | 0.91 | ND | 0.32 | 0.56 | ND | ND | 0.64 | ND | 1 | 1 | |
| 1557519 | 16 | 14.2 | 0.06 | 0.09 | 0.94 | 0.07 | 0.22 | 0.14 | 0.87 | 0.00 | 0.84 | 1 | ||
| 6587216 | 17 | 19.2 | 0.80 | 0.19 | 0.72 | 0.69 | 0.48 | 0.42 | 0.01 | 0.53 | 0.42 | 1 | 1 | |
| 2285750 | 17 | 39.5 | 0.17 | 0.90 | ND | 0.20 | 0.43 | ND | ND | 0.70 | ND | 1 | 1 | |
| 7211306 | 17 | 78.0 | 0.28 | 0.90 | 0.29 | 0.47 | 0.60 | 0.73 | 0.00 | 0.55 | 0.54 | 1 | 1 | |
| 953786 | 18 | 19.9 | 0.20 | 0.79 | 0.99 | 0.36 | 0.60 | 0.68 | 0.80 | 0.52 | 0.21 | 1 | 1 | |
| 17638989 | 19 | 12.5 | 0.56 | 0.01 | ND | 0.46 | 0.31 | ND | ND | 0.54 | ND | 1 | 1 | |
| 1418032 | 20 | 2.0 | 0.27 | 0.98 | ND | 0.43 | 0.59 | ND | ND | 0.65 | ND | 1 | 1 | |
| 6086473 | 20 | 8.4 | 0.76 | 0.16 | ND | 0.62 | 0.51 | ND | ND | 0.53 | ND | 1 | 1 | |
| 293553 | 20 | 30.5 | 0.64 | 0.02 | 0.26 | 0.52 | 0.28 | 0.23 | 0.25 | 0.58 | 0.19 | 1 | 1 | |
| 1689045 | 21 | 16.5 | 0.01 | 0.01 | 0.85 | 0.01 | 0.07 | 0.02 | 0.84 | 0.00 | 0.83 | 1 | ||
| 1475930 | 22 | 21.6 | 0.18 | 0.84 | ND | 0.27 | 0.45 | ND | ND | 0.61 | ND | 1 | 1 | |
| 3747295 | × | 17.5 | 0.06 | 0.43 | 0.93 | 0.15 | 0.29 | 0.37 | 0.86 | 0.33 | 0.45 | 1 | 1 | |
| 1978240 | × | 23.6 | 0.72 | 0.51 | 0.03 | 0.68 | 0.55 | 0.56 | 0.68 | 0.08 | 0.48 | 1 | ||
| 734329 | × | 42.4 | 0.87 | 0.16 | 0.34 | 0.73 | 0.44 | 0.36 | 0.45 | 0.67 | 0.07 | 1 | 1 | |
| 5981813 | × | 74.2 | 0.90 | 0.51 | 0.09 | 0.72 | 0.59 | 0.47 | 0.78 | 0.31 | 0.35 | 1 | 1 | |
| 992864 | × | 110.3 | 0.06 | 0.00 | 0.93 | 0.08 | 0.08 | 0.04 | 0.86 | 0.05 | 0.92 | 1 | ||
| 2380316 | × | 117.3 | 0.81 | 0.11 | 0.06 | 0.63 | 0.34 | 0.29 | 0.72 | 0.65 | 0.00 | 1 | 1 | |
| 1867024 | × | 147.7 | 0.06 | 0.01 | 0.87 | 0.06 | 0.07 | 0.02 | 0.79 | 0.02 | 0.84 | 1 | ||
| 762656 | × | 152.7 | 0.82 | 0.10 | 0.27 | 0.66 | 0.36 | 0.30 | 0.46 | 0.68 | 0.09 | 1 | 1 | |
Allele frequency for SNP allele defined in supplemental Table.
Fst between the different continental populations were calculated using the Weir and Cockerham (1984) algorithm.
The megabase (Mb) position based on HG35 build.
SNP AIMs not previously reported in EURA, AMI, and MAM populations.
SNP AIMs used to examine admixture including WAFR contribution.
SNP AIMs used to examine admixture AMI and EURA admixture.
