Abstract
Background:
Only 40–70% of metastatic colorectal cancers (mCRCs) with wild-type (WT) KRAS oncogene respond to anti-epidermal growth factor receptor (anti-EGFR) antibody treatment. EGFR amplification has been suggested as an additional marker to predict the response. However, improved methods for bringing the EGFR analysis into routine laboratory are needed.
Methods:
The material consisted of 80 patients with mCRC, 54 of them receiving anti-EGFR therapy. EGFR gene copy number (GCN) was analysed by automated silver in situ hybridisation (SISH). Immunohistochemical EGFR protein analysis was used to guide SISH assessment.
Results:
Clinical benefit was seen in 73% of high (⩾4.0) EGFR GCN patients, in comparison with 59% of KRAS WT patients. Only 20% of low EGFR GCN patients responded to therapy. A high EGFR GCN number associated with longer progression-free survival (P<0.0001) and overall survival (P=0.004). Together with KRAS analysis, EGFR GCN identified the responsive patients to anti-EGFR therapy more accurately than either test alone. The clinical benefit rate of KRAS WT/high EGFR GCN tumours was 82%.
Conclusion:
Our results show that automated EGFR SISH, in combination with KRAS mutation analysis, can be a useful and easily applicable technique in routine diagnostic practise for selecting patients for anti-EGFR therapy.
Keywords: colorectal cancer, anti-EGFR therapy, silver in situ hybridisation, immunohistochemistry, predictive marker
The major prognostic determinant for patients with advanced colorectal cancer (CRC) with non-resectable metastases is the response to systemic therapy (Cunningham et al, 2010). For part of these patients, recent advances, including anti-epidermal growth factor receptor (EGFR) therapy have added clinical benefit and extended the median survival time (Cunningham et al, 2004; Douillard et al, 2010; Grothey, 2010; Peeters et al, 2010).
Tumours harbouring activating mutations of KRAS, a signalling molecule downstream of EGFR, do no benefit from the anti-EGFR monoclonal antibodies cetuximab and panitumumab (Linardou et al, 2008; Allegra et al, 2009). In KRAS wild-type (WT) patients, on the other hand, the addition of cetuximab to cytotoxic treatment in first line improves the response rates with 16–24% compared with cytotoxic therapy alone. However, about 40% of the previously untreated (Bokemeyer et al, 2009; Chang et al, 2009; Van Cutsem et al, 2009) and about 60–70% of the previously treated (Moroni et al, 2005; Lievre et al, 2006, 2008; Chang et al, 2009) KRAS WT patients do not respond to anti-EGFR treatment combined with chemotherapy. Consequently, there is a need for predictive markers among the KRAS WT patients. Changes in molecules downstream of EGFR, in particular BRAF gene mutations, PIK3CA mutations and loss of expression of the PTEN tumour-suppressor protein appear to associate with resistance to anti-EGFR treatment (Laurent-Puig et al, 2009; Siena et al, 2009). However, even the combination of these is likely to identify only a minority of non-responsive KRAS WT patients (Laurent-Puig et al, 2009).
Unlike the EGFR protein expression level assessed by immunohistochemistry (IHC) (Cunningham et al, 2004; Saltz et al, 2004; Chung et al, 2005), an increased EGFR gene copy number (GCN) has been associated with a favourable response to anti-EGFR therapy among KRAS WT patients (Moroni et al, 2005; Lievre et al, 2006; Sartore-Bianchi et al, 2007; Cappuzzo et al, 2008). Fluorescence in situ hybridisation (FISH) technique has been used in most previous studies (Moroni et al, 2005; Sartore-Bianchi et al, 2007; Cappuzzo et al, 2008; Personeni et al, 2008; Scartozzi et al, 2009). The FISH results are challenging to interpret and the lack of standardisation of analytical methods and scoring systems may partly explain why the EGFR GCN evaluation has not been incorporated into the clinical practice yet (Martin et al, 2009).
Silver in situ hybridisation (SISH) is a technique that can be applied to automated detection of EGFR GCN and chromosome 7 (Chr-7) number. SISH-based EGFR GCN can be easily performed, because it can be analysed by conventional bright field light microscopy. In addition, the chromogen of SISH is very stable unlike fluorochromes in FISH. The aim of this study was to evaluate the predictive value of EGFR GCN and Chr-7 number assessed by SISH from areas with highest IHC reactivity in patients with metastatic or locally advanced CRC treated with anti-EGFR monoclonal antibody therapy. The correlation between EGFR GCN and EGFR protein expression, as determined by IHC, was also evaluated, since previous reports have been conflicting (Shia et al, 2005; Spindler et al, 2006; Frattini et al, 2007; Hemmings et al, 2009).
Patients and methods
Patients
This retrospective study comprises a series of 80 metastatic or locally advanced CRC patients, 62 of whom were treated with anti-EGFR therapy at the Turku University Hospital. In all, 50% of the patients had metastatic disease at the time of diagnosis. The median age of the patients at diagnosis was 60 years (range, 34–73). Patient characteristics and treatments are presented in Table 1. Ten of the treated patients had a mutation in the KRAS gene, as the anti-EGFR therapy was administered before establishment of the predictive value of KRAS testing. The treatment response could be reliably evaluated for 54 out of 62 (87%) of treated patients. Of those, 25 KRAS WT patients received cetuximab or panitumumab either as single therapy or irinotecan combination therapy in a chemorefractory phase of the disease (⩾third line therapy). The response to anti-EGFR treatment was evaluated by computed tomography or magnetic resonance imaging according to the Response Evaluation Criteria in Solid Tumours (Eisenhauer et al, 2009). The study was conducted in accordance with the Declaration of Helsinki. The clinical data were retrieved and histological samples collected and analysed with the endorsement of the National Authority for Medico-Legal Affairs.
