Abstract
Fast axonal conduction depends on myelin, which is formed by Schwann cells in the PNS. We found that the transcription factor Yin Yang 1 (YY1) is crucial for peripheral myelination. Conditional ablation of Yy1 in the Schwann cell lineage resulted in severe hypomyelination, which occurred independently of altered Schwann cell proliferation or apoptosis. In Yy1 mutant mice, Schwann cells established a 1:1 relationship with axons but were unable to myelinate them. The Schwann cells expressed low levels of myelin proteins and of Egr2 (also called Krox20), which is an important regulator of peripheral myelination. In vitro, Schwann cells that lacked Yy1 did not upregulate Egr2 in response to neuregulin1 and did not express myelin protein zero. This phenotype was rescued by overexpression of Egr2. In addition, neuregulin-induced phosphorylation of YY1 was required for transcriptional activation of Egr2. Thus, YY1 emerges as an important activator of peripheral myelination that links neuregulin signaling with Egr2 expression.
The ability of the nervous system to communicate with the periphery depends on faithful transmission of information to target tissues through peripheral nerves. The speed of propagation of action potentials in these nerves depends on myelin, which is formed by Schwann cells. Impaired differentiation of Schwann cells or damage to myelin results in debilitating peripheral neuropathies1. Given the clinical relevance of PNS myelination, it is not surprising that it has been the focus of several mechanistic studies. Investigation of the molecules at the axon-Schwann cell interface that trigger myelination led to the discovery of type III neuregulin1 (refs. 2–4). This axon-derived signal modulates almost every aspect of Schwann cell development and interacts with erbB2 and erbB3 receptors to initiate a signaling cascade that is essential for modulating the timing and abundance of myelin formation in peripheral nerves2,4–7.
Many transcription factors also modulate Schwann cell differentiation, including Egr2, Pou3f1 (also known as Oct-6), Sox10, Brn1 and Brn2 (refs. 8–13). Among them, a key modulator of the transcriptional program of peripheral myelination is Egr2, a zinc finger transcription factor that is regulated by axonal contact and is induced as Schwann cells begin to myelinate. Analysis of Egr2-deficient mice and correlation of mutations in EGR2 with human peripheral neuropathies have provided compelling evidence that Egr2 is important for myelination of peripheral nerves10,14,15. Gene expression studies have revealed that Egr2 acts as a positive regulator of the myelination process16,17 although the molecular mechanisms that regulate its expression remain only partially understood. Egr2 is regulated by both soluble and membrane-bound neuregulins4,16,18 and its concentration is partially modulated by calcium-dependent events19.
Together these studies have indicated that peripheral myelination is the result of the interplay between extracellular signals and an intricate network of transcription factors, orchestrated by Egr2. However, many of the molecular connections between cell surface receptors and transcription factors that modulate myelination are unknown. We have identified the zinc finger protein YY1 as an important modulator of PNS myelination downstream of neuregulin1 (NRG1) signaling. The MEK-dependent cascade that was initiated by NRG1 treatment was responsible for activation of YY1 and increased expression of Egr2. In addition, Schwann cells that lacked YY1 owing to silencing or genetic ablation had low levels of Egr2 and showed impaired myelin gene expression, a phenotype that could be rescued by overexpression of Egr2.
Results
Severe hypomyelination in sciatic nerves lacking Yy1
We generated mutants with conditional ablation of Yy1 in myelinating cells by crossing Yy1 loxP-flanked mice with the Cnp-cre line as described previously20. Although the mice were viable, the number of survivors decreased with age and dropped markedly after the third postnatal week (Supplementary Fig. 1a). In addition, surviving mice did not gain as much weight as their control siblings (Supplementary Fig. 1b). Heterozygous Yy1 mice (Yy1loxP/+; Cnp-cre+/−) appeared normal and were used as littermate controls. We detected clinical signs of peripheral hypomyelination in the homozygous Yy1 mutants (Yy1loxP/loxP; Cnp-cre+/−) during the second postnatal week; they were characterized by hindlimb weakness, flaccid tail paralysis and abnormal hindlimb posture reflexes (Fig. 1). The onset of clinical signs was consistent with the temporal profile of Yy1 expression in the developing sciatic nerve. Yy1 was expressed at birth, but its transcript levels peaked at postnatal day (P)10 and its expression profile closely resembled that of Egr2 during development10,21 (Fig. 1b). Consistent with its role as transcription factor, we found YY1 in the nuclei of myelinated Schwann cells in wild-type mice (Fig. 1c) but not in the sciatic nerves of Yy1loxP/loxP; Cnp-cre+/− mice (Fig. 1d).
Figure 1.
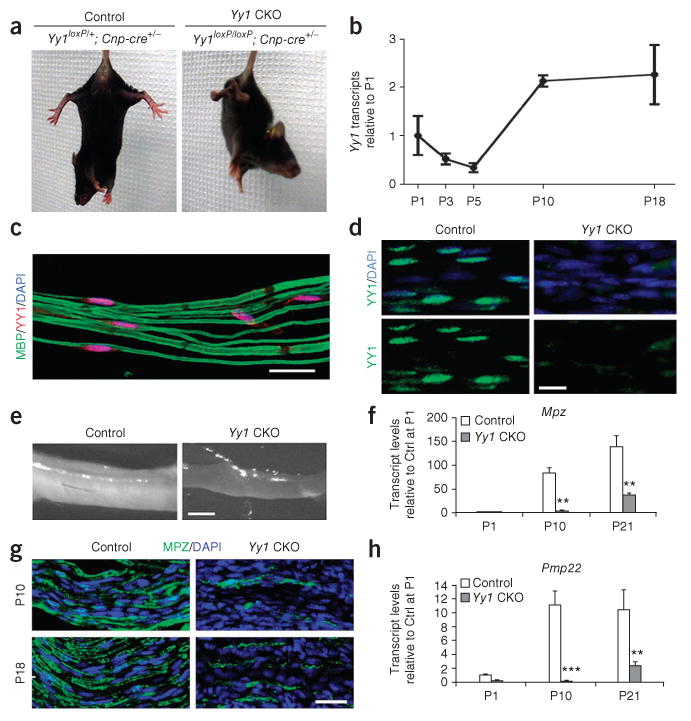
Peripheral nerve hypomyelination in mice with conditional ablation of Yy1. (a) Normal hindlimb postural reflex in a P18 control mouse (Yy1loxP/+; Cnp-cre+/−), characterized by spreading of the limbs, and abnormal reflex in the mutant Yy1loxP/loxP Cnp-cre+/− mouse (CKO), characterized by crossing of the hind limbs. (b) Transcript levels of Yy1 in mouse sciatic nerves during development measured using quantitative reverse transcription PCR (qRT-PCR) n = 3 at each time point). (c) Teased sciatic nerves from P21 wild-type mice stained for YY1 (red) and myelin basic protein (MBP; green) and processed for confocal analysis. Scale bar, 20 μm. (d) Immunohistochemistry for YY1 (green) in the sciatic nerve of controls but not in mutant (Yy1loxP/loxP;Cnp-cre+/−) mice at P18. Cell nuclei were counterstained with DAPI (blue). (e) Gross examination of sciatic nerves dissected from Yy1loxP/loxP;Cnp-cre+/− and control mice at P18 show the white opaque appearance of control nerves and much thinner and more translucent appearance of mutant nerves. (f) qRT-PCR of RNA from sciatic nerves of Yy1loxP/loxP;Cnp-cre+/− mice and control siblings at P1, P10 and P21. The bar graphs represent the transcript levels of Mpz and Pmp22 relative to controls. Error bar, s.d.; **P < 0.01, ***P < 0.001 (n = 3). (g) Longitudinal sections of the sciatic nerves from control and Yy1loxP/loxP; Cnp-cre+/− mice at P10 and P18 were stained for MPZ (green) and nuclei were counterstained with DAPI (blue). Scale bars, 20 μm in c, d, g and 1 mm in e.