Of the 78 AIMs used in the study, 44 were used in three population analyses of the Argentine samples together with European American (EURA), Amerindian (AMI), Mexican, Mexican American (MAM), and West African (AFR) genotyped with the same markers. These 44 markers were highly informative: EURA/AFR, mean Fst = 0.44; EURA/AMI, mean Fst = 0.47; and AMI/AFR, mean Fst = 0.52. They included substantial numbers of AIMs with Fst > 0.6 (EURA/AFR 19 AIMs; EURA/AMI 20 AIMs; and AMI/AFR 20 AIMs). There was no evidence for linkage disequilibrium (LD) between these markers in each of the populations for this set of SNPs (r2 < 0.2 for all marker pairs on the same chromosome) with the exception of rs4936512/rs1648180 (r2 = 0.36 in AMI).
Of the 78 AIMs, 66 AIMs (mean EURA/AMI Fst = 0.63) were used in two population analyses together with European American, Amerindian, and Mexican American samples genotyped with the same markers (Table 1). This analysis did not include those markers that did not distinguish between European and Amerindian ancestry. For this set of SNPs only three marker pair combinations on the same chromosome showed minimal or moderate LD in any of these populations: (rs6601288/rs11778591, r2 = 0.238 in MA; rs4936512/rs1648180, r2 = 0.36 in AMI; rs1418032/rs6086473, r2 = 0.49 EURA, r2 = 0.48 MA, and ARG, r2 = 0.52).
All genotyping was performed using TaqMan assays (Applied Biosciences) using procedures previously described (Collins-Schramm et al., 2004). Allele frequencies for each population are provided in Table 1. All of the AIMs were in Hardy-Weinberg (H-W) equilibrium in each population group.
Data analyses
Admixture proportions
Population admixture proportions were determined using both 1) the weighted least square method of Long (1991) applied in the program ADMIX.PAS, and 2) by utilizing the Bayesian clustering algorithms developed by Pritchard and applied in the program STRUCTURE v2.1 (Pritchard et al., 2000a; Falush et al., 2003). Individual admixture proportions were determined using STRUCTURE 2.1. Population structure was examined using STRUCTURE v2.1. Each STRUCTURE analysis was performed without any prior population assignment and was performed at least five times with similar results using >10,000 replicates and burn-in cycles under the admixture model applying the infer α option with a separate α estimated for each population (where α is the Dirichlet parameter for degree of admixture). Most runs were performed under the λ = 1 option where λ parameterizes the allele frequency prior and is based on the Dirichlet distribution of allele frequencies. The log likelihood of each analysis at varying number of population groups (k) is also estimated in each STRUCTURE analysis. Nearly identical results were observed when markers showing evidence of moderate LD (see above) were excluded or when the linkage option in STRUCTURE was applied.
Fst was determined using Genetix software (Belkhir et al., 2001) that applies the Weir and Cockerham algorithm (Weir and Cockerham, 1984)and H-W examined using Genpop software (http://wbiomed.curtin.edu.au/genepop/). LD was examined using the Genetix software (Belkhir et al., 2001).
RESULTS
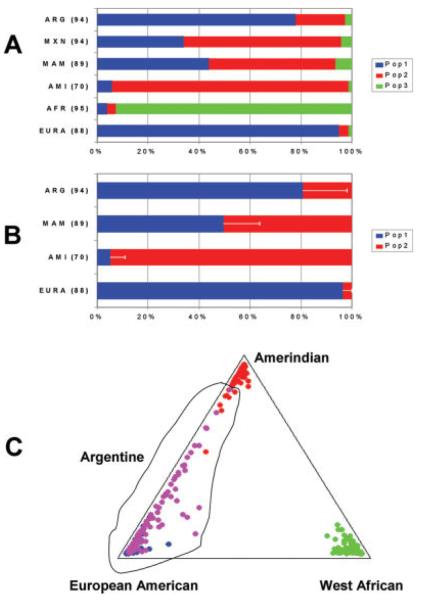
Our initial studies examined Argentine genetic structure using 44 AIMs that distinguish between European, African, and Amerindian origin. The mean contributions of European, African, and Amerindian populations were estimated using previously reported genotypes of these putative parental population groups (Yang et al., 2005). Application of the Bayesian cluster algorithms using STRUCTURE and comparison with Mexican and Mexican American populations are shown in Figure 1A. The mean frequency of the different population groups corresponding to the predominant cluster group in Europe, Amerindian, and Africa were 0.780, 0.195, and 0.025 respectively. Analyses using the weighted least mean squares test based on the allele frequencies in the putative parental populations showed similar results with the following estimated ancestry components in the Argentine subjects: European, 0.802 ± 0.013 (standard error), Amerindian 0.181 ± 0.0132, and African 0.017 ± 0.0077.
