Abstract
The alignment of full-length human cDNA sequences to the finished sequence of the human genome provides a unique opportunity to study the distribution of genes throughout the genome. By analyzing the distances between 23,752 genes, we identified a class of divergently transcribed gene pairs, representing more than 10% of the genes in the genome, whose transcription start sites are separated by less than 1000 base pairs. Although this bidirectional arrangement has been previously described in humans and other species, the prevalence of bidirectional gene pairs in the human genome is striking, and the mechanisms of regulation of all but a few bidirectional genes are unknown. Our work shows that the transcripts of many bidirectional pairs are coexpressed, but some are antiregulated. Further, we show that many of the promoter segments between two bidirectional genes initiate transcription in both directions and contain shared elements that regulate both genes. We also show that the bidirectional arrangement is often conserved among mouse orthologs. These findings demonstrate that a bidirectional arrangement provides a unique mechanism of regulation for a significant number of mammalian genes.
The availability of the finished human genome sequence and the ever-growing collections of full-length cDNA libraries have made genome-scale studies of human gene organization a reasonable endeavor. Computational work by ourselves and others (Adachi and Lieber 2002) has identified a major class of gene pairs that are arranged head-to-head on opposite strands with less than 1000 base pairs separating their transcription start sites. This arrangement of gene pairs has previously been termed “bidirectional.”
Individual examples of bidirectional gene pairs have been known for years, and in some cases it has been shown that a bidirectional promoter regulates the transcription of a gene pair whose levels need to be coordinately expressed for several reasons. For example, some bidirectional promoters serve to maintain a stoichiometric relationship, such as the histone genes (Albig et al. 1997; Ahn and Gruen 1999; Maxson et al. 1983), whereas others regulate the coexpression of genes that function in the same biological pathway (Schmidt et al. 1993; Sugimoto et al. 1994; Momota et al. 1998), or to control expression through different timepoints such as genes involved in the cell cycle (Guarguaglini et al. 1997). Other bidirectional promoters provide coordinated responses to induction signals such as heat shock (Hansen et al. 2003).
Nearly all of the known bidirectional promoters were found in the process of studying a single gene, and Adachi and Lieber (2002) identified bidirectional genes on chromosomes 21 and 22. In the present study, we conducted a genome-wide analysis of gene organization in the human genome and identified a large class of bidirectional genes representing more than 10% of all human genes. To further this analysis, we explored the conservation of the head-to-head arrangements in the mouse genome, and we analyzed a large human microarray data set to see whether the endogenous human transcripts are coordinately regulated in any way. Using experimental approaches, we also functionally characterized the activity of a large sample of these bidirectional promoters. We categorized the functions of the proteins of the bidirectional genes and performed a thorough sequence analysis on the bidirectional promoter sequences.
RESULTS AND DISCUSSION
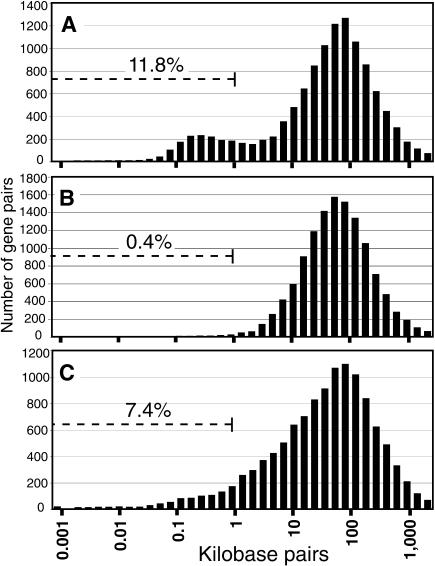
We calculated the distances between the transcription start sites (TSSs) of nearest gene neighbors for each of the 23,752 genes we predicted (see Methods and Supplemental Table 1 available online at www.genome.org). A bimodal distribution is seen for the distances between TSSs of nearest neighbors on opposite strands (Fig. 1A); this type of distribution is not observed for nearest neighbors on the same strand (Fig. 1B) or when the distances are calculated between the 3′ ends of the genes (Fig. 1C). There are 1352 pairs of genes (11% of the total) where members of a pair are located on opposite strands, and their TSSs are separated by less than 1000 base pairs (Suppl. Table 2). We designated the region between the two TSSs a putative bidirectional promoter. Of the 1352 bidirectional gene pairs, 315 pairs (23%) had genes whose transcripts were predicted to overlap at the 5′ ends, whereas 1037 pairs (77% of the bidirectional pairs) are nonoverlapping (Suppl. Fig. 1). Interestingly, the majority (67%) of the 1037 nonoverlapping bidirectional promoters are less than 300 base pairs in length.
