Abstract
A large familyof neural protocadherin (Pcdh) proteins is encoded bythree closelylinked mammalian gene clusters (α, β, and γ). Pcdh α and γ clusters have a striking genomic organization. Specifically, each “variable” exon is spliced to a common set of downstream “constant” exons within each cluster. Recent studies demonstrated that the cell-specific expression of each Pcdh gene is determined bya combination of variable-exon promoter activation and cis-splicing of the corresponding variable exon to the first constant exon. To determine whether there are other similarlyorganized gene clusters in mammalian genomes, we performed a genome-wide search and identified a large number of mammalian genes containing multiple variable first exons. Here we describe several clusters that contain about a dozen variable exons arrayed in tandem, including UDP glucuronosyltransferase (UGT1), plectin, neuronal nitric oxide synthase (NOS1), and glucocorticoid receptor (GR) genes. In all these cases, multiple variable first exons are each spliced to a common set of downstream constant exons to generate diverse functional mRNAs. As an example, we analyzed the tissue-specific expression profile of the mouse UGT1 repertoire and found that multiple isoforms are expressed in a tissue-specific manner. Therefore, this variable and constant genomic organization provides a genetic mechanism for directing distinct cell- and tissue-specific patterns of gene expression.
Various genome sequencing projects are generating vast amounts of sequence data, but at present, our ability to understand and interpret the sequence information is limited. An important aspect of genome annotation is the identification of general patterns of genomic organization that may provide important information about regulatory mechanisms of gene expression. In particular, genomic organization may provide a mechanism for generating transcript diversity through complex alternative splicing. For example, the Drosophila Down syndrome cell adhesion molecule (Dscam) gene contains four alternative cassette exons, each with multiple variants, and can potentially generate more than 38,000 mRNAs through alternative splicing—more than the total number of genes in the fly genome (Schmucker et al. 2000).
The mammalian central nervous system contains billions of cells with trillions of specific connections. These vast numbers of connections are determined by combinatorial interactions between large numbers of cell-surface molecules. Pcdh proteins have been proposed as the prime candidates for specifying neuronal connections in the central nervous system (Kohmura et al. 1998; Wu and Maniatis 1999). We previously identified three closely linked mammalian Pcdh clusters (α, β, and γ) that encode ∼60 diverse mRNAs (Wu and Maniatis 1999; Wu et al. 2001). These mRNAs are expressed in cell-specific patterns in the central nervous system (Kohmura et al. 1998; Wang et al. 2002b).
In both mice and humans, Pcdh α and γ clusters have a striking genomic organization similar to that of the immunoglobulin and T-cell receptor clusters. Within each Pcdh cluster, a tandem array of more than a dozen variable exons spans a large region of ∼200 kb of genomic DNA. Each of these unusually large variable exons encodes all of the six extracellular cadherin domains, a transmembrane segment, and a small part of the cytoplasmic domain (Wu and Maniatis 2000). The remaining cytoplasmic domain is encoded by three constant exons that are located downstream from the variable exon tandem array. Each variable exon is separately spliced to the first constant exon to generate diverse functional mRNAs (as an example, see the mouse Pcdhα genomic organization in Fig. 1A). Recent studies have demonstrated that each variable exon is preceded by a distinct promoter (Tasic et al. 2002; Wang et al. 2002a). Activation of a specific variable region promoter transcribes a long precursor RNA that contains all of the downstream variable exons and the entire constant region. However, only the 5′-most exon is selectively cis-spliced to the first constant exon to generate diverse functional mRNAs. Thus, cell-specific Pcdh expression patterns in the brain are determined by a combination of differential promoter activation and alternative cis-splicing.
Figure 1.
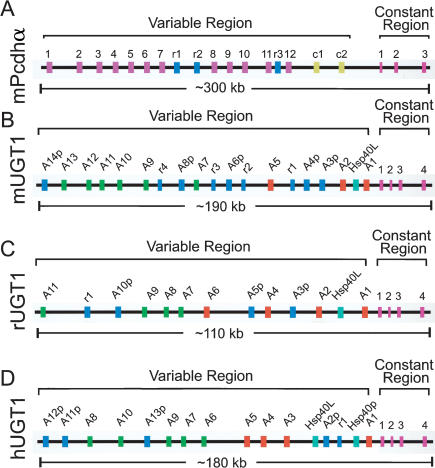
Similar genomic organization of Pcdh and UGT1 gene clusters. Shown are comparisons of the genomic organization of mouse Pcdh (A), and mouse (B), rat (C), and human (D) UGT1 clusters. Each cluster has multiple, highly similar, tandem variable exons followed by one set of constant exons. They are indicated by vertical colored bars: (mauve) mouse Pcdhα variable exons; (yellow) C-type Pcdh variable exons; (green) phenol-type UGT1 variable exons; (orange) bilirubin-type UGT1 variable exons; (blue) pseudogenes or relics (present in both Pcdh and UGT1 clusters); (turquoise) non-UGT1 genes in the UGT1 cluster; (pink) constant exons (present in both Pcdh and UGT1 clusters). The approximate length of each cluster is shown below the corresponding panels. The pseudogenes are represented by a letter “p” following the gene symbol, whereas the relic sequences are represented by a letter “r.” (Pcdh) Protocadherin; (UGT) UDP glucuronosyltransferase; (Hsp40L) heat-shock protein 40kd like gene.
To determine whether there are other genes with a similar unusual organization in mammalian genomes, we searched the GenBank databases and found that the genomic organization of the UGT1 cluster is very similar to that of the Pcdh clusters. About a dozen similar UGT1 variable exons are organized in a tandem array. A common set of four UGT1 constant exons is located downstream from the variable exon tandem array. We cloned eight of the mouse UGT1 genes and characterized their tissue-specific expression profiles. Each variable exon is separately spliced to the first constant exon to generate diverse UGT1 mRNAs encoding distinct protein isoforms. In addition, we provide evidence that each variable exon of the UGT1 cluster is likely preceded by a distinct promoter. We also found that the I-branching β-1,6-N-acetylglucosaminyltransferase (IGnT) cluster has a similar variable and constant genomic organization. Three highly similar variable exons are each separately spliced to a common set of downstream constant exons. Finally, we describe several genes that have more than 10 tandem variable first exons and a common set of downstream constant exons; however, their variable exons do not display sequence similarity. These include plectin, neuronal nitric oxide synthase (NOS1), and glucocorticoid receptor (GR) genes. In all of these cases, about a dozen variable exons are each separately spliced to a common set of downstream constant exons to generate diverse mRNAs. Thus, the unusual genomic organization of variable and constant regions is more prevalent in mammalian genomes than previously realized. The organization of multiple variable first exons has important implications regarding cell- and tissue-specific, as well as developmentally regulated, gene expression.
RESULTS
Identification and Cloning of Mouse UGT1 Genes
The vertebrate animal removes numerous xenobiotic and endobiotic compounds from its body by converting them to more water-soluble glucuronides. UDP glucuronosyltransferase proteins (UGTs) are a superfamily of enzymes that catalyze the glucuronidation of numerous chemical compounds (Tukey and Strassburg 2000). These proteins play an important role in the pharmacokinetic clearance of environmental toxins, endogenous metabolites, and therapeutic drugs. For example, genetic mutations of the human UGT1A1 gene cause jaundice, including Crigler-Najjar syndrome and Gilbert syndrome (Tukey and Strassburg 2000). The UGT1 subfamily of UGT proteins catalyzes the glucuronidation of the donor substrate (UDP-glucuronic acid) to numerous acceptor substrates (aglycone compounds). The N-terminal aglycone-binding domains of UGT1 proteins are distinct, but very similar to each other. In contrast, the C-terminal UDP-binding domains are identical among all the UGT1 proteins. As with Pcdh proteins, the diversity of UGT1 proteins is determined by an unusual genomic organization. The rat and human UGT1 clusters have recently been characterized (Emi et al. 1995; Gong et al. 2001). However, the number of mouse UGT1 genes and their genomic organization are unknown. Here we identify 14 mouse UGT1 genes by a combination of sequence analyses and cloning experiments.
