Abstract
Data analysis sometimes requires the relaxation of parametric assumptions in order to gain modeling flexibility and robustness against mis-specification of the probability model. In the Bayesian context, this is accomplished by placing a prior distribution on a function space, such as the space of all probability distributions or the space of all regression functions. Unfortunately, posterior distributions ranging over function spaces are highly complex and hence sampling methods play a key role. This paper provides an introduction to a simple, yet comprehensive, set of programs for the implementation of some Bayesian non- and semi-parametric models in R, DPpackage. Currently DPpackage includes models for marginal and conditional density estimation, ROC curve analysis, interval-censored data, binary regression data, item response data, longitudinal and clustered data using generalized linear mixed models, and regression data using generalized additive models. The package also contains functions to compute pseudo-Bayes factors for model comparison, and for eliciting the precision parameter of the Dirichlet process prior. To maximize computational efficiency, the actual sampling for each model is carried out using compiled FORTRAN.
Keywords: Bayesian semiparametric analysis, Random probability measures, Random functions, Markov chain Monte Carlo, R
1. Introduction
In many practical situations, a parametric model cannot be expected to coherently describe the chance mechanism generating an observed dataset. Unrealistic features of some common models (e.g., the thin tails of the normal distribution when compared to the distribution of the observed data) can lead to unsatisfactory inferences. Constraining the analysis to a specific parametric form may limit the scope and type of inferences that can be drawn from such models. In these situations, we would like to relax parametric assumptions in order to gain modeling flexibility and robustness against mis-specification of a parametric statistical model. In the Bayesian context such flexible inference is typically achieved by placing a prior distribution on infinite-dimensional spaces, such as the space of all probability distributions for a random variable of interest. These models are usually referred to as Bayesian nonparametric (BNP) or semiparametric (BSP) models depending on whether all or at least one of the parameters is infinite dimensional (see, e.g. Dey, Müller, and Sinha, 1998; Walker, Damien, Laud, and Smith, 1999; Ghosh and Ramamoorthi, 2003; Müller and Quintana, 2004; Hanson, Branscum, and Johnson, 2005).
BNP is a relatively young research area in statistics. First advances were made in the sixties and seventies, and were primarily mathematical formulations. It was only in the early nineties with the advent of sampling based methods, in particular Markov Chain Monte Carlo (MCMC) methods, that substantial progress has been made. Posterior distributions ranging over function spaces are highly complex and hence sampling methods play a key role. The introduction of MCMC methods in the area began with the work of Escobar (1994) for Dirichlet process mixtures. A number of themes are still undergoing development, including issues in theory, methodology and applications. We refer to Walker et al. (1999), Müller and Quintana (2004) and Hanson et al. (2005) for recent overviews.
While BNP and BSP are extremely powerful and have a wide range of applicability, they are not as widely used as one might expect. One reason for this has been the gap between the type of software that many applied users would like to have for fitting models and the software that is currently available. The most general programs currently available for Bayesian inference are BUGS (see, e.g. Gilks, Thomas, and Spiegelhalter, 1994) and OpenBugs (Thomas, O'Hara, Ligges, and Sibylle, 2006). BUGS can be accessed from the publicly available R program (R Development Core Team, 2009), using the R2WinBUGS package (Strurtz, Ligges, and Gelman, 2005). OpenBugs can run on Windows and Linux, as well as from inside R. In addition, various R packages exist that directly fit particular Bayesian models. We refer to Appendix C in Carlin and Louis (2008), for an extensive list of software for Bayesian modeling. Although the number of fully Bayesian programs continues to burgeon, with many available at little or no cost, they generally do not include semiparametric models. An exception to this rule is the R package bayesm (Rossi, Allenby, and McCulloch, 2005; Rossi and McCulloch, 2008), including functions for some models based on Dirichlet process priors (Ferguson, 1973). The range of different Bayesian semiparametric models is huge. It is practically impossible to build flexible and efficient software for the generality of such models.
In this paper we present an up to date introduction to a publicly available R (R Development Core Team, 2009) package designed to help bridging the previously mentioned gap, the DPpackage, originally presented in Jara (2007). Although the name of the package is due to the most widely used prior on the space of the probability distributions, the Dirichlet Process (DP) (Ferguson, 1973), the package includes many other priors on function spaces. Currently, DPpackage includes models considering DP (Ferguson, 1973), mixtures of DP (MDP) (Antoniak, 1974), DP mixtures (DPM) (Lo, 1984; Escobar and West, 1995), linear dependent DP (LDDP) (De Iorio, Müller, Rosner, and MacEachern, 2004; De Iorio, Johnson, Müller, and Rosner, 2009), weight dependent DP (WDDP) (Müller, Erkanli, and West, 1996), hierarchical mixture of DPM of normals (HDPM) (Müller, Quintana, and Rosner, 2004), centrally standardized DP (CSDP) (Newton, Czado, and Chapell, 1996), Polya Trees (PT) (Ferguson, 1974; Mauldin, Sudderth, and Williams, 1992; Lavine, 1992, 1994), mixtures of Polya trees (MPT) (Lavine, 1992, 1994; Hanson and Johnson, 2002; Hanson, 2006; Christensen, Hanson, and Jara, 2008), mixtures of triangular distributions (Perron and Mengersen, 2001), and random Bernstein polynomials (Petrone, 1999a,b; Petrone and Wasserman, 2002). The package also includes models considering Penalized B-Splines (Lang and Brezger, 2004).