Table 1. Baseline characteristics of patients who underwent SISH for EGFR and chromosome 7 and analysis of KRAS gene mutational status (a) and the subgroup of these patients that received anti-EGFR therapy with evaluable treatment response and sufficient follow up data (b).
| (a) Eligible patients for KRAS mutational status analysis, EGFR and chromosome 7 SISH analysis (n=80) |
(b) Patients treated with anti-EGFR therapy (n=54)
|
||
|---|---|---|---|
| KRAS WT and MT, n=80 | KRAS WT, n=44 | KRAS MT, n=10 | |
| n (%) | n (%) | n (%) | |
| Sex | |||
| Female | 34 (42) | 18 (40.9) | 6 (60) |
| Male | 46 (58) | 26 (59.1) | 4 (40) |
| Site of primary tumour | |||
| Colon | 51 (63.8) | 32 (72.7) | 6 (60) |
| Rectum | 28 (35) | 12 (27.3) | 4 (40) |
| Unknown | 1 (1.2) | ||
| Metastatic sites | |||
| Single | 28 (35) | 19 (43.2) | 2 (20) |
| Multiple | 52 (65) | 25 (56.8) | 8 (80) |
| Tumour differentiation grade | |||
| Grade 1 | 11 (13.8) | 6 (13.6) | 1 (10) |
| Grade 2 | 50 (62.5) | 28 (63.7) | 6 (60) |
| Grade 3 | 13 (16.2) | 6 (13.6) | 2 (20) |
| Unknown | 6 (7.5) | 4 (9.1) | 1 (10) |
| Follow-up data of the patients | |||
| Alive with disease | 16 (20) | 10 (22.7) | — |
| Alive and free of disease | 5 (6.2) | 1 (2.3) | — |
| Died of disease | 59 (73.8) | 33 (75) | 10 (100) |
| KRAS mutational status | |||
| KRAS WT | 54 (67.5) | 44 (100) | — |
| KRAS MT | 24 (30) | — | 10 (100) |
| Not evaluable | 2 (2.5) | — | — |
| Anti-EGFR treatment | |||
| Cetuximab | 51 (63.8) | 35 (79.5) | 10 (100) |
| Panitumumab | 10 (12.5) | 8 (18.2) | — |
| Both | 1 (1.2) | 1 (2.3) | — |
| None | 18 (22.5) | — | — |
| Line of therapy | |||
| First | 8 (12.9) | 5 (11.4) | 1 (10) |
| Second | 14 (22.6) | 12 (27.3) | — |
| Third or more | 40 (64.5) | 27 (61.3) | 9 (90) |
| Anti-EGFR combination therapy | |||
| Anti-EGFR combined to IRI | 46 (74.2) | 32 (72.7) | 9 (90) |
| Anti-EGFR combined to OXA | 10 (16.1) | 8 (18.2) | 1 (10) |
| Anti-EGFR combined to CAP | 2 (3.2) | 1 (2.3) | — |
| Single treatment | 4 (6.5) | 3 (6.8) | — |
Abbreviations: CAP=capecitabine; EGFR=epidermal growth factor receptor; IRI=irinotecan; MT=mutated; OXA=oxaliplatin; SISH=silver in situ hybridization; WT=wild type.
Procedures
Formalin-fixed, paraffin-embedded samples with at least 30% of CRC cells were selected and analysed for KRAS point mutations within codons 12 and 13 with the DxS K-RAS mutation kit (DxS Ltd, Manchester, UK).
In all, 3μm sections were stained with two monoclonal antibodies against EGFR (VentanaMedical Systems/Roche Diagnostics, Tucson, AZ, USA). EGFR (clone 3C6) mAb is directed against the extracellular domain of human EGFR, and EGFR (clone 5B7) mAb against the internal domain of human EGFR. All 80 tumour specimens were stained with the 5B7 anti-EGFR antibody and 74 tumour samples with the 3C6 anti-EGFR antibody. Stainings were performed with BenchMark XT (Ventana/Roche) using ultraVIEW Universal DAB Detection Kit (Ventana/Roche). EGFR IHC was scored independently by three observers (OC, JS, and ML) blinded of the clinical information. Three scoring parameters were recorded: the highest (covering at least 10% of the tumour area), the most common staining intensity, and the localisation of staining (membranous, cytoplasmic or both). Four categories of staining intensity were used: 0 (negative), + (weak), ++ (moderate), and +++ (strong, similar to the intensity of the epidermal basal layer). In cases of discordance, a consensus score was used.
EGFR gene was detected from 5μm sections with EGFR DNA Probe (Ventana/Roche) and Chr-7 from parallel sections with Chr-7 oligonucleotide Probe (Ventana/Roche). SISH was performed with the BenchMark XT using ultraVIEW SISH Detection Kit (Ventana/Roche). From each tumour EGFR GCN (number of copies of gene per cell) and Chr-7 number (number of copies of chromosome per cell) were analysed by two observers (ML and JS) from the area of highest IHC reactivity. Forty tumour cells with the highest number of copies were analysed from the EGFR SISH slides. In addition to the average EGFR GCN and Chr-7 number, EGFR/Chr-7 copy number ratio was assessed.
FISH analysis with Vysis EGFR/CEP 7 FISH Probe Kit (Abbott Molecular Inc., Des Plaines, IL, USA) was performed on nine samples selected based on EGFR SISH results (three samples with clusters, three samples with more than four copies, and three samples with normal two copies), using standard protocols.
