Abstract
Chromosome condensation plays an essential role in the maintenance of genetic integrity. Using genetic, cell biological, and biochemical approaches, we distinguish two cell-cycle-regulated pathways for chromosome condensation in budding yeast. From G2 to metaphase, we show that the condensation of the ∼1-Mb rDNA array is a multistep process, and describe condensin-dependent clustering, alignment, and resolution steps in chromosome folding. We functionally define a further postmetaphase chromosome assembly maturation step that is required for the maintenance of chromosome structural integrity during segregation. This late step in condensation requires the conserved mitotic kinase Ipl1/aurora in addition to condensin, but is independent of cohesin. Consistent with this, the late condensation pathway is initiated during the metaphase-to-anaphase transition, supports de novo condensation in cohesin mutants, and correlates with the Ipl1/aurora-dependent phosphorylation of condensin. These data provide insight into the molecular mechanisms of higher-order chromosome folding and suggest that two distinct condensation pathways, one involving cohesins and the other Ipl1/aurora, are required to modulate chromosome structure during mitosis.
Keywords: Condensin, cohesin, mitotic chromosome structure, SMC proteins, IPL1/Aurora
The faithful transmission of eukaryotic chromosomes is orchestrated through several evolutionarily conserved mechanisms (for recent reviews, see Losada and Hirano 2001; Nasmyth 2002). During S phase, identical sister chromatids are paired, thus promoting their bipolar attachment to the mitotic spindle. As chromosomes proceed into mitosis, both euchromatic and repetitive regions undergo dramatic structural changes. This condensation of chromosomes serves two essential functions: (1) the organization of sister chromatids into discrete and separable units, and (2) the shortening of chromosome arms along their linear axis, which prevents bisection by cytokinesis.
Our understanding of chromosome condensation is at an early stage. A key breakthrough came from the isolation of condensin, a five-member complex from Xenopus mitotic extracts required for the in vitro condensation of exogenously added sperm chromatin (Hirano et al. 1997). Condensin is conserved from yeast to man, and is essential for viability. In budding yeast, the five condensin subunits are Smc2p (XCAP-E/hCAP-E/cut14/CeMIX-1), Smc4p (XCAP-C/hCAP-C/cut3/DmSMC4/CeSMC4), Brn1p (hCAP-H/BRRN1/Sp cnd2/DmBarren), Ycg1p (XCAP-G/hCAP-G/cnd4), and Ycs4p (XCAP-D2/Eg7/hCAP-D2/CNAP-1/cnd3). Each subunit is essential for viability and is required for chromosome condensation and transmission during mitosis (Strunnikov et al. 1995; Freeman et al. 2000; Lavoie et al. 2000, 2002; Ouspenski et al. 2000; Bhalla et al. 2002). Inactivation of any of the non-SMCs in vivo precludes both binding of the SMCs to chromatin and also condensation, consistent with the in vitro observation that only the holoenzyme is competent to constrain positive supercoils in DNA (Kimura and Hirano 2000; Lavoie et al. 2002). This energy-dependent supercoiling reaction involves the local wrapping of DNA around the condensin complex (Bazett-Jones et al. 2002). Although this activity is very likely to be critical to condensin function, it has not been straightforward to extrapolate how it contributes to condensation in vivo. In particular, estimates of condensin stoichiometry on chromatin (assuming 1 condensin per 5–10 kb) suggest that for every positive supercoil introduced, up to 25 negative supercoils could be constrained by nucleosomes (Sutani and Yanagida 1997; MacCallum et al. 2002). It follows then that a molecular understanding of condensation will require additional tools to bridge the gap between in vivo and in vitro systems.
One approach toward this goal is to define in vivo intermediates in condensation and correlate chromosome morphology with the activity of known condensation factors. Previous studies monitored morphological changes in whole chromosomes during the onset of mitosis (Hiraoka et al. 1989; Manders et al. 1999). Interpreting the significance of such morphological changes is complicated by the absence of condensin mutants to unambiguously distinguish between bona fide condensation and other cell-cycle-related events like changes in transcription. In addition, classical cytological stains lack the resolution to follow early steps in condensation because these methods do not resolve individual strands of chromatin in interphase or early mitosis. The latter limitation has been overcome in budding yeast through the use of fluorescence in situ hybridization (FISH) to follow the compaction state of defined chromosomal regions (Guacci et al. 1994). In particular, the highly repetitive, ∼1-Mb RDN locus undergoes a condensin-dependent transition from a loosely organized structure in interphase (descriptively called a puff) to a more highly ordered structure in mitosis resembling a line or loop. Recently, an amplified GFP-tagged transgene in CHO cells was shown to undergo a similar morphological transition from interphase to mitosis (Dietzel and Belmont 2001), suggesting that the overall mechanism of chromosome compaction, as well as the condensation machinery, is conserved between eukaryotes. From these studies, new questions arise. Does mitotic chromosome structure arise through a single-step mechanism or by a more complex pathway with multiple intermediates? If there are other intermediates, are they all dependent on condensin function?
The condensin-dependent compaction of chromosomes is likely to be regulated both temporally and spatially, that is, to limit it to mitosis and to ensure the highly regular geometry of mitotic chromosomes. Interestingly, several different mechanisms for the regulation of condensin function have been defined in different organisms (Kimura et al. 1998; Sutani et al. 1999; Giet and Glover 2001). It remains unclear, however, which of these mechanisms have been evolutionarily conserved. For example, in both budding yeast and Sordaria, the establishment and maintenance of condensation require proteins involved in sister-chromatid cohesion including the cohesins (Castano et al. 1996; Guacci et al. 1997; van Heemst et al. 1999; Hartman et al. 2000; Hanna et al. 2001; Lavoie et al. 2002). A universal role for cohesins in condensation has been disputed, however, because in higher eukaryotes, most cohesins are removed from chromosomes in late prophase, and the depletion of cohesins has only subtle effects on chromosome compaction (Losada et al. 1998; Sumara et al. 2000; Waizenegger et al. 2000; Warren et al. 2000; Sonoda et al. 2001). Although these data have inspired models proposing that cohesin removal would be a prerequisite to condensin function in higher eukaryotes, a recent report showed that the persistence of cohesin in Xenopus egg extracts does not preclude condensin loading or function on chromosomes, consistent with the situation in budding yeast (Losada et al. 2002). A similar discrepancy has arisen in studies of the regulation of chromosome condensation by the conserved mitotic kinase Ipl1/aurora B. This kinase mediates histone H3 phosphorylation at serine 10, and is required for chromosome structure and segregation in vivo (Biggins et al. 1999; Hsu et al. 2000; Adams et al. 2001; Giet and Glover 2001; Kang et al. 2001; Hagstrom et al. 2002; Kaitna et al. 2002). In vitro, however, aurora B is not required for condensin loading or function, but functions with the polo-like kinase to promote cohesin dissociation in prophase (Losada et al. 2002; MacCallum et al. 2002).
Clearly, a molecular understanding of condensation will require a rapprochement between the in vitro and in vivo approaches. Toward this goal, we have sought to identify intermediate steps in condensation in highly synchronized cells, during a single cell cycle. In this report, we identify five morphologically and biochemically distinct stages in mitotic chromosome condensation. In addition, we define two distinct pathways for condensin-dependent chromosome folding and show that each pathway is temporally regulated during mitosis. From G2 to metaphase, chromosome condensation is established and maintained through the cohesin-dependent pathway. In anaphase, chromosome structure becomes regulated through the action of the Ipl1/aurora kinase. This maturation step is required for the maintenance of chromosome integrity during segregation (i.e., in the absence of cohesins), but not for the establishment of metaphase chromosome structure. We discuss a model for higher-order chromosome folding.
