Abstract
Background:
The aim of this study is to determine whether immunohistochemical (IHC) assessment of Ki67 and p53 improves prognostication of oestrogen receptor-positive (ER+) breast cancer after breast-conserving therapy (BCT). In all, 498 patients with invasive breast cancer from a randomised trial of BCT with or without tumour bed radiation boost were assessed using IHC.
Methods:
The ER+ tumours were classified as ‘luminal A’ (LA): ER+ and/or PR+, Ki-67 low, p53−, HER2− or ‘luminal B’ (LB): ER+ and/or PR+and/or Ki-67 high and/or p53+ and/or HER2+. Kaplan–Meier and Cox proportional hazards methodology were used to ascertain relationships to ispilateral breast tumour recurrence (IBTR), locoregional recurrence (LRR), distant metastasis-free survival (DMFS) and breast cancer-specific survival (BCSS).
Results:
In all, 73 patients previously LA were re-classified as LB: a greater than four-fold increase (4.6–19.3%) compared with ER, PR, HER2 alone. In multivariate analysis, the LB signature independently predicted LRR (hazard ratio (HR) 3.612, 95% CI 1.555–8.340, P=0.003), DMFS (HR 3.023, 95% CI 1.501–6.087, P=0.002) and BCSS (HR 3.617, 95% CI 1.629–8.031, P=0.002) but not IBTR.
Conclusion:
The prognostic evaluation of ER+ breast cancer is improved using a marker panel, which includes Ki-67 and p53. This may help better define a group of poor prognosis ER+ patients with a greater probability of failure with endocrine therapy.
Keywords: breast cancer, biomarker, Ki67, p53, luminal B
Oestrogen receptor-positive (ER+) breast cancer comprises approximately 75% of all breast cancers and treatments targeting oestrogen synthesis (aromatase inhibitors) or the ER (tamoxifen) are the most effective adjuvant therapies. Gene expression profiling (GEP) studies over the past decade have established molecular subtypes of ER+ luminal disease, which are characterised by differences in outcome and underlying biology, largely now referred to as luminal A (LA) or luminal B (LB), the latter being characterised by increased proliferation and higher grade as well as lower levels of ER related genes (Perou et al, 2000; Sørlie et al, 2001). Despite the successes of endocrine therapy in reducing annual recurrences and death by 41% and 34%, respectively, resistance occurs in about 30% of patients treated with tamoxifen (Early Breast Cancer Trialists’ Collaborative Group (EBCTCG), 2005). Therefore, predicting the likely prognosis in an individual patient before treatment would allow early selection of optimal therapies, the importance of which is highlighted in the most recent St Gallen guidelines for the treatment of early breast cancer (Goldhirsch et al, 2009).
The abundant data derived from GEP studies have clearly identified the significance of genomic grade and proliferation signatures in prognosis and response to endocrine therapy (reviewed in detail in Musgrove and Sutherland, 2009 and Sotiriou and Pusztai, 2009). However, given the current costs of such molecular testing, translating these findings into an economical, reproducible and readily applicable panel for immunohistochemistry (IHC) in a routine pathology setting is a priority. Most previous IHC definitions of LA and LB tumours include ER+ and/or PR+, with HER2 positivity defining LB, creating a population size of approximately 5–10% (Cheang et al, 2008; Nguyen et al, 2008; Millar et al, 2009b; Blows et al, 2010). However, GEP studies have documented the LB population to be larger than this, averaging approximately 16% (ranging from 10 to 21%, reviewed in detail in Sorlie et al, 2003 and Hu et al, 2006), suggesting that this poorer prognosis subtype may be under-represented using this definition. This discrepancy is most likely explained by the fact that only approximately 30% of LB cancers are in fact HER2 positive (Carey et al, 2006). Although proliferation is the key discriminator of luminal tumours, the optimal subclassification of luminal tumours by GEP has yet to be defined (Weigelt et al, 2010b). Several studies have, however, shown that intrinsic subtype as defined by IHC ‘mirrors’ the subtypes identified by GEP and that the IHC subtypes so defined have distinct clinical outcomes (Neilsen et al, 2004; Abd El-Rehim et al, 2005; Cheang et al, 2008, 2009; Blows et al, 2010). Such IHC definitions are now in common clinical usage. Some recent studies have addressed the issue of a more refined definition of good and poor prognosis ER+ cancer, and used a modified IHC definition to include assessment of the proliferation marker Ki-67 (Cheang et al, 2009; Cuzick et al, 2009; Hugh et al, 2009), which results in a larger proportion of LB tumours with independent prognostic power (Cheang et al, 2009). This latter study defined a Ki67 cutpoint (14%) derived from GEP analyses. This set of biomarkers more closely resembles the Oncotype Dx assay of known predictive and prognostic power in ER+, lymph node-negative cancer, which is largely driven by proliferation, HER2- and ER-related genes (Paik et al, 2004). However, a recent head to head comparison of a four IHC biomarker panel of ER, PR, HER2 and Ki-67 (IHC 4) has been shown to provide prognostic information, which is at least equivalent to Oncotype Dx using material from the ATAC trial (Cuzick et al, 2009). This important study identifies the robustness of prognostic data, which can be provided by routine IHC. Some observers support the view that GEP currently offers no more that routine IHC when combined with important morphological features (not assessable by GEP), such as lymphatic vascular invasion and lymph node status (Weigelt and Reis-Filho, 2010). In addition, these routine analyses can be performed at a fraction of the cost of commercially available GEP tests. In addition, it also supports the concept that measurement of a few well chosen protein products can identify clinically significant patient groups (Ring et al, 2006). Histological grade is a key component of routine pathology reporting and of prognostic importance, but may, in some circumstances, be affected by subjectivity, along with problems with inadequate or delayed fixation, which can result in undergrading (Rakha et al, 2010). Incorporation of biomarkers as surrogates for molecular grade into routine reporting may help more reliably define good and poor prognosis patients, most significantly for grade 2 invasive carcinomas, which comprise 37–49% of all breast cancers (Rakha et al, 2010).
To further validate an IHC panel of markers for routine application in a clinical setting, we assessed a new biomarker panel to differentiate good prognosis (LA) and poor prognosis (LB) tumours in a cohort of predominantly ER+ early breast cancer patients enrolled in a randomised clinical trial of conservative surgery, post-operative whole breast radiotherapy and then randomised to an additional cavity boost or not. We previously described the clinical usefulness of a five biomarker panel (Millar et al, 2009b; ER, PR, HER2, CK 5/6 and EGFR) and have further defined luminal tumours by including Ki-67 and p53 status, the latter described in higher grade tumours, overexpressed more frequently within LB (Sorlie, 2004; Jacquemier et al, 2008; Hugh et al, 2009; Carey, 2010; Weigelt et al, 2010b) and as a predictor of endocrine resistance in some studies (Yamashita et al, 2006). These markers have easily available and well-characterised antibodies in current use, which can be immediately applied to clinical practise.
