Abstract
Protein kinase PKR is activated during viral infection and phosphorylates the α subunit of eukaryotic translation initiation factor 2 (eIF2), leading to inhibition of translation and viral replication. We report fast evolution of the PKR kinase domain in vertebrates, coupled with positive selection of specific sites. Substitution of positively selected residues in human PKR with residues found in related species altered sensitivity to PKR inhibitors from different poxviruses. Species-specific differences in sensitivity to poxviral pseudosubstrate inhibitors were identified between human and mouse PKR, which were traced to positively-selected residues near the eIF2α-binding site. Our findings indicate how an antiviral protein evolved to evade viral inhibition while maintaining its primary function. Moreover, the identified species-specific differences in the susceptibility to viral inhibitors have important implications for studying human infections in non-human model systems.
Introduction
Recognition of viral nucleic acids by host proteins is an essential component of the innate immune response against viruses, leading to activation of enzymes that initiate different antiviral responses1,2. One of these sensors is the protein kinase PKR. Binding of dsRNA, which is generated during viral transcription and replication, to the N-terminal dsRNA-binding domains of PKR, triggers kinase dimerization and autophosphorylation3,4. Activated PKR then phosphorylates eIF2α on Ser51, converting eIF2 into an inhibitor of its guanine nucleotide exchange factor eIF2B, and thereby downregulating translation. In addition to PKR, four eIF2α kinases, HRI, PERK (also known as PEK), GCN2 and PKZ, have been identified in vertebrates5,6. The eIF2α kinases share a closely related kinase domain (KD) that is connected to different regulatory domains reflecting the mode of kinase activation by various stresses. While PKR and the fish-specific paralog PKZ sense nucleic acids linked to viral infection, HRI responds to heme deficiency, PERK detects endoplasmic reticulum stress, and GCN2 is activated under conditions of amino acid starvation. By linking various stress signals to increased eIF2α phosphorylation and the attendant changes in global and gene-specific translation, the eIF2α kinases enable cells to adapt their proteomes to their environment7.
PKR plays an important role in the response to different RNA and DNA viruses, which form dsRNA during transcription and replication8. To counteract this host response, viruses have adopted a diverse set of strategies against PKR including: 1) blocking PKR expression, 2) preventing kinase activation, and 3) interfering with eIF2α phosphorylation9. Two of the best-characterized viral PKR inhibitors are the vaccinia virus (vac) proteins K3L and E3L. Orthologs of these proteins are present in many other poxviruses including swinepox virus (swpv) and variola virus (var), the causative agent of smallpox. K3L, which resembles the N-terminus of eIF2α, acts as a pseudosubstrate and competitive inhibitor of PKR10–14. The vacK3L14 and related myxoma virus M156R15 proteins resemble the N-terminal OB-fold domain in eIF2α16,17 with the greatest differences localized to the helix insert region that includes the Ser51 phosphorylation site in eIF2α. Like vacK3L, the related swpvC8L protein (40% amino acid sequence identity to vacK3L) is a potent inhibitor of PKR both in yeast and mammalian cells18. Supporting the notion that the vacK3L protein is a pseudosubstrate inhibitor of PKR, mutations altering residues in the vacK3L OB-fold domain that are conserved in eIF2α impaired K3L inhibition of PKR both in yeast13 and in vitro14. In contrast to the pseudosubstrate inhibitor K3L, vacE3L, which consists of an N-terminal Z-DNA binding domain linked to a C-terminal double-stranded RNA binding domain, interferes with PKR activation and heterodimerizes with the kinase19–21. The identification of two poxvirus proteins using distinct strategies to subvert PKR function indicates the critical role of PKR in suppressing viral replication. Interestingly, both K3L and E3L act as virulence and host-range factors, which has been proposed to reflect varying amounts of PKR and activator dsRNA in different cells22.
The crystal structure of the PKR KD bound to eIF2α revealed a typical protein kinase structure with a smaller N-terminal lobe involved in ATP binding and a larger C-terminal lobe that binds eIF2α23. The PKR KD was dimerized in a back-to-back orientation and the dimer contact residues, which are well conserved among the eIF2α kinases, were restricted to the N-terminal lobe. In contrast, the concave surface of the eIF2α OB-fold domain docked onto helix αG in the C-terminal lobe of both KD protomers, positioning the Ser51 phosphorylation site near the PKR active site cleft between the two lobes of the KD23. Both PKR dimerization and autophosphorylation on Thr446 were shown to be important for eIF2α phosphorylation and for binding to vacK3L4. Interestingly, PKR helix αG is longer and its position is displaced in comparison to other kinases23. Consistent with the notion that helix αG is important for eIF2α recognition, mutation of Thr487 in helix αG impaired eIF2α phosphorylation but not kinase autophosphorylation or phosphorylation of non-specific substrates4. Taken together, the eIF2α binding site on PKR, which is shared by pseudosubstrates like the vacK3L protein, extends from helix αG to the kinase active site.
Previously, we noted that the KDs of PKR genes were more divergent than those of other eIF2α kinases24. Here, using phylogenetic analyses we report accelerated evolution of the PKR KD due to positive selective pressure. Interestingly, swapping the identity of a positively selected residue in helix αG of human and mouse PKR had opposing effects on K3L sensitivity without affecting eIF2α phosphorylation. Our results indicate a potential role for PKR in preventing cross-species infections and reveal a potential limitation in using model systems to study human viral infections.
Results
Accelerated Evolution of the PKR kinase domain
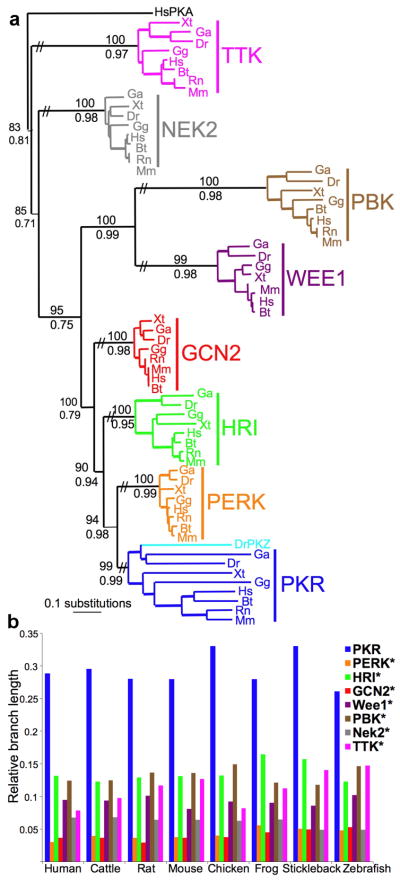
We performed phylogenetic analyses to systematically analyze the evolution of PKR genes, we compared the evolutionary trajectory of the KDs of PKR, the other three eIF2α kinases, HRI, PERK and GCN2 and four related kinases from the same eight vertebrate species: human (Homo sapiens, Hs), cattle (Bos taurus, Bt), mouse (Mus musculus, Mm), rat (Rattus norvegicus, Rn), chicken (Gallus gallus, Gg), frog (Xenopus tropicalis, Xt), stickleback fish (Gasterosteus aculeatus, Ga) and zebrafish (Danio rerio, Dr). Because all paralogous KDs had the same time to evolve, differences in the branch lengths directly reflect differences in the evolutionary pace. Interestingly, PKR branch lengths exceeded those of the other kinases by 2- to 9-fold (Fig. 1 and Supplementary Table 1), indicating a faster rate of evolution of the PKR KDs. A substantial increase in the ratio of nonsynonymous (coding for a different amino acid) to synonymous (non-coding, silent) substitutions was observed for the PKR genes, further supporting a faster rate of evolution (Supplementary Figs. 1,2). Stabilizing selection, which is the conservation of existing residues, was identified for the other eIF2α kinases, as indicated by the low ratios of nonsynonymous to synonymous substitutions. The accelerated evolution of the PKR KD is remarkable given that the different eIF2α kinases all phosphorylate the same substrate, and thus are under strong functional constraints to maintain enzymatic activity and interaction with eIF2α.