Statistical analysis
Statistical analyses were performed with the SAS 9.2 and Enterprise Guide 4.2 programs (SAS Institute Inc., Cary, NC, USA). Frequency table data were analysed with the χ2-test or Fisher's exact test. Spearman correlation coefficients were calculated when correlations were analysed. The optimal cut-off values for EGFR GCN and Chr-7 number were defined with the receiver operating characteristic (ROC) analysis generated on response to treatment (clinical benefit vs progressive disease (PD)). Kaplan–Meier and log-rank tests as well as Cox proportional hazards regression model were used for univariate survival analysis. When analysing progression-free survival (PFS), the survival time was calculated from the onset of anti-EGFR treatment until disease progression. When evaluating the overall survival (OS), the survival time was calculated from the onset of anti-EGFR therapy until death. Multivariate survival analysis was carried out by using Cox's proportional hazards regression model. All statistical tests were two-sided. P-values <0.05 were considered to be statistically significant.
Results
EGFR IHC and EGFR and Chr-7 SISH analysis
Owing to the chromogenic detection method of EGFR GCN and EGFR protein, it was possible to assess both parameters from identical tumour areas and to compare the results. The EGFR protein expression levels and subcellular localisations were examined by two different anti-EGFR antibodies: clone 5B7 against the intracellular domain and clone 3C6 against the extracellular domain, hereafter referred to as intracellular and extracellular domain antibodies, respectively. In general, the intensity and subcellular localisation of IHC reactivity showed considerable intratumoural variation with both antibodies (Figure 1). Therefore, the following parameters were determined: localisation, highest, and most common intensity. The results obtained with the two different antibodies statistically significantly correlated with each other disregarding the parameter used (P<0.0001, Spearman). The most intense areas were scored as moderate (++) in a majority of the tumours, while only one-tenth of the tumours showed areas of strong intensity (+++). The most common EGFR staining intensity was low (+) with both antibodies. The frequencies of these parameters are presented in Table 2.
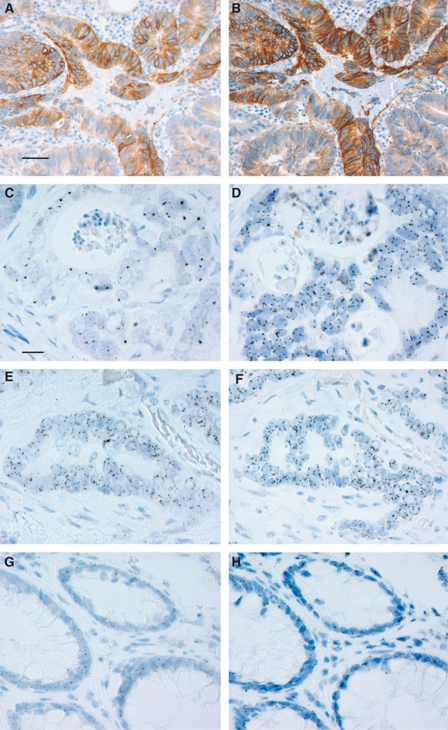
Figure 1.
Epidermal growth factor receptor immunohistochemistry, EGFR, and Chr-7 SISH in colorectal cancer and normal colorectal tissues. Epidermal growth factor receptor IHC with clones 5B7 (A) and 3C6 (B). EGFR SISH revealing gene clusters (C) and the corresponding Chr-7 SISH (D). EGFR SISH with GCN ⩾4.0 (E) and the corresponding Chr-7 SISH (F). EGFR SISH (G) and Chr-7 SISH (H) in normal colorectal tissue. Scale bar 0.05 mm (A, B), 0.02 mm (C–H).
Table 2. EGFR protein expression assessed by anti-EGFR clone 5B7 (n=80) and anti-EGFR clone 3C6 antibodies (n=74).
| 5B7 (H) | 5B7 (C) | 3C6 (H) | 3C6 (C) | |
|---|---|---|---|---|
| Intensity | ||||
| Negative | 0 (0) | 11 (13.8) | 9 (12.2) | 31 (41.9) |
| 1+ | 19 (23.8) | 50 (62.5) | 20 (27.0) | 37 (50.0) |
| 2+ | 53 (66.2) | 19 (23.7) | 38 (51.3) | 6 (8.1) |
| 3+ | 8 (10) | 0 (0) | 7 (9.5) | 0 (0) |
| Localisation | ||||
| Membranous | 23 (28.75) | 11 (13.75) | 24 (32.4) | 11 (14.9) |
| Cytoplasmic | 23 (28.75) | 46 (57.5) | 18 (24.3) | 28 (37.8) |
| Both | 34 (42.5) | 12 (15) | 23 (31.1) | 4 (5.4) |
| Negative | 0 (0) | 11 (13.75) | 9 (12.2) | 31 (41.9) |
Abbreviations: C=most common staining; EGFR=epidermal growth factor receptor; H=highest staining.
Values are given n (%).
The marked variation in EGFR expression as analysed by IHC might reflect an intratumoural variation in the EGFR GCN. Therefore, we assessed the EGFR GCN and Chr-7 number from areas with strongest EGFR staining. The mean EGFR GCN was 5.5 (median 5.5) and the mean Chr-7 number 5.4 (median 5.3).
The optimal cut-off values for EGFR GCN and Chr-7 number as determined with ROC curves were 4.0 (sensitivity 86%, specificity 72%, AUC 83%) and 4.5 (sensitivity 84%, specificity 79%, AUC 85%), respectively. The optimal cut-off value for EGFR GCN was in addition defined with ROC analysis for the selected patients with chemorefractory disease who received anti-EGFR therapy±irinotecan in ⩾third line. The cut-off value proved to be 4.0 (sensitivity 89%, specificity 67%, AUC 84%) in this patient group as well. In all, 51 tumours out of 80 (64%) had an EGFR GCN above cut-off value determined by ROC-analysis (⩾4.0). The EGFR GCN analysis by SISH could not be performed in 2 out of 80 (2.5%) of the cases. Chr-7 number was above the cut-off value (⩾4.5) in 48 out of 80 (60%) of the tumours. The highest EGFR/Chr-7 GCN ratio was 2.8 (mean 1.05, median 1.0). The EGFR FISH results from nine selected tumours correlated with the SISH results.
