Abstract
Background:
The epidermal growth factor receptor-targeted monoclonal antibody cetuximab (Erbitux) was recently introduced for the treatment of metastatic colorectal cancer. Treatment response is dependent on Kirsten-Ras (K-Ras) mutation status, in which the majority of patients with tumour-specific K-Ras mutations fail to respond to treatment. Mutations in the oncogenes B-Raf and PIK3CA (phosphoinositide-3-kinase) may also influence cetuximab response, highlighting the need for a sensitive, accurate and quantitative assessment of tumour mutation burden.
Methods:
Mutations in K-Ras, B-Raf and PIK3CA were identified by both dideoxy and quantitative pyrosequencing-based methods in a cohort of unselected colorectal tumours (n=102), and pyrosequencing-based mutation calls correlated with various clinico-pathological parameters.
Results:
The use of quantitative pyrosequencing-based methods allowed us to report a 13.7% increase in mutation burden, and to identify low-frequency (<30% mutation burden) mutations not routinely detected by dideoxy sequencing. K-Ras and B-Raf mutations were mutually exclusive and independently associated with a more advanced tumour phenotype.
Conclusion:
Pyrosequencing-based methods facilitate the identification of low-frequency tumour mutations and allow more accurate assessment of tumour mutation burden. Quantitative assessment of mutation burden may permit a more detailed evaluation of the role of specific tumour mutations in the pathogenesis and progression of colorectal cancer and may improve future patient selection for targeted drug therapies.
Keywords: K-Ras, mutation, dideoxy sequencing, pyrosequencing, colorectal tumour, personalised medicine
Colorectal (large bowel) cancer is the third most common cause of cancer-related death in the western world, with >36 000 new cases diagnosed annually in the United Kingdom (http://info.cancerresearchuk.org/cancerstats/types/bowel/). Despite recent advances in our understanding of disease pathogenesis and treatment, 5-year survival, particularly for patients presenting with advanced disease (Dukes’ stage C or D tumours) remains <10% (http://info.cancerresearchuk.org/cancerstats/types/bowel/survival/index.htm). Hence, there is a need for new treatment approaches and identification of optimised quantitative patient selection biomarkers for existing treatments.
Colorectal carcinogenesis is a multi-step process resulting in a progression from healthy bowel, through the formation of benign colorectal adenomas, to the development of colorectal tumours and, ultimately, to metastatic disease (Fearon and Vogelstein, 1990). Tumour formation is accompanied by an accumulation of genetic events, including chromosomal abnormalities, mutations in key tumour-suppressor genes, oncogenes and DNA mismatch repair genes, as well as epigenetic changes (Leslie et al, 2003; Söreide et al, 2006). For many years, mutations in a relatively limited number of key genes including APC (adenomatous polyposis coli), Kirsten-Ras (K-Ras) and p53 were considered to have a central role in the development of colorectal cancer, whereas more recent data have identified an increasingly complex network of genes and mutations associated with disease pathogenesis (Fearon and Vogelstein, 1990; Smith et al, 2002; Leslie et al, 2003; Conlin et al, 2005; Suehiro et al, 2008), progression, survival and treatment response (Soong et al, 2000; Smith et al, 2002; Lièvre et al, 2006; Kato et al, 2007).
Colorectal cancer is primarily treated by surgery, followed by adjuvant, usually 5-fluorouracil (5-FU)-based, chemotherapy in patients with adverse pathology following surgical resection (Koopman and Punt, 2009; Des Guetz et al, 2010). However, 5-FU is effective in less than one-third of patients, and it is currently not possible to predict which patients will respond to treatment or will experience severe treatment-associated toxicities (Longley et al, 2003). Similar inter-patient differences in response are seen with additional chemotherapy drugs, including irinotecan and oxaliplatin (Eng, 2009) and with novel drug treatments including bevacizumab, targeted to the vascular endothelial growth factor (Van Meter and Kim, 2010) and cetuximab, a monoclonal antibody targeted to the epidermal growth factor receptor (EGFR) (de Castro-Carpeno et al, 2008; De Roock et al, 2008; Karapetis et al, 2008; Lievre et al, 2008; Nicolantonio et al, 2008; Sartore-Bianchi et al, 2009).
When epidermal growth factor ligands bind to the EGFR, they activate a signalling pathway cascade, mediated by downstream effectors of the mitogen-activated protein kinase (MAPK) pathway and other pathways including the phosphoinositide-3-kinase (PIK3CA)/AKT signalling pathway. These effectors (K-Ras, B-Raf, ERK, MAPK, PIK3CA and AKT) influence cellular proliferation, adhesion, angiogenesis, migration and survival (Wagner and Nebreda, 2009). Blocking EGFR with antibody-based drugs including cetuximab (Erbitux) or panitumumab (Vectibix) inhibits signalling pathways downstream of this receptor. However, mutations in the K-Ras, B-Raf or PIK3CA genes, common in colorectal tumours, result in structural changes in the corresponding proteins, altered effector binding and permanent activation of downstream signalling pathways, independent of EGFR blockade (McCubrey et al, 2006; Scaltriti and Baselga, 2006). Therefore, although the therapeutic benefit of EGFR-targeted therapy in colorectal tumours has been clearly established (Cunningham et al, 2004; Saltz et al, 2004; de Castro-Carpeno et al, 2008), response is preferentially observed in tumours without mutations in K-Ras, whereas patients with tumours carrying K-Ras mutations have response rates below 10% (Lièvre et al, 2006; Di Fiore et al, 2007; Hecht et al, 2007; Amado et al, 2008; De Roock et al, 2008; Karapetis et al, 2008; Lievre et al, 2008; Allegra et al, 2009; Bokemeyer et al, 2009). K-Ras mutations have been reported in between 25 and 37% of colorectal tumours (Smith et al, 2002; Yuen et al, 2002; Calistri et al, 2005; Oliveira et al, 2007), with mutations most commonly described in codons 12 and 13 (Bos, 1989; Smith et al, 2010). A similar differential response to cetuximab has recently been associated with mutations in other EGFR-dependent signalling molecules including B-Raf and PIK3CA (Nicolantonio et al, 2008; Prenen et al, 2009; Sartore-Bianchi et al, 2009). The frequency of B-Raf and PIK3CA mutations in colorectal tumours has been estimated between 10 and 17% (Davies et al, 2002; Smith et al, 2002; Yuen et al, 2002; Calistri et al, 2005; Oliveira et al, 2007) and between 10 and 25% (Samuels et al, 2004; Velho et al, 2005; Nosho et al, 2008), respectively. V600E mutations in B-Raf are the most prevalent and therefore the most commonly analysed mutations in colorectal tumours (Davies et al, 2002; Yuen et al, 2002), whereas exons 9 (codons 542 and 545) and 20 (codons 1023 and 1047) have been shown to harbour ∼80% of all PIK3CA mutations (Samuels et al, 2004). Mutations in K-Ras and B-Raf are considered mutually exclusive (Oliveira et al, 2007) unlike mutations in K-Ras and PIK3CA (Bader et al, 2005).