Figure 1.
Distribution of distances between human genes and their nearest neighbors. (A) The distribution of distances between 5′ ends of genes on opposite strands shows a bimodal distribution, indicating that 11.8% of genes have TSSs separated by less than 1000 bp. (B) Analysis of the distribution of distances between the 5′ ends on the same strand shows that only 0.4% of gene pairs are closer than 1000 bp. (C) Analysis of the distribution of distances between the 3′ ends of genes on opposite strands shows that 7.4% of gene pairs are closer than 1000 bp.
Sequence Analysis of Bidirectional Promoters
A basic analysis of the bidirectional promoter sequences allowed us to identify sequence characteristics unique to this class. Other groups have noted examples of GC-rich promoters that function in both directions (Qvist et al. 1998; Adachi and Lieber 2002; Kawai et al. 2003). This is consistent with our findings that bidirectional promoters have a median GC-content of 66% compared to 53% for nonbidirectional promoters. Furthermore, 77% of the bidirectional promoters are located within a CpG island, compared to 38% of nonbidirectional promoters. We found no difference in the length and sequence characteristics of the CpG islands associated with bidirectional promoters and CpG islands associated with unidirectional promoters. Interestingly, only 8% of the bidirectional promoters contain a strict TATA-box on either strand, which is not significantly different than what one would expect by chance considering the nucleotide frequencies of the bidirectional promoters. In contrast, 28% of the nonbidirectional promoters contain a TATA-box on the forward strand, which is significantly more than the 18% one would expect by chance.
Functional Classes of Bidirectional Genes
It was noted that many DNA-repair genes are arranged head-to-head in the human genome (Adachi and Lieber 2002). By downloading the functional annotation from SOURCE (http://source.stanford.edu/; Diehn et al. 2003) and comparing the proportion of occurrences of “DNA-repair” in the annotated bidirectional genes to the occurrences in the annotated nonbidirectional genes, we also found that DNA-repair genes are more than fivefold overrepresented in the bidirectional class. In addition, we searched for other overrepresented functional categories and gene ontology terms in the bidirectional class of genes (for a complete list of terms and results, see Suppl. Table 3). We found that chaperone proteins are almost threefold overrepresented, mitochondrial genes are more than twofold overrepresented, and a class of DEAD-box RNA-helicases is also twofold overrepresented in the bidirectional genes.
Bidirectional Genes in the Mouse Genome
As evolutionary conservation often indicates functional importance, we estimated the percent of bidirectional gene pairs in the human genome with orthologous bidirectional gene pairs in the mouse genome. To accomplish this, we mapped the exons of each human bidirectional gene to the corresponding location in the mouse genome by using the human/mouse synteny data available at the UCSC Genome Bioinformatics Site. Of the 1037 nonoverlapping human bidirectional pairs, we mapped both transcripts of 335 gene pairs where both of the 5′-most human exons were successfully aligned to the mouse genome. We located an additional 464 orthologous pairs that are close to each other on opposite strands, but we could not calculate the distances between them accurately because the human first exons did not align to the mouse genome. Eighty-one percent of the 335 pairs with both first exons mapped had TSSs within 1000 base pairs of each other in the mouse genome, and 92% were within 2000 base pairs (Suppl. Fig. 2). We wished to know whether selection is acting to conserve this distance or whether there had been inadequate time for the distances between bidirectional gene pairs to be altered by random genome dynamics. To test this, we mapped 389 human gene pairs whose 3′ ends were less than 1000 base pairs apart to the mouse genome. Of these pairs, only 41% were less than 1000 base pairs apart, and 65% were separated by less than 2000 base pairs (Suppl. Fig. 2). A stronger conservation of distances between bidirectional genes located head-to-head compared to the distances between genes located tail-to-tail suggests that selection is acting to maintain bidirectional gene organization. However, a component of this conservation may be accounted for by a higher constraint in general at the 5′ ends of genes compared to the 3′ ends of genes.
