Abstract
If the 5′-proximal AUG triplet in a mammalian mRNA is followed by a short open reading frame (sORF), a significant fraction of ribosomes resume scanning after termination of sORF translation, and reinitiate at a downstream AUG. To examine the underlying mechanism, we examined reinitiation in vitro using a series of mRNAs that differed only in the 5′-untranslated region (UTR). Efficient reinitiation was found to occur only if the eIF4F complex, or at a minimum the central one-third fragment of eIF4G, participated in the primary initiation event at the sORF initiation codon. It did not occur, however, when sORF translation was driven by the classical swine fever virus or cricket paralysis virus internal ribosome entry sites (IRESs), which do not use eIF4A, 4B, 4E, or 4G. A critical test was provided by an mRNA with an unstructured 5′-UTR, which is translated by scanning but does not absolutely need eIF4G and eIF4A: There was efficient reinitiation in a standard reticulocyte lysate, when initiation would be largely driven by eIF4F, but no reinitiation in an eIF4G-depleted lysate. These results suggest that resumption of scanning may depend on the interaction between eIF4F (or the eIF4G central domain) and the ribosome being maintained while the ribosome translates the sORF.
Keywords: Scanning ribosome mechanism, initiation factor eIF4F, initiation factor eIF4G, cricket paralysis virus IRES, classical swine fever virus (pestivirus) IRES
A very large proportion of mammalian cellular mRNAs conform to the straightforward model of initiation site selection by strictly linear ribosome scanning, in which initiation occurs exclusively at the 5′-proximal AUG codon (Kozak 1989). However, there are three circumstances in which linear scanning can result in initiation at downstream AUG codons: (1) if the context of the 5′-proximal AUG is unfavorable so that it is inefficiently recognized by the scanning ribosome (Kozak 1986); (2) if the context of the 5′-proximal AUG is favorable, but it is located very close to the 5′-cap (Kozak 1991a); and (3) if the 5′-proximal AUG is followed by a short open reading frame, or sORF (Kozak 1987). In the last case, it is thought that some, but not all, of the ribosomes that have completed translation of the sORF may resume scanning and (re)initiate translation at an AUG located farther downstream. Because a significant fraction of the ribosomes do not resume scanning, an upstream sORF results in a decrease in translation of the main (downstream) ORF (Kozak 1987).
sORFs are found in a significant proportion of mammalian mRNAs, perhaps as many as 40% (Peri and Pandey 2001), and are particularly common in mRNAs coding for proto-oncogenes, transcription factors, and so forth (Kozak 1991b). In contrast, sORFs are rather rare in yeast mRNAs, notwithstanding the well-known case of GCN4 mRNA, which has four sORFs (Hinnebusch 1997). Moreover, if a random sORF is introduced into a yeast mRNA, the outcome is that reinitiation does not occur (Yun et al. 1996), which is also the case with GCN4 sORF-4, and probably also sORFs-2 and 3 (Hinnebusch 1997). Thus, in yeast, reinitiation after translation of an upstream sORF, as occurs with GCN4 sORF-1, seems to be the exception rather than the rule. Thus there are likely to be significant differences between yeast and mammalian systems in the details of the reinitiation mechanism.
Resumption of scanning and reinitiation in mammalian systems occurs only if the first ORF is short (Luukkonen et al. 1995; Kozak 2001), but it seems to be not so much the length per se that is the critical parameter, but rather the time taken for the ribosome to translate the sORF. Thus an ORF that is short enough to be permissive for reinitiation becomes nonpermissive if it has a pseudoknot structure that would be expected to cause ribosome pausing (Somogyi et al. 1993; Kontos et al. 2001; Kozak 2001). The reason that sORF translation needs to be completed rapidly may be related to another enigmatic aspect of reinitiation. What initiation factors, if any, need to be associated with the 40S subunit to promote its scanning from the sORF termination codon to the downstream initiation site, and how does the 40S subunit acquire the necessary initiation factors? The two possibilities are (1) that such factors are, and have to be acquired de novo, or (2) that the critical initiation factors that promoted scanning of the 40S subunits to the sORF initiation codon remain associated with the ribosome (albeit perhaps loosely associated) for a short time following initiation of sORF translation.
One initiation factor that certainly has to be acquired de novo is eIF2 in the form of an eIF2/GTP/Met-tRNAi ternary complex (Hinnebusch 1997), because the Met-tRNAi in the ternary complex associated with the 40S subunit as it scans to the sORF initiation codon will have been used to initiate sORF translation, and it is presumed that the eIF2 entity will then be released as an eIF2/GDP complex. In the yeast GCN4 system it has been shown that following translation of sORF-1, the average distance traversed by a 40S subunit before it reacquires competence for reinitiation is inversely related to the concentration of available eIF2/GTP/Met-tRNAi ternary complex (Dever et al. 1995; Hinnebusch 1997). This dependency on distance is thought to be really a time-dependent relationship: the lower the intracellular ternary complex concentration, the longer the delay before the small ribosomal subunit acquires Met-tRNAi, and hence the farther it will have traveled along the mRNA after terminating sORF translation. Implicit in this model is the hypothesis that 40S subunits can and do resume scanning initially without an associated ternary complex. Thus if the initiation factors necessary to promote 40S scanning have to be acquired de novo following sORF translation, they must be acquired very much faster than a new ternary complex. This is in sharp contrast with primary initiation when the binding of the ternary complex to 40S subunits is very rapid, much faster than the subsequent steps in initiation (Darnbrough et al. 1973). This contrast strongly suggests the possibility that the factors necessary for resumption of scanning do not have to be (re)acquired de novo, and that the factors that promoted 40S subunit scanning from the 5′-end to the sORF initiation codon might remain associated with the ribosome for a few seconds following initiation, with the result that if the translation of the sORF is completed quickly, they will be suitably located to drive ribosome scanning to the next AUG triplet.
Although a direct study of factor–ribosome interactions that persist for only a few seconds is technically unfeasible, what can be done is to examine whether resumption of scanning/reinitiation is dependent on the precise complement of initiation factors that were involved in promoting primary initiation at the sORF initiation codon. Using this approach, we show here that efficient reinitiation only occurs if the eIF4 family of factors (either eIF4A, 4B, and 4F or eIF4A, 4B, and the central domain of eIF4G) participated in the original initiation event at the sORF AUG codon.
Results
Our assay system is based on the triose phosphate isomerase-CAT (TPI-CAT) fusion described by Zhang and Maquat (1997), which consists of the 5′-untranslated region (UTR) and the first 38 codons of TPI placed immediately upstream of the CAT coding sequence. These investigators reported that when a termination codon was introduced at codon 1 or codon 2 (in their numbering system in which the initiation codon was designated as codon 0, or 0Met), there was reinitiation in an in vitro-coupled transcription–translation system at codon 14 (14Met) at about 40% of the efficiency of the primary initiation at 0Met.
We based our studies on the 2ter version rather than 1ter, because mutation of the codon immediately downstream of the TPI initiation codon might influence the efficiency of initiation at 0Met through context effects (Boeck and Kolakofsky 1994; Grünert and Jackson 1994). We made a number of replacements of the 5′-UTR of this construct (Fig. 1A). The entire TPI 5′-UTR was replaced with one consisting essentially of 19 tandem CAA repeats; this is reported to give a completely unstructured 5′-UTR, and as a consequence initiation can occur, albeit at reduced efficiency, even if eIF4A, 4B, and 4F (or 4G) and ATP are omitted (Pestova and Kolupaeva 2002). In addition, we replaced the entire TPI 5′-UTR with the internal ribosome entry sites (IRESs) of two picornaviruses, encephalomyocarditis virus (EMCV) and human rhinovirus-2 (HRV); with the classical swine fever virus (CSFV) IRES, which promotes initiation independent of initiation factors eIF4A, 4B, 4E, or 4G (Pestova et al. 1998a); or with the intergenic IRES of cricket paralysis virus (CrPV), which does not require any of the canonical initiation factors (Wilson et al. 2000; Pestova and Hellen 2003). For the first three of these IRESs, an exact substitution was made such that the TPI 0Met codon was in the precise position of the viral initiation codon. For the CrPV IRES where initiation occurs without Met-tRNAi and without an AUG codon (Wilson et al. 2000; Jan and Sarnow 2002; Pestova and Hellen 2003), six codons of CrPV coding sequence were included upstream of TPI 0Met (Fig. 1B), and thus the sORF comprised eight sense codons.
