Abstract
The output of a genome assembler generally comprises a collection of contiguous DNA sequences (contigs) whose relative placement along the genome is not defined. A procedure called scaffolding is commonly used to order and orient these contigs using paired read information. This ordering of contigs is an essential step when finishing and analyzing the data from a whole-genome shotgun project. Most recent assemblers include a scaffolding module; however, users have little control over the scaffolding algorithm or the information produced. We thus developed a general-purpose scaffolder, called Bambus, which affords users significant flexibility in controlling the scaffolding parameters. Bambus was used recently to scaffold the low-coverage draft dog genome data. Most significantly, Bambus enables the use of linking data other than that inferred from mate-pair information. For example, the sequence of a completed genome can be used to guide the scaffolding of a related organism. We present several applications of Bambus: support for finishing, comparative genomics, analysis of the haplotype structure of genomes, and scaffolding of a mammalian genome at low coverage. Bambus is available as an open-source package from our Web site.
Large-scale whole-genome shotgun sequencing (WGS) was successfully used in 1995 to sequence, for the first time, the complete genome of a free-living organism, Haemophilus influenzae (Fleischmann et al. 1995), a 1.83 million-base-pair (Mb) bacterium. Previously this technique—pioneered by Fred Sanger in 1982 (Sanger et al. 1982)—had only been used to sequence DNA molecules of at most 200 thousand base pairs (kb). Shotgun sequencing involves breaking up the DNA at random into a collection of small fragments, sequencing those fragments, then using a computer program, called an assembler, to piece back together the original molecule. For Haemophilus, the original assembly contained 140 contiguous pieces of DNA (contigs) which then were joined together through laboratory experiments. The success of the Haemophilus assembly largely hinged upon the use of a “double-ended shotgun” strategy, in which both ends of a collection of DNA fragments of known sizes (2 kb and 15-20 kb) were sequenced. The pairing of sequencing reads from the ends of each fragment allowed the researchers at The Institute for Genomic Research (TIGR) to order and orient the 140 contigs and then quickly fill in the gaps to produce the complete sequence of the genome. This procedure of ordering and orienting a collection of contigs, using paired read information, is called scaffolding (Roach et al. 1995) because it builds a virtual scaffold upon which a genome can be completed.
The success of the Haemophilus sequencing project led TIGR to incorporate scaffolding into all other sequencing projects, using a computer program called “grouper”. Other sequencing centers performed scaffolding in a largely manual fashion, and until recently “grouper” was the only stand-alone scaffolder in use. The successful use of WGS at Celera to assemble the fruit fly genome (Myers et al. 2000) spurred the development of new assembly programs (Huson et al. 2001; Kent and Haussler 2001; Pevzner and Tang 2001; Batzoglou et al. 2002; Jaffe et al. 2003; Mullikin and Ning 2003) specifically designed to handle large genomes. These large-scale WGS assemblers rely heavily on read pairing information, and all of them include a scaffolding module. Moreover, it is now standard practice to report the scaffolds as part of the output of an assembler.
Our group, also faced with the challenge of handling increasingly larger genomes, decided to rewrite the aging “grouper” program. Our extensive experience in finishing genomes had taught us the value of using all available information for scaffolding, not only the information contained in read pairs but also diverse types of independent mapping data. The laboratory procedures for ordering and orienting contigs are much more expensive than generating shotgun reads (Tettelin et al. 1999; Beigel et al. 2001), thus any additional information we can obtain electronically leads to considerable savings. We have developed a flexible scaffolder, Bambus, designed to accommodate large (including mammalian-sized) genomes and to use a variety of sources of linking information. Since October 2002, Bambus has been used successfully in all sequencing projects at TIGR. Bambus is available as an open-source package, freely available for downloading and redistribution, through our Web site at http://www.tigr.org/software/bambus.
The following sections describe in detail the current version of our software (version 2.3) and demonstrate its use in a variety of applications: scaffolding as support for finishing efforts, comparative analysis of multiple strains of Bacillus anthracis, analysis of the haplotype structure of eukaryotic genomes, and the analysis of a mammalian genome at low sequence coverage.
Shotgun Sequencing Overview
A shotgun sequencing project begins with a sample containing large numbers of identical (ideally) double-strand DNA molecules, which are sheared at random into a collection of small fragments. Both ends of each fragment are then sequenced to obtain a collection of sequencing reads as shown in Figure 1. Current sequencing technologies are only able to generate reads between 600 and 1200 bp long, and thus the centers of the fragments usually remain unsequenced.
Figure 1.
Pairing of reads from a fragment.
The reads have a defined orientation, as shown by the arrows in the figure, reflecting the DNA strand from which they were generated. The fragments are selected from a collection of libraries, where each library consists of fragments of roughly equal size that have been inserted into a cloning vector. Reads from opposite ends of the same clone insert are known as clone mates. A typical bacterial sequencing project may use a mixture of libraries of 2-4 kb and 10-12 kb in size, whereas larger projects also use fosmid (30-42 kb) or BAC (50-150 kb) libraries. The reads are then assembled into contiguous pieces (contigs) using an assembler. In all but the simplest cases, repeats and incomplete information prevent the assembler from reconstructing the original DNA molecule as a single contig. The output of the assembler thus consists of a collection of contigs, each contig representing a tiling of reads. Each contig has an implicit orientation, corresponding to the DNA strand being reconstructed. Each read is therefore represented by both its coordinates within the contig and its relative orientation with respect to the chosen orientation of the contig. Virtually all large assemblies contain many clone mates that reside in different contigs. This information, coupled with knowledge of the approximate size of the fragment, allows us to infer the relative placement and distance between the two contigs, as shown in Figure 2.
Figure 2.
Contig adjacency as inferred from clone mates A and B.
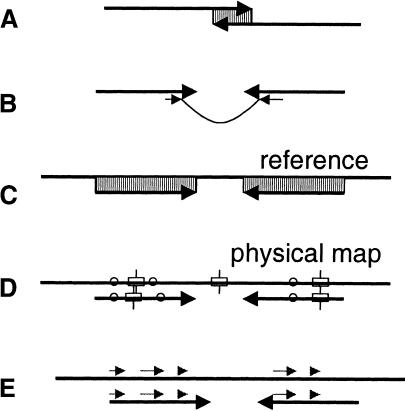
Mated reads from opposite ends of the same fragment have traditionally been the only source of information used in scaffolding. Increasingly, though, other data sources are becoming available that provide equally powerful information for ordering and orienting contigs. One example is an alignment to a completed reference genome (Fig. 3C), a type of data that is becoming increasingly common as the number of sequenced genomes increases. We used this approach in our analysis of the Bacillus anthracis genome (Read et al. 2002), an example of which is described in the Results section. Additional sources of linking information include contig overlaps, physical maps, and information about the conservation of gene synteny (Fig. 3). The first source, contig overlaps, may sound confusing if one assumes that the assembler always merges overlapping contigs. This is not always the case, as for example when a mixture of two divergent haplotypes is being assembled. In regions where the two haplotypes differ, the polymorphisms may be sufficient to prevent the assembler from putting contigs together. Scaffolding software can relax the criteria for detecting contig overlaps and then utilize this information to generate larger contigs.
Figure 3.
Sources of linking information between contigs. (A) overlaps, (B) clone mates, (C) alignments to reference genome, (D) alignments to physical maps, (E) conservation of gene synteny.