Results
Condensation intermediates of the RDN locus
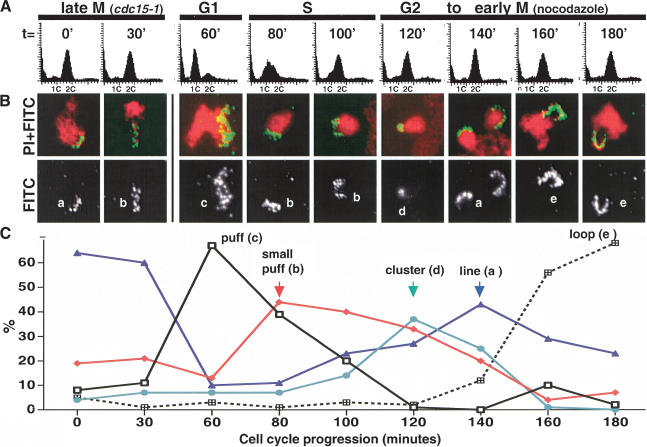
To assess whether in vivo chromosome condensation proceeds through a single-step mechanism or through a progression of intermediates, we monitored chromosome morphology during the cell cycle, focusing on the repetitive RDN locus. This ∼1-MB region on Chromosome XII comprises 100–150 copies of the 9.1-kb rDNA repeat, and is known to require condensin for both mitotic chromosome structure and faithful partitioning. We further reasoned that owing to its size and uniformity in composition, the rDNA would serve as an especially sensitive reporter for condensin function and facilitate the identification of partially folded species. From G1 to M phase, five distinct rDNA species were observed (Fig. 1, a–e). Cells were first synchronized using a temperature-sensitive cdc15-1 allele, which arrests the cell cycle in mitosis following chromosome segregation but prior to mitotic exit. The cells were then synchronously released into an unperturbed G1 phase by lowering the temperature to 23°C and allowed to proceed to the next mitosis. rDNA morphology was correlated with cell cycle state by analyzing samples taken at different time points by FISH versus flow cytometry (Fig. 1) and cell morphology (data not shown). In cdc15-1-arrested cells, the rDNA exhibited a compacted structure clearly distinct from metaphase loops, and which we describe as a line (Fig. 1B, species a). As the rDNA decondenses in telophase and enters into the next cell cycle, chromosome compaction is lost as judged by the more diffuse FISH signal in G1/S phase (puffs, species b, c). During G2/M, when condensin becomes active, a succession of additional rDNA species was observed (Fig. 1B,C). Surprisingly, the rDNA adopted its most compacted morphology at the earlier time points, with the rDNA signal restricted to an intensely stained region (cluster, species d). The next observed rDNA morphology (line, species a) consisted of a more extended staining pattern relative to the cluster, but with the rDNA signal closely associated to the bulk chromosomes. Morphologically, the lines arising preanaphase onset were indistinguishable from those observed postanaphase (at the cdc15-1 arrest point), suggesting that condensation and decondensation may be related processes. Both rDNA clusters and lines were transient, peaking within 1–2 time points, and suggesting that these species reflect intermediate stages of condensation. As the lines disappeared, looped structures two to three times longer extending away from the bulk chromosomes accumulated (loop, species e). At 180 min postrelease into the cell cycle, >70% of the nuclei possessed rDNA loops characteristic of metaphase cells. This preanaphase rDNA morphology is easily distinguished from the rDNA lines because of its position relative to the bulk chromosomal mass as well as its length, and is likely to reflect the most highly ordered (although not compacted) rDNA structure. It is noteworthy that whereas all of these rDNA species were observed in the presence of nocodazole, which was used to provide a defined end point to the experiment, rDNA puffs, clusters, lines, and loops were readily observed in cells released into drug-free medium, indicating that they are not artifacts of the cell cycle arrest (data not shown).
Figure 1.
Cell-cycle-dependent intermediates in rDNA condensation. (A) A cdc15-1 (YBL26c-9a) strain was used to synchronize the cells in M phase. Following release to the permissive temperature to monitor an unperturbed G1, nocodazole was added to the culture 1 h postrelease to prevent cycling beyond the first cell cycle. Time points were taken as shown, and cells were processed for flow cytometric analysis and FISH, using an rDNA probe. (B) Micrographs of FISH of the yeast rDNA (FITC, green) and chromosomes (PI, red). (Bottom panels) The isolated rDNA signal. At each time point, a representative micrograph of the most prominent or newly emerging species is shown. Letters classify the morphologies detected, the quantitation of which is shown in C. At least 100 nuclei were scored per time point.
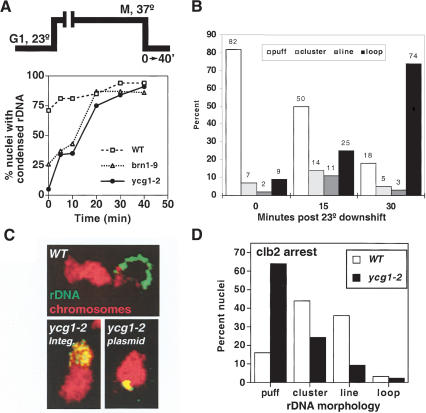
An alternative interpretation of our results is that the observed variations in rDNA structure result from cell-cycle-dependent changes in nucleolar physiology. Although the transcription of the ribosomal RNA has been shown to be independent of cell cycle state in budding yeast (Elliot and McLaughlin 1979) and should therefore have little impact on chromosome morphology, we addressed this issue directly by monitoring condensation independently of cell cycle progression. To do this, a time course of chromosome condensation initiated in cells prearrested in mitosis was performed (Fig. 2). Two previously described condensin mutants, brn1-9 and ycg1-2, which fail to condense chromosomes at the restrictive temperature but efficiently fold chromosomes upon return to 23°C, were used to initiate condensation in mitotically blocked cells (Lavoie et al. 2002). This delayed condensation is functional, as it restores cell viability upon release from the mitotic block. To ensure a complete inactivation of condensin, G1-synchronized cultures were shifted to the restrictive temperature prior to their release into prewarmed medium containing nocodazole, a microtubule-depolymerizing drug. Under these conditions, condensin remained inactive from G1 to M phase. Condensation was then initiated by returning the arrested cells to the permissive temperature. A time course of chromosome condensation (as judged by the appearance of rDNA loops) shows that condensation is complete within ∼20 min (Fig. 2A). rDNA puffs, clusters, lines, and loops were observed, and their kinetics of appearance/disappearance reflect that seen during the cell cycle experiment (Figs. 1 and 2B). It is noteworthy, however, that both the rDNA cluster and short line species were highly transient, with the lines failing to accumulate beyond 15%–20% in any time point. Although it is difficult to determine the time required to reform active condensin after temperature downshift, we estimate from the biphasic kinetics of recondensation that the conversion from rDNA puffs to loops in M-phase cells takes approximately 10 min (data not shown). In contrast, during the cell cycle progression experiment, a similar accumulation of loops (>60%) accumulated after 40–60 min, some four to six times longer, suggesting that the mitotic activation of condensin is a slow step. Whether this apparent increase in condensation kinetics accounts for the rapid disappearance of clusters and lines remains to be determined. Nonetheless, the detection of clusters, lines, and loops during condensation in mitotically arrested cells argues that the mechanism of condensation is conserved and that these rDNA species result from higher-order chromosome folding rather than cell-cycle-dependent changes in nucleolar physiology.
Figure 2.