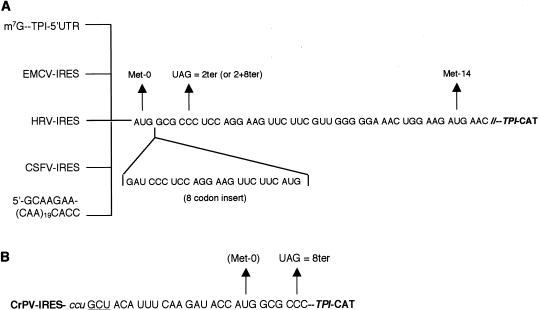
Figure 1.
Constructs used in this investigation. (A) The sequence of the first 16 codons of the ORF of the TPI-CAT fusion construct (TPI/norm). The stop codon mutation introduced in TPI/2ter is shown, as is the site of reinitiation at 14Met. The eight additional codons introduced to generate TPI/+8norm and TPI/2+8ter (and the related constructs with different 5′-UTRs) is given below the site of insertion. The TPI 5′-UTR was replaced by either the EMCV IRES (nt 286–833), human rhinovirus (HRV) IRES (nt 15–610), or CSFV IRES (nt 1–372), such that the initiation codon (0Met) occupied the exact position of the viral initiation codon. The TPI 5′-UTR was also replaced with a synthetic 68-nt 5′-UTR consisting largely of 19 tandem CAA repeats. (B) Sequence of the start of the ORF of the cricket paralysis virus (CrPV) IRES/TPI-CAT fusion constructs, CrPV/norm and CrPV/8ter. All residues up to and including the GAUA motif are derived from the CrPV genome sequence. The lowercase ccu codon is believed to occupy the ribosomal P-site, and the underlined GCU codon is the first codon to be decoded by an aminoacyl-tRNA, with the result that the protein product will have N-terminal alanine (Wilson et al. 2000).
For most of these constructs (except that with the CrPV IRES), we also examined the effect of expanding the sORF by an additional eight codons (Fig. 1A). These constructs will be referred to as 2+8ter, and the control as +8norm. The reason for making this expansion is that, as the ribosome is thought to maintain intimate contact with the mRNA over only 30–35 nucleotides (nt), centered on the codon occupying the P-site (Wolin and Walter 1988; Pestova et al. 1998b), precisely the same mRNA sequence would be in contact with a ribosome that was in the process of translating the sORF termination codon, irrespective of the nature of the 5′-UTR. A further advantage is that when the ribosome is at the 0Met initiation codon, the sORF termination codon would not be in intimate contact with the ribosome, and thus the mutation of the CCC (norm) codon to UAG (ter) could not have any direct influence on initiation efficiency at 0Met.
As explained in detail following, reinitiation efficiency was assessed as the ratio between the yield of slightly truncated reinitiation product translated from the ter mRNAs and the yield of full-length product from the control (norm) mRNA. Thus the validity and accuracy of these determinations obviously depends on two types of premise: (1) that the quantity and `quality' of the two mRNAs in the pair of assays was the same; and (2) that the frequency of initiation at 0Met (and the initiation frequency at downstream AUGs due to leaky scanning) was the same with the ter mRNA as with the matched control (norm) mRNA. Regarding the first presumption, great care was taken to ensure equal concentrations of the two RNAs, and all experiments were repeated several times with at least two independent preparations of each mRNA. The second presumption, which has been inherent in all previous assays of reinitiation efficiency (Luukkonen et al. 1995; Zhang and Maquat 1997; Kozak 2001), is unavoidable because sORF translation products are degraded extremely rapidly in the system, and so are difficult to detect, and their yield cannot give a valid measure of initiation frequency (Hackett et al. 1986). Expansion of the sORF to a length that would give a more stable and quantifiable product is not an option, because this would severely reduce reinitiation efficiency (Luukkonen et al. 1995; Kozak 2001). However, as shown later for the CSFV/norm and CSFV/2ter pair of mRNAs (and also tested with other matched pairs), the addition of G418 to suppress termination at the sORF stop codon can provide strong support that the two mRNAs were very similar in quantity and quality (see Fig. 5B). The G418 controls also confirm that the two RNAs were comparably stable in the system, and we have also verified, using 32P-labeled transcripts, gel electrophoresis, and autoradiography, that the mutation from sense codon to termination codon had no effect on RNA stability (data not shown), and that differential stabilities cannot therefore be the explanation of differences in apparent reinitiation efficiency.
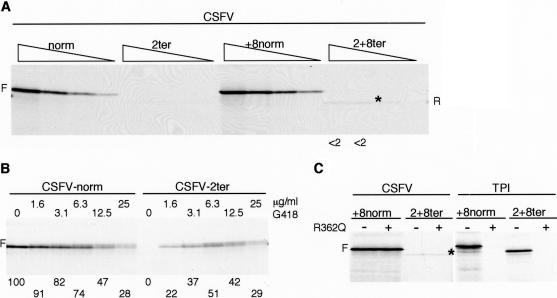
Figure 5.
The CSFV IRES does not support efficient reinitiation. (A) Uncapped CSFV/norm, CSFV/2ter, CSFV/+8norm, and CSFV/2+8ter mRNAs were each translated at 100, 50, 25, and 12.5 μg/mL. The reinitiation product seen with CSFV/2+8ter mRNA is indicated by an asterisk on the autoradiograph, and R in the right-hand margin. Reinitiation efficiency was determined and expressed as in Fig. 2. (B) Uncapped CSFV/norm and CSFV/2ter mRNAs were each translated at 80 μg/mL in the presence of increasing concentrations of G418, as designated. The combined yield of the doublet of full-length products was determined by densitometry and is expressed relative to the yield obtained in the control assay of CSFV/norm, which was set at 100. (C) Uncapped CSFV/+8norm and CSFV/2+8ter mRNAs were each translated at 70 μg/mL in the absence (–) or presence (+) of 300 μg/mL (6.5 μM) R362→Q dominant negative eIF4A (Pause et al. 1994). A parallel control assay was carried out with 20 μg/mL capped TPI/+8norm and TPI/2+8ter mRNAs.
Reinitiation on mRNAs with the TPI 5′-UTR
Figure 2A shows a representative experiment using capped TPI/2ter mRNA and the corresponding control TPI/norm. In all such experiments, the yields of the various translation products were determined either by phosphorimaging or by densitometry of the autoradiographs, using a range of different exposures to ensure that the determinations were made within the linear response range of the film. The raw data were adjusted for the methionine content of each product, with the assumption that the retention or removal the initiating methionine residue would follow the rules deduced by Boissel et al. (1988) and Moerschell et al. (1990).
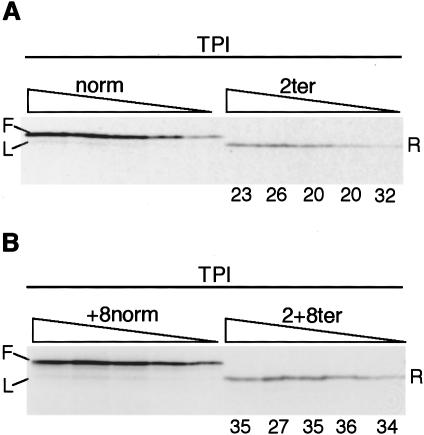
Figure 2.
Reinitiation on the TPI/2ter and TPI/2+8ter mRNAs. (A) Capped TPI/norm and TPI/2ter mRNAs were each translated at 40, 20, 10, 5, and 2.5 μg/mL. Reinitiation efficiency was calculated as described in the text, and is given (as %) below each lane of the assays of TPI/2ter mRNA. (B) Capped TPI/+8norm and TPI/2+8ter mRNAs were translated at the same RNA concentrations as in A, and reinitiation efficiency calculated and presented as in A. The full-length product (F) from the control mRNAs and the product arising from leaky scanning (L) are indicated, as is the reinitiation product (R).