Iterative BLAST searches of public databases identified sequences that span the genomic region of mouse UGT1 genes. Sequence analysis of this genomic DNA revealed the presence of nine variable exons encoding polypeptides very similar to the N-terminal half of human UGT1 proteins. We cloned eight mouse UGT1 genes, two of which match previously cloned cDNAs (S57479 and S64760; Kong et al. 1993). The DNA sequences of each clone contain a single open reading frame of ∼1600 bp, encoding a protein with a divergent N-terminal domain and an identical C-terminal domain. The 5′ regions of cDNA sequences encoding the N-terminal domain are highly similar. Each of these sequences is identical to a different region of ∼850 bp of genomic DNA. Thus, the N-terminal domain of each protein is encoded by a single exon. Fourteen mouse UGT1 variable exons are organized in a tandem array, spanning a region of ∼187 kb of genomic DNA (Fig. 1B). In contrast, the 3′ regions of all mouse UGT1 cDNA sequences are identical and match four segments of genomic DNA located ∼2 kb downstream from the last variable exon. Therefore, the C-terminal domains of all UGT1 proteins are identical and are encoded by four constant exons (Fig. 1B). These four exons span a region of ∼4 kb of genomic DNA sequences. In addition, there is a consensus 5′ splice site immediately downstream from the last nucleotide of each variable exon, and a consensus 3′ splice site immediately upstream of the first nucleotide of the first constant exon (data not shown). Therefore, alternative RNA splicing of each variable exon to the first constant exon generates diverse UGT1 mRNAs. We conclude that the genomic organization of the mouse UGT1 cluster is similar to that of the mouse Pcdhα clusters (Fig. 1).
Like the Pcdh variable cDNA sequences, the variable cDNA coding sequences of the UGT1 cluster are present as uninterrupted exons. In addition, we identified five UGT1 pseudogenes that have significant nucleotide sequence similarity to the nine mouse UGT1 genes; however, their coding regions are interrupted by multiple stop codons. Finally, we identified four UGT1 relics whose sequences have limited similarity to the functional UGT1 genes. The relic sequences are much shorter than the functional genes and pseudogenes.
Tissue-Specific Expression Profile of the Mouse UGT1 Repertoire
Glucuronidation is thought to occur mainly in the liver. Nevertheless, recent studies of human UGT1 genes have shown that some of these genes are expressed in specific extrahepatic tissues (Tukey and Strassburg 2000). However, tissue-specific expression patterns of the entire UGT1 repertoire have not been analyzed to date. Characterization of the whole mouse UGT1 gene repertoire provides an opportunity to systematically investigate its tissue-specific expression patterns.
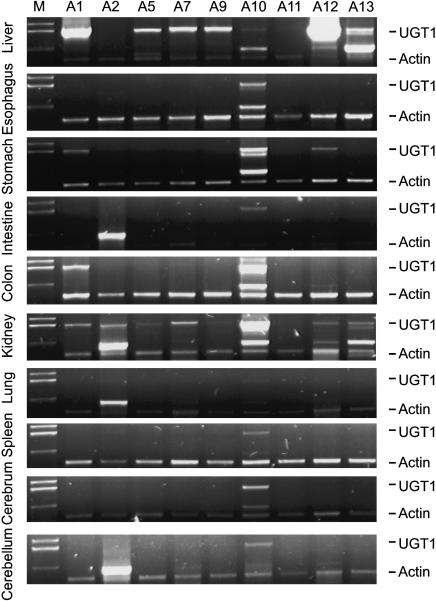
We measured the expression patterns of the entire mouse UGT1 gene repertoire in a wide variety of tissues by using isoform-specific duplex reverse-transcriptase polymerase-chain-reaction (DRT-PCR) methods. These experiments revealed that each UGT1 gene displays a distinct tissue-specific expression pattern (Fig. 2). For example, UGT1A1, the gene mainly responsible for the glucuronidation of bilirubin, is expressed in the stomach, intestine, colon, and kidney, and is expressed most abundantly in the liver. Furthermore, the mouse UGT1A10 gene is expressed in all of the tissues examined thus far, with highest levels in colon and kidney tissues. In addition, the mouse UGT1A12 gene is the most abundantly expressed UGT1 gene in the liver. Finally, we found that the liver expresses almost all UGT1 genes, consistent with its being the main organ for detoxification. Interestingly, the kidney expresses all UGT1 genes except UGT1A11 (Fig. 2). Given that toxic chemicals are eliminated through biliary and renal tissues, these results are consistent with the important role of the UGT1 genes in detoxification.
Figure 2.
Tissue-specific expression profiles of mouse UGT1 gene cluster. The expression of UGT1 was assessed by DRT-PCR using primers specific for each isoform. We used an actin primer pair as an internal control. The tissue sources are shown on the left of each panel. The amplified cDNA products are indicated on the right of each panel. The band slightly above the actin cDNA is a nonspecific PCR product. The UGT1 isoforms are indicated above each lane. (M) Marker.
Comparison of the Mouse, Rat, and Human UGT1 Clusters
The rat UGT1 locus was previously reported to contain seven functional genes and two pseudogenes (Emi et al. 1995). We have annotated the rat UGT1 genomic sequences and found that they have one additional functional gene (A11) and one additional pseudogene (A3p; Fig. 1C). Moreover, we found that there is a relic located between A11 and pseudogene A10p. Based on sequence similarity and substrate-specificity, the rat UGT1 genes have been divided into two groups, the bilirubin group and the phenol group (Emi et al. 1995). The rat UGT1 locus is considerably smaller than the corresponding mouse and human loci primarily because the intergenic sequences are shorter (Fig. 1).
The human UGT1 locus has recently been extensively analyzed and found to contain 13 UGT1 genes (Gong et al. 2001). These variable exons are organized in a tandem array followed by a set of four constant exons (Fig. 1D). Based on sequence similarity, the human UGT1 proteins can also be divided into two groups, namely, UGT1 A1 through A5 (bilirubin group) and UGT1 A6 through A10 (phenol group).
We used the VISTA program (Dubchak et al. 2000) to compare genomic sequences of the human, mouse, and rat UGT1 clusters (data shown as Supplemental Fig. S1, available online at www.genome.org). The variable and constant exons are highly conserved. However, there are almost no highly conserved intergenic sequences in the UGT1 locus, except for one intronic region (position of 187 kb in Supplemental Fig. S1) located between constant exons 1 and 2.
The overall genomic organization patterns of the mouse, rat, and human UGT1 clusters are similar (Fig. 1). For example, the organization of the constant region is highly conserved. Like the constant region of the human and rat UGT1 clusters, the constant region of the mouse UGT1 cluster is also organized into four exons. In addition, these constant exons are highly conserved. The encoded constant-region polypeptide sequences are 90% identical between mice and humans. Furthermore, the length of the first three constant exons is identical among mice, rats, and humans. Finally, as in the human and rat UGT1 clusters, the mouse UGT1 cluster can also be divided into two groups, namely, UGT1 A1 through A5 (bilirubin group) and UGT1 A7 through A13 (phenol group).
The variable exons are organized in a tandem array in mice, rats, and humans. However, their numbers are different in the three species. The mouse UGT1 cluster is predicted to have 14 genes, whereas the human cluster has 13 genes and the rat cluster has 11 genes (Fig. 1). There are five pseudogenes in the mouse UGT1 cluster, whereas there are only four pseudogenes in the human UGT1 cluster and three pseudogenes in the rat UGT1 cluster (Fig. 1). Finally, there are four relic sequences within the variable region of the mouse UGT1 cluster in contrast to one relic each in rats and humans.
