Abstract
Integration of HIV-1 DNA into the genome of the host cell is an essential step in the viral replication cycle that is mediated by the virally encoded integrase protein. We have used atomic force microscopy to study stable complexes formed between HIV-1 integrase and viral DNA and their interaction with host DNA. A tetramer of integrase stably bridges a pair of viral DNA ends, consistent with previous analysis by gel electrophoresis. The intasome, comprised of a tetramer of integrase bridging a pair of viral DNA ends, is highly stable to high ionic strength that would strip more loosely associated integrase from internal regions of the viral DNA. We also observe tetramers of integrase associated with single viral DNA ends; time course experiments suggest that these may be intermediates in intasome assembly. Strikingly, integrase tetramers are only observed in tight association with viral DNA ends. The self-association properties of intasomes suggest that the integrase tetramer within the intasome is different from the integrase tetramer formed at high concentration in solution in the absence of viral DNA. Finally, the integration product remains tightly bound by the integrase tetramer, but the 3′ ends of the target DNA in the complex are not restrained and are free to rotate resulting in relaxation of initially supercoiled target DNA.
Keywords: integrase, intasome, HIV-1, DNA integration, retrovirus
Introduction
Integration of a DNA copy of the viral genome into the DNA of the host cell is an essential step in the replication cycle of HIV-1 and other retroviruses 1,2. After infection, the virion core is uncoated to expose a nucleoprotein complex, termed the reverse transcription complex (RTC)3 that contains two copies of the viral RNA, reverse transcriptase, and integrase along with other viral and cellular proteins. Reverse transcription occurs within the RTC to generate the linear blunt ended viral DNA that is the substrate for integration. The viral DNA remains associated with viral and cellular proteins in a complex called the preintegration complex (PIC) 4. The PIC is then transported to the nucleus where the viral DNA is integrated into the host genome by the viral integrase protein. The PIC is a poorly defined entity that contains a number of proteins in addition to integrase that may play roles in nuclear import and/or auxiliary roles in integration. PICs isolated from infected cells efficiently integrate their viral DNA in vitro5,6,7 and this system has provided a powerful tool for dissecting the integration mechanism. Analysis of the structure of the integration intermediate8,9 showed that integration proceeds in a two- step process. First, two nucleotides are cleaved from each end of the initially blunt-ended viral DNA in a reaction termed 3′ end processing. Then, in the strand transfer step, the 3′ hydroxyls of the terminal adenosine at each end of the viral DNA exposed by the 3′ processing reaction attack a pair of phosphodiester bonds in the target DNA. In the resulting integration intermediate, the 3′ ends of the viral DNA are covalently joined to the 5′ ends of the target DNA at the site of integration. The 5′ ends of the viral DNA and the 3′ ends of the target DNA are not joined in the integration intermediate which must be subsequently repaired by cellular enzymes to complete the integration process. A striking feature of the association of integrase with viral DNA in the PIC is its stability against high-ionic strength which results in dissociation of many other protein DNA complexes; PICs remain integration competent after challenge with ionic strength of greater than 0.5M NaCl10,11,12 implying that integrase remains tightly associated with the viral DNA.
HIV-1 integrase catalyzes the key DNA cutting and joining steps of integration in vitro with simple DNA substrates that mimic the two ends of the viral DNA and either Mg2+ or Mn2+ as a cofactor13,14,15. Under most in vitro reaction conditions the DNA strand transfer reaction products differ from those generated in vivo in that only a single viral DNA end is inserted into a single strand of target DNA, a reaction referred to as half-site integration. In vivo, both ends of viral DNA insert at the site of integration, with a stagger of 5 base pairs separating the sites of integration on the two target DNA strands in the case of HIV-1. The reason behind the sloppiness of the in vitro reaction that results in this uncoupling of insertion of pairs of viral DNA ends is unknown. Half-site integration occurs efficiently under a broad range of reaction conditions in vitro, but under such conditions highly stable complexes that mimic the in-vivo association of integrase with viral DNA in the PIC have not been detected.