This study aimed to define the predictive value of a more refined luminal IHC biomarker signature in those patients who were ER+, with disease relapse and death from breast cancer as end-points.
Materials and methods
Study subjects
Training cohort
Cases were drawn from the St Vincent's Campus Outcome Cohort, which comprised 292 invasive ductal carcinomas treated between February 1992 and August 2002 at St Vincent's Hospital, Sydney, Australia. Ethics approval for use of tissue and clinicopathological data was granted by the Human Research Ethics Committee of St Vincent's Hospital, Sydney (Ref. SVH H94/080 and 00/036). A more detailed description of the clinicopathological characteristics of the cohort is published elsewhere (Millar et al, 2009a; López-Knowles et al, 2010). In summary, 40% of tumours were >20 mm, 45% were grade 3, 43% were lymph node positive, 68% were ER positive, 57% were PR positive and 18% were HER2 fluorescent in situ hybridisation (FISH) positive (HER2:CEP17 ratio >2.2). Median age was 54 years, and patients were treated with endocrine therapy (49%), chemotherapy (38%) or both (24%). Cases were prospectively followed up for a median of 64 months, and the outcome events measured were as follows: recurrence (local or distant; 25%), metastasis (23%) and breast cancer-specific death (18%). This cohort was used to identify differences in expression of several cell cycle and apoptotic markers, including Ki67 and p53 (CM McNeil et al, manuscript in preparation), between LA and B cancers using the following definitions: LA: ER+ and/or PR+ and HER2− and LB: ER+ and/or PR+ and HER2+. Using the median expression levels for Ki67 and p53 as the cutpoints (5% and 10%, respectively), we were able to demonstrate a significant difference in level of expression between LA and LB for these antigens (P=0.0158 and P=0.0061, respectively). Subsequently, we modified our definition of LA and LB to include Ki67 and p53 status as follows: ‘LA’: ER+ and/or PR+ and HER2−, Ki67 low and p53 negative and ‘LB’: ER+ and/or PR+ and/or HER2+ and/or Ki67 high and/or p53+. Kaplan–Meier analysis for breast cancer specific death showed a significant difference in outcome between these two groups of ER+ patients (P=0.0002) using this updated classifier (CM McNeil et al, manuscript in preparation).
Study validation cohort
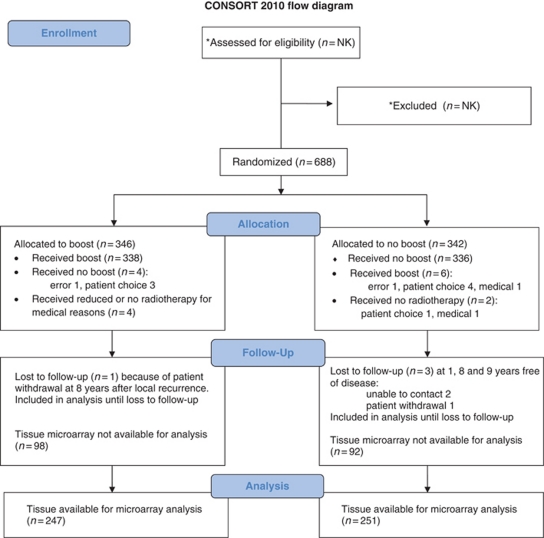
In this biomarker study, tissue was available from 498 patients (from a total of 688) with invasive breast cancer who were enrolled into a randomised clinical trial, which compared the benefit of the addition of a local cavity boost of radiotherapy to breast-conserving therapy (BCT; Clinical Trials Registry NCT00138814). The study was conducted at St George, Wollongong and Liverpool Hospitals, Sydney, New South Wales, Australia between 1996 and 2003 when the trial was closed to accrual. Follow-up for this analysis continued until September 2008. Clinicopathological details are summarised in Supplementary Table 1, and have been previously published in detail Millar et al (2009b). This study was approved by the Human Research Ethics Committee of the St George Hospital, Sydney, Australia (ref. no.: 96/84). The flow of patients through the trial is summarised in a CONSORT flow diagram (Figure 1). Patients were randomised using random blocking sequences set up before commencing of the study. Following patient consent, a person independent of the study both generated the sequence and assigned participants to interventions as below. This was an unblinded study.
Figure 1.
*The trial recruited from three main centres (St George, Wollongong and Liverpool Hospitals). Although the total number of patients assessed for eligibility and excluded for all centres is not known, this data are available for the main recruiting centres at St Geroge Hospital, which contributed the majority of patients in the trial, n=546 (number assessed, n=2046; excluded, n=1500: not meeting criteria, n=943; declined to partcipate, n=235; other reasons, n=322; patients randomised in the trials, n=536).
All patients with invasive carcinoma received local excision and axillary sentinel node biopsy or axillary clearance. Adjuvant chemotherapy (AC or CMF) was given to 23.7% of patients and 44.9% received adjuvant endocrine therapy with tamoxifen. No patients received adjuvant trastuzumab. For patients subsequently classified as modified ‘LA’, 49.5% received endocrine therapy and 13.4% received chemotherapy, and those classified as modified ‘LB’ 55.7% received endocrine therapy and 25% received chemotherapy. Patients were randomised to whole breast radiotherapy of 50 Gy in 25 fractions or whole breast radiotherapy of 45 Gy in 25 fractions plus a tumour bed boost of 16 Gy in eight fractions. Supraclavicular fields were not added unless there were four or more nodes positive. In all, 17 patients had positive margins, 65 had clearance of <1 mm and a further 86 had <2 mm clearance, the remainder being well clear. HER2 status was unknown at the time of treatment.
Study definitions
Patients were assessed at 6 weeks after radiation therapy, 6 monthly for 2 years, then annually thereafter with annual breast imaging. Follow-up time for this biomarker cohort was calculated from the date of the first surgical procedure to the date of the first event, as outlined below, or to the last known confirmed date of breast cancer disease-free status. Median follow-up time was 84 months (range 1–134 months). The primary end point was time to ipsilateral breast tumour recurrence (IBTR). This included any ipsilateral in-breast recurrence (invasive or non-invasive). The secondary end points were locoregional recurrence (LRR: IBTR, axilla, chest wall, internal mammary or supraclavicular fossa lymph nodes) and time to distant metastases and death.
Tissue microarray (TMA) construction, IHC and FISH
TMAs were constructed from formalin-fixed paraffin-embedded tissue blocks, which were available from 498 invasive carcinomas, using 1 mm diameter punches with up to three cores sampled from each tumour. Antibodies used in IHC were Ki-67 (1 : 100, SP6 neomarkers), p53 (1:50, DO-7; Dako, Carpentaria, CA, USA), ER (1:100, 6F11; Dako), PR (1:200, PgR 636; Dako), CK 5/6 (1:80, MAB1602; Chemicon International, Temecula, USA), EGFR (1:100, H11; Dako).