Macroscopic examination of the sciatic nerves showed hypomyelination; the nerves were thick and opaque white in control mice and thin and translucent in Yy1 mutants (Fig. 1e). At the molecular level, the sciatic nerves of Yy1loxP/loxP; Cnp-cre+/− mice had lower levels of myelin gene transcripts than did those of control siblings. The differences in transcripts, including myelin protein zero (Mpz) and peripheral myelin protein 22 (Pmp22), between Yy1loxP/loxP; Cnp-cre+/− and control siblings were not evident at P1, but became statistically significant at P10 and persisted throughout development (Fig. 1f). Immunohistochemical analysis of MPZ confirmed the severe hypomyelination at P10 and P18 in the sciatic nerve of Yy1loxP/loxP; Cnp-cre+/− mice and supported the idea that YY1 is important for myelination from P10 (Fig. 1g). At the ultrastructural level, the sciatic nerves of Yy1loxP/loxP; Cnp-cre+/− mice at P18 had very few myelinated axons (14.3 ± 6.3%, total of 692 axons counted and three mice analyzed) compared with controls (54.7 ± 8.5%, 779 axons counted and three mice analyzed; Fig. 2a). The myelinated axons detected in Yy1loxP/loxP; Cnp-cre+/− mice had thinner myelin sheaths, as indicated by the greater g ratio (ratio of axon diameter to the myelinated fiber diameter) in Yy1 mutants (0.853 ± 0.076) compared with controls (0.691 ± 0.088; Fig. 2b).
Figure 2.
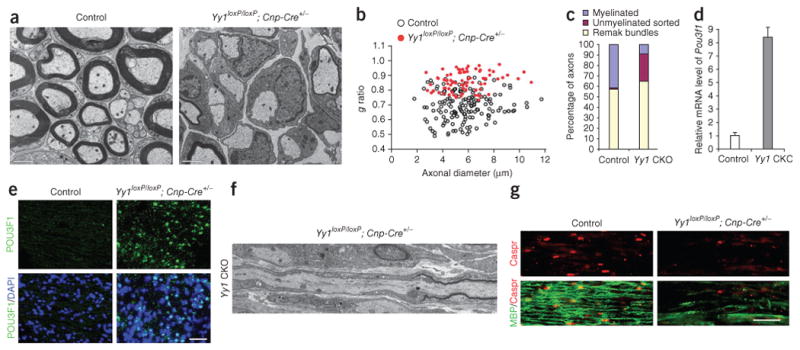
Ablation of Yy1 impairs the ability of Schwann cells to myelinate. (a) Electron micrographs of sciatic nerves of controls and Yy1 mutants show severe hypomyelination in Yy1loxP/loxP; Cnp-cre+/− mice at P18. The Schwann cells in the mutant mice established a 1:1 relationship with large-caliber axons but could not myelinate them. (b) Scatter plot indicating the g ratios of individual fibers as a function of axon diameter (n = 78 axons for Yy1loxP/loxP; Cnp-cre+/− mice; n = 154 axons for controls). (c) Axons were classified into three categories: myelinated (blue bar), unmyelinated-sorted (red bar) and unmyelinated-Remak bundles (yellow bar) and quantified. (d) qRT-PCR revealed elevated Pou3f1 transcripts in the sciatic nerves of Yy1 mutant mice compared to controls. (e) Immunohistochemical validation of Pou3fl expression in longitudinal sections of the sciatic nerves from P18 Yy1loxP/loxP; Cnp-cre+/− mice and control mice. (f) Electron micrographs of longitudinal sections of sciatic nerves in mutant mice showed heminodes, consisting of unpaired paranodal regions, whereas nodal regions were rarely detected. (g) Longitudinal sections of P18 sciatic nerves from control and Yy1loxP/loxP; Cnp-cre+/− mice were stained for MBP (green) and for the paranodal protein Caspr (red). Note the uniform staining of myelinated MBP+ fibers and paired Caspr expression in control animals and the reduced level of MBP and disorganization of Caspr expression in mutant mice. Scale bars, 2 μm in a, 50 μm in e and 20 μm in g.
In Yy1loxP/loxP; Cnp-cre+/− mice we detected Schwann cells in a 1:1 relationship with large caliber axons (>1 μm), but the percentage of unmyelinated large caliber axons was much greater than in controls (Fig. 2c). Because peripheral nerve myelination proceeds in sequential stages that include the association of Schwann cells with clustered axons, followed by radial sorting and 1:1 segregation with the axon1, our data suggested that the sorting process was not affected in Yy1loxP/loxP; Cnp-cre+/− mice and that only the late stages of myelination were impaired. In addition, high levels of Pou3f1 (a marker of pro-myelinating cells) persisted in Yy1loxP/loxP; Cnp-cre+/− mice (Fig. 2d,e). Consistent with impaired developmental myelination, we rarely saw adjacent myelin segments in Yy1loxP/loxP; Cnp-cre+/− sciatic nerves and we noted the prevalence of heminodes (half of a node characterized by the presence of an unpaired paranode; Fig. 2f). The ultrastructural appearance of heminodes in Yy1 mutants at P18 was consistent with the immunohistochemical detection of unpaired clusters of contactin-associated paranode protein (Caspr) labeling (Fig. 2f). This further confirmed the hypomyelinating phenotype.
We also observed defective myelination in the absence of Yy1 in vitro, in dorsal root ganglia (DRG) explants (Supplementary Fig. 2a,b), which contained neurons and Schwann cell precursors22. In explants from control siblings DRG axons were effectively myelinated (18.1 ± 4.7 myelin segments per field; Fig. 3a), whereas myelination was rare in explants from Yy1loxP/loxP; Cnp-cre+/− mice (2.1 ± 1.1 myelin segments per field; Fig. 3b). The lack of myelinated segments correlated with low levels of myelin gene transcripts (Fig. 3c) and severely decreased myelin protein levels, as detected by protein blot (Fig. 3d). To further characterize the effect of YY1 on Schwann cell myelination, we repeated the experiment with DRG explants isolated from Yy1loxP/loxP;Plp-CreERT embryos23 at embryonic day (E)13.5, in which Yy1 was deleted in myelinating cells in a tamoxifen-inducible manner23 (Fig. 3e,f). We determined the efficiency of recombination in myelinating Schwann cells by treating cultures with 10 nM, 100 nM and 1 μM 4-hydroxy-tamoxifen (4OH-TM) for 48 h and assaying the nuclear localization of Cre and the expression of YY1 using immunocytochemistry (Supplementary Fig. 2c). As expected, 4OH-TM induced nuclear translocation of Cre in a dose-dependent fashion only in Schwann cells, but not in fibroblasts and neurons, that continued to express YY1 (Supplementary Fig. 2c). The analysis of myelin segments in explants treated with 1 μM 4OH-TM (Fig. 3f) revealed only 2.2 ± 0.6 myelin segments per field compared to 20.5 ± 4.9 segments in mock-treated explants and 15.3 ± 2.9 segments in Yy1loxP/loxP explants treated with 4OH-TM (Fig. 3e). To confirm that defective myelination in the Yy1loxP/loxP; Cnp-cre+/− explants depended on the Schwann cells and not on the DRG neurons, we generated co-cultures of wild-type rat Schwann cells with DRG neurons from Yy1loxP/loxP; Cnp-cre+/− or control embryos (Fig. 3g). Both cultures showed similar numbers of myelin segments, which supports the idea that the defect in the Yy1loxP/loxP; Cnp-cre+/− mice depended on Schwann cell function.