An increased EGFR GCN and Chr-7 number correlated positively with EGFR IHC analysed by the intracellular domain antibody (Spearman, P=0.01 for both) (Table 3). The correlation remained statistically significant when the staining intensity (IHC) was dichotomised into categories 0 and + vs ++ and +++. A significant correlation between extracellular domain antibody reactivity and an increased Chr-7 number was seen (Spearman, P=0.04), whereas, no correlation was observed between extracellular domain antibody reactivity and EGFR GCN. The subcellular localisation of the EGFR IHC (intracellular and extracellular domain antibodies) did not correlate with EGFR GCN or the Chr-7 number. KRAS mutational status did not correlate either with EGFR and Chr-7 SISH or EGFR IHC results.
Table 3. Correlations of EGFR GCN (SISH), Chr-7 number (SISH), KRAS status and EGFR protein expression (IHC), n=74 (P-values, Spearman).
| KRAS status | EGFR GCN (SISH) continuous variable | Chr-7 (SISH) continuous variable | |
|---|---|---|---|
| Anti-EGFR clone 5B7, intensity | |||
| Highest† | NS | 0.01* | 0.01* |
| Most common† | NS | NS | NS |
| Positive or negative≠ | NS | 0.01* | 0.04* |
| Anti-EGFR clone 3C6, intensity | |||
| Highest† | NS | NS | 0.04* |
| Most common† | NS | NS | NS |
| Positive or negative≠ | NS | NS | NS |
| Localisation | |||
| 5B7§ | NS | NS | NS |
| 3C6§ | NS | NS | NS |
| EGFR GCN (SISH) | |||
| Continuous variable | NS | — | <0.0001* |
| Cut-off 4.0 | NS | — | — |
| Chr-7 number (SISH) | |||
| Continuous variable | NS | <0.0001* | — |
| Cut-off 4.5 | NS | — | — |
Abbreviations: Chr-7=chromosome-7; EGFR=epidermal growth factor receptor; GCN=gene copy number; IHC=immunohistochemistry; NS=not significant; SISH=silver in situ hybridisation; *Significant P-value; †0, 1+, 2+, or 3+ ≠Positive 2+ or 3+, negative 0, or 1+ §Membranous, cytoplasmic, both cytoplasmic and membranous or negative.
EGFR SISH and treatment response
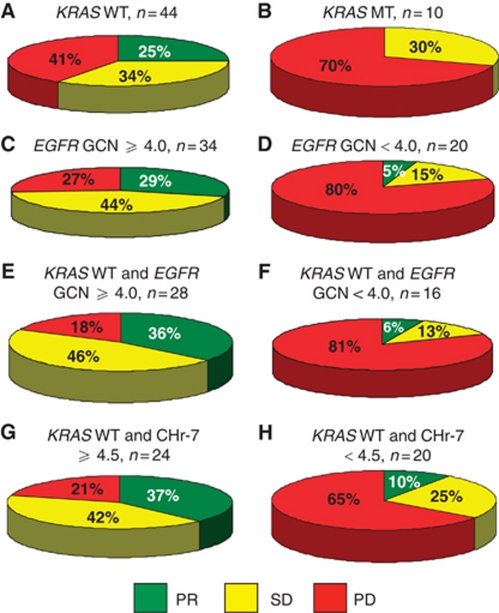
In all, 73% of high EGFR GCN (⩾4.0) patients showed clinical benefit (complete response (CR)+partial response (PR)+stable disease (SD)) from anti-EGFR therapy, whereas only 20% of low EGFR GCN (<4.0) benefited from treatment (Figure 2). In comparison, 59% of the KRAS WT patients showed clinical benefit. In KRAS WT patients with a high EGFR GCN (⩾4.0), clinical benefit was more frequent (82%) than in the overall KRAS WT or high EGFR GCN population. A high Chr-7 number (⩾4.5) was also significantly associated with an improved anti-EGFR treatment response among KRAS WT patients.
Figure 2.
Response to anti-EGFR therapy according to EGFR GCN, Chr-7 number, and KRAS status (A–H).
Anti-EGFR drugs were given as first-line treatment to five KRAS WT patients, four of which (80%) showed an objective response. Interestingly, all four patients had an EGFR GCN ⩾4.0. The fifth KRAS WT patient had an EGFR GCN <4.0 and progressed during therapy. We performed the statistical analyses separately by excluding the five KRAS WT patients who received anti-EGFR therapy as first-line treatment. Improved response rates were still seen in the group of KRAS WT patients with a high EGFR GCN (⩾4.0); an objective response was observed in 25% (6 out of 24), SD in 54% (13 out of 24) and PD in 21% (5 out of 24) of the patients. In the patients with a low EGFR GCN (<4.0), progressive disease was seen in 80% (12 out of 15) of the cases (Fisher's exact test, P=0.002).
In addition, the statistical analyses were performed separately for the KRAS WT chemorefractory CRC patients who received anti-EGFR therapy in ⩾third line, either as single drug therapy (n=3) or in combination with irinotecan (n=22). In all, 84% of the patients with a high EGFR GCN (⩾4.0) achieved either a SD or PR. In contrast, the clinical benefit rate was only 33% for the patients with a low EGFR GCN (<4.0) (Fisher's exact test, P=0.03). Stable disease was the best response recorded for 13 out of 25 patients in this selected patient group and of those 69% (9 out of 13) had a prolonged SD (⩾24 weeks). When excluding the patients with SD duration of <24 weeks from the analysis a significant association between treatment response and EGFR GCN status was still seen in a similar fashion (Fisher's exact test, P=0.02).
