Abstract
The Vpr protein of HIV-1 functions as a vital accessory gene by regulating various cellular functions, including cell differentiation, apoptosis, nuclear factor of κB (NF-κB) suppression and cell-cycle arrest of the host cell. Several reports have indicated that Vpr complexes with the glucocorticoid receptor (GR), but it remains unclear whether the GR pathway is required for Vpr to function1. Here, we report that Vpr uses the GR pathway as a recruitment vehicle for the NF-κB co-activating protein, poly(ADP-ribose) polymerase-1 (PARP-1). The GR interaction with Vpr is both necessary and sufficient to facilitate this interaction by potentiating the formation of a Vpr–GR–PARP-1 complex. The recruitment of PARP-1 by the Vpr–GR complex prevents its nuclear localization, which is necessary for Vpr to suppress NF-κB. The association of GR with PARP-1 is not observed with steroid (glucocorticoid) treatment, indicating that the GR association with PARP-1 is a gain of function that is solely attributed to HIV-1 Vpr. These data provide important insights into Vpr biology and its role in HIV pathogenesis.
A trademark of HIV infection is the diminution of the CD4+ T-cell count of the host, which invariably leads to eventual immunodeficiency2. It is believed that various viral factors contribute to this effect by suppressing both immune activation and T-cell expansion3–6. The 96-amino-acid viral protein R (Vpr), which has a relative molecular mass of 14,000, has been implicated in both the destruction and suppression of potential antigen-specific T cells through multiple mechanisms7. In fact, Vpr is sufficient to suppress mitogen or anti-CD3-dependent proliferation and activation of T cells. Additionally, Vpr is present in the serum of infected patients and can efficiently reactivate viruses from latency8,9. Furthermore, Vpr possesses intrinsic transduction properties, which indicates that there are various viral-induced pathogenesis events that occur within a non-viral infection setting10. Other reported important activities include host cell-cycle arrest at the G2/M stage, nuclear transport of the pre-integration complex, host-cell apoptosis, nuclear herniations and the induction of immune suppression11–20.
Glucocorticoid receptor II (GR-II) has been identified as an in vivo target for Vpr12,20–22. The Vpr–GR interaction is dependent on the signature LXXLL motif, the abrogation of which attenuates the GR-dependent co-activation and transcription that is induced by Vpr. In addition, co-treatment with the GR antagonist mifpristone (Mif) blocks several pathogenic functions of Vpr, including apoptosis and viral transcription12,17,19. However, the mechanism behind nuclear factor of κB (NF-κB) suppression by Vpr currently remains unresolved. Furthermore, the functional deviations between Vpr and glucocorticoid treatments indicates that different mechanisms may occur.
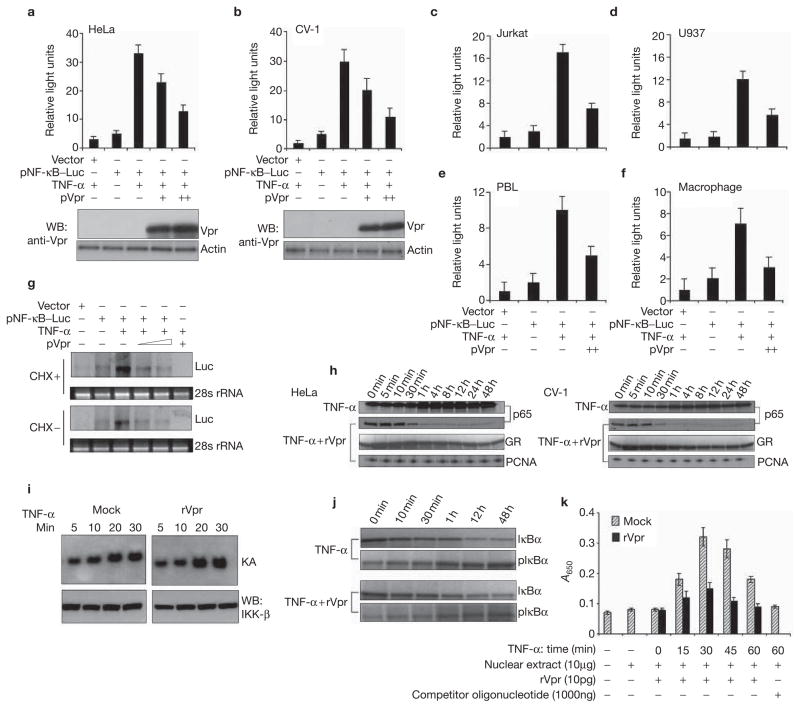
In an effort to understand the role of the GR in Vpr-mediated NF-κB suppression, we compared NF-κB-dependent transcriptional activation in cells with a functional GR and in CV-1 cells, a monkey kidney cell line that expresses an endogenous GR, but is refractory of function23. As shown in Fig. 1a, co-transfection of Vpr but not of a control vector into HeLa cells, is sufficient to inhibit tumour necrosis factor-α (TNF-α)-induced NF-κB transcription. The inhibition was also observed in cells prone to HIV-1 infection, including Jurkat T cells, U937 monocytes and primary peripheral blood leukocyte (PBL) cells and macrophages (Fig. 1c–f). More interestingly, the same inhibitory effect was also observed in CV-1 cells that possess a non-functional GR (Fig. 1b), indicating that GR-mediated transcription is not required for NF-κB suppression, contrary to previous reports that suggested that GR activation leads to an upregulation of inhibitory I-κB12. This was further verified, as shown by the fact that inhibition of de novo protein synthesis via cycloheximide treatment did not attenuate Vpr-mediated NF-κB-dependent transcription (Fig. 1g). Vpr treatment was also accompanied by a reduced nuclear duration of RelA (p65) in both functional GR and non-functional GR cells (Fig. 1h). This result could be due to a failure of the formation of transcriptional complexes, which prevents acetylation and extended presence of RelA within the nucleus, as Vpr did not significantly affect its initial nuclear localization24. As upstream kinase inhibition could manifest the same effect, we next examined the activity of I-κB kinase-β (IKKβ). Vpr treatment did not affect the kinase activity of IKKβ (Fig. 1i) nor did it affect in vivo phosphorylation and turnover of I-κBα (Fig. 1j). However, Vpr potently attenuated the DNA-binding activity of NF-κB (RelA) at both the initial (Fig. 1k) and the later time points, and this effect was specific to RelA and not to other transcriptional factors. Last, co-transfection or Vpr treatment directly attenuated RelA-mediated transcriptional activation (see Supplementary Information, Fig. S1a–d). Taken together, these results indicate that Vpr suppresses NF-κB at the transcriptional level independently of concomitant de novo transcription induced by GR activation.
Figure 1.
Vpr transcriptionally suppresses NF-κB through a pathway that does not require a functional glucocorticoid receptor. Vpr suppresses tumour necrosis factor-α (TNF-α)-induced NF-κB-mediated transcription in HeLa (a), CV-1 (b), Jurkat (c), U937 (d), PBL (e) and macrophage (f) cells. A total of 1 × 106 cells were co-transfected with the indicated expression vectors and luciferase activity was measured. Total proteins were extracted from the HeLa and CV-1 cells, as indicated above, and the presence of Vpr was analysed by western blotting (WB) using specific Vpr antibodies. (g) Vpr suppresses NF-κB-dependent transcription in the absence of new protein synthesis. HeLa cells were treated with or without cycloheximide (CHX; 10 μg ml−1) for 30 min, and transfected with pNF-κB–Luc plasmids (2 μg). After 6 h of transfection, the medium was replaced with or without a CHX-containing medium, and cells were incubated for 24 h before RNA extraction. TNF-α (5 ng ml−1) was added to cells and incubated for 6 h before RNA extraction. Total RNA (20 μg) was resolved on 1.2% formaldehyde gel and analysed by northern blotting using a radiolabelled luciferase DNA as a probe. The loading markers were 28S rRNA level. (h) Nuclear levels of RelA (p65) in both functional GR (HeLa) or non-functional GR (CV-1) cells. Cells were treated with or without rVpr (10 pg) and stimulated with TNF-α (5 ng ml−1). Cells were collected post-treatment, as indicated, and were immunoblotted with a polyclonal antibody against NF-κB (p65). As a control, Vpr effects on GR were also analysed with antibody against GR and anti-PCNA as an internal control for nuclear loading. (i) IKKβ kinase activity in cells treated with rVpr and TNF-α. Serum-starved HeLa cells (1.5 × 106) were treated with TNF-α (5 ng ml−1) at indicated time points with or without rVpr (10 pg) and were then subjected to kinase activity, as described in Methods. Top, kinase assay (KA) revealed by autoradiography; bottom, the immunoprecipitates were also subjected to western blotting (WB) to determine the amount of precipitated proteins. (j) I-κBα phosphorylation and its turnover in rVpr-treated cells was measured from lysates simulated with the conditions described above. (k) HeLa cells were treated with TNF-α at indicated time points in the presence or absence of rVpr (10 pg). Cells were collected at indicated time points and nuclear proteins (10 μg) were incubated in 96-well plates lined with oligonucleotides specific for NF-κB (p65) for the ELISA-based transcription-factor assay. Values and bars represent mean (n = 3) and SD. Scale bars in b, c and i represent 10 μm.
