Abstract
Epithelial-to-mesenchymal transition (EMT) is fundamental to both embryogenesis and tumor metastasis. The Notch intercellular signaling pathway regulates cell fate determination throughout metazoan evolution, and overexpression of activating alleles is oncogenic in mammals. Here we demonstrate that Notch activity promotes EMT during both cardiac development and oncogenic transformation via transcriptional induction of the Snail repressor, a potent and evolutionarily conserved mediator of EMT in many tissues and tumor types. In the embryonic heart, Notch functions via lateral induction to promote a selective transforming growth factor-β (TGFβ)-mediated EMT that leads to cellularization of developing cardiac valvular primordia. Embryos that lack Notch signaling elements exhibit severely attenuated cardiac snail expression, abnormal maintenance of intercellular endocardial adhesion complexes, and abortive endocardial EMT in vivo and in vitro. Accordingly, transient ectopic expression of activated Notch1 (N1IC) in zebrafish embryos leads to hypercellular cardiac valves, whereas Notch inhibition prevents valve development. Overexpression of N1IC in immortalized endothelial cells in vitro induces EMT accompanied by oncogenic transformation, with corresponding induction of snail and repression of VE-cadherin expression. Notch is expressed in embryonic regions where EMT occurs, suggesting an intimate and fundamental role for Notch, which may be reactivated during tumor metastasis.
Keywords: Notch, endocardium, lateral induction, EMT, snail, TGFβ
Epithelial-to-mesenchymal transition (EMT) is fundamental to both normal development and the progression of malignant epithelial tumors (for review, see Thiery 2002). During EMT, epithelial cells undergo sweeping alterations in gene expression to lose apical/basolateral polarity, sever intercellular adhesive junctions, degrade basement membrane components, and become migratory. Several signaling pathways seem to be common to EMT regulation during both development and tumor progression, leading to the notion that developmentally regulated EMT and tumor metastasis are under the control of common molecular mechanisms (Thiery 2002), and raising the hypothesis that tumor metastasis could be regarded as a reactivation of at least some aspects of the embryonic program of EMT.
The snail family of Zinc-finger-containing transcriptional repressors is believed to play a pivotal role in the process of EMT (Nieto 2002). Expression of various snail family members has been tightly associated with cells undergoing both metastatic and developmental EMT (Nieto et al. 1992; Romano and Runyan 2000). One important target of Snail repression is the E-cadherin gene, the primary cadherin that is responsible for homotypic adhesion between members of an epithelial sheet (Batlle et al. 2000; Cano et al. 2000).
A classical example of developmentally regulated EMT occurs during the initial stages of cardiac morphogenesis. At embryonic day 8.5 (E8.5), the mouse heart tube is composed of an outer myocardial layer lined by a monolayer of specialized endothelium, the endocardium. Both are separated by a thick extracellular matrix, the cardiac jelly, secreted primarily by the myocardial cells (Eisenberg and Markwald 1995). At about E9.5, and in response to regionalized myocardial signals, a subset of endocardial cells that overlies the atrio-ventricular (AV) canal and outflow tract regions undergoes EMT (Eisenberg and Markwald 1995). These cells delaminate from the endocardial sheet and invade the cardiac jelly to form the endocardial cushions, which will contribute to cardiac valve development and heart septation. This process likely involves (1) an initial activation of endocardial cells that endows them with the ability to transform; (2) regulation of cell-to-cell adhesion that accompanies the acquisition of a migratory phenotype; and (3) selective recruitment into the cardiac cushions. Numerous functional studies in the chick and mouse indicate that myocardial paracrine and endocardial autocrine production of transforming growth factor-β2 (TGFβ2) and TGFβ3 are required for endocardial cushion formation (see, for example, Camenisch et al. 2002a). Similarly, analysis of NF1 null murine embryos and manipulation of cardiac explants derived from them indicate an important role for Ras signaling during endocardial cushion recruitment (Lakkis and Epstein 1998). Recent data from zebrafish suggest that the evolutionarily conserved Notch signaling pathway may have a role in endocardial EMT, as expression of the receptor Notch 1b localizes onto the endocardium of the presumptive AV valve region prior to and during the stages when EMT occurs (Walsh and Stainier 2001; Hurlstone et al. 2003).
Notch regulates cell fate specification through local cell interactions in invertebrate and vertebrate organisms. Vertebrates have multiple homologs of Notch that are expressed at many sites in the developing vertebrate body. The Notch genes encode transmembrane receptors containing a large extracellular domain composed of a variable number of epidermal growth factor (EGF)-like repeats and an intracellular domain (NIC) with six ankyrin/cdc 10 motifs and a number of nuclear localization signals. The Notch receptors interact at the extracellular side with different membrane-bound ligands of the Delta and Serrate/Jagged families. These ligands contain an extracellular domain with EGF-like repeats and a conserved DSL (Delta/Serrate/LAG2) domain (for review, see Artavanis-Tsakonas et al. 1999). The signal induced by ligand binding is delivered to the nucleus in a process involving proteolytic processing of the receptor by a γ-secretase activity (Kopan 2002) and nuclear translocation of NIC (Kopan 2002). In the nucleus, NIC binds to the transcription factor RBPJK/CBF1/Su(H), forming a complex that activates the expression of target genes. Notch activation during invertebrate and vertebrate neurogenesis has been associated with the maintenance of an uncommitted state (Lewis 1998; Artavanis-Tsakonas et al. 1999), as the major targets of the pathway are genes of the hairy/Enhancer of split (Hes) family, that encode basic helix-loop-helix (bHLH) proteins that repress transcription of pro-neural bHLH genes required for neurogenesis (Artavanis-Tsakonas et al. 1999). Notch signaling also can positively regulate cell fate choices, as it is able to promote the glial fate in rat neural stem cells (Morrison et al. 2000) and to regulate the T/B cell fate choice in lymphoid progenitors (Robey 1999). Truncated versions of the Notch receptor are considered constitutively active (Kopan et al. 1994), and are able to inhibit cellular differentiation in vitro or to bias cell lineage decisions in vivo (Robey 1999). Overexpression of these Notch activating alleles is oncogenic in mammals (reviewed in Artavanis-Tsakonas et al. 1999). In vertebrates, Notch has been shown to be involved in a variety of developmental processes such as neurogenesis, somitogenesis, and angiogenesis (for reviews, see Lewis 1998; Pourquie 2000; Gridley 2001). In addition to the zebrafish data (Walsh and Stainier 2001; Hurlstone et al. 2003), targeted mutations in both murine Notch1 (Swiatek et al. 1994) and RBPJk (Oka et al. 1995) produce severe pericardial edema, suggesting that Notch may play a role in murine cardiac development.
In this study we have addressed the role of Notch in cardiac EMT. We find that Notch acts via lateral induction in the endocardium, and is crucial for endocardial EMT. By the analysis of Notch1 and RBPJk mutant embryos, we demonstrate the absolute requirement for Notch in the promotion of EMT during cardiac valve development, and identify the absence of snail expression and the abnormal maintenance of endocardial VE-cadherin expression. In vitro, cardiac explants of Notch mutants or wild-type embryos treated with the γ-secretase inhibitor N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT; Dovey et al. 2001) show severely impaired EMT. Consistently, transient ectopic expression of activated Notch 1 (N1IC) in zebrafish embryos leads to hypertrophic cardiac valves and DAPT treatment inhibits valve formation. Overexpression of N1IC in immortalized endothelial cells in vitro induces snail, yielding attenuated VE-cadherin expression, loss of contact inhibition, and EMT accompanied by oncogenic transformation. We propose that promotion of EMT may be a general role of Notch both in development and tumor progression.
Results
Notch is active in the embryonic endocardium
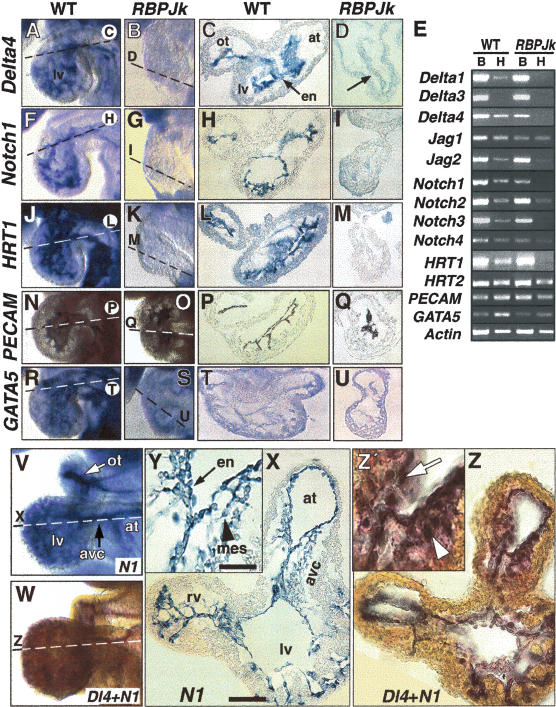
The cardiac expression pattern of Notch signaling components was analyzed in wild-type, RBPJk, and Notch1 mutant embryos at E8.5–E9.5, a period that coincides with endocardial maturation and EMT initiation. The ligand Delta4 is expressed at relatively high levels in the mouse endocardium at E8.5 (Fig. 1A,C; Krebs et al. 2000), and its expression is severely attenuated in RBPJk (Fig. 1B,D) and Notch1 mutants (data not shown), as RT-PCR analysis confirmed (Fig. 1E). The remaining Notch ligands are expressed at moderate levels in the heart from E9.0 onward, with reduced expression in RBPJk and Notch1 mutants (Fig. 1E and data not shown). The receptor Notch1 (Fig. 1F,H; Del Amo et al. 1992; Reaume et al. 1992), and to a lesser extent the three other Notch receptors, are expressed throughout the endocardium of wild-type hearts at E8.5, and their transcription is severely reduced in RBPJk mutants (Fig. 1G,I,E and data not shown). As evidence of Notch activity, we examined the transcription of two putative cardiovascular Notch targets, HRT1/HESR1/HEY1 and HRT2/gridlock/HEY2 (Nakagawa et al. 1999). HRT1 is expressed throughout the endocardium of E8.5–9.0 wild-type embryos (Fig. 1J,L,E), and is significantly reduced in RBPJk (Fig. 1K,M,E) and Notch1 mutants (data not shown). In contrast, myocardial ventricle-specific expression of HRT2 appears to be unaffected (data not shown and Fig. 1E). Thus, HRT1 is an endocardial target of Notch. Marker analysis revealed that CD31/PECAM (Drake and Fleming 2000) is strongly expressed throughout the cardiac endothelium of wild-type (Fig. 1N,P) and RBPJk mutants (Fig. 1O,Q) and, although GATA5 (Morrisey et al. 1997) appears to be slightly reduced by RT-PCR (Fig. 1E), in situ hybridization shows endocardial expression in both wild-type (Fig. 1R,T) and RBPJk mutants (Fig. 1S,U). These results suggest that Notch is not required for early stages of endocardial differentiation.