Coexpression of Endogenous Transcripts of Bidirectional Genes
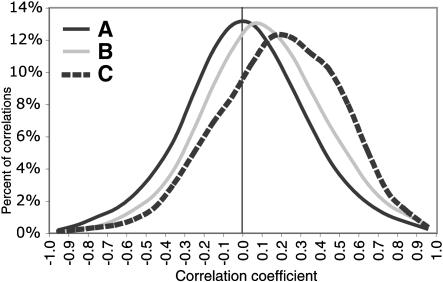
Given their shared cis-regulatory sequences, the transcript levels of the two genes in a bidirectional gene pair may be coordinately regulated (Lee and Song 2000; Ikeda et al. 2002). To test this hypothesis, we assembled a compendium of more than 1500 microarray experiments from the Stanford Microarray Database (http://genome-www5.stanford.edu/MicroArray/SMD/) representing 18 published data sets (Suppl. Table 4). We calculated the Pearson correlation coefficient for each bidirectional gene pair in each of the 18 data sets independently. Overall, the expression of bidirectional gene pairs is more positively correlated than are random pairs across all 18 data sets (Fig. 2), and 17% of all the correlations are significant at P < 0.05. Previous studies in the Drosophila and human genomes showed that genes in the same region of the genome are often coexpressed (Caron et al. 2001; Spellman and Rubin 2002). Figure 2 shows that neighboring genes are more correlated than random, but not as correlated as bidirectional pairs of genes. We also wished to examine the relationship between the length of a bidirectional promoter and the degree of expression correlation; however, we found no significant correlation between these two variables across all of the bidirectional promoters we examined. Previous studies also identified examples of bidirectional genes that are antiregulated (Hirschman et al. 1988; Christoffersen et al. 2001; Nieto et al. 2001; Delpy et al. 2002), and our analysis of the microarray data show that 11% of the significant correlations are negatively correlated. Therefore, the two genes in a bidirectional gene pair are not always coexpressed. Our comprehensive analysis of microarray expression data demonstrates that a bidirectional orientation often provides either a mechanism of coexpression, or for a minority of bidirectional gene pairs, a mechanism of mutually exclusive expression.
Figure 2.
Bidirectional gene pairs show correlated expression patterns in microarray experiments. (A) The frequency distribution of Pearson correlation coefficients of 500,000 random gene pairs averaged over 18 published microarray data sets. (B) The frequency distribution of correlation coefficients of nonbidirectional nearest neighbor gene pairs averaged across the 18 microarray experiments. (C) The frequency distribution of correlation coefficients of bidirectional gene pairs averaged across the 18 microarray experiments.
Functional Activity of Bidirectional Promoter Sequences
To determine whether the bidirectional promoters we predicted function bidirectionally, we assayed their ability to initiate transcription in both directions in a transient transfection assay. We also wished to see whether the class of bidirectional promoters shows directional promoter activity that is different from random promoters of genes not arranged head-to-head in the genome. For 56 random promoters that we identified in a previous study (Trinklein et al. 2003) and for 258 nonoverlapping bidirectional promoters, we cloned each fragment into a luciferase reporter vector in both orientations. We then transfected each construct into one mouse and three human cell lines (see Methods). We measured the activity of each promoter in both directions relative to an internal transfection control and analyzed the results in which a successful transfection was achieved for each promoter in both directions. All raw luciferase data are available in Supplemental Table 6.
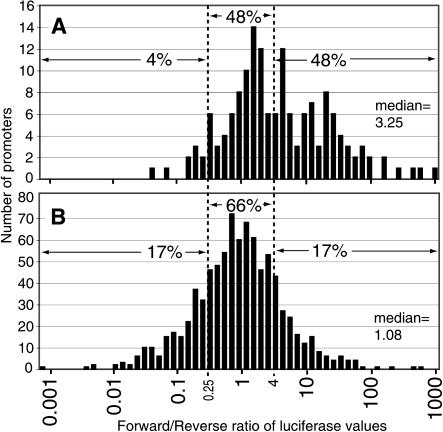
Averaged for the four cell lines, 27% of the random promoters showed significant activity over negative controls in the forward direction and no activity in the reverse, and no random promoters were inactive in the forward direction but active in the reverse. In contrast, 90% of the bidirectional promoters showed significant activity in both directions and 10% were active in one direction but not the other. Figure 3 illustrates that 48% of the random promoters function at least fourfold stronger in the forward direction, and surprisingly, 52% of random promoters showed activity in both directions. We conducted a Kolmogorov-Smirnov test on these two distributions and found them to be significantly different (P < 0.001).