In order to accommodate all such sources of contig adjacency information, Bambus accepts as input abstract contig links characterizing the relative orientation and spacing of adjacent contigs.
METHODS
Background
Scaffolding involves finding an order and orientation of all contigs such that constraints imposed by contig links are satisfied. As described above, contig links may contain a variety of sources of information that imply a particular relative placement of two contigs. When the linking data contain errors, the general problem of globally satisfying all constraints is intractable (Huson et al. 2001; Kent and Haussler 2001). Interestingly, the associated contig orientation problem is also intractable, as is the complementary problem of ordering the contigs when a proper orientation is given. The orientation problem is equivalent to finding a maximum bipartite subgraph, whereas the ordering problem is similar to the Optimal Linear Arrangement problem, both of which are NP-hard (Garey and Johnson 1979). Therefore, it is necessary to pursue approximate solutions in order to obtain solutions in a reasonable amount of compute time.
Prior solutions to the scaffolding problem followed two general frameworks. The first formulates scaffolding as a nonlinear optimization problem under the constraints provided by the contig links (Thayer et al. 1999; Jaffe et al. 2003; J. Vinson, pers. comm.). Due to the complexity of solving such nonlinear systems (typical solutions involve many iterations of complex relaxation steps), practical implementations of this approach are limited to local optimization steps within the scaffolder (Myers et al. 2000; Kent and Haussler 2001; Jaffe et al. 2003). The second framework poses the scaffolding problem in graph theoretic terms. With one exception, the Eulerian graph approach of Pevzner and Tang (2001), all scaffolding algorithms to date construct a graph whose nodes correspond to contigs and whose edges represent the presence of links between the corresponding contigs. In order to reduce the effect of errors, most scaffolders require at least two links between adjacent contigs. They then “bundle” all links between adjacent contigs into a single contig edge and greedily join the contigs into scaffolds. The pathmerging algorithm of Huson et al. (2001) examines the edges in decreasing order of the number of links in the bundle. Whenever the current edge links two distinct scaffolds, the algorithm attempts to merge the scaffolds together in a path-merging step. Arachne (Batzoglou et al. 2002; Jaffe et al. 2003) uses edge weights that depend on both the number of links and the size of the edge, whereas Phusion (Mullikin and Ning 2003) examines edges in order of their lengths, from smallest to largest. This latter approach simplifies the scaffold-merging step by reducing the chances that the scaffolds being merged intertwine. Arachne (Jaffe et al. 2003) and Jazz (Aparicio et al. 2002) incorporate an iterative error-correction step during which scaffolds may be broken and then recombined based on links that were not used during the original greedy step.
Bambus Overview
Most currently existing scaffolders are tightly coupled with a particular assembler, making it difficult or impossible for the user to influence the particular scaffolding algorithm or its parameters. Bambus attempts to overcome these limitations by using a flexible scaffolding algorithm and by providing a general XML-based interface for specifying contig linking information. Conversion scripts can be easily written to allow Bambus to utilize the output of most assembly programs; the current release includes interfaces for TIGR Assembler (Sutton et al. 1995), phrap (Green 1994) or any other assembler producing. ACE files, and Arachne. Abstract contig links inferred from sources other than mate-pair information can be easily provided to Bambus. As an example, the current release of the MUMmer (Delcher et al. 1999, 2002) alignment package contains a utility that converts alignments to a reference genome into Bambus-compatible linking data.
Bambus also provides users with more control over the scaffolding algorithms. The core algorithm is based on a greedy heuristic whose parameters can be specified through a simple configuration file. For example, users can easily modify the minimum number of links from a particular library required to connect two contigs. The order in which links are considered also can be modified. Bambus allows users to specify such an order on the basis of link types or on the basis of the number of links connecting adjacent contigs. Thus, Bambus can emulate both the approach used in Phusion (Mullikin and Ning 2003), where links are considered in order of their lengths, and the approach used by Huson et al. (2001) where links are considered in order of their redundancy.
By design, Bambus does not always generate an unambiguous ordering of contigs. Ambiguities in the scaffold are useful in guiding the finishing of a genome, or when analyzing its haplotype structure. Such examples will be discussed in detail in the Results section. However, when unambiguous scaffolds are required, Bambus can generate them via a separate module that “untangles” the ambiguous scaffolds into a collection of linear scaffolds.
Finally, Bambus documents the decisions taken by its internal algorithms by providing, for every link in the input, one of four possible codes. A valid link is one that was used in a scaffold and that is consistent with all other information; an invalid orientation link is one deemed erroneous due to orientation; an invalid length link is one that violates length constraints; and an unused link is one that was ignored by the program. These codes can be used to validate the output of Bambus, or can highlight problems with the input. For example, large numbers of links marked invalid due to their length may indicate that the library size estimates provided to Bambus were incorrect.
The following sections provide detailed descriptions of the algorithms used by Bambus.
Edge Bundling
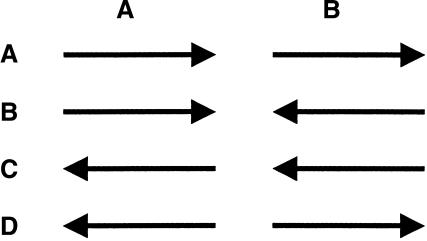
To simplify the task of subsequent scaffolding stages, Bambus starts with a link-bundling step, which combines all links between two contigs into a single edge representing the adjacency relationship. Each individual link, l, can be described in terms of the relative orientation of the adjacent contigs, o, and the range of sizes allowed for the gap between the two contigs [gmin, gmax]. The four possible orientations of two contigs A and B are shown in Figure 4. For each pair of contigs, the bundling procedure starts by partitioning all links l1..k into clusters based on orientation, that is, within each cluster all links imply the same relative orientation of the two contigs. Within each cluster, the bundling procedure finds those links whose size ranges agree, specifically, we identify the largest set of links that are pairwise consistent. This set can be easily identified in linear time because it is sufficient to find the largest clique in the interval graph induced by the inter-contig gap ranges for the links. The links that do not belong to the maximum clique for A and B are given the invalid length tag. The size range for each edge is thus defined by the intersection of size ranges for all links that are pairwise consistent (as described above). Note that the size of a link is defined as a range rather than a (mean, standard deviation) pair. We found this approach to work well in practice; furthermore, the specific distribution of link sizes is difficult or impossible to determine for some types of links, such as those inferred from comparative analyses. Future versions of Bambus will allow the specification of link sizes by mean and standard deviation for those links where such a representation is relevant.
Figure 4.
Possible contig pairings.
At this point, the cluster with the most links is chosen, and all links in other clusters are given the invalid orientation tag. Finally, if the maximum cluster size for A and B is below a certain, user-defined limit (called redundancy), the whole cluster gets removed and no edge is created. The links removed by this final test are tagged as unused and made available to later stages in the algorithm. This approach is similar to that used by most existing scaffolders, which require two mate-pair links between any two contigs, a requirement that dramatically reduces the influence of errors in the link data. Bambus takes the technique one step further by allowing the user to specify different redundancies depending on the confidence in the data (e.g., we could allow single links from 2-kb libraries while requiring at least two BAC-end links).
We would like to bring to the attention of the reader the fact that the heuristics described in the previous paragraph, as well as those used in most other scaffolding programs, are based on the assumption that discrepancies in the linking data are caused by errors alone. This fact is not entirely true; for example, repeats misassembled by the assembler may introduce inconsistencies in the linking data. Although Bambus does not attempt to identify potential repeats, it reports in the output all discrepancies found during the edge-bundling stage in order to allow the users to identify the location of such possible misassemblies.