Condensation intermediates following mitotic arrest. (A) Kinetics of chromosome condensation in M phase. An experimental schematic is shown. Strains CH2523 (WT), CH2524 (brn1-9), and 2153-1 (ycg1-2) were synchronized in G1 with α-factor, shifted to 37°C (0.5 h), and rearrested in mitosis following release into nocodazole-containing media. The cultures were returned to the permissive temperature (23°C), and samples were taken as indicated. rDNA FISH was performed, and the percent nuclei displaying rDNA loops were scored. (B) Quantitation of rDNA species during mitotic condensation. Strain 2153-1 (ycg1-2) was treated as in A, and condensation intermediates were scored by FISH at 0, 15, and 30 min after release to the permissive temperature. The percent nuclei with puff, cluster, line, or loop rDNA morphologies are plotted at each time point. At least 100 nuclei were scored per sample. (C) Accumulation of clusters in a ycg1-2 mutant. Strains YPH499 (WT), YBL02–18b (integrated ycg1-2), and 2125–3a/pBL238-ts2 (ycg1::KAN/CEN6 ARS1 LEU2 ycg1-2) were synchronized in G1, shifted to the restrictive temperature, released, and rearrested in mitosis with nocodazole, followed by rDNA FISH to determine rDNA morphology. Chromosomes are depicted in red with the rDNA signal in green/yellow. (D) Postanaphase rDNA clusters and lines are condensin-dependent. Strains 982–101 (YCG1 GAL1-CLB2Δdb) and 2172–101 (ycg1-2 GAL1-CLB2Δdb) were arrested in late M by overexpression of undegradable Clb2p (see Materials and Methods). The cells were then shifted to the restrictive temperature for 30 min, fixed, and processed for rDNA FISH. Quantitation of rDNA morphologies is shown. More than 100 nuclei were counted per sample.
rDNA intermediates are condensin-dependent
One important question is whether the observed morphologies are dependent on the chromosome condensation machinery. Previously, we generated and analyzed mutants in all three non-SMC subunits of condensin and showed that in the absence of any of these subunits, the rDNA can neither form nor maintain the loops characteristic of metaphase but rather remains in a G1-like (puffed) state (Lavoie et al. 2000, 2002). Thus, condensins are required for the transition from puffs to loops. In addition, condensins are likely to be required for both cluster and line formation because these intermediates occur after puffs but before loops. Additional evidence that clusters are condensin-dependent came from the identification of an allele of condensin that accumulates clusters under certain conditions (Fig. 2C). If chromosome condensation proceeds through multiple steps, a subset of mutants should perform partial reactions and trap intermediates. One such mutant is a temperature-sensitive allele of the Ycg1p subunit of condensin, ycg1-2. When ycg1-2 is expressed from a CEN-based plasmid (which exists in one to three copies per cell), 38% of metaphase-arrested nuclei showed rDNA clusters (versus 53% puffs and 9% loops), suggesting that a partial reaction is supported at the restrictive temperature in a significant proportion of cells (Fig. 2C). In contrast, when the same mutation was integrated into its endogenous chromosomal locus (which allows a tighter copy number control), few clusters were observed, and the rDNA remained in its G1 (puffed) state (<1% clusters). Thus, small twofold to threefold changes in condensin levels may substantially affect the severity of the condensation defects. Importantly, these data indicate that condensin is required not only for the transition of rDNA puff to cluster, but also from cluster to other species, consistent with a multistep pathway for chromosome condensation.
In addition to earlier intermediates in rDNA condensation, we noted that the rDNA in cdc15-1-arrested cells adopted a line-like morphology quite distinct from the puffs seen in G1 cells (Fig. 1; see also Guacci et al. 1994), suggesting that in budding yeast, as in higher eukaryotes, chromosomes remain condensed beyond the metaphase-to-anaphase transition. As the activity of condensin during anaphase had not been directly addressed in any system, we constructed a strain temperature-sensitive for condensin that could be arrested postanaphase onset independently of temperature (ycg1-2 GAL1–CLB2–dbΔ). The overproduction of a nondestructible allele of the Clb2 cyclin causes a mitotic arrest postanaphase onset (Amon et al. 1994). At the permissive temperature, the rDNA exhibited both cluster and line morphologies (Fig. 2D); however, upon inactivation of condensin, the majority of nuclei showed rDNA puffs. Taken together, these data indicate that the maintenance of mitotic rDNA structure is dependent on condensin function from the onset of mitosis until after the metaphase-to-anaphase transition.
Postanaphase condensation is cohesin-independent
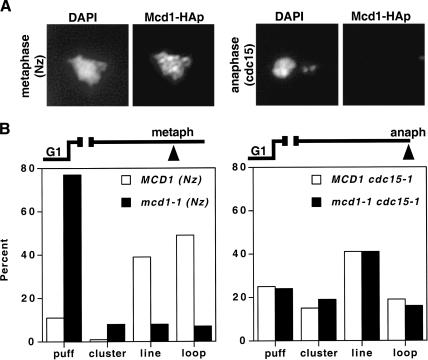
The demonstration that budding yeast chromosomes remain condensed postsegregation poses a potential dilemma for the role of cohesins in condensation. Cohesins are known to be required for both the establishment and maintenance of mitotic chromosome condensation in budding yeast. Paradoxically, it has also been established that the removal of cohesins from chromosomes is required for chromosome segregation at the onset of anaphase, suggesting that little if any cohesin would be available to maintain mitotic chromosome structure (see Uhlmann 2003). To directly address whether cohesins could play a role in condensation during anaphase, we first asked whether cohesin remains associated with chromosomes and/or the rDNA after the chromosomes have segregated to opposite poles but while the rDNA remains condensed (cdc15-1 arrest). For comparison, chromatin spreads using an epitope-tagged cohesin subunit (MCD1-HA) were performed in both preanaphase and postanaphase cells (Fig. 3A). In agreement with published work, Mcd1p was readily detected on metaphase chromosomes but was not observed postanaphase onset under conditions in which the chromosomes remain condensed (cdc15-1; Fig. 3A; Michaelis et al. 1997). We infer from this result that the maintenance of rDNA condensation in anaphase is independent of sister-chromatid cohesion and, indeed, of the intact cohesin complex. One caveat, however, is that the amount of cohesin bound to chromosomes could be underestimated because of epitope masking or may fall below the detection levels of our cytological methods yet prove sufficient to promote condensin function. To address this possibility and eliminate any residual Mcd1p activity, we assayed rDNA structure in metaphase- versus anaphase-arrested cells in a temperature-sensitive mcd1-1 cdc15-1 double mutant (Fig. 3B). Cells were synchronized in G1 and then released at the restrictive temperature in the presence or absence of nocodazole. When cells were arrested in metaphase, little or no rDNA condensation was detected in the absence of Mcd1p function, corroborating published observations that Mcd1p is required for condensation establishment and maintenance prior to anaphase (Guacci et al. 1997). In contrast, both wild-type (WT) and mcd1-1 strains achieved similar levels of condensation when cells were arrested in anaphase (75% and 74% rDNA lines, respectively), indicating that condensation postanaphase onset is not only maintained but can also be established de novo by a cohesin-independent pathway.
Figure 3.
Chromosome condensation in the absence of Mcd1p. (A) Chromatin spreads of Mcd1-HAp in metaphase-versus-anaphase cells. YBL13–2a (MCD1-HA6 cdc15-1) cells were synchronized in G1, released, and rearrested at 37°C either with nocodazole or by allowing the cells to proceed to the cdc15-1 block. Mcd1-HAp was monitored by indirect immunofluorescence using an anti-HA monoclonal antibody (16B12; see Materials and Methods). (B) rDNA condensation in metaphase-versus-anaphase-arrested cells. Cultures of YBL26–5–3a (MCD1 cdc15-1) and YBL111 (mcd1-1 cdc15-1) were synchronized in G1, shifted to the restrictive temperature, and released into prewarmed media plus or minus nocodazole as indicated. Following rearrest in metaphase or anaphase (as indicated), rDNA FISH was performed. Quantitation of rDNA puffs, clusters, lines, and loops is shown. More than 100 nuclei were scored per sample.