All staining was performed using a Dako autostainer following antigen retrieval for all antibodies except for Ki-67, which was performed on a Leica (Wetzlar, Germany)/Bond Max system using ER2 (high pH antigen retrieval). All staining was centrally assessed by one breast Pathologist (EKAM). ER and PR were assessed as positive if a modified ‘H score’ (i.e., percentage × intensity) was >10. CK5/6 and EGFR were considered positive if staining of any intensity was present (i.e., >0). Tumours were considered HER2 positive only if they were HER2 amplified on FISH using a HER2: chromosome 17 ratio >2.2 as positive. p53 and Ki-67 were considered positive if there was >10% positive average nuclear staining of any intensity.
Classification of intrinsic molecular phenotype
Patients were initially subtyped based on the status of their primary tumour as follows: ‘LA’: ER+ and/or PR+ and HER2−, and ‘LB’: ER+ and/or PR+ and HER2+ HER2 enriched: ER− and PR− and HER2+, and basal: ER−, PR−, HER2−, CK 5/6+ and/or EGFR +, unclassified (negative for all five markers). Subsequently they were re-classified as modified ‘LA’: ER+ and/or PR+ and Ki-67 low, p53−, HER2− modified ‘LB’: ER+ and/or PR+ and/or Ki-67 high and/or p53+ and/or HER2+ HER2 enriched: ER− and PR− and HER2+, and basal: ER−, PR−, HER2−, CK 5/6+ and/or EGFR+, unclassified (negative for all five markers).
Statistical analyses
Kaplan–Meier analyses for IBTR, LRR, distant disease-free survival and breast cancer-specific death were estimated for each subtype and compared using the log-rank test. We used Cox proportional hazards univariate analysis to analyse the association between prognostic variables and molecular subtype with IBTR, LRR, metastases and breast cancer-specific death. Multivariate analysis (MVA) was used to construct models identifying those variables which were independently prognostic. Subsequently, step-wise removal of variables was used until resolution. Analyses were performed using Statview 5.0 (Abacus systems, Berkeley, CA, USA) and STATA 10.0 (StataCorp LP, College Station, TX, USA). The ANOVA was used to assess differences in expression of target antigens as continuous variables between intrinsic subtypes.
Results
Assessment of Ki67 and p53 expression between LA and B tumours
Having identified differences in Ki67 and p53 in ER+ tumours in our training cohort, we then assessed the difference between LA and B tumours in expression level of these two antigens in our validation cohort (n=498). Within LB tumours in this cohort, we observed significantly higher levels of Ki-67 and p53 expression (P=0.0008 and 0.0048, respectively). The median average value for both Ki67 and p53 within the validation cohort was 10%. Subsequently, we modified our working definition further for good prognosis modified ‘LA’ as ER+ and/or PR+ and HER2−, Ki67 low and p53− and poor prognosis modified ‘LB’ as ER+ and/or PR+ and/or HER2+ and/or Ki67 high and/or p53+.
Five-year survival rates, univariate analysis of LA and B tumours for IBTR, LRR, distant metastasis-free survival (DMFS) and breast cancer-specific survival (BCSS)
Using these updated definitions, 321 tumours (64.5%) were classified as LA and 96 as LB (19.3%). Thus, 73 previously LA tumours were re-classified as LB (previously only 23 tumours were LB), that is, 4.2-fold increase (4.6–19.3%) with LB now comprising 23% of all ER+ tumours. We then examined the relative contribution of p53 and Ki67 to the updated classification of the 96 LB tumours: 57 of 96 (59.4%) were p53−/Ki67+, 19 (19.7%) were p53+/Ki67−, 12 (12.5%) were p53+/Ki67+ and 8 were p53−/Ki67− (HER2+).
As previously described, no benefit of a tumour bed boost was observed in this group of patients (Millar et al, 2009b). At a median follow-up period of 84 months, the 5-year survival rates for modified LA and modified LB, respectively, using the updated classifier were IBTR 99.3, 96.6% LRR 99.7, 93.4% DMFS 97, 87% and BCSS 99.7, 92.5%. Comparative analyses of the clinicopathological features, crude event rates and univariate analyses of LA and LB groups between the differing definitions are presented in Tables 1 and 2. Univariate Cox proportional hazards were calculated for each measure of outcome for Ki67 and p53 and the modified LA and LB subtypes, which are presented with crude event rates in Table 3. Further crude event rates for modified LA and LB for lymph node negative, lymph node positive and lymphatic vascular invasion are presented in Supplementary Table 2. As expected, the updated classification resulted in increased numbers of events for all outcomes for LB and a reduction for LA. This is mirrored in LB by increases in LVI and LN+ status, with recurrence rates and death rates two to three times that of LA. Univariate analyses showed that modified LA is a significant predictor for all measures of outcome including IBTR (hazard ratio (HR) 0.314, 95% CI 0.136–0.726, P=0.007) where it previously was close to but not statistically significant (P=0.051). Modified LB predicted DMFS and BCSS (P=0.005 and 0.003, respectively) and approached significance for IBTR and LRR (P=0.07 and 0.052, respectively) where previously it was not significant for any outcome measure. Ki67 predicted outcome for all measures (BCSS: HR 4.98, 95% CI 2.530–9.694, P<0.0001). p53+ predicted DMFS and BCSS (HR 3.523, 95% CI 1.731–7.168, P=0.0005) but not IBTR or LRR.
Table 1. Patient tumour characteristics and event rates classified by luminal phenotype.
| Whole cohort, n=498 (%) | Luminal A, n=394 (79.1%) | Luminal B, n=23 (4.6%) | Modified luminal A, n=321(64.5%) | Modified luminal B, n=96 (19.3%) | |
|---|---|---|---|---|---|
| Patient tumour characteristics | |||||
| Size <20 mm | 357 (70.3) | 289 (73.4) | 17 (73.9) | 242 (75.4) | 64 (66.7) |
| LVI+ | 79 (15.8) | 62 (15.7) | 4 (17.4) | 43 (13.4) | 23 (23.9) |
| LN+ | 146 (29.0) | 117 (29.6) | 5 (21.7) | 86 (26.7) | 36 (37.5) |
| Grade 3 | 145 (29.1) | 65 (16.5) | 16 (69.5) | 26 (8.1) | 55 (57.3) |
| EIC+ | 45 (9.0) | 29 (7.4) | 5 (21.7) | 23 (7.2) | 11 (11.5) |
| Median Age | 61 | 62 | 57 | 62 | 61 |
| Events | |||||
| Median follow-up | 84 | 83.5 | 71 | 84 | 78 |
| IBTR | 24 (4.8) | 15 (3.8) | 2 (8.7) | 9 (2.8) | 8 (8.3) |
| LRR | 35 (7.0) | 20 (5.1) | 2 (8.7) | 11 (3.4) | 11 (11.5) |
| DMFS | 47 (9.4) | 30 (7.6) | 2 (8.7) | 16 (4.9) | 16 (16.7) |
| BCSS | 37 (7.4) | 23 (5.8) | 2 (8.7) | 11 (3.4) | 14 (14.6) |
Abbreviations: BCSS=breast cancer-specific survival; DMFS=distant metastasis-free survival; EIC=extensive intraduct carcinoma; ER+ =oestrogen receptor positive; IBTR=ipsilateral breast tumour recurrence; LN=lymph node; LRR=locoregional recurrence; LVI=lymphatic/vascular invasion; PR+ =progesterone receptor positive.