At the end of the bundling stage, the surviving valid links define a set of contig edges. These edges are naturally oriented, in the sense that given a particular orientation for one of the contigs, the relative order of the adjacent contigs is uniquely defined.
Note that two of the possible contig adjacencies (Fig. 4B,D) require that contigs have opposite orientations. We call any such edge in a graph a reversal edge. These edges are essential in understanding the contig ordering problem.
In this graph formulation, scaffolding is equivalent to two separate problems: (1) contig orientation, which requires finding a consistent orientation for all nodes given the constraints imposed by the edges, and (2) contig ordering, which is equivalent to embedding the contigs on a line while preserving the intercontig gaps specified by the edges. Our approaches to solving these problems are presented below.
Contig Orientation
In the absence of errors, the contig orientation problem is trivial because it can be phrased as the problem of coloring a bipartite graph with two colors, the colors corresponding to the two possible orientations of each contig. In the presence of errors, however, the underlying graph may not be bipartite. Undirected cycles that contain an odd number of reversal edges prevent us from assigning a consistent orientation to all nodes in the graph. We are thus faced with an optimization problem: Remove the minimum number of edges (the principle of parsimony suggests that these links are erroneous) such that the remaining graph contains no cycles with an odd number of reversal edges. This optimization problem is NP-hard (Kececioglu and Myers 1995) and so we have opted for a heuristic approach. We greedily assign orientations to contigs ignoring an edge if it conflicts with a previously oriented contig. This simple approach performs very well in practice because the edge-bundling stage removes most erroneous links.
Contig Ordering
Once all contigs have been properly oriented, we need to embed them on a line (or circle in the case of some bacterial chromosomes) by finding an ordering of the contigs that is consistent with the constraints specified by the edges. Again this problem is trivial in the absence of errors, as we can solve it through a simple topological ordering of the nodes in the graph, subject to edge length constraints (see, e.g., Thayer et al. 1999). Errors in the linking data may prevent a consistent solution, though, as for example when constraints propagated along two parallel paths are found to be inconsistent when the two paths meet at the same node. The associated optimization problem that removes the minimum number of edges in order to allow a consistent layout is NP-hard. We use a greedy heuristic, starting with all of the edges at their maximum allowable length (where length corresponds to the fragment size from the DNA library), then assign each contig a coordinate on a linear axis. This procedure starts by anchoring the first unplaced contig, then traverses the graph in a breadth-first manner, placing each contig at the maximum value permitted by its edge constraints. We ignore those links that contradict the placement of an already visited node, specifically, if two parallel paths meet at a node N, the first path to reach N defines its placement. Once a node is placed, its position will not change, and all edges that disagree with its placement are rejected as incorrect. This step is necessary to ensure that the topological ordering of the nodes is consistent; that is, one contig cannot both precede and follow another contig.
Finally, contigs are brought back towards each other as close as possible to the midpoint of the range defined by the length constraints on the edges. The basic rationale behind this “expand-contract” heuristic is that setting the edges at their maximum length provides the opportunity for small contigs to fit in the gaps between other contigs. The subsequent contraction step reduces the gap sizes to the midpoint of the allowable range defined by the edge-bundling procedure. The ordering procedure is repeated until all contigs have been placed in scaffolds.
Note that our approach does not prevent two contigs from occupying the same space. Such a situation may legitimately occur in practice when assembling a mixture of haplotypes. Other scaffolders remove such ambiguity from the data, thus removing information useful in the analysis of the genome. Even in the case when the ambiguous ordering is caused by misassembled repeats, the ambiguous information is useful because it allows finishing experts to identify and resolve these repeats.
Hierarchical Scaffolding
The problems described in the previous sections are computationally intractable when errors are present in the data. We have developed a heuristic framework meant to reduce the effect of errors in the data. Each link presented to Bambus can be assigned a priority reflecting the quality of the link. Scaffolding then proceeds in a hierarchical fashion. The highest-quality links are used to generate a set of scaffolds, then iteratively lower-quality links are added in order to combine already created scaffolds. At each stage, we only use those links that do not conflict with the already computed scaffolds. Note that the quality of a particular link reflects two different factors: (1) the underlying quality of the actual sequencing process—for example, read pairing errors are significantly fewer in small insert libraries than in BAC libraries, and (2) the number of links connecting two contigs—because errors generally occur as independent events, collections of links that are mutually consistent greatly increase the confidence in the contig adjacency information. Note, however, that this latter assumption can be incorrect for misassembled repeats. To reduce the effects of such misassemblies, Bambus includes a mechanism for screening out suspected repeats.
Untangling
As discussed above, the scaffolds produced by Bambus may contain ambiguously placed contigs. Although this situation may be preferred when using the scaffolds to guide finishing efforts, or when analyzing the haplotype structure of a genome, these ambiguous scaffolds cannot be used by analysis software that depends on a single linear molecule, such as gene finders or database search software (e.g., BLAST). To aid such analyses we have developed a program called “untangle” that converts the ambiguous scaffolds into single linear stretches.
The untangling problem is equivalent to finding a path through the graph such that no two nodes on the path overlap. Unfortunately, this problem is NP-complete, essentially equivalent to the “path with forbidden pairs” problem (Garey and Johnson 1979). We implemented a simple heuristic that proceeds in a greedy fashion, traversing the scaffold from left to right and removing links that cause contigs to overlap. The untangler proceeds by iteratively finding a longest non-self-overlapping path, removing all of the nodes on the path and the edges adjacent to them, then repeats the procedure for the remaining nodes.
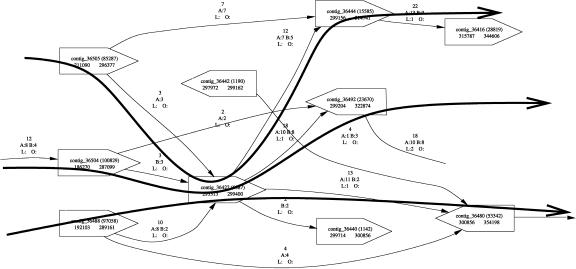
This simple procedure performs fairly well in practice, as will be shown in the next section. It is important to note, however, that tangles such as that shown in Figure 7 (see below) cannot be correctly resolved without splitting the contig corresponding to a collapsed repeat (the contig in the center of the figure) into multiple contigs representing the copies of the repeat. The untangling algorithm described above would choose an arbitrary path through the tangle, then isolate the remaining paths into separate scaffolds. We are in the process of developing a more sophisticated algorithm that correctly identifies and resolves repeats; those results will be presented in a future publication.
Figure 7.
Scaffold “tangled” by a repeat and potential paths through the repeat. This scaffold, from Brucella suis, was resolved by correct placement of three copies of identical ribosomal RNA sequences at distinct locations in the genome.
RESULTS
Genome Finishing
Linking information provided by mated reads from opposite ends of the same fragment has been used to guide the finishing of virtually all genomes sequenced since the completion of H. influenzae. The task of ordering and orienting the contigs is daunting in the absence of linking data. Laboratory techniques exist that help determine the correct ordering of contigs (Tettelin et al. 1999); however, these techniques are expensive in terms of both labor and reagents. Scaffolding greatly reduces the complexity of this task by providing large stretches over which the contigs are correctly ordered and oriented. In this case specialized techniques are only needed to order the scaffolds with respect to each other.