Figure 1. Accelerated evolution of PKR.
(a) Maximum likelihood phylogram of the kinase domains (KDs) of PKR, PERK, HRI, GCN2, WEE1, PBK, NEK2 and TTK from human (Homo sapiens, Hs), cattle (Bos taurus, Bt), mouse (Mus musculus, Mm), rat (Rattus norvegicus, Rn), chicken (Gallus gallus, Gg), frog (Xenopus tropicalis, Xt), stickleback fish (Gasterosteus aculeatus, Ga) and zebrafish (Danio rerio, Dr) [-lnL=12690.22265]. The topology of the Bayesian analysis showed comparable results. Posterior probabilities from the Bayesian analysis and bootstrap values from the maximum likelihood analysis are shown below and above major branches, respectively. (b) Relative branch lengths (from (a)) of KDs in the indicated species. Stars denote significant difference from PKR (P<0.001).
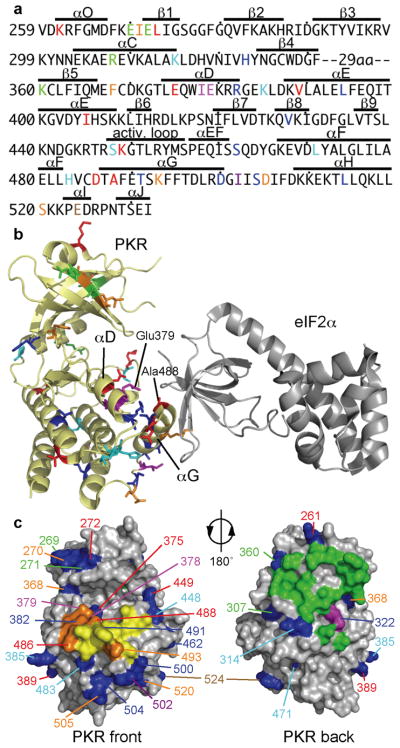
Extending the analysis of PKR evolution, by including sequences of 27 PKR and 3 PKZ genes25, a robust signature of positive diversifying selection was observed on specific branches and sites as determined using the programs PAML, REL, TreeSAAP, and tertiary windowing (Fig. 2a and Supplementary Fig. 3 and Supplementary Table 2). Positive (diversifying) selection describes the process of fixation of selectively advantageous mutations in a population, and can be statistically detected on specific residues in a sequence, or lineages on a phylogenetic tree. In total, 35 sites under positive selection in the PKR KD were identified with high confidence. It is noteworthy that besides the divergent positively selected residues, highly conserved residues are found in the PKR KD, 46 of which are invariant in all PKR genes analyzed (Supplementary Fig. 3). The residues under positive selection were dispersed across the structure of the PKR KD23 with a greater concentration in the C-terminal lobe near the eIF2α binding site (Fig. 2b,c). In addition, all but three (L394, I405 and V428) positively selected residues mapped to the surface of PKR.
Figure 2. Positively selected sites in the PKR kinase domain.
(a) Positively selected residues (PSRs) projected on the sequence of the human PKR (hPKR) kinase domain (KD). PSRs in different lineages are color-coded: vertebrates: red; tetrapods: orange; mammals & birds: blue; primates: azure; rodents: magenta; chicken: purple; xenopus: brown; PKZ: green. Sequences of the highly divergent kinase insert between β4 and β5 have been excluded from the analyses and are not shown. (b) PSRs are depicted as sticks and color-coded as above on a cartoon representation of the hPKR-eIF2α (gray) complex (PDB code 2A19)23. Position of helices αD and αG, which are enriched in PSRs, as well as residues E379 and A488 are indicated. (c) PSRs (blue) are highlighted in a surface representation of the PKR KD. The eIF2α contact site (front, left image) and the dimerization interface (back, right image) are shown. eIF2α contact residues are depicted in yellow; positively selected residues that contact eIF2α are shown in orange. Residues involved in dimerization are colored green; a positively selected residue in the dimer contact region is shown in magenta. Positions of PSRs are color-coded as described in (a).
Altered sensitivity of PKR mutants to poxvirus inhibitors
Why has PKR been subjected to positive selection? Given that PKR has antiviral activity and that many viruses express proteins or RNAs to inhibit PKR activity8,9, it seemed plausible that viruses have driven PKR evolution. We reasoned that viral PKR inhibitors must meet two criteria to be considered as candidates for the driving force behind PKR KD evolution. First, the viral PKR inhibitors must target the PKR KD; and second, the inhibitors must be found in viruses or viral families that infect many vertebrate species. These criteria are met by poxviruses, which include the well-characterized vaccinia virus (vac), swinepox virus (swpv), and variola virus (var), the causative agent of smallpox. All three viruses express two PKR inhibitors: vacK3L (varC3L, swpvC8L) and E3L proteins. Orthologs of K3L and E3L proteins are found in many genera of vertebrate poxviruses (Supplementary Table 3) and, as mentioned above, they act as virulence and host-range factors22.