To calculate reinitiation efficiency, we determined the molar yield (F) of full-length product expressed from the control mRNA (TPI/norm), and the yield (L) of product initiated at 14Met on this mRNA (probably as the result of leaky scanning), as well as the yield (R) of product initiated at 14Met on the termination construct, TPI/2ter mRNA. Reinitiation efficiency was taken as (R–L)/F at each RNA concentration and is given (as %) below each lane in Figure 2A. Thus we are making the precautionary assumption that a small amount of the truncated product translated from the TPI/2ter mRNA originates from leaky scanning from the 5′-end rather than by termination/reinitiation, and that this amount will be equal to the yield of product initiated at 14Met on the control TPI/norm mRNA as a result of leaky scanning. The appropriateness of this adjustment is discussed later. In standard conditions, it makes little difference with the 2+8ter mRNAs, and a rather more significant difference with the 2ter mRNAs, but it has a quite large influence when translation is driven by eIF4G fragments.
As shown in Figure 2A, the efficiency of reinitiation on the capped TPI/2ter mRNA averaged just under 25% (or 34% if no adjustment is made for leaky scanning), but was higher (33%, or 38% if no adjustment is made for leaky scanning) with TPI/2+8ter mRNA (Fig. 2B), for reasons that are not understood. The influence of capping was examined over a range of KCl concentrations, because the optimum K+ for uncapped mRNA translation is lower than for capped, and so we needed to be able to distinguish any influence of capping per se from KCl concentration effects. Figure 3 shows that reinitiation efficiency (on TPI/2+8ter mRNA) is maximal at fairly low K+ concentrations (30–45 mM for capped transcripts, and 15–30 mM for uncapped), and at its maximum, it is slightly higher with uncapped mRNAs than capped.
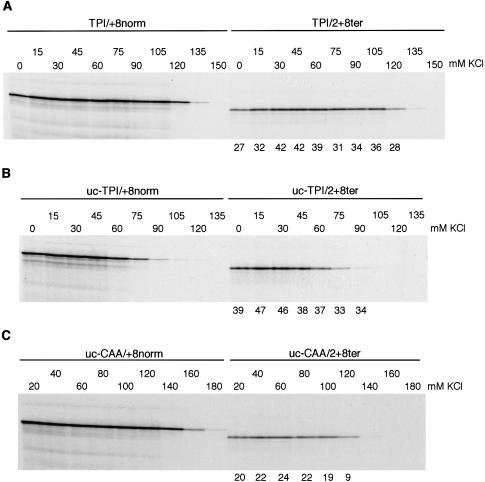
Figure 3.
Effect of capping and KCl concentration on reinitiation efficiency. (A) Capped or (B) uncapped TPI/+8norm and TPI/2+8ter mRNAs were each translated at 25 μg/mL with different concentrations of added KCl as indicated. (C) Uncapped (CAA)19/+8norm and (CAA)19/2+8ter were each translated at 20 μg/mL with the designated concentration of added KCl. Reinitiation efficiency was calculated as in Fig. 2 and is given (as %) below each lane of the assays of TPI/2+8ter or (CAA)19/2+8ter mRNAs.
For both capped and uncapped mRNAs with the TPI 5′-UTR, increasing K+ concentrations are only slightly more inhibitory to reinitiation than to primary initiation at 0Met on the control mRNA (Fig. 3A,B). In contrast, with uncapped (CAA)19 mRNAs, increasing K+ concentrations are much more inhibitory to reinitiation than to primary initiation on the (CAA)19/+8norm mRNA (Fig. 3C). This is not because reinitiation on (CAA)19/2+8ter mRNA is unusually sensitive to inhibition by KCl (in fact it is similar to capped TPI/2+8ter mRNA in this respect and is actually less sensitive than uncapped TPI/2+8ter), but because primary initiation at 0Met on (CAA)19/+8norm mRNA is exceptionally resistant to inhibition by high K+ levels.
A comparison of polyadenylated and nonpolyadenylated capped transcripts showed little influence of a poly(A) tail on reinitiation efficiency in this system (data not shown).
Picornavirus IRESs support reinitiation
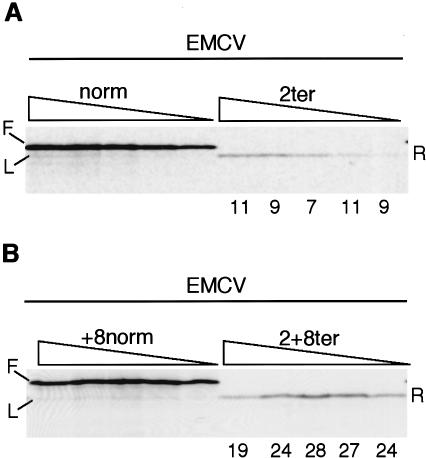
With EMCV/2ter, reinitiation efficiency was about 10%, less than half that seen for the TPI/2ter mRNA (Fig. 4A). However, much higher reinitiation, about 25%, was seen with the EMCV/2+8ter mRNA (Fig. 4B). One possible reason for the lower efficiency with the EMCV/2ter construct may be related to the fact that the EMCV IRES is very potent and recruits ribosomes directly to the initiation codon, without any significant ribosome scanning (Kaminski et al. 1990; Pestova et al. 1996, 1998b). Thus if a ribosome terminates translation just two codons downstream of the initiation codon, there may be a very strong tendency for it to revert back to the 0Met initiation codon rather than scan in a 5′→3′ direction to 14Met, stronger than if the ribosome terminates sORF translation 10 codons downstream of the initiation site, as in EMCV/2+8ter.
Figure 4.
Reinitiation on the EMCV/2ter and EMCV/2+8ter mRNAs. (A) Uncapped EMCV/norm and EMCV/2ter mRNAs, or (B) EMCV/+8norm and EMCV/2+8ter mRNAs were each translated at 40, 20, 10, 5, and 2.5 μg/mL. Reinitiation efficiency was calculated as in Fig. 2, and is given (as %) below each lane of the assays of EMCV/ter mRNAs. The full-length product (F) from the control mRNAs and the product arising from leaky scanning (L) are indicated, as is the reinitiation product (R).
We observed much higher reinitiation with HRV/2ter mRNA than EMCV/2ter (data not shown), in fact higher even than with the TPI/2+8ter construct. The difference between the two picornavirus IRESs may be in part due to the fact that with the HRV IRES, the initial internal entry site is thought to be at AUG576–578, and the ribosomes are then transferred, most likely by scanning, to the correct initiation codon at nt 611 (Borman and Jackson 1992). Thus, with the HRV IRES construct there would probably be even less tendency for the ribosome to revert back to the original entry site after termination at codon 2, which may explain the higher reinitiation efficiency.
The CSFV IRES cannot support efficient reinitiation
With uncapped CSFV/2ter mRNA translated at 100 mM KCl, no reinitiation was observed (Fig. 5A). To ensure that this was not an artifactual result due to poor-quality RNA, or insufficient RNA, we titered G418 into the translation assays of CSFV/2ter mRNA (and the corresponding control) to suppress termination at the end of the sORF. With CSFV/norm, increasing concentrations of G418 caused a progressive inhibition of translation, and instead of a single band of full-length product, a doublet was seen, presumably because of some suppression of termination at the end of the TPI-CAT ORF (Fig. 5B). With CSFV/2ter, the increasing amounts of G418 actually caused a progressive stimulation of translation over the lower end of the concentration range, followed by an inhibition at the upper end (Fig. 5B). The translation product was the same doublet as was seen with CSFV/norm, and at the highest G418 concentrations, the yield of this doublet was precisely the same with CSFV/2ter and CSFV/norm mRNAs (Fig. 5B). We take this as good evidence that the two mRNAs were equivalent in quantity, quality, and stability, and that the frequency of primary initiation at 0Met was very similar on the two templates.
With CSFV/2+8ter, a very faint band corresponding to a reinitiation efficiency of ∼1%–2% was routinely observed (Fig. 5A). The explanation for the lower reinitiation efficiency with CSFV/2ter than with CSFV2+8/ter may be the same as for the corresponding mRNAs with the EMCV IRES, namely, a greater tendency of the ribosome to revert back to the 0Met initiation codon under the influence of the strong IRES when termination occurs just 2 codons, rather than 10 codons, downstream of 0Met. However, it might be prudent to be cautious over drawing too close a parallel between the mechanism of reinitiation on CSFV/2+8ter and other mRNAs, because it is unique among all reinitiation events that we have observed, in that it was completely resistant to the inhibitory effect of R362→Q dominant negative eIF4A mutant (Fig. 5C).