Genome finishing involves targeted sequencing experiments designed to fill in the gaps between the contigs, in order to obtain a single contiguous sequence for each molecule of DNA. Bambus aids this process by providing information about all of the DNA fragments that bridge a gap between two contigs. This information, useful in selecting the most appropriate fragment to be used as a template in a directed sequencing experiment, is provided explicitly by Bambus in a “details” file (Fig. 5). The first line describes the relationship between the two adjacent contigs, namely their estimated coordinates and orientations in the scaffold, as well as a summary of the number of valid (v:) and invalid (l: and o:, wrong length and wrong-orientation, respectively) links. The following lines summarize information about all of the links connecting the two contigs, specifically, the names of the reads, their coordinates and orientation within the contigs, as well as an estimate of the fragment size.
Figure 5.
Detailed information produced by Bambus. Shown are two contigs connected by two valid links (v:2 in the header line), with 1 additional link whose length is outside the estimated range and therefore invalid (l:1). The contigs face away from each other, indicated by the arrows (“→”) in the header. Each pair of linked reads is shown on a separate line, with coordinates indicating the position of the read within its contig. For example, GBRDE74TR is mapped to positions 890-1416 of contig_32, and GBRDE74TF is mapped to positions 413-1207 of contig_38.
Bambus also provides a global overview of the relationships between the contigs. This graphical display is produced with the help of the GraphViz package (Gansner and North 2000). Figure 6 presents an example of the overview image. Each contig's length and coordinates within the scaffold are indicated in the figure. The arrow representing the contig indicates its orientation in the scaffold. The links between the contigs are annotated with information about the number of valid links and the libraries they belong to (represented as single letter codes), as well as information about the number of links deemed invalid due to their length or orientation.
Figure 6.
Graphical output of Bambus.
The situation shown in Figure 6 is a “clean” example, in that the links allow a consistent layout of the contigs along the chromosome. This overview display provides little additional information in the case of such linear scaffolds. The display is, however, extremely useful when analyzing complex scaffolds. As an example, Figure 7 shows the “tangled” structure of a scaffold caused by the presence of a misassembled repeat in the highlighted contig. The image allows us to estimate that at least three copies of this repeat (in this case a ribosomal RNA subunit from Brucella suis; Paulsen et al. 2002) are assembled together in the central contig, because the graph shows at least three possible paths (shown by thick arrows in the figure) through this repeat. Each path corresponds to the location of one of the repeat copies in the genome. This information can be used to design laboratory experiments in order to validate the correct layout of the genome. Moreover, such tangles in the graph are useful in identifying potential misassemblies even in the absence of unusually deep-coverage (a key feature often used to detect misassemblies) regions in the contigs.
Haplotype Detection
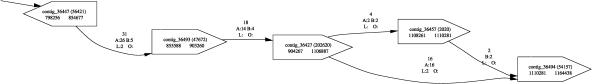
The genomes of most multicellular organisms contain two copies of each chromosome. Though generally similar, the copies, called haplotypes, often differ from each other enough to complicate the assembly process. The differences usually consist of single-base-pair polymorphisms, though longer insertion and deletions are not uncommon. In extreme cases, for example, in the case of the sea-squirt Ciona intestinalis (Dehal et al. 2002), the two copies of a chromosome may differ by up to 15%; moreover, such differences are not uniform across the chromosome. Separating the chromosomes prior to sequencing is usually impossible, leading to a shotgun library that contains a mixture of the chromosomes. Most assembly programs were not specifically developed to handle polymorphic data, and as a result might misassemble these regions. For example, a polymorphic segment might be assembled as two tandem nonidentical copies of the region. In the highly polymorphic regions, assemblers are able to separate the two chromosomes into different contigs. The resulting assembly is thus a mosaic of the two haplotypes. This structure becomes immediately obvious in the output of Bambus. Figure 8 shows such a situation found in the Trypanosoma cruzi genome, a human parasite currently being sequenced at TIGR. The eight contigs in the figure are arranged in a structure typical for polymorphic genomes. The graph can be decomposed into two parallel sections (contigs 2, 3, 5, 6 and contigs 1, 4, 7) that appear to occupy the same place along the chromosome yet whose contigs have not been assembled together. Unlike the case of repeats where the graph appears hopelessly tangled (as in Fig. 7), the scaffolds of polymorphic genomes have structures similar to those in Figure 8 that stretch for long sections of the chromosome. This characteristic “bubble” graph (initially described by Fasulo et al. 2002) is a clear indication of polymorphism. The bubbles—parallel paths—appear in those sections of the genome where the two haplotypes diverge, then rejoin in sections where the two chromosomes are similar (e.g., at contigs 4 and 8). To verify this hypothesis we aligned the contigs to each other using MUMmer (Delcher et al. 2002). The result, shown at the bottom of Figure 8, confirms the existence of two haplotypes that diverge by as much as 17% between each other.
Figure 8.
Haplotype structure in T. cruzi. The top image is Bambus output; the bottom is the haplotype structure as identified by alignments with MUMmer. For each alignment the percent identity is provided in the figure.
We found similar structures in an assembly of the sea-squirt Ciona savignyi that we created from data provided to the Trace Archive (http://www.ncbi.nlm.nih.gov/Traces) by the Whitehead Institute's Center for Genome Research. Similar evidence of widespread polymorphism was found in the related species C. intestinalis (Dehal et al. 2002).
Comparison With Celera Assembler and Arachne
We tested the performance of Bambus on the assembly benchmark data set provided at http://www.tigr.org/tdb/benchmark. These data consist of the shotgun reads from five bacterial genomes: Brucella suis 1330 (Paulsen et al. 2002), Shewanella oneidensis MR1 (Heidelberg et al. 2002), Staphylococcus aureus COL (S. Gill, unpubl. data), Staphylococcus epidermidis RP62A (S. Gill, unpubl. data), and Wolbachia spp (J. Eisen, unpubl. data). All five genomes have been finished to a high degree of accuracy so that they represent reliable references for testing assemblies. We compared the output of Bambus with that produced by the scaffolders used within Celera Assembler (Myers et al. 2000) and Arachne (Batzoglou et al. 2002; Jaffe et al. 2003). It is important to note that the purpose of this comparison was to evaluate the performance of Bambus rather than compare Celera Assembler to Arachne. We separately ran Bambus on the contig sets generated by each assembler, then “untangled” the resulting scaffolds in order to produce a collection of linear scaffolds (for more information on this procedure, see the Methods section). The results are summarized in Table 1. For each set of scaffolds we report the maximum size and span, as well as the N50 size of the scaffolds (The N50 statistic of a set of scaffolds [weighted median] is the minimum scaffold length L such that more than half of the genome is contained in scaffolds of length ≥L). The size of a scaffold represents the sum of the lengths of the contigs contained in the scaffold. The span of a scaffold is an estimate of the length of the section of the genome covered by the scaffolds that includes the estimated distances between linked contigs. Span statistics are of less importance when comparing scaffolds as they are very sensitive to the accuracy of library size estimates. Table 2 describes the overall statistics for each of the five genomes.
Table 1.