We hypothesized that amino acid variations in the PKR KD were fixed during evolution because they provided a selective advantage by altering sensitivity to the viral inhibitors. As such, we predicted that mutations of positively selected residues in PKR will result in altered sensitivity to viral inhibitors. Because we do not know the identity of the PKR inhibitors of ancient viruses that shaped PKR evolution, we employed the best characterized poxvirus inhibitors available: vacK3L, a hyperactive mutant vacK3L-H47R, varC3L and varE3L. To test our hypothesis, we used a heterologous yeast system to analyze wild type and mutant forms of PKR for sensitivity to viral inhibition. Our expectation was that mutation of positively selected residues will confer enhanced sensitivity or resistance to viral inhibitors. In order to test this, we used site-directed mutagenesis to substitute 19 residues that have been under positive selection in human PKR with residues found at the corresponding position in different vertebrate species (Supplementary Table 4). Amongst hundreds of possible mutations, we focused on conspicuous sites, e.g. sites that show evident convergent evolution such as residue 488 (in human PKR) where 7 of the 9 amino acids found at this position evolved independently in different lineages25. The Ala488 residue, which is found in primates, horse, platypus and minnow fish was substituted by Glu, which is found in mouse and Xenopus PKR (Supplementary Fig. 3).
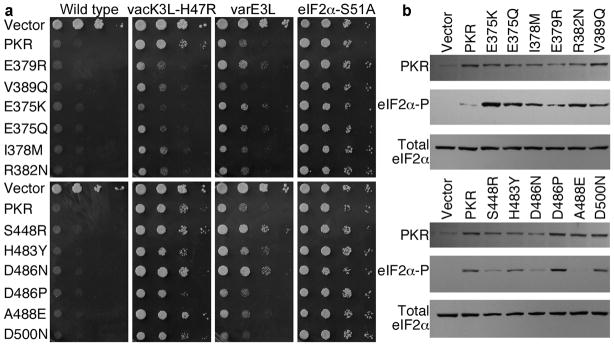
We tested the susceptibility of the PKR mutants to inhibition by vacK3L-H47R and varE3L in Saccharomyces cerevisiae strains that express the respective poxvirus proteins under the control of a galactose-inducible promoter. Expression of human PKR, also under the control of a galactose-inducible promoter, inhibits the growth of yeast13,26,27 (Fig. 3a, 1st panel, row 2). This growth defect is due to eIF2α phosphorylation and the attendant inhibition of protein synthesis, as it is suppressed by an eIF2α-S51A mutation that abolishes the phosphorylation site in eIF2α27 (Fig. 3a, 1st and 4th panel, row 2). Previously, we reported that co-expression of vacK3L or vacE3L suppresses the toxicity associated with expression of PKR in yeast13,20 (Fig. 3a, 2nd and 3rd panels, row 2), and we identified the hyperactive vacK3L-H47R mutant as a more potent inhibitor of PKR13,18. All of the PKR mutants were as toxic as WT PKR when expressed in the WT yeast strain lacking vacK3L (Fig. 3a, 1st panel, and Supplementary Table 4), indicating that the mutations did not alter the ability of PKR to phosphorylate eIF2α.
Figure 3. Altered sensitivity of PKR mutants to poxvirus proteins K3L and E3L.
(a) Yeast growth assays. Plasmids expressing WT or the indicated PKR mutants under the control of a galactose-inducible promoter were introduced into a WT strain or strains expressing vacK3L-H47R, varE3L or non-phosphorylatable eIF2α−S51A. Transformants were grown to saturation and 4-μl serial dilutions (of OD600 = 1, 0.1, 0.01 and 0.001) were spotted on galactose medium and incubated for 3 days at 30°C. (b) Western blot analyses of whole cell extracts from transformants described in (a) of the vacK3L-H47R strain expressing the indicated PKR mutants. Upper panels were probed with anti-Flag antibody to detect human PKR; middle panels were probed with phosphospecific antibodies against Ser51 in eIF2α; bottom panels were probed with polyclonal antiserum against total yeast eIF2α.
Mutations at 6 sites in human PKR (E375K/Q, I378M, E379R, R382N, V389Q, D486P) conferred increased resistance to vacK3L-H47R, restoring PKR toxicity, while 3 mutations (S448R, D486N, and A488E) rendered PKR more sensitive to vacK3L-H47R inhibition (Fig. 3a, 2nd panel, and Supplementary Table 4). Other mutations had no effect. The mutations altering vacK3L-H47R sensitivity mostly clustered in PKR helices αD and αG, which form part of the eIF2α-(and K3L)-docking site on the kinase (Fig. 2b). The concave face of the eIF2α OB-fold domain directly contacts PKR helix αG, while helix αD lies behind and buttresses the position of helix αG23. Genetic tests confirmed that the increased growth defects conferred by the K3L-resistant PKR mutants were due neither to promiscuous phosphorylation of heterologous proteins in yeast nor to kinase hyperactivity. WT PKR and the PKR mutants were non-toxic in a yeast strain expressing non-phosphorylatable eIF2α-S51A (Fig. 3a, 4th panel, and Supplementary Table 4) demonstrating that the growth defects were due to eIF2α phosphorylation. In addition, the PKR mutants conferred a growth defect similar to WT PKR in a yeast strain expressing a desensitized version of eIF2Bα (gcn3-102 allele; Supplementary Table 4) demonstrating that the mutants were not more active than WT PKR. Taken together, these results indicate that the PKR mutations specifically affect the ability of K3L to inhibit PKR. Consistent with this notion, three of the mutations, E375K, E375Q, and E379R, conferred resistance to vacK3L-H47R but increased the sensitivity of PKR to inhibition by varE3L (Fig. 3a).
To further address the mechanism by which the PKR mutations altered sensitivity to vacK3L-H47R inhibition, immunoblotting was used to monitor PKR expression and eIF2α phosphorylation in yeast. Importantly, the PKR mutants were expressed at levels comparable to WT PKR (Fig. 3b), consistent with the observation that the mutants conferred a growth defect similar to WT PKR in the eIF2Bα mutant strain (Supplementary Table 4). Moreover, the PKR helix αD mutants, E375K, E375Q, I378M, E379R, and R382N, that conferred resistance to vacK3L-H47R, were expressed at lower levels than WT PKR (Fig. 3b, upper panel). The impaired expression of these kinases is consistent with the previously reported negative autoregulation of PKR expression in yeast27,28. Thus, enhanced vacK3L-H47R resistance was not due to increased PKR expression for these mutants. As shown in Figure 3b, the level of eIF2α phosphorylation correlated well with the growth observed in the spotting assays. Increased eIF2α phosphorylation was observed in extracts from cells expressing PKR mutants that were resistant to vacK3L-H47R inhibition (see, e.g. Fig. 3b, upper panels, lanes 3–8 versus 2), while reduced levels of eIF2α phosphorylation were observed in extracts from the strains expressing the PKR-S448R, -D486N, -A488E mutants that were more sensitive to vacK3L-H47R inhibition (Fig. 3a,b, lower panels).