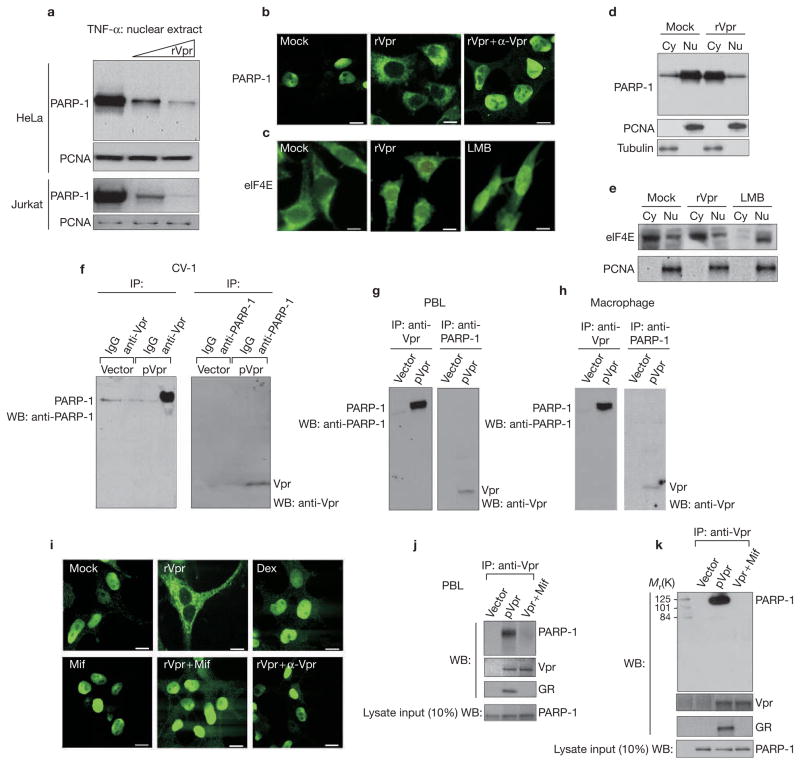
A hypothesis for this inhibition is that Vpr may destabilize the NF-κB transcriptional complex through a direct and/or indirect cross-talk mechanism. Interestingly, recent studies have indicated a role for PARP-1 in co-activating NF-κB-dependent gene expression25–27. Evidence indicates that PARP-1 is linked to the transcriptional potential of NF-κB through formation of a critical transcriptional complex within the nucleus26,27 and, consequently, Parp-1-deficient mice are refractory to toxin-induced sepsis. In view of this, we noticed that treatment of both HeLa and Jurkat cells leads to a dosage-dependent reduction in nuclear localization of PARP-1, as measured by biochemical and immunofluoresence methods (Fig. 2a, b). This effect was specific to PARP-1 and was not associated with other trafficking factors (such as eIF4E) and constitutive nuclear factors (such as PCNA) (Fig. 2c–e). The decrease in nuclear PARP-1 was concomitant with an increase in cytoplasmic PARP-1, indicating a defect in the nuclear or cytoplasmic shuttling of PARP-1 (Fig. 2d). Co-staining of Vpr–GFP (green fluorescent protein) with both PARP-1 and GR revealed significant colocalization within the cytoplasmic and perinuclear regions (see Supplementary Information, Fig. S2a), whereas GFP alone failed to affect PARP-1 nuclear localization.
Figure 2.
Vpr interacts with PARP-1 and inhibits its nuclear localization through a GR interaction-dependent pathway. (a) Vpr prevents the nuclear migration of poly(ADP-ribose) polymerase-1 (PARP-1). HeLa or Jurkat T cells were stimulated with tumour necrosis factor-α (TNF-α) with or without rVpr (10 pg and 1 pg). PARP-1 localization was detected by protein blotting with an anti-PARP-1 polyclonal antibody. Immunoblotting was also performed using an anti-PCNA antibody as an internal control for nuclear loading. (b–c) Cellular localization of PARP-1. HeLa cells were treated with or without rVpr (10 pg) in the presence or absence of anti-Vpr and were stimulated with or without of TNF-α for 3 h. Cells were fixed and stained with an anti-PARP-1 antibody, followed by FITC-conjugated goat anti-rabbit secondary antibody, as described in Methods (b). Localization of eIF4E was also performed to show the specificity of Vpr towards PARP-1. The third panel is eIF4E staining in leptomycin B (LMB; 50 nM)-treated cells to show trafficking (c). Scale bars, 10 μM. (d–e) Biochemical fractionation of nuclear (Nu) versus cytoplasmic (Cy) contents were performed from TNF-α or TNF-α + Vpr-treated cells. Protein samples were separated by SDS–PAGE and analysed by western blot with a polyclonal antibody against PARP-1 or eIF4E antibody for cytosolic versus nuclear expression. (f–h) Vpr and PARP-1 form a complex in multiple cells. CV-1, peripheral blood leukocyte (PBL) and macrophage cells were transfected with a vector control (pDNA) or with pVpr (4 μg) and analysed for co-immunoprecipitation (IP) with respective antibodies. WB, western blot. (i) Dexamethasone is insufficient to regulate PARP-1, whereas the ability of Vpr to regulate PARP-1 localization is sensitive to mifpristone. HeLa cells were treated with Mock, rVpr (10 pg), dexamethasone (Dex; 1 μM), mifpristone (Mif; 1 μM), rVpr (10 pg) + mifpristone (1 μM) or Vpr (10 pg) + anti-Vpr (1:200). Twenty-four hours post-treatment, cells were stimulated with or without 5 ng ml−1 of TNF-α for 3 h, and immunofluorescence was performed using an antibody against PARP-1 followed by FITC-conjugated goat anti-rabbit secondary antibody. Scale bars, 10 μM. (j–k) The interaction of Vpr with PARP-1 is sensitive to mifpristone. HeLa or PBL cells were transfected with vector or pVpr (4 μg) in the presence or absence of mifpristone (1 μM). Protein lysates were immunoprecipitated with an anti-Vpr polyclonal antibody and analysed via protein blotting with anti-PARP-1, glucocorticoid receptor (GR) and Vpr antibodies. Western blots were also performed for PARP-1 expression for the input cell lysate that was extracted from the vector or Vpr-transfected samples.