Comparison of Bambus Using Celera Assembler and Arachne on Five Benchmark Genomes
| Max size | N50 Size | Max span | # Errors | ||
|---|---|---|---|---|---|
| Brucella sius 1330 | Ca | 2,090,920 | 2,090,920 | 2,100,248 | 0 |
| CA Bambus | 1,556,915 | 1,100,469 | 1,578,082 | 0 | |
| CA Bambus norep. | 1,556,915 | 610,258 | 0 | ||
| Arachne | 1,804,830 | 1,804,830 | 1,800,825 | 0 | |
| Arachne Bambus | 1,214,328 | 511,873 | 1,217,772 | 5 | |
| Arachne Bambus norep. | 1,100,196 | 688,090 | 0 | ||
| Shewanella oneidensis MR1 | CA | 228,793 | 62,460 | 230,298 | 0 |
| CA Bambus | 228,793 | 69,276 | 231,668 | 11 | |
| CA Bambus norep. | 228,793 | 55,775 | 5 | ||
| Arachne | 244,815 | 80,523 | 251,981 | 10 | |
| Arachne Bambus | 286,184 | 81,273 | 297,842 | 26 | |
| Arachne Bambus norep. | 286,184 | 81,273 | 26 | ||
| Staphylococcus aureus COL | CA | 65,596 | 10,146 | 66,028 | 0 |
| CA Bambus | 88,011 | 15,661 | 89,215 | 18 | |
| CA Bambus norep. | 88,011 | 15,402 | 10 | ||
| Arachne | 113,689 | 18,139 | 112,139 | 7 | |
| Arachne Bambus | 113,689 | 16,614 | 114,249 | 4 | |
| Arachne Bambus norep. | 113,689 | 15,574 | 7 | ||
| Staphylococcus epidermidis RP62A | CA | 1,090,358 | 781,570 | 1,094,683 | 0 |
| CA Bambus | 776,834 | 610,961 | 791,183 | 2 | |
| CA Bambus norep. | 1,086,408 | 780,057 | 0 | ||
| Arachne | 770,082 | 734,638 | 796,757 | 0 | |
| Arachne Bambus | 1,087,230 | 768,256 | 1,141,607 | 0 | |
| Arachne Bambus norep. | 1,087,230 | 768,256 | 0 | ||
| Wolbachia spp | CA | 256,979 | 83,636 | 262,188 | 0 |
| CA Bambus | 277,395 | 117,944 | 281,045 | 0 | |
| CA Bambus norep. | 277,395 | 119,900 | 0 | ||
| Arachne | 374,494 | 255,964 | 393,785 | 0 | |
| Arachne Bambus | 376,400 | 319,924 | 383,768 | 1 | |
| Arachne Bambus norep. | 376,400 | 194,340 | 0 |
CA, Celera Assembler output; Arachne, Arachne output; CA Bambus and Arachne Bambus, Bambus scaffolds based on Celera Assembler and Arachne contigs; norep., Bambus scaffolds after repeats were masked. The first two columns indicate the maximum and N50 sizes and spans (see main text). The “# errors” column indicates the number of contigs that were incorrectly scaffolded with respect to the reference genome.
Table 2.
Overall Assembly Statistics
| # Contigs | N50 Contig size | Total contig size | # Scaffolds | N50 Scaffold span | Total scaffold span | ||
|---|---|---|---|---|---|---|---|
| Brucella suis | CA | 50 | 100,829 | 3,290,467 | 2 | 2,100,248 | 3,305,486 |
| Arachne | 77 | 79,320 | 3,308,265 | 15 | 1,800,825 | 3,302,830 | |
| Shewanella oneidensis | CA | 392 | 22,444 | 4,792,524 | 166 | 62,400 | 4,827,793 |
| Arachne | 924 | 10,247 | 4,812,904 | 177 | 86,353 | 5,028,099 | |
| Staphylococcus aureus | CA | 429 | 6,496 | 2,114,874 | 252 | 10,739 | 2,178,030 |
| Arachne | 602 | 7,776 | 2,155,916 | 198 | 24,050 | 2,324,505 | |
| Staphylococcus epidermidis | CA | 24 | 539,290 | 2,620,329 | 10 | 782,602 | 2,627,124 |
| Arachne | 81 | 112,603 | 2,662,917 | 35 | 768,373 | 2,793,522 | |
| Wolbachia sp | CA | 194 | 111,803 | 1,524,267 | 139 | 86,177 | 1,553,563 |
| Arachne | 677 | 9,540 | 1,946,056 | 483 | 229,859 | 2,116,116 |
Before discussing the results of the comparison, we would like to point out a particular feature of Celera Assembler that affects the interpretation of the data. Celera Assembler does not incorporate all contigs into scaffolds. Some contigs are considered to have poor quality (based on arrival rate statistics; Myers et al. 2000) and are ignored by the scaffolder. Because these contigs were available to Celera Assembler's scaffolder, we provided them as input to Bambus. Bambus is required to place all contigs in scaffolds and will thus generally create more scaffolds than Celera Assembler. The number of scaffolds should therefore be discounted when comparing Celera Assembler with Bambus.
The results of our comparison show that Bambus performs well when compared to both Celera Assembler and Arachne. Based on size statistics, Bambus outperformed Celera Assembler on three of the five data sets: S. oneidensis, S. aureus, and Wolbachia. Bambus outperformed Arachne on a different set of three genomes: S. oneidensis, S. epidermidis, and Wolbachia. In a fourth genome, S. aureus, Bambus' performance was very slightly worse than Arachne's, with both scaffolders generating the same largest scaffold. Bambus had poor performance (with respect to the other two scaffolders) in only one case: the Brucella suis genome. Closer inspection revealed that Bambus was unable to resolve the layout of the genome around the three rRNA genes (large, 100% identical regions) present in Brucella. Both Celera Assembler and Arachne incorrectly collapsed these genes into a single 6-kbp contig. Removing this contig allowed Bambus to build scaffolds comparable to those produced by Celera Assembler and Arachne.
The overall statistics shown in Table 1 can be misleading, mainly because scaffolds are sometimes much larger because they are incorrect. We aligned all of the contigs to the reference for each genome, then compared each scaffold with the order and orientation of the contigs defined by these alignments. Both Arachne and Celera Assembler produced some incorrect contigs which could not be consistently aligned to the reference. For the purposes of the comparison, we used only those contigs that could be unambiguously located within the reference sequence. A summary of the contigs ignored by the analysis is shown in Table 3. The last column in Table 1 reports the numbers of contigs that were incorrectly placed in scaffolds; that is, those contigs whose order or orientation did not agree with that implied by an alignment to the reference. In the case of a scaffold that contained errors, we identified the largest number of contigs that agreed with the reference and marked the remaining contigs as errors.
Table 3.
Alignment Statistics
|
Celera assembler
|
Arachne
|
|||||
|---|---|---|---|---|---|---|
| Good | Mis-asm. | No match | Good | Mis-asm. | No match | |
| B. suis | 65 | 3 | 3 | 76 | 0 | 1 |
| S. oneidensis | 626 | 12 | 94 | 907 | 7 | 10 |
| S. aureus | 717 | 1 | 62 | 588 | 1 | 19 |
| S. epidermidis | 47 | 6 | 19 | 69 | 2 | 10 |
| Wolbachia | 83 | 24 | 958 | 139 | 5 | 533 |
Good, contigs that confirmed the reference; mis-asm, disagreements between a particular assembly and the reference—these are likely mis-assemblies in the assembly; no match, the number of contigs that did not align to the reference—most likely contaminants (e.g., Wolbachia is known to have a large amount of Drosophila contamination) or small mis-assembled contigs.