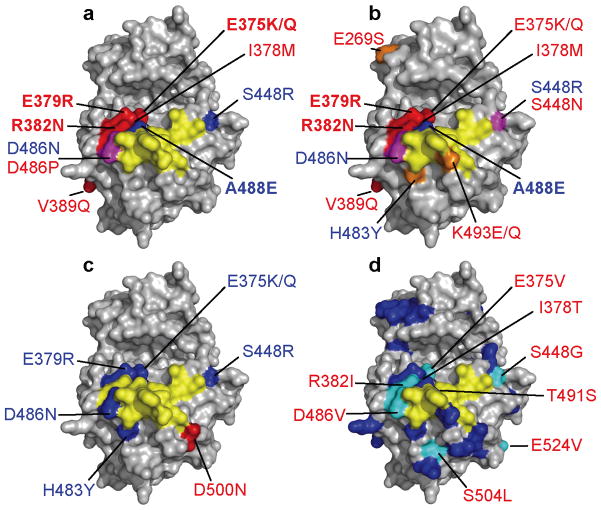
We suspect that multiple poxviruses contributed to the rapid evolution and positive selection in PKR. K3L orthologs from different poxviruses are very diverse, some of which displaying less than 30% sequence identity at the amino acid level (see Supplementary Table 5). The variola virus (var) C3L protein shares 80% amino acid sequence identity with vacK3L. In order to analyze whether K3L orthologs were positively selected during evolution, we calculated dN/dS rates of the K3L orthologs using PAML and REL as implemented in HyPhy. Overall, a weak signature of positive selection was detected, possibly influenced by the short sequence length of K3L orthologs (<100 amino acids), which makes detection of positive selection difficult. In general, the mutations that altered PKR sensitivity to vacK3L-H47R resulted in a similar change in PKR sensitivity to varC3L (Fig. 4 and Supplementary Table 4). However, four mutations (E269S, S448N, K493E and K493Q), that had no effect on the sensitivity of PKR to inhibition by vacK3L-H47R or varE3L, resulted in increased resistance to varC3L. On the other hand, E375Q and I378M mutations in PKR had only minor effects on varC3L sensitivity, while strongly increasing sensitivity to vacK3L-H47R. More PKR mutants affected vacK3L/varC3L than varE3L sensitivity (with only a single mutation D500N conferring greater varE3L resistance), consistent with the notion that pseudosubstrate inhibitors have played a greater role in evolution of the PKR KD. The different responses of these mutants to the various poxvirus inhibitors is consistent with the notion that a variety of viral inhibitors have shaped PKR evolution.
Figure 4. Positively selected residues (PSRs) in PKR that altered sensitivity to poxvirus inhibitors.
(a–d) eIF2α contact residues are depicted in yellow; PSRs that when mutated altered sensitivity to poxvirus inhibitors vacK3L-H47R (a), varC3L (b) and varE3L (c) are shown in red (increased resistance), blue (increased sensitivity) and magenta (distinct sets of mutants showed increased resistance or sensitivity). Mutations that had the strongest effect are shown in bold. (d) PKR mutations (labeled) identified in a random screen as conferring resistance to vacK3L-H47R31. PSRs are colored blue. 9 of 12 random mutations conferring resistance to vacK3L-H47R occurred at positively selected sites (cyan). The remaining three PKR mutations, which altered residues that were not identified to be under positive selection, and the PSR I405 are located in the interior of the protein and cannot be seen.
Differential sensitivity of human and mouse PKR to inhibition by K3L
To extend the analysis of the PKR mutants to human cells, we used a HeLa cell line in which the endogenous PKR was stably knocked down (PKRkd) by RNA interference29. To express PKR in these cells, we introduced synonymous mutations in the shRNA target sequence to obtain knock down resistant PKR (PKRkd-res) (Supplementary Fig. 5a). Following transfection, PKRkd-res was expressed much better and repressed activity of a co-transfected luciferase reporter stronger than wild type PKR (Supplementary Fig. 5b,c). Co-transfection of PKRkd-res with vacK3L, vacK3L-H47R and the swinepox virus (swpv) K3L ortholog C8L, which displays ca. 40% sequence identity on the protein level with vacK3L, partially restored luciferase activity compared to co-transfection with an empty vector (Supplementary Fig. 6a). We next tested two of the PKR mutants that in the yeast assays showed either resistance (PKR-E379R) or increased sensitivity (PKR-A488E) to pseudosubstrate inhibition. As observed in yeast, PKR-E379R was more resistant, and PKR-A488E was more sensitive, than WT PKR to vacK3L inhibition (Fig. 5a). In addition, PKR-E379R was also resistant to swpvC8L (Fig. 5a). Thus, the PKR mutants displayed similar activities in the yeast and mammalian cell assays.
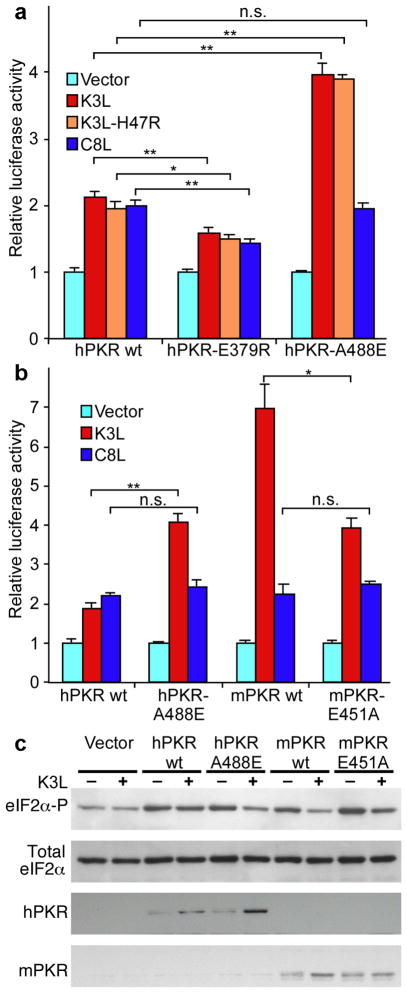
Figure 5. Effects of PKR variants on sensitivity to pseudosubstrate inhibition in HeLa cells.
(a) Human HeLa PKRkd cells were co-transfected with expression vectors for luciferase, knock-down resistant derivatives of human (h)PKR, hPKR-E379R or hPKR-A488E and control, vacK3L, vacK3L-H47R or swpvC8L. After 40 hours, cells were harvested, lysed, and samples were assayed for luciferase activity. Luciferase activity was normalized to control transfections lacking PKR inhibitors. Standard deviations are indicated for three independent transfections. Significant differences are indicated: * = p<0.005; ** = p<0.001; n.s. = not significant (p > 0.05). (b) Co-transfection of luciferase, hPKR, hPKR-A488E, mouse (m)PKR or mPKR-E451A and vacK3L or swinepox C8L. (c) Immunoblot analyses of whole cell extracts from cells transfected with the indicated expression vectors in the absence (−) or presence (+) of vacK3L. The blots were probed with antiserum specific for phosphorylated Ser51 in eIF2α (upper panel), total human eIF2α (second panel), hPKR (third panel), or mPKR (bottom panel).