Luminal A: ER+ and/or PR+, HER2− Luminal B: ER+ and/or PR+, HER2+ modified luminal A: ER+ and/or PR+, Ki67 low and p53− and HER2− modified luminal B: ER+ and/or PR+ and/or Ki67 high and/or p53+ and/or HER2+.
Table 2. Comparative 5 and 10 year event rates for luminal A and B.
|
IBTR
|
LRR
|
DM
|
BCSD
|
|||||
|---|---|---|---|---|---|---|---|---|
| 5 Year (%) | 10 Year (%) | 5 Year (%) | 10 Year (%) | 5 Year (%) | 10 Year (%) | 5 Year (%) | 10 year (%) | |
| Whole cohort (n=498) | 12/498 (2.4) | 23/498 (4.6) | 21/498 (4.2) | 35/498 (6.8) | 34/498 (6.8) | 47/498 (9.4) | 18/498 (3.6) | 37/498 (7.4) |
| 12/24 (50) | 23/24 (95.8) | 21/35 (60) | 35/35 (100) | 34/47 (72.3) | 47/47 (100) | 18/37 (48.6) | 37/37 (100) | |
| Luminal A (n=394) | 4/394 (1) | 14/394 (3.6) | 8/394 (2) | 19/394 (4.8) | 19/394 (4.8) | 30/394 (7.6) | 7/394 (1.8) | 23/394 (5.8) |
| 4/15 (26.6) | 14/15 (93.3) | 8/20 (40) | 19/20 (95) | 19/30 (63.3) | 30/30 (100) | 7/23 (30.4) | 23/23 (100) | |
| Luminal B (n=23) | 1/23 (4.3) | 2/23 (8.7) | 1/23 (4.3) | 2/23 (8.6) | 2/23 (8.6) | 2/23 (8.6) | 1/23 (4.3) | 2/23 (8.6) |
| 1/2 (50) | 2/2 (100) | 1/2 (50) | 2/2 (100) | 2/2 (100) | 2/2 (100) | 1/2 (50) | 2/2 (100) | |
| Modified luminal A (n=321) | 2/321 (0.6) | 9/321 (2.8) | 3/321 (0.9) | 11/321 (3.4) | 9/321 (2.8) | 16/321 (4.9) | 1/321 (0.3) | 11/321 (3.4) |
| 2/9 (22.2) | 9/9 (100) | 3/11 (27) | 11/11 (100) | 9/16 (56.3) | 16/16 (100) | 1/11 (9.1) | 11/11 (100) | |
| Modified luminal B (n=96) | 3/96 (3.1) | 7/96 (7.3) | 6/96 (6.3) | 10/91 (10.9) | 12/96 (12.5) | 16/96 (16.7) | 7/96 (7.3) | 14/96 (14.6) |
| 3/8 (37.5) | 7/8 (87.5) | 6/11 (54.5) | 10/11 (90.1) | 12/16 (75) | 16/16 (100) | 7/14 (50) | 14/14 (100) | |
Abbreviations: BCSD=breast cancer-specific death; DM=distant metastasis; ER+ =oestrogen receptor positive; IBTR=ipsilateral breast tumour recurrence; LRR=locoregional recurrence; PR+ =progesterone receptor positive.
Modified luminal A: ER+ and/or PR+, Ki-67 low, p53−, HER2− modified luminal B: ER+ and/or PR+ and/or Ki-67 high and/or p53+ and/or HER2+. In the top row of each box, the denominator is the total number of patients within that patient group or subtype; in the bottom row of each box, the denominator is the total number of events for each group or subtype.
Table 3. Univariate crude rates and hazard ratio (Cox) for biomarkers and luminal phenotype.
|
IBTR (n=24)
|
LRR (n=35)
|
DDFS (n=47)
|
BCSS (n=37)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CR | HR (95% CI) | P | CR | HR (95% CI) | P | CR | HR (95% CI) | P | CR | HR (95% CI) | P | |
| Ki67 high | 12/129 | 3.126 (1.390–7.029) | 0.0008 | 19/129 | 3.759 (1.923–7.340) | 0.0001 | 24/129 | 3.436 (1.926–6.130) | <0.0001 | 22/129 | 4.948 (2.530–9.674) | <0.0001 |
| p53+ | 3/57 | 1.067 (0.315–3.629) | 0.916 | 5/57 | 1.290 (0.497–3.350) | 0.601 | 11/57 | 2.566 (1.303–5.056) | 0.006 | 11/57 | 3.523 (1.731–7.168) | 0.0005 |
| LA | 15/394 | 0.433 (0.186–1.005) | 0.051 | 20/394 | 0.333 (0.169–0.655) | 0.002 | 30/394 | 0.446 (0.246–0.810) | 0.008 | 23/394 | 0.414 (0.213–0.816) | 0.009 |
| LB | 2/23 | 2.132 (0.500–9.098) | 0.307 | 2/23 | 1.365 (0.327–5.697) | 0.669 | 2/23 | 0.963 (0.233–3.971) | 0.958 | 2/23 | 1.258 (0.302–5.234) | 0.753 |
| Modified LA | 9/321 | 0.314 (0.136–0.726) | 0.007 | 11/321 | 0.233 (0.113–0.478) | <0.0001 | 16/321 | 0.263 (0.144–0.481) | 0.0001 | 11/321 | 0.218 (0.108–0.441) | <0.0001 |
| Modified LB | 8/96 | 2.217 (0.0.945–5.200) | 0.07 | 11/96 | 2.036 (0.995–4.167) | 0.052 | 16/96 | 2.351 (1.285–4.300) | 0.005 | 14/96 | 2.733 (1.406–5.314) | 0.003 |
Abbreviations: CI=confidence interval; CR=crude rate; DMFS=distant metastasis-free survival; ER+ =oestrogen receptor positive; HR=hazard ratio; IBTR=ipsilateral breast tumour recurrence; LA=luminal A; LB=luminal B; LRR=locoregional recurrence; PR+ =progesterone receptor positive.
LA: ER+ and/or PR+ and HER2− LB: B ER+ and/or PR+ and HER2+ modified LA: ER+ and/or PR+, Ki-67 low, p53−, HER2− modified LB: ER+ and/or PR+ and/or Ki-67 high and/or p53+ and/or HER2+.
Bold typescript indicates statistical significance.