K-Ras mutation testing is now mandated by the regulatory authorities in the United States and in Europe (Allegra et al, 2009; van Krieken and Tol, 2009) and is routinely used as a patient selection biomarker for cetuximab prescription in colorectal cancer patients. Current mandatory K-Ras mutation testing is limited to ‘hotspot’ codons 12 and 13, although K-Ras mutations have also been described at codon 61, and we have recently described several additional mutations, one of which results in an alanine-to-threonine amino-acid substitution at codon 146, occurs as frequently as previously described codon 13 mutations and seems to have a similar transforming phenotype (Smith et al, 2010). Analysis of these additional mutations, together with a novel amplification of the K-Ras gene that we have described in ∼2% of colorectal tumours (Smith et al, 2010), would increase the K-Ras mutation burden by more than one-third, and the current K-Ras mutation testing protocols may therefore mis-classify a significant number of patients. In addition, the majority of current mutation analyses simply classify tumours as K-Ras ‘wild type’ or ‘null’, and do not therefore consider the phenotypic consequences of inter-tumour differences in mutation burden.
It is also important to note that not all patients currently classified as ‘wild type’ for K-Ras, B-Raf and PIK3CA respond to cetuximab treatment (Lièvre et al, 2006; Di Fiore et al, 2007; Hecht et al, 2007; De Roock et al, 2008; Lievre et al, 2008; Bokemeyer et al, 2009). Although there are many reasons for this, it is possible that the limited sensitivity of conventional dideoxy sequencing-based methods of mutation assessment may fail to detect low-abundance oncogene mutations. Therefore, improved sensitive and quantitative methods for assessing mutation burden are essential, particularly for the assessment of response in biomarker-defined clinical trials. To address this issue, we have developed novel quantitative pyrosequencing-based methods for the analysis of oncogene mutation burden in colorectal tumours, and demonstrated that a significant number of tumours contain mutations in key oncogenes, which were not detected by conventional dideoxy sequencing analysis.
Materials and methods
Patient recruitment
Unselected Caucasian patients with a histologically confirmed diagnosis of colorectal cancer (ICD-9-CM 153. 1–4, 153.6–9, 154.0 and 154.1), undergoing surgery at the Ninewells Hospital, Dundee (n=102, 50 women, 52 men, age range 42–93 years) were recruited by the Tayside Tissue Bank between January 2005 and April 2007. All tumour samples used in this study were selected and dissected by an experienced pathologist, and were quality controlled by frozen section to ensure that tumour cells were present in least 60% of the sample. This is the same standard currently applied to diagnostic samples used for clinical estimation of K-Ras mutation status. Patient demographics are summarised in Table 1. Written informed consent was obtained from all patients, and the study was approved by the Tayside Tissue Banks Ethics Committee, a sub-committee of the Tayside Committee on Medical Research Ethics. All tumours were classified by the Dukes’ staging system in which Dukes’ A tumours were confined to the bowel wall, Dukes’ B tumours extended locally beyond the bowel and Dukes’ C tumours involved lymph-node metastases (Dukes, 1932). Tumour pathology was additionally classified using TNM (tumour, node, metastasis) staging (Greene, 2002), and the extent of differentiation was assessed by an experienced pathologist.
Table 1. Patient demographics.
| Female | Male | Total | |
|---|---|---|---|
| No. of patients | 50 | 52 | 102 |
| Age (median (range)) | 74.4 (42–93) | 70.3 (43–87) | 72.3 |
| Dukes's stage | |||
| A | 4 (8%) | 12 (22.2%) | 16 (15.7%) |
| B | 22 (44%) | 16 (30.8%) | 40 (37.2%) |
| C | 24 (48%) | 24 (44.4%) | 48 (47.0%) |
| D | 0 | 0 | 0 |
| TNM stage | |||
| I | |||
| T1N0MX | 2 (4%) | 4 (7.4%) | 6 (5.9%) |
| T2N0MX | 3 (6%) | 8 (14.8%) | 12 (11.8%) |
| II | |||
| T3N0MX | 17 (34%) | 14 (26.9%) | 31 (30.4%) |
| T4N0MX | 5 (10%) | 2 (3.7%) | 7 (6.9%) |
| III | |||
| T2N1MX | 0 | 1 (1.9%) | 1 (0.99%) |
| T3N1MX | 6 (2%) | 11 (20.3%) | 17 (16.7%) |
| T3N2MX | 10 (20%) | 7 (12.9%) | 17 (16.7%) |
| T4N1MX | 4 (8%) | 3 (5.6%) | 7 (6.9%) |
| T4N2MX | 3 (6%) | 2 (3.7%) | 5 (4.9%) |
| Tumour localization | |||
| Colon | 37 (74.0%) | 35 (67.3%) | 72 (70.6%) |
| Rectum | 14 (26%) | 17 (32.7%) | 29 (29.4%) |
| Differentiation | |||
| Moderate | 39 (78.0%) | 48 (88.5%) | 87 (83.3%) |
| Poor | 11 (22.0%) | 6 (11.5%) | 17 (16.7%) |
Abbreviation: TNM=tumour, node, metastasis;
Tissue processing
Tumour samples were taken directly from the operating theatre to the pathology department, where an experienced pathologist selected tumour tissues. Samples were then snap frozen in liquid nitrogen and stored at −80°C in the Tayside Tissue Bank until further processing. Genomic DNA was isolated from each tumour sample using a Wizard SV Genomic Purification System (Promega, Southampton, UK) according to the manufacturer's instructions, and DNA concentrations were assessed using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Loughborough, UK).
Mutation detection
Dideoxy sequencing
Mutations in exons 1, 2 and 3 of K-Ras, including the mutation hotspot codons 12, 13, and 61, the mutation codon 146, the B-Raf codon 600 and exons 9 and 20 of PIK3CA were detected by direct sequencing. PCR amplification was performed using the primers and reaction conditions summarised in Tables A and B, Supplementary Information. Dideoxy sequencing was performed by the DNA Analysis Facility at the Ninewells Hospital, Dundee. The software 4Peaks (http://mekentosj.com) was used to visualise and analyse the DNA sequences; mutations were identified based on automated sequence calls made by the analysis software, which were not overruled by the operator to avoid potential subjectivity of assessment of mutation burden.
Generation of pyrosequencing standards
A set of plasmid standards was developed for each K-Ras genotype. PCR products were amplified from cell lines or from tumour tissues of known genotype (PCR and reaction conditions are summarised in Table C, Supplementary Information) and purified using a GFX PCR DNA and Gel Band Purification Kit (GE Healthcare Life Sciences, Little Chalfont, UK). Purified PCR products were then sub-cloned into the pGEMTeasy Vector System I (Promega), and transformed into JM109 high-efficiency competent cells (Promega) following the manufacturer's instructions. Single colonies were grown in Luria-Bertoni broth+100 mg l−1 ampicillin, and plasmids were purified using the GenElute HP Plasmid Miniprep Kit (Sigma-Aldrich, Gillingham, UK). Sequences of plasmid inserts were verified by dideoxy sequencing (DNA Sequencing Facility, University of Dundee). For each mutation tested, a set of standards was created with the following proportions of the wild-type:mutant allele 0 : 100% 5 : 95%, 10 : 90%, 25 : 75%, 50 : 50%, 75 : 25% and 100 : 0%.