The article is organized as follows. Section 2 reviews the general syntax and design philosophy. Although the material in this section was presented in Jara (2007), its inclusion here is necessary in order to make the paper self-contained. In Section 3 the available functions are described in detail. In Section 4 the main features and usages of DPpackage are illustrated by means of simulated and real life data analyses. We conclude with additional comments and discussion in Section 5.
2. Design philosophy and general syntax
The design philosophy behind DPpackage is quite different from the one of a general purpose language. The most important design goal has been the implementation of model-specific MCMC algorithms. A direct benefit of this approach is that the sampling algorithms can be made dramatically more efficient than in a generic environment.
Fitting a model in DPpackage begins with a call to an R function, for instance, DPmodel, or PTmodel. Here “model” denotes a descriptive name for the model being fitted. Typically, the model function will take a number of arguments that control the specific MCMC sampling strategy adopted. In addition, the model(s) formula(s), data, and prior parameters are passed to the model function as arguments. The common elements in any model function are:
prior: an object list which includes the values of the prior hyper-parameters.
mcmc: an object list which must include the integers nburn giving the number of burn-in scans, nskip giving the thinning interval, nsave giving the total number of scans to be saved, and ndisplay giving the number of saved scans to be displayed on screen: the function reports on the screen when every ndisplay iterations have been carried out and returns the process's runtime in seconds. For some specific models, one or more tuning parameters for Metropolis steps may be needed and must be included in this list. The names of these tuning parameters are explained in each specific model description in the associated help files.
state: an object list giving the current value of the parameters, when the analysis is the continuation of a previous analysis, or giving the starting values for a new Markov chain, which is useful to run multiple chains starting from different points.
status: a logical variable indicating whether it is a new run (TRUE) or the continuation of a previous analysis (FALSE). In the latter case the current value of the parameters must be specified in the object state.
Inside the R model function the inputs are organized in a more useable form, the MCMC sampling is performed by calling a shared library written in a compiled language, and the posterior sample is summarized, labeled, assigned into an output list, and returned. The output list includes:
state: a list of objects containing the current value of the parameters.
save.state: a list of objects containing the MCMC samples for the parameters. This list contains two matrices randsave and thetasave which contain the MCMC samples of the variables with random distribution (errors, random effects, etc.) and the parametric part of the model, respectively.
In order to exemplify the extraction of the output elements, consider the abstract model fit:
fit <– DPmodel (…, prior, mcmc,
state, status, ….)
The lists can be extracted using the following code:
fit$state fit$save.state$randsave fit$save.state$thetasave
Based on these output objects, it is possible to use, for instance, the boa (Smith, 2007) or the coda (Plummer, Best, Cowles, and Vines, 2006) R packages to perform convergence diagnostics. For illustration, we consider the coda package here. It requires a matrix of posterior draws for relevant parameters to be saved as a mcmc object. Assume that we have obtained fit1, fit2, and fit3, by independently running a model function three times, specifying different starting values each. To compute the Gelman-Rubin convergence diagnostic statistic for the first parameter stored in the thetasave object, the following commands may be used:
library(coda) coda.obj <– mcmc.list( chain1=mcmc(fit1$save.state$thetasave[,1]), chain2=mcmc(fit2$save.state$thetasave[,1]), chain3=mcmc(fit3$save.state$thetasave[,1])) gelman.diag(coda.obj, transform = TRUE)
Note that the second command line saves the results as a mcmc.list object of class and the third command line computes the Gelman-Rubin statistic from these three chains.
Generic R functions such as print, plot, summary, and anova have methods to display the results of the DPpackage model fit. The function print displays the posterior means of the parameters in the model, and summary displays posterior summary statistics (mean, median, standard deviation, naive standard errors, and credibility intervals). By default, the function summary computes the 95% HPD intervals using the Monte Carlo method proposed by Chen and Shao (1999). The user can display the order statistic estimator of the 95% credible interval by using the following code,
summary(fit, hpd=FALSE)
The plot function displays the trace plots and a kernel-based estimate of the posterior distribution for the parameters of the model. Similarly to summary, the plot function displays the 95% HPD regions in the density plot and the posterior mean. The same plot but considering the the 95% credible region can be obtained by using,
plot(fit, hpd=FALSE)
The anova function computes simultaneous credible regions for a vector of parameters from the MCMC sample using the method described by Besag, Green, Higdon, and Mengersen (1995). The output of the anova function is an anova-like table containing the pseudo-contour probabilities for each of the factors included in the linear part of the model.
3. Implemented Models
In this section we describe in detail the functions available in version 1.0-8 of DPpackage.
3.1. Marginal density estimation
DPdensity, PTdensity, TDPdensity, and BDPdensity functions implement models for marginal density estimation using DPM of normals, MPT, triangular-Dirichlet, and a Bernstein-Dirichlet prior, respectively. The first two functions allow the user to fit uni- and multi-variate models. We next introduce the notation used for each model along with the associated computational approaches used to fit the models.
Dirichlet Process Mixtures of Normals
The DPdensity function considers the multivariate extension of the univariate DPM of normals model presented in Escobar and West (1995). Let yi be a k-dimensional vector of measurements for the ith subject, i = 1,…, n. The model assumes
and
where, the baseline distribution, Go, corresponds to the conjugate normal-inverted-Wishart distribution
To complete the model specification, the following independent hyper-priors are assumed,
and
Note that the inverted-Wishart prior, W | ν, Ψ ~ IWk (ν, Ψ), is parameterized such that .