Phosphorylation of condensin correlates with the cohesin-independent pathway
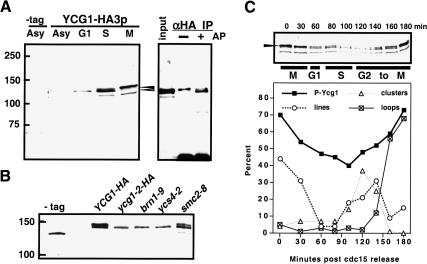
The existence of a second, Mcd1p-independent stage of condensation correlates with the disappearance of cohesins at the metaphase-to-anaphase transition. How does this pathway become activated? The simplest model predicts that chromatin or the condensin complex itself could be modified prior to or during anaphase, and this would permit cohesin-independent condensation. Consistent with this idea, we observed that the condensin subunit Ycg1p is phosphorylated in mitosis, as judged by a mobility shift in SDS-PAGE that is sensitive to alkaline phosphatase treatment (Fig. 4). To better assess the significance of Ycg1p phosphorylation, we undertook a candidate approach to identifying the responsible kinase. The accumulation of phospho-Ycg1p is dependent on the mitotic kinase IPL1/aurora (Fig. 4B), suggesting that in budding yeast as in other organisms, the Ipl1 kinase is poised to play a key role in mitotic chromosome structure (Wei et al. 1998; Giet and Glover 2001; Morishita et al. 2001; Hagstrom et al. 2002; Kaitna et al. 2002).
Figure 4.
Mitotic phosphorylation of the condensin subunit Ycg1p. (A) Western blot analysis of Ycg1–HA3p (2131a) from asynchronously growing (Asy) and synchronized cells (G1, S, and M phase). Whole-cell extracts were separated by SDS-PAGE (6.5% resolving gel), transferred to nitrocellulose, and probed with a mouse monoclonal anti-HA antibody (16B12), which recognizes a doublet of Ycg1p (arrows) and an unrelated 125-kD polypeptide just below. The slower migrating form is sensitive to alkaline phosphatase treatment (right panel). Ycg1–HAp was immunoprecipitated from nocodazole-arrested cells using anti-HA antibodies and protein A Sepharose, and treated plus and minus alkaline phosphatase. Arrows indicate both forms of Ycg1–HAp. (B) Western blot of Ycg1–HAp phosphorylation in IPL1+ (YBL55–9b) and ipl1-321 (YBL55–10b) strains. Cells were synchronized in G1, shifted to 37°C, and released into warm medium containing nocodazole to rearrest the cells in mitosis. G1-versus-M-phase extracts are shown. Arrows mark Ycg1p phospho-forms. (C) Correlation of Ycg1p phosphorylation with rDNA condensation. Following cell synchronization in a cdc15-1 background, cells were released into the cell cycle at 23°C in the presence of nocodazole, and aliquots were taken at each time point for rDNA FISH and FACS analysis (see Fig. 1) as well as for protein analysis by Western blot. Under- and overphosphorylated forms of Ycg1p are indicated (arrows). The amount of Ycg1p shifted to the slower migrating band is plotted over time (lower panel). The temporal succession of rDNA species is redrawn from Fig. 1 for comparison.
One possibility is that Ipl1p could be involved in the loading of condensin onto chromatin as suggested in several in vivo studies (Giet and Glover 2001; Hagstrom et al. 2002; Kaitna et al. 2002; Petersen and Hagan 2003). In budding yeast, however, as in Xenopus extracts, chromatin binding of condensin was not dependent on Ipl1 activity (Losada et al. 2002; data not shown), suggesting a later role for Ipl1p in the condensation process. To further address this, we next asked whether the Ipl1-dependent Ycg1p phosphorylation correlated with condensin activation at the onset of mitosis or occurred later in the condensation process. Figure 4C shows a time course correlating the appearance of phospho-Ycg1p with chromosome condensation. Strikingly, although low levels of Ycg1p phosphorylation can be detected in early mitosis, it peaks ∼40–60 min later with the accumulation of rDNA loops (T = 160–180 min postrelease from the cdc15-1 block). Thus, the phosphorylation of Ycg1p seems unlikely to be required for activating condensin at the onset of mitosis; rather, its timing is consistent with a later role possibly in modulating the cohesin-independent condensation pathway.
Condensation postanaphase onset requires the Ipl1 kinase
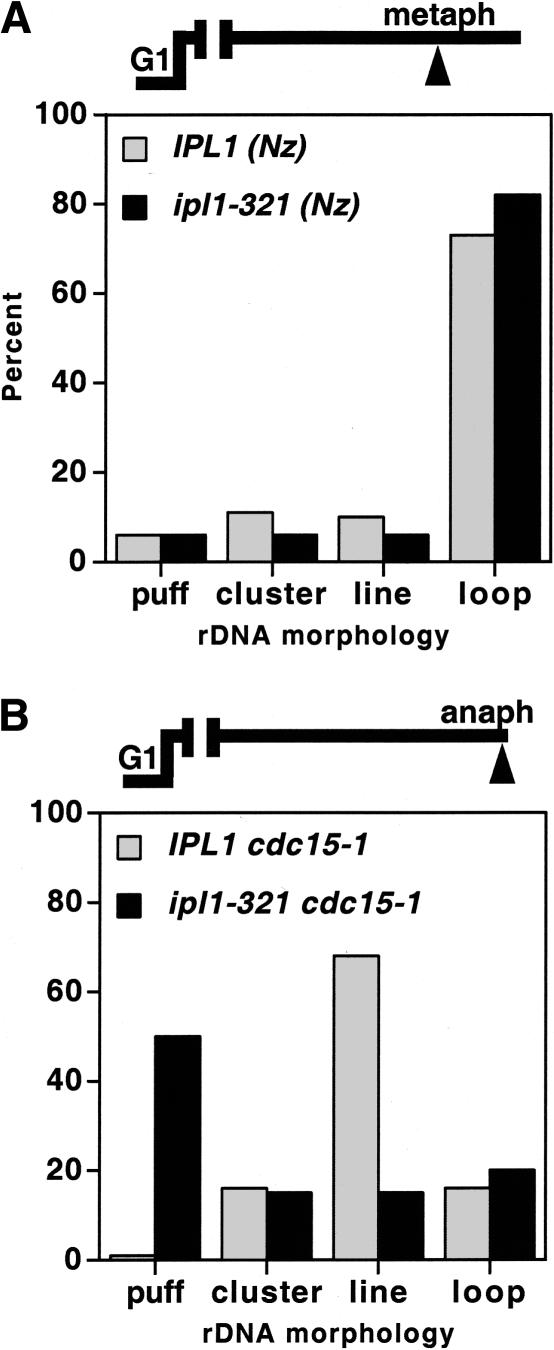
To more directly assess whether IPL1 could be involved in a transition from cohesin-dependent to -independent condensation, we asked whether Ipl1 kinase is required for rDNA condensation in metaphase- versus anaphase-arrested cells. rDNA FISH was performed on chromosomes from cells arrested at either cell cycle stage, plus and minus Ipl1 kinase activity. As shown in Figure 5, chromosome condensation was established and maintained at similar levels in metaphase-arrested wild-type or ipl1-321 cells. In contrast, rDNA condensation in anaphase cells could not be maintained in the absence of Ipl1 function. Rather, the anaphase chromosomes reverted prematurely to G1-like puffs, consistent with the loss and/or misregulation of condensin function. This inhibition of condensin function is unlikely to result from a failure to arrest in anaphase, because the ip1l-321 mutant progresses through mitosis (Biggins and Murray 2001) and, in the cdc15-1 background, accumulated large budded cells with divided nuclei at the restrictive temperature (data not shown). We next asked whether the loss of anaphase condensation in ipl1-321 cells could be attributed to a precocious loss of condensin from chromatin. This does not appear to be the case as no significant change in condensin binding to chromatin was observed in the presence or absence of Ipl1 function (data not shown). Thus, given the persistence of condensin on chromosomes, our data suggest that in the absence of Ipl1 kinase, the cohesin-independent pathway fails to be activated resulting in a disruption of chromosome condensation as cells progress into anaphase.