Concerted integration, also called full-site integration, of pairs of viral DNA ends can be catalyzed by HIV-1 integrase in vitro but the reaction conditions are far more restrictive than for half-site integration16,17,18,19. The viral DNA substrates must be much longer than the 20 base pairs which are sufficient for near maximal efficiency of half-site integration. Concerted integration also requires the presence of high concentrations of the crowding agent polyethylene glycol (PEG) in the reaction mixture and is exquisitely sensitive to both the concentration and stoichiometry of integrase and DNA substrates. Under reaction conditions that promote concerted integration, stable complexes of integrase with viral DNA are formed that mimic the association of integrase with viral DNA in the PIC20,21. These stable synaptic complexes (SSCs) are intermediates on the integration reaction pathway and are rapidly converted into strand transfer complexes (STCs) in the presence of target DNA. In the STC, a pair of viral DNA ends is integrated into the target DNA with integrase remaining tightly associated with the integration product at the site of insertion. Although SSCs are intermediate products in the integration reaction they can be trapped by preventing DNA strand transfer20. This can be accomplished by using a selective inhibitor of DNA strand transfer or carrying out the reaction with pre-processed viral DNA ends terminating with dideoxy adenosine and therefore lacking the 3′-OH that is the nucleophile for attacking the phophodiester bond in the target DNA. A third way to trap the SSC is to use an integrase enzyme with an active site mutation in conjunction with pre-processed viral DNA ends.
We have previously analyzed HIV-1 SSCs by gel electrophoresis in combination with crosslinking and footprinting studies and demonstrated that the viral DNA ends are stably synapsed by a tetramer of integrase that protects approximately 20 base pairs of terminal viral DNA sequence22. Here, we have used atomic force microscopy (AFM) to directly visualize nucleoprotein complexes formed by HIV-1 integrase and DNA substrate under conditions that favor concerted DNA integration. The major species are SSCs in which, a pair of viral DNA ends are bridged by integrase. However, we also observe other species that were not detected in previous analysis by gel electrophoresis, either because they were not stable during electrophoresis or did not enter the gel. A significant fraction of the viral DNA molecules, especially at early time points of assembly, consist of a tetramer of integrase bound to a single viral DNA end. Kinetic studies suggest these are an intermediate on the pathway to the SSC. Strikingly, viral DNA ends bound to integrase dimers were essentially undetectable. The SSCs self -associate through the integrase tetramers suggesting that the integrase tetramer within the SSC is different from free tetramers of integrase in solution that do not self-associate under the same conditions. Finally, the target DNA associated with the SSCs is relaxed even though it was initially supercoiled prior to the reaction. Thus, despite being tightly associated with an integrase tetramer, the 3′ ends of the target DNA strands at the site of integration must be free to rotate before integrase is dissociated. This has implications for the repair of the viral DNA within the STC that must occur to complete the integration process.
Results
Visualization of SSCs by atomic force microscopy
Stable synaptic complexes of HIV-1 integrase and viral DNA substrate were assembled and visualized by atomic force microscopy (AFM) as described in Materials and Methods. The viral DNA substrate contained 32 bases of HIV-1 U5 terminal DNA sequence and non-specific flanking sequence22. Stable synaptic complexes (SSCs) are expected to be visualized as two DNA substrate molecules bridged by a multimer of integrase. In order to prevent DNA strand transfer, the viral DNA substrate was “pre-cleaved” and terminated with 3′ dideoxyadenosine on the transferred strand. Only large uninterruptible aggregates were seen when the low salt assembly mixture was deposited directly onto the derivatized mica. However, when the assembly buffer was exchanged to 500 mM NaCl buffer prior to deposition discrete nucleoprotein complexes were observed. Fig. 1A shows a typical field of view. The majority of the complexes contained a pair of viral DNA ends bridged by integrase (SSCs); a collage of individual complexes at higher magnification is shown in Fig. 1B. The size of the integrase oligomer bridging the viral DNA ends appeared to be relatively uniform and larger than free oligomers of integrase in the absence of DNA; we will refer this oligomer of integrase and the terminal viral DNA sequence as the intasome. When deposited at 500 mM NaCl, viral DNA external to the intasome was largely devoid of bound integrase, although some smaller units were sparcely bound on some DNA molecules. In addition to SSCs, complexes consisting of only a single viral DNA with integrase at the terminus, Single End Complexes (SECs), were also observed (Fig. 1C).
Figure 1.

AFM images of Stable Synaptic Complexes (SSCs) on APS mica. (A) Panoramic view of a 3 μm region. Arrows point to integrase bridging a pair of viral DNA DNA ends within SSCs. (B) Close-ups (340 nm squares) of individual SSCs consisting of two linear 1kb “viral” DNA and an integrase tetramer. (C) Close-ups (340 nm squares) of Single End Complexes (SECs) consisting of one linear 1kb “viral” DNA and an integrase tetramer. Dimensions are indicated by scale bars.