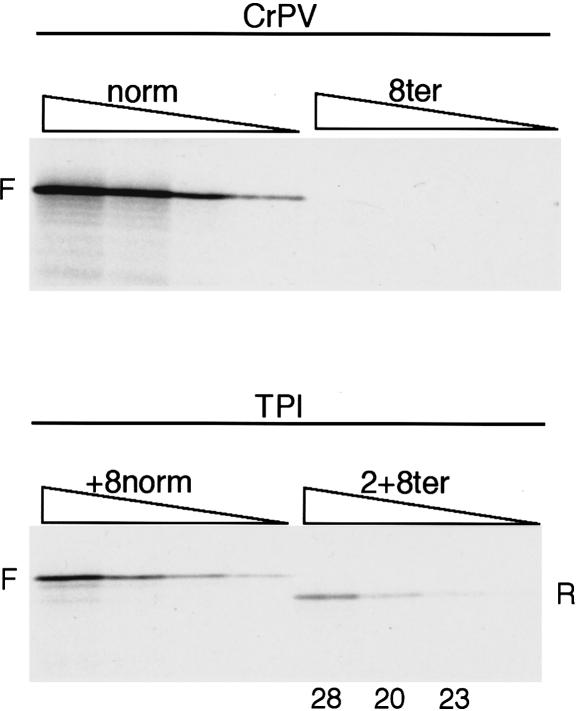
The cricket paralysis virus IRES cannot support reinitiation
Unlike all of the IRESs discussed so far, initiation on this IRES does not occur at an AUG codon, does not use Met-tRNAi, and probably does not require any of the canonical initiation factors (Wilson et al. 2000; Jan and Sarnow 2002; Pestova and Hellen 2003). Instead, the IRES is believed to consist of a triple pseudoknot structure, with one of the pseudoknots acting as a tRNA anticodon loop/codon mimic, which is inserted into the P-site of the ribosome (Kanamori and Nakashima 2001; Jan and Sarnow 2002). The aminoacyl-tRNA corresponding to the N-terminal amino acid residue (Ala) then enters the A-site and is transferred to the P-site by a pseudo-translocation event (Wilson et al. 2000; Pestova and Hellen 2003). Because it is not known whether codons immediately downstream of this initiation site exert any influence on initiation efficiency, we decided to retain some of these codons. The sequence in our reinitiation construct was ccu GCU ACA UUU CAA GAU ACC AUG GCG UAG (Fig. 1B), in which the ccu triplet is part of the pseudoknot that is thought to enter the P-site, and the GCU triplet encodes the N-terminal amino acid residue.
We found that the function of the CrPV IRES was quite strongly influenced by the K+ level: the lower the KCl concentration, the lower was the yield of product of the size expected for initiation at the GCU codon, and the greater the yield of two smaller products that appeared to have been initiated at the next two AUG codons downstream of the GCU triplet. We presume that a low K+ concentration destabilizes the triple pseudoknot structure (Kanamori and Nakashima 2001; Jan and Sarnow 2002), which would have the dual effect of not only compromising the internal initiation mechanism, but also facilitating ribosome scanning through the less structured 5′-UTR. It was found necessary to use 130 mM KCl in order to prevent all initiation at the downstream AUGs on CrPV/norm mRNA and thus to obtain exclusively IRES-driven initiation. Controls with TPI/2+8ter showed that reinitiation could still be observed at this elevated KCl level, albeit at the reduced efficiency of ∼24% (average) rather than ∼33%, but, in sharp contrast, no reinitiation whatsoever was seen with the CrPV/8ter mRNA (Fig. 6).
Figure 6.
The CrPV IRES does not support reinitiation. Uncapped CrPV/norm and CrPV/8ter mRNAs were each translated at 60, 30, 15, and 7.5 μg/mL. The concentration of added KCl was 130 mM. As a control, capped TPI/+8norm and TPI/2+8ter mRNAs were translated at the same KCl concentration at 40, 20, 10, and 5 μg/mL RNA. Reinitiation efficiency was calculated as in Fig. 2 and is given (as %) below each lane of the assays of TPI/2+8ter mRNA.
The central domain of eIF4G can promote reinitiation
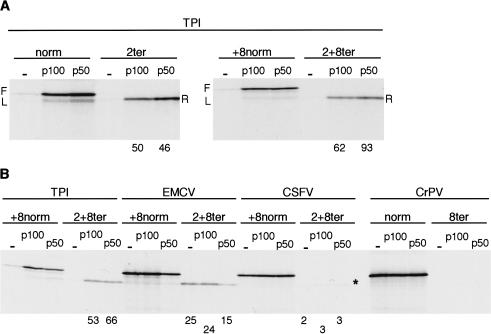
At 100 mM added KCl, the translation of uncapped mRNAs by the scanning mechanism is very inefficient (Fig. 3B), but it can be greatly stimulated by addition of either the C-terminal two-thirds fragment (p100) or the central one-third (p50) of eIF4G (De Gregorio et al. 1998; Ali et al. 2001; Ali and Jackson 2002). Figure 7A shows that although these conditions, especially with p50, give rise to leaky scanning on the TPI/norm control and, to a lesser extent, on TPI/+8norm mRNA, it is nevertheless clear that p100 and p50 promoted even more efficient reinitiation on these uncapped mRNAs than is seen with capped or uncapped TPI/ter mRNAs in the standard lysate system (Figs. 2, 3), when primary initiation would be driven by intact eIF4F complex. Addition of p100 or p50 also appeared to increase reinitiation on capped TPI/ter mRNAs (data not shown), but as in vitro capping is never 100% efficient (Dasso and Jackson 1989), we cannot be sure whether there was a real increase in reinitiation on the majority capped species, rather than just on the uncapped contaminants.
Figure 7.
Addition of p50 (central one-third) or p100 (C-terminal two-thirds) fragments of eIF4G strongly promotes reinitiation on uncapped TPI/ter mRNAs, but not on mRNAs with the CSFV, CrPV, or EMCV IRESs. (A) Uncapped TPI/norm, TPI/2ter, TPI/+8norm, and TPI/2+8ter mRNAs, as indicated, were each translated at 15 μg/mL with 100 mM added KCl, in the absence or presence of 100 nM p100, or 200 nM p50. (B) Uncapped mRNAs as designated were translated at 20 μg/mL (TPI), 15 μg/mL (EMCV), 60 μg/mL (CSFV), or 25 μg/mL (CrPV). Added KCl was 100 mM (135 mM for CrPV/mRNAs), and translation was carried out in the presence or absence of 100 nM p100, or 200 nM p50. Reinitiation efficiency was calculated as in Fig. 2, and is given (as %) below each lane of the assays in which quantifiable reinitiation occurred.
One can envisage two alternative scenarios that could explain the high reinitiation efficiency seen in Figure 7A. One is that the reinitiation is actually being driven by the same p50 or p100 molecules that carried out the primary initiation event, but that this is more efficient than with intact eIF4F, for reasons discussed following. The alternative scenario is that at some stage around the time of termination of sORF translation, ribosomes can actually acquire p50 or p100 de novo (or reacquire these factors) more efficiently than they can acquire intact eIF4F, perhaps because of conformational differences between the fragments and intact eIF4G, and/or perhaps because p50 and p100 were added at a higher concentration (200 and 100 nM, respectively) than the concentration of endogenous eIF4F in the lysate, which we have estimated as ∼25 nM (Ali et al. 2001). If this second explanation is the correct one, a firm prediction is that addition of p50 or p100 should potentiate reinitiation even on CSFV/2+8ter or CrPV/8ter mRNAs where primary initiation is completely independent of, and uninfluenced by, eIF4G or fragments of eIF4G. Figure 7B shows that this was decidedly not the case; nor was reinitiation on EMCV/2+8ter increased. We therefore conclude that there is no evidence for ribosomes being able to acquire p50 or p100 at some point after the primary initiation event, for example, at termination of sORF translation. A further reason for doubting whether such delayed acquisition of p50 or p100 can occur is that if it did one might reasonably expect to see reinitiation even after long ORFs.
Thus the reinitiation seen in Figure 7A is most likely to have been actually driven by the p50 or p100 that promoted the primary initiation event, and the unusually high efficiency of this reinitiation suggests that the interaction of p50 or p100 with the ribosome (via eIF3 as intermediary) may be more stable and persist for longer than the interaction of intact eIF4F with the ribosome. It is possible, certainly with capped mRNAs, that intact eIF4F is tethered quite strongly to the 5′-cap (via the eIF4E/cap interaction), and that this opposes prolonged maintenance of the eIF4G/eIF3/ribosome interactions, whereas p50 or p100 would not have such a strong interaction with the 5′-end.