Comparison of the susceptibility of human and mouse PKR to pseudosubstrate inhibition revealed that mouse (m) PKR was much more sensitive to vacK3L inhibition, while sensitivity to swpvC8L was comparable to hPKR (Fig. 5b). Substitution of A488 in hPKR by E, as found at the corresponding position 451 in mPKR, rendered hPKR more susceptible to vacK3L inhibition, and the converse mutation of E451 in mPKR to A, as found in hPKR, resulted in increased resistance to vacK3L. Interestingly, these mutations had no effect on swpvC8L sensitivity. Thus, single amino acid substitutions at position 488/451 in helix αG of h/mPKR functionally switched kinase sensitivity to pseudosubstrate inhibition. Phosphorylation levels of eIF2α correlated well with the observed inhibition of luciferase activity after co-transfection of PKR and vacK3L (Fig. 5c). While expression of WT and mutant forms of PKR was comparable in control-transfected cells, stronger expression of hPKR-A488E vs. hPKR and mPKR vs. mPKR-E451A was observed after co-transfection with vacK3L, likely due to relieved translational auto-inhibition30. Thus, altered sensitivity to vacK3L is not due to differences in PKR expression.
Discussion
We have shown that the KD of PKR evolved much faster than those of the other eIF2α kinases throughout the vertebrate lineage. This accelerated evolution is due to positive selected pressure. We identified 35 positively selected residues in the PKR KD. In order to test whether this rapid evolution altered PKR sensitivity to viral inhibition, we substituted positively selected residues in human PKR with residues found in related species and tested for sensitivity to inhibition by poxvirus proteins. More than half of the positively selected residues tested altered sensitivity to viral pseudosubstrate inhibitors of the K3L family. Moreover, the finding that some PKR mutants showed differential sensitivity to vacK3L and varC3L, which display 80% sequence identity on the protein level, suggests that pseudosubstrates from different poxviruses exerted virus-specific selective pressure. This interpretation is further supported by the finding that human and mouse PKR showed markedly different sensitivity to vacK3L, while no difference was observed in the sensitivity to swpvC8L.
Independent support for our hypothesis that viruses have driven the rapid evolution of the PKR family comes from an unbiased screen for PKR mutants resistant to K3L inhibition. We randomly mutated human PKR and screened for mutants that were toxic in a yeast strain expressing vacK3L-H47R. Interestingly, 9 of the 12 single amino acid substitutions identified in this screen altered residues that our phylogenetic analyses revealed as under positive selection during evolution (Fig. 4d)31 , supporting our bioinformatic analyses. Like many of the mutations described in this report, the PKR mutations identified in the screen did not affect eIF2α phosphorylation, but instead specifically impaired vacK3L inhibition of PKR. Biochemical studies of the helix αG PKR-D486V mutation identified in the mutagenic screen revealed no change in the Km for eIF2α phosphorylation, but nearly a 15-fold decrease in K3L binding affinity31. We proposed that these random PKR mutants, like the directed mutants examined in this study, differentially affected substrate and pseudosubstrate binding owing to differences in the rigidity of eIF2α and the K3L protein. The overlap in PKR residues identified in the molecular genetic screen and the evolutionary studies provides additional support for our primary hypothesis that viruses, including poxviruses, have driven the rapid evolution of the PKR family.
Mutations that altered the sensitivity of PKR to inhibition by the K3L orthologs were enriched in helices αD and αG. While mutations in helix αG might directly affect K3L binding, we propose that mutations in other parts of PKR, including helix αD, might indirectly affect pseudosubstrate interactions by reposition helix αG. The question arises then as to how repositioning of helix αG has a greater impact on K3L inhibition than on eIF2α phosphorylation. It is noteworthy that the greatest structural differences between eIF2α and the K3L protein are restricted to the helix insert region that includes the Ser51 phosphorylation site in eIF2α. The well-ordered (low relative B-factors) and apparently rigid helix insert region in the K3L protein consists of a four turn α-helix and a 3/10 helix 14. In contrast, the helix insert region in eIF2α contains two 3/10 helices and an intervening linker containing Ser5116. Although the helix insert region was resolved in the structure of free yeast eIF2α16, it is likely to be quite flexible (high relative B-factors16,17). In further support of this inherent flexibility in the eIF2α helix insert, this region was disordered in the PKR-eIF2α complex structure14. Finally, docking the structure of free eIF2α onto the PKR-eIF2α complex structure revealed that the Ser51 residue must move ~17 Å to access the PKR active site. As both eIF2α and the K3L protein contact PKR through helix αG and the kinase active site, mutations that cause movement of helix αG might weaken binding of the rigid K3L protein by impairing productive contacts near the kinase active site. In contrast, the flexible nature of eIF2α, especially in the vicinity of the Ser51 phosphorylation site, might compensate for altered helix αG positioning and facilitate eIF2α phosphorylation by the PKR helix αD mutants.
While our data indicate that poxvirus K3L orthologs contributed to PKR evolution, it is not clear specifically which of these pseudosubstrates directly contributed to the evolution of PKR. Most likely, a variety of poxviruses, including some that are now extinct, shaped the evolution of PKR genes from different species. Poxviruses infect many different vertebrate species. While variola virus, which was restricted to humans and caused smallpox before its eradication, displays strict host-specificity, other poxviruses show much broader host ranges32. In general, poxvirus tropism appears to be influenced by the manipulation of intracellular signaling cascades after viral entry into a cell32,33. Many different poxviruses can cause zoonotic infections in humans. These infections are usually self-limiting, but occasionally they can cause life-threatening disease, especially in immunocompromised people34. Vaccinia virus, the most extensively studied orthopoxvirus, was used as a vaccine in the eradication of smallpox. While the natural host of vaccinia virus is unknown, the virus can infect a wide range of animals and can occasionally cause serious systematic infection in humans. In addition to poxvirus K3L orthologs, other viral gene products might have exerted positive selective pressure on the PKR KD. Among these other candidates are the double-stranded RNA binding protein E3L and vIF2α, an eIF2α analog found in fish and amphibian iridoviruses that has been suggested to function as an inhibitor of PKR35,36. The notion that other viruses have exerted positive selective pressure on PKR is consistent with the observation that mutations at 8 out of 19 sites under positive selection did not alter sensitivity to vacK3L, varE3L or varC3L.
Importantly, K3L was a more potent inhibitor of mouse PKR than of human PKR. Swapping a single positively-selected amino acid at the corresponding positions in helix αG of human and mouse PKR had opposite effects on the K3L resistance of the kinases. Introducing the mouse Glu in place of A488 in human PKR increased K3L sensitivity, while introducing human Ala in place of mouse E451 conferred greater resistance to K3L inhibition. Accordingly, we would predict that K3L will have a greater impact on infection of mouse cells than of human cells. Consistent with this prediction, it was previously shown that K3L-deleted vaccinia virus is impaired for replication in mouse L929 cells10 but not in human HeLa cells22. Our results provide a new and interesting interpretation of this apparent host-range activity of K3L. Previously it was proposed that K3L and E3L contribute to vaccinia virus host range due to differing amounts of PKR or dsRNA activators in different cells22. Based on our PKR evolution studies, we propose an alternate model in which the altered sensitivity mouse and human PKR to K3L inhibition, in part, underlies the host range effect of K3L.