Pyrosequencing analysis
PCR templates for pyrosequencing analysis were amplified from 10 ng gDNA (or 0.1 pg plasmid standards) using Hotstar Taq Mastermix (Qiagen, Crawley, UK) and 5 pmol of each primer in a total reaction volume of 25 μl (PCR reaction and cycling conditions are summarised in Appendix 1, Supplementary Information). In all, 1 μl of each PCR reaction was analysed on an Agilent 2100 Bioanalyzer (Agilent, Edinburgh, UK) using a DNA 1000 kit, and pyrosequencing was carried out on 0.15–0.5 pmol of each PCR product using the PyroMark MD System (Qiagen) following the manufacturer's instructions, with sequencing primers and assay parameters specific to each mutation (Appendix 1, Supplementary Information). Resulting pyrograms were analysed using the PyroMark MD 1.0 software in ‘AQ mode’. For each assay, duplicate pyrosequencing analysis of tumour samples was performed, and the average of these was taken to represent the identified percentage burden of the mutant allele. The cutoff value, discriminating between the mutant and wild-type sequence, was arbitrarily assigned as 10% mutant allele burden.
Statistical analysis
Two-sided Fisher's exact tests were used to evaluate associations between tumour mutations and age, Dukes’ staging, gender and tumour location. A P-value <0.05 was nominally considered to be statistically significant.
Results
Mutation analysis
Dideoxy sequencing analysis
Genomic DNA was extracted from each tumour (n=102) and processed as described in the ‘Materials and methods’ section. Tumour DNA was then analysed by dideoxy sequencing for mutations in K-Ras exons 1, 2 and 3 (including the hotspot codons 12 and 13 (exon 1), 61 (exon 2) and codon 146 (exon 3)), PIK3CA (exons 9 and 20) and B-Raf (V600E).
K-Ras mutations were identified in 26.4% of tumours, and B-Raf and PIK3CA mutations in 8.8% of tumours, when automatic base calling software was used to assign mutation status (Table 2). The majority of K-Ras mutations were found in codon 12 (18.6%), with a smaller number in codon 13 (5.9%). Consistent with our previous analysis of K-Ras mutation burden in colorectal tumours, no mutations were found in codon 61 (Smith et al, 2002). In addition, a single tumour had a mutation in codon 22 (a C to A transversion substituting glutamine (CAG) for lysine (AAG)), which had been reported previously (Tsukuda et al, 2000), whereas a novel 3 bp in-frame insertion on the boundary of codons 14 and 15 was also found in a single tumour. Two tumours had mutations in K-Ras codon 146. The V600E B-Raf mutation was found in 9 tumours (8.8% of tumours analysed), whereas the majority of PIK3CA mutations were found in codons 542 (3.9%) and 543 (2.9%), whereas only single tumours had mutations in codons 546 and 1047. No mutations were found in PIK3CA codon 1023.
Table 2. Summary of mutation frequencies in K-Ras, B-Raf and PIK3CA as analysed by dideoxy and pyrosequencing.
|
Frequency (%)
|
||||
|---|---|---|---|---|
| Mutation | Nucleotide change | Amino-acid change | Dideoxy sequencing | Pyrosequencing |
| K-Ras | 26.4% | 32.4% | ||
| Codon 12 | G34A | Gly12Ser | 1/102 (1%) | 3/102 (2.9%) |
| G34T | Gly12Cys | 1/102 (1%) | 4/102 (3.9%) | |
| G35T | Gly12Val | 8/102 (7.8%) | 9/102 (8.8%) | |
| G35C | Gly12Arg | 1/102 (1%) | 1/102 (1%) | |
| G35A | Gly12Asp | 8/102 (7.8%) | 8/102 (7.8%) | |
| Codon 13 | G38A | Gly13Asp | 6/102 (5.9%) | 6/102 (5.9%) |
| Codon 61 | None detected | None detected | 0/102 | 0/102 |
| Codon 146 | G436A | Ala146Thr | 2/102 (1.9%) | 2/102 (1.9%) |
| Codon 14 | Ins 41–44 | 14Gly15 | 1/102 (1%) | 1/102 (1%) |
| Codon 22 | C65A | Gln22Lys | 1/102 (0.6%) | 1/102 (0.6%) |
| B-Raf | 8.8% | 11.5% | ||
| Codon 600 | T1798A | Val600Glu | 9/102 (8.8%) | 12/102 (11.5%) |
| PIK3CA | 8.8% | 13.70% | ||
| Codon 542 | G1624A | Glu542Lys | 4/102 (3.9%) | 4/102 (3.9%) |
| Codon 545 | G1633A | Glu545Lys | 3/102 (2.9%) | 7/102 (6.8%) |
| Codon 546 | A1637C | Gln546Pro | 1/102 (1%) | 1/102 (1%) |
| Codon 1023 | None detected | None detected | 0/102 | 0/102 |
| Codon 1047 | A3140G | His1047Arg | 1/102 (1%) | 2/102 (1.9%) |
Pyrosequencing analysis
Pyrosequencing assays for each codon were optimised to include calibration curves generated from titrated proportions of the wild-type:mutant allele, derived from cloned plasmids as described in the ‘Materials and methods’ section. Assay performance was formally assessed and is summarised in Table 3. Duplicate calibration curves were constructed for each tumour panel assessment and were used to derive adjusted percentage mutation burden calls for each genotype in duplicate. Although individual assay performances indicated accuracy at mutation burdens below 10%, we chose to use a conservative cutoff of 10% as the lower limit of quantitation for the assignment of mutation status calls.