Fig. 1.
Estimation of ancestry contributions to the Argentine population using by a Bayesian analysis of population genetic structure. In panel A the results of STRUCTURE analysis (k = 3) using 44 AIMs selected for European, Amerindian, and African information (see methods) are shown. In panel B the results of STRUCTURE analysis using 66 European/Amerindian AIMs (k = 2) is shown. For both panel A and B, the mean population group assignment is shown by color code and the number of subjects in each group is shown in parenthesis. The population groups were European American (EURA), Amerindian (AMI), West African, (AFR), Mexican (MXN), Mexican American (MAM), and Argentine (ARG). In Panel B, the standard deviation is shown for the Amerindian assignment of the individuals. The large standard deviation (SD) observed in the admixed populations are due to the large variation in the individual members of these population groups. The variation in the means between different STRUCTURE runs is <0.5%. In Panel C the same results as panel A are shown for 88 individual EURA subjects (blue), 70 AMI subjects (red), 95 AFR subjects (green), and 94 ARG subjects (magenta) in a triangle plot of the color coded cluster groups corresponding to self identified population affiliations.
The relative contribution of the three putative parental populations in each individual showed a large variation in the ancestral origins and provided an illustration of the relatively small African contribution in the individual Argentine subjects (Fig. 1C). There were only four of 94 Argentine subjects with >10% estimated African contribution and 83 of these Argentine subjects had <5% estimated African contribution. The number of estimated populations was also examined using the STRUCTURE algorithms (see Fig. 2). The estimated probabilities further suggested that there were only two major parental population groups that contribute to the majority of current Argentine individuals.
Fig. 2.
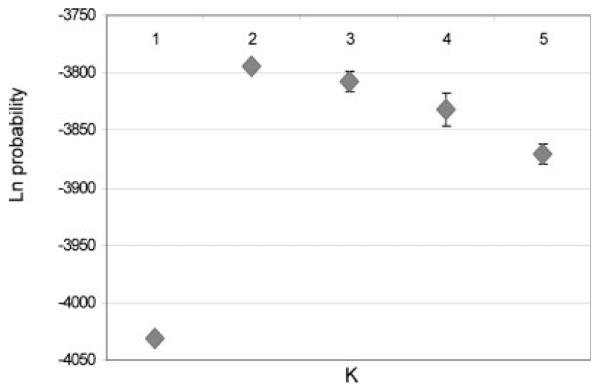
Probability estimations for the number cluster groups (“ancestral or founder populations”) present using AIMs. The ordinate shows the Ln probability (±SD from 5 separate runs) corresponding to the number of clusters (K) when Argentine samples alone are examined with the 44 AIMs selected for distinguishing between European, Amerindian, and African population groups.
To further define the European and Amerindian contribution an additional 34 AIMs were genotyped in the Argentine samples. Analyses were then performed using these 34 markers and 32 markers from the initial set of 44 markers (12 AIMs that did not distinguish between European and Amerindian populations were excluded from this analysis). Analysis using these 66 European/Amerindian AIMs (mean Fst = 0.63) shows an average Amerindian contribution of 19.3% (Fig. 1B). Consistent with these results the estimation by weighted least mean square analysis shows an Amerindian contribution of 18.4%. The results were also similar when the Argentine subjects without evidence of substantial African contribution (<5%) were examined: Amerindian 19.2% using STRUCTURE and 18.3% using the weighted least mean square analysis.