Taken together, our findings indicate that virus tropism might be influenced by species-specific differences in the sensitivity of PKR (and possibly other innate immune system components) to viral inhibitors. We suggest that such differences should be taken into account when interpreting the results of studies on pathogen-host interactions using non-human cell lines or animals as models for human infections. As an extension of this idea, we propose that an important function of PKR might be to thwart inter-species transmission of a virus from a natural host reservoir, against which its PKR inhibitors were optimized, to a genetically more distant and only occasionally infected host.
Methods
Evolutionary analyses
Kinase sequences were obtained through database searches in GenBank. Sequences of 27 PKR and PKZ sequences used in this work have been previously described25. Accession numbers of all sequences used are shown in Supplementary Table 6.
Kinase domain sequences were aligned in MAFFT 637, using iterative refinement methods with weighted sum-of-pairs scores and consistency scores as implemented in the L-INS-I algorithm. Poorly aligned regions were removed with Gblocks38. Alignments were analyzed in PhyML to compute heuristic maximum likelihood (ML) trees from amino acids, and nucleotides, using the JTT and the GTR model including a gamma correction (eight categories of evolutionary rates, an estimated parameter and an estimated proportion of invariant sites), respectively39. Nodal support was estimated by bootstrapping of 100 replications of the original dataset. Additionally, Bayesian inference was implemented (MrBayes), using the previously mentioned models, with 3 independent runs at 2,000,000 generations sampled every 100 generations and four chains.
For each kinase domain, lineage branch lengths (= number of nucleotide substitutions per site) were compared. The total branch length was calculated by adding the length values stemming from the point of speciation per kinase domain paralog and leading to the terminal taxa. Their average length was then obtained by dividing the summed branch lengths by the number of branches associated with the nodes involved within each assemblage. A two-group t-test was employed to assess the significance of the differences in mean branch length between paralogous kinase domains for the eight species per group.
The dN/dS rates of the kinase domains in the evolutionary trajectory from their ancestral sequence to the extant representatives were estimated using four different approaches:
PAML. Analyses were carried out using the inferred unrooted tree topology from the ML search. Branch lengths for this tree were estimated by PAML 440 with the M0 model. All subsequent analyses were run twice to check for consistency. Branch-specific selection was tested on each branch using free-ω-ratio models. Furthermore, likelihood of evolution was compared under models prohibiting (M1, M7, M8a) versus allowing positive selection (M2, M8). Site-specific selection per branch was studied by allowing ω (dN/dS) to vary both among lineages and among sites (Model A: model = 2; Nsites = 2; ω = fixed (1) versus ω = estimated; d.f. = 1).
HyPhy. Evidence of positive selection was sought using a codon-based random effects likelihood (REL) approach, as implemented in HyPhy41. The REL approach allows the synonymous substitution rate to vary among codons, and pinpoints codons under positive or negative selection on a branch. Since this dataset is relatively small, and type I errors might be more common, random effects likelihood (Bayes Factor = 50) was used. Sites identified at Bayes factor 50, and 10 are reported.
Tertiary Windowing. This analysis employs windowing in tertiary structure, and is based on the assumption that co-selected sites might be close in a folded protein, but not in primary sequence space42. The program implementation was modified from the published per window dS to using global dS calculation (http://www.wyomingbioinformatics.org/WebServices/3D-Windowing/index.htm). Using the known structure file of kinase domains from the PDB database (pdb:2A1A), windows of 10 angstrom starting from a central codon were constructed.
TreeSAAP. TreeSAAP43 uses calculations of expected random distributions of possible amino acid changes based on fixed differences between residues. It measures selective influences on a set of structural and biochemical amino acid properties along a phylogenetic tree. TreeSAAP uses Yang’s baseml module44 to calculate ancestral sequences from a specified tree. In this study, we specified 20 categories of change, and a sliding window of 20 bp. Greater than expected numbers of replacements (relative to the neutral model) in categories 18–20 indicate positive diversifying selection.
Structural models were generated using PDB database coordinates (2A1A) of the PKR-eIF2α complex23 using PyMOL software (http://pymol.sourceforge.net/).
Plasmids and yeast strains
Construction of the human PKR expression plasmid using the vector pYX113 (R&D systems) has been described25. In this construct human PKR is tagged at the C-terminus with His6 and Flag epitope tags. For expression in human cells, human and mouse PKR were cloned into the vector pSG5 (Stratagene). Site-specific mutations were generated by PCR mutagenesis using Pfu polymerase (Stratagene) and primers containing the desired mutations following the QuickChange protocol (Stratagene). To obtain knock-down resistant human PKR, we introduced synonymous mutations in the shRNA target sequence using the QuickChange protocol (Stratagene) and primers: F(5'-CTT AAT ACA TAC CGT CAG AAa CAa GGc GTg GTc tTg AAg TAT CAA GAA CTG CCT AAT TC-3') and R(5'-GAA TTA GGC AGT TCT TGA TAc TTc Aag ACc ACg CCt TGt TTC TGA CGG TAT GTA TTA AG-3'); mutated sites are indicated by small letters. All inserts were completely sequenced to confirm desired mutations and exclude additional mutations.
Yeast strains expressing poxvirus inhibitors were generated in three steps. First, cDNAs encoding vacK3L-H47R, varC3L, and varE3L were subcloned in the vector pEMBLyex445 under the control of a yeast GAL-CYC1 hybrid promoter. Second, the GAL-CYC1 promoter and poxviral protein coding sequence from each of the three plasmids was subcloned into the yeast LEU2 integrating plasmid pRS305. Third, the resultant plasmids were digested with EcoRV and each linear fragment was used to transform the yeast strain H2557 (MATα ura3-52 leu2-3 leu2-112 trp1-Δ63 gcn2Δ) generating derivatives in which the GAL-CYC1 construct was integrated at the LEU2 locus. The resulting strains were named J674 (GAL-CYC1-K3L-H47R at LEU2), J664 (GAL-CYC1-C3L at LEU2), and J659 (GAL-CYC1-E3L at LEU2). The WT control strain was constructed by integrating the empty vector pRS305 at the LEU2 locus of strain H2557 generating strain J673. Strain J223 (MATα ura3-52 leu2-3 leu2-112 trp1-Δ63 gcn2Δ SUI2-S51A) was described previously4. Strain J234 (MATα gcn3-102 gcn2::KanMX leu2-3 leu2-112 ura3-52) is a derivative of strain H17 in which the GCN2 gene has been deleted. The gcn3-102 allele encodes eIF2Bα-E44D, which renders eIF2B less sensitive to inhibition of phosphorylated eIF2α27.