Evolutionary Relationship Among Members of the Mouse, Rat, and Human UGT1 Genes
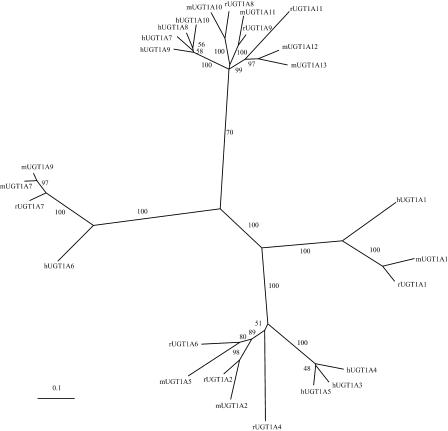
The variable regions of the mouse, rat, and human UGT1 proteins are similar and of almost the same length, and can be divided into bilirubin and phenol groups. Within the bilirubin group, the human, mouse, and rat UGT1A1 genes are orthologs and form an orthologous branch in the phylogenetic tree (Fig. 3). There is an 86% sequence identity between rat and mouse UGT1A1 at the amino acid level, whereas both have a 68% amino acid sequence identity to the human UGT1A1 protein. Other members of this group display a complex evolutionary relationship. For example, the human UGT1A3, UGT1A4, and UGT1A5 genes are paralogous, and they form an orthologous branch with the mouse UGT1A2 and UGT1A5, and rat UGT1A2, UGT1A4, and UGT1A6 genes in the phylogenetic tree (Fig. 3). The average amino acid sequence identity among members of this branch is ∼69%.
Figure 3.
Phylogenetic tree of human, mouse, and rat UGT1 gene clusters. The tree was reconstructed based on variable region polypeptides by using the neighbor-joining method of the CLUSTAL W package. The tree branches are labeled with the percentage support for that partition based on 1000 bootstrap replicates. The scale bar equals a distance of 0.1.
The phenol group also displays two branches in the phylogenetic tree (Fig. 3). In the smaller branch, mouse UGT1A7 and UGT1A9 are paralogous, and they are orthologous to rat UGT1A7 and human UGT1A6. The mouse UGT1A7 and UGT1A9 variable polypeptides have 95% identity but only have 88% and 70% identity to the rat and human orthologs, respectively. This observation indicates that the mouse UGT1A7 and A9 genes were duplicated after speciation. In the larger branch, human UGT1A7, A8, A9, and A10 are paralogous. Mouse UGT1A12 and A13 are paralogous and have one rat ortholog, UGT1A11. Mouse UGT1A10 and rat UGT1A8, and mouse UGT1A11 and rat UGT1A9 are orthologous. The average amino acid sequence identity among members of this branch is ∼73%. These observations indicate that distinct members of the UGT1 cluster are selectively expanded in mice and humans but not in rats.
Identification of a Non-UGT1 Gene Within the Variable Region of the UGT1 Cluster
In the Pcdh locus, there are two noncadherin genes, ORNT2 and TAFII55, between the Pcdh β and γ clusters. Both are single-exon genes and are transcribed from the opposite strand (Wu et al. 2001). Here we identified an Hsp40/DnaJ-like gene within the variable region of the mouse, rat, and human UGT1 clusters (Fig. 1; Supplemental Figs. S1, S2). The Hsp40-like (Hsp40L) gene is located on the opposite strand between UGT1 A1 and A2 in both mice and rats. In humans, this gene appears to be duplicated, with one (HSP40P) located between UGT1A1 and pseudogene A2p and another (HSP40L) between pseudogene A2p and UGT1A3. However, the former appears to be a pseudogene because it has a stop codon in the putative coding region. This gene is transcribed because it has numerous mouse, rat, and human EST matches.
Evidence for Multiple Promoters in the UGT1 Variable Region
In Pcdh clusters, the position of each variable exon corresponds to the location of a CpG island (Wu et al. 2001). We have used the CpGplot program (Larsen et al. 1992) to obtain the distribution of CpG dinucleotides in the UGT1 genomic sequences (http://bioweb.pasteur.fr/seqanal/interfaces/cpgplot.html; Supplemental Fig. S2). The ratio of observed-to-expected CpG dinucleotide frequency peaks at the location of almost every human UGT1 variable exon. It is known that the mouse genome lost some CpG dinucleotides after the divergence of mice and humans (Antequera and Bird 1993). Consistent with this, we note that the ratio is slightly lower in mice and rats than in humans. Nevertheless, this distribution indicates that each variable exon is associated with a promoter.
In the variable regions of the Pcdh α and γ clusters, the sequences immediately upstream of each variable exon are highly conserved between orthologs (Wu et al. 2001). Likewise, there are also conserved sequences upstream of orthologous variable exons in the UGT1 variable region (Supplemental Fig. S1; data not shown). We were not able to find a common motif upstream of all of the UGT1 variable exons using a Gibbs sampler program (Lawrence et al. 1993; http://bayesweb.wadsworth.org/gibbs/gibbs.html). We reasoned that the motif may be different for the two groups of UGT1 variable exons. Indeed, we found a conserved motif upstream of variable exons in the bilirubin group (Supplemental Fig. S3A,B) and also a distinct conserved motif upstream of variable exons in the phenol group (Supplemental Fig. S3C,D). Although the functional significance of these motifs remains to be established, their locations in the loci indicate that they may play a role in the regulation of UGT1 gene expression (Supplemental Fig. S3). The transcription start sites have been mapped immediately upstream of several human UGT1 variable exons (Gong et al. 2001). The sequences that precede a noncoding exon upstream of a rat UGT1 variable exon have been shown to contain promoter activities (Emi et al. 1996). Taken together, we conclude that each UGT1 variable exon is preceded by a promoter, and suggest that the tissue-specific expression patterns of UGT1 genes are determined by a combination of variable-exon promoter activation and cis-splicing of the corresponding cap-proximal exon to the first constant exon.
The I-Branching β-1,6-N-Acetylglucosaminyltransferase (IGnT) Cluster
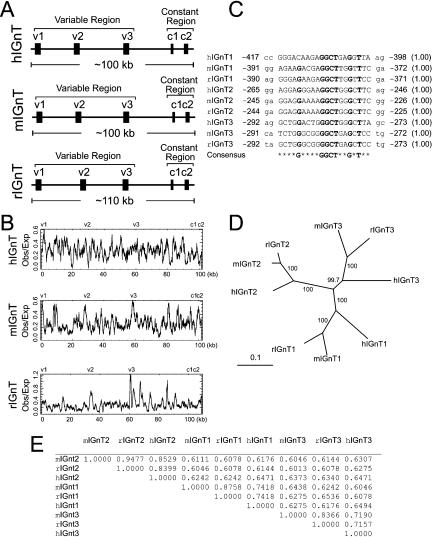
The acetylglucosaminyltransferase proteins are a family of type II transmembrane proteins that play an important role in the synthesis of oligosaccharide structures on glycoproteins. The IGnT genes encode a subfamily of proteins that convert linear carbohydrate chains to branched ones, and are essential for human blood group I antigen formation during development. By performing a genome-wide search, we found that, similar to the Pcdh and UGT1 clusters, the mouse IGnT locus is organized into both variable and constant regions (Fig. 4A). The variable exons are highly similar to each other and are all about the same length. In contrast to the Pcdh and UGT1 clusters, however, the IGnT locus contains only three variable exons (Fig. 4A). The genomic organization of the IGnT locus is conserved among humans, mice, and rats (Fig. 4A). Very recent studies of the human IGnT genes confirm that each variable exon is separately spliced to the first constant exon (Inaba et al. 2003; Yu et al. 2003). Different IGnT isoforms have distinct cell-specific expression profiles. For example, human IGnT3 is expressed in reticulocytes, whereas human IGnT2 is expressed in lens-epithelium cells. Mutations of these isoforms may cause the adult i phenotype and congenital cataracts, respectively (Yu et al. 2003). There is a CpG island corresponding to the position of each variable exon in human, mouse, and rat IGnT genomic DNA sequences (Fig. 4B). In addition, there is a conserved motif upstream of each variable exon in all three species (Fig. 4C). These observations strongly imply that each variable exon is preceded by a separate promoter. Phylogenetic analysis demonstrates that the three variable exons display an orthologous relationship, indicating that they were duplicated before the divergence of humans, mice, and rats (Fig. 4D). A pairwise comparison of the variable polypeptides reveals that the sequence identities between orthologs are significantly higher than those between paralogs (Fig. 4E). A comparison of the genomic sequences of the IGnT locus by using the VISTA program (Dubchak et al. 2000) shows that the variable and constant coding regions are highly conserved (Supplemental Fig. S4). In summary, the genomic organization of the IGnT cluster is similar to that of the Pcdh and UGT1 clusters; the expression pattern of the IGnT genes is likely determined by a combination of activation of a variable-exon promoter and cis-splicing of the corresponding variable exon to the first constant exon.