Next, we hypothesized whether Vpr may be interacting with PARP-1 and, therefore, facilitating its failed nuclear localization. Therefore, plasmid (p) Vpr was transfected into CV-1, PBL cells and macrophages with appropriate controls; they were immunoprecipitated with either anti-Vpr or PARP-1 antibodies and were blotted for the other. As shown in Fig. 2f–h, Vpr was sufficient to co-immunoprecipitate with PARP-1, indicating that the two reside together in a complex in CV-1 and primary HIV-1 target cells. We then asked whether this requires a Vpr–GR interaction, which we have shown previously to be a cellular receptor for Vpr. As shown in Fig. 2i and Supplementary Information, Fig. S2b, the GR antagonist mifpristone is sufficient to reverse the cellular accumulation of PARP-1, indicating that the interaction between Vpr and GR is necessary for Vpr to affect PARP-1 localization. Not surprisingly, the interaction between Vpr and PARP-1 was also sensitive to mifpristone in PBL and HeLa cells (Fig. 2j, k), which indicates that the Vpr–GR complex may facilitate the interaction with PARP-1, and therefore regulate its localization. Paradoxically, steroid treatment (dexamethasone) alone is insufficient to relocate PARP-1 from the nucleus to the cytoplasm (Fig. 2i; and see Supplementary Information, Fig. S2b). In fact, recombinant protein (r) Vpr and dexamethasone exhibit competing effects on PARP-1 localization (see Supplementary Information, Fig. S2c). Therefore, although the Vpr interaction with GR was required for PARP-1 interaction, GR activation by steroids alone is insufficient, implying a molecular distinction between Vpr and steroid-receptor activation.
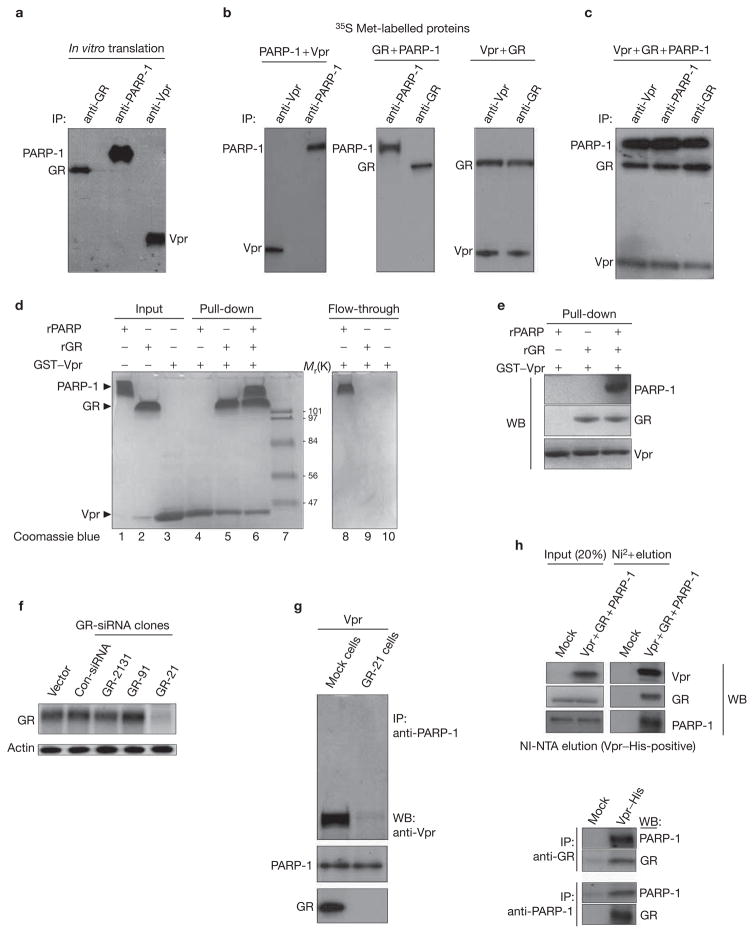
Next, we examined the molecular nature of the Vpr–PARP-1 complex. We in vitro translated PARP-1, Vpr and GR with 35S-Met and performed co-precipitation experiments. As shown in Fig. 3a–c, although Vpr directly interacts with GR as previously shown20, Vpr was unable to directly interact with PARP-1. In addition, as is consistent with dexamethasone experiments, GR is unable to interact with PARP-1. PARP-1 was only sufficient to complex with Vpr when it was complexed with GR (Fig. 3c). This was also observed in glutathione S-transferase (GST) pull-down assays and confirmed by western blot analysis of the pull-down (Fig. 3d, e). Taken together, these results indicate that the Vpr–GR interaction is necessary and sufficient to recruit PARP-1 in vitro.
Figure 3.
The GR interaction with Vpr is both necessary and sufficient to recruit PARP-1. (a–c) Expression plasmids (Vpr, glucocorticoid receptor (GR) and poly(ADP-ribose) polymerase-1 (PARP-1)) (2 μg) were used for coupled in vitro transcription/translation reactions and were carried out as described in Methods. (a) Immunoprecipitation (IP) of the in vitro-translated proteins were performed with anti-Vpr, anti-GR and anti-PARP-1 antibodies. Equal amounts of in vitro-translated proteins were mixed in the following groups: (b) Vpr + PARP-1, GR + PARP-1, Vpr + GR or (c) Vpr + GR + PARP-1, and interactions were performed as described in Methods. (d–e) A glutathione S-transferase (GST) pull-down assay with recombinant forms of each protein was performed as described in Methods. (d) Represents a Coomassie blue staining of the gels, whereas (e) represents the western blot (WB) analysis of the pull-down assay with the indicated antibodies. Input represents 25% of total protein used for pull-down; flow-through represents 10% of unbound material. K, relative molecular mass in thousands. (f) Small interfering RNA (siRNA)-mediated knockdown of GR. Three different GR target siRNA expression vectors were constructed as described in Methods. (g) GR is required for Vpr and PARP-1 interaction in cells. GR-21 stable clone cells were established as described in Methods and transfected with pVpr (4 μg) and co-immunoprecipitation assays were performed with the indicated antibodies. (h) Vpr, PARP-1 and GR reside as a single complex in vivo. Equal amounts of total protein in whole-cell extracts that were prepared from HeLa cells were transfected with mock or pVpr–His + pGR + pPARP-1, respectively, and were bound to Ni-NTA columns, eluted and immunoprecipitated for GR and PARP-1 with Vpr–His-tagged eluted protein (bottom). Vpr–His-tagged eluted proteins were immunoprecipitated with anti-PARP or anti-GR antibodies and blotted back for the other.
To determine whether this can be extrapolated in vivo, we reduced the endogenous levels of GR via small interfering RNA (siRNA). Of the specific clones tested, GR-21 specifically and efficiently knocked down GR protein levels (Fig. 3f). Subsequently, transfection of Vpr into these cells failed to co-immunoprecipitate PARP-1 compared with siRNA control cells, indicating that Vpr requires GR to interact with PARP-1 in vivo (Fig. 3g). Biochemical fractionation of His-tagged Vpr through a nickel column and subsequent co-immunoprecipitation of PARP-1 or GR reveals that the three proteins exist in one complex rather than mutually exclusive complexes in vivo (Fig. 3h).
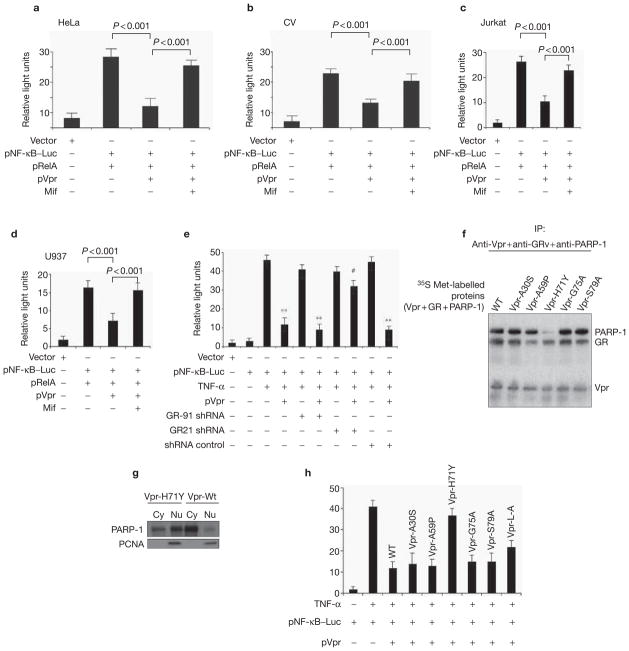
If Vpr recruitment of PARP-1 via GR interaction is, in fact, regulating transcriptional suppression, then prevention of either the Vpr interaction with GR or with PARP-1 should be sufficient to reverse the NF-κB suppression that is induced by Vpr. Accordingly, co-treatment of mifpristone, which abrogates the Vpr–GR interaction, was sufficient to significantly (P < 0.001) recover the transcriptional suppression of co-transfected RelA in HeLa and CV-1 cells, respectively (Fig. 4a, b). Similar results were also obtained in HIV-1 target cells, including Jurkat T cells and U937 monocytes (Fig. 4c, d). In a similar fashion, loss of GR function via siRNA also exhibited an inability for Vpr to suppress NF-κB, again indicating that GR is necessary for Vpr to inhibit NF-κB (Fig. 4e).