EGFR SISH and survival
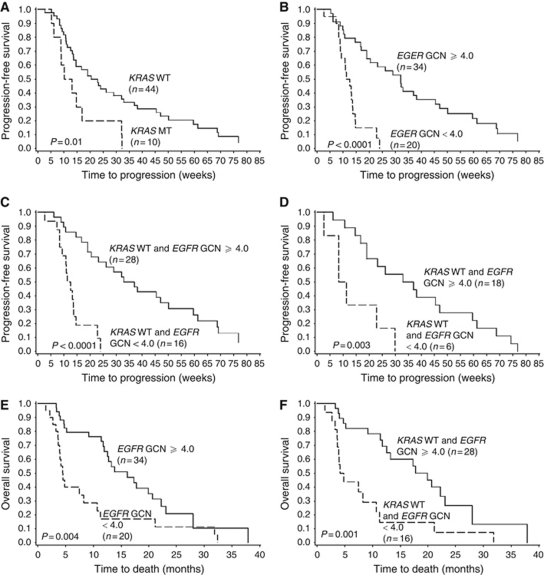
In the entire treated population, the EGFR GCN associated significantly with an improved PFS when using the ROC-curve based cut-off value of 4.0. Interestingly, the PFS time of the KRAS WT patients with EGFR GCN <4.0 was indifferent from those with KRAS mutation. The median PFS time of KRAS WT/EGFR GCN ⩾4.0 was 35 weeks compared with only 12 weeks of the KRAS WT/EGFR GCN <4.0 patients. The PFS remained significantly longer in the KRAS WT patient population with a high EGFR GCN when analysing only the patients treated with anti-EGFR therapy in second line or more (log-rank test, P<0.0001). Furthermore, in the cohort of chemorefractory patients treated either with single panitumumab or cetuximab±irinotecan in ⩾third line (n=25), the median PFS time was significantly longer in the KRAS WT/EGFR GCN ⩾4.0 patients than in the KRAS WT/EGFR GCN <4.0 patients; 35 vs 10 weeks (log-rank test, P=0.003; Cox test, P=0.007, HR: 0.22, 95% CI: 0.08–0.66). Similar results were obtained when excluding the patients with a short SD duration (<24 weeks) from the analysis (log-rank test, P=0.0008; Cox test, P=0.003, HR: 0.15, 95% CI: 0.04–0.53; PFS time 42 vs 8 weeks). Other factors associated with improved PFS in the entire group of anti-EGFR treated patients were tumour differentiation grade (log-rank test, P=0.001) and the absence of KRAS gene mutation (log-rank test, P=0.01).
The EGFR GCN ⩾4.0 associated significantly with improved OS (log-rank test, P=0.004) in the entire treated population and in the subgroup of KRAS WT patients (log-rank test, P=0.001). The Chr-7 number did not associate with OS. The median OS time for patients with KRAS WT/EGFR GCN ⩾4.0 tumours was 85 weeks compared with 19 weeks for those with KRAS WT/EGFR GCN below the cut-off value. When excluding the patients treated with anti-EGFR therapy in first line the OS was still significantly higher in the patients with an EGFR GCN ⩾4.0 (log-rank test, P=0.001). In the selected patient group treated with anti-EGFR antibodies±irinotecan in ⩾third line an EGFR GCN ⩾4.0 predicted a prolonged OS as well; 74 vs 16 weeks in the group of patients with a low EGFR GCN (log-rank test, P=0.0005; Cox test, P=0.003, HR: 0.13, 95% CI: 0.03–0.49). The results remained significant when the patients with a short SD duration (<24 weeks) were excluded from the analysis (log-rank test, P=0.0003; Cox test, P=0.004, HR: 0.08, 95% CI: 0.01–0.44; PFS time 89 vs 14 weeks).
The responses, survival times and P-values are summarised in Table 4, survival curves shown in Figure 3.
Table 4. Tumour response of patients with KRAS WT (n=54) and KRAS MT (n=10) metastatic or locally advanced colorectal cancer treated with anti-EGFR therapy according to ROC curve based cut-off values of EGFR GCN and chromosome 7 number evaluated by SISH.