Figure 3.
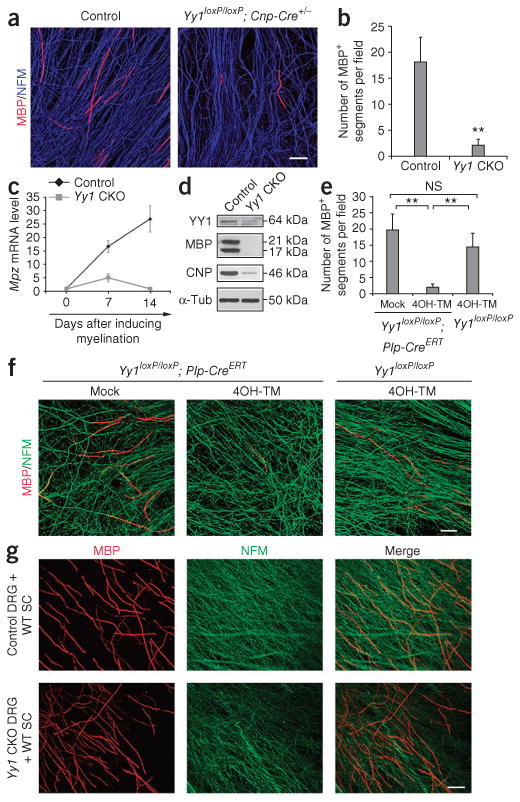
Effect of Yy1 ablation on the ability of Schwann cells to myelinate in vitro. (a) In vitro explant DRG cultures from E13.5 control and Yy1loxP/loxP; Cnp-cre+/− embryos were immunostained for MBP (red) and neurofilament medium chain (NFM, blue). Note the few myelin segments in cultures from the Yy1loxP/loxP; Cnp-cre+/− mice compared to controls. (b) Bar graph shows the number of myelin segments shown in a. (c) qRT-PCR of mRNA levels of Mpz in myelinating cultures established from Yy1loxP/loxP; Cnp-cre+/− and control DRGs and examined at days 0, 7 and 14 after induction of myelination. (d) Protein blot analysis of MBP and CNPase in myelinating cocultures derived from Yy1loxP/loxP; Cnp-cre+/− and control DRGs kept in culture for 21 d. Full-length blots are shown in Supplementary Figure 5. (e,f) Immunofluorescence of cocultures of DRG neurons and Schwann cells from E13.5 transgenic embryos (Yy1loxP/loxP; Plp-creERT) treated with 1 μM Tamoxifen (4OH-TM) for 2 d and then cultured for additional 12 d to induce myelination. MBP (red) and neurofilament (NFM, green). (g) Immunofluorescence of Yy1loxP/loxP; Cnp-cre+/− and control DRG neurons cultured with wild-type rat Schwann cells (WT SC) for 14 d and then stained for MBP (red) and NFM (green). Scale bars, 50 μm. Error bars, s.d.; **P < 0.01.
YY1 is an important upstream regulator of Egr2
We reasoned that the mechanism of action of YY1 in Schwann cells might involve Egr2 because the two transcripts shared a similar expression profile in the developing sciatic nerve and because of the marked similarities between the phenotype of the Yy1loxP/loxP; Cnp-cre+/− mice and those of Egr2Lo/Lo mutants10,14 and Egr2-binding protein Nab1 (NGFI-A binding protein 1)−/−; Nab2 (NGFI-A binding protein 2)−/− double mutants17. In all three mutant mice, similar clinical signs developed during the second postnatal week and included tremor and weakness of the hind limbs. The macroscopic appearance of the sciatic nerves was identical (thin and translucent) and the Schwann cells in the three mutants showed successful completion of radial sorting and defective myelination10,14,17. Because Egr2Lo/Lo (refs. 10,14,24) and Nab1−/−; Nab2−/− double mutants17 showed increased proliferation and apoptosis compared to control mice, we investigated whether similar features occurred in the Yy1loxP/loxP; Cnp-cre+/− mice. We used BrdU incorporation to measure S-phase entry and performed immunohistochemical analysis of the sciatic nerve of Yy1 mutant and control mice at P1, P10 and P18 (Fig. 4a,b). Although we detected no difference at the early developmental time points, similar to the Egr2Lo/Lo mice14, we found a higher percentage of BrdU+ cells in the sciatic nerves of Yy1loxP/loxP; Cnp-cre+/− mice than in those of controls at P10 and P18 (Fig. 4b). The detection of a proliferative phenotype at p10, but not at p1, in Yy1loxP/loxP; Cnp-cre+/− mice was consistent with the developmental pattern of Yy1 expression in the sciatic nerve and correlated with the downregulation of the cell cycle inhibitor Cdkna1 (also called p21), which is an important regulator of the G1-S phase transition (Fig. 4c). We detected similar results in DRG co-cultures from Yy1loxP/loxP; Cnp-cre+/− mice and controls (Supplementary Fig. 3). We next examined apoptosis in the sciatic nerves. Although we found no difference at P2 or P10, we detected a transient and modest increase in TUNEL+ cells at P18 in Yy1loxP/loxP; Cnp-cre+/− mice compared with controls (Fig. 4d). This was associated with increased transcripts of genes involved in apoptosis, such as Jun and Tgfb1 (refs. 25,26; Fig. 4e). Together, these data suggested that there might be a genetic interaction between YY1 and Egr2.
Figure 4.
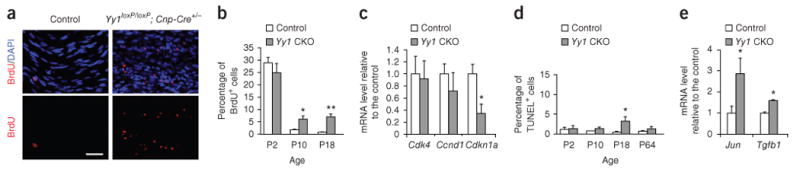
Ablation of Yy1 modulates Schwann cell S-phase entry during the second week of development. (a) Longitudinal sections of the sciatic nerves from P18 Yy1loxP/loxP; Cnp-cre+/− and control mice stained with anti-BrdU (red) antibodies after 2 h pulse labeling of BrdU in vivo. Scale bar, 20 μm. (b) The fraction of BrdU+ cells in the sciatic nerves at multiple developmental time points was quantified and referred to the percentage of DAPI+ cells. (c) qRT-PCR of genes involved in proliferation (Cdk4, cyclin D1 (Ccnd1) and P21 (Cdkn1a)) in the sciatic nerves of mutant mice and control littermates at P10. (d) TUNEL assay of the sciatic nerves from mutant mice and control littermates at the indicated time points. The fraction of TUNEL+ cells in the sciatic nerves was quantified and referred to the percentage of DAPI+ cells. (e) qRT-PCR of genes involved in apoptosis (Jun and Tgfb1) in the sciatic nerves of mutant mice and control littermates at P21. Error bar, s.d.; *P < 0.05, **P < 0.01. n = 3 for each genotype at P2, P10, P18 and P21 and n = 2 at P64.