Figure 4.
Recruitment of PARP-1 to the Vpr–GR complex is necessary for Vpr-mediated transcriptional suppression of NF-κB. HeLa (a), CV-1 (b), Jurkat (c) and U937 (d) cells were transfected with the nuclear factor of κB (NF-κB)-dependent reporter plasmid with RelA plasmids and with pCMVLacZ (1 μg) with pVpr (4 μg) in the presence or absence of mifpristone (Mif; 1 μM), and luciferase assays were conducted as described in Methods. (e) Glucocorticoid receptor (GR) is necessary for the transcriptional suppression induced by Vpr. HeLa cells were transfected with the NF-κB-dependent reporter plasmid (2 μg), with 5 μg of short hairpin RNA (shRNA) pGR-91 or shRNA pGR-21 or shRNA control plasmid with or without Vpr (4 μg). Luciferase activity was measured as described in Methods. Asterisks (**) over a bar indicates statistically significant results (p < 0. 005) whereas the hash symobl (#) indicates results that are not statistically significant (p < 0.01). (f) Vpr H71Y interacts with GR but fails to recruit poly(ADP-ribose) polymerase-1 (PARP-1). Expression plasmids (Vpr and different Vpr mutants, GR and PARP-1) (2 μg) were used for coupled in vitro transcription/translation reactions and interaction action assays were carried out as described in Methods. IP, immunoprecipitation; WT, wild type. (g) The H71Y Vpr mutant fails to prevent PARP-1 nuclear localization. Total nuclear (Nu) and cytosol (Cy) protein extracts of HeLa cells were prepared 48 h post-transfection from wild-type Vpr (Vpr-Wt) or mutant Vpr (Vpr-H71Y)-transfected cells and were western blotted for PARP-1 antibody and anti-PCNA as an internal control for nuclear loading. (h) Analysis of Vpr and differing mutants for NF-κB-mediated transcription was measured as described in Methods. HeLa cells were transfected with pLuc–NF-κB (5 μg), pCMVLacZ (1 μg), with or without Vpr-Wt and Vpr mutants (2 μg), and luciferase activity in whole-cell lysates was assayed as described in Methods.
The role of PARP-1 in regulating NF-κB suppression seems to be entirely structural and not associated with its enzymatic properties, as DPQ (a nicotinic acid analogue) and a PARP-1 enzymatic inhibitor, did not significantly repress TNF-α-induced NF-κB transcription28. Meanwhile, small interfering RNA (siRNA)-mediated knockdown of PARP-1 had a more pronounced effect on NF-κB-dependent transcription relative to knockdown efficiency (see Supplementary Information, Fig. S3a–c). To determine whether recruitment of PARP-1 to the Vpr–GR complex was required for Vpr to suppress NF-κB, we screened previously described mutants29 for their ability to interact with GR and/or PARP-1. As shown in Fig. 4f, a point mutation of Vpr at H71 to tyrosine (Y) exhibited binding to GR but, importantly, failed to coprecipitate PARP-1. Consistently, transfection of this mutant into HeLa cells exhibited an obvious attenuation of cytoplasmic localization of PARP-1 compared with wild-type Vpr (Fig. 4g). Last, transfection of this mutant, but not others, exhibited a significant reduction in its ability to repress NF-κB-dependent transcription (Fig. 4h). Taken together, these results indicate that Vpr recruitment to the GR complex is required for efficient NF-κB transcription. It also indicates that, to a large extent, the Vpr–GR interaction is merely a priming effect to potentiate the recruitment of PARP-1, which is necessary for transcriptional suppression.
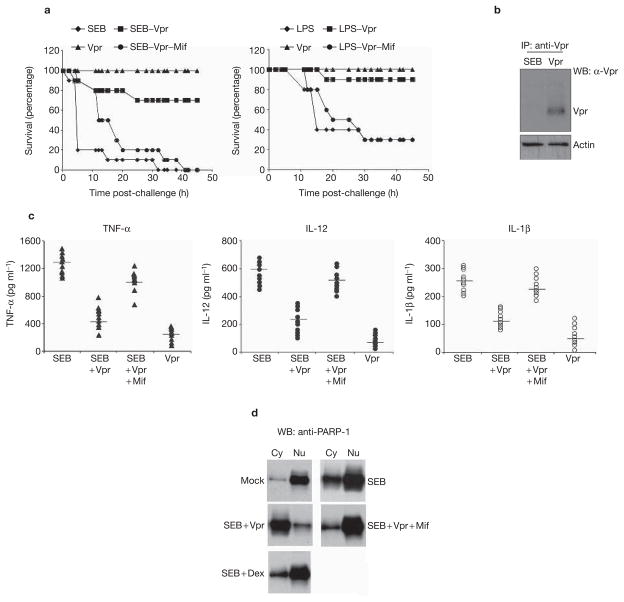
To determine whether the interaction between PARP-1 and Vpr is relevant in vivo, we next challenged mice with lipopolysaccharide (LPS) or staphyloccal enterotoxin B (SEB) and measured the ability of Vpr to suppress immune activation and PARP-1 nuclear localization. SEB and LPS have been extensively used as toxin models to study the effects on immune cells19,30. Knockout mice that are deficient in the Parp-1 gene are refractory to the toxic effects of LPS challenge25. Using the 3-galactos-mine-sensitized mouse model30, we investigated the protective activity of recombinant Vpr that was delivered intravenously (Fig. 5a) against SEB and LPS challenge. Injection of Vpr into mice effectively repressed the hyperinflammation (TNF-α, interleukin (IL)-12 and IL-1β) that was induced by SEB (Fig. 5c). The splenocytes from mice injected with mock, SEB, SEB + Vpr, SEB + Dex or SEB + Vpr + mifpristone were then extracted and lysed. The proteins were fractionated between cytoplasmic and nuclear extracts and blotted for PARP-1 expression. Mice that were injected with SEB alone demonstrated significant localization of PARP-1 to the nucleus, which supports its role as a co-activator for NF-κB. However, mice that were co-injected with SEB and Vpr failed to localize PARP-1 into the nucleus (Fig. 5d). This effect could be reversed by co-injection with mifpristone, indicating that the Vpr–GR interaction is necessary in vivo for preventing nuclear localization of PARP-1. By contrast, SEB- and dexamethasone-injected mice failed to prevent the localization of Parp-1 into the nucleus. These results support the in vitro finding that failed PARP-1 nuclear localization is specific to Vpr and is not manifested by a traditional steroid effect. Furthermore, immunoprecipitation of Vpr within the splenic lysates reveals its presence within this lymphoid organ post-intravenous injection (Fig. 5b). Taken together, these results indicate that the inhibition of nuclear localization of PARP-1 by Vpr is a relevant phenomenon both in vitro and in vivo.
Figure 5.
The PARP-1–Vpr–GR interaction is relevant in vivo. (a) Groups of 10 female, Balb/C mice for each group aged 6–12 weeks were sensitized by intravenous (i.v) injection with 20 mg D-galactosamine for 30 min prior to challenge with 10 μg of LPS or SEB toxin. Mice were challenged with rVpr (100 μg) in the presence or absence of 10 μM mifpristone (Mif), 1 h after SEB introduction via an intravenous method. We examined the survival of the SEB-challenged animals. (b) Immunoblot analysis of Vpr in the spleen post-rVpr injection. Total cellular proteins were prepared from the splenocytes challenged with SEB or SEB + Vpr. Protein lysates were immunoprecipitated with anti-Vpr antibody, followed by western blot (WB) analysis with an anti-Vpr antibody blotted back for Vpr. Lysates from the splenocytes were also loaded and blotted for actin as an internal control for immunoprecipitation. (c) Vpr-mediated suppression of cytokine secretion in mice challenged with SEB, SEB + rVpr or SEB + rVpr + mifpristone. Serum from ten mice per group were harvested prior to SEB treatment and serum from mice in the tested groups was also harvested. Serum tumour necrosis factor-α (TNF-α), interleukin (IL)-12 and IL-1β levels were determined by standard cytokine ELISA, as described in Methods. Results are expressed as pg ml−1 ± SD of individual determinants. (d) Vpr prevents poly(ADP-ribose) polymerase-1 (PARP-1) nuclear localization in vivo. Total nuclear (Nu) and cytosolic (Cy) protein extracts were prepared from the challenged splenocytes and analysed by western blot with a polyclonal antibody against PARP-1 for cytosolic versus nuclear location. Dex, dexamethasone.