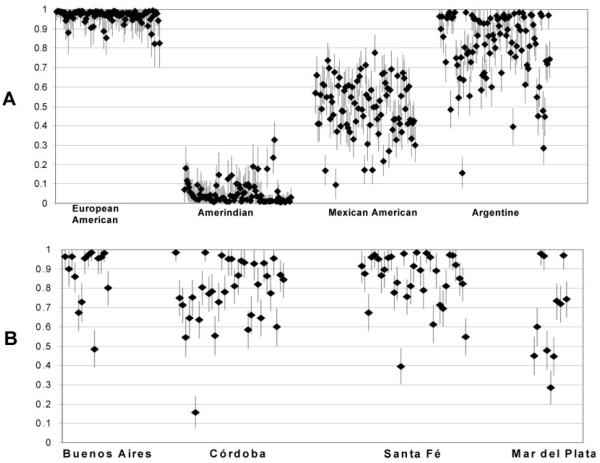
When individual admixture is considered, a large variation in the Amerindian and European contribution is evident by the STRUCTURE analysis (see Fig. 3). The putative Amerindian contribution in individual Argentine subjects varied between 1.5 and 84.5%. For this analysis using STRUCTURE, the mean 90% Bayesian confidence limits for individual assignments was 15% (Fig. 3A) demonstrating that the individual Amerindian/European admixture can be clearly distinguished among these individuals. We also examined that whether there were differences in the Amerindian and European contribution in the largest four recruitment regions used in the current study (Fig. 3B.). Although there was considerable variation in the individual admixture, the mean Amerindian and European contribution varied in these recruitment regions: Buenos Aires, 12.2% Amerindian and 87.8% European; Santa Fé, 15.5% Amerindian and 84.5% European; Córdoba, 22.8% Amerindian and 77.2% European; and Mar del Plata, 33.1% Amerindian and 66.9% European. Using a nonparametric test (two-tailed Wilcoxon-Mann-Whitney U test), the admixture contribution (Amerindian vs. European) was significantly different between Córdoba and Buenos Aires (p = 0.0072), Córdoba and Santa Fé (p = 0.0266), Mar del Plata and Santa Fé (p = 0.0439), and Buenos Aires and Mar del Plata (p = 0.0414).
Fig. 3.
A, Analysis of population genetic structure in individuals of European, Amerindian, Argentine, and Mexican American origin. Each symbol represents an individual examined with 66 selected AIMs and analyzed using the STRUCTURE program under the condition of two populations. The position of the symbol on the Y-axis indicates the most probable admixture from Amerindian (proximity to 0) or the European population (proximity to 1.0) and the error bars show 90% Bayesian confidence limits, e.g., the Argentine subject with the largest Amerindian contribution. In panel B, the Argentine results are shown separated by regional recruitment areas and included Buenos Aires (15 subjects), Córdoba (33 subjects), Santa Fé (33 subjects), and Mar del Plata (11 subjects). Two individuals recruited from La Plata were not included (subject 1, 0.967 European; subject 2, 0.948 European).
DISCUSSION
In this study the admixture characteristics in the current Argentine nonindigenous population were examined using nuclear genome AIMs. The Argentine population like other predominant populations in much of the “New World” is composed of members with varying contributions from three continental groups: European, Amerindian, and African. Similar to Mexican American and Mexican populations the current nonindigenous Argentine population is primarily composed of individuals with substantial admixture components from European and Amerindian ancestry. Our results consistent with another recent study (Fejerman et al., 2005) showed limited African admixture within this population. The vast majority, 83 of the 94 subjects had less than a 5% African contribution. Interestingly, our 94 individuals come from all of Argentina while those of Fejerman et al. (2005) are from Buenos Aires: the similar results show that in all of Argentina there is very low African ancestry inclusion and not just in Buenos Aires where the population has always been considered more European. Using two different analytic methods, one applying Bayesian clustering algorithm and the other a weighted least square, the Amerindian contribution was in agreement (19.5% versus 18.1%). These estimates are somewhat greater than that found in the single previous study (Avena et al., 2001) that used a limited number of blood banking antigens in which the Amerindian contribution was estimated at 15.9%. The current estimates using SNP AIMs distributed throughout the genome provide both a more accurate estimate as well as the ability to examine individual admixture. As shown in the current study the relative contribution of Amerindian ancestry varies greatly between different individuals. The variance of the Amerindian contribution was 3.9% in the current study. Although these conclusions are based on putative representatives of the parental populations, our previous studies showing small variation in AIMs allele frequency distribution in disparate subpopulations suggests that these markers selected for very high frequency differences between continental populations provide a reasonable assessment of admixture despite the uncertainties reflecting the original parental population groups.