Figure 1.
Endocardial expression of Notch and marker genes. Whole-mount and transverse sections of E8.5–9.5 hearts. (A–D) Delta4 mRNA is expressed in the endocardium of wild-type embryos (A, C), and is severely reduced in RBPJk mutants (B, D). (E) Semiquantitative RT-PCR analysis in E8.5–9.0 wild-type and RBPJk mutants. (B) Body; (H) heart. (F–I) Notch1 is strongly expressed in the endocardium of wild-type embryos (F, H) and is strongly reduced in RBPJk mutants (G, I). (J–M) Endocardial HRT1 expression in wild-type (J, L) is strongly reduced in RBPJk mutants (K, M). (N–Q) PECAM1protein is present in the endocardium of wild-type (N, P) and RBPJk (O, Q) embryos. (R–U) GATA5 is expressed in the endocardium and myocardium of wild-type (R, T) and RBPJk (S, U) embryos. (V–Z) E9.5 wild-type hearts. (V, X, Y) Notch1 expression. (V, Z, Z′) Double in situ with Delta4 (red) and Notch1 (blue). (X, Y) Notch1 expression in endocardium (arrow in Y) and mesenchyme (arrowhead in Y); (Z, Z′) Delta4 and Notch1 coexpression. Coincident signal produces black/purple coloration. Dashed lines indicate plane of section. (en) Endocardium; (ot) outflow tract; (avc) atrio-ventricular canal; (at) atrium; (lv) left ventricle, (rv) right ventricle; (mes) mesenchyme. Note the defective looping of RBPJk mutants. Scale bar, 25 μm in M, O; 100 μm in L, N.
At E9.5, when endocardial EMT begins, Notch1 is expressed throughout the endocardium (Fig. 1V,X) and in cushion mesenchyme (Fig. 1Y), coinciding precisely with Delta4 expression, as double label in situ hybridization reveals (Fig. 1W,Z,Z′). Thus, specific Notch genes are expressed in an embryonic field comprising the endocardium and cushion mesenchyme. Unlike observations of Notch1b expression in zebrafish (Walsh and Stainier 2001), we have not detected any regionalization of Notch1 expression in the E9.5–E10.5 endocardium (data not shown).
Targeted mutations in Notch pathway elements prevent endocardial EMT
Serial sections of E8.5–E10.5 embryos were analyzed for cardiac developmental abnormalities in Notch pathway mutants. The diagrams in Figure 2, A and B, indicate the approximate plane of the sections shown in Figure 2, E–P; the different cardiac chambers appear color-coded. Chamber identity appears to be unaffected in RBPJk mutants, as indicated by the expression of Tbx5 (Fig. 2C,D; Bruneau et al. 1999). Sections were analyzed at planes that demonstrated good AV and outflow tract morphology for embryos of each genotype.
Figure 2.
Notch pathway mutants initiate endocardial EMT. (A–B) Diagrams showing a lateral view of E9.5 wild-type (A) and RBPJk mutant (B) hearts with the cardiac chambers depicted in color. The Roman numbers indicate the levels of the sections shown in panels E–P. (E–H) H&E-stained E9.5 heart transverse sections at the atrio-ventricular canal level. (E, F) Wild type. (F) Mesenchymal cells (arrowhead) invading the cushion. (G, H) RBPJk mutants collapsed endocardium (arrow). (I–P) TEM of E9.5 atrio-ventricular canal endocardia. (I) Wild-type rounded endocardial cells with processes directed toward the cardiac jelly (arrows), among flattened nonmigratory endothelial cells (arrowhead). (J, L) RBPJk mutant endocardial cells extending pseudopodia (arrow in L). Lipid vesicles (asterisks) are found in RBPJk mutant endocardial cells (K, L) and in wild-type migratory mesenchymal cells (N). Adherens junctions (arrowheads in O, P) are conspicuous in RBPJk mutants. Scale bar, 25 μm in F, H; 100 μm in E, G.
At E9.5, wild-type endocardial cells undergo EMT and can be visualized within the cardiac cushions (Fig. 2E,F, and data not shown). In contrast, RBPJk mutants exhibit a collapsed endocardium and lack mesenchymal cushion cells (Fig. 2G,H). A similar phenotype was observed in Notch1 embryos (not shown), suggesting that endocardial EMT is defective in Notch pathway mutants. Following elegant ultrastructural studies that define cellular features of endocardial EMT (Markwald et al. 1977), the endocardium of E9.5 RBPJk mutant embryos was examined by transmission electron microscopy (TEM). An early sign of EMT in the AV canal is the appearance of loosely associated and hypertrophied endocardial cells that extend long pseudopodia and filopodia (Fig. 2I) to initiate invasion of the cardiac jelly (Markwald et al. 1977). In RBPJk mutants, endocardial cells of this region remain tightly associated, although they extend pseudopodia toward the jelly (Fig. 2J,L). Wild-type transforming endocardial and migratory cells that invade the cushion (Fig. 2I,M,N) accumulate a high density of lipid vesicles (detail in Fig. 2N), as do RBPJk mutant endocardial cells (Fig. 2J,K,L). The junctional complexes that maintain the physical integrity of the endocardium (Fig. 2M,O) must be locally disassembled for EMT to occur in the AV canal and outflow tract regions; such complexes remain prominent throughout the entire endocardium of RBPJk mutants (Fig. 2P). These data indicate that cushion-associated endocardium in Notch pathway mutants shows features of activated, premigratory cells (Markwald et al. 1977), suggesting that Notch mediates a later step in the progression of EMT.
Notch pathway mutants demonstrate impaired expression of TGFβ signaling components
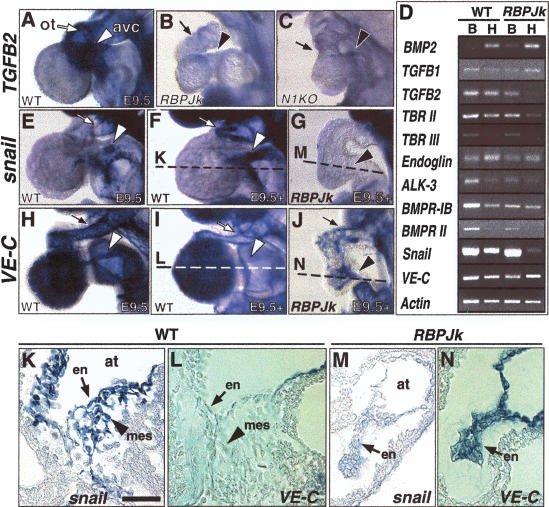
We next examined the expression of TGFβ family members that are thought to be part of the paracrine, myocardial signals that promote endocardial EMT (Nakajima et al. 2000). At E9.5, TGFβ2 expression is restricted to the outflow tract and AV canal myocardium of wild-type embryos (Fig. 3A and Dickson et al. 1993), but TGFβ2 transcription is severely reduced in a majority of RBPJk (9 of 12; Fig. 3B) and Notch1 (7 of 9; Fig. 3C) mutants. This effect is specific for TGFβ2. Thus, expression of BMP2, which is also restricted to the outflow tract and AV canal myocardium (Lyons et al. 1990), or endocardial TGFβ1 expression (Akhurst et al. 1990), remains unaffected (data not shown and Fig. 3D). Notch receptors are not expressed in the myocardium during these developmental stages and Notch1 and RBPJk mutants show normal expression of a variety of myocardial markers (data not shown), suggesting that myocardial differentiation is not impaired. Thus, Notch activity may be generating a novel endocardial signal that promotes regional myocardial TGFβ2 expression. The endocardial receptors for TGFβ2, TBRI-TBRII and TBRIII (Boyer and Runyan 2001), exhibit reduced expression in RBPJk mutants (Fig. 3D and data not shown), whereas expression of the co-receptor endoglin and of the BMP-related receptors ALK3, BMPR-1B, and BMPRII (Kingsley 1994) appears unaffected (data not shown and Fig. 3D). These results imply that endocardial Notch activity is specifically required for the expression of multiple TGFβ2 pathway elements.
Figure 3.