Figure 5.
Ipl1 kinase requirement for chromosome structure postanaphase onset. FISH analysis of rDNA condensation in IPL1 (YBL26–5–3a; IPL1 cdc15-1) versus ipl1-321 (YBL76–9b; ipl1-321 cdc15-1) strains in metaphase (A) versus anaphase cells (B). Following synchronization in G1, cells were released at 37°C and rearrested in metaphase using nocodazole (A) or allowed to proceed to the anaphase block (cdc15-1; B). After 2.5 h, cells were fixed and processed for rDNA FISH. rDNA morphologies were scored as indicated. More than 100 nuclei were counted per sample.
Discussion
rDNA condensation proceeds through defined intermediates
In this study, we have defined five distinct species for the mitotic condensation of the RDN locus during the course of a single cell cycle: large and small puffs in G1–S versus clusters, lines, and loops in G2/M phase. These in vivo rDNA species likely reflect progressive levels of chromosome organization. Indeed, rDNA clusters are significantly more compacted than the premitotic puffs, yet do not represent end products. Rather, with time, rDNA further aligns into short lines and finally loops, which are clearly resolved from the bulk of yeast chromosomes. We suggest that these different rDNA morphologies reflect in vivo kinetic intermediates inherent to the condensation process. Consistent with this, a similar succession of the rDNA species is observed both in cycling as well as mitotically arrested cells, and the transition between species is dependent on condensin, an established component of the condensation machinery.
Intriguingly, the earliest intermediate of chromosome condensation we detected is the most highly compacted (cluster), which over time resolves into larger, and presumably more ordered, lines and loops (Fig. 6A). The simplest interpretation is that these intermediates reflect the two essential functions of condensation: chromosome compaction and resolution. At first glance, it may seem counterintuitive that chromosome condensation would initiate through an apparently more compact structure. One possible interpretation is that clusters represent misfolded, off-pathway products. This seems unlikely because rDNA clusters, which accumulate in the presence of the plasmid-borne ycg1-2 allele, are converted to loops with similar kinetics to uncondensed rDNA puffs upon return to the permissive temperature (data not shown). Alternatively, the progression of rDNA intermediates from clusters into lines/loops may reflect an ordered series of reactions that are required for mitotic chromosome folding. Such a multistep process is reminiscent of other higher-order nucleoprotein complex assembly reactions. For instance, the formation of the phage Mu transposome, which promotes the transposition of the Mu genome into new DNA sites, involves interactions between multiple proteins bound to three distant DNA regions, the two Mu ends and the transpositional enhancer (Chaconas and Harshey 2002). At an early step of the reaction, all three DNA regions are brought together into a compact, yet transient three-site synaptic complex. This unstable complex precedes the formation of a series of progressively more stable transpososomes in which protein and DNA composition, as well as DNA topology, are altered.
Figure 6.
Condensation is a multistep process and can proceed through two distinct pathways. (A) Model of chromosome condensation. As cells enter into the G2/early M phase of the cell cycle, condensation of the chromosomes proceeds through three distinct stages: clustering, alignment, and resolution. Both condensins (gray balls) and cohesins (black bars) are required for the establishment and maintenance of condensation until metaphase. Resolution of the more compacted cluster to lines and finally loops could result from protein–protein interactions between condensin molecules, in a manner reminiscent of higher-order nucleoprotein assembly pathways (see Discussion). At the metaphase-to-anaphase transition, however, a different condensation pathway operates. The Ipl1 kinase promotes a further maturation of chromosomes necessary for the maintenance of structural integrity in the absence of cohesins. This maturation event may in part be accomplished through the phosphorylation of condensin, namely, through its Ycg1p subunit. Condensin would thus have two distinct active forms specific to each condensation pathway: a foldase that promotes cohesin-dependent condensation and an Ipl1-dependent maturase required postanaphase onset. (B) Conservation of condensation pathways in budding yeast versus higher eukaryotes. In S. cerevisiae, the Ipl1/aurora-independent (cohesin-dependent) pathway is activated in G2/M and persists to metaphase (white bar). Prior to the metaphase-to-anaphase transition (indicated by an arrow), the Ipl1/aurora kinase pathway (black bar) is activated, such that chromosome structural integrity is maintained during anaphase. In higher eukaryotes, condensin function initiates in prophase in an aurora-independent manner and during which time cohesins are still present on chromosomes. In late prophase, aurora stimulates the loading of condensin on chromatin, coincident with the removal of bulk cohesins, and suggesting that the Ipl1/aurora pathway is activated significantly before the metaphase-to-anaphase transition.
In a similar manner, condensin-dependent intermediates can be modeled as steps in higher-order nucleoprotein complex assembly (Fig. 6A). First, condensin binds its substrate, presumably wrapping chromatin around a protein core as has been observed in vitro with naked DNA (Bazett-Jones et al. 2002). Given the repetitive nature of the rDNA, it seems likely that condensin would be regularly distributed throughout the array, as suggested by protein localization studies (Freeman et al. 2000; Bhalla et al. 2002). Second, higher-order complex formation would involve interactions between distant, condensin-bound DNA sites. This early assembly step could be stimulated by cohesins bound within each rDNA repeat (Laloraya et al. 2000), consistent with the proposal that cohesins serve as cis-acting factors in condensation (Lavoie et al. 2002). Once a rudimentary architecture is generated, further resolution of the chromosomes would occur generating line and loop morphologies, possibly involving protein–protein interactions between condensin molecules. If the higher-order assembly of condensins were robust (sterically constrained, high affinity and/or low dissociation), then condensin function might also provide rigidity to the chromosomes as suggested by recent studies in Caenorhabditis elegans (Stear and Roth 2002). Consistent with this idea, the transition we observe from rDNA lines to loops involves the formation of a more extended rDNA structure, which bulges away from the nucleus rather than folding randomly over itself. These features of loops strongly suggest that the chromosomes become rigid.
It remains to be determined whether the condensation intermediates of the RDN locus of Chromosome XII and the ensuing model for condensation will prove a faithful model for euchromatic chromosomes in yeast and other organisms. Although budding yeast condensation uses the same machinery as all other eukaryotes, the rDNA consists of specialized substrate consisting of an ∼1 Mb of repeated structure. The uniformity and size of the RDN locus are likely to facilitate the detection of intermediate species yet to be reported at euchromatic sites in yeast and other organisms. One caveat, however, is that additional levels of regulation may make aspects of these intermediates specific to this locus or to repetitive DNA. Indeed, the rDNA appears to remain condensed later in mitosis than euchromatic sites (Guacci et al. 1994). This difference may reflect that specific regions of chromosomes condense at different rates. Alternatively, it could also derive from the increased sensitivity of the rDNA assay. Although it is not presently possible to distinguish between these two nonexclusive possibilities, the unequivocal requirement for condensin for both rDNA and euchromatic chromosome condensation argues that the fundamental mechanism of condensation is likely to be conserved. Because the enzymatic/structural role for condensin in condensation is unlikely to change substantially with different DNA substrates, an understanding of mitotic rDNA structure is likely to provide significant insight into the fundamental mechanism of condensin function in vivo that will be applicable to euchromatic sequences in yeast and other organisms.