Table 3. Performance assessment of pyrosequencing assays.
|
Assay performance
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
(Percentage point bias at expected allele frequency)
|
||||||||||||
| Gene | Nucleotide | Codon | Substitution | R 2 | Slope | 0% | 5% | 10% | 25% | 50% | 75% | 100% |
| BRAF | 1799 | 600 | T>A, Val>Glu | 0.998 | 0.918±0.0058 | 1.140 | 2.19 | 1.74 | 0.40 | 1.83 | 4.06 | 6.30 |
| KRAS | 12 | 34 | G>T, Gly>Ser | 0.997 | 0.956±0.0092 | 0.74 | 2.03 | 1.80 | 1.11 | 0.04 | 1.20 | 2.35 |
| KRAS | 12 | 34 | G>A, Gly>Cys | 0.997 | 0.930±0.0089 | 0 | 0.08 | 0.30 | 1.42 | 3.30 | 5.18 | 7.06 |
| KRAS | 12 | 35 | G>A, Gly>Asp | 0.995 | 0.901±0.0102 | 2.48 | 2.61 | 2.07 | 0.42 | 2.31 | 5.05 | 7.79 |
| KRAS | 12 | 35 | G>T, Gly> Val | 0.997 | 0.956±0.0092 | 0.74 | 2.03 | 1.80 | 1.11 | 0.04 | 1.20 | 2.35 |
| KRAS | 12 | 35 | G>C, Gly> Ala | 0.990 | 0.970±0.0157 | 0.62 | 4.52 | 4.36 | 3.89 | 3.10 | 2.31 | 1.53 |
| KRAS | 13 | 38 | G>A, Gly> Asp | 0.990 | 0.883±0.0112 | 0.58 | 0.49 | 0.18 | 2.17 | 5.50 | 8.82 | 12.15 |
| KRAS | 61 | 182 | T>A, Lys>Gln | 0.997 | 0.938±0.0073 | 3.44 | 1.93 | 1.60 | 0.61 | 1.04 | 2.68 | 4.33 |
| KRAS | 436 | 146 | G>A, Ala>Thr | 0.997 | 0.979±0.0069 | 0 | 2.1 | 2.65 | 4.4 | 4.35 | 3.45 | 1.65 |
| PIK3CA | 1624 | 542 | G>A, Glu>Lys | 0.992 | 0.972±0.0120 | 0 | 3.18 | 3.33 | 3.76 | 4.49 | 5.22 | 5.95 |
| PIK3CA | 1634 | 545 | A>G, Glu>Gly | 0.996 | 0.932±0.0101 | 0.08 | 2.06 | 1.69 | 0.59 | 1.25 | 3.09 | 4.93 |
| PIK3CA | 1633 | 545 | G>A, Glu>Lys | 0.994 | 0.949±0.0119 | 0.75 | 3.49 | 3.22 | 2.41 | 1.07 | 0.27 | 1.61 |
| PIK3CA | 1637 | 546 | A>C, Gln>Pro | 0.978 | 0.894±0.0217 | 0 | 1.18 | 1.78 | 3.56 | 6.53 | 9.50 | 12.47 |
| PIK3CA | 3140 | 1047 | A>G, His>Arg | 0.996 | 0.968±0.0084 | 2.15 | 3.83 | 3.66 | 3.16 | 2.33 | 1.49 | 0.66 |
R2, correlation co-efficient of best-fit dose–response line; Slope, slope of best–fit dose-response line.
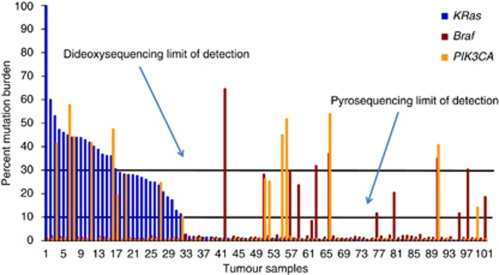
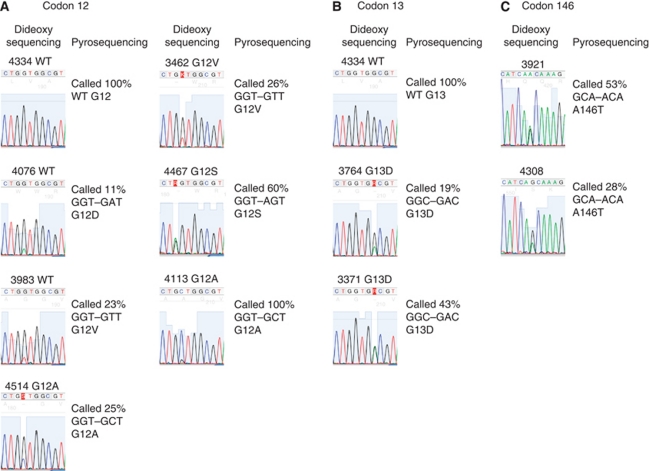
Pyrosequencing analysis revealed mutation frequencies of 32.4, 11.5 and 13.7% for K-Ras, B-Raf and PIK3CA, respectively, thus significantly increasing the number of tumours with mutations in K-Ras, B-Raf or PIK3CA (Table 2). In total, an additional 14 tumours carrying K-Ras, B-Raf or PIK3CA mutations were identified by pyrosequencing analysis, including all mutations previously identified by dideoxy sequencing analysis. In addition, K-Ras, B-Raf and PIK3CA mutation burden ranged from 11 to 99%, 12 to 65% and 14 to 54% of cells, respectively, highlighting marked inter-tumour heterogeneity in mutation burden (Figure 1). In addition to providing quantitative assessment of mutation burden, the increased sensitivity of pyrosequencing analysis allowed us to identify low-frequency mutations (mutation burden <30%) in a subset of tumours, which had not been identified by automated base calling analysis of our dideoxy sequencing data. A comparison of K-Ras mutation calls assessed by dideoxy and pyrosequencing is illustrated in Figure 2 – similar data were obtained for B-Raf and PIK3CA (data not shown). All of the additional K-Ras, B-Raf and PIK3CA mutations identified by pyrosequencing analysis were retrospectively manually confirmed in the dideoxy sequencing traces (e.g., samples 3983 and 4076, Figure 2).
Figure 1.
Inter-tumour variation in K-Ras, B-Raf and PIK3CA mutation burden Quantitative mutation detection was performed by pyrosequencing analysis, as described in the ‘Materials and methods’ section. Inter-tumour differences in mutation burden for K-Ras (blue), B-Raf (red) and PIK3CA (yellow) is illustrated, where each bar represents a different tumour sample. Tumours with K-Ras mutations are grouped to the left, with additional B-Raf and PIK3CA mutations highlighted. Arbitrary limits of detection for pyrosequencing (10% mutation burden) and dideoxy sequencing (30% mutation burden) are illustrated, highlighting the additional mutations identified by pyrosequencing analysis.
Figure 2.
Mutation analysis of K-Ras codons 12, 13 and 146. Mutation detection was performed by dideoxy and pyrosequencing analyses, as described in the ‘Materials and methods’ section. Mutation status in samples analysed by dideoxy sequencing was assigned by automated base calling using 4Peaks software, and is shown in comparison with quantitative analysis of mutation burden, assessed by pyrosequencing. Representative analyses are illustrated.
In confirmation of previous reports, K-Ras and B-Raf mutations were mutually exclusive in our tumour series (Yuen et al, 2002; Suehiro et al, 2008), whereas mutations in K-Ras and PIK3CA, in and B-Raf and PIK3CA were found together, but occurred in only 7.8 and 2.9% of tumours, respectively. A mutation in at least one of these genes, previously associated with response to EGFR-targeted antibody therapies, was found in 57.8% of all tumours analysed by pyrosequencing, an increase in K-Ras mutation burden of 27.4% compared with current mandatory analysis of K-Ras mutations restricted to codons 12 and 13, and a 33.3% increase when mutations in B-Raf and PIK3CA were additionally considered.