The computation implementation is based on the marginalized version of the model where the random probability measure G is integrated out. Although the baseline distribution, G0, is a conjugate prior in this model specification, the algorithms with auxiliary parameters described in MacEachern and Müller (1998) and Neal (2000) are adopted. Specifically, the no-gaps algorithm of MacEachern and Müller (1998) and algorithm 8 of Neal (2000), with m = 1, are considered. The default method is algorithm 8 of Neal (2000).
Mixtures of Polya trees
The current implementation of the PTdensity function considers a MPT model as in Hanson (2006). As in the previous section, let yi be a k-dimensional vector of measurements for the ith subject, i = 1,…, n. The model assumes
and
where M is the maximum level of the partition to be updated (the default value is M = ∞), Πμ,Σ = {πj}j≥0 is a set of partitions of ℝk, indexed by μ and Σ, and Aα is a family of non-negative vectors controlling the variability of the process indexed by α. Following Hanson (2006), the PT is centered around the Nk (μ, Σ) distribution by taking
with γα(j, r) = αj21k, and further taking each level j of the sequence of partitions in Πμ,Σ, as the sets arising from a location-scale transformation μ + Σ1/2z of the Cartesian products of intervals obtained as quantiles from the standard univariate normal distribution, where Σ1/2 is the Cholesky decomposition of Σ. Notice that we consider a different parameterization than the one considered by Hanson (2006), were Σ1/2 is taken to be the unique symmetric square root of Σ. The base sets for level j are given by
for vectors p = (p1,…,pk) with pi ∈ {1,…, 2j}, i = 1,…, k. The location-scale transformation applied to each base set yields the final sets B(j, p) = {μ + Σ1/2z : z ∈ B0(j, p}, such that πj = {B(j,p) : p ∈ {1,…, 2j}k}.
The model specification is completed by assuming the following hyper-priors
and
As noticed by Jara, Hanson, and Lesaffre (2009), the PT prior specification is dependent on the square root of the centering covariance matrix considered to define the partitions sets. Indeed, in the Nk (μ, Σ)-centered multivariate extension considered by Hanson (2006), the direction of the sets are completely defined through the decomposition of the covariance matrix by the unique symmetric square root. In the context of multivariate random effects distributions, Jara et al. (2009) proposed a novel mixture of PT priors where the effect of the partitions is smoothed over by mixing over the decomposition of the centering covariance matrix (see, Section 3.2). This option will be considered in future version of the package.
For univariate analyses using a finite (M < ∞) PT, a full version of the model is considered where the Dirichlet vectors are updated during the MCMC scheme. For univariate analysis with a fully specified PT (M = ∞) and for multivariate analyses, a marginalized version of the model is considered, where the random probability measure G is integrated out. The baseline parameters μ and Σ, and the precision parameter α are updated using Metropolis-Hastings (MH) steps (Tierney, 1994).
Bernstein-Dirichlet prior
The function BDPdensity consider density estimation using a Bernstein-Dirichlet prior (BDP) proposed by Petrone (1999a,b). For a continuous cdf G on (0,1], the associated Bernstein polynomial (BP) is defined as
which is a mixture of beta distributions. Its density is given by
where β(x | j,k − j +1) stands for a beta density with parameters j and k − j + 1. Petrone (1999a,b) proposed a hierarchical prior, called the Bernstein polynomial prior (BPP), where the random density f(·) is given by the following mixture of beta densities,
| (1) |
where wj,k = G(j/k) − G((j − 1)/k), k as probability mass function ρ(·), and given k, wk = (w1,k,…,wk,k) has distribution Hk(·) on the k-dimensional simplex
Petrone (1999a,b) called expression (1) the Bernstein polynomial density with parameters k and wk, and shows that to assume wk = (w1,k, … ,wk,k) ~ Dirichlet(ζ1,k,… ,ζk,k), with ζj,k = α(G0(j/k) − G0((j − 1)/k)), j = 1,…, k, G0 a probability distribution on (0,1] and α a positive constant, is equivalent to assume that G | α, G0 ~ DP(αG0). Petrone (1999a,b) refers to this as the Bernstein-Dirichlet prior (BDP) and discussed MCMC algorithms to scan the posterior distribution.
Our MCMC implementation is similar to the one described by Petrone (1999a,b) but adds the resampling step described by Bush and MacEachern (1996) for Dirichlet process mixture models. The function BDPdensity considers
and
where yi is the data transformed to lie in (0,1] and G0 = Beta(a0, b0). It is further assumed that
and
where DU(A) refers to the discrete uniform distribution on the set A. Although BDP are naturally defined as probability models for distributions on the unit interval (0,1], different measurable mappings could be considered to transform the data when the support is not the unit interval. With this aim we consider the uniform CDF on the range of the data.
Mixtures of triangular distributions
The TDPdensity function considers a triangular-Dirichlet prior (TDP) for univariate density estimation. The logic behind the TDP is similar to the BDP construction but replaces the beta kernels in the mixture model by triangular distributions as proposed by Perron and Mengersen (2001). The model is given by
and
and
where yi is the data transformed to lie in (0,1], kmax is the upper limit of the discrete uniform prior or the number of components in the mixture of Triangular distributions, α is the total mass parameter of the Dirichlet process component, and G0 is the centering distribution of the DP. The centering distribution corresponds to a G0 = Beta(a0, b0) distribution.