The intasome contains a tetramer of integrase
The volumes of integrase oligomers unambiguously synapsing a pair of viral DNA molecules were calculated as described in Materials and Methods and plotted as a histogram (Fig. 2A). The predominant volume for the SSC is about 220 nm3 (> 60% of particles). The volume distribution, as seen in the histogram, appears rather broad. This is commonly observed with AFM-estimated protein volumes. The complex electrostatic interactions between the protein and the substrate, as well as those between the protein and the AFM probe typically result in significant broadening of measured particle volume distributions, even after correcting for the finite tip size. However, the center of an AFM-based volume histogram can be assigned with a high certainty and robustness. In the case of SSC, this volume of 220 nm3, which according to Eq. (1) (see Materials and Methods) corresponds to a molecular mass of between 115 and 130 kDa depending on the hydration ratio of the intasome. An integrase tetramer would have a mass of 129 kDa, so we conclude that the majority of the complexes contain a tetramer of integrase consistent with previous crosslinking experiments. The distribution is skewed to the right with the presence of larger protein oligomers, most likely dimers and trimers of the basic tetrameric unit. The latter might be either small aggregates of 2–3 SSCs, all but one of which have lost their DNAs upon adsorption to the substrate, or aggregates of a single SSC with unbound IN. The long term stability of the SSC (data not shown) points to a highly stable association and argues for the second scenario. It is striking that integrase tetramers were never observed in the absence of DNA, bound to DNA at locations internal from the ends, or both bound at both ends of the viral DNA substrate. Assembly of the tetramer therefore requires sequence-specific interactions with the viral DNA end. The latter conclusion is confirmed by the similar core volume distribution observed for SEC (Fig. 2B). The mean is only slightly lower than that of the SSC but still closer to the tetramer than any other oligomer. Again, the distribution is expectedly broad for the same reasons described for the SSC, with the possible additional factor of a different bias in the preferred protein adsorption orientation when it is “anchored” by only one DNA.
Figure 2.
(A) Histogram of the volume distribution of the intasomes in the SSCs. Only complexes containing two viral DNA ends exiting from the integrase multimer were included in the analysis. A fit to three Gaussian components is superimposed on the histogram. (B) Histogram of the volume distribution of SECs
DNA chain exit angles and attachment orientations
We measured the apparent exit angles between the two DNA chains in intasomes and the results are plotted in Fig. 3, together with a double Gaussian fit of the data; the fit excludes the zero degree complexes which appear to comprise a discrete sub-population in which the exiting DNAs are intertwined. We speculate that the observed distribution reflects two preferred orientations of adhesion of the complexes to the mica surface. Consistent with this interpretation, SSCs with angles closer to the 120° region of the distribution are lower in height (~3.3 nm ) compared to those closer to 70° (~4 nm). In addition, the preferred orientation of the elongated intasome with respect to the DNA chains is visibly different between complexes with the ~70° and the ~120° DNA angles (data not shown). Since projection of the angle between the DNAs onto the two-dimensional mica surface can only decrease the apparent angle, the real angle is likely to be closer to the higher end of the observed distribution.
Figure 3.
The SSC “projections” onto a substrate plane typically show a broad distribution of predominantly obtuse angles at which the two DNA exit the integrase core. Acute angles <60° are only rarely seen, while all the observed 0° angles correspond to the two DNAs winding around each other near the protein core (left). The orientation of the elongated intasome relative to the DNA is different between the approximately 70° and 120° complexes (compare middle and right images).
Intasomes self-associate
Although integrase tetramers bridging a pair of viral DNA ends were the predominant species when deposited on the mica at 0.5 M NaCl, we also observed higher order aggregates of such complexes. Self-association of SSCs is also observed in gel electrophoresis experiments (Fig 4A). When complexes are assembled with a mixture of viral DNAs of two different lengths, the single SSC splits into three bands (Fig 4B, Group 1), corresponding to short paired with short, short paired with long, and long paired with long DNAs as previously derscribed22. The next complex in the ladder series (Fig. 4B, Group 2) splits into five bands demonstrating that it consists of two SSCs; beyond the third complex in the series the number of bands cannot be unambiguously counted but they clearly increase in number, consistent with a simple series of SSCs. AFM visualization can add more detailed information about the aggregation process. First it appears that lower concentrations of NaCl increase both the number and size of aggregates (Fig. 4C) although it is clear that the constituent complexes of these aggregates are the same as seen individually at higher salt. Strikingly, aggregation is mediated by self-association of the intasomes in the SSCs and the rest of the DNA length is largely devoid of bound integrase. This is clearly seen in complexes of a small number of SSCs as shown in Fig. 4C. Under the same conditions, HIV-integrase in solution does not form oligomers greater than tetramers (data not shown). Although, self-association of intasomes cannot be biologically relevant because PICs contain only a single pair of viral DNA ends and can therefore make only one intasome, it implies that the integrase tetramer within the intasome is different from the integrase tetramer in solution, because the later does not form higher order oligomers under the same conditions. Integrase tetramers associated with a single viral DNA end also associate with intasomes, as evidenced by odd numbers of DNA molecules exiting some complexes, suggesting that at least part of the integrase oligomer (perhaps the monomers that bind the single DNA) in these complexes may be in the same configuration as in the intasome.