Reduced TGFβ2 and Snail expression with persistent VE-cadherin expression associate to an impaired EMT in Notch pathway mutants. E9.5 hearts whole-mount and transverse sections. (A–C) TGFβ2 expression in wild-type (A), RBPJk (B), and Notch1 (C) mutants. (D) Semiquantitative RT-PCR in E9.5–10.0 embryos; (B) body; (H) heart. (E–G, K, M) snail transcription in E9.5 versus E9.5–10.0 hearts. (E, F, K) Wild type. (G, M) RBPJk mutants. (H–J, L, N) VE-cadherin expression in E9.5 versus E9.5–10.0 hearts. (H, I, L) Wild type. (J, N) RBPJk mutants. Scale bar, 50 μm.
Snail is expressed in endocardial cells undergoing EMT and is absent in Notch pathway mutants
Slug is a putative target of TGFβ2 during chick endocardial EMT (Romano and Runyan 2000). We find that Slug is not expressed in the murine heart, but its homolog snail is detected in the outflow tract and atrio-ventricular canal endocardium at the onset of EMT (Fig. 3E), and its expression increases progressively over time (Fig. 3F,K). E9.5 RBPJk mutants show severely reduced snail expression (Fig. 3G,M). Interestingly, snail is also detected in cushion mesenchyme (Fig. 3K), in a pattern exactly matching that of Delta4 (Fig. 1Z), Notch 1 (Fig. 1Y), Notch2, and Notch3 (data not shown). Thus Notch may act via snail to promote and maintain the cardiac mesenchymal phenotype. This is consistent with the requirement for snail in the production of mesenchyme from epiblast cells during mouse gastrulation (Carver et al. 2001), and with the ability of snail to repress E-cadherin expression and induce EMT in epithelial tumor cells (Batlle et al. 2000; Cano et al. 2000). Vascular endothelial cadherin (VE-cadherin), the predominant adhesion molecule of the endothelial adherens junction (Lampugnani et al. 1992), is transcribed throughout the endocardium of E8.5 wild-type embryos (data not shown). By E9.5 (Fig. 3H), it shows a striking expression pattern reciprocal to that of snail: VE-cadherin expression is reduced specifically in the outflow tract and AV canal endocardium, where snail expression becomes strongest as EMT proceeds (Fig. 3, cf. H,I,L and E,F,K). In contrast, RBPJk and Notch1 mutants maintain intense endocardial VE-cadherin expression (Fig. 3J,N and data not shown). Thus, the impairment of EMT in RBPJk and Notch1 mutants may in part be due to their inability to reduce endocardial cellular adhesion.
Notch pathway mutants show impaired EMT in vitro
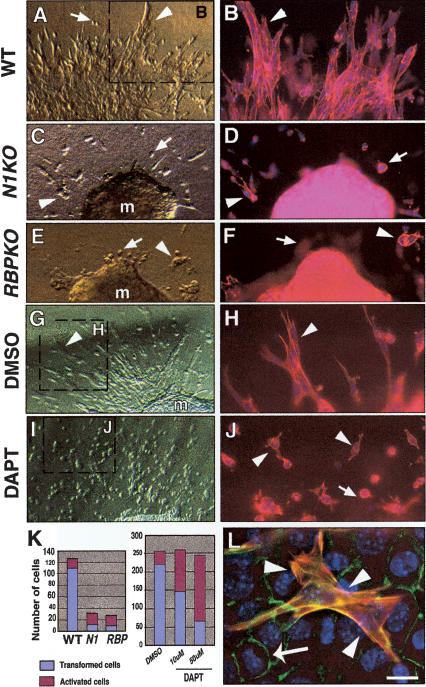
As Notch1 and RBPJk mutant embryos die at around E10.5 (Swiatek et al. 1994; Oka et al. 1995), preventing any further characterization in vivo, we used the Markwald and Runyan explant model of endocardial EMT (Runyan and Markwald 1983) to extend our observations to later developmental stages. E9.5 wild-type and mutant hearts were microdissected and explanted onto type I collagen gels. Twenty-seven of 32 (85%) explants became attached to the gel and exhibited spontaneous beating of the associated myocardium for at least 4 d. Phalloidin-TRITC staining after 73 h revealed that 24 of 27 (89%) wild-type explants exhibited elongated individual mesenchymal cells positive for α-smooth muscle actin (Camenisch et al. 2002b), which migrated through the collagen gel (around 110 cells/explant, 85% of total cells of endocardial origin in the explant; Fig. 4A,B,K left panel,L), and a few rounded cells of “intermediate phenotype” that represent a cell population that is activated but does not transform (Camenisch et al. 2002b) were also observed (around 25 cells/explant, 15% total; Fig. 4A,K left panel). In contrast, a majority of Notch1 (9 of 10; Fig. 4C,D) and RBPJk (8 of 9; Fig. 4E,F) mutant explants transform very poorly (around 10 cells/explant, 30% total; Fig. 4K left panel), and rounded cells can be readily observed (around 20 cells/explant, 70% total; Fig. 4C–F,K left panel). This result indicates that Notch1 and RBPJk mutants suffer a severe impairment of endocardial EMT rather than a simple developmental delay. This phenotype is not rescued by the addition of TGFβ2, TGFβ3, or both (data not shown) to the explant media, suggesting a cell-autonomous defect consistent with the reduced expression of TBRII and TBRIII in these mutant embryos (Fig. 3D and data not shown).
Figure 4.
Impaired EMT in Notch1, RBPJk, and DAPT-treated cardiac explants cultures. E9.5 atrio-ventricular canal explants cultured for 72 or 96 h. (A, B) Wild type. (C, D) Notch1 mutant. (E, F) RBPJk mutant. (G, H) Wild-type explant cultured with DMSO. (I, J) Wild-type explant cultured with 10 μM DAPT. (B, D, F, H, J) Phalloidin-TRITC staining. Transformed mesenchymal cells invadingthe collagen gel are indicated by arrowheads, and activated but not transformed cells by arrows. (K) Quantitative analysis of explants. (Left table) The number of transformed mesenchymal cells is significantly reduced in explants from Notch1 and RBPJk mutants. (Right table) Wild-type DAPT-treated explants show a dose-dependent reduction in transformed mesenchymal cells and an increase in activated but not transformed cells. (m) Myocardium. (L) Wild-type explant with transformed cells expressing α-SMA-Cy3 (arrowheads) surrounded by nontransformed endothelial cells stained with Phalloidin-FITC. Scale bar, 15 μm in A, C, E, G, I; 5 μm in B, D, F, H, J; and 2 μm in L.
To independently substantiate the requirement of Notch for endocardial EMT, we cultured E9.5 wild-type explants in collagen gels and cultured them in the presence of DAPT, a well-characterized and highly specific γ-secretase inhibitor (Dovey et al. 2001). After overnight culture, DAPT was added at concentrations that have been proven to inhibit γ-secretase activity and interfere with Notch signaling in zebrafish (Geling et al. 2002) and in chick cultured embryos (Dale et al. 2003). After 4 d of culture, control explants (dimethylsulphoxide [DMSO], n = 26) show a high number of transformed mesenchymal cells (around 220 cells/explant, 88% of total cell number; Fig. 4G,H,K right panel). In contrast, explants treated with DAPT show a dose-dependent reduction in the number of transformed cells: 10 μM DAPT (n = 22) produced about 150 cells/explant, (56% of total) with a parallel increase in cells of intermediate phenotype (Fig. 4I,J,K right panel), and 50 μM DAPT (n = 42) produced an even more drastic reduction in the number of transformed mesenchymal cells (90 cells/explant, 36%) and a larger increase in cells of intermediate phenotype (Fig. 4K right panel). These results indicate that in DAPT-treated explants, the endocardial cells accumulate in an “intermediate state” (Camenisch et al. 2002b) and are unable to progress through EMT. This phenotype is reminiscent of although less severe than that observed in Notch pathway mutants, probably because the γ-secretase inhibitor was added with a 24-h periodicity and its effective concentration would oscillate with time. E9.5 wild-type posterior embryonic regions comprising the presomitic mesoderm (PSM) were cultured in media containing DMSO or 10 μM DAPT for 4.5 h and hybridized with a probe for Lunatic Fringe (L-Fng), a Notch target gene (Barrantes et al. 1999). Only PSM regions cultured with DAPT show a drastic reduction in L-Fng expression (data not shown), indicating that these DAPT concentrations efficiently block Notch signaling.
Manipulation of Notch signaling activity perturbs valve development in zebrafish embryos
To test whether hyperactivation of Notch signaling could promote excessive EMT, capped mRNA encoding the constitutive active form of mouse Notch1 (N1IC) was injected into groups of one- to two-cell stage zebrafish embryos, transiently inducing Notch activity. Development was monitored throughout cardiac valve formation, from 48 h postfertilization (hpf) to 5 d post-fertilization (Stainier 2001). A large percentage (around 80%) of injected embryos died before reaching 48 hpf, mainly owing to serious gastrulation defects, alterations in brain formation, and severe antero-posterior axis deformities secondary to somitogenesis defects, consistent with previous reports where the Notch pathway is constitutively active (Dornseifer et al. 1997; Raya et al. 2003a). We reasoned that the variability in phenotype severity could be the consequence of the injected mRNA being distributed randomly. Indeed, detection of the injected N1IC by in situ hybridization revealed a wide variation in both the intensity and the distribution of the mRNA among the individual embryos (see Supplementary Table 1). Notably, cardiac expression of mouse N1IC RNA persisted in ∼15% of embryos for at least 3 d of development (n = 32; Fig. 5A,B, Supplementary Table 1). In turn, the vast majority (92 of 128 embryos, 71%) of embryos that survived beyond 48 hpf developed normally compared with wild type (Fig. 5C) or with embryos injected with control mRNA (eGFP, not shown). In contrast, 17 embryos (14%) showed pericardial distension indicative of cardiac or circulatory malfunction (Fig. 5D). The remaining embryos (19, 15%) exhibited a number of gross developmental defects and were excluded from this study. Histological analysis revealed the AV valves with two well-developed cusps in 5-day-old wild-type embryos (Fig. 5E), and 15 of 17 (88%) embryos with pericardial distension exhibited enlarged AV valves and hypertrophic endocardial cushions (Fig. 5F). Injection of similar amounts of mRNA encodinge GFP did not result in any alterations in the AV valves or endocardial cushions (data not shown).