Kaplan–Meier analysis of intrinsic subtype
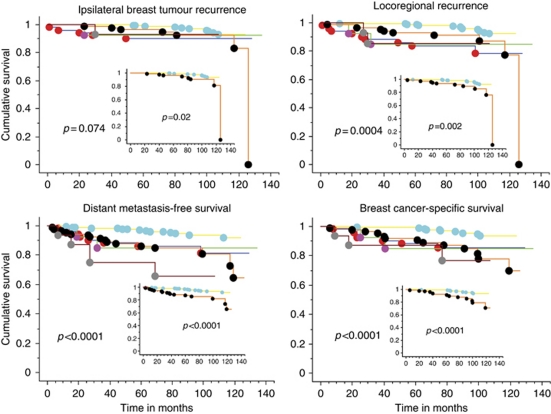
Kaplan–Meier analysis (log-rank test) comparing modified LA and LB alone was significant for all measures of outcome IBTR P=0.02, LRR P=0.002, DMFS and BCSS both P<0.0001 (Figure 2 inserts). This classifier also showed improvement in the degree of statistical significance between all molecular subtypes compared with the previously reported five biomarker panel, which was observed for LRR P=0.0004 (previously 0.012), DMFS P<0.0001 (previously 0.0035) and BCSS P=0.0001 (previously 0.048) but not for IBTR (P=0.074, previously 0.346, Figure 2). Although LA had an excellent prognosis, LB had adverse survival, similar to basal, HER2-enriched and unclassified subtypes.
Figure 2.
Kaplan–Meier estimates for ipsilateral breast tumour recurrence, locoregional recurrence, distant metastasis-free survival and breast cancer-specific survival for all intrinsic subtypes and for luminal A vs luminal B (inserts). Luminal A  n=321, luminal B
n=321, luminal B  n=96, basal
n=96, basal  n=52, HER2 enriched
n=52, HER2 enriched  n=13, unclassified
n=13, unclassified  n=16.
n=16.
MVA for IBTR, LRR, DMFS and BCSS
We then constructed multivariable models of clinicopathological features and intrinsic subtype to assess predictive value and compare HRs between intrinsic subtypes, using modified LA as a reference group.
Ispilateral breast tumour recurrence
Only margin status (HR 3.158, 95% CI 1.067–9.348, P=0.378) and grade (HR 3.13, 95% CI 1.4–7.012, P=0.0055) independently predicted recurrence with no other prognostic variable or intrinsic subtype reaching statistical significance.
Locoregional recurrence
Luminal B (HR 3.612, 95% CI 1.555–8.340, P=0.003), basal, unclassified and extensive intraduct carcinoma positive were independent predictors of outcome in the final resolved model (Table 4).
Table 4. Cox proportional hazards multivariate models.
| Variable | HR | 95% CI | P |
|---|---|---|---|
| Locoregional recurrence | |||
| Grade 3 | 1.938 | 0.823–4.568 | 0.130 |
| Size>20 mm | 0.861 | 0.408–1.817 | 0.694 |
| LN+ | 2.188 | 1.054–4.542 | 0.036 |
| LVI | 1.286 | 0.546–3.026 | 0.564 |
| EIC+ | 3.136 | 1.328–7.405 | 0.009 |
| Subtype | |||
| Modified LA (reference) | 1.0 | ||
| Modified LB | 2.483 | 0.982–6.281 | 0.055 |
| Basal | 3.939 | 1.281–12.114 | 0.017 |
| HER2 | 1.931 | 0.382–9.754 | 0.426 |
| Unclassified | 4.471 | 0.926–21.59 | 0.062 |
| Resolved model | |||
| EIC+ | 2.476 | 1.070–5.730 | 0.034 |
| Modified LB | 3.612 | 1.555–8.340 | 0.003 |
| Basal | 5.541 | 2.279–13.47 | <0.001 |
| HER2 | 3.549 | 0.764–16.51 | 0.106 |
| Unclassified | 4.913 | 1.077–22.42 | 0.040 |
| Distant metastasis free survival | |||
| Grade 3 | 1.100 | 0.529–2.287 | 0.879 |
| Size>20 mm | 1.372 | 0.742–2.540 | 0.313 |
| LN+ | 3.822 | 2.036–7.175 | <0.001 |
| LVI | 1.832 | 0.960–3.499 | 0.067 |
| Subtype | |||
| Modified LA (reference) | 1.0 | ||
| Modified LB | 2.872 | 1.326–6.222 | 0.007 |
| Basal | 3.273 | 1.139–9.396 | 0.028 |
| HER2 | 1.825 | 0.386–8.639 | 0.448 |
| Unclassified | 9.902 | 3.269–29.99 | <0.001 |
| Resolved model | |||
| LN+ | 4.013 | 2.154–7.477 | <0.001 |
| LVI | 2.011 | 1.075–3.764 | 0.029 |
| Modified LB | 3.023 | 1.501–6.089 | 0.002 |
| Basal | 3.902 | 1.657–9.191 | 0.002 |
| HER2 | 2.064 | 0.472–9.026 | 0.336 |
| Unclassified | 10.87 | 3.882–30.461 | <0.001 |
| Breast cancer specific death | |||
| Grade 3 | 1.307 | 0.570–2.997 | 0.527 |
| Size>20 mm | 1.879 | 0.927–3.807 | 0.080 |
| LN+ | 4.535 | 2.153–9.553 | <0.001 |
| LVI | 2.085 | 1.030–4.223 | 0.041 |
| Subtype | |||
| Modified LA (ref) | 1.0 | ||
| Modified LB | 3.084 | 1.280–7.431 | 0.012 |
| Basal | 3.780 | 1.155–12.37 | 0.028 |
| HER2 | 2.095 | 0.412–10.65 | 0.373 |
| Unclassified | 8.167 | 1.997–33.40 | 0.003 |
| Resolved model | |||
| LN+ | 4.906 | 2.353–10.22 | <0.001 |
| LVI | 2.518 | 1.267–5.004 | 0.008 |
| Modified LB | 3.617 | 1.629–8.031 | 0.002 |
| Basal | 5.715 | 2.173–15.03 | <0.001 |
| HER2 | 2.907 | 0.641–13.17 | 0.166 |
| Unclassified | 10.37 | 2.801–38.42 | <0.001 |
Abbreviations: CI=confidence interval; EIC+ =extensive intraduct component of DCIS=ductal carcinoma in situ; HR=hazard ratio; LA=luminal A; LB=luminal B; LN=lymph node; LVI=lymphatic vascular invasion.
Bold typescript indicates statistical significance.
DMFS and BCSS
Luminal B was an independent predictor of adverse outcome for both metastases and breast cancer-specific death in the final resolved models (LB DMFS: HR 3.023, 95% CI 1.501–6.089, P=0.002; BCSS: HR 3.617, 95% CI 1.629–8.031, P=0.002), along with LVI, LN positivity, basal and unclassified (Table 4).