Correlation with pathological data and patient details
The mutation status of K-Ras, B-Raf and PIK3CA, based on pyrosequencing assessment of mutation burden, was then correlated with patient demographics and various clinico-pathological parameters, as the increased sensitivity of pyrosequencing analysis allowed us to more accurately evaluate these correlations. Mutations in K-Ras codons 14 and 22 were excluded from this analysis, as their phenotypes have not yet been fully characterised.
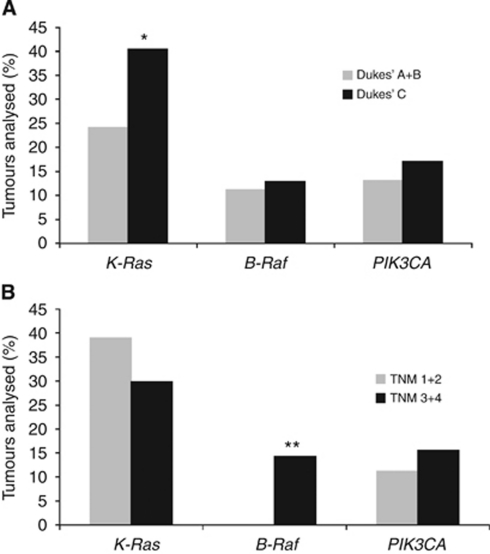
No differences in mutation frequencies comparing gender or median age were observed in our patient cohort (Table 4). In agreement with previous literature (Smith et al, 2002), a significantly higher proportion of rectal tumours harboured K-Ras mutations (40.0 vs 27.7%, P=0.04), whereas no significant differences in mutation frequencies were found for B-Raf (6.7 vs 13.9%, P=0.07) or PIK3CA (13.9 vs 16.7%, P=0.79). In addition, B-Raf mutation burden was significantly inversely correlated with differentiation status, in which 8.2% of moderately differentiated tumours had a B-Raf mutation compared with 29.4% of poorly differentiated tumours (P=0.0002). An additional significant correlation was found between Dukes’ stage and K-Ras mutation status, in which K-Ras mutations were more common in Dukes’ C than in Dukes’ A and B tumours (P=0.01) (Figure 3). This observation is consistent with our previous report (Smith et al, 2002), and associations between K-Ras mutation and poorer prognosis and time to relapse (Andreyev et al, 1998; Conlin et al, 2005). There were also significant correlations between T stage and the presence of a B-Raf mutation (P=0.00002), in which B-Raf mutations were restricted to more advanced tumours (T stages 3 and 4). Similarly, K-Ras mutations were overrepresented in tumours with lymph-node metastasis (N1 and 2) compared with lymph node-negative (N0) tumours (38.3 vs 25.5%, P=0.03).
Table 4. Associations between mutation status and various clinico-pathological parameters.
| N | K-Ras mutant | B-Raf mutant | PIK3CA mutant | |
|---|---|---|---|---|
| Male | 52 | 17 (32.6%) | 6 (11.5%) | 9 (17.3%) |
| Female | 50 | 15 (30.0%) | 6 (12.0%) | 6 (12%) |
| Age median | 72.4 years | 71.4 years | 75.3 years | 72.9 years |
| Colon | 72 | 20 (27.7%) | 10 (13.9%) | 10 (13.9%) |
| Rectum | 30 | 12 (40.0%)§ | 2 (6.7%) | 5 (16.7%) |
| Dukes’ A+B | 54 | 13 (24.1%) | 6 (11.1%) | 7 (13.0%) |
| Dukes’ C | 47 | 19 (40.4%)$ | 6 (12.8%) | 8 (17.0%) |
| T stage 1+2 | 18 | 7 (38.9%) | 0 | 2 (11.1%) |
| T stage 3+4 | 84 | 25 (29.8%) | 12 (14.2%)† | 13 (15.5%) |
| N stage 0 | 55 | 14 (25.5%) | 6 (10.9%) | 8 (14.5%) |
| N stage 1/2 | 47 | 18 (38.3%)* | 6 (12.8%) | 7 (14.8%) |
| Moderate differentiation | 85 | 27 (31.8%) | 7 (8.2%) | 12 (14.1%) |
| Poor differentiation | 17 | 5 (29.4%) | 5 (29.4%)# | 3 (17.6%) |
§P=0.04, $P=0.01, †P=0.00002, *P=0.03, #P=0.0002. The (paired) bold values highlight significant results.
Figure 3.
Distribution of K-Ras, B-Raf and PIK3CA mutations according to Dukes’ and TNM stage. The presence of mutations in K-Ras, B-Raf, and PIK3CA was determined by pyrosequencing analysis as described in the ‘Materials and methods’ section. Tumours were categorised according to (A) Dukes’ and (B) TNM staging, and further sub-divided by the presence of mutations in K-Ras, B-Raf, and PIK3CA. *P=0.03, **P=0.00002.
Discussion
Mutations in oncogenes including K-Ras, B-Raf and PIK3CA confer an important growth advantage to cancer cells (Vogelstein et al, 1988) and are found in more than one-third of all tumours. In colorectal tumours, K-Ras mutations have been associated with a more aggressive tumour phenotype (Smith et al, 2002) and reduced patient survival (Andreyev et al, 1998, 2001; Conlin et al, 2005), whereas B-Raf and PIK3CA mutations have also been associated with both disease pathogenesis and prognosis (Davies et al, 2002; Yuen et al, 2002; Calistri et al, 2005; McCubrey et al, 2006; Nicolantonio et al, 2008; Sartore-Bianchi et al, 2009; Baldus et al, 2010). However, the majority of current analyses of mutations in these genes are usually restricted to single amino-acid mutation hotspots, and mutation reporting is limited to a simple binary ‘wild-type’ or ‘mutant’ classification.
Several recent clinical reports provide compelling evidence that only a minority of colorectal tumours with K-Ras, B-Raf or PIK3CA mutations respond to novel EGFR-targeted monoclonal antibody therapies including cetuximab and panitumimab (Lièvre et al, 2006; Nicolantonio et al, 2008; Allegra et al, 2009; Sartore-Bianchi et al, 2009). K-Ras mutation testing is now mandatory before the prescription of these drugs, and it is therefore essential that analysis of mutation burden is as comprehensive and quantitative as possible.
We have previously described K-Ras mutations with previously described ‘hotspot’ codons, which significantly increase the K-Ras mutation burden in human colorectal tumours (Smith et al, 2010). Our current data, resulting from the analysis of an independent patient series, confirm the presence of K-Ras codon 146 mutations in colorectal tumours and report additional K-Ras mutations in codons 14 and 22. The codon 14 insertion, resulting in an in-frame creation of an additional glycine residue, has not previously been reported. Therefore, it is particularly interesting to note that a similar insertion mutation, K-Ras10Gly11, results in a hyperactive form of K-Ras (Bollag et al, 1996) – the phenotypic consequences of the codon 14 insertion are currently being evaluated in our laboratory. In contrast, the point mutation in codon 22 has been reported before (Tsukuda et al, 2000; Simi et al, 2008), although the resulting phenotype has not been fully characterised.