Interestingly the European and Amerindian contribution appeared to vary in part with recruitment location. A lower Amerindian contribution and higher European contribution was observed in Buenos Aires and Santa Fe that are provinces with a high incidence of European (mainly Italian and Spaniard) immigration. Córdoba which had a higher Amerindian contribution was an important city during the “Spanish conqueror age” (year 1500) and “mestizos or criollos” are very common in this province. On the other hand, the most important immigrant stream in Santa Fé was of Italian people during the first part of the 20 century (1900–1950). With respect to Mar del Plata, which also showed a higher Amerindian contribution, this settlement was very large and actually called Sierra de los Padres for the Padres, who converted Amerindians to Christianity and there remain many Amerindian surnames in usage (i.e., Millapan, Quitrupán). There is also large migration of people from the neighboring and more Amerindian countries such as Bolivia or Paraguay and the Province of Corrientes. However, it must be cautioned that although some of these differences (e.g., Buenos Aires vs. Córdoba, and Santa Fé vs. Córdoba) were statistically significant, larger sample sizes will be necessary to confirm these regional differences. These studies further suggest that additional sample of other regions within Argentina will be needed to provide a more accurate estimation of the admixture variation and accurate representation of the Argentine population.
In contrast to the Mexican and Mexican American populations the predominant contribution to this Argentine population comes from Europe. However, the Amerindian contribution (~20%) and substantial variation in the Amerindian contribution when individual subjects are examined clearly indicates that admixture must be considered when evaluating traits and candidate genes for traits in this population. The current study provides a list of AIMs that can be used for this purpose.
The present report adds to the number of admixed populations in the “New World” that have been examined for relative continental contributions using a genome-wide panel of AIMs. Previous studies have examined African Americans, Puerto Ricans, Mexican Americans, and Mexicans (Collins-Schramm et al., 2004; Yang et al., 2005). The different frequency of certain diseases or disease endophenotypes in Amerindian or Hispanic populations compared with European populations suggests that these various “Hispanic” populations may be particularly informative for deciphering complex genetic diseases including rheumatoid arthritis, asthma, systemic lupus erythematosus, Type 1 diabetes, Type 2 diabetes, and certain malignancies (Del Puente et al., 1989; Erlich et al., 1993; Molokhia and McKeigue, 2000; Williams et al., 2000; Silman and Pearson, 2002; Collado-Mesa et al., 2004; Pons-Estel et al., 2004; Gonzalez Burchard et al., 2005; Salari et al., 2005). For systemic lupus erythematosus, a recent study of several Hispanic populations suggests that the “Mestizo” populations in both Mexico and Argentina have a higher risk for particular phenotypes seen in this disease including renal disease (Pons-Estel et al., 2004). Although specific studies utilizing methods such as those described in the current will be necessary to define whether ancestry is linked to these traits, the small African contribution to the Mexican and Argentine populations (see Fig. 1) suggests that Amerindian ancestry may be the common factor for these two populations. For asthma, a lower severity of disease is associated with Amerindian ancestry in Mexican Americans (Salari et al., 2005). For Type 1 diabetes there is a much lower incidence of disease in Mexican and Amerindian populations than in European populations (Collado-Mesa et al., 2004), whereas the opposite is suggested for rheumatoid arthritis and Type 2 diabetes(Del Puente et al., 1989; Molokhia and McKeigue, 2000; Williams et al., 2000; Silman and Pearson, 2002). Although environmental, socioeconomic, and other differences will need to be considered in such studies, the difference in ancestry is at least another candidate for elucidating the differences in diseases and disease phenotypes in these different populations.
The large variation in Amerindian contribution suggests that the Argentine population is quite suitable for admixture mapping studies to examine the chromosomal location of various disease phenotypes. Thus, if differences in susceptibility or disease manifestations are in fact linked to ancestry, admixture mapping may help identify the actual gene variations responsible. The difference in relative admixture between Argentine and Mexican or Mexican American populations could also provide an opportunity to examine gene/gene interactions that may differ depending on additional epistatic interactions with the background genetic make-up. Further studies will also be necessary to examine the substructure differences within the European and Amerindian contributions that may also underlie particular phenotypic differences or confound clinical epidemiology and candidate gene studies. However, the major population genetic differences that can result in hidden stratification or ancestry linkage to traits within this population should be discernable using AIMs such as those used in the current study.