On average, Bambus produces slightly more erroneous scaffolds than Arachne and Celera Assembler. This is not surprising given that this version of Bambus was designed as a support tool for finishing efforts, by providing a consistent view of all linking information between contigs. The software used to generate linear scaffolds is confused by misassembled repeats, leading to the observed errors. We would like to point out that the input to Bambus consisted of only the location of reads within the contigs and information about the pairing of the reads. Scaffolders that are tightly coupled with a particular assembly package can usually utilize additional information, such as the location of repeats and potential misassemblies, and therefore can have a distinct advantage over general-purpose scaffolders such as Bambus. We provided Bambus with the capability to screen out those links anchored in user-specified sections of the genome. This feature allowed us to reduce the influence of repeats on Bambus output. We identified potential repeats in the genome using the Closure Repeat Finder (open-source package available from the TIGR software page http://www.tigr.org/software). This package, based on “reputer” (Kurtz and Schleiermacher 1999), identifies nearly identical (>97.5% identity) repeats of more than 200 bp. Such repeats often confuse assembly programs, leading to ambiguous linking information. Bambus can ignore all reads anchored in such repeats, thus limiting the effects of potential misassemblies. After this repeat-screening step, Bambus was able to generate better scaffolds. A similar phenomenon can be observed in our analysis of low-coverage dog genome data. With the exception of S. oneidensis and S. aureus, the repeat-masked scaffolds contained no errors. Whereas in most cases the removal of repeat-induced links led to smaller scaffolds (as evidenced by a decrease in the N50 scaffold sizes), in S. epidermidis the absence of ambiguous data allowed Bambus to generate significantly larger scaffolds. For the same genome, Bambus generated larger scaffolds than Arachne, all of them correct. Similarly, in Wolbachia Bambus outperforms Celera Assembler and generates scaffolds similar to Arachne's.
Comparative Genomics
An ever-increasing number of genomes are becoming available to researchers, providing opportunities for comparative genomic analyses. Although for some genomes a complete sequence is available, for many others the only available data are shotgun sequencing reads and a collection of contigs from a preliminary assembly of these reads. In the latter case, the use of scaffold data is essential for comparative genomic applications by providing long-range relationships between individual contigs. Here we describe the use of Bambus in two different comparative projects.
The first project is an analysis of multiple strains of Bacillus anthracis. The project started at TIGR after the bio-terror attacks of October 2001 with an analysis of the differences between the strain used in the attacks and a common laboratory strain (Read et al. 2002). These differences can be used to design highly precise strain identification assays. The genome of the Ames strain of B. anthracis was completed at TIGR (Read et al. 2003) and provides a reference for comparisons with other related strains. We have sequenced the genomes of Bacillus cereus, B. anthracis strain Kruger B, and an ancestral Ames strain (with more strains underway). We assembled each of the individual strains using Celera Assembler. In order to take advantage of the reference sequence provided by the completed Ames genome, we aligned the contigs from each assembly to the reference using MUMmer. The “showtiling” program of the MUMmer package was used to convert the alignments into a set of Bambus-compatible links between the contigs. We then used Bambus to produce scaffolds in a hierarchical manner. We first used the mate-pair information, then added the links inferred from the alignment data to link together the resulting scaffolds. In all but the case of B. cereus, Bambus was able to generate a single scaffold spanning the entire main chromosome (5.23 Mb). Because B. cereus is a more distant relative of B. anthracis, the nucleotide-level comparisons did not yield sufficient information to produce a single scaffold. We therefore aligned the B. cereus contigs to the B. anthracis reference at the protein level using “promer,” another program packaged with MUMmer. The protein-level links allowed us to generate a correct scaffold spanning most of the main chromosome of B. cereus. Note that the alignment data were only used in places where mate-pair data were either nonexistent or insufficient. This method can thus be used for scaffolding even when some rearrangements have occurred between the two genomes. Giving priority to the alignment data would limit our ability to detect such rearrangements.
The second comparative project is an assembly of the Drosophila pseudoobscura genome using data provided by the Human Genome Sequencing Center at Baylor College of Medicine. D. pseudoobscura was sequenced with the specific goal of making a comparison with Drosophila melanogaster and using the comparison to improve gene prediction in Drosophila species. We assembled the shotgun data using Celera Assembler, obtaining a 146-Mb assembly (available from ftp://ftp.tigr.org/pub/data) comprised of 4653 scaffolds. As is clear from Tables 1 and 2, Celera Assembler is particularly conservative when generating scaffolds, a feature that allows it to produce few errors, sometimes at the expense of scaffold size. In order to obtain larger scaffolds, we used Bambus to build upon the Celera Assembler scaffolds using mate-pair information that did not conflict with the layout imposed by Celera Assembler. In addition, we allowed Bambus to utilize “degenerate” contigs from Celera Assembler (contigs that the assembler concludes are repetitive or otherwise unreliable, and that it does not use further) that could help bridge the gap between two scaffolds. The result was a collection of 4349 scaffolds that were 3 kb longer on average. Using these larger scaffolds, we identified 912 potential large-scale rearrangements between the genomes of D. pseudoobscura and D. melanogaster. This number correlates well with other estimates of the number of chromosomal rearrangements that have occurred since the divergence of D. melanogaster and D. pseudoobscura (Gonzalez et al. 2002).
Analysis of Low-Coverage Shotgun Data From the Dog Genoem
Due to the large costs involved in completely sequencing a mammalian genome, funds are currently available for sequencing only those organisms considered of strategic importance to biomedical research. In order to assess the usefulness of lowcoverage data, researchers at TIGR and Celera Genomics sequenced the genome of a standard poodle to 1.5 × coverage (Kirkness et al. 2003). The analysis of these data can pave the way for similar low-pass sequencing projects meant to provide the scientific community with genomic data from multiple mammals, even when funds are not available for the completion of these genomes.
The data, consisting of 6.22 million reads from two genomic libraries with 2-kb and 10-kb insert sizes respectively, were assembled using Celera Assembler. The assembly consisted of 1.09 million contigs of mean length 1393 bases, and 0.85 million singleton reads. At 1.5 × coverage, few contigs can be linked by multiple clones, and scaffolds of double-linked dog contigs incorporated only ∼660 Mb of the assembled sequence. Scaffold data are especially important for such low-coverage genomes, as they provide long-range linking information between specific areas of interest within the genome. One particular use of these data was to identify regions of the genome that were located near known genetic markers.
In order to incorporate more contigs within scaffolds, Bambus was set to allow single links when generating scaffolds and rerun over the entire data set. To reduce the effect of errors in the read-pairing data, Bambus was run in a hierarchical fashion, first building a collection of double-linked scaffolds, then combining the scaffolds using the single-link data that did not conflict with the already generated scaffolds. Due to the large number of repeats present in the dog genome, the resulting scaffolds were considerably “tangled” as described above. We used the “untangle” program to disambiguate these scaffolds.
To further aid the disambiguation process, we ignored the links of any contig that connected to six or more other contigs, as well as the links of any contig containing any sequence covered by more than 10 reads. Such contigs usually represent misassemblies and can confuse the untangling algorithm. We thus removed the linking information for 17,318 contigs before attempting the disambiguation of the scaffolds. The statistics of these three scaffolding approaches are presented in Table 4.
Table 4.