Figure 5.
Manipulation of Notch signaling activity perturbs valve development in zebrafish. (A, B) In situ hybridization showing mouse N1IC expression in the heart (arrowhead) of 56- and 72-hpf injected zebrafish embryos. (C, D) Embryos at 5 d postfertilization. (C) Wild type. (D) N1IC mRNA-injected embryo exhibiting pericardial edema. Arrowhead points to pericardial sac in C, D. (E, F) H&E sagittal sections; arrowheads indicate the leaflets of the bicuspid atrio-ventricular valves. (E) Wild-type embryo. (F) Representative N1IC-injected embryo with pericardial distention. Note the abnormally large size of the atrio-ventricular valves (arrowheads). The asterisk labels the large pericardial space in the injected embryos. (G, H) Mitotic cells labeled with anti-phospho-histone H3 (red) at 4 d postfertilization in sagittal sections of control-(G) and N1IC-injected (H) embryos. Note the presence of a mitotic cell (arrowhead) in the endocardial cushion of the N1IC-injected embryo (H). DAPI (blue) stains cell nuclei. The position of the atrio-ventricular valve is indicated by asterisks. (I, J) Detection of zebrafish snail1 transcripts by whole-mount in situ hybridization in 3 d postfertilization control-(I) and N1IC-injected (J) embryos. Note the up-regulation of snail1 transcripts in the heart region of N1IC-injected embryos (arrow in J). (K–M) H&E sagittal sections of 5 d postfertilization embryos treated with DMSO (K), or DAPT at final concentrations of 50 (L) or 100 (M) μM. Arrowheads indicate the leaflets of the bicuspid atrio-ventricular valves. Note the valve atrophy in embryos in which Notch activity was down-regulated (K, M). The asterisk labels the pericardial space. In all of the images, embryo view is lateral, dorsal to the top, anterior to the left. (at) Atrium; (pe) pericardium; (v) ventricle. Scale bar, 50 μm.
To better understand the cellular dynamics leadin to valve hypertrophy induced by Notch activation, we analyzed the status of cell proliferation in sections of control- and N1IC-injected embryos. Because the number of cells in mitosis (detected by immunostaining with anti-phospho histone H3 antibodies) is extremely low, we had to analyze serial sections from 20 control embryos to detect one mitotic cell in the valve region (Fig. 5G). Interestingly, we detected the presence of mitotic cells in the endocardial cushion in 5 of 14 N1IC-injected embryos (Fig. 5H), suggesting that expression of N1IC may cause an increase in proliferation. We also analyzed whether Notch activation in zebrafish resulted in changes in snail expression. For this purpose, we injected N1IC into transgenic zebrafish embryos expressing eGFP under the control of the myosin light chain 2a (mlc2a) promoter (Raya et al. 2003b), which allowed us to select for embryos displaying valve hypertrophy (see Supplementary Movies 1 and 2). Whole-mount in situ hybridization analyses revealed increased transcripts of snail1 in the heart region of N1IC-injected (Fig. 5J), compared with uninjected embryos (Fig. 5I), whereas snail2 was not expressed in either group (data not shown). Taken together, these results indicate that constitutive Notch activation induces excessive endocardial EMT. For the converse experiment, that is, down-regulation of Notch activity, we treated zebrafish embryos with DAPT, a treatment previously reported to inhibit Notch activity in zebrafish embryos in vivo (Geling et al. 2002). Zebrafish embryos were incubated in the presence of different concentrations of DAPT or the vehicle alone (DMSO) from 36 hpf until 4 or 5 d postfertilization. Remarkably, treatment with 50 or 100 μM DAPT resulted in noncompetent AV valves (Supplementary Movie 3). Histological analyses of these embryos (n = 20 for each treatment) revealed atrophic endocardial cushions compared with DMSO-treated embryos (Fig. 5K–M). These results support the notion that Notch activity is critical for endocardial EMT.
Notch promotes snail induction and EMT in immortalized endothelial cells in vitro
To test whether endocardial cells are unique in this response to Notch signaling, subclones of the spontaneously immortalized pig aortic endothelial cell line (PAE), expressing either N1IC and GFP (PAE-N1IC) or GFP alone (PAE-GFP), were established. All 12 PAE-N1IC clones express severely attenuated levels of VE-cadherin, relative to the 5 PAE-GFP clones and the parental non-transfected line (Fig. 6A). PAE-GFP cells grow evenly on plastic to become contact inhibited on confluence (Fig. 6B). PAE-N1IC clones, however, lose contact inhibition, exhibit a less endothelial-like, less elongated morphology, and undergo focus formation and morphological transformation (Fig. 6C). VE-cadherin attenuation is a relatively rapid response to Notch activity, as ectopic expression of N1IC inhibited the VE-cadherin promoter (Gory et al. 1999) to about 40% of control values, whereas Ets-1 activated the reporter modestly as previously reported, and NFkB-p50 and empty vector produced little effect at 24 h in transient luciferase assays (Fig. 6D). The VE-cadherin promoter contains two consensus snail-binding sequences (CAGGTG), and transient expression of snail induced potent promoter repression to about 10% of control values (Fig. 6D). Thus, N1IC might act via snail induction to attenuate VE-cadherin expression, as these data and analysis of the Notch mutant endocardia suggested. To address this possibility, we constructed a mutant version of the murine snail molecule that lacks 122 amino-terminal amino acids that are required for interaction with the intracellular transcriptional repression systems. This truncated snail molecule (DN-Snail) retains the ability to bind snail DNA target sequences and functions as a dominant negative by direct competition for binding with endogenous Snail at specific promoter sites (Aybar et al. 2003). In cotransfection studies, we found that DN-Snail could relieve the Notch-mediated repression of the VE-Cadherin promoter in a dose-dependent fashion (Fig. 6E). Furthermore, semiquantitative RT-PCR on the PAE-N1IC and PAE-GFP cell lines using degenerate snail primers (Supplementary Table 2) revealed that Snail mRNA was readily amplified from PAE-N1IC but not from PAE-GFP expressing clones, whereas VE-cadherin expression was dramatically reduced in PAE-N1IC when compared with PAE-GFP clones (Fig. 6F). Similarly, N1IC was able to transiently activate a snail promoter luciferase reporter (Tan et al. 2001) up to sevenfold in a dose-dependent fashion in wild-type PAE cells and wild-type mouse fibroblasts, but not in RBPJk-deficient fibroblasts (Fig. 6G), demonstrating that the effect of Notch on snail induction is N1IC /RBPJK dependent. The lack of RBPJK consensus binding sites in the published snail promoter suggests that the effect may be indirect. In addition, 20 of 26 neomycin-resistant PAE subclones expressing ectopic snail (PAE-sna) also produced attenuated VE-cadherin expression and a transformed phenotype identical to that of N1IC stable clones (Fig 6H and data not shown), indicating that snail may be the major effector of Notch-mediated transformation in this system. Induction of the snail repressor and loss of VE-cadherin expression indicates that cells have undergone conversion from endothelia/epithelia to mesenchyme as an intimate part of Notch-mediated oncogenic transformation. Accordingly, in collagen gels, PAE-GFP cells interact and form tubules (Fig. 6I), whereas PAE-N1IC clones form disorganized masses of colonies surrounded by wide halos of cells independently migrating through the matrix and exhibiting filamentous actin staining (Fig. 6J,K). Xenograft results mirror those of the collagen gels: PAE-N1IC clones injected in matrigel carrier solution produce large subcutaneous tumors that grow rapidly to expand on average sevenfold (volumetrically) over a 32- to 48-d period, readily infiltratingthe matrigel and adjacent normal tissue. In contrast, PAE-GFP xenograft size did not significantly increase, and cells that were recovered remained in intimate contact, were unable to colonize the matrigel matrix, and only slightly penetrated surrounding tissue (see Supplementary Fig. 1).
Figure 6.
Constitutive Notch signaling produces an invasive phenotype via snail induction and VE-cadherin repression in PAE cells. (A) PAE-N1IC clones express little VE-cadherin. (B, C) Cells on plastic (10×). (B) PAE-GFP (wild type). (C) PAE N1IC. (D) Snail and N1IC significantly repress VE-cadherin promoter activity. (E) DN-snail suppresses VE-cadherin repression mediated by N1IC. (F) RT-PCR analysis of snail and VE-cadherin expression in PAE-N1IC clones 1 and 2 and in PAE-GFP clone 3. Snail is induced in N1IC-expressing clones, and VE-cadherin is down-regulated. (G) The snail promoter is activated by N1IC in a dose-dependent manner in wild-type PAE (black bars) and MEFs (blue bars), but not in RBPJk mutant MEFs (red bars). (H) Expression analysis of PAE-snail subclones. (I–K) Cells in collagen I matrix. (I) PAE-GFP (10×). (J, K) PAE N1IC (10×, 60×). (L) Notch1-IC does not induce SMAD activity in PAE cells. (M) PAE cells express different TBRIII levels irrespective of N1IC. (N) Activation of the PAI-Luc reporter correlates with TBRIII expression levels but not with N1IC. (N1IC) myc-epitope tagged N1IC; (VE-C) VE-cadherin; (Actin) β-actin; (Snail) HA epitope-tagged Snail; (p50) NF-kB-p50; (pCDNA3) empty vector; (GAPDH) glyceraldehyde-3-phosphate dehydrogenase.