Yeast growth assays
Yeast strains were transformed with the PKR expression plasmids using standard methods. For each transformation, four single colonies were selected and colony purified. Purified transformants were streaked on SGal medium (inducing conditions: yeast minimal complete medium containing 10% (w/v) galactose and all amino acids except uracil) and grown at 30°C for 3–4 days. Additionally, representative transformants were additionally grown to saturation in SD medium, and 4-μl of serial dilutions (of OD600 = 1, 0.1, 0.01, and 0.001) were spotted on SGal medium and incubated for 3 days at 30°C. For western blot analysis, yeast transformants were grown and whole cell extracts (WCEs) were prepared as described25 with the exception that cells were grown for 4 hrs under inducing conditions before harvesting.
Cell line, transient transfections, luciferase activity and western blot
HeLa control and HeLa PKR knock-down cells were kindly provided by Charles Samuel29 and were grown in DMEM medium containing 10% FCS. Cells were seeded the day before transfection. For luciferase assays, 3×104cells were seeded and were co-transfected using 2 μl GenJet for HeLa cells (SignaGen Laboratories) with 0.05 μg luciferase plasmid pGL3promoter (Promega), 0.1 μg PKR expression vector and 0.9 μg of the vacK3L, vacK3L-H47R or swinepox C8L expression vectors18. In control experiments the empty pSG5 vector was used. For each plasmid triplicate transfections were performed. After 40 hours cells were harvested and luciferase activity was determined using a luciferase detection kit (Promega) luminometer (Pharmingen, Monolight 3010). Experiments were performed at least three times and representative experiments are shown. Statistical significance was determined using Student's t-test.
For western blot analysis, 1×105 cells were seeded on 6-well plates and were co-transfected using 4 μl GenJet for HeLa cells (SignaGen Laboratories) with 0.2 μg PKR expression vector and 1.8 μg of the vacK3L, vacK3L-H47R or swinepox C8L expression vectors. 24 hours after transfection, whole cell extracts (WCEs) were obtained by SDS lysis and sonication. Protein content was measured using the Biorad Protein Assay (Biorad).
Immunoblot analysis
6 μg of WCEs were subjected to SDS-PAGE and proteins were transferred to nitrocellulose membranes. For immunoblot analysis, the lower part of the membrane was probed with rabbit phosphospecific antibodies directed against Ser51 in eIF2α (BioSource International). The membranes were stripped and re-probed with polyclonal antibodies against yeast eIF2α46 or human eIF2α (Santa Cruz Biotechnology). The upper part of the membranes was probed with antibodies against human PKR (71/10), and then stripped and re-probed with antibodies against mouse PKR (Santa Cruz Biotechnology). Flag-tagged proteins were detected with anti-Flag Antibody (Applied Biological Materials).
Supplementary Material
Acknowledgments
We are grateful to Alan Hinnebusch, Michael Yu and Loubna Tazi for helpful discussion and for careful reading of the manuscript; Frank Sicheri, Madhusudan Dey, David McClellan, Anthony Furano and Christian Tellgren-Roth for helpful discussion; and Charles Samuel (University of California, Santa Barbara) for kindly providing the PKR-knock-down cells, and Makiko Kawagishi-Kobayashi and Madhusudan Dey (NICHD) for yeast strains. This work was supported by funding from the Intramural Research Program of the National Institutes of Health, NICHD to TED.
Footnotes
Authors contributions:
This study was initiated by SR and experiments were conceived and designed by SR, KD, JSG and TED. Sequences were compiled by SR and evolutionary analyses performed by KD. Yeast and cell line experiments were performed by SR. C3L- and E3L-expressing yeast strains were constructed by EJS. The paper was written by SR, KD and TED.
Supplementary information accompanies this manuscript.
References
- 1.Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 2.Ishii KJ, Akira S. Innate immune recognition of nucleic acids: beyond toll-like receptors. Int J Cancer. 2005;117:517–523. doi: 10.1002/ijc.21402. [DOI] [PubMed] [Google Scholar]
- 3.Kaufman RJ. Double-stranded RNA-activated protein kinase PKR. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2000. pp. 503–527. [Google Scholar]
- 4.Dey M, et al. Mechanistic link between PKR dimerization, autophosphorylation, and eIF2α substrate recognition. Cell. 2005;122:901–913. doi: 10.1016/j.cell.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 5.Barber GN. The dsRNA-dependent protein kinase, PKR and cell death. Cell Death Differ. 2005;12:563–570. doi: 10.1038/sj.cdd.4401643. [DOI] [PubMed] [Google Scholar]
- 6.Dever TE, Dar AC, Sicheri F. The eIF2α Kinases. In: Mathews MB, Sonenberg N, Hershey JW, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; 2007. pp. 319–344. [Google Scholar]
- 7.Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- 8.Toth AM, Zhang P, Das S, George CX, Samuel CE. Interferon action and the double-stranded RNA-dependent enzymes ADAR1 adenosine deaminase and PKR protein kinase. Prog Nucleic Acid Res Mol Biol. 2006;81:369–434. doi: 10.1016/S0079-6603(06)81010-X. [DOI] [PubMed] [Google Scholar]
- 9.Langland JO, Cameron JM, Heck MC, Jancovich JK, Jacobs BL. Inhibition of PKR by RNA and DNA viruses. Virus Res. 2006;119:100–110. doi: 10.1016/j.virusres.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 10.Beattie E, Tartaglia J, Paoletti E. Vaccinia virus-encoded eIF-2α homolog abrogates the antiviral effect of interferon. Virology. 1991;183:419–422. doi: 10.1016/0042-6822(91)90158-8. [DOI] [PubMed] [Google Scholar]
- 11.Davies MV, Elroy-Stein O, Jagus R, Moss B, Kaufman RJ. The vaccinia virus K3L gene product potentiates translation by inhibiting double-stranded-RNA-activated protein kinase and phosphorylation of the α subunit of eukaryotic initiation factor 2. J Virol. 1992;66:1943–1950. doi: 10.1128/jvi.66.4.1943-1950.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll K, Elroy-Stein O, Moss B, Jagus R. Recombinant vaccinia virus K3L gene product prevents activation of double-stranded RNA-dependent, initiation factor 2α-specific protein kinase. J Biol Chem. 1993;268:12837–12842. [PubMed] [Google Scholar]