Figure 3.
Functional activity of bidirectional and random promoters. (A) The distribution of the ratios of promoter strength in the forward and reverse directions for 56 random nonbidirectional promoters. (B) The distribution of the ratios of promoter strength in the forward and reverse directions for 258 bidirectional promoters. Based on a Kolmogorov-Smirnov test, these two distributions are significantly different (P < 0.001).
These results imply that more than half of all human promoters do not exhibit strong directionality in transcript initiation. Our observation that bidirectional promoters contain very few TATA-boxes is in agreement with previous studies (Qvist et al. 1998; Dong et al. 2000; Kawai et al. 2003) and suggests that the TATA element regulates the directionality of transcription. Nevertheless, we see that some TATA-less promoters show strong directional activity, and some TATA-containing promoters show activity in both directions. Therefore, the direction of transcription initiation is not always explained by the presence of a TATA-box.
Although the majority of bidirectional promoters show activity in both directions, 10% function only in one direction. Taken together with the anticorrelated expression data, these results support the hypothesis that the majority of bidirectional promoters regulate the coexpression of a bidirectional gene pair, whereas a minority of bidirectional promoters induce the transcription initiation of one gene while inhibiting transcription in the other direction.
We also looked at the activity of these promoters across the four different cell lines. If the strength of a promoter was four times greater in one direction than the other in one cell line, we call that “unidirectional activity” in that cell line, and if the strengths in both directions were within fourfold of each other, we call that “bidirectional activity.” Of all the random promoters, 33% showed bidirectional activity in half of the cell lines and unidirectional activity in the other half, 38% showed unidirectional activity in the same direction in all four cell lines, and 29% showed bidirectional activity in all four cell lines. Of the bidirectional promoters we assayed, 22% showed bidirectional activity in half of the cell lines and unidirectional activity in the other half, 21% showed unidirectional activity in the same direction in all four cell lines, and 57% showed bidirectional activity in all four cell lines. Interestingly, one bidirectional promoter showed unidirectional activity in the forward direction in one cell line, and unidirectional activity in the opposite direction in a different cell line. Taken together, these results suggest that the directionality of promoter activity may be regulated to some degree in a cell type-specific manner.
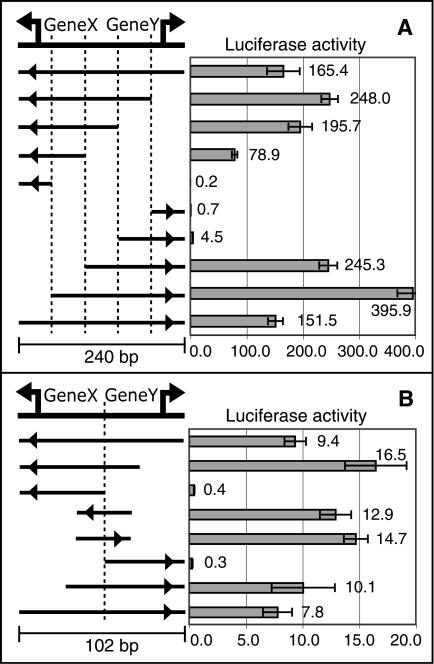
To further investigate the transcription initiation mechanism of bidirectional promoters, we measured the transcriptional activity of deletion constructs of 10 randomly chosen bidirectional promoters. Our aim was to determine whether these promoters share elements necessary for transcription initiation in both directions or whether they function as two independent promoters. For each of the 10 pairs, we observed that a shared fragment was necessary for full promoter activity in both directions (see Fig. 4A for one example, Suppl. Fig. 3 for all data), suggesting that most bidirectional promoters share at least some regulatory elements. In one case, we localized full promoter activity in both directions to a 30-bp sequence (Fig. 4B). In nine of 10 cases, we saw that deletion of the TSS of one transcript increased transcriptional activity in the opposite direction, indicating some degree of competition for determining the direction of transcription initiation. These results demonstrate that most bidirectional promoters act as inseparable functional units that coordinately regulate the transcription of both genes.