TGFβ signaling is critical for endocardial cushion entry (Nakajima et al. 2000), and TGFβ signals can cooperate with ras to induce experimental tumor cell line metastasis in vitro (Oft et al. 2002). Similarly, TGFβ signalingis correlated with the induction of the snail homolog slug (Romano and Runyan 2000). Thus, we expected that Notch would function to induce snail and EMT at least in part by activation of the TGFβ system in PAE cells and in the endocardium of murine embryos. Surprisingly, we were unable to find evidence of TGFβ/SMAD activity, either in the stable N1IC expressing lines, or by transient transfection of N1IC into wild-type PAE cells. Accordingly, the PAI-1 (plasminogen activator inhibitor-1) promoter, a potent indicator of TGFβ/SMAD-directed transcriptional activity, is not activated in response to N1IC, nor the irrelevant NFkBp50 molecule, although it is responsive to treatment with TGFβ1 within 24 h, indicating that the TGFβ signaling system is intact (Fig. 6L). Comparable results were obtained by treatment with porcine TGFβ2 (data not shown). Similarly, overexpression of a constitutively active version of the SMAD2 molecule does not repress the VE-cadherin promoter (Fig. 6E). We tested whether the PAE-N1IC clones expressed higher levels of the TBRIII molecule, and found that the expression level of TBRIII is not correlated with NIC expression (Fig. 6M). We were unable to assess the expression levels of other TBR molecules, probably due to their porcine origin. One indication of more TBR expression by NIC-PAE cells would be an increased sensitivity to TGFβ treatment in the PAE-N1IC versus PAE-GFP cells. However, we found that the magnitude of TGFβ induction of the PAI-1 promoter correlated with the expression levels of the TBRIII molecule in the PAE subclones, but not with the presence or absence of N1IC (Fig. 6N). Finally, subclones of PAE cells were derived that stably express constitutively active SMAD2. These lines maintain VE-cadherin expression and do not lose contact inhibition in vitro on plastic (data not shown). In summary, we have not found evidence for TGFβ/SMAD activity in the repression of the VE-cadherin promoter or the transformation of the PAE cells by N1IC, suggesting that Notch may induce Snail either by TGFβ-dependent, SMAD-independent means, or via novel mechanisms.
A recent report has shown that Notch1 deletion in the epidermis leads to increased levels of free β-catenin (Nicolas et al. 2003); PAE-N1IC expressing cells do not show alterations in β-catenin levels or subcellular localization (data not shown), suggesting that Notch does not affect Wnt signaling in this endothelial setting.
Discussion
Endocardial Notch functions by lateral induction
Notch has been proposed to control cellular fates according to two different mechanisms, lateral inhibition and lateral induction (Artavanis-Tsakonas et al. 1999). A key feature of lateral inhibition is the down-regulation of Delta genes on Notch activation through a negative feedback loop that drives neighboring cells into different developmental pathways (Lewis 1998). A typical example of lateral inhibition mediated by Notch is the control of neurogenesis in Drosophila and vertebrates (for review, see Beatus and Lendahl 1998). In contrast, during lateral induction, Notch activation promotes production of Notch ligands: Cells expressing high levels of ligand stimulate their neighbors to do the same, giving rise to a field of cells that simultaneously express high levels of both ligand and receptor. All cells in this signaling field are stimulated to make their cell fate choices cooperatively. This mechanism is at work during Drosophila wing margin development (Panin et al. 1997). Although not well understood, Notch is thought to function by lateral induction during vertebrate somite boundary formation (Lewis 1998) and some aspects of inner ear development (Eddison et al. 2000).
Our work demonstrates that specific Notch pathway elements are coexpressed in an embryonic field comprising endocardial and cardiac mesenchymal cells between E8.5 and 9.5 of murine embryogenesis. Delta4 may be the most relevant ligand at this stage, as it has the strongest expression throughout the endocardium, and Delta1 and Jagged2 are respectively expressed in ventricular and atrial endocardium from E9.0 onward (data not shown). Similarly, Notch1 is the receptor with strongest endocardial expression, and the three receptors Notch1–Notch3 are expressed in transforming mesenchymal cells of the outflow tract and AV canal (Fig. 1M and data not shown). The similarity between the phenotypes of Notch1 and RBPJk mutant embryos indicates that Notch1 may be the most relevant Notch receptor in the endocardium. Coexpression of both Notch ligands, receptors, and transcriptional targets such as HRT1/HESR1/HEY1 in cells that form a one-layer endocardium suggests that these cells are able to simultaneously send and receive lateral signals and therefore behave as a field, adopting cellular fates cooperatively. Inactivation of endocardial Notch signaling leads to attenuated expression of Notch ligands and receptors (Fig. 1B,D,G,I), indicating that Notch pathway element expression is regulated by a positive feedback loop that is consistent with lateral induction. During Drosophila wing margin development, a typical example of lateral induction, the activity of the glycosyltransferase Fringe modulates Notch receptiveness to ligands, positioning and restricting Notch activation to the developing wing margin (Panin et al. 1997). Similarly, Notch activity modulated by Lunatic Fringe is crucial for the establishment of boundaries in the presomitic mesoderm (Evrard et al. 1998; Zhang and Gridley 1998). In contrast, we have not detected endocardial expression of any Fringe-related gene between E8.5 and E10.5 (data not shown), indicating that Fringe is likely not involved in the establishment of a boundary of distinct Notch activation in the endocardium versus the rest of the endothelium, making Notch-mediated lateral induction in the endocardium a phenomenon different from those previously characterized (Lewis 1998).
Notch activity and endocardial competence
Vascular and cardiac endothelium have a different developmental history (Liao et al. 1997), and the earliest expression of Notch pathway genes is detected in both endothelia at around E7.5 (data not shown and Del Amo et al. 1992; Mailhos et al. 2001), after emergence of both lineages has occurred. Thus it is unlikely that Notch is involved in the establishment of cardiac versus vascular endothelial cell fate. Differential expression of Notch signaling elements in the developing vasculature has also been associated with arterial versus venous fate choices (Krebs et al. 2000). Because Notch ligand and receptor expression in the endocardia of wild-type embryos overlaps at the cellular level, it is doubtful that loss of Notch signaling is promoting a “more arterial” or “more venous” cell fate choice. Thus the role of endocardial Notch is most likely to confer “competence” to the endocardial territory, allowing response to local developmental cues and the implementation of specific developmental programs, such as EMT. Similarly, Notch signaling does not seem to be involved in cardiac chamber identity specification, as Tbx5 expression is correctly regionalized in the myocardium of RBPJk (Fig. 2D) or Notch1 (data not shown) mutant embryos. Overexpression of activated Notch in zebrafish embryos produces hypercellular cushions, suggesting that Notch, like Ras activity, is tightly regulated during development to produce “just enough” cushion entry and mesenchymal conversion.
Notch promotes EMT during endocardial cushion formation
We have shown that targeted inactivation of Notch in mice impairs endocardial EMT in vivo and in explant assays in vitro, the latter phenotype being reproduced in wild-type explants cultured with the γ-secretase inhibitor DAPT (Dovey et al. 2001). Histological analysis of E9.5 mutant AV canal endocardia reveals that cells remain in close association, abnormally maintaining adherens junctions, and do not invade the cardiac jelly, despite exhibiting features of activated premigratory endocardial cells. These observations correlate with a specific reduction of TGFβ2 expression in the AV canal and outflow tract myocardium of Notch1 and RBPJk mutant embryos. TGFβ2 has been proposed to induce expression of the transcription factor slug in the chick heart (Romano and Runyan 2000), and Notch1 and RBPJk mutant embryos have a severe reduction in snail transcription, the mammalian slug homolog expressed in the heart at this stage. Concomitantly, VE-cadherin expression remains abnormally stabilized in the AV canal and outflow tract endocardium of the mutants, suggesting that the lack of snail repressor expression prevents the down-regulation of VE-cadherin in this tissue, thus blocking endocardial EMT. These findings demonstrate that Notch activity is required for endocardial EMT. Supporting this idea are gain-of-function experiments in zebrafish, whereby transient overexpression of N1IC in the heart leads to the formation of hypertrophic AV valves, whereas Notch inhibition impairs valve development.
Antibody blocking experiments in chick and murine cardiac explants indicate that TGFβ2 signaling is required for endocardial cushion entry (Camenisch et al. 2002a). The regionalized myocardial TGFβ2 expression is severely reduced in Notch pathway mutants, suggesting that Notch activity may lead to the production of a novel retrograde signal from endocardium to myocardium, because Notch receptors are not expressed by cardiac myocytes. This would result in the coordinate induction of the myocardial TGFβ2 signal and endocardial snail expression, leading to regionalized EMT and cushion entry.
Notch induction of the snail repressor and EMT is not restricted to specialized cardiac endothelial cells
Further insight into the mechanism of Notch function in EMT is based on our in vitro experiments with immortalized PAE cells: N1IC-expressing PAE clones show increased snail transcription and decreased VE-cadherin expression, adopting a transformed and motile phenotype, indicating that they have converted into mesenchyme. This is consistent with the findings that snail or slug overexpression can drive some epithelial tumor cells through mesenchymal conversion in vitro (Savagner et al. 1997; Batlle et al. 2000; Cano et al. 2000), and that in patient tumor samples, expression of snail is highly correlated with invasive carcinomas (Blanco et al. 2002). Transient transfection studies in PAE cells indicate that N1IC and snail are able to repress the VE-cadherin promoter, and that N1IC activates the snail promoter in PAE cells and in wild-type mouse embryonic fibroblasts. This activation is lost in RBPJk mutant fibroblasts, indicating that the effect of N1IC on the snail promoter is RBPJK dependent. Recent studies have shown that Notch can function as a Ras downstream effector in epithelial tumor cells (Weijzen et al. 2002), and that Ras is able to activate the snail promoter (Peinado et al. 2003). Because Ras activity also modulates endocardial EMT (Lakkis and Epstein 1998), we suggest that Notch signaling may participate in a Ras-mediated snail activation in the endocardium. The simplest interpretation of our cardiac cushion recruitment data is that endocardial Notch activity is required for TGFβ2 signaling that in turn induces snail to repress VE-cadherin. However, the PAE results indicate that this epistasis relationship may be more complex, as VE-cadherin repression occurs in a TGFβ/SMAD-independent fashion in PAE cells (Fig. 6L,N). Taken together, our data indicate that Notch represses VE-cadherin expression in two different endothelial settings, and can function in a manner independent of TGFβ signaling, most likely via snail induction.