To investigate this possibility, we compared gene profiling of the sciatic nerves from control and Yy1loxP/loxP; Cnp-cre+/− mice at P21 and identified 1,575 upregulated genes and 1,151 downregulated genes. We then compared this set of 2,725 genes with published gene profiling datasets from Egr2Lo/Lo and Nab1−/−; Nab2−/− mice17. Among the genes whose expression changed in Yy1 mutants, 36.67% were also changed in Egr2Lo/Lo mice and 24.60% were also altered in Nab1−/−; Nab2−/− mice. Of the regulated genes, 351 overlapped in all three mutants (Fig. 5a). The shared subsets of genes that had decreased expression included the myelin genes Pmp22, Mpz and Mbp, and those encoding lipid metabolism enzymes such as stearoyl-coenzyme A desaturase 2 (Scd2), fatty acid desaturase 2 (Fads2), 24-dehydrocholesterol reductase (Dhcr24) and HMG coenzyme A reductase (Hmgcr; Fig. 5b), and this is consistent with the hypomyelinating phenotype and with the involvement of these genes in several human neuropathies. Additional transcripts that were significantly decreased in all three mutants included several growth factors such as fibroblast growth factor 1 (Fgf1), Fgf4 and bone morphogenetic protein 7 (Bmp7), and receptors such as Frizzled-related protein 3 (Frzb) and G protein-coupled receptor family C group 5 (Gprc5b). The transcripts that were upregulated during the third postnatal week in all three mutants included the transcription factors Sox4, Sox5, Egr1, Pou3f1, Btg1, Id2 and Id4 (refs. 27,28; Fig. 5b) and also genes related to the cell cycle such as cyclin B1 (Ccnb1), cyclin D1 (Ccnd1), cell division cycle associated 3 (Cdca3) and pituitary tumor-transforming 1 (Pttg1).
Figure 5.
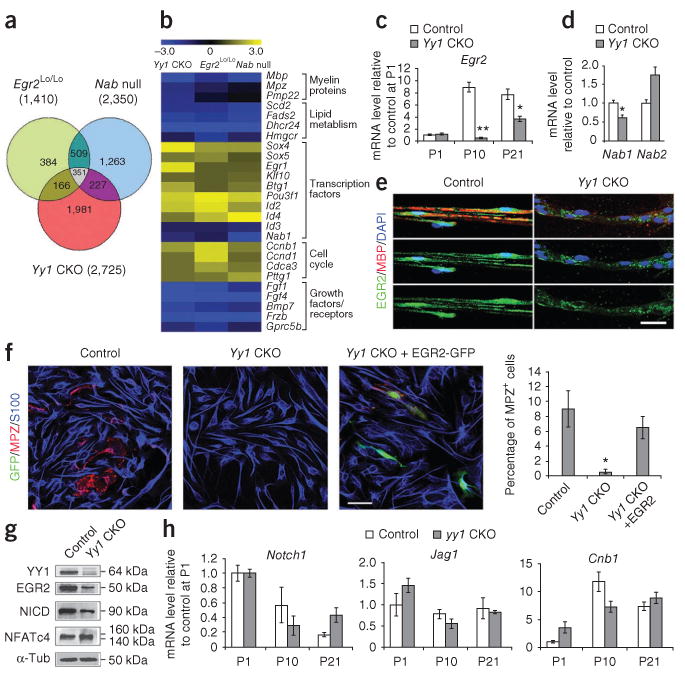
YY1 regulates Egr2 expression. (a) Intersection of genes altered in the sciatic nerves of Yy1loxP/loxP; Cnp-cre+/− mice and of Egr2Lo/Lo and Nab1−/−;Nab2−/− mice17. (b) Comparison of gene expression in Yy1loxP/loxP; Cnp-cre+/−, Egr2Lo/Lo and Nab1−/−;Nab2−/− mice. Similar gene expression signatures in the sciatic nerves of the three mutants suggested that these molecules might share a common signaling pathway. The expression of Nab1, Nab2 (c) and Egr2 (d) was measured by qRT-PCR in the sciatic nerves at P21. (e) Immunohistochemistry showed decreased Egr2 (green) immunoreactivity in teased sciatic nerve fibers of Yy1loxP/loxP; Cnp-cre+/− mice. (f) Purified mouse Yy1loxP/loxP; Cnp-cre+/− Schwann cells cultured in myelination medium for 3 d showed normal S100 (blue) expression but low immunoreactivity for MPZ (red), and ectopic expression of Egr2 in these cells rescued the phenotype as indicated by the detection of MPZ in GFP+ transfected cells. Bar graph shows quantification of the fraction of MPZ+ cells among S100+ cells from two experiments. (g) Protein blot of myelinating cocultures from Yy1loxP/loxP; Cnp-cre+/− and control DRGs maintained for 21 d in vitro showed low Egr2, whereas the NICD was only mildly affected. Full-length protein blots are shown in Supplementary Figure 5. (h) qRT-PCR of Notch1, Jag1 and CnB1 in the sciatic nerves of Yy1loxP/loxP; Cnp-cre+/− and control mice at distinct developmental time points. Error bar, s.d.; *P < 0.05, **P < 0.01 (n = 3). Scale bars, 20 μm.
The similarity between the gene expression signatures in the sciatic nerves of Egr2Lo/Lo mice and Yy1loxP/loxP; Cnp-cre+/− mice was consistent with a possible interaction between YY1 and the Egr2-Nab regulatory network. As the levels of Yy1 transcript changed only slightly in Egr2Lo/Lo mice (1.2-fold) and in Nab1−/−; Nab2−/− mice (0.95-fold)14,17, it was unlikely that Yy1 was downstream of the Egr2–Nab complex. An alternative possibility was that Yy1 was upstream of Egr2. Indeed, levels of Egr2 transcripts in the sciatic nerve of Yy1loxP/loxP; Cnp-cre+/− mice were significantly lower than control values at P10 and reached 38 ± 8% of control levels at P21 (Fig. 5c). Levels of Nab1 transcript were less affected in Yy1loxP/loxP; Cnp-cre+/− mice (62 ± 6.3% of the level of the controls, n = 3) and Nab2 transcripts were increased (70% more than controls), which suggests that the effect of Yy1 on myelination was unlikely to be mediated by the modulation of Nab1 and Nab2 levels (Fig. 5d). The decrease in Egr2 transcripts in Yy1loxP/loxP; Cnp-cre+/− mice was consistent with the detection of decreased levels of Egr2 protein by immunohistochemistry (Fig. 5e) and protein blot (Fig. 5f). To prove that Egr2 was downstream of YY1, we overexpressed Egr2 in Yy1loxP/loxP; Cnp-cre+/− Schwann cells; this overexpression rescued the expression of MPZ (Fig. 5f). Together these data provide strong evidence that YY1 is an upstream regulator of Egr2.
To investigate whether the hypomyelinating phenotype of Yy1 mutants resulted from changes in the expression of genes that negatively affect postnatal peripheral myelination, we searched the microarray database for enrichment in genes in the Notch signaling pathway29. We also searched for genes related to the neuregulin-calcineurin-NFATcy pathway, which modulates Schwann cell differentiation19. Pathway analysis conducted with the DAVID Gene Functional Classification Tool did not reveal any enrichment of genes associated with these two signaling pathways (data not shown). To test further whether the decrease in Egr2 transcripts in Yy1loxP/loxP; Cnp-cre+/− mice resulted from altered Notch signaling, we performed protein blot analysis and quantitative real-time PCR. The decreased levels of the intracellular domain of Notch (NICD) detected in Yy1loxP/loxP; Cnp-cre+/− mice (Fig. 5g), together with the similar levels of Notch1 and its ligand Jagged1 throughout development (Fig. 5h), ruled out the involvement of this pathway in the hypomyelinating phenotype of the mutants. Similarly, the pattern of expression of calcineurin B (Cnb1, also known as Ppp3r1; Fig. 5h) and the lack of similarities between the phenotypes of Yy1loxP/loxP; Cnp-cre+/− and Ppp3r1 mutant mice suggested that YY1 acts as upstream regulator of Egr2, independent of calcineurin B or Notch signaling.