Figure 4.
Characteristics of the IGnT cluster. (A) Organization of the human, mouse, and rat IGnT clusters. Variable and constant exons are indicated. (B) Distribution of CpG islands in the genomic sequences of the human, mouse, and rat IGnT clusters. Shown are ratios of observed to expected CpG dinucleotide frequency of an 800-bp sliding window for the genomic sequences of the IGnT cluster in the three species. (C) A conserved motif and its relative position (negative numbers flanking the motif) to the translation start codon in the human, mouse, and rat IGnT clusters. The probability of finding the motif within 500 nt upstream of the translation start codon is shown within parentheses at the right. (D) Phylogenetic tree of the human, mouse, and rat IGnT variable protein sequences. (E) Percentage identities of a pairwise comparison between two human, mouse, or rat IGnT variable protein sequences.
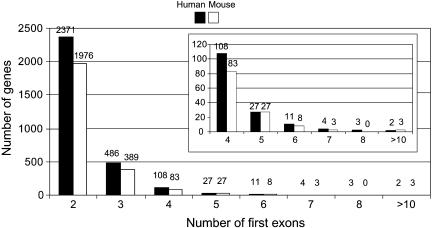
A Very Large Number of Mammalian Genes Contain Multiple Variable First Exons
We performed a genome-wide search for genes or gene clusters that contain multiple variable exons. We mapped all human, mouse, and rat mRNAs to their respective genomes and found a surprisingly large number of genes containing multiple variable first exons. There are >3000 such genes found in the human genome and >2000 in the mouse genome (Fig. 5). Extensive analyses showed that, besides the immunoglobulin and T-cell receptor clusters, only the variable exons of the Pcdh, UGT1, and IGnT clusters display sequence similarity. The variable exons of all the other genes do not have any sequence similarity. Although it is not known whether each variable exon of these genes is preceded by a separate promoter, we suggest that there are multiple promoters in these genes.
Figure 5.
Genome-wide distribution of human and mouse genes that have more than one first exon. The numbers are shown above each histogram. The inset shows an enlargement of the distribution of genes with more than three first exons.
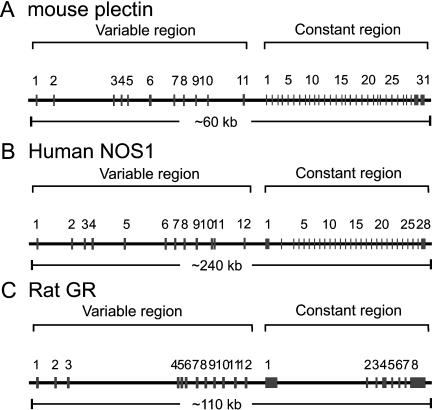
The majority of the genes have two alternative first exons (Fig. 5). The number of genes containing four or more variable exons decreases significantly; however, this number may be underestimated because there could be many more mRNAs that have not yet been sequenced (Fig. 5). Several of these genes have more than 10 variable exons, including the plectin, NOS1, and GR genes. Below we describe their genomic organization (Fig. 6) in detail.
Figure 6.
Genomic organization of mouse plectin (A), human NOS1 (B), and rat GR (C) genes. Each gene contains a tandem array of multiple first exons in the variable region, each of which is separately spliced to a common set of downstream constant exons. The approximate length of each gene is shown below the corresponding panels.
The Plectin Gene
Plectin is a versatile cytolinker protein that interacts directly with diverse membrane skeleton proteins and various intermediate filament proteins (Janda et al. 2001). Plectin isoforms are expressed in a wide variety of tissues, including muscle, brain, lung, spleen, and heart. It has been reported that the mouse plectin gene has an unusual 5′-transcript complexity and its transcripts exhibit distinct tissue-specific expression patterns (Fuchs et al. 1999). Similar to Pcdh and UGT1, the mouse plectin gene is also organized into variable and constant regions. Eleven variable exons are organized in a tandem array spanning a region of ∼40 kb of genomic DNA. Downstream from the tandem array, there are 31 constant exons spanning a region of ∼20 kb of genomic DNA (Fig. 6A). Each variable exon is separately spliced to the first constant exon to generate diverse plectin mRNAs.
There are only four known first exons in the rat plectin gene and two known first exons in the human plectin gene. We analyzed the genomic DNA sequences of the rat and human plectin variable regions and found seven and six new variable exons, respectively. A VISTA comparison of mouse sequences with the rat genomic DNA sequences revealed that all of the 11 variable exon sequences are conserved (Supplemental Fig. S5). In addition, a VISTA comparison of mouse and human genomic DNA sequences revealed that eight of the 11 variable exons are conserved (Supplemental Fig. S5). Eight of the 11 variable exons have putative coding regions, potentially encoding distinct plectin isoforms. The coding regions of variable first exons are all in the same open reading frame, and the encoded polypeptides are highly conserved among the three species (Supplemental Fig. S6). This indicates that each variable polypeptide has a distinct function. In contrast to the highly similar variable exons of the Pcdh and UGT1 clusters, plectin variable exons are of different lengths and do not display sequence similarity. Moreover, we cannot identify a conserved motif upstream of each variable exon by a Gibbs sampler program. We analyzed the CpG island distribution of human, mouse, and rat plectin variable regions and found that positions of almost all variable exons correspond to locations of CpG islands (Supplemental Fig. S7). Interestingly, there are additional upstream exons that are reported to splice to variable exon 1 in mice (Fuchs et al. 1999). Consistently, there is a CpG island upstream of variable exon 1 corresponding to the position of the upstream exons (Supplemental Fig. S7). These results imply that there are multiple promoters in the plectin variable region. Although the mechanism of tissue-specific plectin expression is unknown, it is very likely that there are multiple promoters in the plectin variable region, and that the tissue-specific expression patterns are determined by a combination of differential promoter activation and alternative cis-splicing.
The NOS1 Gene
Neuronal nitric oxide synthase (NOS1) has been implicated in diverse physiological and pathological processes, such as neurotransmission, muscle contraction, and sexual function (Forstermann et al. 1998). The NOS1 gene is expressed in a tissue- and cell-specific, as well as developmentally regulated, fashion (Lee et al. 1997). The genomic organization of the human NOS1 gene is similar to that of the Pcdh clusters, containing variable and constant regions. Nine exon 1 variants have been reported thus far (Wang et al. 1999). We identified three additional human NOS1 variable exons by comparing cDNA and EST sequences with genomic DNA sequences. These 12 variable exons are organized in a tandem array spanning a region of ∼120 kb of genomic DNA. Downstream from the variable exon tandem array, there are 28 constant exons spanning a region of another 120 kb of genomic DNA (Fig. 6B). Each variable exon is separately spliced to the first constant exon to generate diverse NOS1 mRNAs.