Reinitiation is impaired if the primary scanning-dependent initiation event occurs without the involvement of eIF4A and eIF4G
The results described so far point unambiguously to the conclusion that efficient reinitiation occurs only if the eIF4 family of initiation factors (eIF4A plus either eIF4F or the central domain of eIF4G) participated in the primary initiation event, the delivery of the 40S subunit to 0Met. For a more rigorous test of this proposition, we used an mRNA with a 5′-UTR consisting essentially of 19 repeats of the triplet CAA, which is thought to be completely unstructured (Pestova and Kolupaeva 2002). Initiation on this RNA can occur by a scanning mechanism, albeit at reduced efficiency, in the complete absence of eIF4A, 4B, and 4F (or 4G) or ATP, or in the presence of the dominant negative R362→Q eIF4A mutant (Pause et al. 1994; Pestova and Kolupaeva 2002). Under these conditions, eIF1, eIF1A, and eIF3 are absolutely required for successful scanning, and this scanning is highly sensitive to secondary structure: Even a hairpin with just 4 A-U pairs centrally located in the 5′-UTR completely inhibited initiation (Pestova and Kolupaeva 2002). When the eIF4 family of factors is present, it is not quite clear whether both modes of initiation (dependent or independent of the eIF4 factors) occur concurrently, or whether the eIF4-dependent mechanism dominates.
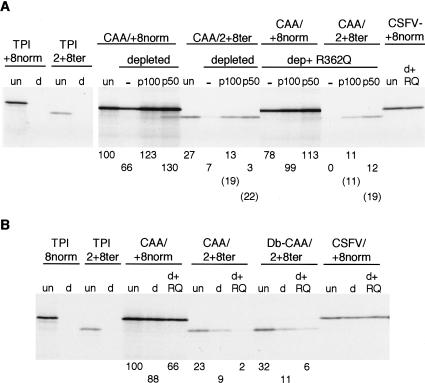
Uncapped mRNA was used for these experiments because it has the advantage that the mRNA population is homogeneous with respect to the nature of the 5′-end, whereas any preparation of capped transcripts invariably includes some uncapped species (Dasso and Jackson 1989). The uncapped (CAA)19/2+8ter and its matched control, (CAA)19/+8norm, were translated at 100 mM added KCl, either in standard lysate, or in the eIF4G-depleted system (∼95% depletion) we have described recently (Ali et al. 2001; Ali and Jackson 2002), which was either supplemented directly with recombinant p100 or p50 (Fig. 8A), or first preincubated with R362→Q dominant negative eIF4A (to ensure that any residual traces of eIF4F, or 4G, would be completely inactivated) and then supplemented with p50 or p100 (Fig. 8A).
Figure 8.
Reinitiation on (CAA)19/2+8ter occurs only if eIF4G (or the p100 or p50 fragments of eIF4G) was involved in the primary initiation event at 0Met. (A) Uncapped (CAA)19/+8norm and (CAA)19/2+8ter mRNAs were each translated at 20 μg/mL and with 100 mM added KCl, in either normal (nondepleted) lysate (un), or in eIF4G-depleted lysate, or in eIF4G-depleted lysate that had been preincubated for 10 min with 150 μg/mL (3.3 μM) R362→Q dominant negative eIF4A, as indicated. Where indicated, the assays were supplemented with either 100 nM of p100 or p50. The translation efficiency of the (CAA)19/+8norm mRNA under these different conditions is given under the corresponding lane, and is expressed relative to the yield in the nondepleted lysate (un), which was set at 100. Reinitiation efficiency was calculated as for Fig. 2, and is given under each lane of the (CAA)19 /2+8ter mRNA translation assays. Numbers in parentheses are the calculated reinitiation efficiency if no adjustment is made for the yield of product resulting from putative leaky scanning on the (CAA)19 /+8norm mRNA; that is, these numbers represent the R/F yield ratio rather than (R–L)/F. (B) Insertion of tandem CAA codons between the sORF termination codon and the reinitiation site does not facilitate efficient eIF4G-independent reinitiation. Uncapped (CAA)19/+8norm, (CAA)19/2+8ter, and Double-CAA/2+8ter (see text) mRNAs were each translated at 20 μg/mL in standard (nondepleted) lysate, or in eIF4G-depleted lysate that had been preincubated for 10 min with 7.5 μM R362→Q dominant negative eIF4A, as indicated. The efficiency of translation of (CAA)19/+8norm mRNA in the depleted lysate is expressed relative to that observed in standard lysate, which was set at 100. Apparent reinitiation efficiency was determined from the yield of reinitiation product relative to the yield of full-length product from (CAA)19/+8norm mRNA translated under the same conditions, and is given below each lane for assays of (CAA)19/2+8ter and Double-CAA/2+8ter RNAs. Data for positive and negative controls are also included for each panel: capped TPI/+8norm and TPI/2+8ter mRNAs were each translated at 20 μg/mL in nondepleted lysate (un) or eIF4G-depleted lysate (d); uncapped CSFV/+8norm was translated at 60 μg/mL in nondepleted lysate (un), or eIF4G-depleted lysate (d), or eIF4G-depleted lysate that had been preincubated with R362→Q dominant negative eIF4A (d+RQ).
As expected from the results of Pestova and Kolupaeva (2002), primary initiation at 0Met on the control (CAA)19/+8norm mRNA was only slightly lower (∼70%) in the depleted lysate than in the standard lysate, even if the depleted lysate had been preincubated with the dominant negative eIF4A mutant (Fig. 8A). In the standard lysate system, the reinitiation efficiency on (CAA)19/2+8ter was typically about 25% (Fig. 8A). Reinitiation efficiency in the eIF4G-depleted lysate was severely reduced, and was zero if this depleted lysate had been preincubated with R362→Q dominant negative eIF4A (Fig. 8A), or if it was simply supplemented with the mutant eIF4A without any preincubation step (data not shown). Addition of p100 rescued reinitiation to about half that seen in the standard (nondepleted) lysate (Fig. 8A). The ability of p100 (and p50) to rescue reinitiation in the depleted lysate that had been preincubated with R362→Q dominant negative eIF4A is especially remarkable when one considers that the added p100 or p50 would be quite rapidly inactivated or `poisoned' by the dominant negative eIF4A (Pause et al. 1994), which was present in 33-fold molar excess over p100 or p50.
The calculated reinitiation efficiency would be even higher (numbers in parentheses in Fig. 8A) if no adjustment were made for the yield of product initiated at 14+8Met on (CAA)19/+8norm mRNA as a result of presumed leaky scanning. This prompts the question of whether this adjustment is actually appropriate and valid. One consideration that suggests that it might not be appropriate is that ribosomes are known to pause at termination codons (Wolin and Walter 1988). A ribosome paused at the termination codon of the sORF would not only block any following ribosome from reaching 14+8Met by leaky scanning, but, by stalling this following ribosome in the vicinity of the 0Met codon, might also promote even more exclusive use of this codon as the initiation site. These arguments suggest that there would be less leaky scanning to 14+8Met on the (CAA)19/2+8ter mRNA than on (CAA)19/+8norm, and thus the real reinitiation efficiency is likely to approach the values given in parentheses in Figure 8A.
Previously we noted that primary initiation at 0Met on (CAA)19/+8 norm mRNA was unusual in its strong resistance to increasing KCl concentration (Fig. 3C), whereas the response of reinitiation on (CAA)19/2+8ter to increasing KCl was not at all unusual in comparison with TPI/2+8ter. With hindsight it seems likely that the ability of primary initiation to operate at high KCl may reflect the fact that it can be supported by the combination of eIF1, eIF1A, and eIF3, without the need for eIF4A, 4B, 4E, and 4G (Pestova and Kolupaeva 2002), whereas efficient reinitiation has a very strong requirement that the primary initiation event at 0Met was promoted by eIF4F or eIF4G/4A (and presumably eIF4B).