NRG1-dependent YY1 phosphorylation regulates Egr2
The expression of Egr2 expression is crucial for peripheral nerve myelination and is regulated by axonal contact and soluble neuregulin4,17. Treatment of cultured Schwann cells17,18 with soluble NRG1 for 1 h was sufficient to induce upregulation of Egr2 transcripts (Fig. 6a). To determine whether YY1 was part of the signaling pathway that was responsible for the upregulation of Egr2 in response to NRG1, we silenced its expression using short hairpin RNA (shRNA) plasmids transfected into primary Schwann cells (Fig. 6b) and measured the transcript levels (Fig. 6c). Yy1 silencing significantly decreased the NRG1-mediated expression of Egr2 transcripts (Fig. 6c) whereas the levels of Nab1, Nab2 and Pou3f1 transcripts remained unchanged (Fig. 6d).
Figure 6.
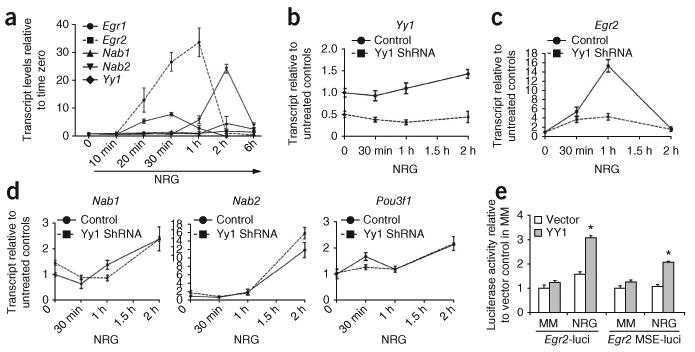
YY1 regulates Egr2 expression in response to NRG1. (a) Purified rat Schwann cells were cultured in minimal medium for 18 h and then exposed to NRG1. The transcript levels of Egr1, Egr2, Nab1, Nab2 and Yy1 at each time point were analyzed by qRT-PCR. Egr2 transcripts increased at 20 min, peaked at 1 h and then gradually returned to basal levels by 2 h, whereas the expression of Yy1 increased only moderately. (b-d) Transcript levels of Yy1, Egr2, Nab1 and Nab2 in Schwann cells either mock-transfected or transfected with Yy1-specific shRNAs and cultured in the absence or presence of NRG1 for 30 min, 1 h or 2 h. The expression of each gene in control cells at time zero was arbitrarily set as 1. (e) Luciferase activity measured in NRG1-treated Schwann cells transfected with reporter constructs containing either the Egr2 promoter or the myelinating Schwann cell enhancer (MSE) of Egr2, together with pCX-Yy1 or empty vector. Values were referred to the readings obtained in untreated cells transfected with empty vector. Error bars, s.d.; *P < 0.05, **P < 0.01.
The role of YY1 in regulating the NRG-induced expression of Egr2 was consistent with the presence of multiple YY1 binding sites in the promoter and conserved regulatory region (MSE, myelinating Schwann cell element) of Egr2 (Supplementary Fig. 4). To test the effect of YY1 on the activity of the Egr2 promoter and MSE, we performed a series of cotransfection experiments with YY1 constructs and luciferase reporter genes, driven either by the mouse Egr2 promoter or by the MSE region upstream of a minimal promoter19. We treated cotransfected Schwann cells with NRG1 and measured reporter activity. Consistent with the role of YY1 as an NRG1-dependent downstream activator of Egr2, we detected increased reporter activity in cotransfected cells only after treatment with NRG1 (Fig. 6e).
To determine whether YY1 directly modulated the activity of the Egr2 promoter and MSE in living cells, we performed chromatin immunoprecipitation (ChIP). Chromatin was cross-linked from Schwann cells before or after exposure of Schwann cells to NRG1 for 1 or 12 h. We then tested the recruitment of YY1 to precise chromatin loci using immunoprecipitation with antibodies to YY1 followed by quantitative ChIP with primers specific for the Egr2 locus (Fig. 7a). YY1 was recruited to the promoter and MSE region of Egr2 only after treatment with NRG1 for 1 h (Fig. 7b) and was consistent with the kinetics of Egr2 transcription in response to NRG1 (Fig. 6a).
Figure 7.
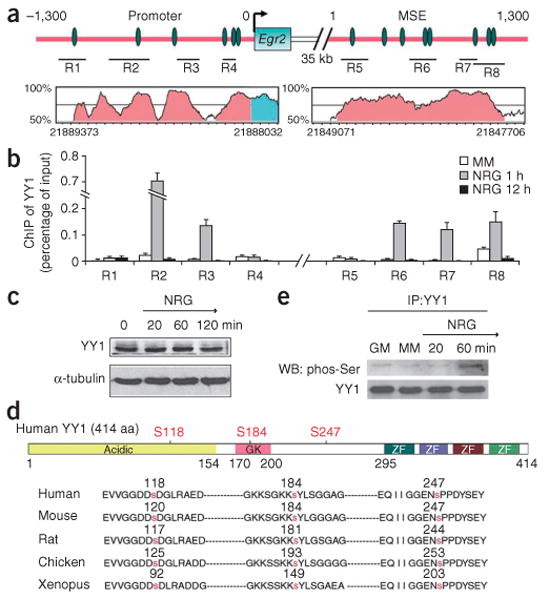
YY1 binds to chromatin at the Egr2 locus only in Schwann cells treated with NRG1. (a) Schematic diagram of the promoter and MSE of Egr2. YY1 consensus binding sequences (green ovals) and the regions (R1–R8) analyzed by chromatin immunoprecipitation are indicated. The plot in pink shows highly conserved sequences between rat and human. Scale marks on x axis indicate 50 bp. (b) Chromatin from Schwann cells cultured in minimal medium (MM, white bar) or treated with NRG1 for 1 h (gray bars) or 12 h (black bars) was immunoprecipitated (ChIP) with antibodies against YY1 and the regions indicated in a were amplified. The results are expressed as percent of input. YY1 was recruited to multiple regions of the Egr2 promoter and MSE after 1 h NRG1 treatment. (c) Protein blot analysis of YY1 in Schwann cells after NRG1 treatment. (d) Schematic diagram of human YY1 protein including the acidic N terminus, the glycine-lysine–rich central domain (GK) and the C-terminal DNA-binding domain, which is composed of four zinc fingers (ZF). Three highly conserved serine residues are marked in red. (e) Co-immunoprecipitation of protein lysates from rat Schwann cells kept in growth medium (GM) or minimal medium (MM), or treated with NRG1 for 20 min or 1 h. After immunoprecipitation (IP) with anti-YY1 antibodies, the protein blot (WB) was probed with anti-phospho-serine antibodies. Full-length blots are presented in Supplementary Figure 5.
We next investigated how NRG1 treatment promoted binding of YY1 to the Egr2 locus. Because neither the levels nor the subcellular localization of YY1 changed upon NRG1 treatment (Fig. 7c), we investigated whether its activity was modulated by post-translational modifications induced by NRG1. Analysis of the amino acid sequence of YY1 revealed several potential phosphorylation sites that were highly conserved among species (Fig. 7d). These sites were consistent with the results of proteomic studies in which YY1 was found to be phosphorylated on S118, S184 and S247 in Hela cells30,31. Analysis of the sequences that surrounded these serine residues, using the Group-based Prediction System32, indicated that these residues were potential MEK phosphorylation sites. Because activation of the MEK-MAPK cascade has been identified as part of NRG1 signaling, we defined the time course of serine phosphorylation of YY1 in Schwann cells exposed to NRG1. Protein extracts from Schwann cells treated with NRG1 for 20 min or 1 h were immunoprecipitated with YY1 antibodies and processed for protein blot analysis using anti-phospho-serine antibodies. We detected an immunoreactive band in the 1-h treatment sample, which indicated YY1 phosphorylation (Fig. 7e).