Supplementary Material
ACKNOWLEDGMENTS
We thank Adriana I. Scollo, Armando M. Perichon y Mariano C.R. Tenaglia, CEDIM, Diagnóstico Molecular y Forense SRL. Rosario, Argentina for their help in DNA preparation of the Argentine samples. The participants in the collection of Argentine samples included: Pilar C. Marino, M.D., Estela L. Motta, M.D., Servicio de Reumatología, Hospital Interzonal General de Agudos “Dr. Oscar Alende”, Mar del Plata, Argentina; Cristina Drenkard, M.D., Emilia Menso, M.D. Servicio de Reumatología de la UHMI 1, Hospital Nacional de Clínicas, Universidad Nacional de Córdoba, Córdoba, Argentina; Guillermo A. Tate, M.D., Organización Médica de Investigación, Buenos Aires, Argentina; Jose L. Presas, M.D., Hospital General de Agudos Dr. Juán A. Fernandez, Buenos Aires, Argentina; Marcelo Abdala, M.D., Mariela Bearzotti, Ph.D., Facultad de Ciencias Medicas, Universidad Nacional de Rosario y Hospital Provincial del Centenario, Rosario, Argentina; Francisco Caeiro, M.D., Ana Bertoli, M.D., Servicio de Reumatología, Hospital Privado, Centro Medico de Córdoba, Córdoba, Argentina; Susana Roverano, M.D., Hospital José M. Cullen, Santa Fe, Argentina; Cesar E. Graf, M.D., Griselda Buchanan, Ph.D., Estela Bertero, Ph.D., Hospital San Martín, Paraná, Hospital Felipe Heras, Concordia, Entre Ríos, Argentina; Sebastian Grimaudo, Ph.D., Jorge Manni, M.D., Departamento de Inmunología, Instituto de Investigaciones Médicas “ lfredo Lanari”, Buenos Aires, Argentina; Enrique R. Soriano, M.D., Carlos D. Santos, M.D., Sección Reumatología, Servicio de Clínica Medica, Hospital Italiano de Buenos Aires y Fundación Dr. Pedro M. Catoggio para el Progreso de la Reumatología, Buenos Aires, Argentina; Fernando A. Ramos, M.D., Sandra M. Navarro, M.D., Servicio de Reumatología, Hospital Provincial de Rosario, Rosario, Argentina; Marisa Jorfen, M.D., Elisa J. Romero, Ph.D., Servicio de Reumatología Hospital Escuela Eva Perón. Granadero Baigorria, Rosario, Argentina; Juan C. Marcos, M.D., Ana I. Marcos, M.D., Servicio de Reumatología, Hospital Interzonal General de Agudos General San Martín, La Plata; Alicia Eimon, M.D. Centro de Educación Médica e Investigaciones Clínicas (CEMIC), Buenos Aires, Argentina; Cristina G. Battagliotti, M.D., Hospital de Niños Dr. Orlando Alassia, Santa Fe, Argentina.
Grant sponsor: NIH; Grant number: R01 DK071185; Grant sponsors: Torsten and Ragnar Söderbergs Stiftelse, the Swedish Research Council, the Marcus Borsgtröms Foundation, the Swedish Rheumatism Association, the Gustav V: 80-year Jubilee, and Royal Swedish Academy of Sciences.
Footnotes
This article contains supplementary material available via the Internet at http://www.interscience.wiley.com/jpages/0002-9483/suppmat
LITERATURE CITED
- Avena SA, Goicoechea AS, Dugoujon JM, Slepoy MG, Slepoy ES, Carnese FR. Analisis antropogenetico de los aportes indigena y Africano en Muestras Hospitalaras de la ciudad de Buenos Aires. Rev Argent Antropol Biol. 2001;3:79–99. [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX, software under Windows™ for the genetic of populations. Laboratory Genome, Populations, Interactions CNRS UMR 5000, University of Montpellier II; Montpellier, France: 2001. [Google Scholar]
- Bonilla C, Shriver MD, Parra EJ, Jones A, Fernandez JR. Ancestral proportions and their association with skin pigmentation and bone mineral density in Puerto Rican women from New York city. Hum Genet. 2004;115:57–68. doi: 10.1007/s00439-004-1125-7. [DOI] [PubMed] [Google Scholar]
- Burchard EG, Ziv E, Coyle N, Gomez SL, Tang H, Karter AJ, Mountain JL, Perez-Stable EJ, Sheppard D, Risch N. The importance of race and ethnic background in biomedical research and clinical practice. N Engl J Med. 2003;348:1170–1175. doi: 10.1056/NEJMsb025007. [DOI] [PubMed] [Google Scholar]
- Collado-Mesa F, Barcelo A, Arheart KL, Messiah SE. An ecological analysis of childhood-onset type 1 diabetes incidence and prevalence in Latin America. Rev Panam Salud Publica. 2004;15:388–394. doi: 10.1590/s1020-49892004000600004. [DOI] [PubMed] [Google Scholar]