Our representation is equivalent to the mixture of triangular distributions proposed by Perron and Mengersen (2001), with random weights following a Dirichlet prior. However, in this function we exploit the underlying DP structure, thus avoiding the use of Reversible-Jump algorithms (Green, 1995). In fact, the same MCMC algorithm considered for the BDP prior is implemented in the TDPdensity function.
3.2. Nonparametric random effects distributions in mixed effects models
Assume that for each of m experimental units the regression data (Yij, xij, zij), 1 ≤ i ≤ m, 1 ≤ j ≤ ni, is recorded, where Yij is a response variable, and xij ∈ ℝp and zij ∈ ℝq are vectors of p and q explanatory variables, respectively. Let Y i = (Yn1, …, Yini)T, Xi = (xi1, …, xini)T, and Zi = (zi1, …, zini)T, i = 1, …, m. The observations are assumed to be conditionally independent with exponential family distribution,
The means μij = E (Yij | ϑij, τ) and variances are related to the canonical ϑij and dispersion parameter τ via μij = b′ (ϑij) and , respectively. The means μij are related to the p-dimensional and q-dimensional “fixed” effects vectors βF and βR, respectively, and the q-dimensional “random” effects vector bi via the link relation
| (2) |
where, h(·) is a known monotonic differentiable link function, and ηij is called the linear predictor. Due to software limitations, the analyses are often restricted to the setting in which the random effects follow a multivariate normal distribution, . In this context, Bayesian nonparametric extensions incorporate a probability model for the random effects distribution in order to better represent the distributional uncertainty and to avoid the effects of the miss-specification of an arbitrary parametric random effects distribution. Bush and MacEachern (1996) and Kleinman and Ibrahim (1998b) describe Bayesian semiparametric versions of the linear mixed model considering DP prior for the random effects distribution. Under this approach the DP prior is centered at a normal base mesure with zero mean. Similar approaches were considered by Mukhopadhyay and Gelfand (1997) and Kleinman and Ibrahim (1998a) in the context of GLMM. In order to avoid the discrete nature of the DP realizations, Müller and Rosner (1997) consider a DPM of normals model in the context of a normal nonlinear mixed model. Alternatively, Walker and Mallick (1997) and Hanson (2006) consider PT and mixtures of PT priors in random intercept models. Jara et al. (2009) propose a novel mixture of multivariate PT priors to define flexible nonparametric models for multivariate distributions that reduces the undesirable sensitivity to the choice of the partitions associated with the PT constructions. Under these approaches, the parametric assumption is relaxed by considering
and
where H is one of the previously mentioned probability models for probability distributions. We will specify the nonparametric priors in more detail next, but first it is necessary to discuss some important issues regarding the specification of the semiparametric model. Specifically, it is important to stress that under parametrization (2), βR represents the mean of random effects, and bi represents the subject-specific deviation from the mean. It follows that fixing the mean of the normal prior distribution for the random effects b at zero in the parametric context corresponds to an identification restriction for the model parameters (see e.g., Newton, 1994; San Martín, Jara, Rolin, and Mouchart, 2007). Equivalently, the random probability measure must be appropriately restricted in a semiparametric GLMM specification. In our settings, the location of G is “confounded” with the parameters βR. Although such identification issues present no difficulties to a Bayesian analysis in the sense that a prior is transformed into a posterior using the sampling model and the probability calculus, if the interest focuses on a “confounded” parameter then such formal assurances have little practical value. Furthermore, as more data become available, the posterior mass will not concentrate on a point in the model, making asymptotic analysis difficult. As pointed out by Newton (1994), from a computational point of view, identification problems imply ridges in the posterior distribution and MCMC methods can be difficult to implement in these situations.
Following Jara et al. (2009), we consider the following re-parameterization of the model
and
where β = βF, and θi = βR + bi, and we center the nonparametric priors for G at a Nq (μ, Σ) distribution. Notice that samples under the original parameterization can be obtained in a straightforward manner from MCMC samples as explained in Jara et al. (2009) for PT priors. For DP or DPM priors the ∊–DP approximation proposed by Muliere and Tardella (1998) is considered, with ∊ = 0.01. The latter is similar to the approach proposed by Gelfand and Kottas (2002) who considered a fixed truncation to the DP. When a DP or DPM prior is used to model the random effects distribution, Dunson, Yang, and Baird (2007a) and Li, Müller, and Lin (2007) proposed alternative strategies to avoid the identifiability problem described above but these approaches are not implemented in the current version of DPpackage.
The functions DPlmm, DPglmm, and DPolmm implement mixed effects models using a DP prior for G such that
The functions DPMlmm, DPMglmm, and DPolmm consider a DPM of normals prior for G such that
and
The functions PTlmm, PTglmm, and PTolmm consider a multivariate PT prior for G such that
where O is a q × q orthogonal matrix defining the “direction” of the partition sets. The models are completed by assuming the following prior distributions:
and
where Γ and IW refers to the Gamma and inverted Wishart distributions, respectively. As before, the inverted Wishart prior is parameterized such that E(Σ) = T−1/(ν0 − q − 1).
The DPlmm, DPMlmm and PTlmm functions consider the normal sampling distribution with an identity link. The DPglmm, DPMglmm, and PTglmm functions include the following sampling distributions (link): binomial (logit and probit), Poisson (log) and gamma (log). The DPolmm, DPMolmm and PTolmm consider a multinomial sampling distribution and an ordered-probit link function.
In all functions, a marginalized version of the semiparametric GLMM is considered where the random probability distribution G is integrated out. For the multinomial and probit-binomial models, the latent variable approach of Albert and Chib (1993) is considered.