Notch has not previously been suspected to play a role in the regulation of EMT, nor to modulate either TGFβ signaling nor cellular adhesion, although its activity coincides with processes that involve selective changes in cellular adhesion, as, for example, neural crest formation (Endo et al. 2002), somitogenesis (Pourquie 2000), and angiogenesis (Gridley 2001). Our findings illuminate this novel function of Notch and demonstrate that it can occur in both embryonic contexts and during Notch-mediated transformation in vitro. Notch3 has been tightly associated with a metastatic expression signature in analysis of a wide variety of solid tumor types in vivo (Ramaswamy et al. 2003), and our data suggest that Notch may play a causal role in this process. Similarly, angiogenesis requires transient endothelial EMT, and Notch1; Notch4 double mutant embryos exhibit severe angiogenic abnormalities (Krebs et al. 2000), suggesting that Notch-mediated EMT may underlie this defect. These findings broaden our understanding of developmentally regulated EMT processes including the occurrence of congenital cardiac valve abnormalities (Srivastava 2001), and open possible new avenues for cancer therapeutic intervention via inhibition of Notch-mediated angiogenesis and/or tumor metastasis.
Materials and methods
Embryo genotyping
Notch1 (Conlon et al. 1995) and RBPJk (Oka et al. 1995) homozygous mutant embryos were obtained by mating males and females heterozygous for these targeted mutations. Embryos were genotyped by PCR analysis of the yolk sacs, as described.
Histology, in situ hybridization, and immunohistochemistry
Hematoxylin and eosin staining was performed by standard methods. Single- and double-color whole-mount in situ hybridization and sectioning were performed according to standard protocols. The probes used in this study were Delta4 (600-bp 3′ fragment), Notch1, GATA5, HRT1 and HRT2, TGFβ2, and snail that were kindly provided by Janet Rossant, Ed Morrisey, D. Srivastava, Rosemary Akhurst, and Angela Nieto; the VE-C probe was a 1.6-kb 5′ fragment. The probe for zebrafish snail1 was kindly provided by Alex Schier. A fragment representing the publicly available sequence of zebrafish snail2 was cloned by PCR. In double-color in situs, the Delta4 probe was developed in red with Fast Red (Sigma), and the Notch1 probe in blue using NBT/BCIP solution (Roche). Overlapping staining produces a dark brown color. For whole-mount immunohistochemistry, embryos were fixed overnight in methanol: DMSO (4:1), bleached in methanol:DMSO: H2O2 (4:1:1), rehydrated, and processed accordingto standard protocols. Embryos were incubated with 10 μg/mL of anti-mouse CD31 (platelet endothelial-cell adhesion molecule or PECAM; PharMingen). For the detection of mitotic cells, zebrafish embryos were fixed in 4% paraformaldehyde for 2 h and 14-μm cryostat sections incubated with anti-phospho histone H3 (Upstate) 1:100. DAPI was used to visualize nuclei.
Transmission electron microscopy
Embryos were fixed overnight (2.5% glutaraldehyde, 2% paraformaldehyde in 0.1 M phosphate buffer at pH 7.4), postfixed (1% osmium tetroxide in 0.1 M phosphate buffer), washed, dehydrated, and infiltrated with Epon-araldite. Ultrathin sections were imaged on a Hitachi H-800MT transmission electron microscope.
RNA isolation
E8.5–E9.5 wild-type and RBPJk mutant embryos were dissected in ice cold PBS. RNA from bodies and hearts was purified separately (Trizol, Invitrogen) according to manufacturer instructions. In the case of PAE clones, RNA was extracted from ∼30 × 106 cells with Trizol.
Semiquantitative RT-PCR
First-strand cDNA synthesis was performed accordingto the manufacturer directions (Amersham). One microgram of total RNA was used per reaction. For primers and conditions, see Supplementary Table 2. For cDNA synthesis from PAE cells, poly(A)+ RNA was obtained using PolyATtract kit (Promega) and 1 μg was used per reaction. Amplified PCR products were cloned in the PCRII-TOPO (Invitrogen) vector and sequenced. PCR products were quantitated by phosphorimager analysis (BioRad). The GenBank accession number for porcine snail is AY263371.
Explant system for in vitro mesenchymal transformation and γ-secretase inhibitor treatment
Rat tail collagen type I (BD Biosciences 354236) was prepared according to manufacturer instructions. The solution was dispensed in four-well microculture dishes (NUNC) and allowed to solidify inside a 37°C, 5% CO2 incubator. Subsequently, collagen gels were washed several times with media containing DMEM, 5% FCS, 0.1% insulin, transferrin, and selenium (ITS) plus antibiotics, and drained. AV tissue was harvested in sterile PBS from E9.5 embryos. AV explants were placed on drained collagen gels with the endocardium facing down and allowed to attach for 5 h at 37°C, 5% CO2. Media (0.5 mL/well) was added and the cultures reincubated for up to 3 d. In rescue experiments, collagen gel was conditioned with TGFβ2, TGFβ3 (Peprotech, 10 ng/mL), or both prior to addition of explants, and was changed daily. Explants were fixed and processed for phalloidin-TRITC staining (Sigma) to stain the cytoskeleton or α-smooth muscle actin (α-SMA-Cyan, Sigma) to detect mesenchymal cells, accordingto standard protocols (Camenisch et al. 2002b), and mounted onto excavated slides with Vectashield media containing DAPI. For the γ-secretase inhibitor treatment, DAPT (Calbiochem, γ-secretase inhibitor IX, Cat. 565770) was diluted in DMSO and added to the media at 10 or 50 μM final concentration, maintaining a maximal DMSO concentration of 0.2%. Wild-type AV tissue was harvested and handled as described earlier. After overnight incubation at 37°C, 5% CO2, explants have started transformation, and media containing DMSO as control, or 10 or 50 μM DAPT was added to the explant. Every 24 h, fresh media with DMSO or DAPT was added for a period of 4 d and during this time, explants maintained a strong myocardial beating. After this time, selected control and DAPT-treated explants were stained with Tripan blue to asses viability, giving similar results. Subsequently, explants were stained with phalloidin-TRITC as described earlier. For quantification, two different independent observers naive to the experimental conditions of AV explants enumerated mesenchymal and not transformed endothelial cells numbers, and statistical analysis was carried out using an ANOVA one-way model.
Manipulation of zebrafish embryos
Wild-type zebrafish (AB line) or mlc2a-eGFP (Raya et al. 2003) embryos were maintained at 28.5°C by standard methods (Westerfield 2000). Capped mouse N1IC RNA was transcribed using the mMessage mMachine kit (Ambion). Injections were performed according to standard protocols. One hundred picograms of mRNA was injected into the yolks of one- or two-cell stage embryos that were allowed to develop from 48 hpf up to 5 d, classified, and analyzed by histology or in situ hybridization. DAPT was directly added to the water of 36 hpf embryos from a 50-mM stock in DMSO, at final concentrations of 10, 50, or 100 μM. Controls received amounts of DMSO equivalent to embryos treated with 100 μM DAPT. The incubations proceeded until 4–5 d postfertilization.
Stable cell line production
Plasmids encoding GFP and neomycin resistance (pCDNA3-GFP), or pCDNA3-GFP plus myc-N1IC, or murine HA-Snail with neomycin resistance, or constitutively activated SMAD2 plus neomycin resistance were linearized and transfected (Lipofectamine 2000) into 12-well or 6-well plates seeded with 5 × 104 or 2 × 105 PAE cells. G418 was added 12 h later to 1 μg/mL and emergent clones were isolated for analysis.
Cell culture and type I collagen assay
Cells were maintained in media with or without 1 μg/mL G418. Collagen solution was prepared as described earlier, cells added, and dispensed in 250-μL aliquots with 1–2 × 104 cells into 24-well plates. Polymerization occurred at room temperature, 10–20 min. Wells were rinsed twice and cultured with 500 μL complete media at 37°C, 5% CO2. To initially place cells in one focal plane, we allowed 250 μL of collagen solution to polymerize, washed it twice, and added 1–2 × 104 cells in 500 μL complete media to each well and incubated it as described earlier. After several hours, media was carefully removed and 250 μL of collagen solution was gently overlaid, polymerized, and washed, and the plates were incubated. Media was replaced every second day.
Western blotting
Whole-cell lysates were prepared in the presence of protease inhibitors (Complete mini tablet Roche 1–836–153). Western blotting was performed with 60 μg of each extract and antisera specific for VE-cadherin (Santa Cruz Biotechnology C-19, 1:200), myc epitope tag (Sigma, 1:1000), HA epitope tag(Sigma, 1:1000), Snail (Santa Cruz clone T18; 1:200), β actin (Sigma, 1:5000) in TBST/0.5% milk followed by anti-rabbit-HRP (Amersham, 1:2000), anti-mouse HRP (Amersham, 1:2000), or anti-goat-HRP (Santa Cruz Biotech, 1:500). Immunoreactive proteins were detected by chemiluminescence (ECL Plus, Amersham) and autoradiography.