The Ipl1/aurora, a second pathway for condensation in budding yeast
Our study demonstrates that condensins mediate higher-order chromosome structure from G2 through anaphase, and supports the model that a single basic machinery is responsible for condensation throughout mitosis. Previous work in both yeast and Sordaria has also established the involvement of cohesins in chromosome condensation, at least up to the metaphase (Guacci et al. 1997; van Heemst et al. 1999; Hartman et al. 2000). Although the mechanism of cohesin involvement is not fully understood, we have recently proposed that cohesins (but not sister-chromatid cohesion) serve as cis-factors for condensation in early M-phase cells by delimiting domains of condensin function (Lavoie et al. 2002). We now extend this study and show that in contrast to condensation up to and including metaphase, the rDNA condensation in anaphase is independent of the essential cohesin subunit Mcd1p/Scc1p and can occur de novo. This result is consistent with the observed destruction of Mcd1p/Scc1p, which occurs at the metaphase-to-anaphase transition (Uhlmann et al. 1999), and suggests that a second, cohesin-independent condensation pathway is required for chromosome condensation postanaphase onset (this study). This second condensation pathway is characterized by its requirement for the conserved mitotic kinase Ipl1/aurora. A previous study of aurora-depleted Drosophila cells had observed defects in chromosome condensation in anaphase (Adams et al. 2001), suggesting that this pathway may be conserved among eukaryotes.
How could Ipl1/aurora B promote a second condensation pathway? One possibility suggested by recent experiments in Xenopus extracts is that the aurora B and polo-like kinases are required for cohesin release from chromosomes, and either the kinases or the release of cohesins are required for sister-chromatid resolution although not chromosome compaction (Losada et al. 2002). Likewise in budding yeast, the timing of the Ipl1-dependent condensation pathway also coincides with the removal of cohesins and the resolution of sisters (albeit in anaphase rather than earlier in mitosis; see Fig. 6). This removal of cohesins is not sufficient for anaphase condensation, however, as the premature inactivation of cohesins using a temperature-sensitive mutant fails to support condensation in the absence of Ipl1 function. Rather, we propose that the aurora kinase acts as a maturase, converting the maintenance of condensation from a cohesin-dependent to an -independent mechanism.
Ipl1/aurora could act as a maturase by modifying the condensation machinery or the substrate chromatin. Indeed, one target of Ipl1/aurora is histone H3. However, H3 phosphorylation is unlikely to promote chromosome maturation in budding yeast, as unphosphorylatable mutants show no significant condensation defects, either in metaphase or in anaphase (Lavoie et al 2002; data not shown). Even in other organisms the importance of H3 phosphorylation for condensation has been contested (Adams et al. 2001). Another potential target of aurora is the condensin complex. Consistent with this, we observe Ipl1-dependent phosphorylation of the Ycg1 subunit of condensin. Although the functional significance of this phosphorylation remains to be determined, aurora B has been shown to affect multiple aspects of chromosome structure in other species. Ipl1/aurora promotes condensin association with chromatin in some organisms (Giet and Glover 2001; Petersen et al. 2001; Hagstrom et al. 2002; Kaitna et al. 2002). However, this stimulation of condensin loading cannot wholly explain the role of aurora in condensation, as it does not strictly correlate with condensin function in either C. elegans (Kaitna et al. 2002), or in Xenopus laevis and budding yeast, where condensin binding is independent of aurora function (Losada et al. 2002; MacCallum et al. 2002; this study). Based on its time of action and its phenotype of tangled chromosomes, we suggest that Ipl1/aurora directly or indirectly modifies condensin to ensure chromatid resolution and thus sister separation during anaphase (Fig. 6).
Aurora kinase and cohesins: two independent pathways for condensation?
An important question that remains is how the cell orchestrates the transition between both condensation pathways. Temporally, overlap between the pathways is likely to occur. Indeed, the Ipl1 kinase is known to phosphorylate several in vivo yeast targets prior to anaphase, including histone H3 and the kinetochore component Ask1p (Hsu et al. 2000; Cheeseman et al 2002). Similarly, the Ipl1-dependent phosphorylation of condensin peaks prior to anaphase onset, but after significant chromosome condensation is observed (this study). In Saccharomyces cerevisiae, cohesins remain associated with chromosomes until the onset of anaphase, whereas in higher eukaryotes, the majority of cohesins are removed by prometaphase/metaphase (Michaelis et al. 1997; Losada et al. 1998, 2000; Darwiche et al. 1999; Sumara et al. 2000; Waizenegger et al. 2000; Warren et al. 2000), a time when aurora B is already associated with chromosomes (Adams et al. 2001; Giet and Glover 2001). We suggest that the early removal of cohesins in these systems could be compensated by an earlier activation of aurora B function as a maturase (Fig. 6B). In addition, this earlier activation could drive condensation in experiments in which cohesins are inactivated.
Because both the Ipl1 and cohesin pathways can independently drive condensation, is either pathway sufficient for the biological function of condensation in chromosome segregation? This question is difficult to address experimentally because of the essential functions of both aurora and cohesins in other aspects of chromosome segregation like kinetochore function and sister-chromatid cohesion. Nonetheless, it seems likely that both pathways will be required because of constraints imposed by chromosome segregation. Prior to anaphase, condensation serves to compact sister chromatids and also partition them into two distinct domains, facilitating their subsequent segregation during anaphase. In budding yeast, the cohesin-dependent condensation pathway serves this function as condensation in the absence of cohesins leads to misfolded/tangled chromosomes (Lavoie et al. 2002). After the onset of anaphase, condensation is further needed to prevent sister-chromatid bridging and/or lagging chromosomes in the plane of cytokinesis and may also facilitate encapsulation in the reforming or dividing nucleus. The cohesin-dependent pathway cannot provide this function because cohesins must be inactivated for anaphase to occur. Thus, although both pathways could in principle establish and maintain condensation, we propose that a division of labor occurs such that chromosome compaction is first established by ordered folding (cohesin pathway) followed by a chromosome resolution step to preclude chromosome bridging and maintain condensation once cohesins are removed (aurora pathway). Together both pathways orchestrate a temporal and structural hierarchy of chromosome folding, coordinating the multiple functions of condensation throughout mitosis.
Materials and methods
Media and reagents
Yeast cultures were grown in rich media (Guthrie and Fink 1991) with 2% dextrose (YPD) or 2% raffinose for gal inductions (YPRaf). Raffinose, galactose, α-factor, and nocodazole were from Sigma. Restriction enzymes were from Boehringer Mannheim. The rDNA FISH probe was as previously described (Guacci et al. 1994). The antibodies used were mAb 16B12 (anti-HA; BabCO), mouse anti-DIG and pig-anti-goat-FITC (Boehringer Mannheim), goat anti-mouse-FITC (BabCO), and goat anti-mouse HRP (Jackson Immunolabs). ECL reagents were from NEN. G418 was from GIBCO BRL.
Yeast strains and plasmids
Yeast strains (Table 1) were constructed using standard techniques (Guthrie and Fink 1991). Conditional temperature-sensitive condensin alleles are as previously described (Lavoie et al. 2000, 2002). The plasmid-borne ycg1-2ts allele is in YCplac111, a CEN6 ARS1 LEU2 vector (Gietz and Sugino 1988) to generate pBL238-ts2. The cdc15-1 mutation used for cell cycle arrests postanaphase was obtained from strain 4536-151 (originally from the Hartwell collection) and backcrossed 10 times to an S288c background (YPH derivation). To generate C-terminal triple HA-tagged YCG1 strains (YCG1-HA), single-step gene replacement used the Schizosaccharomyces pombe his5+ gene to complement a ura3 deficiency. p473 was used as a source for PCR of 3XHA:his5+ as described previously (Lavoie et al. 2002). Colony PCR and Western blotting verified integration into the correct site. Galactose-inducible Clb2Δdb strains were generated by integration of p185 (URA3: GAL1-CLB2Δdb in YIplac211; a generous gift from A. Amon, MIT) into 982 (WT) and 2172 (ycg1-2), generating 982–101 and 2172–101, respectively.
Table 1.