In contrast to variable exons of the Pcdh, UGT1, and IGnT clusters, and similar to the plectin cluster, the variable exons of human NOS1 have different lengths and do not display sequence similarity. Moreover, all of the variable exons are noncoding. Thus, these exons only generate transcript diversity; they will not generate different NOS1 protein isoforms. In addition, we cannot identify a conserved DNA sequence motif upstream of each variable exon. Nevertheless, the 5′-untranslated region of NOS1 mRNAs, which contain distinct variable exon sequences, has specific effects on the translational efficiency of NOS1 mRNAs (Wang et al. 1999). We analyzed the CpG dinucleotide distribution in the human NOS1 variable region genomic DNA sequences by using the CpGplot program. We found that the positions of variable exons 1, 5, and 12 correspond to a CpG island, and that variable exons 2-4 and exons 6-11 are clustered in two distinct CpG islands (Supplemental Fig. S8). Previous studies have demonstrated that variable exons 6 and 7 are transcribed from separate promoters (Xie et al. 1995). These exons and their promoters do not display significant sequence similarity. Therefore, they do not appear to be paralogous. Taken together, we conclude that there are at least six distinct promoters in the human NOS1 variable region, and that promoter activation and alternative cis-splicing may establish the tissue-specific expression patterns of diverse NOS1 mRNAs.
Interestingly, although the organization of the NOS1 constant region is highly conserved between mice and rats, we can only identify one mouse NOS1 variable exon in the EST database and three putative NOS1 variable exons in the mouse genomic sequences (draft mouse genomic sequences of the February 2003 freeze). These four variable exons are weakly conserved between mice and humans. All of the other human NOS1 variable exons do not have mouse orthologs. In addition, we can only identify one variable exon in the rat NOS1 genomic region (the rat January 2003 freeze; http://genome.ucsc.edu/), although three rat NOS1 variable exons have been reported previously (Lee et al. 1997). These observations indicate that the organization of the NOS1 variable exons is different in these mammalian genomes and that the NOS1 gene may be differentially expressed in different species.
The Glucocorticoid Receptor Gene
The glucocorticoid receptor gene (GR) belongs to the nuclear receptor superfamily. It encodes a transcription factor that regulates genes involved in a wide variety of physiological and pathophysiological processes, such as glucose homeostasis, bone turnover, lung maturation, and inflammation. The transcripts of GR genes display very complex expression patterns in a tissue- and cell-specific, as well as an environment-dependent, manner (Yudt and Cidlowski 2002). The genomic organization of the rat GR gene contains 12 variable exons and eight constant exons (Fig. 6C; McCormick et al. 2000). These variable exons do not display any sequence similarity to each other, and their upstream regions do not share a conserved motif. However, there is a consensus 5′ splice site immediately downstream from each variable exon. Thus, each variable exon is separately spliced to the first constant exon to generate diverse GR mRNAs (McCormick et al. 2000).
We mapped all variable exons to the rat draft genomic sequence except variable exons 2 and 3, which may be located within several sequence gaps between exons 1 and 4. Alignment of the variable region of rat GR genes with the corresponding human and mouse genomic DNA sequences revealed that many, but not all, of the variable exons are conserved (Supplemental Fig. S9). The sequence similarity of the GR variable exons between mice and rats is >75%, but is lower when either is compared with humans. We noted that the position of variable exon 1 corresponds to the location of a CpG island (data not shown). Interestingly, variable exons 4 to 12 are clustered within a 2-kb CpG-rich region. It is very likely that there are multiple promoters in the variable region of the GR gene. Indeed, the human GR gene has been shown to have at least three promoters, corresponding to variable exons 1, 6, and 10, whereas the mouse GR gene has four promoters, corresponding to variable exons 1, 5, 6, and 11 (Chen et al. 1999; Breslin et al. 2001). Like the NOS1 variable exons, all variable exons of the GR gene are noncoding. Therefore, the diverse GR mRNAs encode identical proteins. It has been shown, however, that the diverse GR mRNAs are expressed in a tissue-specific fashion in the hippocampus, liver, and thymus (McCormick et al. 2000). These tissue-specific expression patterns may be regulated by a combination of differential variable promoter activation and alternative cis-splicing.
DISCUSSION
Multiple Variable First Exons Generate Protein/Transcript Diversity
The Pcdh clusters have an unusual genomic organization in which multiple variable exons are each spliced to a common set of downstream constant exons. To determine whether there are additional mammalian genes or gene clusters that display a similar genomic organization, we conducted a genome-wide search in human, mouse, and rat genomic sequences, and found more than 3000 genes containing multiple first exons. We described in detail the organization of several examples, including UGT1, NOS1, plectin, and GR genes. In each case, more than a dozen variable exons are organized in a tandem array followed by a common set of downstream constant exons. This genomic organization provides genetic multiplicity for generating diversity. In the cases of Pcdh, UGT1, and IGnT genes, the variable exons are very similar to each other and encode diverse polypeptides. In the cases of NOS1 and GR genes, the variable exons are noncoding. These genes only display mRNA transcript diversity. Thus, these multiple variable exons provide purely genetic diversity for gene regulation. The plectin gene, however, exemplifies an interesting intermediate type. Some of its variable exons are coding, but others are noncoding. However, similar to the NOS1 and GR genes, the variable exons of the plectin gene do not display sequence similarity to each other.
Pcdh proteins are proposed to play an important role in specifying diverse neuronal connections in the CNS. Both detoxification and neuronal connections require tremendous diversity. A tandem array of Pcdh and UGT1 variable exons, encoding highly similar but distinct polypeptides, provides a means for generating such diversity. Both Pcdh and UGT1 proteins are typical type I transmembrane proteins, that is, they have a single transmembrane segment located close to the C terminus. The oligomer structure of UGT1 proteins may provide diverse substrate specificity (Iyanagi et al. 1998). Similarly, Pcdh proteins may provide nearly unlimited diversity for specific neuronal connections by forming combinatorial oligomers. In the case of Pcdh genes, each variable exon transcript without constant exons may encode a short-form Pcdh protein (Wu and Maniatis 1999) with a characteristic Pcdh structure. In the case of UGT1 genes, each variable exon only encodes the N-terminal half of the protein, without a transmembrane segment. In contrast, the constant region encodes the C-terminal half of the protein, including the transmembrane segment (Iyanagi et al. 1998). Thus, a single variable exon itself does not appear to encode a functional UGT1 protein. In addition, by using the BLAST program we searched the human, mouse, and rat EST and cDNA databases with intron sequences immediately downstream from each variable exon, but could not find any transcripts that contain a variable exon and its immediately downstream intron sequences. A similar search for the Pcdh clusters reveals many such transcripts. Therefore, unlike Pcdh clusters, the UGT1 cluster does not have variable-only forms of diversity.