The computational implementation associated to the functions DPMlmm and DPMolmm, and to the probit-Bernoulli model included in the DPMglmm function, is based on the use of MCMC methods for conjugate priors for a collapsed state of MacEachern (1998). For the poisson, Gamma, and logit-binomial models included in the DPglmm and DPMglmm functions, MCMC methods for non-conjugate priors are used. Specifically, algorithm 8 of Neal (2000), with m = 1, is considered. In this case, a MH step with the iterative weighted least square (IWLS) normal proposal of Gamerman (1997) is used to update fixed and random effects.
For the functions DPlmm and DPolmm, and the probit-Bernoulli model included in DPglmm, the MCMC strategy described by Bush and MacEachern (1996) is employed. Finally, for the PTlmm, PTgmm and PTomm the modified IWLS proposal normal proposal described by Jara et al. (2009) is considered for sampling the random effects. In these functions, IWLS normal proposal of Gamerman (1997) is used to update the fixed effects in the nonconjugate case. The PT centering and precision parameters are updated using adaptive MCMC algorithms as described by Jara et al. (2009).
3.3. Semiparametric IRT-type models
Item response theory (IRT) models are widely used in educational measurement (see e.g., De Boeck and Wilson, 2004). Rasch-type models (Rasch, 1960) are typical examples of this class and can be viewed as a particular case of GLMM (see e.g., De Boeck and Wilson, 2004). In Rasch-type models, the linear predictor ηij depends on two parameters in an additive way ηij = θi − βj, where θi ∈ ℝ corresponds to the ability of subject i, i = 1, …, m, and βj ∈ ℝ corresponds to the difficulty of probe/item j, j = 1, …, p. The difficulty and ability parameters are interpreted as “fixed” and “random” effects, respectively. Two versions of the model are considered here: the Rasch model (RM) and the Rasch Poisson count model (RPCM). In the RM, Yij represents a binary variable coding the correct answer of individual i to the item j, such that
where Ψ(x) = exp(x)/(1 + exp(x)). In the RPCM the sampling distribution is given by
where Yij is an “unbounded” count variable, typically representing the number of miss-reading / miss-copying for the subject i in the text j. We consider semiparametric versions of the models where the abilities distribution G is modeled using DP, PT and DPM priors. To avoid identification problems in the semiparametric specification of the model (see, San Martín et al., 2007), we fixed the first difficulty parameter at 0 and consider a normal prior for the remaining elements in the vector
The functions DPrasch and DPraschpoisson implement semiparametric versions of the RM and RPCM, respectively, where
and
In a similar way, the functions FPTrasch and FPTraschpoisson implement semiparametric versions of the RM and RPCM, respectively, using a finite PT prior,
where, the PT is centered around a N(μ, σ2) distribution, by taking each m level of the partition Πμ, σ2 to coincide with the k/2m, k = 0, …, 2m quantiles of the N(μ, σ2) distribution. The family 𝒜α = {α∊ : ∊ ∈ E*}, where and Em is the m-fold product of E = {0,1}, was specified as α∊1 … ∊m = αm2. For the DP and PT priors, the model is completed by assuming
and
The functions DPMrasch and DPMraschpoisson consider DPM of normals priors for the abilities distribution in a RM and RPCM, respectively, given by
and
where . We further assume that
and
In all functions, the difficulty and ability parameters are updated using a MH step with the IWLS normal proposal of Gamerman (1997). The computational implementation in the DPrasch and DPraschpoisson functions is based on the marginalization of the DP and on the use of algorithm 8 of Neal (2000), with m = 1. The DPM implementations of functions DPMrasch and DPMraschpoisson are based on the finite approximation for DP proposed by Ishwaran and James (2002). Finally, the functions using finite PT priors for the abilities distribution, FPTrasch and FPTraschpoisson, fit a full version of the models where the PT conditional probabilities are updated during the MCMC scheme. In this case, the abilities, centering and precision parameters are updated using slice sampling (Neal, 2003).
3.4. Semiparametric meta-analysis models
The DPmeta, DPMmeta and PTmeta functions implement random (mixed) effects univariate metaanalysis models using a MDP, DPM of normals, and MPT prior for the random effects, respectively. In this case, the conditional model is given by
where the variances are known, Xi is a p-dimensional design vector, excluding an intercept term, and
The DPmeta function assumes that
and
The PTmeta function, replaces the latter assumption by a PT prior,
where the PT prior is centered around a N(μ, σ2) distribution. The PTmeta function can also center the PT prior around a N(0, σ2) distribution for the median-0 model described by Branscum and Hanson (2008). This model is fitted if the option frstlprob is set equal to TRUE in the model prior object. In this case, the design vector xi includes an intercept term and the associated regression coefficient represents the median effect. The computational implementation of the DPmeta and PTmeta functions are based on the marginalization of the DP and PT, respectively. In both cases, the model specification is completed by assuming
and
The the average effect in the DPmeta function is sampled using the method of composition and the ∊–DP approximation proposed by Muliere and Tardella (1998), with ∊ = 0.01. For the PTmeta function, the mean effect is sampled using the finite PT approximation described by Jara et al. (2009).
The DPMmeta function considers a location DPM of normals priors for the study effects
and
where . This function further assumes that
and
The computational implementation of the model is based on the marginalization of the DP and on the use of MCMC methods for conjugate priors for a collapsed state, as presented in MacEachern (1998). The average effect is also sampled using the method of composition and the ∊–DP approximation proposed by Muliere and Tardella (1998), with ∊ = 0.01.