Figure 4.
Self-association of intasomes. (A) Self-association of SSCs is evident as a ladder of bands in gel electrophoresis. The extent of self-association is variable, but increases with incubation time and is greater after incubation at low ionic strength. (B) The ladder is due to self-association of assembled SSCs and not recruitment of additional protomers of integrase. When SSCs are assembled from a mixture of “short” and “long” viral DNA molecules, the first band in the ladder series splits into three (Group 1) as a result of pairing of short with short, short with long, and long with long DNA molecules. The next band in the ladder series splits into five (Group 2), consistent with self-association of two SSCs. The number of bands in the higher order members of the series cannot be unambiguously counted. (C) AFM shows that self-association occurs through the intasomes and does not involve bridging of internal regions of the viral DNA by integrase. The images on the left and middle were from a sample deposited at 500 mM NaCl, while the image on the right was from a sample deposited at 150 mM NaCl.
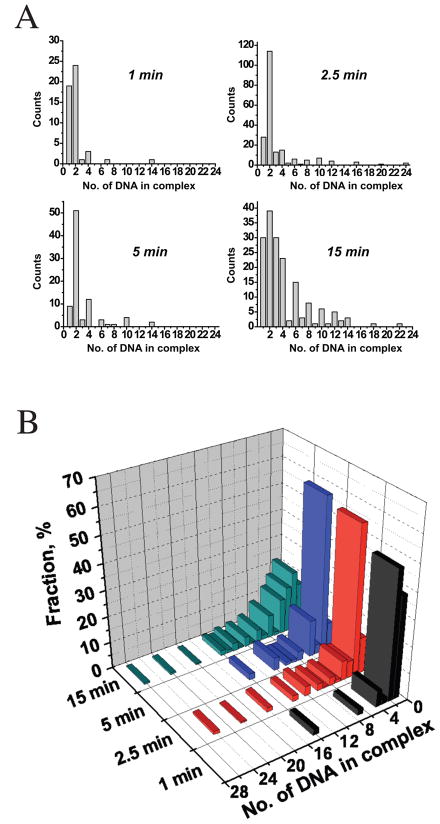
Time course of complex formation
We carried out a time course of complex assembly to investigate the temporal relationship between the appearance the SECs, SSCs, and the higher order aggregates. For this purpose, we counted the number of DNAs exiting all complexes for which the DNA molecules could be unambiguously counted, excluding larger aggregates for which the number of DNA molecules could not be counted with certainty. Figure 5A shows histograms of the number of DNAs exiting from complexes after four different assembly times and the same data is represented as a three dimensional histogram in Fig. 5B. At the earliest time assembly time point of 1 min SECs and SSCs predominated and were almost equal in abundance. By 2.5 min, complete SSCs with a pair of viral DNAs exiting a single intasome were the dominant species. As the incubation time was further lengthened, SSCs and SECs self-associated to form higher order aggregates. Moreover, the SSC aggregates with even numbers of DNA dominate and follow a trend characteristic of an indefinite association (see Supplemental Material) while the number of SSC aggregates with odd numbers of DNA remains small and does not change with the size of the SSC aggregates. It is also notable that after prolonged storage, the ratio of SECs to SSCs remains essentially unchanged (data not shown). The protein volume distribution for individual SSC remained unchanged with the reaction time, as well as with prolonged storage. In the absence of DNA strand transfer, SSCs appeared to be extremely stable, as we observed noticeable SSC decomposition only after months of storage.
Figure 5.
Time course of complex formation. The number of DNA chains exiting integrase clusters (tetramers and higher order units resulting from self-association) were plotted for various times of complex assembly. Larger clusters for which the DNA chains could not be unambiguously counted were ignored. (A) Histograms of the distribution of the number of DNAs per cluster after 1 min, 2.5 min, 5 min, and 15 min assembly times. (B) The same data represented as a three dimensional histogram.
The 3′ ends of the target DNA are unrestrained in the STC
SSCs were trapped using viral DNA ends with a 3′ dideoxyadenosine, which prevents DNA strand transfer. When reactions are carried out in the presence of Mg2+ with normal viral DNA ends terminating with an adenosine with a 3′ hydroxyl, together with a plasmid DNA as the target, strand transfer complexes (STC) are the predominant complexes observed in non-denaturing electrophoresis22. We found that the electrophoretic mobility of STCs is the same regardless of whether the target DNA is initially supercoiled or nicked (Fig. 6A), a result that would not be expected if the supercoiling of the target DNA is maintained in the STC. AFM imaging of STCs confirmed that the target DNA is relaxed in the STC even when it was initially supercoiled (Fig 6B); this is not due to contaminating nuclease activity because the majority of the unreacted plasmid DNA molecules remained supercoiled (Fig. 6B and data not shown). We conclude that the 3′ ends of the cut target DNA are not restrained in the STC and are free to rotate resulting in relaxation of the target DNA.