To identify the kinase that phosphorylated YY1 in response to NRG1, we tested the functional consequences of pharmacologically blocking the three main pathways downstream of NRG1 (refs. 3,19). We used inhibitors of Akt activity (LY294002), MEK kinase (U0126 and PD98059) and the phospholipase C (PLC)γ-calcineurin pathway (cyclosporin A (CsA)). Of these compounds, the MEK inhibitors most severely impaired the NRG1-mediated increase in Egr2 expression (Fig. 8a) and blocked YY1 phosphorylation (data not shown). Using ChIP, we also showed that the recruitment of YY1 to the MSE regions of the Egr2 locus in response to NRG1 treatment was impaired by treatment with U0126 but not with CsA (Fig. 8b).
Figure 8.
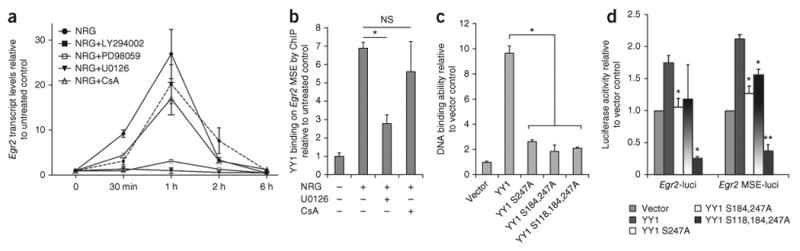
The regulation of Egr2 by phosphorylated YY1 is mediated by NRG1-dependent MEK activation. (a) Rat Schwann cells were treated with NRG and the PI(3)K inhibitor LY294002, with the MEK inhibitors PD98059 or U0126, or with the calcineurin inhibitor CsA. The transcript levels of Egr2 were assessed by qRT-PCR. (b) ChIP of samples from Schwann cells either untreated or treated with NRG1 alone or with the MEK inhibitor U0126 or the calcineurin inhibitor CsA for 1 h and immunoprecipitated with antibodies to YY1. Note the decreased binding to region 7 of the Egr2 MSE in cells treated with U0126 but not CsA, in response to NRG stimulation. (c) Luciferase assay of Schwann cells cotransfected with YY1 Ser→Ala mutation at position 118, 184, 247 and with Yy1 translucent reporter. Note the decreased ability of mutant YY1 to activate a YY1 binding sequence–driven reporter after NRG treatment. (d) Luciferase assay of Schwann cells treated with NRG1 and cotransfected with the indicated reporter constructs and the mutant Yy1 constructs. Error bar, s.d.; *P < 0.05. **P < 0.01.
To determine the functional role of NRG1-induced phosphorylation of YY1, we generated multiple Ser→Ala mutantions at position 118, 184 and 247 of YY1 and tested the consequences for the transcriptional activity of YY1 using a YY1 TransLucent Reporter20. Mutation of one, two or three serine residues impaired the ability of YY1 to activate a luciferase reporter driven by a cis-acting enhancer element containing multiple YY1 binding sites upstream of a minimal promoter after NRG treatment (Fig. 8c). Although single Ser→Ala substitutions only moderately affected the ability of YY1 to transactivate luciferase reporters driven by the Egr2 regulatory elements (Fig. 8d), the mutant construct with three amino acid substitutions produced the most significant impairment (Fig. 8d). Therefore, phosphorylation of YY1 is a key signal downstream of NRG1 that is required for the upregulation of Egr2.
Our results identify YY1 as a crucial regulator of peripheral nerve myelination that functions as an activator upon NRG1-dependent phosphorylation of key serine residues.
Discussion
YY1 is a key regulator of postnatal myelination in the PNS
The notable peripheral nerve hypomyelination that we detected in mice with conditional ablation of Yy1 provides the first in vivo genetic evidence that YY1 has a crucial role in peripheral nerve myelination. The expression of YY1 in the developing sciatic nerve paralleled that of the key transcriptional regulator Egr2, which has been extensively characterized for its role in promoting the attainment of a myelinating phenotype10,21. The temporal expression profile of YY1 suggested that it might be involved in the late postnatal development of peripheral nerves. Consistent with this prediction, at birth the sciatic nerves of Yy1loxP/loxP; Cnp-cre+/− mice and control mice were similar in appearance. The proliferation rates, numbers and gene expression profiles of Schwann cells were also similar between wild-type and Yy1loxP/loxP; Cnp-cre+/− mice during the first neonatal days. Early Schwann cell development was therefore unperturbed in the Yy1loxP/loxP; Cnp-cre+/− mice and the detection of a correct 1:1 relationship with the axons confirmed that Schwann cells underwent radial sorting and axonal segregation. We observed severe hypomyelination during the second postnatal week, characterized by decreased levels of myelin proteins and of Egr2.
Notably, the presence of hypomyelination after normal segregation, increased proliferation and compensatory apoptosis at P10 resembled the phenotype of Egr2 mutants10,14. A comparison between the gene expression profiles of Yy1 deficient mice and Egr2 hypomorphs revealed marked similarities in the patterns of gene expression in the peripheral nerves of these mutants14. The ability of Egr2 to rescue the ability of cultured Yy1 mutant Schwann cells to express myelin proteins suggested that Egr2 is downstream of Yy1. This conclusion was supported by evidence that overexpression of Yy1 activated the expression of luciferase reporter genes driven by the Egr2 promoter and regulatory elements and that YY1 is physically bound to these elements in the chromatin of cultured Schwann cells exposed to NRG1.
YY1 activity is regulated by NRG1-dependent phosphorylation
Myelination of the PNS depends on the interplay between extracellular signals and a complex transcriptional network that modulates Schwann cell maturation. Previous work has shown that NRG1 type III is the axonal signal that binds to its receptors erbB2 and erbB3 and regulates the timing and amount of myelination as well as myelin sheath thickness2,4. Besides its role in myelination, NRG1 also regulates multiple stages of Schwann cell development, including the early commitment of neural crest cells and precursor migration1,6, and several groups have attempted to decipher the signaling events downstream of the activation of erbB2 and erbB3 receptors.