|
Treatment response
|
PFS
|
OS
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total no. of patients | PR | SD | PD | P-value Fisher's exact test | PFS time median (days) | P-value log-rank test | P-value Cox testa | HR | 95% CI | OS time median (days) | P-value log-rank test | P-value Cox testa | HR | 95% CI | |
| KRAS WT and MT patients | 54 | ||||||||||||||
| KRAS status | |||||||||||||||
| KRAS WT | 44 | 11 (25) | 15 (34.1) | 18 (40.9) | NS | 151 | 0.01 | 0.01 | 0.40 | 0.20–0.84 | 352 | 0.3 | 0.3 | 0.67 | 0.32–1.38 |
| KRAS MT | 10 | 0 | 3 (30) | 7 (70) | 81 | 249 | |||||||||
| EGFR GCN status | |||||||||||||||
| EGFR GCN ⩾4.0 | 34 | 10 (29.4) | 15 (44.1) | 9 (26.5) | 0.0006 | 224 | <0.0001 | <0.0001 | 0.21 | 0.10–0.43 | 483 | 0.004 | 0.006 | 0.41 | 0.22–0.77 |
| EGFR GCN <4.0 | 20 | 1 (5) | 3 (15) | 16 (80) | 84 | 134 | |||||||||
| KRAS WT patients | |||||||||||||||
| EGFR GCN | |||||||||||||||
| ⩾4.0 | 28 | 10 (35.7) | 13 (46.4) | 5 (17.9) | 0.0002 | 244 | <0.0001 | <0.0001 | 0.17 | 0.07–0.39 | 598 | 0.001 | 0.002 | 0.32 | 0.16–0.66 |
| <4.0 | 16 | 1 (6.2) | 2 (12.5) | 13 (81.3) | 84 | 134 | |||||||||
| Chromosome 7 number | |||||||||||||||
| ⩾4.5 | 24 | 9 (37.5) | 10 (41.7) | 5 (20.8) | 0.009 | 214 | 0.2 | 0.2 | 0.67 | 0.35–1.29 | 520 | 0.1 | 0.1 | 0.56 | 0.28–1.13 |
| <4.5 | 20 | 2 (10) | 5 (25) | 13 (65) | 94 | 225 | |||||||||
| KRAS MT patients | |||||||||||||||
| EGFR GCN | |||||||||||||||
| ⩾4.0 | 6 | 0 | 2 (33.3) | 4 (66.7) | NS | 94 | NS | 369 | NS | ||||||
| <4.0 | 4 | 0 | 1 (25) | 3 (75) | 77 | 134 | |||||||||
| Chromosome 7 number | |||||||||||||||
| ⩾4.5 | 6 | 0 | 2 (33.3) | 4 (66.7) | NS | 94 | NS | 369 | NS | ||||||
| <4.5 | 4 | 0 | 1 (25) | 3 (75) | 77 | 134 | |||||||||
Abbreviations: CI=confidence interval; CR=complete response; EGFR=epidermal growth factor receptor; GCN=gene copy number; HR=hazards ratio; MT=mutated; NS=not significant; OS=overall survival; PD=progressive disease; PFS=progression-free survival; PR=partial response; ROC= receiver operating characteristic; SD=stable disease; SISH=silver in situ hybridisation; WT=wild type.
Cox proportional hazards regression model. Treatment response values are given n (%). Significant P-values are shown in bold type.
Figure 3.
Kaplan–Meier curves for PFS (A–D) and OS (E–F). Progression-free survival in anti-EGFR treated patients by (A) KRAS and (B) EGFR gene copy number (GCN). (C) Progression-free survival in KRAS WT patients (n=44) according to EGFR GCN. (D) Progression-free survival according to EGFR GCN in selected chemorefractory KRAS WT patients treated with anti-EGFR therapy±irinotecan in ⩾third line (n=25). (E) Overall survival by EGFR GCN. (F) Overall survival in KRAS WT patients according to EGFR GCN.
Multivariate survival analysis
Variables that in univariate survival analysis significantly associated with PFS and OS in the anti-EGFR treated patient group were included in the Cox's multivariate analysis. The multivariate analysis for PFS included EGFR GCN, tumour differentiation grade, and KRAS status. EGFR GCN (P=0.0003, HR: 0.22, 95% CI: 0.09–0.50), tumour differentiation grade (P=0.02, HR: 0.38, 95% CI: 0.16–0.88), and KRAS (P=0.04, HR: 0.44, 95% CI: 0.20–0.97) proved to be independent predictors of PFS. When the KRAS WT patients were analysed separately, only EGFR GCN (P=0.0003, HR: 0.16, 95% CI: 0.06–0.43) independently predicted PFS. EGFR GCN and tumour differentiation grade were entered for OS analysis. Both variables predicted OS: EGFR GCN (P=0.02, HR: 0.44, 95% CI: 0.22–0.86), tumour differentiation grade (P=0.046, HR: 0.43, 95% CI: 0.19–0.99). In the KRAS WT subgroup of patients, EGFR GCN remained as a statistically significant predictor of OS (P=0.01, HR: 0.35, 95% CI: 0.16–0.78).
Discussion
This study shows that EGFR GCN analysis, when performed from areas with highest EGFR expression, is a highly promising method for predicting the efficacy of anti-EGFR therapy in locally advanced or metastatic CRC. Together with KRAS analysis EGFR GCN identifies the responsive patients more accurately than either test alone. In all, 73% of patients with a high EGFR GCN (⩾4.0) responded to anti-EGFR therapy, whereas a clear majority (80%) of the patients with a low EGFR GCN were non-responders. In comparison, 41% of the KRAS WT patients did not respond to treatment.
Previous reports, in which chromogenic ISH (CISH) and FISH were used to evaluate the EGFR GCN and/or Chr-7, have provided evidence for the association of increased EGFR GCN and response to anti-EGFR treatment. However, the predictive value of our study seems to be better than those (Moroni et al, 2005; Lievre et al, 2006; Frattini et al, 2007; Sartore-Bianchi et al, 2007; Cappuzzo et al, 2008; Personeni et al, 2008). What could be the explanation for this difference? One potential factor may be the use of IHC to guide the selection of the area for in situ analysis. The EGFR expression showed marked intratumoural variation and therefore, IHC was used to indicate the strongest EGFR immunoreactivity for evaluation of the EGFR GCN and Chr-7 number by SISH. This protocol might also explain why the EGFR GCN values were higher in our study than in most other studies reported. Another possible explanation could be the usage of a different EGFR probe. However, as the FISH analyses of nine selected cases were in concordance with SISH results, this is an unlikely explanation.
Methodological difficulties as well as reproducibility concerns have until now prevented the usage of EGFR GCN as a predictive marker in the clinic. The fully automated SISH technique offers several advantages compared with manually performed FISH and CISH. Automation improves reproducibility and compared with FISH, SISH enables morphological identification of the analysed tissue, which facilitates the interpretation (Dietel et al, 2007).