In this study, we have elucidated the mechanistic role for GR in mediating NF-κB suppression by Vpr. Although the presence of the structural GR protein is required, its transcriptional property is not. The structural GR, following interaction with Vpr, induces the recruitment of PARP-1, and the GR–Vpr interaction alone is sufficient to complex with PARP-1 in vitro. Accordingly, we propose that the ‘gain of function’ of the Vpr–GR interaction is a priming event for PARP-1 recruitment and hence prevents its nuclear accumulation. Although the molecular nature of the Vpr–Gr–PARP-1 complex remains unknown, the interaction of Vpr with GR may induce conformational changes within the complex that expose binding sites for the PARP-1 interaction. Consistently, steroid treatment is insufficient to recruit PARP-1 and interact with GR. Future work will identify the details of this regulation, such as the specific importin that may be involved or the role of the nuclear-cytoplasmic shuttling properties of the Vpr.
Although our results reveal a mechanism by which Vpr can suppress NF-κB, we propose that prevention of PARP-1 nuclear localization may have greater consequences for HIV infection. First, PARP-1 has recently been shown to be a modulator of chromatin structure by binding to nucleosomes. In vivo, PARP-1 binding to nucleosomes has been correlated with transcriptionally repressed chromatin domains31. This indicates that Vpr has a potent ability to transcriptionally alter gene expression, which may also be associated with failed PARP-1 nuclear localization. Second, PARP-1 has been shown to be a negative regulator of TAT-mediated HIV transcription by directly competing for the TAT RNA32. Therefore, retaining PARP-1 within the cytoplasm could potentially augment TAT-mediated transcription and viral infectivity. Consistently, it has been observed that Vpr can augment TAT-mediated viral production33,34.
In conclusion, the present results help to clarify a controversy in the Vpr literature, but at the same time raise several important questions regarding Vpr biology. We observe that the GR complex is a central target of Vpr in vitro and in vivo. The Vpr–GR complex is a gain-of-function interaction that represses NF-κB transcription through the additional recruitment of PARP-1, a crucial regulator of NF-κB. It is likely that Vpr targeting of PARP-1 could add to the T-cell depletion and immunosuppressive effects that are observed during HIV-1 infection, illustrating the complexity of HIV-1 infection. These studies indicate that understanding the relationship of Vpr to PARP-1 activation may reveal new insights into the important roles of Vpr, such as cell-cycle arrest and apoptosis, in host-cell pathogenesis.
METHODS
Cell culture
The human CD4+ T-cell line Jurkat, the monocyte line U937, HeLa cells and the African green monkey cell line (CV-1) (non-functional GR-expressing cells)23 were obtained from the American Type Culture Collection (Rockville, MD). Cells were passaged in RPMI-1640 or DMEM (Gibco-BRL, Carlsbad, CA) supplemented with 10% fetal bovine serum, 2 mM 3-glutamine, 100 U ml−1 penicillin-G and 100 μg ml−1 streptomycin. Cells were maintained at 37°C and 5% CO2, and were tested routinely to certify that they were Mycoplasma-negative. Leukopacks from individual donors were obtained from the Centre for AIDS research clinical core facilities at the University of Pennsylvania School of Medicine (Philadelphia, PA) in order to isolate PBL cells, as well as macrophages. PBL cells were stimulated with 5 μg ml−1 phytohaemagglutinin (PHA) (Sigma, St Louis, MO) before the addition of 5 U ml−1 of human recombinant interleukin-2 (hrIL-2) (R&D Systems, Minneapolis, MN)16. For isolation of macrophages, CD14+ cells were purified from peripheral blood mononuclear cells by positive enrichment using autoMACS (Miltenyi Biotec, Auburn, CA) according to the manufacturer’s instructions. Adherent macrophages were cultured in complete RPMI-1640 medium with 500 U ml−1 of rhIL-4 and 1,000 U ml−1 of rhGM-CSF (R&D Systems) per 106 cells. The purity of the monocyte cell populations that were isolated was >98%, as determined by fluorescence-activated cell sorting staining for CD14 (ref. 17).
Plasmid constructions
The PARP-1 expression vector was generously provided by Zhao-Qi Wang (IARC, Lyon, France) and was subcloned into the mammalian expression vector pVax1 (Invitrogen, Carlsbad, CA). Human GR DNA was amplified using gene-specific primers and cloned into KpnI and XhoI sites of the mammalian expression vector pVax1 vector (Invitrogen). Oligonucleotides encoding HIV-1 Vpr and point mutations within the Vpr regions were introduced using the QuickChange Kit (Stratagene, La Jolla, CA) using specific primers and cloned into the pVax1 vector (Invitrogen). Plasmids encoding GST–Vpr were generated by polymerase chain reaction (PCR) and cloned into BamHI and XhoI sites of the pGEX-4T-3 vector (Amersham Biosciences, Piscataway, NJ) for bacterial expression. For plasmids encoding Vpr–His, Vpr was generated by PCR and cloned into a pcDNA3.1/His vector (Invitrogen), as described previously16. The p65/RelA expression construct was a gift from Douglas R. Green (La Jolla Institute for Allergy and Immunology, San Diego, CA). The pTAL–Luc (luciferase) and pNF-κB–Luc vectors were purchased from BD Biosciences (San Jose, CA). All constructs were verified by automated DNA sequencing. Highly purified (>95%) recombinant Vpr protein (pTYB4-Vpr) was prepared by using the IMPACT-CN system (New England Biolabs, Ipswich, MA) as described previously17. Recombinant human TNF-α was obtained from R&D Systems, whereas mifpristone, SEB and dexamethasone were obtained from Sigma.
Transfection and luciferase assay
Cells (0.5 × 106 per well) were seeded on 60-mm plates the day before the transfection. Transfections were conducted with the FuGENE 6 transfection reagent (Roche Applied Science, Indianapolis, IN) or using a Nucleofector Device (Amaxa, Gaithersburg, MD). The total amounts of DNA and the equal molar ratios of promoters were kept constant in all set-ups by using empty vectors19. Due to differences in transfection efficiencies, an expression plasmid (pCMV β-galactosidase) was co-transfected as a transfection efficiency control, and luciferease activities were normalized based on β-galactosidase activity with the chemiluminescent β-gal reporter gene assay kit (Roche, NJ). Cells were harvested, washed three times with phosphate-buffered saline (PBS) and lysed in 100 μl of the Reporter lysis buffer (RLB) according to the manufacturer’s instructions (Roche). Cell debris was removed by centrifugation and the supernatant was used in the luciferase assay using LUMAT LB9501 (Berthold, TN).