Evolutionary Origin of the Variable and Constant Genomic Organization
The variable exons arrayed in tandem raise interesting implications regarding their evolution. The three closely linked Pcdh clusters appear to result from gene duplication followed by diversification (Wu and Maniatis 1999). The organization of both the Pcdh α and γ constant regions is identical and is conserved among humans, mice, and rats (Wu et al. 2001; data not shown), whereas the Pcdh β cluster does not have a constant region. The three clusters may be duplicated from a common ancestral gene. The sequences of the constant exon of the Pcdh α and γ clusters diverged to encode different cytoplasmic domains, potentially functioning in distinct intracellular signal transduction pathways, whereas the Pcdh β cluster lost its constant region during evolution. The extensive tandem array of variable exons within each cluster appears to result from duplications of ancestral variable exons within each cluster. For the UGT1 cluster, the variable exons of the bilirubin and phenol groups appear to be duplicated separately from two ancestral variable exons (Fig. 1), which are themselves duplicated from a single ancestral variable exon. Some variable exons, for example, mouse UGT1A12 and A13, appear to result from species-specific expansion of a variable exon. On the other hand, the duplication of the IGnT variable exons occurred before the separation of the human, mouse, and rat lineages, because the three variable exons display strictly orthologous relationships (Fig. 4).
For variable exons that display close similarities, the unit of duplication appears to include both the coding sequence and its upstream regulatory region. For Pcdh, UGT1, and IGnT, within a given cluster, the coding regions of multiple variable exons not only share sequence similarity, but also have similar lengths. In fact, there are conserved sequence motifs located upstream from each variable exon of these clusters (Wu et al. 2001; Fig. 4; Supplemental Fig. S3). However, most of the other regulatory sequences appear to have been diversified. For example, the sequences flanking the highly conserved motifs are different among distinct variable exons. This diversification provides a genetic basis for cell- or tissue-specific gene expression in response to developmental programs or environmental stimuli. In a few cases, however, for example, Pcdh γb6 and γb7, the promoter sequences are highly conserved. These could be the result of either selection pressure for similar gene expression patterns or simply not enough time for diversification after duplication.
Interestingly, in all cases the transcriptional direction for variable exons is the same, that is, toward the constant region. Even in the cases of pseudogenes or relics, the remnant codons are always on the same strand. This may reflect an exon duplication mechanism that can only result in a parallel tandem array. In addition, this may also reflect that the parallel arrangement of variable exons and their promoters ensures that the functional mRNAs can be generated by cis-splicing mechanisms.
Several models have been proposed to explain the preservation of duplicated genes. The classic model for the evolution of duplicate genes posited that paralogous genes arise by acquiring new adaptive functions (Ohno 1970). Recently, the duplication-degeneration-complementation (DDC) model hypothesized that paralogous genes accomplish complementary subfunctions of the ancestral gene (Force et al. 1999). The organization of Pcdh, UGT1, and IGnT is unusual in that only the first exon was duplicated. Nevertheless, this organization supports aspects of both models. The vast expansion of variable exons by arrayed arrangements in the UGT1 and Pcdh clusters may reflect purifying selection for the enormous diversity required for detoxification or neuronal connectivity. On the other hand, the prototypical UGT1 or Pcdh genes may have been expressed ubiquitously and had much broader functions.
For variable exons that do not display sequence similarity, such as in the plectin, NOS1, and GR genes, we can only speculate about their evolutionary origin. One scenario is that their variable first exons are the result of exon duplication, with high selection pressure to quickly diversify after duplication for tissue-specific and development-dependent regulation. Another possibility is that tandem variable first exons are the result of fortuitous transcriptions. Initially, there may have been many transcription start sites for each of these genes. During evolution, these genes may have been under selection pressure for diverse tissue-specific promoter activation. Selective splicing that joins a cap-proximal 5′ splice site to the 3′ splice site of the first constant exon removes all the intervening sequences. Hence, specific promoter activation and cap-proximal cis-splicing may have evolved for these multiple regulatory strategies. This scenario, and the duplication of Pcdh and UGT1 variable exons, would be convergent evolution for tandem arrays of multiple first exons. Whatever the origin of tandem variable first exons, it is very likely that this organization provides a genetic mechanism for the diverse gene regulation required for complex mammalian development and adaptation.
Implication of the Variable and Constant Genomic Organization for Gene Regulatory Complexity
Many mammalian genes are expressed in a cell- or tissue-specific manner. This is especially prominent in the nervous system. The nervous system is extremely heterogeneous in that a large repertoire of different neuronal cell types makes specific connections, and these vast numbers of cell types are defined by specific gene expression. How is this specific expression achieved? The variable and constant genomic organization can afford the specific expression of diverse mRNAs that may translate into a variety of protein isoforms with distinct functions. In addition, a repertoire of distinct 5′ sequences may influence posttranscriptional gene regulation such as mRNA processing, export, stability, and translation. For example, different 5′-UTR sequences of the NOS1 mRNAs have been implicated in the regulation of their translation (Wang et al. 1999). A combination of differential promoter activation and alternative cis-splicing may provide a powerful means for regulating gene expression in response to a wide variety of intrinsic and extrinsic signals.
It is important to note that the organization of multiple variable first exons is very different from that of the single promoter with multiple transcription start sites. In the latter case, the multiple transcripts share common sequences at the 3′ portion of the first exon and use the same 5′ splice site to splice to the second exon. In the case of multiple variable first exons, different variable first exons do not share a common 5′ splice site, and each variable first exon is likely preceded by a distinct promoter.
The variable and constant region genomic organization provides a framework for differential and complex patterns of gene expression. The flexibility to generate complex patterns of gene expression is possible because the promoters preceding each variable exon are distinct, although the promoters are clearly related in the case of the Pcdh clusters. Different strengths of 5′ splice sites of variable exons may also contribute to the specific patterns of gene expression. This gene expression plasticity may play an important role in the molecular and cellular adaptation to environmental cues.
How Many Variable and Constant Genomic Organizations Exist in Mammalian Genomes?
Large-scale genomic sequencing has outpaced our ability to annotate sequences. To date, sequencing cDNA clones is still the most reliable method to identify genes. Although EST sequencing has provided valuable information about genome annotation, it usually does not provide enough information about the 5′-end of genes because most ESTs are derived from the 3′-end. Recently, the Institute of Physical and Chemical Research (Japan; RIKEN) has sequenced 60,770 full-length mouse cDNA clones (http://fantom2.gsc.riken.go.jp/; Okazaki et al. 2002). Even this large cDNA collection provides limited information about multiple variable exons.
It will be interesting to see how many genes containing a tandem array of variable exons exist in a single mammalian genome. Most of these genes are likely expressed in a tissue- or cell-specific manner. Furthermore, they may be expressed at very low levels. Therefore, it is extremely difficult to identify genes with a tandem variable exon array. In light of our identification of gene clusters containing a tandem array of variable exons, undoubtedly many other genes will be found to contain multiple variable first exons in the future.
METHODS
Identification of Gene Clusters That Have Variable and Constant Genomic Organization
To identify genes that have a genomic organization similar to the Pcdh clusters, we first collected all human, mouse, and rat transcripts from various sources, including MGC, Refseq, RIKEN, and GenBank. Next we identified the genomic regions spanning the transcripts by mapping them onto the public genomes (http://genome.ucsc.edu/; human April 2003 freeze, mouse February 2003 freeze, and rat November 2002 freeze) using the BLAT program (Kent 2002). A transcript was considered mapped to the genome if it shares >95% sequence identity with a genomic region over 95% of the transcript's length. If a transcript maps to multiple regions on the genome, the region it matches best is retained and all others are discarded.
Because EST sequences are a rich source of terminal exons, we also retrieved ESTs for each species from dbEST and mapped them onto the respective genomes. Because of their poor sequence quality and possible contamination, ESTs were first cleaned by removing vector sequences, low-complexity regions, and repeats. The parameters for mapping ESTs onto genomes were also relaxed to >95% sequence identity, with the genomic region over 90% length of the EST, while allowing up to 15 bp mismatches at the 5′-end.