Several studies have indicated that EGFR IHC does not predict the response to EGFR-targeted therapies (Cunningham et al, 2004; Saltz et al, 2004; Chung et al, 2005; Cappuzzo et al, 2008). In addition, the correlation between EGFR IHC and EGFR GCN has been poor (Shia et al, 2005; Spindler et al, 2006; Frattini et al, 2007). Here, EGFR IHC with intracellular domain 5B7 antibody showed a significant correlation with the EGFR GCN and Chr-7 number. Our results may be due to the properties of the antibodies used. The novel 5B7 antibody detects the functionally active intracellular domain of EGFR, whereas other commercially available antibodies bind to the external domain of the EGFR. However, also IHC scoring method may have a role, the highest intensity assessment providing the best correlation with EGFR GCN. Typically, a constant intensive membranous staining correlated with areas of EGFR amplification. Consequently, although IHC does not predict treatment response, it is important for guiding SISH analysis, that is, indicating tumour areas with highest degree of EGFR GCN.
Currently, patients with metastatic CRC are screened for KRAS status and only those with KRAS WT tumours receive anti-EGFR therapy. This selection is not absolute and about half of the patients with KRAS WT tumours will receive the anti-EGFR monoclonal antibodies unnecessarily. Although, BRAF, PIK3CA/PTEN, and NRAS alterations explain a fraction of unresponsiveness (Laurent-Puig et al, 2009; Bardelli and Siena, 2010; De Roock et al, 2010) the search for further predictive markers in this setting is feasible. Improved predictive testing would minimise the risk of exposing the patients to harmful side-effects caused by EGFR targeted therapies and at the same time reduce the healthcare costs.
Our results suggest that cetuximab and panitumumab should not be offered to KRAS WT patients with EGFR GCN <4.0. Furthermore, according to our results, the analysis of EGFR GCN by SISH could in certain cases be used as a substitute for KRAS analysis, for example, when only a small biopsy of the tumour has been taken and the amount of tumour tissue is insufficient for KRAS analysis.
Acknowledgments
We thank Sinikka Kollanus for preparing sections for immunohistochemistry, Tero Vahlberg for assistance with the statistical analyses, Jaakko Liippo for help with art work, Sakari Knuutila for KRAS analysis and Ventana Medical Systems, Inc. for reagents and valuable comments. This study was funded by the Finnish Medical Foundation, the Finnish Cancer Foundation, the National Graduate School of Clinical Investigations, and Turku University Hospital EVO grant.
References
- Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL (2009) American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 27: 2091–2096 [DOI] [PubMed] [Google Scholar]
- Bardelli A, Siena S (2010) Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol 28: 1254–1261 [DOI] [PubMed] [Google Scholar]
- Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, de Braud F, Donea S, Ludwig H, Schuch G, Stroh C, Loos AH, Zubel A, Koralewski P (2009) Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol 27: 663–671 [DOI] [PubMed] [Google Scholar]
- Cappuzzo F, Finocchiaro G, Rossi E, Janne PA, Carnaghi C, Calandri C, Bencardino K, Ligorio C, Ciardiello F, Pressiani T, Destro A, Roncalli M, Crino L, Franklin WA, Santoro A, Varella-Garcia M (2008) EGFR FISH assay predicts for response to cetuximab in chemotherapy refractory colorectal cancer patients. Ann Oncol 19: 717–723 [DOI] [PubMed] [Google Scholar]
- Chang DZ, Kumar V, Ma Y, Li K, Kopetz S (2009) Individualized therapies in colorectal cancer: KRAS as a marker for response to EGFR-targeted therapy. J Hematol Oncol 2: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KY, Shia J, Kemeny NE, Shah M, Schwartz GK, Tse A, Hamilton A, Pan D, Schrag D, Schwartz L, Klimstra DS, Fridman D, Kelsen DP, Saltz LB (2005) Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol 23: 1803–1810 [DOI] [PubMed] [Google Scholar]
- Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, Starling N (2010) Colorectal cancer. Lancet 375: 1030–1047 [DOI] [PubMed] [Google Scholar]
- Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, Chau I, Van Cutsem E (2004) Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med 351: 337–345 [DOI] [PubMed] [Google Scholar]
- De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, Penault-Llorca F, Rougier P, Vincenzi B, Santini D, Tonini G, Cappuzzo F, Frattini M, Molinari F, Saletti P, De Dosso S, Martini M, Bardelli A, Siena S, Sartore-Bianchi A, Tabernero J, Macarulla T, Di Fiore F, Gangloff AO, Ciardiello F, Pfeiffer P, Qvortrup C, Hansen TP, Van Cutsem E, Piessevaux H, Lambrechts D, Delorenzi M, Tejpar S (2010) Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 11: 753–762 [DOI] [PubMed] [Google Scholar]
- Dietel M, Ellis IO, Höfler H, Kreipe H, Moch H, Dankof A, Kölble K, Kristiansen G (2007) Comparison of automated silver enhanced in situ hybridisation (SISH) and fluorescence ISH (FISH) for the validation of HER2 gene status in breast carcinoma according to the guidelines of the American Society of Clinical Oncology and the College of American Pathologists. Virchows Arch 451: 19–25 [DOI] [PubMed] [Google Scholar]
- Douillard JY, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, Rivera F, Kocákova I, Ruff P, Błasińska-Morawiec M, S˘makal M, Canon JL, Rother M, Oliner KS, Wolf M, Gansert J (2010) Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) vs FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol 28: 4697–4705 [DOI] [PubMed] [Google Scholar]
- Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J (2009) New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 45: 228–247 [DOI] [PubMed] [Google Scholar]
- Frattini M, Saletti P, Romagnani E, Martin V, Molinari F, Ghisletta M, Camponovo A, Etienne LL, Cavalli F, Mazzucchelli L (2007) PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. Br J Cancer 97: 1139–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grothey A (2010) EGFR antibodies in colorectal cancer: where do they belong? J Clin Oncol 28: 4668–4670 [DOI] [PubMed] [Google Scholar]
- Hemmings C, Broomfield A, Bean E, Whitehead M, Yip D (2009) Immunohistochemical expression of EGFR in colorectal carcinoma correlates with high but not low level gene amplification, as demonstrated by CISH. Pathology 41: 356–360 [DOI] [PubMed] [Google Scholar]
- Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet JB, Lecomte T, Rougier P, Lievre A, Landi B, Boige V, Ducreux M, Ychou M, Bibeau F, Bouche O, Reid J, Stone S, Penault-Llorca F (2009) Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol 27: 5924–5930 [DOI] [PubMed] [Google Scholar]
- Lievre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, Ychou M, Bouche O, Landi B, Louvet C, Andre T, Bibeau F, Diebold MD, Rougier P, Ducreux M, Tomasic G, Emile JF, Penault-Llorca F, Laurent-Puig P (2008) KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol 26: 374–379 [DOI] [PubMed] [Google Scholar]
- Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Cote JF, Tomasic G, Penna C, Ducreux M, Rougier P, Penault-Llorca F, Laurent-Puig P (2006) KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res 66: 3992–3995 [DOI] [PubMed] [Google Scholar]
- Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, Kosmidis P, Papadimitriou CA, Murray S (2008) Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol 9: 962–972 [DOI] [PubMed] [Google Scholar]
- Martin V, Mazzucchelli L, Frattini M (2009) An overview of the epidermal growth factor receptor fluorescence in situ hybridisation challenge in tumour pathology. J Clin Pathol 62: 314–324 [DOI] [PubMed] [Google Scholar]
- Moroni M, Veronese S, Benvenuti S, Marrapese G, Sartore-Bianchi A, Di Nicolantonio F, Gambacorta M, Siena S, Bardelli A (2005) Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol 6: 279–286 [DOI] [PubMed] [Google Scholar]
- Peeters M, Price TJ, Cervantes A, Sobrero AF, Ducreux M, Hotko Y, André T, Chan E, Lordick F, Punt CJ, Strickland AH, Wilson G, Ciuleanu TE, Roman L, Van Cutsem E, Tzekova V, Collins S, Oliner KS, Rong A, Gansert J (2010) Randomized phase III study of panitumumab with fluorouracil, leucovorin, and irinotecan (FOLFIRI) compared with FOLFIRI alone as second-line treatment in patients with metastatic colorectal cancer. J Clin Oncol 28: 4706–4713 [DOI] [PubMed] [Google Scholar]
- Personeni N, Fieuws S, Piessevaux H, De Hertogh G, De Schutter J, Biesmans B, De Roock W, Capoen A, Debiec-Rychter M, Van Laethem JL, Peeters M, Humblet Y, Van Cutsem E, Tejpar S (2008) Clinical usefulness of EGFR gene copy number as a predictive marker in colorectal cancer patients treated with cetuximab: a fluorescent in situ hybridization study. Clin Cancer Res 14: 5869–5876 [DOI] [PubMed] [Google Scholar]
- Saltz LB, Meropol NJ, Loehrer Sr PJ, Needle MN, Kopit J, Mayer RJ (2004) Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol 22: 1201–1208 [DOI] [PubMed] [Google Scholar]
- Sartore-Bianchi A, Moroni M, Veronese S, Carnaghi C, Bajetta E, Luppi G, Sobrero A, Barone C, Cascinu S, Colucci G, Cortesi E, Nichelatti M, Gambacorta M, Siena S (2007) Epidermal growth factor receptor gene copy number and clinical outcome of metastatic colorectal cancer treated with panitumumab. J Clin Oncol 25: 3238–3245 [DOI] [PubMed] [Google Scholar]
- Scartozzi M, Bearzi I, Mandolesi A, Pierantoni C, Loupakis F, Zaniboni A, Negri F, Quadri A, Zorzi F, Galizia E, Berardi R, Biscotti T, Labianca R, Masi G, Falcone A, Cascinu S (2009) Epidermal growth factor receptor (EGFR) gene copy number (GCN) correlates with clinical activity of irinotecan-cetuximab in K-RAS wild-type colorectal cancer: a fluorescence in situ (FISH) and chromogenic in situ hybridization (CISH) analysis. BMC Cancer 9: 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shia J, Klimstra DS, Li AR, Qin J, Saltz L, Teruya-Feldstein J, Akram M, Chung KY, Yao D, Paty PB, Gerald W, Chen B (2005) Epidermal growth factor receptor expression and gene amplification in colorectal carcinoma: an immunohistochemical and chromogenic in situ hybridization study. Mod Pathol 18: 1350–1356 [DOI] [PubMed] [Google Scholar]
- Siena S, Sartore-Bianchi A, Di Nicolantonio F, Balfour J, Bardelli A (2009) Biomarkers predicting clinical outcome of epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer. J Natl Cancer Inst 101: 1308–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindler KL, Lindebjerg J, Nielsen JN, Olsen DA, Bisgard C, Brandslund I, Jakobsen A (2006) Epidermal growth factor receptor analyses in colorectal cancer: a comparison of methods. Int J Oncol 29: 1159–1165 [PubMed] [Google Scholar]
- Van Cutsem E, Kohne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D’Haens G, Pinter T, Lim R, Bodoky G, Roh JK, Folprecht G, Ruff P, Stroh C, Tejpar S, Schlichting M, Nippgen J, Rougier P (2009) Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med 360: 1408–1417 [DOI] [PubMed] [Google Scholar]