In vitro binding assay
Using the TNT-coupled in vitro transcription/translation system (Promega Corp., WI), 35S-labelled protein products were generated using plasmids containing Vpr wild type, Vpr mutants, GR or PARP-1. The reaction mix was prepared according to the instructions that were supplied by the manufacturer, and the reaction was carried out at 30 °C for 1 h as described previously22,29. Equal amounts (25 μl) of in vitro-translated 35S-labelled proteins were mixed and incubated for 60 min at 4 °C in a binding buffer (25 mM Hepes (pH 7.9), 150 mM KCl, 0.1% Nonidet P-40, 5% glycerol, 0.5 mM dithiothreitol and 0.4 mM phenylmethylsulfonyl fluoride). Proteins were pre-cleared with protein G-Sepharose 4 Fas Flow beads (Amersham Biosciences) and goat immunoglobulin G (IgG) (Santa Cruz Biotechnology, Santa Cruz, CA) at 4°C for 1 h. The Sepharose beads were washed four times with 5 ml of protein lysis buffer containing 0.5 M NaCl and once more with protein lysis buffer. Proteins were immunoprecipitated using anti-PARP-1 (1:500; Cell Signaling Technology, Danvers, MA), anti-GR (1:1000; Active Motif, Carlsbad, CA) and anti-Vpr (1:200; NIH AIDS Research & Reference Program, Germantown, MD) antibodies. Approximately 5 mg of protein G-Sepharose beads were added to each immunoprecipitation reaction from a freshly prepared stock solution (100 mg ml−1), and the samples were incubated at 4 °C for 90 min in a rotating shaker. The beads were washed three times with binding buffer containing high salt and bovine serum albumin (BSA) and finally suspended in 2× SDS sample buffer. The immunoprecipitated protein complexes were eluted from the Sepharose beads by being briefly boiled and then were resolved using 4–15% SDS–PAGE gels. The gel was fixed, treated with a 1 M sodium salicylate solution and dried in a gel drier (Bio-Rad, Hercules, CA). The dried gel was exposed overnight to X-ray film and developed using the automated developer (Kodak, Rochester, NY).
Cell extracts, SDS–PAGE and western blotting
Subcellular fractions were extracted, as described previously, with some modifications35. Briefly, experimental cells were collected by centrifugation at 300 g for 10 min, washed twice with ice-cold PBS, and then once with washing buffer (10 mM Hepes-KOH, pH 7.3, 110 mM KOAc, 2 mM Mg[OAc]2, 2 mM DTT). After resuspension in one volume of lysis buffer (5 mM Hepes-KOH, pH 7.3, 10 mM KOAc, 2 mM −1 each of leupeptin, pepstatin Mg[OAc]2, 2 mM DTT, 1 mM PMSF, and 1 μg ml and aprotinin (Sigma)) and swelling for 10 min on ice, the cells were lysed in a stainless steel Dounce homogenizer. After centrifugation at 1,500 g for 15 min, the supernatant was cleared by centrifugation at 120,000 g for 1 h and then dialysed overnight against transport buffer (20 mM Hepes-KOH, pH 7.3, 110 mM KOAc, 2 mM Mg[OAc]2, 1 mM EGTA). The resulting cytosol was frozen in liquid nitrogen and stored at −80 °C.
To prepare a nuclear extract, the pellet of the 1,500 g spin was washed in 4 ml of lysis buffer and resuspended in 10 ml of 20 mM Hepes-KOH, pH 7.9, 25% glycerol, 0.5 M NaCl, 1.5 M MgCl2, 0.2 mM EDTA, 1 mM PMSF and 1 mM DTT. The nuclei were lysed by ten strokes in a stainless steel Dounce homogenizer, left on ice for 15 min and then centrifuged at 35,000 g for 30 min. The resulting supernatant was dialysed overnight against transport buffer and then centrifuged again at 100,000 g for 30 min. The final supernatant was frozen in liquid nitrogen and stored at −80 °C. The protein concentration was determined with a BCA-200 protein assay kit (Pierce, Rockford, IL) following the manufacturer’s instructions. Proteins were separated by SDS–PAGE and blotted onto to PVDF transfer membrane using standard methods16. Blots were blocked with 5% BSA in TBS-T20 (10 mM Tris-HCl, pH 8, 150 mM NaCl, 0.05% Tween-20) for 1–2 h. Proteins were detected with the specific anti-PARP-1, anti-GR, anti-Vpr polyclonal (AIDS Research and Reference Reagent Program); anti-RelA/p65 (C-20, sc-372; Santa Cruz Biotechnology); anti-actin, anti-PCNA (BD Biosciences); anti-tubulin and Tom20 (Santa Cruz Biotechnology) antibodies in 3% BSA in TBS-Tween-20 and with horseradish-peroxide-coupled goat anti-mouse or anti-rabbit IgG (Sigma) in 1:5,000 dilution. Last, bands were visualized by autoradiography using Amersham ECL system (Amersham Biosciences).
ELISA-based transcription-factor activity assay and EMSA assay
HeLa cells (2 × 106) were treated with TNF-α (5 ng ml−1) with or without rVpr (10 pg). Nuclear proteins were extracted from experimental cell lysates at indicated time points, as described above, and quantified using the protein assay dye reagent. Enzyme-linked immunosorbent assay (ELISA)-based transcription-factor activity assays were performed following the manufacturer’s protocol using BD Mercury TransFactor kits (BD Biosciences). Briefly, 10 μg of nuclear extracts were incubated within 96-well format chambers coated with oligonucleotides containing consensus binding sequences of p65 NF-κB, ATF-2, CREB-1 or c-Fos. Bound transcription factors were then detected by specific antibodies, followed by a horseradish-peroxidase-conjugated secondary antibody, which was used to detect the bound primary antibody. For competition assays, excess competitor oligonucleotides were co-incubated with nuclear protein. For the p65 transcription-factor activity assays, 1,000-ng competitor oligonucleotides were used. Samples were run in triplicate, and all experiments were performed on at least three separate occasions. For electrophoretic mobility shift assays (EMSAs), nuclear extracts were prepared from cells that were untreated or rVpr cells in the presence or absence of rTNF-α stimulation; 10 μg of experimental samples was incubated with 32P-labelled oligonucleotide probes (5′-GTAGGGGACTTTCCGAGCTCGAGA TCCTATG-3′; NF-κB binding site is underlined) in binding buffer. Protein–DNA complexes were resolved on a 5% acrylamide gel using 0.5% TBE buffer, dried and visualized by autoradiography12.
IKK kinase assay
IKK activity was detected by immunoprecipitation of IKK and addition of radiolabelled ATP to recombinant I-κBβ (Biosource, Camarillo, CA). Briefly, for immune-complex kinase assays, serum-starved HeLa cells (2 × 106) were treated with TNF-α with or without recombinant Vpr. The treated cells were then lysed in an immunoprecipitation–kinase buffer (50 mM HEPES (pH 8.0), 150 mM NaCl, 25 mM EGTA, 1 mM EDTA, 0.1% Tween-20 and 10% glycerol) containing a cocktail of protease inhibitors (20 μg ml−1 soybean trypsin inhibitor, 2 μg ml−1 aprotinin, 5 μg ml−1 leupeptin and 100 μg ml−1 PMSF) and phosphatase inhibitors (50 mM NaF and 0.1 mM Na3VO4). To analyse IKK activity, 100 μg protein extracts that were prepared using the procedures described for immunoblot analysis were incubated with 1 μg IKKβ polyclonal antibodies (Santa Cruz Biotechnology) and 50 μg Protein A/G agarose-conjugated beads (Santa Cruz Biotechnology) for 3 h at 4 °C. After washing with buffer (50 mM HEPES (pH 7.0), 250 mM NaCl, 1 mM EDTA and 0.1% NP-40) twice and kinase buffer once, the beads were incubated with 20 μl kinase buffer. The kinase activity was determined at 30 °C for 30 min in a 30 μl reaction mixture containing 50 mM HEPES (pH 8.0), 10 mM MgCl2, 2.5 mM EGTA, 1 mM DTT, 10 μM glycerophosphate, 1 mM NaF, 0.1 mM Na3VO4, 0.1 mM PMSF, 10 μM ATP, 1 μg of I-κBβ substrate (Biosource), and 185 kBq [32P-γ] ATP (222 TBq mmol−1; NEN-Perkin Elmer, Norwalk, CT). Reaction products were separated by SDS–PAGE, and phosphorylated proteins were detected by autoradiography. The immunoprecipitates were also subjected to western blotting to determine the amount of precipitated proteins.