A crucial event in myelination is the upregulation of Egr2. The levels of Egr2 are increased during the transition to myelinating Schwann cells and this increase has been attributed to axonal contact as well as to membrane-bound or soluble neuregulins4,17. Despite the evidence that Egr2 is crucial for peripheral nerve myelination, there is limited information on the NRG1-dependent pathways that regulate Egr2 expression. Three signaling pathways have been shown to be activated by erbB2–erbB3 receptor complexes in response to ligand binding: phosphatidylinositol-3-OH kinase (PI(3)K), PLCγ and MEK19,33–35. Although many reports have focused on the activation of the PI(3)K pathway, a recent study has implicated the PLCγ pathway, in cooperation with Sox10, in activating the transcription of Egr2 and Mpz19. It has been proposed that Ca2+ influx, mediated by the PLCγ pathway and followed by activation of calcineurin and dephosphorylation of NFATc4, activates Egr2 and Mpz. However the analysis of sciatic nerves from mice with mutations in Ppp3r1 showed substantial levels of Egr2 transcripts and therefore suggested that there are alternative pathways of activation of Egr2 in the absence of calcineurin B19. We have provided several lines of evidence that YY1 is a key regulator of Egr2. First, we have defined the in vivo kinetics of binding of YY1 to the chromatin in the promoter and MSE regulatory region of Egr2 upon exposure of Schwann cells to NRG1. Second, we have shown that silencing of Yy1 prevents the NRG1-dependent increase in Egr2, without affecting the levels of Pou3f1, Nab1 or Nab2. Third, we have shown that the NRG1-dependent serine phosphorylation of YY1 is crucial for the recruitment of YY1 to the Egr2 locus and that it depends on the activation of the MEK cascade (Supplementary Fig. 6). Previous studies had identified the downstream MAPKs as mediators of the proliferative and migratory effects of NRG1 in Schwann cell precursors35,36. Our data are not inconsistent with this possibility, because we focused our analysis at the postnatal stage of development, when YY1 is increased, rather than at earlier embryonic stages. Downstream of MEK kinase, at least two pathways with opposing effects have been identified. The activation of the Ras-MAPK pathway, for instance, has been associated with lack of differentiation34,37,38, whereas activation of p38 kinase has been associated with myelination37,39. However, these effects were dependent on the concentrations of soluble NRG and the strength of MAPK activation. Together these data suggest that in Schwann cells, as in other experimental systems, exposure to signaling molecules (NRG1) determined an outcome that depended on the strength of the signal, the enzymes that were activated (Akt, PLCγ, MEK), the levels and kinetics of activation of each enzyme and the specificity of the cellular substrates, which may differ at distinct developmental stages. Future studies might be needed to determine the molecular identity and functional significance of NRG-dependent activation at distinct stages in Schwann cell development and in response to exposure to axon-bound or soluble ligands.
The role of YY1 is unique and cell-type specific
In oligodendrocytes, the myelin-forming cells of the CNS, YY1 forms repressive complexes with HDAC1 and HDAC2 that downregulate transcriptional inhibitors of myelin gene expression, such as Tcf7l2 and Id4, and favor progression to a myelinating phenotype20. In other words, YY1 modulates oligodendrocyte differentiation by promoting a de-repression mechanism (inhibiting the inhibitors). Considering that both oligodendrocytes and Schwann cells can produce myelin and that their differentiation is similarly impaired in the absence of Yy1, the simplest explanation would suggest that YY1 has a similar role in myelin-forming cells of the PNS and CNS. However, we have shown that the role of YY1 in the PNS is quite different from its role in the CNS and that it acts as transcriptional activator of Egr2, a key regulator of Schwann cell myelination. The ability of YY1 to activate the expression of Egr2 is not constitutive, but rather is modulated by phosphorylation events that can be induced by exposure of Schwann cells to NRG1. Therefore, even though the peripheral and central phenotypes of Yy1 mutant mice are similar (hypomyelination), the YY1-dependent mechanisms in the CNS and PNS are distinct and cell specific. In oligodendrocytes, YY1 is part of repressive complexes that contain histone deacetylases and the repression of transcriptional inhibitors depends on its acetylation state40. By contrast, in Schwann cells YY1 functions as activator of gene expression whose activity depends on the phosphorylation state of specific serine residues, consistent with early studies on the signals that modulate the yin and yang functions of this zinc-finger protein41.
The essential function of YY1 in Schwann cell myelination, as revealed by the phenotype of Yy1 conditional knockout mice, does not exclude the possibility that YY1participates in earlier stages of Schwann cell development. Yy1 deletion studies in Xenopus have identified several target genes that are involved in neural crest development, such as slug, snail and Otx2 (refs. 42,43). It will be interesting to determine whether YY1 has distinct functions at different stages of Schwann cell lineage development and to identify the post-translational modifications of this transcription factor that might integrate the cross-talk among distinct signaling pathways.
Methods
Methods and any associated references are available in the online version of the paper at http://www.nature.com/natureneuroscience/.
Supplementary Material
Acknowledgments
We thank Y. Shi, K.A. Nave and B. Popko for mouse lines; G. Crabtree, D. Meijer and M. Grumet for plasmids and antibodies; H. Kim and P. Maurel for advice on Schwann cell and myelination cultures; C. Krier for assistance with microarrays; J. Salzer, K. Jessen, P. Brophy and C. Taveggia for discussions; and N. Kuo, J. Li and R. Srinivasan for technical assistance. This work was supported by grant numbers 5R01NS042925-08, R01NS052738-04 and NS04295-08S ARRA supplement and NMSS RG-4134 to P.C. and in part by the NJ Commission on Spinal Cord Research (08B-010-SCR3 to Y.H.). Electron microscopy was performed at the VCU Department of Anatomy and Neurobiology Microscopy Facility and supported, in part, by funding from a US National Institutes of Health Natinal Institute of Neurological Disorders and Stroke Center core grant (5P30NS047463-02).
Footnotes
Note: Supplementary information is available on the Nature Neuroscience website.
Author Contributions: Y.H. conducted the majority of the experiments and analyzed the data. J.Y.K. generated the YY1 point mutation plasmids and performed the immuno-precipitation and western blots. J.D. performed the ultrastructural analysis of the sciatic nerve. C.M.-V. and A.T. helped with the DRG co-culture experiments and conducted part of the in vitro myelination studies. J.S. performed the conservation analysis and contributed to the analysis of gene expression in the three mutants. P.C. supervised the project, analyzed the data, formatted the figures, uploaded microarray data and wrote the manuscript.