Importantly, our use of quantitative pyrosequencing analysis allowed us to identify mutations in K-Ras, B-Raf and PIK3CA with mutation frequencies ranging from 10 to 30%, which were not detected by automatic base calling software, routinely used in the analysis of dideoxy sequencing traces. These findings are in agreement with previous pyrosequencing studies, which have described a mutation detection threshold of 5–10% of mutant cells (Ogino et al, 2005; Dufort et al, 2009). Our data highlight an overall 13.7% increase in mutation burden (comparing the results of dideoxy and pyrosequencing analyses), and identifies a sub-set of tumours which would be erroneously classified as ‘wild type’ by conventional sequencing analysis, with potentially important implications for the prescription of EGFR-targeted therapies.
Although clinical response to cetuximab and related drugs is clearly dependent on K-Ras status, only one in two K-Ras ‘wild-type’ patients respond to treatment, based on current limited analysis of K-Ras mutation status (Karapetis et al, 2008; Lievre et al, 2008; De Roock et al, 2010). Although there are many complex factors which will inevitably contribute to variability in response, our data suggest that a significant proportion of non-responder patients may be mis-classified, either because of the presence of an additional oncogene mutation which influences cetuximab response or because of the presence of a relatively low-frequency mutation which is not detected by conventional dideoxy sequencing. Intra- and inter-tumour heterogeneity in mutation burden are also likely to be significant determinants of treatment response, and are not routinely considered in current binary ‘wild-type/mutant’ tumour classifications. In general, only a single piece of tumour is analysed for each patient, although previous studies have reported differences in mutation burden, for example, comparing tumour centres and invasion fronts (Baldus et al, 2010). Therefore, each individual ‘tumour’ sample may have a different normal/tumour cell ratio or a different proportion of infiltrating lymphocytes or other contaminating cell types, each of which can influence the apparent mutation burden. It is currently not possible to determine whether the marked inter-tumour variability in mutation burden observed in our patient cohort results from tumour sampling bias, or represents true differences in clonality, wherein some tumour cells contain mutations and other do not. This issue is central to the interpretation of mutation data, and we highlight the need for additional studies, for example, using laser capture micro-dissected material to address this issue. It is also clearly important that we are able to better predict the tumour phenotypes arising from varying proportions of wild-type and mutant cells – for example, should patients with 50% K-Ras mutant alleles be treated with cetuximab? In current testing protocols, these tumours would be classified as ‘mutant’, whereas a significant proportion of tumour cells have retained the ability to respond to EGFR inhibitors. Studies to address this issue using regulatable plasmids to vary the relative proportion of wild-type and mutant K-Ras are currently underway in our laboratory.
Recent data from our own laboratory and from that of De Roock et al (2010) additionally highlight the need to consider marked differences in phenotypes associated with individual oncogene mutations. Our data, in which wild-type and various mutant forms of K-Ras were transiently expressed in NIH3T3 cells, revealed significant differences in gene expression induced following the introduction of individual K-Ras mutations, suggesting that the K-Ras genotype may be a significant determinant of chemotherapy response (Smith et al, 2010). Consistent with this hypothesis, in a recent meta-analysis of cetuximab clinical trial data, De Roock et al (2010) clearly demonstrated that patients with colorectal tumours with a G13D mutation were significantly more likely to respond to cetuximab treatment than other K-Ras mutant tumours and survived longer. In additional in vitro experiments, these authors further demonstrated that cells expressing K-Ras G13D were phenotypically more similar to wild-type K-Ras than to cells containing alternative K-Ras mutations. These findings are consistent with previous reports highlighting differences in transforming potential, comparing K-Ras codon 12 and codon 13 mutations (Guerrero et al, 2000, 2002; Cespedes et al, 2006), and again highlights the need to extend clinical studies in this area.
Quantitative pyrosequencing-based analysis of mutation burden has also allowed us to more accurately investigate correlations between mutation burden and key clinico-pathological parameters. As would be expected from previous reports, K-Ras and B-Raf mutations were mutually exclusive in our tumour cohort, and were associated with tumours located in the rectum and colon, respectively (Yuen et al, 2002; Suehiro et al, 2008). Our data additionally confirmed our own previous report of increased K-Ras mutation burden in advanced Dukes’ C tumours (Smith et al, 2002) and identified a novel association associating K-Ras mutations with the presence of lymph-node metastases, consistent with the hypothesis that the K-Ras mutation is associated with a more aggressive tumour phenotype. Similarly, and consistent with the report of Baldus et al (2010), we found B-Raf mutation status to be inversely correlated with tumour differentiation. Like K-Ras, tumours with B-Raf mutations were restricted to more advanced tumours (T stages 3 and 4).
Our experimental approach has obvious application to the analysis of additional tumour types, for example, pancreatic tumours, in which K-Ras mutations are present in the majority (>90%) of tumours analysed (Bos, 1989), and can be easily extended to other mutation and tumour targets. In colorectal cancer, pyrosequencing-based mutation detection methods may also prove to be a powerful approach in the analysis of mutation burden in pre-malignant lesions, for example, colorectal adenomas to identify individual patients at the highest risk of disease progression and in metastatic disease for example tumours which have metastasised to liver or lymph nodes, the primary targets for adjuvant chemotherapy.
In conclusion, therefore, the use of sensitive pyrosequencing-based methods for mutation detection facilitates the identification of low-frequency tumour mutations and permits a quantitative assessment of intra- and inter-tumour differences in mutation burden. Quantitative assessment of oncogene mutation burden using pyrosequencing or other quantitative technologies including Sequenom MassArray (Sequenom, Hamburg, Germany) and next-generation sequencing methods may permit a more detailed evaluation of the role of specific tumour mutations in the pathogenesis and progression of colorectal cancer and may improve future patient selection for targeted drug therapies.