Saccharomyces cerevisiae strains
| Strain | Plasmid | Mating type | Genotype | Background | Reference |
|---|---|---|---|---|---|
| 2125-3a | pBL238-ts2 | MATa | ura3-52 lys2-801 ade2-101 trp1Δ63 leu2Δ1 his3Δ200 ycg1::KAN/ycg1-2 LEU2 CEN6 ARS1 | S288c | This study |
| 2131a | MATa | ura3-52 lys2-801 ade2-101 trp1Δ63 leu2Δ1 his3Δ200 YCG1-HA3:his5+ | S288c | This study | |
| 2153-1 | MATa | bar1 ura3 leu2 trp1 ade2 his3Δ200 gal1 ycg1-2:KAN arg- | A364A | This study | |
| 2172-101 | MATa | ura3 leu2 ycg1-2:KAN MCD1-HA6 can1 gal1 URA3:GAL-CLB2Δdb | A364A | This study | |
| 982-101 | MATa | bar1 trp1 ura3 URA3:GAL-CLB2Δdb | A364A | This study | |
| CH2523 | MATa | leu2Δ1 trp1Δ63 ura3-52 BRN1:TRP1 | S288c | Lavoie et al. 2000 | |
| CH2524 | MATa | brn1-9:TRP1 ura3-52 leu2Δ1 trp1Δ63 | S288c | Lavoie et al. 2000 | |
| YBL02-18b | MATa | ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2-801 ade2-101 ycgl-2:KAN | S288c | This study | |
| YBL111 | MATa | cdc15-1 ura3-52 trp1Δ63 mcd1-1 | S288c | This study | |
| YBL13-2a | MATa | MCD1-HA6 leu2 BRN1:TRP1 TRP? cdc15-1 leu2 ade3 can?1 his7 | Other | This study | |
| YBL26-5-3a | MATa | cdc15-1 ura3-52 trp1Δ63 | S288c | This study | |
| YBL26c-9a | MATa | leu2 ade2 trp1 ura-cdc15-1 YCG1-HA3:his5+ | Other | This study | |
| YBL55-10b | MATa | ipl1-321 YCG1-HA3:his5+ ura3 leu2 lys2 ade2 trp1-1:256LacO:TRP1 his3, 11:pCUP1-GFP12-LacI12:HIS3 bar1 | Other | This study | |
| YBL55-9b | MATa | IPL1+ YCG1-HA3:his5+ ura3 leu2 lys2 ade2 bar1 trp1Δ63 his3 | Other | This study | |
| YBL76-9b | MATa | ipl1-321 cdc15-1 his3Δ200 trp1Δ63 | S288c | This study | |
| YPH499 | MATa | ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 | S288c | Sikorski and Hieter 1989 |
Cell cycle synchronization
For the cell cycle release experiment (cdc15-1), an early log phase culture of YBL26c-9a (MATa cdc15-1) was shifted from 23°C to 37°C for 3 h (89% large budded cells). The cells were then synchronously released into the cell cycle by placing the culture in a 23°C waterbath. Time points were taken as indicated, and cell cycle progression was monitored by light microscopy, flow cytometry, and FISH analysis. For G1 synchrony-release experiments, early log phase cultures were first arrested in G1 using 3 μM final α-factor for 2.5 h at 23°C (85%–95% schmoos). The cultures were then shifted to 37°C in a waterbath for 0.5 h and released by washing the cells three times with 37°C YPD plus 0.1 mg/mL pronase E (Sigma). The final resuspension medium also contained 20 μg/mL nocodazole, and the cells were incubated for another 2.5 h, at which time >90% of cells were large budded with an undivided nucleus. For FISH, cells were fixed with 0.36% formaldehyde for 1.5 h. Rescue of condensation experiments was carried out similarly except that cells were shifted back to 23°C as indicated after 2 h at 37°C to maintain the mitotic arrest. For Clb2Δdb-induced synchrony, cells were grown in YP + 2% raffinose, blocked, and released from a G1 (α-factor) block except that 2% galactose was added to the cells immediately upon release into the cell cycle. After 3 h, the cells were shifted to the restrictive temperature for 30 min and the extent of condensation determined by FISH.
Flow cytometry, chromatin spreads, and FISH
Determination of DNA content in cycling cells was by flow cytometry as previously described (Lavoie et al. 2000). Chromatin spreads were as described (Michaelis et al. 1997; Lavoie et al. 2002). Both mouse anti-HA and goat anti-mouse-FITC antibodies were at 1/500. DNA was stained with DAPI. FISH was performed as in Guacci et al. (1994) and Lavoie et al. (2000, 2002), using digoxigenin-labeled rDNA probes and FITC-conjugated secondary and tertiary antibodies. Chromosomes were counter-stained with propidium iodide in antifade mounting medium (Intergen). Images were obtained using a Zeiss epifluorescence microscope, and were recorded digitally with the use of a Princeton charge coupled device with Signal Analytics processing software, which allows image superimposition. For Western blots, both the 16B12 as well as the goat-anti-mouse-HRP antibodies were used at 1/2500 in 2% milk/PBST.
Acknowledgments
We are indebted to Angelika Amon (MIT) for the CLB2–Δdb overexpressor. We thank A. Amon, Yixian Zheng, Terence Murphy, Jim Wilhelm, and members of our laboratories for useful discussions and critical reading of the manuscript. This work was funded by grants from the Canadian Institutes of Health Research (B.L.) and the Howard Hughes Medical Institute (D.K.). B.L. is a CIHR New Investigator.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1150404.
References
- Adams R.R., Maiato, H., Earnshaw, W.C., and Carmena, M. 2001. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153: 865–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon A., Irniger, S., and Nasmyth, K. 1994. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell 77: 1037–1050. [DOI] [PubMed] [Google Scholar]
- Bazett-Jones D.P., Kimura, K., and Hirano, T. 2002. Efficient supercoiling of DNA by a single condensin complex as revealed by electron spectroscopic imaging. Mol. Cell 9: 1183–1190. [DOI] [PubMed] [Google Scholar]
- Bhalla N., Biggins, S., and Murray, A.W. 2002. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol. Biol. Cell 13: 632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S. and Murray, A.W. 2001. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes & Dev. 15: 3118–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Severin, F.F., Bhalla, N., Sassoon, I., Hyman, A.A., and Murray, A.W. 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes & Dev. 13: 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano I., Brzoska, P., Sadoff, B., Chen, H., and Christman, M. 1996. Mitotic chromosome condensation in the rDNA requires TRF4 and DNA topoisomerase I in S. cerevisiae. Genes & Dev. 10: 2564–2576. [DOI] [PubMed] [Google Scholar]
- Chaconas G. and Harshey, R.M. 2002. Transposition of phage Mu DNA. In Mobile DNA II (eds. N.L. Craig, A. Lambowitz, M. Gellert, and R. Craigie), pp. 384–402. American Society for Microbiology, Washington, DC.