Competing Financial Interests: The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Jessen KR, Mirsky R. The origin and development of glial cells in peripheral nerves. Nat Rev Neurosci. 2005;6:671–682. doi: 10.1038/nrn1746. [DOI] [PubMed] [Google Scholar]
- 2.Michailov GV, et al. Axonal neuregulin-1 regulates myelin sheath thickness. Science. 2004;304:700–703. doi: 10.1126/science.1095862. [DOI] [PubMed] [Google Scholar]
- 3.Nave KA, Salzer JL. Axonal regulation of myelination by neuregulin 1. Curr Opin Neurobiol. 2006;16:492–500. doi: 10.1016/j.conb.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Taveggia C, et al. Neuregulin-1 type III determines the ensheathment fate of axons. Neuron. 2005;47:681–694. doi: 10.1016/j.neuron.2005.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan R, Hardy WR, Dankort D, Laing MA, Muller WJ. Modulation of Erbb2 signaling during development: a threshold level of Erbb2 signaling is required for development. Development. 2004;131:5551–5560. doi: 10.1242/dev.01425. [DOI] [PubMed] [Google Scholar]
- 6.Garratt AN, Voiculescu O, Topilko P, Charnay P, Birchmeier C. A dual role of erbB2 in myelination and in expansion of the Schwann cell precursor pool. J Cell Biol. 2000;148:1035–1046. doi: 10.1083/jcb.148.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen S, et al. Neuregulin 1-erbB signaling is necessary for normal myelination and sensory function. J Neurosci. 2006;26:3079–3086. doi: 10.1523/JNEUROSCI.3785-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaegle M, et al. The POU proteins Brn-2 and Oct-6 share important functions in Schwann cell development. Genes Dev. 2003;17:1380–1391. doi: 10.1101/gad.258203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaegle M, et al. The POU factor Oct-6 and Schwann cell differentiation. Science. 1996;273:507–510. doi: 10.1126/science.273.5274.507. [DOI] [PubMed] [Google Scholar]
- 10.Topilko P, et al. Krox-20 controls myelination in the peripheral nervous system. Nature. 1994;371:796–799. doi: 10.1038/371796a0. [DOI] [PubMed] [Google Scholar]
- 11.Britsch S, et al. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15:66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedrich RP, Schlierf B, Tamm ER, Bosl MR, Wegner M. The class III POU domain protein Brn-1 can fully replace the related Oct-6 during schwann cell development and myelination. Mol Cell Biol. 2005;25:1821–1829. doi: 10.1128/MCB.25.5.1821-1829.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryu EJ, et al. Misexpression of Pou3f1 results in peripheral nerve hypomyelination and axonal loss. J Neurosci. 2007;27:11552–11559. doi: 10.1523/JNEUROSCI.5497-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le N, et al. Analysis of congenital hypomyelinating Egr2Lo/Lo nerves identifies Sox2 as an inhibitor of Schwann cell differentiation and myelination. Proc Natl Acad Sci USA. 2005;102:2596–2601. doi: 10.1073/pnas.0407836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warner LE, et al. Mutations in the early growth response 2 (EGR2) gene are associated with hereditary myelinopathies. Nat Genet. 1998;18:382–384. doi: 10.1038/ng0498-382. [DOI] [PubMed] [Google Scholar]
- 16.Svaren J, Meijer D. The molecular machinery of myelin gene transcription in Schwann cells. Glia. 2008;56:1541–1551. doi: 10.1002/glia.20767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le N, et al. Nab proteins are essential for peripheral nervous system myelination. Nat Neurosci. 2005;8:932–940. doi: 10.1038/nn1490. [DOI] [PubMed] [Google Scholar]
- 18.Murphy P, et al. The regulation of Krox-20 expression reveals important steps in the control of peripheral glial cell development. Development. 1996;122:2847–2857. doi: 10.1242/dev.122.9.2847. [DOI] [PubMed] [Google Scholar]
- 19.Kao SC, et al. Calcineurin/NFAT signaling is required for neuregulin-regulated Schwann cell differentiation. Science. 2009;323:651–654. doi: 10.1126/science.1166562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He Y, et al. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007;55:217–230. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zorick TS, Syroid DE, Arroyo E, Scherer SS, Lemke G. The transcription factors SCIP and Krox-20 mark distinct stages and cell fates in Schwann cell differentiation. Mol Cell Neurosci. 1996;8:129–145. doi: 10.1006/mcne.1996.0052. [DOI] [PubMed] [Google Scholar]
- 22.Päiväläinen S, et al. Myelination in mouse dorsal root ganglion/Schwann cell cocultures. Mol Cell Neurosci. 2008;37:568–578. doi: 10.1016/j.mcn.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 23.Doerflinger NH, Macklin WB, Popko B. Inducible site-specific recombination in myelinating cells. Genesis. 2003;35:63–72. doi: 10.1002/gene.10154. [DOI] [PubMed] [Google Scholar]
- 24.Zorick TS, Syroid DE, Brown A, Gridley T, Lemke G. Krox-20 controls SCIP expression, cell cycle exit and susceptibility to apoptosis in developing myelinating Schwann cells. Development. 1999;126:1397–1406. doi: 10.1242/dev.126.7.1397. [DOI] [PubMed] [Google Scholar]
- 25.Parkinson DB, et al. Krox-20 inhibits Jun-NH2-terminal kinase/c-Jun to control Schwann cell proliferation and death. J Cell Biol. 2004;164:385–394. doi: 10.1083/jcb.200307132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parkinson DB, et al. Transforming growth factor beta (TGFbeta) mediates Schwann cell death in vitro and in vivo: examination of c-Jun activation, interactions with survival signals, and the relationship of TGFbeta-mediated death to Schwann cell differentiation. J Neurosci. 2001;21:8572–8585. doi: 10.1523/JNEUROSCI.21-21-08572.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mager GM, et al. Active gene repression by the Egr2. NAB complex during peripheral nerve myelination. J Biol Chem. 2008;283:18187–18197. doi: 10.1074/jbc.M803330200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart HJ, et al. Helix-loop-helix proteins in Schwann cells: a study of regulation and subcellular localization of Ids, REB and E12/47 during embryonic and postnatal development. J Neurosci Res. 1997;50:684–701. doi: 10.1002/(SICI)1097-4547(19971201)50:5<684::AID-JNR6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 29.Woodhoo A, et al. Notch controls embryonic Schwann cell differentiation, postnatal myelination and adult plasticity. Nat Neurosci. 2009;12:839–847. doi: 10.1038/nn.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beausoleil SA, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci USA. 2006;103:5391–5396. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou FF, Xue Y, Chen GL, Yao X. GPS: a novel group-based phosphorylation predicting and scoring method. Biochem Biophys Res Commun. 2004;325:1443–1448. doi: 10.1016/j.bbrc.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Maurel P, Salzer JL. Axonal regulation of Schwann cell proliferation and survival and the initial events of myelination requires PI 3-kinase activity. J Neurosci. 2000;20:4635–4645. doi: 10.1523/JNEUROSCI.20-12-04635.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrisingh MC, et al. The Ras/Raf/ERK signaling pathway drives Schwann cell dedifferentiation. EMBO J. 2004;23:3061–3071. doi: 10.1038/sj.emboj.7600309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meintanis S, Thomaidou D, Jessen KR, Mirsky R, Matsas R. The neuron-glia signal beta-neuregulin promotes Schwann cell motility via the MAPK pathway. Glia. 2001;34:39–51. [PubMed] [Google Scholar]
- 36.Kim HA, DeClue JE, Ratner N. cAMP-dependent protein kinase A is required for Schwann cell growth: interactions between the cAMP and neuregulin/tyrosine kinase pathways. J Neurosci Res. 1997;49:236–247. [PubMed] [Google Scholar]
- 37.Fragoso G, et al. Inhibition of p38 mitogen-activated protein kinase interferes with cell shape changes and gene expression associated with Schwann cell myelination. Exp Neurol. 2003;183:34–46. doi: 10.1016/s0014-4886(03)00101-8. [DOI] [PubMed] [Google Scholar]
- 38.Zanazzi G, et al. Glial growth factor/neuregulin inhibits Schwann cell myelination and induces demyelination. J Cell Biol. 2001;152:1289–1299. doi: 10.1083/jcb.152.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haines JD, Fragoso G, Hossain S, Mushynski WE, Almazan G. p38 Mitogen-activated protein kinase regulates myelination. J Mol Neurosci. 2008;35:23–33. doi: 10.1007/s12031-007-9011-0. [DOI] [PubMed] [Google Scholar]
- 40.He Y, Sandoval J, Casaccia-Bonnefil P. Events at the transition between cell cycle exit and oligodendrocyte progenitor differentiation: the role of HDAC and YY1. Neuron Glia Biol. 2007;3:221–231. doi: 10.1017/S1740925X08000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Becker KG, Jedlicka P, Templeton NS, Liotta L, Ozato K. Characterization of hUCRBP (YY1, NF-E1, delta): a transcription factor that binds the regulatory regions of many viral and cellular genes. Gene. 1994;150:259–266. doi: 10.1016/0378-1119(94)90435-9. [DOI] [PubMed] [Google Scholar]
- 42.Kwon HJ, Chung HM. Yin Yang 1, a vertebrate polycomb group gene, regulates antero-posterior neural patterning. Biochem Biophys Res Commun. 2003;306:1008–1013. doi: 10.1016/s0006-291x(03)01071-4. [DOI] [PubMed] [Google Scholar]
- 43.Morgan MJ, Woltering JM, In der Rieden PM, Durston AJ, Thiery JP. YY1 regulates the neural crest-associated slug gene in Xenopus laevis. J Biol Chem. 2004;279:46826–46834. doi: 10.1074/jbc.M406140200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