Once all mRNAs and ESTs were mapped onto the genome, we determined the exon/intron structure by comparing a transcript sequence with its corresponding genomic region sequence using the Sim4 program (Florea et al. 1998). We first put all transcripts whose second exons have the same coordinates into a group, that is, they have a common second exon. We then identified all the groups whose transcripts have more than one distinct (i.e., nonoverlapping) first exon. We filtered out members of immunoglobulin and T-cell receptor gene clusters. If the variable exons were located close to each other, we then searched for other novel variable exons in the same genomic region.
We annotated the UGT1 cluster by using the BLAST program (Altschul et al. 1997) and the GCG sequence analysis package. The UGT1 genomic sequences were downloaded from the UCSC database (http://genome.ucsc.edu/). The potential coding sequences were aligned by using the multiple sequence alignment program Pileup. The 5′ splice sites were identified manually by inspecting the alignment and were confirmed by comparing the genomic sequences to the corresponding cDNA sequences. The putative translation start codon was determined by inspecting the translated signal peptide sequences.
The genomic organization of mouse plectin was determined by comparing the cDNA sequences (AF180006-AF180023; Fuchs et al. 1999) with the mouse draft genomic sequences (Waterston et al. 2002; http://genome.ucsc.edu/). The organization of human and rat plectin genes was determined by a sequence comparison between species. The genomic organization of the human NOS1 gene was determined by comparing the cDNA sequences (AF446131-AF446140; AF049712-AF049720; U11422; NM000620) and the human genomic sequences (Lander et al. 2001; http://www.ncbi.nlm.nih.gov/). Interestingly, the junction between variable exons 10 and 11 is a dual acceptor/donor splice site. The only other known example of such a combined splice site is in the IRF3 gene (Karpova et al. 2000). The genomic organization of the rat GR constant region was determined by comparing the cDNA sequences (NM_012576) with the rat draft genomic sequences (http://www.hgsc.bcm.tmc.edu/) using the BLAT program (Kent 2002). The 5′-region organization was previously reported to contain 11 variable exons (McCormick et al. 2000), and we identified an additional variable exon by an EST database search. The organization of mouse and human GR genes was determined by sequence comparison between species.
Cloning and Expression Profiling of Mouse UGT1 Genes
Nine forward and one reverse PCR primers for the mouse UGT1 cluster were designed based on the genomic DNA sequences (AC087780 and AC087801) identified by iterative BLAST searches. The nine forward primer sequences are: AGCTTGAG TAGATCTCTGGCAG (UGT1A1), TACAGCCCTCAGCATTCAG GAG (UGT1A2), TGAGGAAACCTGACTCTTCTGC (UGT1A5), CAGTCACTGAGTGTGTGTGTGG (UGT1A7), GTCGGTGCT GAGTTTGTAAGTAG (UGT1A9), ATTGCCTTAGGCTG CACTTCTG (UGT1A10), ATTCTCCTGGGCTGTGTTTCTC (UGT1A11), GACAGGATTACTCTGGGCTCTG (UGT1A12), and GACTGGATTGCTCTGGACTTTG (UGT1A13). The reverse primer is CCACTTCTCAATGGGTCTTGGA. The mouse UGT1 cDNAs were amplified from total RNA by reverse-transcriptase polymerase chain reaction (RT-PCR). The first-strand cDNA was synthesized from total RNA by using Superscript reverse transcriptase (Invitrogen) with either a random primer or a UGT1-specific primer. The subsequent PCR reactions were performed using each forward primer paired with the same reverse primer. The total RNA was isolated from a wide variety of mouse tissues by Trizol (Invitrogen) according to the manufacturer's instructions. The PCR products were cloned and sequenced for both DNA strands by using fluorescent dye terminators on an automated DNA sequencer. We cloned all mouse UGT1 genes except UGT1A11. We noted that the sequences of mouse UGT1A11 from the whole genome shotgun (WGS) method have multiple stop codons, in contrast to the UGT1A11 sequences from the BAC clone (AC087801), which do not contain any stop codons. We performed PCR experiments to amplify the mouse UGT1A11 genomic DNA. By sequencing the cloned PCR products, we found that there were no nonsense mutations in the genomic sequences. However, we were unable to detect UGT1A11 expression in a wide variety of tissues from the same mouse by RT-PCR method. Therefore, the UGT1A11 may be a pseudogene.
The possibility of DNA contamination of total RNA was excluded by RT-PCR amplification of mouse β-actin with primers TGTTACCAACTGGGACGACATG and TCTTGATCTTCATGGT GCTAGG spanning exon 4-intron 4-exon 5-intron 5-exon 6 junctions. The product is 761 bp when a cDNA template is present, and will be 1311 bp if there is genomic DNA contamination.
We detected expression profiles of UGT1 genes in mouse tissues by using semiquantitative DRT-PCR with isoform-specific primers. Briefly, the cDNAs were synthesized in a 40-μL reaction with 80 picomoles of random 10-mer oligonucleotides and 2 μg of total RNAs by using Reverse Transcriptase (Invitrogen) according to the manufacturer's protocol. Then, 2 μL of the resulting cDNAs was used for one PCR reaction with each isoform-specific variable region forward primer and a constant region reverse primer. A pair of mouse β-actin primers was added to each PCR reaction after six or 10 cycles and continued for a total of 35 cycles.
Phylogenetic Analysis of UGT1 and IGnT Genes
The cloned or predicted UGT1 and IGnT variable-region coding sequences were translated, and the resulting polypeptides were aligned using CLUSTAL W (version 1.82) with default parameters (Thompson et al. 1994). A phylogenetic tree was reconstructed by using the Neighbor-Joining algorithm in the CLUSTAL W package. Gaps in the alignment were ignored in the tree-building process. The robustness of the tree partitions was evaluated by bootstrap analysis.
Acknowledgments
We thank M. Capecchi and T. Maniatis for advice, M. Hobbs, C. Thummel, and R. Weiss for critical reading of the manuscript, members of the Wu lab for discussion, and three anonymous reviewers for helpful suggestions. Q.W. is a recipient of the Basil O'Connor Award from the March of Dimes. Supported by a grant from the American Cancer Society (to Q.W.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1225204. Article published online before print in December 2003.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data described have been submitted to the GenBank/EMBL/DDDJ data libraryunder accession nos. AY227194-AY227201, AY435128-AY435153 and AY480022-AY480051.]