- 13.Kawagishi-Kobayashi M, Silverman JB, Ung TL, Dever TE. Regulation of the protein kinase PKR by the vaccinia virus pseudosubstrate inhibitor K3L is dependent on residues conserved between the K3L protein and the PKR substrate eIF2α. Mol Cell Biol. 1997;17:4146–4158. doi: 10.1128/mcb.17.7.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dar AC, Sicheri F. X-ray crystal structure and functional analysis of vaccinia virus K3L reveals molecular determinants for PKR subversion and substrate recognition. Mol Cell. 2002;10:295–305. doi: 10.1016/s1097-2765(02)00590-7. [DOI] [PubMed] [Google Scholar]
- 15.Ramelot TA, et al. Myxoma virus immunomodulatory protein M156R is a structural mimic of eukaryotic translation initiation factor eIF2α. J Mol Biol. 2002;322:943–954. doi: 10.1016/s0022-2836(02)00858-6. [DOI] [PubMed] [Google Scholar]
- 16.Dhaliwal S, Hoffman DW. The crystal structure of the N-terminal region of the α subunit of translation initiation factor 2 (eIF2α) from Saccharomyces cerevisiae provides a view of the loop containing serine 51, the target of the eIF2α-specific kinases. J Mol Biol. 2003;334:187–195. doi: 10.1016/j.jmb.2003.09.045. [DOI] [PubMed] [Google Scholar]
- 17.Nonato MC, Widom J, Clardy J. Crystal structure of the N-terminal segment of human eukaryotic translation initiation factor 2α. J Biol Chem. 2002;277:17057–17061. doi: 10.1074/jbc.M111804200. [DOI] [PubMed] [Google Scholar]
- 18.Kawagishi-Kobayashi M, Cao C, Lu J, Ozato K, Dever TE. Pseudosubstrate inhibition of protein kinase PKR by swine pox virus C8L gene product. Virology. 2000;276:424–434. doi: 10.1006/viro.2000.0561. [DOI] [PubMed] [Google Scholar]
- 19.Chang HW, Watson JC, Jacobs BL. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc Natl Acad Sci U S A. 1992;89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romano PR, et al. Inhibition of double-stranded RNA-dependent protein kinase PKR by vaccinia virus E3: role of complex formation and the E3 N-terminal domain. Mol Cell Biol. 1998;18:7304–7316. doi: 10.1128/mcb.18.12.7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharp TV, et al. The vaccinia virus E3L gene product interacts with both the regulatory and the substrate binding regions of PKR: implications for PKR autoregulation. Virology. 1998;250:302–315. doi: 10.1006/viro.1998.9365. [DOI] [PubMed] [Google Scholar]
- 22.Langland JO, Jacobs BL. The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology. 2002;299:133–141. doi: 10.1006/viro.2002.1479. [DOI] [PubMed] [Google Scholar]
- 23.Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2α by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 24.Rothenburg S, et al. A PKR-like eukaryotic initiation factor 2α kinase from zebrafish contains Z-DNA binding domains instead of dsRNA binding domains. Proc Natl Acad Sci U S A. 2005;102:1602–1607. doi: 10.1073/pnas.0408714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothenburg S, Deigendesch N, Dey M, Dever TE, Tazi L. Double-stranded RNA-activated protein kinase PKR of fishes and amphibians: varying number of double-stranded RNA binding domains and lineage-specific duplications. BMC Biol. 2008;6:12. doi: 10.1186/1741-7007-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chong KL, et al. Human p68 kinase exhibits growth suppression in yeast and homology to the translational regulator GCN2. Embo J. 1992;11:1553–1562. doi: 10.1002/j.1460-2075.1992.tb05200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dever TE, et al. Mammalian eukaryotic initiation factor 2α kinases functionally substitute for GCN2 protein kinase in the GCN4 translational control mechanism of yeast. Proc Natl Acad Sci U S A. 1993;90:4616–4620. doi: 10.1073/pnas.90.10.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romano PR, Green SR, Barber GN, Mathews MB, Hinnebusch AG. Structural requirements for double-stranded RNA binding, dimerization, and activation of the human eIF-2α kinase DAI in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:365–378. doi: 10.1128/mcb.15.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang P, Samuel CE. Protein kinase PKR plays a stimulus- and virus-dependent role in apoptotic death and virus multiplication in human cells. J Virol. 2007;81:8192–8200. doi: 10.1128/JVI.00426-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barber GN, et al. Translational regulation by the interferon-induced double-stranded-RNA-activated 68-kDa protein kinase. Proc Natl Acad Sci USA. 1993;90:4621–4625. doi: 10.1073/pnas.90.10.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seo EJ, et al. Protein kinase PKR mutants resistant to the poxvirus pseudosubstrate K3L protein. Proc Natl Acad Sci U S A. 2008;105:16894–16899. doi: 10.1073/pnas.0805524105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McFadden G. Poxvirus tropism. Nat Rev Microbiol. 2005;3:201–213. doi: 10.1038/nrmicro1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Werden SJ, Rahman MM, McFadden G. Poxvirus host range genes. Adv Virus Res. 2008;71:135–171. doi: 10.1016/S0065-3527(08)00003-1. [DOI] [PubMed] [Google Scholar]
- 34.Lewis-Jones S. Zoonotic poxvirus infections in humans. Curr Opin Infect Dis. 2004;17:81–89. doi: 10.1097/00001432-200404000-00003. [DOI] [PubMed] [Google Scholar]
- 35.Essbauer S, Bremont M, Ahne W. Comparison of the eIF-2α homologous proteins of seven ranaviruses (Iridoviridae) Virus Genes. 2001;23:347–359. doi: 10.1023/a:1012533625571. [DOI] [PubMed] [Google Scholar]
- 36.Majji S, et al. Rana catesbeiana virus Z (RCV-Z): a novel pathogenic ranavirus. Dis Aquat Organ. 2006;73:1–11. doi: 10.3354/dao073001. [DOI] [PubMed] [Google Scholar]
- 37.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 39.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 40.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 41.Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 42.Berglund AC, Wallner B, Elofsson A, Liberles DA. Tertiary windowing to detect positive diversifying selection. J Mol Evol. 2005;60:499–504. doi: 10.1007/s00239-004-0223-4. [DOI] [PubMed] [Google Scholar]
- 43.Woolley S, Johnson J, Smith MJ, Crandall KA, McClellan DA. TreeSAAP: selection on amino acid properties using phylogenetic trees. Bioinformatics. 2003;19:671–672. doi: 10.1093/bioinformatics/btg043. [DOI] [PubMed] [Google Scholar]
- 44.Yang Z. Phylogenetic analysis using parsimony and likelihood methods. J Mol Evol. 1996;42:294–307. doi: 10.1007/BF02198856. [DOI] [PubMed] [Google Scholar]
- 45.Cesareni G, Murray JAH. Plasmid vectors carrying the replication origin of filamentous single-stranded phages. In: Setlow JK, Hollaender A, editors. Genetic engineering: principals and methods. Vol. 9. Plenum Press; New York, N.Y: 1987. pp. 135–154. [Google Scholar]
- 46.Dever TE, et al. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.