- Cheeseman I.M., Anderson, S., Jwa, M., Green, E.M., Kang, J., Yates III, J.R., Chan, C.S., Drubin, D.G., and Barnes, G. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111: 163–172. [DOI] [PubMed] [Google Scholar]
- Darwiche N., Freeman, L.A., and Strunnikov, A. 1999. Characterization of the components of the putative mammalian sister chromatid cohesion complex. Gene 233: 39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzel S. and Belmont, A.S. 2001. Reproducible but dynamic positioning of DNA in chromosomes during mitosis. Nat. Cell Biol. 3: 767–770. [DOI] [PubMed] [Google Scholar]
- Elliot S.G. and McLaughlin, C.S. 1979. Regulation of RNA synthesis in yeast: Synthesis during the cell cycle. Mol. Gen. Genet. 169: 237–243. [DOI] [PubMed] [Google Scholar]
- Freeman L., Aragon-Alcaide, L., and Strunnikov, A. 2000. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell Biol. 149: 811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R. and Glover, D.M. 2001. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 152: 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D. and Sugino, A. 1988. New yeast—Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534. [DOI] [PubMed] [Google Scholar]
- Guacci V., Hogan, E., and Koshland, D. 1994. Chromosome condensation and sister chromatid pairing in budding yeast. J. Cell Biol. 125: 517–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V., Koshland, D., and Strunnikov, A. 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91: 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C. and Fink, G.R. 1991. Guide to yeast genetics and molecular biology. In Methods in enzymology. Academic Press, New York. [PubMed]
- Hagstrom K.A., Holmes, V.F., Cozzarelli, N.R., and Meyer, B.J. 2002. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes & Dev. 16: 729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J.S., Kroll, E.S., Lundblad, V., and Spencer, F.A. 2001. S. cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell. Biol. 21: 3144–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman T., Stead, K., Koshland, D., and Guacci, V. 2000. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in S. cerevisiae. J. Cell Biol. 151: 613–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Kobayashi, R., and Hirano, M. 1997. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell 89: 511–521. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y., Minden, J.S., Swedlow, J.R., Sedat, J.W., and Agard, D.M. 1989. Focal points for chromosome condensation and decondensation revealed by three-dimensional in vivo time-lapse microscopy. Nature 342: 293–296. [DOI] [PubMed] [Google Scholar]
- Hsu J.Y., Sun, Z.W., Li, X., Reuben, M., Tatchell, K., Bishop, D.K., Grushcow, J.M., Brame, C.J., Caldwell, J.A., Hunt, D.F., et al. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102: 279–291. [DOI] [PubMed] [Google Scholar]
- Kaitna S., Pasierbek, P., Jantsch, M., Loidl, J., and Glotzer, M. 2002. The Aurora B kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous chromosomes during meiosis. Curr. Biol. 12: 798–812. [DOI] [PubMed] [Google Scholar]
- Kang J., Cheeseman, I.M., Kallstrom, G., Velmurugan, S., Barnes, G., and Chan, C.S. 2001. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J. Cell Biol. 155: 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K. and Hirano, T. 2000. Dual roles of the 11S regulatory subcomplex in condensin functions. Proc. Natl. Acad. Sci. 97: 11972–11977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Hirano, M., Kobayashi, R., and Hirano, T. 1998. Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science 282: 487–490. [DOI] [PubMed] [Google Scholar]
- Laloraya S., Guacci, V., and Koshland, D. 2000. Chromosomal addresses of the cohesin component Mcd1p. J. Cell Biol. 151: 1047–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie B.D., Tuffo, K.M., Oh, S., Koshland, D., and Holm, C. 2000. Mitotic chromosome condensation requires Brn1p, the yeast homologue of Barren. Mol. Biol. Cell 11: 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie B.D., Hogan, E., and Koshland, D. 2002. In vivo dissection of the chromosome condensation machinery: Reversibility of condensation distinguishes contributions of condensin and cohesin. J. Cell Biol. 156: 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A. and Hirano, T. 2001. Shaping the metaphase chromosome: Coordination of cohesion and condensation. Bioessays 23: 924–935. [DOI] [PubMed] [Google Scholar]
- Losada A., Hirano, M., and Hirano, T. 1998. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes & Dev. 12: 1986–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A., Yokochi, T., Kobayashi, R., and Hirano, T. 2000. Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J. Cell Biol. 150: 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A., Hirano, M., and Hirano, T. 2002. Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes & Dev. 16: 3004–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum D.E., Losada, A., Kobayashi, R., and Hirano, T. 2002. ISWI remodeling complexes in Xenopus egg extracts: Identification as major chromosomal components that are regulated by INCENP-aurora B. Mol. Biol. Cell 13: 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manders E.M., Kimura, H., and Cook, P.R. 1999. Direct imaging of DNA in living cells reveals the dynamics of chromosome formation. J. Cell Biol. 144: 813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C., Ciosk, R., and Nasmyth, K. 1997. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell 91: 35–45. [DOI] [PubMed] [Google Scholar]
- Morishita J., Matsusaka, T., Goshima, G., Nakamura, T., Tatebe, H., and Yanagida, M. 2001. Bir1/Cut17 moving from chromosome to spindle upon the loss of cohesion is required for condensation, spindle elongation and repair. Genes Cells 6: 743–763. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. 2002. Segregating sister genomes: The molecular biology of chromosome separation. Science 297: 559–565. [DOI] [PubMed] [Google Scholar]
- Ouspenski I.I., Cabello, O.A., and Brinkley, B.R. 2000. Chromosome condensation factor Brn1p is required for chromatid separation in mitosis. Mol. Biol. Cell 11: 1305–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J. and Hagan, I.M. 2003. S. pombe aurora kinase/survivin is required for chromosome condensation and the spindle checkpoint attachment response. Curr. Biol. 13: 590–597. [DOI] [PubMed] [Google Scholar]
- Petersen J., Paris, J., Willer, M., Philippe, M., and Hagan, I.M. 2001. The S. pombe aurora-related kinase Ark1 associates with mitotic structures in a stage dependent manner and is required for chromosome segregation. J. Cell Sci. 114: 4371–4384. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter, P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in S. cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E., Matsusaka, T., Morrison, C., Vagnarelli, P., Hoshi, O., Ushiki, T., Nojima, K., Fukagawa, T., Waizenegger, I.C., Peters, J.M., et al. 2001. Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev. Cell 1: 759–770. [DOI] [PubMed] [Google Scholar]
- Stear J.H. and Roth, M.B. 2002. Characterization of HCP-6, a C. elegans protein required to prevent chromosome twisting and merotelic attachment. Genes & Dev. 16: 1498–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov A.V., Hogan, E., and Koshland, D. 1995. SMC2, an S. cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes & Dev. 9: 587–599. [DOI] [PubMed] [Google Scholar]
- Sumara I., Vorlaufer, E., Gieffers, C., Peters, B.H., and Peters, J.M. 2000. Characterization of vertebrate cohesin complexes and their regulation in prophase. J. Cell Biol. 151: 749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutani T. and Yanagida, M. 1997. DNA renaturation activity of the SMC complex implicated in chromosome condensation. Nature 388: 798–801. [DOI] [PubMed] [Google Scholar]
- Sutani T., Yuasa, T., Tomonaga, T., Dohmae, N., Takio, K., and Yanagida, M. 1999. Fission yeast condensin complex: Essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes & Dev. 13: 2271–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F. 2003. Chromosome cohesion and separation: From men and molecules. Curr. Biol. 13: R104–R114. [DOI] [PubMed] [Google Scholar]
- Uhlmann F., Lottspeich, F., and Nasmyth, K. 1999. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400: 37–42. [DOI] [PubMed] [Google Scholar]
- van Heemst D., James, F., Poggeler, S., Berteaux-Lecellier, V., and Zickler, D. 1999. Spo76p is a conserved chromosome morphogenesis protein that links the mitotic and meiotic programs. Cell 98: 261–271. [DOI] [PubMed] [Google Scholar]
- Waizenegger I.C., Hauf, S., Meinke, A., and Peters, J.M. 2000. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 103: 399–410. [DOI] [PubMed] [Google Scholar]
- Warren W.D., Steffensen, S., Lin, E., Coelho, P., Loupart, M., Cobbe, N., Lee, J.Y., McKay, M.J., Orr-Weaver, T., Heck, M.M., et al. 2000. The Drosophila RAD21 cohesin persists at the centromere region in mitosis. Curr. Biol. 10: 1463–1466. [DOI] [PubMed] [Google Scholar]
- Wei Y., Mizzen, C.A., Cook, R.G., Gorovsky, M.A., and Allis, C.D. 1998. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proc. Natl. Acad. Sci. 95: 7480–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]