Figure 4.
Bidirectional promoters function as inseparable units. Normalized luciferase activity is plotted next to each construct, with arrows indicating the direction of transcription. (A) Loss of upstream promoter sequence leads to loss of transcriptional activity, indicating that there are shared elements necessary for full promoter activity in each direction. (B) A similar series of deletion constructs illustrates that a shared 30-bp fragment is able to confer full activity in both directions.
Conclusions
Although the detailed mechanism of transcription initiation on a genome-wide scale is largely unknown, our work has characterized the bidirectional activity of a large sample of human promoters. Furthermore, the catalog of human genes is still incomplete, so the number of bidirectional genes in the genome will likely increase as more genes are discovered. The full implications of this mode of regulation in mammalian genomes will be an interesting area for further investigation. All data and Supplemental material are available for download at http://www.stanford.edu/∼dschroed/bidirectional/index.shtml.
METHODS
Identifying Bidirectional Gene Pairs
We analyzed the UCSC BLAT alignments of Refseq, MGC, and DBTSS cDNA sequences to the Nov. 2002 freeze of the human genome and filtered out incomplete alignments and likely processed pseudogenes. Using a combination of Unigene assignments and exon overlaps, we collapsed cDNAs aligning to the same region in the genome into gene clusters. We defined the transcription start site coordinate and the transcript end coordinate for each gene as the 5′-most boundary and the 3′-most boundary of each gene cluster, respectively. We then compiled a list of the TSS coordinates of 23,752 gene clusters and sorted the list by the TSS on each chromosome. We also compiled a sorted list for the 3′ end of gene clusters. We then separately calculated the distances between the 5′ ends of nearest neighbor genes and the distances between 3′ ends of nearest neighbor genes and recorded the strand of each gene's location.
Sequence Analysis
We downloaded the sequence of each bidirectional promoter plus 50 bp downstream of each transcription start site. We also downloaded the sequence -550 to +50 relative to the TSS of each nonbidirectional promoter. We then determined the nucleotide frequency within each class and calculated the probability of finding at least one occurrence of the TATA element, (C G A)TATA(T A)(T A), per promoter. Next, we searched both the bidirectional and nonbidirectional classes of promoters to find the observed number of promoter fragments with at least one TATA element. We used a χ2 test to determine whether the observed frequencies differed from the expected at a significance cut-off of P < 0.05. Promoters located in CpG islands had TSSs between the start and stop coordinates of an entry in the UCSC cpgIsland table (http://genome.ucsc.edu/). These are genomic regions that start and end with CG-dinucleotides while also meeting traditional CpG-island requirements (Gardiner-Garden and Frommer 1987).
Identification of Mouse Orthologs of Human Bidirectional Genes
We mapped human bidirectional gene coordinates from the April 2003 human freeze to the February 2003 release of the mouse genome using the mouse/human BLASTZ alignment data at UCSC. We required a 20-bp minimum overlap between the human cDNA exon coordinates and the human coordinates that mapped to the mouse sequence in the axtTight alignment file.
Correlations of Microarray Expression Data for Bidirectional Gene Pairs
We downloaded microarray data from 18 separate publications from SMD (http://genome-www5.stanford.edu/MicroArray/smd/). For each of the 18 data sets (Suppl. Table 4), we subtracted the median Cy3/Cy5 ratio of each spot across all of the arrays in the data set, and we averaged the ratios of spots with identical Unigene IDs. We then calculated the Pearson correlation coefficients for the expression ratios of bidirectional gene pairs in each of the data sets separately. We calculated correlations on individual data sets instead of all of the experiments so that a positive correlation in one experiment would not cancel out a negative correlation in another experiment. To calculate the significance of each bidirectional correlation, we also calculated the correlations of 500,000 random pairs of genes within each of the 18 data sets.