Transient luciferase assays, reporters, and constructs
One microgram of total DNA containing 300 ng VE-cadherinluciferase or PAI-1 Luciferase plasmids, and 35 ngp lasmid encoding modifier genes, indicated in the figure legends, was transfected in triplicate (Lipofectamine Plus) according to the manufacturer's instructions into 24-well tissue culture plates that were seeded 12 h earlier with 2 × 104 wild-type PAE cells. For the Snail-Luciferase assays, 0.5–3 μg of N1IC and 0.5 μg of Snail-Luciferase plasmids were transfected into 2 × 104 wild-type PAE cells, mouse embryonic fibroblasts, or RBPJk-deficient embryonic fibroblasts (E. Bertrán and J.L. de la Pompa, in prep.). After 24 h, lysates were made and luciferase activity measured. Luciferase values were normalized for βgalactosidase activity. Experiments were performed at least three times. Porcine TGFβ1 and TGFβ2 were added (R and D Systems), as indicated. A truncated version of the murine snail gene was created by proofreading-PCR to encode a start methionine, followed by the murine snail zinc finger DNA binding domain and an HA epitope tag, using the HA epitope-tagged full-length murine snail gene as template. The sequences of the oligos are CGC GGA TCC ACC ATG GCC GAG GCC TTC ATC GCC TCC and the commercially available pCDNA3.1/BGH Reverse primer (Invitrogen). The resultant PCR fragment was cloned into the parental CMV expression vector, sequenced, and tested for expression by Western blot. CMV-based expression vector encoding a constitutively active murine SMAD2 molecule was obtained from the laboratory of Dr. Alan Balmain.
Acknowledgments
We thank A.García de Herreros and P. Huber for the snail and VE-cadherin cDNAs and promoter fragments; A. Balmain for the constitutively active SMAD2 expression construct; and A. Bigas, F. J. Diaz-Benjumea, A. García de Herreros, F. Giraldez, D. Henrique, R. Muñoz-Chapuli, A. Nieto, and M. Torres for critical reading of the manuscript. This work was funded by grant BFI-2002–01560 from the Spanish Ministry of Science and Technology to J.L.dlP.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.276304.
References
- Akhurst R.J., Lehnert, S.A., Faissner, A., and Duffie, E. 1990. TGF beta in murine morphogenetic processes: The early embryo and cardiogenesis. Development 108: 645–656. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Rand, M.D., and Lake, R.J. 1999. Notch signaling: Cell fate control and signal integration in development. Science 284: 770–776. [DOI] [PubMed] [Google Scholar]
- Aybar M.J., Nieto, M.A., and Mayor, R. 2003. Snail precedes slug in the genetic cascade required for the specification and migration of the Xenopus neural crest. Development 130: 483–494. [DOI] [PubMed] [Google Scholar]
- Barrantes I.B., Elia, A.J., Wunsch, K., De Angelis, M.H., Mak, T.W., Rossant, J., Conlon, R.A., Gossler, A., and de la Pompa, J.L. 1999. Interaction between Notch signalling and Lunatic fringe during somite boundary formation in the mouse. Curr. Biol. 9: 470–480. [DOI] [PubMed] [Google Scholar]
- Batlle E., Sancho, E., Franci, C., Dominguez, D., Monfar, M., Baulida, J., and Garcia De Herreros, A. 2000. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2: 84–89. [DOI] [PubMed] [Google Scholar]
- Beatus P. and Lendahl, U. 1998. Notch and neurogenesis. J. Neurosci. Res. 54: 125–136. [DOI] [PubMed] [Google Scholar]
- Blanco M.J., Moreno-Bueno, G., Sarrio, D., Locascio, A., Cano, A., Palacios, J., and Nieto, M.A. 2002. Correlation of Snail expression with histological grade and lymph node status in breast carcinomas. Oncogene 21: 3241–3246. [DOI] [PubMed] [Google Scholar]
- Boyer A.S. and Runyan, R.B. 2001. TGFbeta Type III and TGFbeta Type II receptors have distinct activities during epithelial-mesenchymal cell transformation in the embryonic heart. Dev. Dyn. 221: 454–459. [DOI] [PubMed] [Google Scholar]
- Bruneau B.G., Logan, M., Davis, N., Levi, T., Tabin, C.J., Seidman, J.G., and Seidman, C.E. 1999. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev. Biol. 211: 100–108. [DOI] [PubMed] [Google Scholar]
- Camenisch T.D., Molin, D.G., Person, A., Runyan, R.B., Gittenberger-de Groot, A.C., McDonald, J.A., and Klewer, S.E. 2002a. Temporal and distinct TGFbeta ligand requirements during mouse and avian endocardial cushion morphogenesis. Dev. Biol. 248: 170–181. [DOI] [PubMed] [Google Scholar]
- Camenisch T.D., Schroeder, J.A., Bradley, J., Klewer, S.E., and McDonald, J.A. 2002b. Heart-valve mesenchyme formation is dependent on hyaluronan-augmented activation of ErbB2-ErbB3 receptors. Nat. Med. 8: 850–855. [DOI] [PubMed] [Google Scholar]
- Cano A., Perez-Moreno, M.A., Rodrigo, I., Locascio, A., Blanco, M.J., del Barrio, M.G., Portillo, F., and Nieto, M.A. 2000. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2: 76–83. [DOI] [PubMed] [Google Scholar]
- Carver E.A., Jiang, R., Lan, Y., Oram, K.F., and Gridley, T. 2001. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol. Cell Biol. 21: 8184–8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon R.A., Reaume, A.G., and Rossant, J. 1995. Notch1 is required for the coordinate segmentation of somites. Development 121: 1533–1545. [DOI] [PubMed] [Google Scholar]
- Dale J.K., Maroto, M., Dequeant, M.L., Malapert, P., McGrew, M., and Pourquie, O. 2003. Periodic notch inhibition by lunatic fringe underlies the chick segmentation clock. Nature 421: 275–278. [DOI] [PubMed] [Google Scholar]
- Del Amo F.F., Smith, D.E., Swiatek, P.J., Gendron-Maguire, M., Greenspan, R.J., McMahon, A.P., and Gridley, T. 1992. Expression pattern of Motch, a mouse homolog of Drosophila Notch, suggests an important role in early postimplantation mouse development. Development 115: 737–744. [DOI] [PubMed] [Google Scholar]
- Dickson M.C., Slager, H.G., Duffie, E., Mummery, C.L., and Akhurst, R.J. 1993. RNA and protein localisations of TGF beta 2 in the early mouse embryo suggest an involvement in cardiac development. Development 117: 625–639. [DOI] [PubMed] [Google Scholar]
- Dornseifer P., Takke, C., and Campos-Ortega, J.A. 1997. Overexpression of a zebrafish homologue of the Drosophila neurogenic gene Delta perturbs differentiation of primary neurons and somite development. Mech. Dev. 63: 159–171. [DOI] [PubMed] [Google Scholar]
- Dovey H.F., John, V., Anderson, J.P., Chen, L.Z., de Saint Andrieu, P., Fang, L.Y., Freedman, S.B., Folmer, B., Goldbach, E., Holsztynska, E.J., et al. 2001. Functional gamma-secretase inhibitors reduce beta-amyloid peptide levels in brain. J. Neurochem. 76: 173–181. [DOI] [PubMed] [Google Scholar]
- Drake C.J. and Fleming, P.A. 2000. Vasculogenesis in the day 6.5 to 9.5 mouse embryo. Blood 95: 1671–1679. [PubMed] [Google Scholar]
- Eddison M., Le Roux, I., and Lewis, J. 2000. Notch signaling in the development of the inner ear: Lessons from Drosophila. Proc. Natl. Acad. Sci. 97: 11692–11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg L.M. and Markwald, R.R. 1995. Molecular regulation of atrioventricular valvuloseptal morphogenesis. Circ. Res. 77: 1–6. [DOI] [PubMed] [Google Scholar]
- Endo Y., Osumi, N., and Wakamatsu, Y. 2002. Bimodal functions of Notch-mediated signaling are involved in neural crest formation during avian ectoderm development. Development 129: 863–873. [DOI] [PubMed] [Google Scholar]
- Evrard Y., Lun, Y., Aulehla, A., Gan, L., and Johnson, R. 1998. Lunatic fringe is an essential mediator of somite segmentation and patterning. Nature 394: 377–381. [DOI] [PubMed] [Google Scholar]
- Geling A., Steiner, H., Willem, M., Bally-Cuif, L., and Haass, C. 2002. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 3: 688–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gory S., Vernet, M., Laurent, M., Dejana, E., Dalmon, J., and Huber, P. 1999. The vascular endothelial-cadherin promoter directs endothelial-specific expression in transgenic mice. Blood 93: 184–192. [PubMed] [Google Scholar]
- Gridley T. 2001. Notch signaling during vascular development. Proc. Natl. Acad. Sci. 98: 5377–5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlstone A.F., Haramis, A.P., Wienholds, E., Begthel, H., Korving, J., Van Eeden, F., Cuppen, E., Zivkovic, D., Plasterk, R.H., and Clevers, H. 2003. The Wnt/beta-catenin pathway regulates cardiac valve formation. Nature 425: 633–637. [DOI] [PubMed] [Google Scholar]
- Kingsley D.M. 1994. The TGF-beta superfamily: New members, new receptors, and new genetic tests of function in different organisms. Genes & Dev. 8: 133–146. [DOI] [PubMed] [Google Scholar]