Figure 6.
(A) Strand transfer complexes (STCs) were assembled with 1 kb viral DNA and either supercoiled (S) or nicked (N) pBR322 plasmid DNA as the target. The products were electrophoresed with or without deproteinization. The mobilities of the STCs assembled with supercoiled or nicked target DNA are indistinguishable. (B) AFM image showing an STC with 1 kb viral DNA and unreacted target DNA molecules. (C) Close-up image of an STC made with 355 bp viral DNA.
Discussion
Bridging of viral DNA ends by a tetramer of integrase
Our results demonstrate that a tetramer of integrase stably synapses a pair of viral DNA ends in agreement with previous analysis of SSCs by gel electrophoresis22. This stable binding of integrase to the viral DNA ends mimics the stable association of integrase with PICs that retain integration activity at high ionic strength. Volume measurements confirm the stably bound unit of integrase to be tetrameric. Strikingly, apart from the termini associated with the integrase tetramer, the rest of the viral DNA appeared devoid of integrase highlighting the stability of the SSC compared to simple non-specific binding of integrase to DNA. Integrase protomers bound sparsely to internal regions of the viral DNA were monomeric or dimeric (data not shown) in contrast to the tetramers specifically associated with the viral DNA termini. It remains possible that additional more weakly associated protomers contribute to integration efficiency, but are stripped under the high ionic strength conditions used for sample preparation. At lower ionic strength the SSCs aggregate (discussed below) making it difficult to visualize the entire length of the DNA in individual complexes.
DNA chains exit the intasome at an obtuse angle
The angle at which the two DNA molecules exit the intasome cannot be accurately determined by AFM because we visualize a two dimensional projection of a three dimensional structure. Judging from the orientation of the DNA chains of the SSC, however, it appears that the intasome attaches to the hydrophobic substrate stronger than the DNA. This is based on the occasional observation whereby the multiple DNA chains of an SSC aggregate tend to orient in similar directions, apparently forced by the rinsing water, but kept immobilized by the protein complexes. Interestingly, the protein in the absence of DNA does not adsorb easily to the APS substrate whereas the IN of an SSC does. It is therefore reasonable to assume that the orientation of the complex on the mica will largely be determined by the tetramer orientation that facilitates its adsorption to the surface. This may well result in preferred attachment orientations and hence different apparent DNA exit angles. It follows that the orientation of the complexes on the mica are not expected to be random. Inspection of SSC images such as those shown in Fig. 1 reveals that the angle distribution for the two DNA chains is not straightforward to interpret. Based on the height differences between the intasomes with acute and obtuse DNA angles, and the corresponding relative orientations of the elongated intasome with respect to the DNA chains, we believe that we are observing two different populations of SSCs distinguished by their attachment orientation. The true angle is likely closer to the higher end of the observed distribution, because projection onto the two dimensional surface can only result in an apparent decrease. In the recently published crystal structure of the closely related prototype foamy virus intasome 23, the axes of the two oligonucleotide DNAs make an angle close to 90° with one another. This is fully consistent with our results, although longer DNAs may make additional contacts so the angle of exit from the intasome need not be equivalent to the angle made by the DNA helices in the vicinity of the active site.
Evidence for a concerted integration intermediate with a single viral DNA end stably bound to a tetramer of integrase
The evidence that concerted integration is mediated by a tetramer of integrase is compeling22,24. With assembly times of 30 min to 1 h, the majority of complexes consisted of a pair of viral DNA ends associated with an integrase tetramer (SSCs) as shown in Fig 1A. However, other species were also observed at lower frequency. A minor but significant fraction of DNA molecules were SECs consisting of a tetramer of integrase bound to only a single viral DNA end. Other complexes contained three of more DNA molecules associated with integrase larger than a tetramer (ranging from octameric to large aggregates). The SEC species is particularly interesting and potentially represents an intermediate in assembly of the SSC or a breakdown product formed after assembly. We have not observed the SEC in gel electrophoresis experiments (data not shown), suggesting that it may be less stable than the SSC and not survive gel electrophoresis. To gain insight into whether the SEC may be an intermediate on the pathway to the SSC we carried out a time course of assembly (Fig. 5). The proportion of SECs relative to SSCs (including those in aggregates) was greatest at the earliest time point and comparable to the SSCs in abundance. At subsequent time points the proportion of SECs decreased whereas the SSCs and aggregates progressively increased in number. No increase in SECs was observed at later time points or after extended storage of the assembled complexes. These results are consistent with the SEC being an intermediate on the pathway to assembly of the SSC.