Immunofluorescence
HeLa cells were maintained in poly-L-lysine-coated glass cover slides at a density of 1 × 104/slide chamber. Experimental cells were fixed in 4% paraformaldehyde and permeabilized for 60 min in PBS containing 40 μg ml−1 digitonin (Sigma). Cells were washed with PBS and blocked with PBS containing 1% BSA + 1.5% normal goat serum, followed by incubation with antibodies against PARP-1 (BD-Pharmingen) or eIF4E (Abcam, Cambridge, MA) for 1 h at room temperature and detected with Alexa-Fluor-488-conjugated secondary antibody (Molecular Probes, Carlsbad, CA). For triple labelling, HeLa cells were transfected with either Vpr–GFP or GFP alone using a Nucleofector Device (Amaxa). After experimental conditions, 4% paraformaldehyde fixed cells were immunostained for GR and PARP-1 using polyclonal anti-GR antibody (Activ Motif) (secondary antibody; Alexa-Fluor-568 goat anti-rabbit IgG) and anti-PARP monoclonal antibody (secondary antibody; Alexa-Fluor-647 goat anti-mouse IgG), respectively. Confocal imaging was performed using a Bio-Rad Radiance 2000 imaging system equipped with a Kr/Ar-ion and red diode laser source connected to a microscope (Nikon TE300 model) with a 60× oil objective35.
Cytokines detection by ELISA
The levels of cytokines in experimental animals were analysed by a capture ELISA. We measured IL-12, IL-1β or TNF-α collected from the challenged mice (36 h post-challenge). Briefly, experimental serum was added to the wells in triplicate at 1:1 dilution and incubated at 37 °C for 2–3 h, followed by washing and incubation with detection antibodies for 1 h. Bound antibodies were developed by the addition of TMB peroxide substrate and detected at 450 nm in an ELISA plate reader according to the manufacturer’s protocol (R&D Systems).
siRNA-mediated knockdown of GR and PARP-1
Three sets of hGRα (GenBank X03225) siRNA oligonucleotides and two sets of human PARP-1 (BC037545) were designed using web resources from Ambion Research, Inc (Austin, TX). For each set, top- and bottom-strand oligonucleotides were synthesized separately and annealed together. The primers for GR–siRNA corresponding to the coding region were made to correspond to the sequence 21, 91, 2131 as follows. 21-top, 5′-gatccCTCCTGGTAGAGAAGAAAATTcaagagaTTTTCTTCTCTACCAGGAGTTTTTTGGAAA-3′, and 21-bottom, 5′-AGCTTTTCCAAAAAACTCCTGGTAGAGAAGAAAAtctcttgAATTTTCTTCTCTACCAGGAGg-3′; 91-top, 5′-gatccAACCCTAAGAGGAGGAGCTTTcaagagaAGCTCCTCCTCTTAGGGTTTTTTTTggaaa-3′, and 91-bottom, 5′-AGCTTTTCCAAAAAAAACCCTAAGAGGAGGAGCTtctcttgAAAGCTCCTCCTCTTAGGGTTg-3′; and 2131-top, 5′-gatccCTGGCAGCGGTTTTATCAATTcaagagaTTGATAAAACCGCTGCCAGTTTTTTggaaa-3′, and 2131-bottom, 5′-AGCTTTTCCAAAAAACTGGCAGCGGTTTTATCAAtctcttgAATTGATAAAACCGCTGCCA Gg-3′. The primers for the human PARP-1 siRNA corresponding to the coding region were made to correspond to the sequences 303 and 618 as follows: 303-top, 5′-gatccGTTGGTAGCAAGGCAGAGAATTcaagagaTTCTCTGCCTTGCTACCAATTTTTTggaaa-3′, and 303-bottom, 5′-AGCTTTTCCAAAAAATTGGTAGCAAGGCAGAGAAtctcttgAATTCTCTGCCTTGCTACCAACG-3′; and 618-top, 5′-gatccAGAGAAAAGGCGATGAGGTTTcaagagaACCTCATCGCCTTTTCTCTTTTTTTggaaa-3′, and 618-bottom, 5′-AGCTTTTCCAAAAAAAGAGAAAAGGCGATGAGGTtctcttgAAACCTCATCGCCTTTTCTCTG-3′.
The selection of siRNA sequences followed the criteria provided by Ambion Research, Inc. The BamHI restriction site was included on each top strand at its 5′ end; similarly, on the bottom strand, the HindIII site was included at the 3′ end to facilitate cloning into the pSilencer 2.0-U6 neo vector (Ambion). In the top strand, the uppercase nucleotides correspond to the nucleotide open reading frame of the mRNA sequence. Following seven bases (CAAGAGA), spacer sequences were included to facilitate the formation of a small hairpin loop. The second stretch of upper-case nucleotides is the reverse complement of the first. The two oligonucleotides were annealed and inserted into a pSilencer 2.0-U6 neo vector (Ambion that had been linearized with BamHI and HindIII enzyme. Verification of the inserted sequences was performed by automated DNA sequencing. The pSilencer 2.0-U6 neo vector, which contains a 21-base scrambled sequence and has no significant homology to mouse, rat or human gene sequences, was used as a negative control to transfect cells.
For stable clone selection, HeLa cells were maintained in DMEM medium with 10% FBS. Cells were seeded into 100 mm-diameter culture dishes. A total of 20 μg of the GR-21 siRNA plasmid was transfected as described above. Twenty-four hours after transfection, the cells were split and cultured in G418 selection in DMEM (1 mg ml−1). Culture medium was replaced every 3 d until colonies were formed. Individual colonies were isolated and seeded into six-well plates. After the cells grew to confluence, their GR suppression was tested by immunoblotting analysis using anti-GR antibody.
GST–Vpr pull-down assay and purification of Vpr–His proteins by Ni-NTA Superflow columns
GST–Vpr proteins were expressed in Escherichia coli BL21 and purified using GST glutathione-Sepharose (Amersham Biosciences), followed by elution with reduced glutathione. The eluted GST fusion proteins were dialysed against dialysis buffer (20 mM Tris-HCl (pH 7.5), 100 mM KCl, 0.1 mM EDTA, 0.1 mM DTT and 10% glycerol) and stored at −20 °C. Recombinant GR and PARP-1 proteins were purchased from Activ Motif and BioMol (Plymouth Meeting, PA), respectively. Approximately 2 μg of GST–Vpr, 1 μg of GR and 1 μg of PARP-1 were used in the pull-down assay, as described by the manufacturer’s instructions. In the assays, 2 μg of GST–Vpr was incubated with 1 μg of the purified GR and PARP-1 in 100 μl of binding buffer (50 mM HEPES, 10 mM MgCl2, × protease 10 mM MnCl2, 1 mM DTT, 1 mM β-glycerophosphate, 1 mM NaF and 1 inhibitor cocktail (EDTA-free; Roche)) supplemented with 10 μg of BSA at 4 °C for 1 h. Glutathione Sepharose (5 μl) was added and the mixture was incubated for 1 h at 4 °C. Complexes were washed four times with the binding buffer supplemented with 0.2% NP-40 and eluted in loading buffer. The precipitates were resolved by SDS–PAGE and analysed by western blotting. Vpr–His protein was purified on Ni-NTA columns according to the manufacturer’s instructions (Qiagen, Valencia, CA). Purified proteins were eluted under mild conditions by adding 200 mM imidazole, as per the manufacturer’s instructions. Fractions were collected and analysed by SDS–PAGE and immunoblotting, as described above.
Northern blot analysis
Twenty micrograms of total RNA were separated by 1.2% formaldehyde-agarose gel electrophoresis and transferred to a Nylon membrane (Schleicher & Schuell, Inc., Keene, NH). The membrane was hybridized with the a32P-labelled-specific cDNA probes for pLuc in Quickhyb hybridization solution (Stratagene) for 12 h at 65 °C, washed twice for 15 min at room temperature with 2× SSC/0.1% SDS, then washed once for 30 min at 60 °C with 0.1× SSC/0.1% SDS. The membrane was exposed to K-type imaging screen and visualized using Molecular Imaging System FX (Bio-Rad).
Supplementary Material
Acknowledgments
PARP-1 expression vector was generously provided by Z.-Q. Wang (IARC, Lyon, France) and p65/RelA expression constructs were gifts from D.R. Green (La Jolla Institute for Allergy and Immunology, San Diego, CA). We thank M.A. Chattergoon and D.J. Laddy for their useful comments, and M.J. Merva for administrative assistance. Support from the National Institutes of Health (NIH) AIDS Research and Reference Reagents program and Centers for AIDS Research (CFAR), University of Pennsylvania, is also acknowledged. This work was supported by grants from the NIH to D.B.W.