The function DPmultmeta implements a multivariate extension of the no-covariate model considered in the DPmeta function, given by
and
where the covariance matrices Σi are known. To complete the model specification, independent hyperpriors are assumed,
and
The computational implementation is similar to the one employed for the DPmeta function.
3.5. Accelerated failure time modeling for interval-censored data
The DPsurvint function implements the algorithm described by Hanson and Johnson (2004) for semiparametric accelerated failure time (AFT) models. The AFT regression model is given by
and
where LN (v| μ, σ2) refers to a log-normal distribution with location and scale parameter μ and σ2, respectively. The model is completed by assuming independent hyperpriors,
and
The likelihood in the AFT model for interval censored data involves the product of indicator functions , where Ai is an interval in the sample space. This fact gives rise to algorithmic possibilities which are unavailable or very difficult to implement under standard hierarchical models with uncensored data. As described in Hanson and Johnson (2004), the DPsurvint function partially sample G, in order to sample (V1, …, Vn, Vn+1, β, α) with perfect accuracy. This can be performed by using the properties of DP. Specifically, the following representation of the process is considered
where j indexes the intervals that define a finite partition of the sample space {B1, …, Bm}, Gj = G(Bj), and Gj(·) = G(· | Bj), with the Gj's being Dirichlet distributed random variables and the Gj's being independent Dirichlet processes. Therefore, G can be updated by first updating {Gj} using Ferguson's definition of DP and then by updating each Gj | {Gj}, … using the Sethuraman (1994) stick-breaking representation of DP (see, e.g. Doss, 1994; Hanson and Johnson, 2004). Based on this, a MH step is used to update the regression coefficients, followed by updates of V1, …, Vn+1.
The function predict.DPsurvint can be used to extract posterior information about the survival curve based on the MCMC output. Given a sample of the parameters of size J, a sample of the survival curve for a given x is drawn as follows. For the jth MCMC scan of the posterior distribution, j = 1, …, J, the survival function evaluated at t is sampled from where
where
and b(j) (t) = α(j) + n − a(j) (t).
3.6. Binary regression with nonparametric link
Consider binary regression data, (Yi, xi), 1 ≤ i ≤ n, where Yi is a binary response variable (Yi ∈ {0,1}) and xi ∈ ℝp is a vector of p explanatory variables. Parametric versions of this model are characterized by the following assumption
where Fφ is a distribution function on ℝ, called the inverse link function in the context of generalized linear models, known up to a Euclidean parameter φ, and m(·) is a known function, called the index function, parameterized by β. Popular parametric versions include a linear index function, , and where Fφ is considered to be a known cumulative distribution function, i.e. with φ = φ0, thus allowing relatively simple treatment of the finite regression parameters, θ = β. The function Pbinary implements parametric versions of this model considering the logit, probit, cloglog, and Cauchy link functions.
The DPbinary, FPTbinary, and CSDPbinary functions replace the parametric inverse link function Fφ by a general distribution G and placing a DP prior,
a finite PT where the first and second quartiles are fixed (Hanson, 2006),
and a CSDP (Newton et al., 1996),
on G, respectively. Newton et al. (1996) described the CSDP as a prior distribution on the space of the probability distribution with fixed location and scale in order to assure sampling identification. The reasoning behind their construction is presented here for completeness. The following definition is a slight modification of the one given by Newton et al. (1996). Let G0 and H be two probability measures on ℝ and (0, d), respectively, such that for all d > 0, G0 ((−∞, −d)) > 0 and G0 ((d, ∞)) > 0. Let θ ~ h, where h is the density of H with respect to Lebesgue measure. Given θ, define the following partition of the real line, A1(θ) = (−∞, θ − d], A2(θ) = (θ − d, 0], A3(θ) = (0, θ], and A1(θ) = (θ, ∞). Finally, suppose that for each θ ∈ (0, d), the random probability measures φ1, φ2, φ3, and φ4 follow conditionally independent DP priors, , i = 1, …, 4. The random probability measure G on (ℝ, ℬ) is said to follow CSDP prior with parameter (α, G0, p, d, h), written G ~ CSDP(αG0, p, d, h), if,
In all cases, the functions allows for misclassified binary responses with known misclassification parameters and the model specification is completed by assuming
and
The DPbinary function allows the user to center the DP around a logistic, normal or Cauchy distribution. The CSDPbinary function takes H ≡ U(0, d) distribution and G0 as the standard logistic distribution. In both functions, a latent variable representation
and
is used, along with a MH step to update the regression coefficients. In the computational implementation of this model, G is considered as latent data and sampled partially with sufficient accuracy to be able to generate V1, …, Vn+1 such that are exactly iid random variables from G, as proposed by Doss (1994). Both Ferguson's definition of DP and the Sethuraman (1994)'s representation of the process are used. As in Bush and MacEachern (1996), an extra step which moves the clusters in such a way that the posterior distribution is still the stationary distribution, is performed in order to improve the mixing of the chain.
The FPTbinary function creates the partition sets based on the logistic distribution. In the computational implementation of the model, MH steps are used to update the regression coefficients and the precision parameter, as described in Hanson (2006).