The fact that tandem CAA triplets in the 5′-UTR allowed primary initiation via scanning in the absence of eIF4G or eIF4F raised the question of whether reinitiation would become eIF4G independent if there is a similar sequence between the sORF termination codon and the reinitiation site. We therefore replaced these 11 codons in (CAA)19/2+8ter and (CAA)19/+8norm by (CAA)10ACC to generate what will be called `Double-CAA' constructs (Db-CAA). We encountered a significant problem in that the tandem CAA codons in the Double-CAA/norm mRNA were highly restrictive to translation (data not shown), no doubt due to sequestration of all the tRNAGln in ribosomal P- and E-sites. Although addition of extra tRNA partly relieved this restriction, we could not be confident that this relief was sufficiently complete that the yield of full-length product from Double-CAA/norm was a valid reference for calculating reinitiation efficiency. It seemed better (or less unsatisfactory) to determine the `apparent' reinitiation efficiency on Double-CAA/2+8ter, using the yield of full-length product from (CAA)19/+8norm as the reference. Figure 8B shows that the apparent reinitiation efficiency on Double-CAA/2+8ter was higher in standard lysate than with (CAA)19/2+8ter, and was also somewhat higher in eIF4G-depleted lysate. However, it is obvious that even with the Double-CAA/2+8ter mRNA, reinitiation efficiency is drastically reduced by depletion of eIF4G and addition of dominant negative eIF4A.
We also made the same replacement in CSFV/2+8ter, and found that the apparent efficiency of reinitiation, using CSFV/+8norm as reference, in standard (nondepleted) lysate increased to an average (over the four RNA concentrations) of 5.1% from an average of 2.0% observed with CSFV/2+8ter (data not shown). Clearly, placing tandem CAA codons between the sORF termination codon and the reinitiation site can result in some increase in reinitiation in systems lacking eIF4G, or with mRNAs, such as CSFV/2+8ter, which do not use eIF4G in the primary initiation process, but the increase is fairly modest and the unstructured segment clearly does not allow eIF4G-independent reinitiation to occur at an efficiency approaching that observed when primary initiation is driven by either intact eIF4G (in the complete eIF4F complex), or p50/eIF4A, or p100/eIF4A.
A reduction in the rate of elongation results in reduced reinitiation efficiency
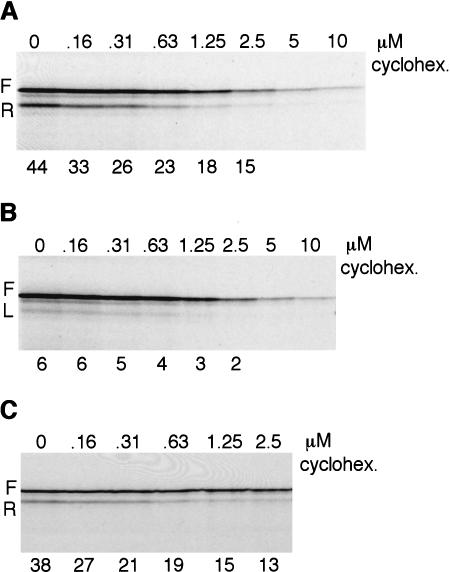
Finally, we have revisited the question of whether the speed of translation of the sORF influences reinitiation efficiency. To this end, we used low concentrations of cycloheximide to reduce the rate of elongation in an assay in which the TPI/+8norm and TPI/2+8ter were cotranslated, to ensure that the two mRNAs really were translated at the same cycloheximide concentrations (Fig. 9A). Controls with TPI/+8norm RNA on its own showed that cycloheximide reduced leaky scanning somewhat (Fig. 9B), probably because a decrease in the rate at which a ribosome translates the coding segment will reduce the probability that the following ribosome may scan past the 5′-proximal AUG. Visual inspection of Figure 9A suggested that cycloheximide reduced reinitiation efficiency, but to make this more transparent, we adjusted the gel loadings to obtain equal yields in all lanes of the full-length product translated from TPI/+8norm mRNA; the decrease in yield of the reinitiation product with increasing cycloheximide now becomes patently obvious (Fig. 9C). The efficiency of reinitiation, adjusted for leaky scanning, fell from ∼38% in the absence of cycloheximide to 13% at the highest cycloheximide level, which reduced the rate of translation by slightly less than sixfold.
Figure 9.
Reinitiation efficiency decreases if the rate of elongation is decreased. (A) Capped TPI/+8norm and TPI/2+8ter mRNAs were cotranslated at 15 μg/mL each and (B) capped TPI/+8norm mRNA was translated at 15 μg/mL in the presence of cycloheximide at the designated concentrations. The apparent reinitiation efficiency, unadjusted for leaky scanning (i.e., R/F), is shown below each lane in A, and the percentage of leaky scanning (L/F) below each lane in B. (C) The gel loadings of the same samples as in A were adjusted so that the yield of full-length product (F) from the TPI/+8norm mRNA would be approximately the same in each lane. The reinitiation efficiency, adjusted for leaky scanning, is given below each lane.
Previous estimates put the rate of elongation on exogenous mRNAs in reticulocyte lysates at ∼60 codons per minute (Jackson 1989). Thus the time taken to translate a 10-codon sORF in the absence of cycloheximide would be expected to be 10 sec plus the time of the pauses that are known to occur after initiation and at termination (Wolin and Walter 1988), but which have not been quantified. A concentration of cycloheximide that would increase the actual elongation time to about 60 sec plus the combined times of the postinitiation and termination pauses results in almost a threefold decrease in reinitiation efficiency. This has strong parallels with the observation of Kozak (2001) that reinitiation efficiency is severely reduced by the presence of a pseudoknot in the sORF, which would be expected to cause ribosome pausing (Somogyi et al. 1993; Kontos et al. 2001).
Discussion
In this work we have been concerned with reinitiation at downstream sites, and have not directly addressed the controversial question of whether there is any backward scanning and reinitiation at upstream sites, as was suggested some years ago (Peabody et al. 1986; Thomas and Capecchi 1986). However, these claims have been questioned on the grounds that the experiments could not distinguish between backward scanning from the termination codon as opposed to leaky scanning from the 5′- end of the mRNA (Kozak 2001). In recently reported in vitro assays, no backward scanning was seen when there were 10 nt between the termination codon and the upstream AUG, but if the separation was only 4 nt, a little backward scanning did occur, though at a level too low for accurate quantitation (Kozak 2001).
Analysis of reinitiation in prokaryotic systems suggests that following termination the ribosome (or more likely just the small subunit) undergoes random, but limited diffusion on the mRNA, and may reinitiate anywhere in a window of 40 nt on either side of the stop codon, with slight preference for downstream sites, and strong preference for the nearest potential reinitiation site (Adhin and van Duin 1990). However, the reinitiation was not very efficient (only 5% of the terminating ribosomes reinitiated), and it required an acceptable Shine-Dalgarno/initiation codon tandem, presumably to stabilize the interaction of the ribosome with the mRNA until it acquired initiator tRNA. If, as seems quite plausible, eukaryotic 40S subunits can also carry out bidirectional diffusion following termination, the absence of stabilizing interactions akin to the S-D/16S rRNA interaction is likely to result in even more rapid dissociation from the mRNA, and hence even lower reinitiation efficiency than in prokaryotes. It is possible that the very low level of reinitiation (albeit at a downstream site) that we saw with CSFV/2+8ter (Fig. 5C), which was atypical in that it was resistant to the dominant negative R362→Q mutant eIF4A (Pause et al. 1994), occurred by this type of random diffusion process.
However, the reinitiation seen with conventional mRNAs such as TPI/2ter and TPI/2+8ter is clearly different (1) because it is much more efficient, and (2) because the bias in favor of reinitiation at downstream rather than upstream sites is extremely strong (Kozak 2001). Given that eukaryotic 40S subunits don't even bind to conventional mRNAs (let alone scan such RNAs) in the absence of initiation factors (Pestova and Kolupaeva 2002), it is intuitive that such efficient and highly directional resumption of scanning and reinitiation must require the participation of initiation factors. The question is how and when does the 40S subunit acquire the necessary factors; and which factors are required? Scanning seems to resume extremely rapidly following termination, starting appreciably before a new eIF2/GTP-Met-tRNAi ternary complex is acquired (Kozak 1987, 2001; Hinnebusch 1997), despite the fact that ternary complex acquisition seems to be a rapid event in primary initiation (Darnbrough et al. 1973). The apparent speed with which scanning resumes raises suspicions that the necessary initiation factors may not need to be acquired de novo, but may already be in place at the time when termination of sORF translation occurs, perhaps because they are the self-same factors as promoted primary initiation at the sORF start codon.