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website.
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Muthumani K, et al. Human immunodeficiency virus type 1 (HIV-1) Vpr-regulated cell death: insights into the mechanism. Cell Death Differ. 2005;12:962–970. doi: 10.1038/sj.cdd.4401583. [DOI] [PubMed] [Google Scholar]
- 2.Gandhi RT, Walker BD. Immunologic control of HIV-1. Annu Rev Med. 2002;53:149–172. doi: 10.1146/annurev.med.53.082901.104011. [DOI] [PubMed] [Google Scholar]
- 3.Ho DD, et al. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 4.Xu XN, et al. Evasion of cytotoxic T lymphocyte (CTL) responses by Nef-dependent induction of Fas ligand (CD95L) expression on simian immunodeficiency virus-infected cells. J Exp Med. 1997;186:7–16. doi: 10.1084/jem.186.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Letvin NL. Progress in the development of an HIV-1 vaccine. Science. 1998;280:1875–1880. doi: 10.1126/science.280.5371.1875. [DOI] [PubMed] [Google Scholar]
- 6.Geleziunas R, et al. HIV-1 Nef inhibits ASK1-dependent death signalling providing a potential mechanism for protecting the infected host cell. Nature. 2001;410:834–838. doi: 10.1038/35071111. [DOI] [PubMed] [Google Scholar]
- 7.Muthumani K, et al. Mechanism of HIV-1 viral protein R-induced apoptosis. Biochem Biophys Res Commun. 2003;304:583–592. doi: 10.1016/s0006-291x(03)00631-4. [DOI] [PubMed] [Google Scholar]
- 8.Levy DN, et al. Serum Vpr regulates productive infection and latency of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1994;91:10873–10877. doi: 10.1073/pnas.91.23.10873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy DN, Refaeli Y, Weiner DB. Extracellular Vpr protein increases cellular permissiveness to human immunodeficiency virus replication and reactivates virus from latency. J Virol. 1995;69:1243–1252. doi: 10.1128/jvi.69.2.1243-1252.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sherman MP, et al. HIV-1 vpr displays natural protein transducing properties: implications for viral pathogenesis. Virology. 2002;302:95–105. doi: 10.1006/viro.2002.1576. [DOI] [PubMed] [Google Scholar]
- 11.Jowett JB, et al. Human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol. 1995;69:6304–6313. doi: 10.1128/jvi.69.10.6304-6313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ayyavoo V, et al. HIV-1 Vpr suppresses immune activation and apoptosis through regulation of nuclear factor κB. Nature Med. 1997;3:1117–1123. doi: 10.1038/nm1097-1117. [DOI] [PubMed] [Google Scholar]
- 13.Stewart SA, et al. Human immunodeficiency virus type 1 Vpr induces apoptosis following cell cycle arrest. J Virol. 1997;71:5579–5592. doi: 10.1128/jvi.71.7.5579-5592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goh WC, et al. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nature Med. 1998;4:65–71. doi: 10.1038/nm0198-065. [DOI] [PubMed] [Google Scholar]
- 15.Jacotot E, et al. Control of mitochondrial membrane permeabilization by adenine nucleotide translocator interacting with HIV-1 viral protein R and Bcl-2. J Exp Med. 2001;193:509–519. doi: 10.1084/jem.193.4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muthumani, et al. HIV-1 Vpr induces apoptosis through caspase 9 in T cells and peripheral blood mononuclear cells. J Biol Chem. 2002;277:37820–37831. doi: 10.1074/jbc.M205313200. [DOI] [PubMed] [Google Scholar]
- 17.Muthumani K, et al. HIV-1 Vpr inhibits the maturation and activation of macrophages and dendritic cells in vitro. Int Immunol. 2005;17:103–116. doi: 10.1093/intimm/dxh190. [DOI] [PubMed] [Google Scholar]
- 18.de Noronha CM, et al. Dynamic disruptions in nuclear envelope architecture and integrity induced by HIV-1 Vpr. Science. 2001;294:1105–1108. doi: 10.1126/science.1063957. [DOI] [PubMed] [Google Scholar]
- 19.Muthumani K, et al. HIV-1 viral protein-R (Vpr) protects against lethal superantigen challenge while maintaining homeostatic T cell levels in vivo. Mol Ther. 2005;12:910–921. doi: 10.1016/j.ymthe.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Refaeli Y, Levy DN, Weiner DB. The glucocorticoid receptor type II complex is a target of the HIV-1 vpr gene product. Proc Natl Acad Sci USA. 1995;92:3621–3625. doi: 10.1073/pnas.92.8.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kino T, et al. The HIV-1 virion-associated protein vpr is a coactivator of the human glucocorticoid receptor. J Exp Med. 1999;189:51–62. doi: 10.1084/jem.189.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramanathan MP, et al. Carboxyl terminus of hVIP/mov34 is critical for HIV-1-Vpr interaction and glucocorticoid-mediated signaling. J Biol Chem. 2002;277:47854–47860. doi: 10.1074/jbc.M203905200. [DOI] [PubMed] [Google Scholar]
- 23.Doppler W, et al. Expression level-dependent contribution of glucocorticoid receptor domains for functional interaction with STAT5. Mol Cell Biol. 2001;21:3266–3279. doi: 10.1128/MCB.21.9.3266-3279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen LF, Greene WC. Shaping the nuclear action of NF-kappaB. Nature Rev Mol Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 25.Oliver FJ, et al. Resistance to endotoxic shock as a consequence of defective NF-κB activation in poly(ADP-ribose) polymerase-1 deficient mice. EMBO J. 1999;18:4446–4454. doi: 10.1093/emboj/18.16.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassa PO, et al. The enzymatic and DNA binding activity of PARP-1 are not required for NF-κB coactivator function. J Biol Chem. 2001;276:45588–45597. doi: 10.1074/jbc.M106528200. [DOI] [PubMed] [Google Scholar]
- 27.Chang WJ, Alvarez-Gonzalez R. The sequence-specific DNA binding of NF-κB is reversibly regulated by the automodification reaction of poly (ADP-ribose) polymerase 1. J Biol Chem. 2001;276:47664–47670. doi: 10.1074/jbc.M104666200. [DOI] [PubMed] [Google Scholar]
- 28.Hassa PO, et al. The enzymatic and DNA binding activity of PARP-1 are not required for NF-κB coactivator function. J Biol Chem. 2001;276:45588–45897. doi: 10.1074/jbc.M106528200. [DOI] [PubMed] [Google Scholar]
- 29.Mahalingam S, et al. Nuclear import, virion incorporation, and cell cycle arrest/differentiation are mediated by distinct functional domains of human immunodeficiency virus type 1 Vpr. J Virol. 1997;71:6339–6347. doi: 10.1128/jvi.71.9.6339-6347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arad G, et al. Superantigen antagonist protects against lethal shock and defines a new domain for T-cell activation. Nature Med. 2000;6:414–421. doi: 10.1038/74672. [DOI] [PubMed] [Google Scholar]
- 31.Kim MY, et al. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 32.Parent M, et al. Poly(ADP-ribose) polymerase-1 is a negative regulator of HIV-1 transcription through competitive binding to TAR RNA with Tat-positive transcription elongation factor b (p-TEFb) complex. J Biol Chem. 2005;280:448–457. doi: 10.1074/jbc.M408435200. [DOI] [PubMed] [Google Scholar]
- 33.Sawaya BE, et al. Cooperative interaction between HIV-1 regulatory proteins Tat and Vpr modulates transcription of the viral genome. J Biol Chem. 2000;275:35209–35214. doi: 10.1074/jbc.M005197200. [DOI] [PubMed] [Google Scholar]
- 34.Kashanchi F, et al. Cell cycle-regulated transcription by the human immunodeficiency virus type 1 Tat transactivator. J Virol. 2000;74:652–660. doi: 10.1128/jvi.74.2.652-660.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zong WX, et al. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 2004;18:1223–1226. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.