Dog Genome Scaffolding Statistics
| # Scaffolds | Mean length | N50 Length | Mean span | N50 Span | |
|---|---|---|---|---|---|
| hierarchical | 522,102 | 3,769 | 23,902 | 8,641 | 28,410 |
| hierarchical untangled | 636,575 | 3,091 | 13,431 | 8,855 | 21,846 |
| repeats removed | 663,171 | 2,972 | 12,178 | 8,542 | 37,905 |
To ascertain the quality of these scaffolds, we used an independently generated 512-kb reference sequence (consisting of four overlapping BACs with GenBank accession numbers AC114891, AC114332, AC113570, AC114890). After removing known repeats, we identified a set of 222 contigs that unambiguously mapped to the reference sequence. For the purpose of this analysis, we ignored any singleton sequences, as they provide little scaffolding information; moreover, their placement is very sensitive to library size estimates. We then validated the scaffolds by comparing the order and orientation of the contigs inferred from the mapping to the reference with that produced by Bambus. We analyzed only those scaffolds that contained two or more mapped contigs. For each such scaffold we identified the largest set of contigs that agreed with the order and orientation specified by the reference sequence. The remaining contigs in the scaffold were marked as “incorrectly placed”. The results are summarized in Table 5. The analysis clearly shows that the removal of suspected repeats improved the scaffolding. When repeats were removed, only two contigs were incorrectly placed by the scaffolder, as opposed to 16 when all contigs were used.
Table 5.
Validation of Scaffolding in a Low Sequence-Coverage Genome
| Total | Correct | Incorrect | N/A | |
|---|---|---|---|---|
| hierarchical untangled | 222 | 175 | 16 | 31 |
| repeats removed | 222 | 182 | 2 | 38 |
N/A, those contigs whose placement could not be validated due to insufficient linking data.
The results of this experiment are a clear testament to Bambus' ability to provide correct scaffolding information, even in the particularly difficult case of a highly repetitive genome at low sequence coverage. Even without removing known repeats from consideration, Bambus was able to correctly scaffold more than 90% of the contigs.
DISCUSSION
The diverse applications described in this paper demonstrate that Bambus is a versatile, general-purpose scaffolder. We have used it together with our collaborators to build scaffolds on data ranging from simple bacterial genomes to complex mammalian genomes. Bambus performs well in comparison to the scaffolders packaged with Celera Assembler and Arachne—assemblers with a proven track record in assembling large whole-genome shotgun data sets. Although Bambus has certain limitations in generating linear scaffolds, it is the only scaffolder able to highlight the haplotype structure of a genome. Furthermore, Bambus is the only scaffolder that provides the detailed information required during the finishing stages of a sequencing project. Some of this information is already available as part of the output of sequence assemblers; however, specialized software is usually needed to extract it in a format suitable for use during finishing. Furthermore, we would like to emphasize the fact that the current version of Bambus does not attempt to replace, or compete with, the specialized scaffolders included in most assemblers. These scaffolders have access to more detailed information about the contigs produced by the assembler and contain sophisticated algorithms in order to correctly navigate through tangles caused by repeats. Often, such scaffolders are used as part of an iterative process that produces increasingly better assemblies by interleaving assembly and scaffolding routines. Bambus was designed to complement such assemblers by providing users with the means for analyzing the relationships between contigs and scaffolds that could not be unambiguously determined by the assembler. This task is particularly important, especially as the specific assumptions made by the assemblers are often not clearly described in their documentation; moreover, most assemblers attempt to resolve only a small subset of the scaffolding problems described above. As an example, the haplotype problem is virtually ignored by most state-of-the-art assemblers.
Feature Summary
The previous section described examples of a wide variety of applications for Bambus, ranging from simple bacterial genomes to complex mammals. The features that are key to Bambus' ability to handle such diverse data sets are:
A flexible and extensible XML interface that allows arbitrary linking data to be provided;
Comprehensive output including both a graphical overview as well as detailed information about each scaffold;
Priorities that can be assigned to linking data on the basis of individual libraries or specific redundancies (number of links required between adjacent contigs);
The ability to screen out specified sections of the genome (e.g., repeats);
Flexibility in specifying scaffolding parameters through a simple configuration file;
The ability to use data produced by most commonly-used assemblers.
Implementation Details
Bambus consists of a collection of C++ and Perl programs that interact through a simple XML-based interface. The main scaffolding engine, “grommit,” is written entirely in C++ and operates on abstract contig links. A collection of Perl scripts collect the linking data from multiple sources, convert it to the input format for “grommit,” then process the output to generate a wide variety of reports. An image of the scaffold layout is generated with the use of the GraphViz (Gansner and North 2000) graphdrawing package from AT&T. An additional set of Perl scripts called “bamboo” provide an interface between the Bambus output and a relational database, and a collection of Web pages provide easy in-house access to scaffold data as relevant to finishing teams.
Bambus accepts input directly from the output of TIGR Assembler, Phrap, and from the common. ACE file format produced by several other assemblers. Scripts are included to convert Arachne and Celera Assembler output to Bambus input. In addition, contig linking data can be specified through an XML-based interface, allowing the integration of Bambus into other software packages.
Timing and Memory Usage
To evaluate Bambus' performance we scaffolded the data from three genomes: Bacillus anthracis, Drosophila pseudoobscura, and Canis familiaris. The results are summarized in Table 6. All of the jobs were executed on a Compaq Alpha machine with four 500-MHz processors, and 32 GB of RAM running Tru64 Unix version 5.1. We reported both the overall execution statistics and those for just the C++ component of our code in order to allow an accurate evaluation of the true computational complexity. The Perl component of Bambus uses hash tables—datastructures known to be inefficient in the current versions of Perl. Also note that the dog (Canis familiaris) data set is particularly complex because it represents a 1.5 × coverage assembly containing singleton contigs (normally excluded by most assemblers). This data set is therefore a worst-case scenario for mammalian-sized genomes.
Table 6.
Timing and Memory Usage
| Genome | Genome size | # of contigs | # of mates | Scaffolder component | Execution time | Maximum memory |
|---|---|---|---|---|---|---|
| Bacillus anthracis | 5.2 Mbases | 4,739 | 39,489 | ALL | 2m46s | 74 MB |
| C++ | 19s | 74 MB | ||||
| Drosophila pseudoobscura | 140 Mbases | 45,872 | 1,018,385 | ALL | 59m22s | 1.4 GB |
| C++ | 2m56s | 465 MB | ||||
| Canis familiaris | 2900 Mbases | 1,944,009 | 2,696,416 | ALL | 4h32m16s | 6.2 GB |
| C++ | 55m27s | 4.2 GB |
The data are separated into ALL—aggregate for a complete run of Bambus and C++-data for the C++ component of the code. The dog data is a worst-case scenario due to the low coverage (1.5 ×) and the fact that singleton contigs were included in the analysis.
Availability
The current version of Bambus (version 2.3) is available under an open-source license from http://www.tigr.org/software/bambus. The code has been tested on three commonly used computing platforms: Intel PCs running Linux, Sun/Solaris, and Alpha/Ultrix computers. We continue to improve the algorithms used in Bambus in order to overcome the challenges posed by complex genome projects. Bambus is also part of the open-source whole-genome assembly package AMOS currently being developed jointly by TIGR, the University of Maryland, and other research groups.
Additional Data
Dog Genome Data
GenBank accessions AACN010000001-AACN011089636 and CE000001-CE853796.
Benchmark Data Sets (Trace Archive Accessions)
Brucella suis 1330: TI 185503887-185523886 and TI 18533567-185551438.
Shewanella oneidensis MR1: TI 202843116-202899853.
Staphylococcus aureus COL: TI 175102203-175183597.