Our results show unambiguously that efficient reinitiation occurs only if eIF4F (or at least the central domain of eIF4G in conjunction with eIF4A and likely eIF4B) participated in the primary initiation event at the sORF initiation codon. Particularly significant are the results for the mRNA with the (CAA)19 5′-UTR in the eIF4G-depleted lysate, where no reinitiation occurred, yet primary initiation at the sORF initiation codon was only slightly reduced relative to what happens in the standard lysate. This observation shows that even though eIF1, eIF1A, and eIF3 may be important for reinitiation, they cannot be sufficient in the absence of an involvement of eIF4G/eIF4A in the primary initiation event.
It is important to emphasize that the mere presence of eIF4F in the system is not sufficient, as shown by the fact that efficient reinitiation did not occur when the CSFV and CrPV IRESs were used to drive primary initiation in the normal lysate that contains eIF4F in relative abundance. Nor does addition of high concentrations of p50 or p100 promote reinitiation on these mRNAs (Fig. 7B). Efficient reinitiation specifically requires that eIF4F must have participated in the delivery of the primed 40S subunit to the sORF initiation codon. The implication of these results is that association between eIF4G/eIF4A (or the eIF4F complex) and the 40S subunit (an association that is perhaps bridged by eIF3) is likely to persist, not just throughout the whole process of 40S subunit scanning through the 5′-UTR, as we have speculated previously (see Fig. 4 of Jackson 2000), but is likely to be maintained for a short time after primary initiation at the 5′-proximal AUG. It is quite probable that the interactions between the small ribosomal subunit, eIF3, and eIF4G are rearranged in some way following ribosome subunit joining, and that in this rearranged state they are weaker than previously and thus will not persist for very long, probably only for a few seconds. We postulate that if sORF translation (including the termination step) is completed before these interactions are disrupted, the ribosome will resume scanning and can reinitiate at a downstream AUG codon. No reinitiation will occur, however, if these interactions are disrupted before sORF translation is complete, or if such interactions never existed in the first place, as is the case with the CSFV/2ter and CrPV/8ter mRNAs, or with the (CAA)19/2+8ter mRNA in the eIF4G-depleted lysate. In such circumstances, the ribosome probably dissociates from the mRNA immediately following termination, due to the absence of associated initiation factors to stabilize its interaction with the RNA.
Although this model may seem speculative given that we have not shown directly that initiation factors, in particular eIF4G and likely also eIF3, remain associated with the ribosome during translation of the sORF, the hypothesis is entirely consistent with all our results. Moreover, there is no obvious viable alternative model that is consistent both with the results and with the fact that efficient reinitiation occurs only if the upstream ORF is short. While direct evidence would clearly be very desirable, the difficulty is that if the postulated interactions between the ribosomes, eIF3 and eIF4G, persist for only a short time after initiation of sORF translation, as is implied by the facts that a reduction in elongation rate decreases reinitiation efficiency (Fig. 9) and a pseudoknot (which would cause ribosome pausing) completely abolishes it (Kozak 2001), a direct demonstration of these interactions would probably be impossible with current technology. In principle, a biochemical approach could be used, which would start by forming 80S initiation complexes, necessarily over a brief time of about 60 sec (Darnbrough et al. 1973), and then attempt to fix any ribosome-associated factors by using a chemical cross-linking reagent which would have to be potentially reversible but give a high cross-linking yield within 30–60 sec yet without any artifactual cross-links. However, one has to also consider the problem that although any eIF4G in such fixed 80S complexes might be present because of the postulated eIF4G/eIF3/ribosome interaction relay, it could equally well be there by virtue of interaction of eIF4G with the mRNA, either indirectly via eIF4E as an intermediary binding the 5′-cap or via direct interaction of the eIF4G central domain with the mRNA. To ensure that only eIF4G, which is interacting with the ribosome either directly or via eIF3 as an intermediary, will be in the fixed 80S initiation complexes it would be necessary to disrupt the direct and indirect interactions between eIF4G and the mRNA. This obviously can be done only after the 80S initiation complexes have formed, but, for practical reasons, probably has to be done before the fixation step, thus seriously risking that the eIF4G/eIF3/ribosome interactions would be spontaneously disrupted in the meantime. There appears to be no satisfactory biochemical approach to demonstrating a short-term persistence of these interactions, and there is currently no way of studying such interactions in real time (e.g., via fluorescence stop-flow methods). For these reasons we have had to rely on an indirect strategy. Fortunately it has given a set of results that are remarkably self-consistent and that collectively make a compelling case for the model we have suggested.
Although the sequence of the majority of sORFs seems to have little influence on the degree to which they decrease translation of the main ORF, in a handful of cases sequence is critical. The results of mutagenesis indicate that it is the amino acid sequence of the encoded peptide (rather than the nucleotide sequence), and in some cases also the exact position of the termination codon, that is of paramount importance (for review, see Morris and Geballe 2000). Sequence-dependent sORFs of this type cause ribosomes to pause on the sORF, and it has been suggested that this inhibits translation of the main (downstream) ORF by preventing ribosomes accessing the initiation codon by leaky scanning (Morris and Geballe 2000). Although this is very plausible, our results and those of Kozak (2001) show that the pause would also have the effect of making the stalled ribosome incapable of subsequent resumption of scanning after it had overcome the barrier and completed translation of the sORF.
Materials and methods
Plasmids
All constructs were in a background of pSP73 vector (Promega) modified by insertion of an 85-residue poly(A) tract between the SmaI and BglII sites. The TPI constructs have a 5′-UTR consisting of 25 nt of residual pSP73 polylinker followed by 43 nt of TPI 5′-UTR. This 5′-UTR was replaced by either the EMCV IRES (R-strain, nt 286–833) from pEMCV L-VP0–(6A; Kaminski and Jackson 1998), or the CSFV IRES (nt 1–372) from the C/Cs cDNA construct described by Fletcher et al. (2002), or the HRV IRES (nt 15–610) from pJ 10–610 described by Borman and Jackson (1992). In all three cases, the initiation codon of the TPI-CAT ORF was in the exact position of the viral initiation codon, but in the EMCV construct its context was changed from wild-type AUAAUG to ACCAUG, and in the CSFV construct from CACAUG to CCCAUG. The 5′-UTR of TPI/norm and TPI/2ter was also replaced with the CrPV intergenic IRES (nt 6025–6232) from pGEM3-CrPV-Luc (Wilson et al. 2000), a segment that includes six codons of coding sequence downstream of the IRES. Details of plasmid construction strategy are given in the Supplemental Material.
Transcription reactions and translation assays
Prior to transcription, all constructs were linearized with SmaI, except when polyadenylated RNA (85 A residues) was required, in which case linearization was with BglII. Capped and uncapped RNAs were synthesized using bacteriophage T7 RNA polymerase, and the RNA product was isolated and quantitated exactly as described by Dasso and Jackson (1989).
In vitro translation reactions were carried out in nuclease-treated rabbit reticulocyte lysates as described previously (Kaminski et al. 1990). In brief, the reactions contained 60%–75% (by volume) nuclease-treated reticulocyte lysate (Promega), with 500 μCi/mL [35S]methionine (Amersham International, SJ1515, >1000 Ci/mmole), and incubation was at 30°C for 60 min. Added KCl was at 100 mM unless otherwise stated. For constructs with the HRV IRES, assays were carried out with 70 mM added KCl, and the reticulocyte lysate was replaced by an 80/20 (v/v) mixture of reticulocyte lysate and HeLa cell high-salt postribosomal supernatant as defined in Hunt and Jackson (1999). Translation products were separated by 20% polyacrylamide gel electrophoresis and visualized by autoradiography with Hyperfilm βmax (Amersham) or Biomax (Kodak). Quantitation was carried out by densitometry of the autoradiograms using Phoretix software or by phosporimaging and volumetric analyses using Imagequant software (Molecular Dynamics).
The preparation of eIF4G-depleted reticulocyte lysate was carried out exactly as described by Ali et al. (2001). His-tagged p100 fragment of eIF4GI (amino acids 621–1560 in the 1560 amino acid eIF4GI isoform) was overexpressed and purified exactly as described in Ali et al. (2001), and the same approach was used to obtain His-tagged p50 fragment (eIF4GI643–1076). Recombinant His-tagged R362–Q dominant negative eIF4A was purified as described by Pestova et al. (1998a).