Absence of stable association of viral DNA ends with integrase dimers
Models of the nucleoprotein complex that carries out concerted DNA integration require at least a tetramer of integrase. Assuming the catalytic core domain dimer observed in all crystal structures to date is preserved in the active complex, a tetramer or higher multimer is required to position a pair of active sites with the correct spacing for concerted integration. However, it has been suggested that a dimer may be sufficient for integration of single viral DNA ends and that assembly of a dimer on each viral DNA may be an intermediate in the assembly of the SSC that mediates concerted integration24,25,26. Our results do not support this hypothesis. In fact, in our AFM studies integrase dimers on the ends of the viral DNA were essentially absent, even at the 1 min assembly time point where SECs were seen in abundance. Therefore if dimers do bind to DNA ends these dimers must either be unstable and be dissociated under our conditions for AFM or, once a dimer binds the DNA signal sequence a second dimer binds so fast that a dimer bound to the DNA is never seen. In contrast, tetramers assembled on single DNA ends were readily observed, the latter fact being consistent with the SEC protein volume distribution (Fig. 2B).
Integrase tetramer in the SSC differs from the tetramer in the absence of DNA
The vital unit of integrase in the SSC that carries out concerted integration is a tetramer. Integrase also forms tetramers in solution as well as monomers and dimers. Integrase aggregates also form at low ionic strength. The question arises as to whether the solution tetramer is the same as the tetramer in the SSC. The aggregation properties of the SSC persuade us that they are different. SSCs tend to form a ladder of bands in gel electrophoresis (Fig. 4A). Experiments in which SSCs are formed with DNAs of different length indicate this ladder is a series of SSCs, formed from units of individual SSCs each comprising a tetramer of integrase and two DNAs (Fig. 4B). Our AFM studies show that the self-association of SSCs occurs directly through contacts of tetramers bound to DNA and not through integrase-DNA contacts involving DNA not sequestered within the intasome. Self-association of SSCs is greater at longer time points and is greatly promoted by lowering the ionic strength below 0.5M NaCl during sample preparation. Even at 0.5M, some of the complexes are self-associated and at 0.1M NaCl the aggregates become so large that individual SSCs are difficult to locate. Once formed, SSCs remain aggregated under conditions where integrase protein alone is readily solubilized by high ionic strength. Moreover, judging from the orientation of the DNA chains of the SSC it appears that the protein oligomer attaches to the hydrophobic substrate stronger than the DNA. This is based on the frequent observation whereby the multiple DNA chains of an SSC aggregate tend to orient in similar directions, apparently forced by the rinsing water but kept immobilized by the intasomes. Interestingly, the protein in the absence of DNA does not adsorb easily to the APS substrate. Based on the above observations, we propose that the integrase tetramer within the SSC differs from that in solution resulting in its greater propensity to aggregate.
In aggregates containing only a few SSCs the integrase tetramers are usually readily resolved in AFM as shown in Fig. 4C. However as the numbers of SSCs in the aggregate increases, counting the SSCs becomes problematic. In contrast, counting the number of DNA molecules within the aggregated complexes is much easier. The distribution of the number of DNA molecules in the aggregates is revealing. The histogram in Fig. 5 shows that even numbers of DNAs predominate over odd numbers. This reinforces our conclusion that the aggregates result from self-association of intasomes and not non-specific aggregation of integrase and DNA. In addition to the predominant single SSCs and SECs observed when samples are prepared at 0.5M NaCl, we also observe association of SSCs and SECs, each with their own clearly resolved integrase tetramer. Odd numbers of DNA molecules, such as three DNAs exiting from two integrase tetramers, are seen with these complexes. Counting the DNAs exiting from the larger aggregates indicates they also contain SECs. However, SECs must be a minor component of the aggregates because of the preponderance of aggregates containing even numbers of DNA molecules. Furthermore, the ratio of SECs to SSCs remains essentially constant over long storage times. These observations also support our proposition that SECs are an intermediate and their failure to accumulate during the time course is not due to their consumption into aggregates. We conclude that the self-association properties of SSCs and SECs are indistinguishable, but differ from integrase tetramers in the absence of DNA.