Functional Assays of Bidirectional Promoters
We PCR amplified over 300 bidirectional promoters ranging from 50–1000 base pairs (list of primers is available as Suppl. Table 5). BglII and MluI restriction sites were included at the 5′ end of the forward and reverse primers, respectively, to direct the promoter orientation while cloning into the luciferase reporter vector. We ligated each digested PCR product upstream of the luciferase gene in the pGL3-basic vector (Promega). The PCR primers were designed so that the amplified products would not contain the AUG translation start of the endogenous gene; therefore, translation is ensured to be initiated by the AUG of the luciferase reporter gene. We also modified pGL3-basic by flipping the multiple cloning site, so that we could ligate each fragment in the flipped orientation on a separate plasmid. We made minipreps (QIAGEN) of each construct and diluted each to a concentration of 50 ng/μL. To control for transfection efficiency, we cotransfected 50 ng of each experimental luciferase plasmid with 8 ng of the renilla-containing pRL-TK control plasmid (Promega) into Hela, WI-38 human primary fibroblasts, HT1080, and mouse embryonic fibroblasts (ATCC). After 24 h, we prepared lysates, and assayed luciferase and renilla activity in a 96-well plate luminometer (Wallace). We eliminated data from failed transfections and determined the promoter strength of each DNA fragment by calculating the luciferase-to-renilla ratio. For each cell type, we calculated the standard deviation of 11 negative control DNA fragments and set our threshold for a positive signal at three standard deviation units above the mean of the negatives within each cell type.
We randomly chose 10 of the cloned bidirectional promoters from which we made deletion constructs. We designed four PCR primers evenly spaced throughout the cloned bidirectional fragment to PCR-amplify truncated versions of the bidirectional promoter (see Suppl. Fig. 4). We designed only two primers for the bidirectional promoters shorter than 150 bp. We then cloned these truncated fragments into the luciferase reporter vector and assayed their transcriptional activity as described above.
Acknowledgments
We thank the members of the Myers Lab for helpful discussion and support. We also thank Arend Sidow for comments regarding the mouse conservation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1982804.
Footnotes
[Supplemental material is available online at www.genome.org and http://www-shgc.stanford.edu/myerslab].
References
- Adachi, N. and Lieber, M.R. 2002. Bidirectional gene organization: A common architectural feature of the human genome. Cell 109: 807-809. [DOI] [PubMed] [Google Scholar]
- Ahn, J. and Gruen, J.R. 1999. The genomic organization of the histone clusters on human 6p21.3. Mamm. Genome 10: 768-770. [DOI] [PubMed] [Google Scholar]
- Albig, W., Kioschis, P., Poustka, A., Meergans, K., and Doenecke, D. 1997. Human histone gene organization: Nonregular arrangement within a large cluster. Genomics 40: 314-322. [DOI] [PubMed] [Google Scholar]
- Caron, H., van Schaik, B., van der Mee, M., Baas, F., Riggins, G., van Sluis, P., Hermus, M.C., van Asperen, R., Boon, K., Voute, P.A., et al. 2001. The human transcriptome map: Clustering of highly expressed genes in chromosomal domains. Science 291: 1289-1292. [DOI] [PubMed] [Google Scholar]
- Christoffersen, C.A., Brickman, T.J., Hook-Barnard, I., and McIntosh, M.A. 2001. Regulatory architecture of the iron-regulated fepD-ybdA bidirectional promoter region in Escherichia coli. J. Bacteriol. 183: 2059-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpy, L., Decourt, C., Le Bert, M., and Cogne, M. 2002. B cell development arrest upon insertion of a neo gene between JH and Emu: Promoter competition results in transcriptional silencing of germline JH and complete VDJ rearrangements. J. Immunol. 169: 6875-6882. [DOI] [PubMed] [Google Scholar]