3.7. ROC curve estimation
The DProc function performs a ROC curve analysis based on DPM of normals models for density estimation. Let x1, …, xn and y1, …, ym be the diagnostic marker measurements for the healthy and diseased subjects, respectively. The model is given by
and
where, the baseline distributions, Gz0, z = {x, y}, correspond to the conjugate normal-inverted-Wishart distribution
To complete the model specification, the model is extended by assuming independent hyper-priors,
The survival and ROC curves are estimated by using a Monte Carlo approximation to the posterior means E(Gx|x1, …, xn) and E(Gy|y1 …, ym), which is based on MCMC samples from posterior predictive distribution for a future observation. The optimal cut-off point is based on the efficiency of the test and is built on Cohen's kappa as defined in Kraemer (1992).
3.8. Median regression modeling
Consider regression data (yi, xi), i = 1, …, n, where yi is the response and xi is a p-dimensional vector of predictors. By default, the PTlm function fits a median regression model using a scale MPT prior for the distribution of the errors (Hanson and Johnson, 2002),
and
where, the PT is centered around a N(0, σ2) distribution, by taking each m level of the partition Πσ2 to coincide with the k/2m, k = 0, …, 2m quantiles of the N(0, σ2) distribution. The family 𝒜α = {α∊ : ∊ ∈ E*}, where and Em is the m-fold product of E = {0,1}, was specified as α∊1 … ∊m = αm2. To complete the model specification, independent hyperpriors are assumed,
Optionally, if frstlprob=FALSE (the default value is TRUE) is specified, a mean regression model is considered. In this case, the following PT prior is considered
where, the PT is centered around a N(μ, σ2) distribution. In this case, the intercept term is automatically excluded from the model and the hyperparameters for the normal prior for μ must be specified. The normal prior is given by
In the computational implementation of the model, random-walk Metropolis steps are used to update the regression coefficients and hyperparameters.
3.9. Models for related distributions
The current version of DPpackage considers models for related random probability distributions based on particular implementations of the dependent DP (DDP) proposed by MacEachern (1999, 2000), a natural generalization of the approach discussed by Müller et al. (1996) for nonparametric regression to the context of conditional density estimation, and the hierarchical mixture of DPM models (HDPM) proposed by Müller et al. (2004). These approaches and the associated functions are described next.
Linear dependent Dirichlet process
MacEachern (1999, 2000), proposes the DDP as an approach to define a prior model for an uncountable set of random measures indexed by a single continuous covariate, say x, {Gx : x ∈ χ ⊂ ℝ}. The key idea behind the DDP is to create an uncountable set of DPs (Ferguson, 1973) and to introduce dependence by modifying the Sethuraman (1994)'s stick-breaking representation of each element in the set. If G follows a DP prior with precision parameter α and base measure G0, denoted by G ~ DP(αG0), then the stick-breaking representation of G is
| (3) |
where B is a measurable set, δa(·) is the Dirac measure at and ωl = Vl Πj<l(1 − Vj), with . MacEachern (1999, 2000) generalizes (3) by assuming the point masses θ(x)l, l = 1, …, to be dependent across different levels of x, but independent across l.
De Iorio et al. (2004) and De Iorio et al. (2009) proposed a particular version of the DDP where the component of the atoms defining the location in a DDP mixture model follows a linear regression model , where x is a p-dimensional design vector. An advantage of this model for related random probability measures, referred to as the Linear DDP (LDDP), is that it can be represented as DPM of linear (in the coefficients) regression models. This approach is implemented in the LDDPdensity function, where for the regression data (yi, xi), i = 1, …, n, the following model is considered
and
where G0 ≡Np(β|μb, Sb) Γ (σ−2|τ1/2, τ2/2). The LDDP model specification is completed with the following hyper-priors
and
The LDDPsurvival function implements this model in the context of survival data. Now let yi the time to event for the ith subject. The LDDP mixture of survival models is given by
with the same hierarchical specification given above for the LDDPdensity function. Note that this function can deal with censored observations by using a data-augmented approach.
Finally, the LDDPrach and LDDPrachpoisson functions consider this modeling strategy in a Rasch and Rasch Poisson model context, respectively, as in Fariña, Quintana, San Martín, and Jara (2009). Here the linear predictor is given by ηij = θi − βj, where the abilities follow a LDDP mixture of normals model based on subject-specific covariates included in xi,
These functions fit a marginalized version of the models where the random probability measure G is integrated out. Full inference on the conditional density, and survival and hazard functions in the case of the LDDPsurvival function, at covariate level are obtained using the ∊–DP approximation proposed by Muliere and Tardella (1998), with ∊ = 0.01.
Weight dependent Dirichlet process
Let , where zi is a p-dimensional vector of continuous predictors. The LDDP of the previous section defines a mixture model where the weights are independent of the predictors z, given by
where the weights ωl follow a stick-breaking construction and . Motivated by regression problems with continuous predictors different extensions have been proposed by making the weights dependent on covariates (see, e.g. Griffin and Steel, 2006; Duan, Guindani, and Gelfand, 2007; Dunson, Pillai, and Park, 2007b; Dunson and Park, 2008), such that
| (4) |
An earlier approach that is related to the latter references and that also induces a weight-dependent DP model, as in expression (4), was discussed by Müller et al. (1996). These authors fitted a “standard” DPM of multivariate Gaussian distributions to the complete data di = (yi, zi)′, i = 1, …, n, and looked at the induced conditional distributions. Although Müller et al. (1996) focused on the mean function only, m(z) = E(y|z), their method can be easily extended to provide inferences for the conditional density at covariate level z, i.e. a “density regression” model in the spirit of Dunson et al. (2007b). The extension of the approach of Müller et al. (1996) for related probability measures is implemented in the DPcdensity function, where the model is given by
and
where k = p + 1 is the dimension of the vector of complete data di, the baseline distribution G0 is the conjugate normal-inverted-Wishart (IW) distribution . To complete the model specification, the following hyper-priors are assumed
and
This model induce a weight dependent mixture models, as in expression (4), where the components are given by
and
where the weights ωl follow a DP stick-breaking construction and the remaining elements arise from the standard partition of the vectors of means and (co)variance matrices given by
respectively.