Discussion
Oestrogen receptor-positive early breast cancer is the commonest form of the disease and tailoring treatment to individual patients is a priority. It is important to identify ER+ patients with a good prognosis who will receive most benefit from endocrine therapy and receive little or no benefit from chemotherapy, and, therefore, avoid any toxicity. In addition, it is also beneficial to identify patients who will have little or no benefit from endocrine therapy. GEP studies have consistently identified at least two groups of ER+ tumours; the less favourable LB group being characterised by higher histological grade and higher expression of proliferation and HER2-related genes, such as MKI67, MYBBL2, CCNB1, HER2 and GRB7, and lower levels of ER-related genes. Although there is some consistency in the recognition of these differing subgroups between GEP studies, there is some doubt as to the stability of the classifiers used by different single sample predictors (Weigelt et al, 2010b) and most assays are not yet ready for routine clinical use (De Ronde et al, 2010). As a result, a simple and relatively cheap test using IHC surrogates would be easier to transfer into clinical practise. Various combinations of markers have been assessed to develop a robust IHC panel for routine pathology reporting, most recently adding Ki67 to ER, PR, HER2 to better assess proliferative luminal tumours (Cheang et al, 2009; Hugh et al, 2009). Assessing ER+ tumours with surrogates for molecular grade may strengthen patient selection as histological grade can be compromised in some specimens because of sub-optimal fixation.
Using an independent discovery cohort of 292 patients, we identified a significant difference in expression in Ki-67 and p53 within ER+ cancers, which was associated with differences in clinical outcomes (breast-cancer specific death; CM McNeil et al, manuscript in preparation). These findings were subsequently validated in a detailed analysis of 498 early breast cancer patients, in which we compared good and poor prognosis ‘LA’ and ‘LB’ IHC signatures, which included Ki-67 and p53 in addition to ER, PR and HER2. This updated definition provided superior predictive power and better discrimination between the two groups of luminal tumours for all measures of outcome. In all, 73 previously LA tumours were reclassified as LB, increasing the size of the ‘LB’ group by >four-fold from 4.6 to 19.7% of the cohort, better reflecting GEP estimates of the size of the LB population. Using this definition, ‘LB’ was an independent predictor of poor prognosis in MVA for LRR, DMFS and BCSS but not for IBTR for the whole cohort. As well as demonstrating its superior predictive power over the most frequently used classifier or ER, PR, HER2 alone, we also performed additional analyses to make a comparison with ER+ breast cancer classified by hormone receptor (HR) status alone (data not shown). Some studies have shown a significant difference in outcome between double-positive (i.e., ER+ PR+) and single-receptor positive HR status (i.e., ER+ PR− or ER− PR+, Rakha et al, 2007). This latter group may correspond to the LB subtype (Rakha et al, 2009). Our further analyses of these subgroups demonstrated that HR status alone was inferior to our updated five biomarker classifier: in univariate analysis good prognosis double-positive status (ER+ PR+) was only statistically predictive for distant metastases and death (not IBTR or LRR) and single-positive status (i.e., poor prognosis ‘LB’) was not predictive for any measure of outcome in univariate analysis.
Our updated classification of ER+ disease also improves the statistical significance in survival between all intrinsic subtypes, where the adverse survival and HR of our poor prognosis ‘LB’ group is three times that of ‘LA’ and closer to that of HER2-enriched and basal subtypes. One limitation of this study is that recurrence rates may be over estimated for LB, as the prognosis of HER2-positive LB tumours (24% of all LB tumours) would currently be modified by the benefits of Herceptin treatment (which was not used in this study) and an underestimate for LA, as only 44.9% of patients received adjuvant tamoxifen. An additional limitation of this study is the difference in cut points used for Ki67 positivity where the training cohort median was 5% and the validation cohort median was 10%. Although we have identified good and poor prognostic groups with our signature, the relatively wide confidence intervals, which reflect the small numbers of events, strongly suggests the importance of further independent validation. Further analyses in a larger data set with a greater number of events may provide narrower confidence intervals, which along with assessment of the hazard ratio will determine the likely clinical significance derived from this panel of markers.
These findings suggest a potential role for this biomarker panel in better defining groups of ER+ cancer of low and high molecular grade, allowing better selection of patients for endocrine therapy alone or with AC. Although Ki67 alone identifies approximately 60% of LB tumours, p53 adds a further 20% of cases, 12% are positive for both markers, 8% are negative for both but HER2 positive. This study builds upon previous work (Cheang et al, 2009) using a cut point for optimal determination of ‘high’ Ki-67 proliferation rate at 14% through correlation with the PAM50 classifier using RT–PCR. They identified a LB population, which was 42% of the cohort (includes their LB and luminal HER2 cases). Although the cut point of 14% correlates with GEP estimates it may, in practical terms, be difficult to discern by IHC. Ki67 has long been analysed in breast cancer cohorts with varied results in terms of its predictive value. A recent review has recommended its inclusion as a routine biomarker in breast cancer (Yerushalmi et al, 2010), but its application as a stand alone biomarker has been debated (Stuart-Harris et al, 2008). Therefore, its inclusion in a panel to help define molecular grade and better subtype ‘LA’ and ‘LB’ cancers is independently prognostic and valuable. However, its role as a predictive marker appears less certain. A pre- and post-biopsy analysis of endocrine treated breast cancer has demonstrated that only the post-treatment tumour Ki67 (at 2 weeks) was predictive of response to endocrine therapy, whereas baseline Ki67 was not (Dowsett et al, 2007). High Ki67 status in BIG 1–98 suggested a potential benefit in selecting letrozole over tamoxifen in post-menopausal patients (Viale et al, 2008). Most recently a significant study identified that the prognostic information provided by ‘IHC4’ (ER, PR, HER2 and Ki-67) was at least equivalent to Oncotype Dx (Cuzick et al, 2009) and highlights the relevance of these readily available routine pathology markers in the clinical management of breast cancer.
p53 overexpression in breast cancer assessed by IHC is, rather over simplistically, assumed to act as a surrogate for TP53 mutations and is associated with higher tumour grade and responsiveness to radiotherapy, chemotherapy and endocrine therapy (Thompson and Lane, 2010). Although the p53 pathway is undoubtedly highly complex, its assessment by IHC does appear to provide meaningful information. p53 mutations are more frequent in the LB group compared with LA (Weigelt et al, 2010a), being described in 71% of LB tumours but only 16% of LA (Sorlie, 2004). p53 currently features as one of five antibodies in the Mammostrat (Clarient, Inc., Aliso Viejo, CA, USA) IHC test shown to be of predictive value in ER+, tamoxifen-treated early breast cancer (Ring et al, 2006; Bartlett et al, 2010). Mammostrat uses a five IHC panel (p53, HTF9C, CEACAM5, NDRG1, SLC7A5) with an algorithm that is independent of ER and PR status to identify low-, medium- and high-risk groups. The initial published study (Ring et al, 2006) demonstrated HRs of 1.8 and 2.3 (training and validation cohorts, respectively) for high risk compared with the low and medium risks for disease recurrence. Elevated expression of p53 was observed by IHC in our cohorts and appeared to be a useful classifier and was included in the updated definition of poor prognosis ‘LB’ cancer.