The 3′ ends of the cleaved target DNA are not constrained within the STC
When reactions are carried out with wild type integrase, viral DNA ends with 3′ hydroxyls, and a circular target DNA, strand transfer complexes (STCs) form efficiently and are readily visualized by AFM (Fig. 6). Volume measurements confirm that integrase within the STC is tetrameric. It is notable that SSC self-association does not prevent the formation of STCs. Also, even when the circular target DNA is supercoiled, the target DNA segment associated with the SSC is always relaxed. This is not the result of nuclease contamination because a large majority of unreacted target DNA molecules remain supercoiled (Fig. 6B and data not shown). This result is confirmed when products are analyzed by gel electrophoresis; the STC migrates at the same position regardless of whether the target DNA is initially supercoiled or nicked (Fig. 6A). We conclude that the 3′ ends of cleaved target DNA in the STC are not sequestered within the complex but are free to rotate and allow relaxation of the target DNA. This may have important consequences for integration in vivo where cellular enzymes must complete the integration process, taking over after formation of the integration intermediate by integrase. Our results suggest that the 3′ ends of the cleaved host DNA may be accessible to cellular repair enzymes even before dissociation of the integrase tetramer.
Materials and Methods
Preparation of SSCs and STCs
Viral DNA substrates and HIV-1 integrase protein were prepared as described20. The integrase used was the W235F mutant because of its reduced aggregation propensity. Except as otherwise stated in figure legends, complexes were assembled using 1 kb precleaved viral DNAs terminating with a 3′ dideoxyadenosine. Strand transfer complexes were made using blunt end viral DNA and supercoiled pUC18 as the target. Typical reaction mixtures for assembly of SSCs (25 μl final volume) were assembled by incubating 400 nM integrase on ice in 20 mM HEPES pH 7.5, 12% DMSO, 5 mM DTT, 10% PEG-6000, 10 mM MgCl2, 20 μM ZnCl2, and 200 mM NaCl (final), followed by addition of 50 nM viral DNA substrate. These components were preincubated on ice for 0.5 hr and the reaction was then initiated by transfer to 37°C and incubation was continued for 2 hrs. The reactions were stopped by addition of 10 mM EDTA and incubated at 30°C for 30 min. The reaction mixture was centrifuged at 10,000 g for 40 min, the supernatant was removed, and the pellet was resuspended in 10 μl suspension buffer (20 mM Hepes, pH 7.5, 500mM NaCl). The solution was then passed through a P30 column equilibrated with suspension buffer. The DNA concentration was measured using a NanoDrop spectrophotometer. For AFM analysis the solutions were further diluted to the desired concentration.
The sample preparation procedure was modified as follows for the time-course experiments. The assembly reaction was stopped by adding 10 times volume of ice-cold suspension buffer and incubation was continued for another 15 min on ice. The sample was briefly centrifuged at 15,000 g for 5 min and the supernatant was diluted to desired concentration.
AFM imaging
For AFM imaging, the SSC or STC stock solution was diluted in 20 mM HEPES, 0.5 M NaCl buffer. To achieve similar DNA base-pair concentrations, the final concentrations ranged from 1 nM to 30 nM, depending on the DNA lengths used. Typically, a 7 μl drop was placed onto a freshly cleaved 12 mm mica disk (Ted Pella, Inc.) pre-treated with 1-(3-aminopropyl)silatrane (APS) solution according to the protocol described previously27. After 10 min incubation, the mica was washed with 400 μl of deionized water and dried in argon gas flow. Commercial AFM instruments (Picoforce Multimode with Nanoscope V controller and E scanner head and a Bioscope with Nanoscope V controller and Hybrid SZ-scanner head by Veeco, CA, USA) were used for imaging. All AFM imaging was performed in the tapping mode using supersharp probes (TESP-SS by Veeco, CA, USA) with nominal radii of 2–5 nm and nominal spring constant of 42 N/m, in air under controlled humidity.
The raw AFM images were preprocessed with Nanoscope software and analyzed with the NIH ImageJ image analysis software package.
Volume calculations
The volumes of the protein core particles of SSC were measured on the assumption of their ellipsoidal shape. The axes of each ellipsoid were approximated taking into account the AFM probe shape, using a simple geometric model for convolution of the spherical tip and ellipsoidal molecule. We estimated the probe dimensions based on the DNA width at half height, measured in the same image. The volume of a protein, Vpr, in air is estimated by including the residual hydration water of the molecule 28,
| Eq. 1 |
where Mpr is the molecular mass of the protein, Vp, the partial specific volume of the protein (~0.73–0.74 cm3/g), d is the hydration ratio in grams of water per gram of protein, Vw is the partial specific volume of water (1 cm3/g) and Na is Avogadro’s number. The true hydration ratio depends on protein composition and conformation but for a variety of proteins it has been measured to be in the range of 0.3–0.4.
Supplementary Material
Acknowledgments
This work was supported by the intramural research programs of the NIDDK and NIBIB of the National Institutes of Health and by the NIH AIDS Targeted Antiviral Program.