Acknowledgments
We thank Lynne Maquat for the TPI-CAT fusion constructs; Peter Sarnow for the cricket paralysis virus IRES; Simon Morleyand Christopher Hellen for initiation factor expression constructs; Will Turner for carrying out pilot experiments; Iraj Ali for advice and for recombinant initiation factor preparations; and Rosemary Farrell for technical support. This work was supported by a Wellcome Trust Programme Grant.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.276504.
References
- Adhin M.R. and van Duin, J. 1990. Scanning model for translational reinitiation in eubacteria. J. Mol. Biol. 213: 811–818. [DOI] [PubMed] [Google Scholar]
- Ali I.K. and Jackson, R.J. 2002. The translation of capped mRNAs has an absolute requirement for the central domain of eIF4G but not for the cap-binding initiation factor eIF4E. Cold Spring Harbor Symp. Quant. Biol. 66: 377–387. [DOI] [PubMed] [Google Scholar]
- Ali I.K., McKendrick, L., Morley, S.J., and Jackson, R.J. 2001. Truncated initiation factor eIF4G lacking an eIF4E binding site can support capped mRNA translation. EMBO J. 20: 4233–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeck R. and Kolakofsky, D. 1994. Position +5 and +6 can be major determinants of the efficiency of non-AUG initiation codons for protein synthesis. EMBO J. 13: 3608–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissel J-P., Kasper, T.J., and Bunn, H.F. 1988. Cotranslational amino-terminal processing of cytosolic proteins. J. Biol. Chem. 263: 8443–8449. [PubMed] [Google Scholar]
- Borman A. and Jackson, R.J. 1992. Initiation of translation of human rhinovirus RNA: Mapping the internal ribosome entry site. Virology 188: 685–696. [DOI] [PubMed] [Google Scholar]
- Darnbrough C.H., Legon, S., Hunt, T., and Jackson, R.J. 1973. Initiation of protein synthesis: Evidence for messenger RNA independent binding of methionyl transfer RNA to 40S ribosomal subunits. J. Mol. Biol. 76: 379–403. [DOI] [PubMed] [Google Scholar]
- Dasso M.C. and Jackson, R.J. 1989. On the fidelity of mRNA translation in the nuclease-treated rabbit reticulocyte lysate system. Nucleic Acids Res. 17: 3129–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E., Preiss, T., and Hentze, M.W. 1998. Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′ end dependent. RNA 4: 828–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T.E., Yang, W., Åström, S., Byström, A.S., and Hinnebusch, A.G. 1995. Modulation of tRNAi, eIF-2 and eIF-2B expression shows that GCN4 translational control is inversely coupled to the level of eIF-2/GTP/Met-tRNA ternary complexes. Mol. Cell. Biol. 15: 6351–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher S.P., Ali, I.K., Kaminski, A., Digard, P., and Jackson, R.J. 2002. The influence of viral coding sequences on pestivirus IRES activity reveals further parallels with translation initiation in prokaryotes. RNA 8: 1558–1571. [PMC free article] [PubMed] [Google Scholar]
- Grünert S. and Jackson, R.J. 1994. The immediate downstream codon strongly influences the efficiency of utilisation of eukaryotic translation initiation codons. EMBO J. 13: 3618–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett P.B., Petersen, R.B., Hensel, C.H., Albericio, F., Gunderson, S.I., Palmenberg, A.C., and Barany, G. 1986. Synthesis in vitro of a 7 amino acid peptide encoded in the leader RNA of Rous sarcoma virus. J. Mol. Biol. 190: 45–57. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A.G. 1997. Translational regulation of yeast GCN4: A window on factors that control initiator-tRNA binding to the ribosome. J. Biol. Chem. 272: 21661–21664. [DOI] [PubMed] [Google Scholar]
- Hunt S.L. and Jackson, R.J. 1999. Polypyrimidine tract binding protein is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA 5: 344–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J. 1989. An unusual co-ordinated cleavage event in the processing of encephalomyocarditis virus polypeptides. Virology 172: 363–366. [DOI] [PubMed] [Google Scholar]
- ____. 2000. A comparative view of initiation site selection mechanisms. In Translational control of gene expression (eds. N. Sonenberg, J.W.B. Hershey, and M.B. Mathews), pp. 127–183. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Jan E. and Sarnow, P. 2002. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 324: 889–902. [DOI] [PubMed] [Google Scholar]
- Kaminski A. and Jackson, R.J. 1998. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA 4: 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski A., Howell, M.T., and Jackson, R.J. 1990. Initiation of encephalomyocarditis virus RNA translation: The authentic initiation site is not selected by a scanning mechanism. EMBO J. 9: 3753–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori Y. and Nakashima, N. 2001. A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA 7: 266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontos H., Napthine, S., and Brierley, I. 2001. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol. Cell. Biol. 21: 8657–8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44: 283–292. [DOI] [PubMed] [Google Scholar]
- ____. 1987. Effects of intercistronic length on the efficiency of reinitiation by eukaryotic ribosomes. Mol. Cell. Biol. 7: 3438–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1989. The scanning model for translation–An update. J. Cell Biol. 108: 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1991a. A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr. 1: 111–115. [PMC free article] [PubMed] [Google Scholar]
- ____. 1991b. An analysis of verterbrate messenger RNA sequences–Intimations of translational control. J. Cell Biol. 115: 887–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2001. Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 29: 5226–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luukkonen B.G.M., Tan, W., and Schwartz, S. 1995. Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J. Virol. 69: 4086–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerschell R.P., Hosokawa, Y., Tsunasawa, S., and Sherman, F. 1990. The specificities of yeast methionine aminopeptidase and acetylation of amino-terminal methionine in vivo. J. Biol. Chem. 265: 19638–19643. [PubMed] [Google Scholar]
- Morris D.R. and Geballe, A.P. 2000. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 20: 8635–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A.N., Méthot, N., Svitkin, Y., Merrick, W.C., and Sonenberg, N. 1994. Dominant negative mutants of mammalian translation initiation factor eIF4A define a critical role for eIF4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13: 1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody D.S., Subramani, S., and Berg, P. 1986. Effect of upstream reading frames on translation efficiency in simian virus 40 recombinants. Mol. Cell. Biol. 6: 2704–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peri S. and Pandey, A. 2001. A reassessment of the translation initiation codon in vertebrates. Trends Genet. 17: 685–687. [DOI] [PubMed] [Google Scholar]
- Pestova T.V. and Hellen, C.U.T. 2003. Translation elongation after assembly of ribosomes on the cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes & Dev. 17: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V. and Kolupaeva, V.G. 2002. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes & Dev. 16: 2906–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Hellen, C.U.T., and Shatsky, I.N. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16: 6859–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Shatsky, I.N., Fletcher, S.P., Jackson, R.J., and Hellen, C.U.T. 1998a. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes & Dev. 12: 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Borukhov, S.I., and Hellen, C.U.T. 1998b. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394: 854–859. [DOI] [PubMed] [Google Scholar]
- Somogyi P., Jenner, A.J., and Brierley, I. 1993. Ribosomal pausing during translation of an RNA pseudoknot. Mol. Cell. Biol. 13: 6931–6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas K.R. and Capecchi, M.R. 1986. Introduction of homologous DNA sequences into mammalian cell lines induces mutations in the cognate gene. Nature 324: 34–38. [DOI] [PubMed] [Google Scholar]
- Wilson J.E., Pestova, T.V., Hellen, C.U.T., and Sarnow, P. 2000. Initiation of protein synthesis from the A site of the ribosome. Cell 123: 511–520. [DOI] [PubMed] [Google Scholar]
- Wolin S.L. and Walter, P. 1988. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 7: 3559–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun D.F., Laz, T.M., Clements, J.M., and Sherman, F. 1996. mRNA sequences influencing translation and the selection of AUG initiator codons in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 19: 1225–1239. [DOI] [PubMed] [Google Scholar]
- Zhang J. and Maquat, L.E. 1997. Evidence that translation reinitiation abrogates nonsense-mediated mRNA decay in mammalian cells. EMBO J. 16: 826–833. [DOI] [PMC free article] [PubMed] [Google Scholar]