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antequera, F. and Bird, A. 1993. Number of CpG islands and genes in human and mouse. Proc. Natl. Acad. Sci. 90: 11995-11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslin, M.B., Geng, C.D., and Vedeckis, W.V. 2001. Multiple promoters exist in the human GR gene, one of which is activated by glucocorticoids. Mol. Endocrinol. 15: 1381-1395. [DOI] [PubMed] [Google Scholar]
- Chen, F., Watson, C.S., and Gametchu, B. 1999. Multiple glucocorticoid receptor transcripts in membrane glucocorticoid receptor-enriched S-49 mouse lymphoma cells. J. Cell. Biochem. 74: 418-429. [PubMed] [Google Scholar]
- Dubchak, I., Brudno, M., Loots, G.G., Pachter, L., Mayor, C., Rubin, E.M., and Frazer, K.A. 2000. Active conservation of noncoding sequences revealed by three-way species comparisons. Genome Res. 10: 1304-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emi, Y., Ikushiro, S., and Iyanagi, T. 1995. Drug-responsive and tissue-specific alternative expression of multiple first exons in rat UDP-glucuronosyltransferase family 1 (UGT1) gene complex. J. Biochem. (Tokyo) 117: 392-399. [DOI] [PubMed] [Google Scholar]
- ———. 1996. Xenobiotic responsive element-mediated transcriptional activation in the UDP-glucuronosyltransferase family 1 gene complex. J. Biol. Chem. 271: 3952-3958. [DOI] [PubMed] [Google Scholar]
- Florea, L., Hartzell, G., Zhang, Z., Rubin, G.M., and Miller, W. 1998. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 8: 967-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force, A., Lynch, M., Pickett, F.B., Amores, A., Yan, Y.L., and Postlethwait, J. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstermann, U., Boissel, J.P., and Kleinert, H. 1998. Expressional control of the `constitutive' isoforms of nitric oxide synthase (NOS I and NOS III). FASEB J. 12: 773-790. [PubMed] [Google Scholar]
- Fuchs, P., Zorer, M., Rezniczek, G.A., Spazierer, D., Oehler, S., Castanon, M.J., Hauptmann, R., and Wiche, G. 1999. Unusual 5′ transcript complexity of plectin isoforms: Novel tissue-specific exons modulate actin binding activity. Hum. Mol. Genet. 8: 2461-2472. [DOI] [PubMed] [Google Scholar]
- Gong, Q.H., Cho, J.W., Huang, T., Potter, C., Gholami, N., Basu, N.K., Kubota, S., Carvalho, S., Pennington, M.W., Owens, I.S., et al. 2001. Thirteen UDP glucuronosyltransferase genes are encoded at the human UGT1 gene complex locus. Pharmacogenetics 11: 357-368. [DOI] [PubMed] [Google Scholar]
- Inaba, N., Hiruma, T., Togayachi, A., Iwasaki, H., Wang, X.H., Furukawa, Y., Sumi, R., Kudo, T., Fujimura, K., Iwai, T., et al. 2003. A novel I-branching β-1,6-N-acetylglucosaminyltransferase involved in human blood group I antigen expression. Blood 101: 2870-2876. [DOI] [PubMed] [Google Scholar]
- Iyanagi, T., Emi, Y., and Ikushiro, S. 1998. Biochemical and molecular aspects of genetic disorders of bilirubin metabolism. Biochim. Biophys. Acta 1407: 173-184. [DOI] [PubMed] [Google Scholar]
- Janda, L., Damborsky, J., Rezniczek, G.A., and Wiche, G. 2001. Plectin repeats and modules: Strategic cysteines and their presumed impact on cytolinker functions. Bioessays 23: 1064-1069. [DOI] [PubMed] [Google Scholar]
- Karpova, A.Y., Howley, P.M., and Ronco, L.V. 2000. Dual utilization of an acceptor/donor splice site governs the alternative splicing of the IRF-3 gene. Genes & Dev. 14: 2813-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT—The BLAST-like alignment tool. Genome Res. 12: 656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohmura, N., Senzaki, K., Hamada, S., Kai, N., Yasuda, R., Watanabe, M., Ishii, H., Yasuda, M., Mishina, M., and Yagi, T. 1998. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron 20: 1137-1151. [DOI] [PubMed] [Google Scholar]
- Kong, A.N., Ma, M., Tao, D., and Yang, L. 1993. Molecular cloning of two cDNAs encoding the mouse bilirubin/phenol family of UDP-glucuronosyltransferases (mUGTBr/p). Pharm. Res. 10: 461-465. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Larsen, F., Gundersen, G., Lopez, R., and Prydz, H. 1992. CpG islands as gene markers in the human genome. Genomics 13: 1095-1107. [DOI] [PubMed] [Google Scholar]
- Lawrence, C.E., Altschul, S.F., Boguski, M.S., Liu, J.S., Neuwald, A.F., and Wootton, J.C. 1993. Detecting subtle sequence signals: A Gibbs sampling strategy for multiple alignment. Science 262: 208-214. [DOI] [PubMed] [Google Scholar]
- Lee, M.A., Cai, L., Hubner, N., Lee, Y.A., and Lindpaintner, K. 1997. Tissue- and development-specific expression of multiple alternatively spliced transcripts of rat neuronal nitric oxide synthase. J. Clin. Invest. 100: 1507-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick, J.A., Lyons, V., Jacobson, M.D., Noble, J., Diorio, J., Nyirenda, M., Weaver, S., Ester, W., Yau, J.L., Meaney, M.J., et al. 2000. 5′-Heterogeneity of glucocorticoid receptor messenger RNA is tissue specific: Differential regulation of variant transcripts by early-life events. Mol. Endocrinol. 14: 506-517. [DOI] [PubMed] [Google Scholar]
- Ohno, S. 1970. Evolution by gene duplication. Springer Verlag, Heidelberg, Germany.
- Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563-573. [DOI] [PubMed] [Google Scholar]
- Schmucker, D., Clemens, J.C., Shu, H., Worby, C.A., Xiao, J., Muda, M., Dixon, J.E., and Zipursky, S.L. 2000. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101: 671-684. [DOI] [PubMed] [Google Scholar]
- Tasic, B., Nabholz, C.E., Baldwin, K.K., Kim, Y., Rueckert, E.H., Ribich, S.A., Cramer, P., Wu, Q., Axel, R., and Maniatis, T. 2002. Promoter choice determines splice site selection in protocadherin α and γ pre-mRNA splicing. Mol. Cell 10: 21-33. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukey, R.H. and Strassburg, C.P. 2000. Human UDP-glucuronosyltransferases: Metabolism, expression, and disease. Annu. Rev. Pharmacol. Toxicol. 40: 581-616. [DOI] [PubMed] [Google Scholar]
- Wang, X., Su, H., and Bradley, A. 2002a. Molecular mechanisms governing Pcdh-γ gene expression: Evidence for a multiple promoter and cis-alternative splicing model. Genes & Dev. 16: 1890-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Newton, D.C., Robb, G.B., Kau, C.L., Miller, T.L., Cheung, A.H., Hall, A.V., VanDamme, S., Wilcox, J.N., and Marsden, P.A. 1999. RNA diversity has profound effects on the translation of neuronal nitric oxide synthase. Proc. Natl. Acad. Sci. 96: 12150-12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Weiner, J.A., Levi, S., Craig, A.M., Bradley, A., and Sanes, J.R. 2002b. γ protocadherins are required for survival of spinal interneurons. Neuron 36: 843-854. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Wu, Q. and Maniatis, T. 1999. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell 97: 779-790. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Large exons encoding multiple ectodomains are a characteristic feature of protocadherin genes. Proc. Natl. Acad. Sci. 97: 3124-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Q., Zhang, T., Cheng, J.F., Kim, Y., Grimwood, J., Schmutz, J., Dickson, M., Noonan, J.P., Zhang, M.Q., Myers, R.M., et al. 2001. Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res. 11: 389-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, J., Roddy, P., Rife, T.K., Murad, F., and Young, A.P. 1995. Two closely linked but separable promoters for human neuronal nitric oxide synthase gene transcription. Proc. Natl. Acad. Sci. 92: 1242-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, L.C., Twu, Y.C., Chou, M.L., Reid, M.E., Gray, A.R., Moulds, J.M., Chang, C.Y., and Lin, M. 2003. The molecular genetics of the human I locus and molecular background explain the partial association of the adult i phenotype with congenital cataracts. Blood 101: 2081-2088. [DOI] [PubMed] [Google Scholar]
- Yudt, M.R. and Cidlowski, J.A. 2002. The glucocorticoid receptor: Coding a diversity of proteins and responses through a single gene. Mol. Endocrinol. 16: 1719-1726. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://genome.ucsc.edu; UCSC Genome Bioinformatics.
- http://www.ncbi.nlm.nih.gov; NCBI.
- http://www.hgsc.bcm.tmc.edu; Baylor College of Medicine HGSC.
- http://fantom2.gsc.riken.go.jp; RIKEN FANTOM2.
- http://bioweb.pasteur.fr/seqanal/interfaces/cpgplot.html; CpG plot.
- http://bayesweb.wadsworth.org/gibbs/gibbs.html; Gibbs sampler.