Acknowledgments
We gratefully acknowledge the Tayside Tissue Bank for help with patient recruitment and sample processing and Vincent Rao for technical assistance. This work was funded by a project grant from the Translational Medicine Research Collaboration, Cancer Research UK (C4639/A5661) and a Strategic Research Development Grant from the Scottish Funding Council.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL (2009) American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 27: 2091–2096 [DOI] [PubMed] [Google Scholar]
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD (2008) Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 26: 1626–1634 [DOI] [PubMed] [Google Scholar]
- Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA (1998) Kirsten ras mutations in patients with colorectal cancer: the multicenter ‘RASCAL’ study. J Natl Cancer Inst 90: 675–684 [DOI] [PubMed] [Google Scholar]
- Andreyev HJN, Norman AR, Cunningham D, Oates JR (2001) Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer 85: 692–69611531254 [Google Scholar]
- Bader AG, Kang S, Zhao L, Vogt PK (2005) Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer 5: 921–929 [DOI] [PubMed] [Google Scholar]
- Baldus SE, Schaefer K-L, Engers R, Hartleb D, Stoecklein NH, Gabbert HE (2010) Prevalence and heterogeneity of KRas, BRaf, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastasis. Clin Cancer Res 16: 790–799 [DOI] [PubMed] [Google Scholar]
- Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, de Braud F, Donea S, Ludwig H, Schuch G, Stroh C, Loos AH, Zubel A, Koralewski P (2009) Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol 27: 663–671 [DOI] [PubMed] [Google Scholar]
- Bollag G, Adler F, elMasry N, McCabe PC, Conner Jr E, Thompson P, McCormick F, Shannon K (1996) Biochemical characterization of a novel KRAS insertion mutation from a human leukemia. J Biol Chem 271: 32491–32494 [DOI] [PubMed] [Google Scholar]
- Bos JL (1989) Ras oncogenes in human cancer: a review. Cancer Res 49: 4682–4689 [PubMed] [Google Scholar]
- Calistri D, Rengucci C, Seymour I, Lattuneddo A (2005) Mutation analysis of p53, K-ras, and BRAF genes in colorectal cancer progression. J Cell Phys 204: 484–488 [DOI] [PubMed] [Google Scholar]
- Cespedes MV, Sancho FJ, Guerrero S, Parreno M, Casanova I, Pavon MA, Marcuello E, Trias M, Cascante M, Capella G, Mangues R (2006) K-ras Asp12 mutant neither interacts with Raf, nor signals through Erk and is less tumorigenic than K-ras Val12. Carcinogenesis 27: 2190–2200 [DOI] [PubMed] [Google Scholar]
- Conlin A, Smith G, Carey FA, Wolf CR, Steele RJ (2005) The prognostic significance of K-ras, p53, and APC mutations in colorectal carcinoma. Gut 54: 1283–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, Chau I, Van Cutsem E (2004) Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med 351: 337–345 [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S (2002) Mutations in the BRaf gene in human cancer. Nature 417: 949–952 [DOI] [PubMed] [Google Scholar]
- de Castro-Carpeno J, Belda-Iniesta C, Casado Saenz E, Hernandez Agudo E, Feliu Batlle J, Gonzalez Baron M (2008) EGFR and colon cancer: a clinical view. Clin Transl Oncol 10: 6–13 [DOI] [PubMed] [Google Scholar]
- De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, Penault-Llorca F, Rougier P, Vincenzi B, Santini D, Tonini G, Cappuzzo F, Frattini M, Molinari F, Saletti P, De Dosso S, Martini M, Bardelli A, Siena S, Sartore-Bianchi A, Tabernero J, Macarulla T, Di Fiore F, Gangloff AO, Ciardiello F, Pfeiffer P, Qvortrup C, Hansen TP, Van Cutsem E, Piessevaux H, Lambrechts D, Delorenzi M, Tejpar S (2010) Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 11: 753–762 [DOI] [PubMed] [Google Scholar]
- De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, Lamba S, Arena S, Frattini M, Piessevaux H, Van Cutsem E, O’Callaghan CJ, Khambata-Ford S, Zalcberg JR, Simes J, Karapetis CS, Bardelli A, Tejpar S (2010) Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA 304: 1812–1820 [DOI] [PubMed] [Google Scholar]
- De Roock W, Piessevaux H, De Schutter J, Janssens M, De Hertogh G, Personeni N, Biesmans B, Van Laethem JL, Peeters M, Humblet Y, Van Cutsem E, Tejpar S (2008) KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Ann Oncol 19: 508–515 [DOI] [PubMed] [Google Scholar]
- Des Guetz G, Uzzan B, Morere JF, Perret G, Nicolas P (2010) Duration of adjuvant chemotherapy for patients with non-metastatic colorectal cancer. Cochrane Database Systematic Review, (1): Article number: CD007046 [DOI] [PMC free article] [PubMed]
- Di Fiore F, Blanchard F, Charbonnier F, Le Pessot F, Lamy A, Galais MP, Bastit L, Killian A, Sesboüé R, Tuech JJ, Queuniet AM, Paillot B, Sabourin JC, Michot F, Michel P, Frebourg T (2007) Clinical relevance of KRAS mutation detection in metastatic colorectal cancer treated by cetuximab plus chemotherapy. Br J Cancer 96: 1166–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufort S, Richard MJ, de Fraipont F (2009) Pyrosequencing method to detect KRAS mutation in formalin-fixed and paraffin-embedded tumor tissues. Anal Biochem 391: 166–168 [DOI] [PubMed] [Google Scholar]
- Dukes CE (1932) The classification of cancer of the rectum. J Pathol Bacteriol 35: 323–332 [Google Scholar]
- Eng C (2009) Toxic effects and their management: daily clinical challenges in the treatment of colorectal cancer. Nat Rev Clin Oncol 6: 207–218 [DOI] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B (1990) A genetic model for colorectal tumorigenesis. Cell 61: 759–767 [DOI] [PubMed] [Google Scholar]
- Greene FL (ed) (2002) AJCC Cancer Staging Manual. Springer: New York [Google Scholar]
- Guerrero S, Casanova I, Farre L, Mazo A, Capella G, Mangues R (2000) K-ras codon 12 mutation induces higher level of resistance to apoptosis and predisposition to anchorage-independent growth than codon 13 mutation or proto-oncogene overexpression. Cancer Res 60: 6750–6756 [PubMed] [Google Scholar]
- Guerrero S, Figueras A, Casanova I, Farre L, Lloveras B, Capella G, Trias M, Mangues R (2002) Codon 12 and codon 13 mutations at the K-ras gene induce different soft tissue sarcoma types in nude mice. FASEB J 16: 1642–1644 [DOI] [PubMed] [Google Scholar]
- Hecht JR, Patnaik A, Berlin J, Venook A, Malik I, Tchekmedyian S, Navale L, Amado RG, Meropol NJ (2007) Panitumumab monotherapy in patients with previously treated metastatic colorectal cancer. Cancer 110: 980–988 [DOI] [PubMed] [Google Scholar]
- Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR (2008) K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 359: 1757–1765 [DOI] [PubMed] [Google Scholar]