Although the number of events was small, additional exploratory multivariate analyses for patients treated with tamoxifen alone (n=169, 10 events) showed that the poor prognosis ‘LB’ definition retained independent prognostic significance in the final resolved model for breast cancer specific death (HR 5.361, 95% CI 1.418–20.25, P=0.013). This finding suggests that ‘LB’ has five times the risk of death compared with ‘LA’ in patients treated with endocrine therapy. The predictive value of this classification would however require further testing within the setting of a randomised trial of endocrine therapy.
Our updated definition of ER+ cancer translates into an IBTR-free survival at 5 years of 99.3% for LA and 96.6% LB, LRR-free survival 99.7 and 93.4%. A similar recent study using ER, PR and Ki67 in the definition for LA and LB found local recurrence-free rates at 10 years of 92% for LA and 90% for LB (Voduc et al, 2010). Importantly, our findings further support the observations of this group, who found that LB was associated with increased risk of LRR. These results highlight the role of proliferation and grade, mirrored by the Oncotype Dx assay (Mamounas et al, 2005), as a predictor of locoregional recurrence, and may help further refine patient selection regarding therapy for optimal locoregional control. A subsequent study analysed patterns of metastases and found both LA and LB had a predilection for bone as a metastatic site and found that LB had a distant relapse rate similar to basal tumours at 15 years (Kennecke et al, 2010). In summary, this study suggests that good and poor prognosis ER+ breast cancers can be reliably and easily discriminated using Ki67 and p53 in addition to ER, PR and HER2 in routine pathology IHC. This definition greatly enhances the detection of poor prognosis ER+ ‘LB’ breast cancers, with an outcome closer to that of basal and HER2-enriched tumours. This approach may help more reliably define groups of ER+ patients with an excellent prognosis and identify those at risk of early relapse who may benefit from more frequent follow-up and early intervention with alternative therapies and/or chemotherapy. Further, larger studies in randomised clinical trials of endocrine therapy are required to assess the clinical utility of this classification and its value as a predictor of therapeutic responsiveness.
Acknowledgments
We thank the National Health and Medical Research Council of Australia (Program Grant 535903, Project Grant 535947, the Fellowship 427601 RLS), the Cancer Institute New South Wales (Translational Program Grant 10/TPG/1-04), the Cancer Australia (Project Grant 626201), the Petre Foundation and the RT Hall Trust.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Abd El-Rehim DM, Ball G, Pinder SE, Rakha E, Paish C, Robertson JF, Macmillan D, Blamey RW, Ellis IO (2005) High throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA analyses. Int J Cancer 116: 340–350 [DOI] [PubMed] [Google Scholar]
- Bartlett JMS, Thomas J, Ross DT, Seitz RS, Ring BZ, Beck RA, Pedersen HC, Munro A, Kunkler IH, Campbell FM, Jack W, Kerr GR, Johnstone L, Cameron DA, Chetty UL (2010) Mammostrat as a tool to stratify breast cancer patients at risk of recurrence during endocrine therapy. Breast Cancer Res 12: R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blows FM, Driver KE, Schmidt MK, Broeks A, van Leeuwen FE, Wesseling J, Cheang MC, Gelmon K, Nielsen TO, Blomqvist C, Heikkilä P, Heikkinen T, Nevanlinna H, Akslen LA, Bégin LR, Foulkes WD, Couch FJ, Wang X, Cafourek V, Olson JE, Baglietto L, Giles GG, Severi G, McLean CA, Southey MC, Rakha E, Green AR, Ellis IO, Sherman ME, Lissowska J, Anderson WF, Cox A, Cross SS, Reed MW, Provenzano E, Dawson SJ, Dunning AM, Humphreys M, Easton DF, García-Closas M, Caldas C, Pharoah PD, Huntsman DL (2010) Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med 7: e1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey LA (2010) Through a glass darkly: advances in understanding breast cancer biology, 2000–2010. Clin Breast Cancer 10: 188–195 [DOI] [PubMed] [Google Scholar]
- Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, Deming SL, Geradts J, Cheang MC, Nielsen TO, Moorman PG, Earp HS, Millikan RC (2006) Race, breast cancer subtypes and survival in the Carolina breast cancer study. JAMA 295: 2492–2502 [DOI] [PubMed] [Google Scholar]
- Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, Watson M, Davies S, Bernard PS, Parker JS, Perou CM, Ellis MJ, Nielsen TO (2009) Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst 101: 736–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, Perou CM, Nielsen TO (2008) Basal like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res 14: 1368–1376 [DOI] [PubMed] [Google Scholar]
- Cuzick J, Dowsett M, Wale C, Salter J, Quinn E, Zabaglo L, Howell A, Buzdar A, Forbes JF (2009) Prognostic value of a combined ER, PgR, Ki67, HER2 immunohistochemical (IHC4) score and comparison with the GHI recurrence score – results from TransATAC. Cancer Res 69: 503S (abstract) [Google Scholar]
- De Ronde J, Wessels L, Wesseling J (2010) Molecular subtyping ready to use? Lancet Oncol 11: 306–307 [DOI] [PubMed] [Google Scholar]
- Dowsett M, Smith IE, Ebbs SR, Dixon JM, Skene A, A’Hern R, Salter J, Detre S, Hills M, Walsh G, IMPACT Trialists Group (2007) Prognostic value of Ki67 expression after short term presurgical endocrine therapy for primary breast cancer. J Natl Cancer Inst 99: 167–170 [DOI] [PubMed] [Google Scholar]
- Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) (2005) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 365: 1687–1717 [DOI] [PubMed] [Google Scholar]
- Goldhirsch A, Ingle JN, Gelber RD, Coates AS, Thürlimann B, Senn HJ (2009) Thresholds for therapy: highlights of the international expert consensus on the primary therapy of early breast cancer. Ann Oncol 20: 1319–1329 [DOI] [PubMed] [Google Scholar]
- Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, Nobel A, Parker J, Ewend MG, Sawyer LR, Wu J, Liu Y, Nanda R, Tretiakova M, Ruiz Orrico A, Dreher D, Palazzo JP, Perreard L, Nelson E, Mone M, Hansen H, Mullins M, Quackenbush JF, Ellis MJ, Olopade OI, Bernard PS, Perou CM (2006) The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics 7: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugh J, Hanson J, Cheang MC, Nielsen TO, Perou CM, Dumontet C, Reed J, Krajewska M, Treilleux I, Rupin M, Magherini E, Mackey J, Martin M, Vogel C (2009) Breast cancer subtypes and response to docetaxel in node-positive breast cancer: use of an immunohistochemical definition in the BCIRG 001 trial. J Clin Oncol 27: 1168–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemier J, Charafe-Jauffret E, Monville F, Esterni B, Extra JM, Houvenaeghel G, Xerri L, Bertucci F, Birnbaum D (2008) Association of GATA3, p53, Ki67 status and vascular peritumoral invasion are strongly prognostic in luminal breast cancer. Breast Cancer Res 11: R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennecke H, Yerushalmi R, Woods R, Cheang MC, Voduc D, Speers CH, Nielsen TO, Gelmon K (2010) Metastatic behaviour of breast cancer subtypes. J Clin Oncol 28: 3271–3277 [DOI] [PubMed] [Google Scholar]