Abbreviations
- HIV-1
human immunodeficiency virus type 1
- SSC
stable synaptic complex
- STC
strand transfer complex
References
- 1.Brown PO. Integration. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press; 1997. pp. 161–203. [PubMed] [Google Scholar]
- 2.Craigie R. Retroviral DNA Integration. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. ASM Press; Washington, D.C: 2002. pp. 613–630. [Google Scholar]
- 3.Warrilow D, Tachedjian G, Harrich D. Maturation of the HIV reverse transcription complex: putting the jigsaw together. Reviews in Medical Virology. 2009;19:324–337. doi: 10.1002/rmv.627. [DOI] [PubMed] [Google Scholar]
- 4.Bowerman B, Brown PO, Bishop JM, Varmus HE. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989;3:469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- 5.Brown PO, Bowerman B, Varmus HE, Bishop JM. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 6.Ellison V, Abrams H, Roe T, Lifson J, Brown P. Human immunodeficiency virus integration in a cell-free system. J Virol. 1990;64:2711–2715. doi: 10.1128/jvi.64.6.2711-2715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farnet CM, Haseltine WA. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc Natl Acad Sci USA. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujiwara T, Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 9.Brown PO, Bowerman B, Varmus HE, Bishop JM. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci USA. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee MS, Craigie R. Protection of retroviral DNA from autointegration: involvement of a cellular factor. Proc Natl Acad Sci USA. 1994;91:9823–9827. doi: 10.1073/pnas.91.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farnet CM, Bushman FD. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 12.Wei SQ, Mizuuchi K, Craigie R. A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J. 1997;16:7511–7520. doi: 10.1093/emboj/16.24.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bushman FD, Fujiwara T, Craigie R. Retroviral DNA integration directed by HIV integration protein in vitro. Science. 1990;249:1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- 14.Sherman PA, Fyfe JA. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci USA. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 16.Hindmarsh P, Leis J. Reconstitution of concerted DNA integration with purified components. Advances in Virus Research. 1999;52:397–410. doi: 10.1016/s0065-3527(08)60308-5. [DOI] [PubMed] [Google Scholar]
- 17.Sinha S, Pursley MH, Grandgenett DP. Efficient concerted integration by recombinant human immunodeficiency virus type 1 integrase without cellular or viral cofactors. J Virol. 2002;76:3105–3113. doi: 10.1128/JVI.76.7.3105-3113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, Craigie R. Processing the viral DNA ends channels the HIV-1 integration reaction to concerted integration. J Biol Chem. 2005;280:29334–29339. doi: 10.1074/jbc.M505367200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinha S, Grandgenett DP. Recombinant human immunodeficiency virus type 1 integrase exhibits a capacity for full-site integration in vitro that is comparable to that of purified preintegration complexes from virus-infected cells. J Virol. 2005;79:8208–8216. doi: 10.1128/JVI.79.13.8208-8216.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M, Craigie R. Nucleoprotein complex intermediates in HIV-1 integration. Methods. 2009;47:237–242. doi: 10.1016/j.ymeth.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandey KK, Bera S, Zahm J, Vora A, Stillmock K, Hazuda D, Grandgenett DP. Inhibition of human immunodeficiency virus type I concerted integration by strand transfer inhibitors which recognize a transient structural intermediate. J Virol. 2007;81:12189–12199. doi: 10.1128/JVI.02863-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, Mizuuchi M, Burke TR, Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464:232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guiot E, Carayon K, Delelis O, Simon F, Tauc P, Zubin E, Gottikh M, Mouscadet JF, Brochon JC, Deprez E. Relationship between the oligomeric status of HIV-1 integrase on DNA and enzymatic activity. J Biol Chem. 2006;281:22707–22719. doi: 10.1074/jbc.M602198200. [DOI] [PubMed] [Google Scholar]
- 25.Faure A, Calmels C, Desjobert C, Castroviejo M, Caumont-Sarcos A, Tarrago-Litvak L, Litvak S, Parissi V. HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res. 2005;33:977–986. doi: 10.1093/nar/gki241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bera S, Pandey KK, Vora AC, Grandgenett DP. Molecular Interactions between HIV-1 Integrase and the Two Viral DNA Ends within the Synaptic Complex that Mediates Concerted Integration. J Mol Biol. 2009;389:183–198. doi: 10.1016/j.jmb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shlyakhtenko LS, Gall AA, Filonov A, Cerovac Z, Lushnikov A, Lyubchenko YL. Silatrane-based surface chemistry for immobilization of DNA, protein-DNA complexes and other biological materials. Ultramicroscopy. 2003;97:279–287. doi: 10.1016/S0304-3991(03)00053-6. [DOI] [PubMed] [Google Scholar]
- 28.Cantor CR, Schimmel PR. Biophysical Chemistry: Part II: Techniques for the Study of Biological Structure and Function. W. H. Freeman & Co; San Francisco: 1980. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.