- Kopan R. 2002. Notch: A membrane-bound transcription factor. J. Cell Sci. 115: 1095–1097. [DOI] [PubMed] [Google Scholar]
- Kopan R., Nye, J.S., and Weintraub, H. 1994. The intracellular domain of mouse Notch: A constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development 120: 2385–2396. [DOI] [PubMed] [Google Scholar]
- Krebs L.T., Xue, Y., Norton, C.R., Shutter, J.R., Maguire, M., Sundberg, J.P., Gallahan, D., Closson, V., Kitajewski, J., Callahan, R., et al. 2000. Notch signaling is essential for vascular morphogenesis in mice. Genes & Dev. 14: 1343–1352. [PMC free article] [PubMed] [Google Scholar]
- Lakkis M.M. and Epstein, J.A. 1998. Neurofibromin modulation of ras activity is required for normal endocardial-mesenchymal transformation in the developing heart. Development 125: 4359–4367. [DOI] [PubMed] [Google Scholar]
- Lampugnani M.G., Resnati, M., Raiteri, M., Pigott, R., Pisacane, A., Houen, G., Ruco, L.P., and Dejana, E. 1992. A novel endothelial-specific membrane protein is a marker of cell-cell contacts. J. Cell Biol. 118: 1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. 1998. Notch signalling and the control of cell fate choices in vertebrates. Semin. Cell Dev. Biol. 9: 583–589. [DOI] [PubMed] [Google Scholar]
- Liao W., Bisgrove, B.W., Sawyer, H., Hug, B., Bell, B., Peters, K., Grunwald, D.J., and Stainier, D.Y. 1997. The zebrafish gene cloche acts upstream of a flk-1 homologue to regulate endothelial cell differentiation. Development 124: 381–389. [DOI] [PubMed] [Google Scholar]
- Lyons K.M., Pelton, R.W., and Hogan, B.L. 1990. Organogenesis and pattern formation in the mouse: RNA distribution patterns suggest a role for bone morphogenetic protein-2A (BMP-2A). Development 109: 833–844. [DOI] [PubMed] [Google Scholar]
- Mailhos C., Modlich, U., Lewis, J., Harris, A., Bicknell, R., and Ish-Horowicz, D. 2001. Delta4, an endothelial specific notch ligand expressed at sites of physiological and tumor angiogenesis. Differentiation 69: 135–144. [DOI] [PubMed] [Google Scholar]
- Markwald R.R., Fitzharris, T.P., and Manasek, F.J. 1977. Structural development of endocardial cushions. Am. J. Anat. 148: 85–119. [DOI] [PubMed] [Google Scholar]
- Morrisey E.E., Ip, H.S., Tang, Z., Lu, M.M., and Parmacek, M.S. 1997. GATA-5: A transcriptional activator expressed in a novel temporally and spatially-restricted pattern during embryonic development. Dev. Biol. 183: 21–36. [DOI] [PubMed] [Google Scholar]
- Morrison S.J., Perez, S.E., Qiao, Z., Verdi, J.M., Hicks, C., Weinmaster, G., and Anderson, D.J. 2000. Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell 101: 499–510. [DOI] [PubMed] [Google Scholar]
- Nakagawa O., Nakagawa, M., Richardson, J.A., Olson, E.N., and Srivastava, D. 1999. HRT1, HRT2, and HRT3: A new subclass of bHLH transcription factors marking specific cardiac, somitic, and pharyngeal arch segments. Dev. Biol. 216: 72–84. [DOI] [PubMed] [Google Scholar]
- Nakajima Y., Yamagishi, T., Hokari, S., and Nakamura, H. 2000. Mechanisms involved in valvuloseptal endocardial cushion formation in early cardiogenesis: Roles of transforming growth factor (TGF)-beta and bone morphogenetic protein (BMP). Anat. Rec. 258: 119–127. [DOI] [PubMed] [Google Scholar]
- Nicolas M., Wolfer, A., Raj, K., Kummer, J.A., Mill, P., van Noort, M., Hui, C.C., Clevers H., Dotto, G.P., and Radtke, F. 2003. Notch1 functions as a tumor suppressor in mouse skin. Nat. Genet. 33: 416–421. [DOI] [PubMed] [Google Scholar]
- Nieto M.A. 2002. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell Biol. 3: 155–166. [DOI] [PubMed] [Google Scholar]
- Nieto M.A., Bennett, M.F., Sargent, M.G., and Wilkinson, D.G. 1992. Cloning and developmental expression of Sna, a murine homologue of the Drosophila snail gene. Development 116: 227–237. [DOI] [PubMed] [Google Scholar]
- Oft M., Akhurst, R.J., and Balmain, A. 2002. Metastasis is driven by sequential elevation of H-ras and Smad2 levels. Nat. Cell Biol. 4: 487–494. [DOI] [PubMed] [Google Scholar]
- Oka C., Nakano, T., Wakeham, A., de la Pompa, J.L., Mori, C., Sakai, T., Okazaki, S., Kawaichi, M., Shiota, K., Mak, T.W., et al. 1995. Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development 121: 3291–3301. [DOI] [PubMed] [Google Scholar]
- Panin V.M., Papayannopoulos, V., Wilson, R., and Irvine, K.D. 1997. Fringe modulates Notch-ligand interactions. Nature 387: 908–912. [DOI] [PubMed] [Google Scholar]
- Peinado H., Quintanilla, M., and Cano, A. 2003. Transforming growth factor beta 1 induces snail transcription factor in epithelial cell lines. Mechanisms for epithelial-mesenchymal transitions. J. Biol. Chem. 278: 21113–21123. [DOI] [PubMed] [Google Scholar]
- Pourquie O. 2000. Segmentation of the paraxial mesoderm and vertebrate somitogenesis. Curr. Top. Dev. Biol. 47: 81–105. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S., Ross, K.N., Lander, E.S., and Golub, T.R. 2003. A molecular signature of metastasis in primary solid tumors. Nat. Genet. 33: 49–54. [DOI] [PubMed] [Google Scholar]
- Raya A., Kawakami, Y., Rodriguez-Esteban, C., Buscher, D., Koth, C.M., Itoh, T., Morita, M., Raya, R.M., Dubrova, I., Bessa, J.G., et al. 2003a. Notch activity induces Nodal expression and mediates the establishment of left-right asymmetry in vertebrate embryos. Genes & Dev. 17: 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya A., Koth, C.M., Buscher, D., Kawakami, Y., Itoh, T., Raya, R.M., Sternik, G., Tsai, H.J., Rodriguez-Esteban, C., and Izpisua-Belmonte, J.C. 2003b. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc. Natl. Acad. Sci. (Suppl. 1) 100: 11889–11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaume A.G., Conlon, R.A., Zirngibl, R., Yamaguchi, T.P., and Rossant, J. 1992. Expression analysis of a Notch homologue in the mouse embryo. Dev. Biol. 154: 377–387. [DOI] [PubMed] [Google Scholar]
- Robey E. 1999. Regulation of T cell fate by Notch. Annu. Rev. Immunol. 17: 283–295. [DOI] [PubMed] [Google Scholar]
- Romano L.A. and Runyan, R.B. 2000. Slug is an essential target of TGFbeta2 signaling in the developing chicken heart. Dev. Biol. 223: 91–102. [DOI] [PubMed] [Google Scholar]
- Runyan R.B. and Markwald, R.R. 1983. Invasion of mesenchyme into three-dimensional collagen gels: A regional and temporal analysis of interaction in embryonic heart tissue. Dev. Biol. 95: 108–114. [DOI] [PubMed] [Google Scholar]
- Savagner P., Yamada, K.M., and Thiery, J.P. 1997. The zinc-finger protein slug causes desmosome dissociation, an initial and necessary step for growth factor-induced epithelial-mesenchymal transition. J. Cell Biol. 137: 1403–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D. 2001. Genetic assembly of the heart: Implications for congenital heart disease. Annu. Rev. Physiol. 63: 451–469. [DOI] [PubMed] [Google Scholar]
- Stainier D.Y. 2001. Zebrafish genetics and vertebrate heart formation. Nat. Rev. Genet. 2: 39–48. [DOI] [PubMed] [Google Scholar]
- Swiatek P.J., Lindsell, C.E., del Amo, F.F., Weinmaster, G., and Gridley, T. 1994. Notch1 is essential for postimplantation development in mice. Genes & Dev. 8: 707–719. [DOI] [PubMed] [Google Scholar]
- Tan C., Costello, P., Sanghera, J., Dominguez, D., Baulida, J., de Herreros, A.G., and Dedhar, S. 2001. Inhibition of integrin linked kinase (ILK) suppresses beta-catenin-Lef/Tcf-dependent transcription and expression of the E-cadherin repressor, snail, in APC-/- human colon carcinoma cells. Oncogene 20: 133–140. [DOI] [PubMed] [Google Scholar]
- Thiery J.P. 2002. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2: 442–454. [DOI] [PubMed] [Google Scholar]
- Walsh E.C. and Stainier, D.Y. 2001. UDP-glucose dehydrogenase required for cardiac valve formation in zebrafish. Science 293: 1670–1673. [DOI] [PubMed] [Google Scholar]
- Weijzen S., Rizzo, P., Braid, M., Vaishnav, R., Jonkheer, S.M., Zlobin, A., Osborne, B.A., Gottipati, S., Aster, J.C., Hahn, W.C., et al. 2002. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat. Med. 8: 979–986. [DOI] [PubMed] [Google Scholar]
- Westerfield M. 2000. The zebrafish book: A guide for the laboratory use of zebrafish (Danio rerio). University of Oregon Press, Eugene, OR.
- Zhang N. and Gridley, T. 1998. Defects in somite formation in lunatic fringe deficient mice. Nature 394: 374–377. [DOI] [PubMed] [Google Scholar]