- Kato S, Iida S, Higuchi T, Ishikawa T, Takagi Y, Yasuno M (2007) PIK3CA mutation is predictive of poor survival in patients with colorectal cancer. Int J Cancer 121: 1771–1778 [DOI] [PubMed] [Google Scholar]
- Koopman M, Punt CJ (2009) Chemotherapy, which drugs and when. Eur J Cancer 45(Suppl 1): 50–56 [DOI] [PubMed] [Google Scholar]
- Leslie A, Pratt NR, Gillespie K, Sales M, Kernohan NM (2003) Mutations in APC, K-Ras and p53 are associated with specific chromosomal aberrations in colorectal adenocarcinoma. Cancer Res 63: 4656–4661 [PubMed] [Google Scholar]
- Lievre A, Bachet J-B, Boige V, Cayre A, Le Corre D, Buc E, Ychou M, Bouché O, Landi B, Louvet C, André T, Bibeau F, Diebold MD, Rougier P, Ducreux M, Tomasic G, Emile JF, Penault-Llorca F, Laurent-Puig P (2008) KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol 26: 374–379 [DOI] [PubMed] [Google Scholar]
- Lièvre A, Bachet J-B, Corre DL, Boige V, Landi B, Emile J-F, Cote J-F, Tomasic G, Penna C, Ducreux M, Rougier P, Penault-Lorca F, Laurent-Puig P (2006) KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res 66: 3992–3995 [DOI] [PubMed] [Google Scholar]
- Longley DB, Harkin DP, Johnston PG (2003) 5-Fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer 3: 330–338 [DOI] [PubMed] [Google Scholar]
- McCubrey JA, Steelman LS, Abrams SL, Lee JT, Chang F, Bertrand FE, Navolanic PM, Terrian DM, Franklin RA, D’Assoro AB, Salisbury JL, Mazzarino MC, Stivala F, Libra M (2006) Roles of the RAF/MEK/ERK and PI3K/PTEN/AKT pathways in malignant transformation and drug resistance. Adv Enzyme Regul 46: 249–279 [DOI] [PubMed] [Google Scholar]
- Nicolantonio FD, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, Dosso SD, Maxxucchelli L, Frattini M, Siena S, Bardelli A (2008) Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol 26: 5705–5712 [DOI] [PubMed] [Google Scholar]
- Nosho K, Kawasaki T, Ohnishi M, Suemoto Y, Kirkner GJ, Zepf D, Yan L, Longtine JA, Fuchs CS, Ogino S (2008) PIK3CA mutation in colorectal cancer: relationship with genetic and epigenetic alterations. Neoplasia 10: 534–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Kawasaki T, Brahmandam M, Yan L, Cantor M, Namgyal C, Mino-Kenudson M, Lauwers GY, Loda M, Fuchs CS (2005) Sensitive sequencing method for KRAS mutation detection by pyrosequencing. J Mol Diagn 7: 413–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira C, Velho S, Moutinho C, Ferreira A, Preto A, Domingo E, Capelinha AF, Duval A, Hamelin R, Machado JC, Schwartz Jr S, Carneiro F, Seruca R (2007) KRAS and BRAF oncogenic mutations in MSS colorectal carcinoma progression. Oncogene 26: 158–163 [DOI] [PubMed] [Google Scholar]
- Prenen H, De Schutter J, Jacobs B, De Roock W, Biesmans B, Claes B, Lambrechts D, Van Cutsem E, Tejpar S (2009) PIK3CA mutations are not a major determinant of resistance to the epidermal growth factor receptor inhibitor cetuximab in metastatic colorectal cancer. Clin Cancer Res 15: 3184–3188 [DOI] [PubMed] [Google Scholar]
- Saltz LB, Meropol NJ, Loehrer Sr PJ, Needle MN, Kopit J, Mayer RJ (2004) Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol 22: 1201–1208 [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J (2004) High frequency of mutations of the PIK3CA gene in human cancers. Science 304: 554. [DOI] [PubMed] [Google Scholar]
- Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, Di Nicolantonio F, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A (2009) PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res 69: 1851–1857 [DOI] [PubMed] [Google Scholar]
- Scaltriti M, Baselga J (2006) The epidermal growth factor receptor pathway: a model for targeted therapy. Clin Cancer Res 12: 5268–5272 [DOI] [PubMed] [Google Scholar]
- Simi L, Pratesi N, Vignoli M, Sestini R, Cianchi F, Valanzano R, Nobili S, Mini E, Pazzagli M, Orlando C (2008) High-resolution melting analysis for rapid detection of KRAS, BRAF, and PIK3CA gene mutations in colorectal cancer. Am J Clin Pathol 130: 247–253 [DOI] [PubMed] [Google Scholar]
- Smith G, Bounds R, Wolf H, Steele RJ, Carey FA, Wolf CR (2010) Activating K-Ras mutations outwith ‘hotspot’ codons in sporadic colorectal tumours – implications for personalised cancer medicine. Br J Cancer 102: 693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G, Carey FA, Beattie J, Wilkie MJV, Lightfood TJ (2002) Mutations in APC, Kirsten-Ras, and p53-alternative genetic pathways to colorectal cancer. Proc Natl Acad Sci USA 99: 9433–9438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soong R, Powell B, Elsaleh H, Gnanasampanthan G, Smith DR (2000) Prognostic significance of TP53 gene mutations in 995 cases of colorectal carcinoma: influence of tumour site, stage, adjuvant chemotherapy and type of mutation. Eur J Cancer 36: 2053–2060 [DOI] [PubMed] [Google Scholar]
- Söreide K, Janssen EAM, Söiland H, Körner H, Baak JPA (2006) Microsatellite instability in colorectal cancer. Br J Surg 93: 395–406 [DOI] [PubMed] [Google Scholar]
- Suehiro Y, Wong CW, Chirieac LR, Kondo Y, Shen L, Webb CR, Chan YW, Chan AS, Chan TL, Wu TT, Rashid A, Hamanaka Y, Hinoda Y, Shannon RL, Wang X, Morris J, Issa JP, Yuen ST, Leung SY, Hamilton SR (2008) Epigenetic-genetic interactions in the APC/WNT, RAS/RAF, and P53 pathways in colorectal carcinoma. Clin Cancer Res 14: 2560–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda K, Tanino M, Soga H, Shimizu N, Shimizu K (2000) A novel activating mutation of the K-ras gene in human primary colon adenocarcinoma. Biochem Biophys Res Commun 278: 653–658 [DOI] [PubMed] [Google Scholar]
- van Krieken H, Tol J (2009) Setting future standards for KRAS testing in colorectal cancer. Pharmacogenomics 10: 1–3 [DOI] [PubMed] [Google Scholar]
- Van Meter ME, Kim ES (2010) Bevacizumab: current updates in treatment. Curr Opin Oncol 22: 586–591 [DOI] [PubMed] [Google Scholar]
- Velho S, Oliveira C, Ferreira A, Ferreira AC, Suriano G (2005) The prevalence of PI3KCA mutations in gastric and colon cancer. Eur J Cancer 41: 1649–1654 [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC (1988) Genetic alterations during colorectal tumour development. N Engl J Med 319: 525–532 [DOI] [PubMed] [Google Scholar]
- Wagner EF, Nebreda AR (2009) Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer 9: 537–549 [DOI] [PubMed] [Google Scholar]
- Yuen ST, Davies H, Chan TL, Ho JW, Bignell GR, Cox C, Stephens P, Edkins S, Tsui WW, Chan AS, Futreal PA, Stratton MR, Wooster R, Leung SY (2002) Similarity of the phenotypic patterns associated with BRAF and KRAS mutations in colorectal Neoplasia. Cancer Res 62: 6451–6455 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.