Staphylococcus epidermidis RP62A: TI 175142192-175162191 and TI 175294503-175314500 and TI 175183598-175204295.
Wolbachia sp: TI 184661597-184671587 and TI 185523887-185533553.
Data Sets Used in This Paper
Acknowledgments
We thank Art Delcher for his careful review and suggestions. We also thank the anonymous reviewers for their detailed comments. This work was supported in part by the NIH under grant R01-LM06845.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1536204.
References
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301-1310. [DOI] [PubMed] [Google Scholar]
- Batzoglou, S., Jaffe, D.B., Stanley, K., Butler, J., Gnerre, S., Mauceli, E., Berger, B., Mesirov, J.P., and Lander, E.S. 2002. ARACHNE: A whole-genome shotgun assembler. Genome Res. 12: 177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel, R., Alon, N., Apaydin, M.S., Fortnow, L., and Kasif, S. 2001. An optimal procedure for gap closing in whole genome shotgun sequencing. In Proceedings of the Fifth Annual International Conference on Computational Biology (RECOMB), pp. 22-30. ACM, New York.
- Dehal, P., Satou, Y., Campbell, R.K., Chapman, J., Degnan, B., De Tomaso, A., Davidson, B., Di Gregorio, A., Gelpke, M., Goodstein, D.M., et al. 2002. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science 298: 2157-2167. [DOI] [PubMed] [Google Scholar]
- Delcher, A.L., Kasif, S., Fleischmann, R.D., Peterson, J., White, O., and Salzberg, S.L. 1999. Alignment of whole genomes. Nucleic Acids Res. 27: 2369-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher, A.L., Phillippy, A., Carlton, J., and Salzberg, S.L. 2002. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 30: 2478-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasulo, D., Halpern, A., Dew, I., and Mobarry, C. 2002. Efficiently detecting polymorphisms during the fragment assembly process. Bioinformatics 18: S294-S302. [DOI] [PubMed] [Google Scholar]
- Fleischmann, R.D., Adams, M.D., White, O., Clayton, R.A., Kirkness, E.F., Kerlavage, A.R., Bult, C.J., Tomb, J.F., Dougherty, B.A., Merrick, J.M., et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269: 496-512. [DOI] [PubMed] [Google Scholar]
- Gansner, E.R. and North, S. 2000. An open graph visualization system and its applications to software engineering. Software—Practice and Experience 30: 1203-1233. [Google Scholar]
- Garey, M.R. and Johnson, D.S. 1979. Computers and intractability. W.H. Freeman, New York.
- Gonzalez, J., Ranz, J.M., and Ruiz, A. 2002. Chromosomal elements evolve at different rates in the Drosophila genome. Genetics 161: 1137-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, P. 1994. PHRAP documentation: ALGORITHMS. http://www.phrap.org
- Heidelberg, J.F., Paulsen, I.T., Nelson, K.E., Gaidos, E.J., Nelson, W.C., Read, T.D., Eisen, J.A., Seshadri, R., Ward, N., Methe, B., et al. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20: 1118-1123. [DOI] [PubMed] [Google Scholar]
- Huson, D.H., Reinert, K., and Myers, E. 2001. The greedy path—Merging algorithm for sequence assembly. In Proceedings of the Fifth Annual International Conference on Computational Biology (RECOMB), pp. 157-163.
- Jaffe, D.B., Butler, J., Gnerre, S., Mauceli, E., Lindblad-Toh, K., Mesirov, J.P., Zody, M.C., and Lander, E.S. 2003. Whole-genome sequence assembly for Mammalian genomes: Arachne 2. Genome Res. 13: 91-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kececioglu, J.D. and Myers, E.W. 1995. Combinatorial algorithms for DNA sequence assembly. Algorithmica 13: 7-51. [Google Scholar]
- Kent, W.J. and Haussler, D. 2001. Assembly of the working draft of the human genome with GigAssembler. Genome Res. 11: 1541-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkness, E.F., Bafna, V., Halpern, A.L., Levy, S., Remington, K., Rusch, D.B., Delcher, A.L., Pop, M., Wang, W., Fraser, C.M., et al. 2003. The dog genome: Survey sequencing and comparative analysis. Science 301: 1898-1903. [DOI] [PubMed] [Google Scholar]
- Kurtz, S. and Schleiermacher, C. 1999. REPuter: Fast computation of maximal repeats in complete genomes. Bioinformatics 15: 426-427. [DOI] [PubMed] [Google Scholar]
- Mullikin, J.C. and Ning, Z. 2003. The phusion assembler. Genome Res. 13: 81-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, E.W., Sutton, G.G., Delcher, A.L., Dew, I.M., Fasulo, D.P., Flanigan, M.J., Kravitz, S.A., Mobarry, C.M., Reinert, K.H., Remington, K.A., et al. 2000. A whole-genome assembly of Drosophila. Science 287: 2196-2204. [DOI] [PubMed] [Google Scholar]
- Paulsen, I.T., Seshadri, R., Nelson, K.E., Eisen, J.A., Heidelberg, J.F., Read, T.D., Dodson, R.J., Umayam, L., Brinkac, L.M., Beanan, M.J., et al. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. 99: 13148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevzner, P.A. and Tang, H. 2001. Fragment assembly with double-barreled data. Bioinformatics (Suppl.) 17: S225-233. [DOI] [PubMed] [Google Scholar]
- Read, T.D., Salzberg, S.L., Pop, M., Shumway, M., Umayam, L., Jiang, L., Holtzapple, E., Busch, J.D., Smith, K.L., Schupp, J.M., et al. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296: 2028-2033. [DOI] [PubMed] [Google Scholar]
- Read, T.D., Peterson, S.N., Tourasse, N., Baillie, L.W., Paulsen, I.T., Nelson, K.E., Tettelin, H., Fouts, D.E., Eisen, J.A., Gill, S.R., et al. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423: 81-86. [DOI] [PubMed] [Google Scholar]
- Roach, J.C., Boysen, C., Wang, K., and Hood, L. 1995. Pairwise end sequencing: A unified approach to genomic mapping and sequencing. Genomics 26: 345-353. [DOI] [PubMed] [Google Scholar]
- Sanger, F., Coulson, A.R., Hong, G.F., Hill, D.F., and Petersen, G.B. 1982. Nucleotide sequence of bacteriophage λ DNA. J. Mol. Biol. 162: 729-773. [DOI] [PubMed] [Google Scholar]
- Sutton, G.G., White, O., Adams, M.D., and Kerlavage, A.R. 1995. TIGR Assembler: A new tool for assembling large shotgun sequencing projects. Genome Sci. Technol. 1: 9-19. [Google Scholar]
- Tettelin, H., Radune, D., Kasif, S., Khouri, H., and Salzberg, S.L. 1999. Optimized multiplex PCR: Efficiently closing a whole-genome shotgun sequencing project. Genomics 62: 500-507. [DOI] [PubMed] [Google Scholar]
- Thayer, E.C., Olson, M.V., and Karp, R.M. 1999. Error checking and graphical representation of multiple-complete-digest (MCD) restriction-fragment maps. Genome Res. 9: 79-90. [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.ncbi.nlm.nih.gov/Traces; NCBI Trace Archive.
- http://www.tigr.org/tdb/benchmark; Shotgun sequence assembly benchmark data.
- http://www.tigr.org/software/; TIGR software page.
- http://www.tigr.org/software/bambus;, Bambus Web page.