- Diehn, M., Sherlock, G., Binkley, G., Jin, H., Matese, J.C., Hernandez-Boussard, T., Rees, C.A., Cherry, J.M., Botstein, D., Brown, P.O., et al. 2003. SOURCE: A unified genomic resource of functional annotations, ontologies, and gene expression data. Nucleic Acids Res. 31: 219-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, S., Lester, L., and Johnson, L.F. 2000. Transcriptional control elements and complex initiation pattern of the TATA-less bidirectional human thymidylate synthase promoter. J. Cell Biochem. 77: 50-64. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden, M. and Frommer, M. 1987. CpG islands in vertebrate genomes. J. Mol. Biol. 196: 261-282. [DOI] [PubMed] [Google Scholar]
- Guarguaglini, G., Battistoni, A., Pittoggi, C., Di Matteo, G., Di Fiore, B., and Lavia, P. 1997. Expression of the murine RanBP1 and Htf9-c genes is regulated from a shared bidirectional promoter during cell cycle progression. Biochem. J. 325 (Pt 1): 277-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, J.J., Bross, P., Westergaard, M., Nielsen, M.N., Eiberg, H., Borglum, A.D., Mogensen, J., Kristiansen, K., Bolund, L., and Gregersen, N. 2003. Genomic structure of the human mitochondrial chaperonin genes: HSP60 and HSP10 are localised head to head on chromosome 2 separated by a bidirectional promoter. Hum. Genet. 112: 71-77. [DOI] [PubMed] [Google Scholar]
- Hirschman, J.E., Durbin, K.J., and Winston, F. 1988. Genetic evidence for promoter competition in Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 4608-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda, S., Ayabe, H., Mori, K., Seki, Y., and Seki, S. 2002. Identification of the functional elements in the bidirectional promoter of the mouse O-sialoglycoprotein endopeptidase and APEX nuclease genes. Biochem. Biophys. Res. Commun. 296: 785-791. [DOI] [PubMed] [Google Scholar]
- Kawai, Y., Asai, K., Miura, Y., Inoue, Y., Yamamoto, M., Moriyama, A., Yamamoto, N., and Kato, T. 2003. Structure and promoter activity of the human glia maturation factor-γ gene: A TATA-less, GC-rich and bidirectional promoter. Biochim. Biophys. Acta 1625: 246-252. [DOI] [PubMed] [Google Scholar]
- Lee, S.G. and Song, K. 2000. Identification and characterization of a bidirectional promoter from the intergenic region between the human DDX13 and RD genes. Mol. Cells 10: 47-53. [DOI] [PubMed] [Google Scholar]
- Maxson, R., Cohn, R., Kedes, L., and Mohun, T. 1983. Expression and organization of histone genes. Annu. Rev. Genet. 17: 239-277. [DOI] [PubMed] [Google Scholar]
- Momota, R., Sugimoto, M., Oohashi, T., Kigasawa, K., Yoshioka, H., and Ninomiya, Y. 1998. Two genes, COL4A3 and COL4A4 coding for the human α3(IV) and α4(IV) collagen chains are arranged head-to-head on chromosome 2q36. FEBS Lett. 424: 11-16. [DOI] [PubMed] [Google Scholar]
- Nieto, C., Puyet, A., and Espinosa, M. 2001. MalR-mediated regulation of the Streptococcus pneumoniae malMP operon at promoter PM. Influence of a proximal divergent promoter region and competition between MalR and RNA polymerase proteins. J. Biol. Chem. 276: 14946-14954. [DOI] [PubMed] [Google Scholar]
- Qvist, H., Sjostrom, H., and Noren, O. 1998. The TATA-less, GC-rich porcine dipeptidylpeptidase IV (DPPIV) promoter shows bidirectional activity. Biol. Chem. 379: 75-81. [PubMed] [Google Scholar]
- Schmidt, C., Fischer, G., Kadner, H., Genersch, E., Kuhn, K., and Poschl, E. 1993. Differential effects of DNA-binding proteins on bidirectional transcription from the common promoter region of human collagen type IV genes COL4A1 and COL4A2. Biochim. Biophys. Acta 1174: 1-10. [DOI] [PubMed] [Google Scholar]
- Spellman, P.T. and Rubin, G.M. 2002. Evidence for large domains of similarly expressed genes in the Drosophila genome. J. Biol. 1: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, M., Oohashi, T., and Ninomiya, Y. 1994. The genes COL4A5 and COL4A6, coding for basement membrane collagen chains α5(IV) and α6(IV), are located head-to-head in close proximity on human chromosome Xq22 and COL4A6 is transcribed from two alternative promoters. Proc. Natl. Acad. Sci. 91: 11679-11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein, N.D., Aldred, S.J., Saldanha, A.J., and Myers, R.M. 2003. Identification and functional analysis of human transcriptional promoters. Genome Res. 13: 308-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www-shgc.stanford.edu/myerslab; Supplemental information.
- http://genome-www5.stanford.edu/MicroArray/SMD; Stanford Microarray database.
- http://source.stanford.edu; SOURCE.
- http://genome.ucsc.edu; UCSC Genome Browser.