- López-Knowles E, O’Toole SA, McNeil CM, Millar EKA, Qiu MR, Crea P, Musgrove EA, Sutherland RL (2010) PI3K pathway activation in breast cancer is associated with the basal-like phenotype and cancer-specific mortality. Int J Cancer 126(5): 1121–1131 [DOI] [PubMed] [Google Scholar]
- Mamounas E, Tang G, Bryant J (2005) Association between the 21-gene recurrence score assay (RS) and risk of locoregional failure in node-negative, ER-positive breast cancer: results from NSABP B-14 and NSABP B-20. 28th Annual San Antonio Breast Cancer Symposium December 8–11 2005; San Antonio, TX (abstract 29)
- Millar EK, Anderson LR, McNeil CM, O’Toole SA, Pinese M, Crea P, Morey AL, Biankin AV, Henshall SM, Musgrove EA, Sutherland RL, Butt AJ (2009a) BAG-1 predicts patient outcome and tamoxifen responsiveness in ER-positive invasive ductal carcinoma of the breast. Br J Cancer 100: 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar EK, Graham PH, O’Toole SA, McNeil CM, Browne L, Morey AL, Eggleton S, Beretov J, Theocharous C, Capp A, Nasser E, Kearsley JH, Delaney G, Papadatos G, Fox C, Sutherland RL (2009b) Prediction of local recurrence, distant metastases, and death after breast-conserving therapy in early-stage invasive breast cancer using a five-biomarker panel. J Clin Oncol 27: 4701–4708 [DOI] [PubMed] [Google Scholar]
- Musgrove EA, Sutherland RL (2009) Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer 9: 631–643 [DOI] [PubMed] [Google Scholar]
- Neilsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, Akslen LA, Ragaz J, Gown AM, Gilks CB, van de Rijn M, Perou CM (2004) Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast cancer. Clin Cancer Res 10: 5367–5374 [DOI] [PubMed] [Google Scholar]
- Nguyen PL, Taghian AG, Katz MS, Niemierko A, Abi Raad RF, Boon WL, Bellon JR, Wong JS, Smith BL, Harris JR (2008) Breast cancer subtype approximated by estrogen receptor, progesterone receptor, and HER-2 is associated with local and distant recurrence after breast conserving therapy. J Clin Oncol 26: 2373–2378 [DOI] [PubMed] [Google Scholar]
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N (2004) A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351: 2817–2826 [DOI] [PubMed] [Google Scholar]
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D (2000) Molecular portraits of human breast tumours. Nature 406: 747–752 [DOI] [PubMed] [Google Scholar]
- Rakha EA, El Sayed ME, Green AR, Paish EC, Powe DG, Gee J, Nicholson RI, Lee AHS, Robertson JFR, Ellis IO (2007) Biologic and clinical characteristics of breast cancer with single hormone receptor-positive phenotype. J Clin Oncol 25: 4772–4778 [DOI] [PubMed] [Google Scholar]
- Rakha EA, Reis-Filho JS, Baehner F, Dabbs DJ, Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR, Palacios J, Richardson AL, Schnitt SJ, Schmitt FC, Tan PH, Tse GM, Badve S, Ellis IO (2010) Breast cancer prognostic classification in the molecular era: the role of histological grade. Breast Cancer Res 12: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakha EA, Reis-Filho JS, Ellis IO (2009) Combinatorial biomarker expression in breast cancer. Breast Can Res Treat 120: 293–308 [DOI] [PubMed] [Google Scholar]
- Ring BZ, Seitz RS, Beck R, Shasteen WJ, Tarr SM, Cheang MC, Yoder BJ, Budd GT, Nielsen TO, Hicks DG, Estopinal NC, Ross DT (2006) Novel prognostic immunohistochemical biomarker panel for estrogen receptor-positive breast cancer. J Clin Oncol 24: 3039–3047 [DOI] [PubMed] [Google Scholar]
- Sorlie T (2004) Molecular portraits of breast cancer: tumour subtypes as distinct disease entities. Eur J Cancer 40: 2667–2675 [DOI] [PubMed] [Google Scholar]
- Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lønning P, Børresen-Dale AL (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98: 10869–10874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lønning PE, Brown PO, Børresen-Dale AL, Botstein D (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 100: 8418–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou C, Pusztai L (2009) Gene-expression signatures in breast cancer. N Engl J Med 360: 790–800 [DOI] [PubMed] [Google Scholar]
- Stuart-Harris R, Caldas C, Pinder SE, Pharoah P (2008) Proliferation markers and survival in early breast cancer: a systematic review and meta-analysis of 85 studies in 32 825 patients. Breast J 17: 323–334 [DOI] [PubMed] [Google Scholar]
- Thompson AM, Lane DP (2010) p53 transcriptional pathways in breast cancer: the good, the bad and the complex. J Pathol 220: 401–403 [DOI] [PubMed] [Google Scholar]
- Viale G, Giobbie-Hurder A, Regan MM, Coates AS, Mastropasqua MG, Dell’Orto P, Maiorano E, MacGrogan G, Braye SG, Ohlschlegel C, Neven P, Orosz Z, Olszewski WP, Knox F, Thürlimann B, Price KN, Castiglione-Gertsch M, Gelber RD, Gusterson BA, Goldhirsch A, Breast International Group Trial 1–98 (2008) Prognostic and predictive value of centrally reviewed Ki-67 labelling index in postmenopausal women with endocrine-responsive breast cancer: results from Breast International Group trial 1–98 comparing adjuvant tamoxifen with letrozole. J Clin Oncol 26: 5569–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voduc KD, Cheang MCU, Tyldesley S, Gelmon K, Nielsen TO, Kennecke H (2010) Breast cancer subtypes and the risk of local and regional relapse. J Clin Oncol 28: 1684–1691 [DOI] [PubMed] [Google Scholar]
- Weigelt B, Baehner FL, Reis-Filho JS (2010a) The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade. J Pathol 220: 263–280 [DOI] [PubMed] [Google Scholar]
- Weigelt B, Mackay A, A’hern R, Natrajan R, Tan DS, Dowsett M, Ashworth A, Reis-Filho JS (2010b) Breast cancer molecular profiling with single sample predictors: a retrospective analysis. Lancet Oncol 11: 339–349 [DOI] [PubMed] [Google Scholar]
- Weigelt B, Reis-Filho JS (2010) Molecular profiling currently offers no more than tumour morphology and basic immunohistochemistry. Breast Cancer Res 12(Suppl 4): 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita H, Toyama T, Nishio M, Ando Y, Hamaguchi M, Zhang Z, Kobayashi S, Fujii Y, Iwase H (2006) p53 protein accumulation predicts resistance to endocrine therapy and decreased post-relapse survival in metastatic breast cancer. Breast Cancer Res 8: R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerushalmi R, Woods R, Ravdin PM, Hayes MM, Gelmon KA (2010) Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol 11: 174–183 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.