- Collins-Schramm HE, Chima B, Morii T, Wah K, Figueroa Y, Criswell LA, Hanson RL, Knowler WC, Silva G, Belmont JW, Seldin MF. Mexican American ancestry-informative markers: examination of population structure and marker characteristics in European Americans, Mexican Americans, Amerindians and Asians. Hum Genet. 2004;114:263–271. doi: 10.1007/s00439-003-1058-6. [DOI] [PubMed] [Google Scholar]
- Del Puente A, Knowler WC, Pettitt DJ, Bennett PH. High incidence and prevalence of rheumatoid arthritis in Pima Indians. Am J Epidemiol. 1989;129:1170–1178. doi: 10.1093/oxfordjournals.aje.a115238. [DOI] [PubMed] [Google Scholar]
- Erlich HA, Zeidler A, Chang J, Shaw S, Raffel LJ, Klitz W, Beshkov Y, Costin G, Pressman S, Bugawan T, Rotter JI. HLA class II alleles and susceptibility and resistance to insulin dependent diabetes mellitus in Mexican-American families. Nat Genet. 1993;3:358–364. doi: 10.1038/ng0493-358. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejerman L, Carnese FR, Goicoechea AS, Avena SA, Dejean CB, Ward RH. African ancestry of the population of Buenos Aires. Am J Phys Anthropol. 2005;128:164–170. doi: 10.1002/ajpa.20083. [DOI] [PubMed] [Google Scholar]
- Gonzalez Burchard E, Borrell LN, Choudhry S, Naqvi M, Tsai HJ, Rodriguez-Santana JR, Chapela R, Rogers SD, Mei R, Rodriguez-Cintron W, Arena JF, Kittles R, Perez-Stable EJ, Ziv E, Risch N. Latino populations: a unique opportunity for the study of race, genetics, and social environment in epidemiological research. Am J Public Health. 2005;95:2161–2168. doi: 10.2105/AJPH.2005.068668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds DA, Stokowski RP, Patil N, Konvicka K, Kershenobich D, Cox DR, Ballinger DG. Matching strategies for genetic association studies in structured populations. Am J Hum Genet. 2004;74:317–325. doi: 10.1086/381716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, McKeigue PM. Control of confounding of genetic associations in stratified populations. Am J Hum Genet. 2003;72:1492–1504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggart CJ, Shriver MD, Kittles RA, Clayton DG, McKeigue PM. Design and analysis of admixture mapping studies. Am J Hum Genet. 2004;74:965–978. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittles RA, Chen W, Panguluri RK, Ahaghotu C, Jackson A, Adebamowo CA, Griffin R, Williams T, Ukoli F, Adams-Campbell L, Kwagyan J, Isaacs W, Freeman V, Dunston GM. CYP3A4-V and prostate cancer in African Americans: causal or confounding association because of population stratification? Hum Genet. 2002;110:553–560. doi: 10.1007/s00439-002-0731-5. [DOI] [PubMed] [Google Scholar]
- Long JC. The genetic structure of admixed populations. Genetics. 1991;127:417–428. doi: 10.1093/genetics/127.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeigue PM. Prospects for admixture mapping of complex traits. Am J Hum Genet. 2005;76:1–7. doi: 10.1086/426949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molokhia M, McKeigue P. Risk for rheumatic disease in relation to ethnicity and admixture. Arthritis Res. 2000;2:115–125. doi: 10.1186/ar76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montana G, Pritchard JK. Statistical tests for admixture mapping with case-control and cases-only data. Am J Hum Genet. 2004;75:771–789. doi: 10.1086/425281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR, Hauser SL, Smith MW, O’Brien SJ, Altshuler D, Daly MJ, Reich D. Methods for high-density admixture mapping of disease genes. Am J Hum Genet. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons-Estel BA, Catoggio LJ, Cardiel MH, Soriano ER, Gentiletti S, Villa AR, Abadi I, Caeiro F, Alvarellos A, Alarcon-Segovia D. The GLADEL multinational Latin American prospective inception cohort of 1,214 patients with systemic lupus erythematosus: ethnic and disease heterogeneity among “Hispanics”. Medicine (Baltimore) 2004;83:1–17. doi: 10.1097/01.md.0000104742.42401.e2. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000a;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am J Hum Genet. 2000b;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Patterson N, De Jager PL, McDonald GJ, Waliszewska A, Tandon A, Lincoln RR, DeLoa C, Fruhan SA, Cabre P, Bera O, Semana G, Kelly MA, Francis DA, Ardlie K, Khan O, Cree BA, Hauser SL, Oksenberg JR, Hafler DA. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat Genet. 2005;37:1113–1118. doi: 10.1038/ng1646. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Salari K, Choudhry S, Tang H, Naqvi M, Lind D, Avila PC, Coyle NE, Ung N, Nazario S, Casal J, Torres-Palacios A, Clark S, Phong A, Gomez I, Matallana H, Perez-Stable EJ, Shriver MD, Kwok PY, Sheppard D, Rodriguez-Cintron W, Risch NJ, Burchard EG, Ziv E. Genetic admixture and asthma-related phenotypes in Mexican American and Puerto Rican asthmatics. Genet Epidemiol. 2005;29:76–86. doi: 10.1002/gepi.20079. [DOI] [PubMed] [Google Scholar]
- Satten GA, Flanders WD, Yang Q. Accounting for unmeasured population substructure in case-control studies of genetic association using a novel latent-class model. Am J Hum Genet. 2001;68:466–477. doi: 10.1086/318195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin MF, Morii T, Collins-Schramm HE, Chima B, Kittles R, Criswell LA, Li H. Putative ancestral origins of chromosomal segments in individual African Americans: implications for admixture mapping. Genome Res. 2004;14:1076–1084. doi: 10.1101/gr.2165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriver MD, Parra EJ, Dios S, Bonilla C, Norton H, Jovel C, Pfaff C, Jones C, Massac A, Cameron N, Baron A, Jackson T, Argyropoulos G, Jin L, Hoggart CJ, McKeigue PM, Kittles RA. Skin pigmentation, biogeographical ancestry and admixture mapping. Hum Genet. 2003;112:387–399. doi: 10.1007/s00439-002-0896-y. [DOI] [PubMed] [Google Scholar]
- Silman AJ, Pearson JE. Epidemiology and genetics of rheumatoid arthritis. Arthritis Res. 2002;4(Suppl 3):S265–S272. doi: 10.1186/ar578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Quertermous T, Rodriguez B, Kardia SL, Zhu X, Brown A, Pankow JS, Province MA, Hunt SC, Boerwinkle E, Schork NJ, Risch NJ. Genetic structure, self-identified race/ethnicity, and confounding in case-control association studies. Am J Hum Genet. 2005;76:268–275. doi: 10.1086/427888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B, Cockerham C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Williams RC, Long JC, Hanson RL, Sievers ML, Knowler WC. Individual estimates of European genetic admixture associated with lower body-mass index, plasma glucose, and prevalence of type 2 diabetes in Pima Indians. Am J Hum Genet. 2000;66:527–538. doi: 10.1086/302773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Li H, Criswell LA, Gregersen PK, Alarcon-Riquelme ME, Kittles R, Shigeta R, Silva G, Patel PI, Belmont JW, Seldin MF. Examination of ancestry and ethnic affiliation using highly informative diallelic DNA markers: application to diverse and admixed populations and implications for clinical epidemiology and forensic medicine. Hum Genet. 2005;118:382–392. doi: 10.1007/s00439-005-0012-1. [DOI] [PubMed] [Google Scholar]
- Zhang C, Chen K, Seldin MF, Li H. A hidden Markov modeling approach for admixture mapping based on case-control data. Genet Epidemiol. 2004;27:225–239. doi: 10.1002/gepi.20021. [DOI] [PubMed] [Google Scholar]
- Zhu X, Cooper RS, Elston RC. Linkage analysis of a complex disease through use of admixed populations. Am J Hum Genet. 2004;74:1136–1153. doi: 10.1086/421329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Luke A, Cooper RS, Quertermous T, Hanis C, Mosley T, Gu CC, Tang H, Rao DC, Risch N, Weder A. Admixture mapping for hypertension loci with genome-scan markers. Nat Genet. 2005;37:177–181. doi: 10.1038/ng1510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.