The DPcdensity function fits a marginalized version of the model where the random probability measure G is integrated out. Full inference on the conditional density at covariate level z is obtained using the ∊–DP approximation proposed by Muliere and Tardella (1998), with ∊ = 0.01.
Hierarchical mixture of Dirichlet process mixture of normals
The HDPMdensity function considers the hierarchical mixture of DPM of normal models for density estimation presented in Müller et al. (2004). Let yij be the q-dimensional vector of responses for the jth observation, j = 1, …, ni, for the ith group, i = 1, …, I. The model assumes that
where Fi is assumed to arise as a mixture model Fi = ∊H0 + (1 − ∊)Hi of one common distribution H0 and a distribution Hi that is specific or idiosyncratic to the ith group. The random probability measures Hi, i = 0,1, …, I in turn are given a DPM of normal prior,
with
The model specification is completed by assuming the following hyper-priors,
and
where δc represents the Dirac measure at c, and β(a, b) represents the beta distribution with parameters a and b.
The HDPMcdensity function considers the extension of the previously described approach to the inclusion of continuos predictors z. This functions fits the HDPM model to the complete data di = (yi, zi)′, i = 1, …, n, and reports the induced conditional distributions.
3.10. Generalized additive models
The PSgam function fits a generalized additive model (see, e.g. Hastie and Tibshirani, 1990) using Penalized splines (see e.g., Eilers and Marx, 1996; Lang and Brezger, 2004). The linear predictors ηi, i = 1, …, n, are modeled in an additive way. Let xi be a p-dimensional design vector and zi be a q-dimensional vector of continuous predictors. Then, the model is given by
where the effect fj of the a covariate zj is approximated by a polynomial spline with equally spaced knots, written in terms of a linear combination of B-spline basis functions. Specifically, the function fj is aproximated by a spline of degree l with r equally spaced knots within the domain of zj,
where are B-spline basis function of degree l, and bjm represents the associated B-spline coefficients. For the parametric component of the model, a normal prior distribution is assumed,
For the vector of basis coefficients bj = (bj1, …, bj(l+r))T, independent Gaussian smoothness priors (Lang and Brezger, 2004) are assumed
The precision matrix acts as a penalty matrix to enforce smoothness and is defined through , where Dj is a first or second order difference matrix for adjacent B-spline coefficients. The variance (or inverse smoothing) parameter controls the amount of smoothness. Note that the log-penalty corresponds exactly to the penalty term introduced by Eilers and Marx (1996) in a frequentist penalized likelihood setting. For the variance parameters, we assume independent inverse gamma priors
Finally, for the gamma and Gaussian models, an inverse gamma prior is assumed for the dispersion parameter σ2,
The computational implementation of the model is model-specific. For the Poisson, gamma, and binomial (logit) models, fixed and random effects are updated using MH steps with a IWLS normal proposal (see, West, 1985; Gamerman, 1997). For the probit-Bernoulli model, the latent variable representation of the binary responses is used, leading to conjugate normal updates.
3.11. Additional tools
Additional functions included in the package are DPelicit and PsBF. The DPelicit function implements methods for eliciting the DP prior using exact and approximated formulas for the mean and variance of the number of clusters given the total mass parameter and the number of subjects (see, e.g. Jara, García-Zattera, and Lesaffre, 2007). The PsBF function computes pseudo-Bayes factors for model comparison.
The practical implementation of models based on DP priors with a random precision parameter requires adopting values for the hyperparameters a0 and b0. The discrete nature of the DP realizations leads to their well-known clustering properties. The choice of a0 and b0 needs some careful thoughts, as the parameter α directly controls the number of distinct components. Kottas, Müller, and Quintana (2005), referred to as the KMQ approach, and Jara et al. (2007), referred to as the JGL approach, proposed strategies for the specification of these hyperparameters.
The KMQ approach is based on approximations of the conditional mean and conditional variance of the number of clusters, given the precision parameter α (see e.g., Liu, 1996). Specifically, denoting by n the number of elements associated to the DP prior, and n* the number of resulting clusters, their approach relies on
| (5) |
| (6) |
Using the fact that a priori and , the resulting expressions for the prior mean and variance of n* are
| (7) |
and
| (8) |
On the other hand, the JGL approach is based on the exact value of conditional mean and conditional variance of the number of clusters given the precision parameter α. They noted that the approximations given by the expression (5) and expression (6) may be dangerous when α is considered a function of n. For instance, (5) gives 0 instead of 1 with . Better approximations may be obtained by noticing that
and
where ψ0(·) and ψi(·) represents the digamma and trigamma function, respectively. Using these results, an approximation based on a first-order Taylor series expansion, and the fact that a priori and we get
and
These expressions could be used in order to evaluate the robustness of the model to the specification of prior distribution for the precision parameter. The function DPelicit computes either the expected value and the standard deviation of the number of clusters, given the values of the parameters of the Gamma prior for the precision parameter, a0 and b0, or the value of the parameters a0 and b0 of the Gamma prior distribution for the precision parameter, α, given the prior judgement for the expected number and the standard deviation of the number of clusters. With this objective in mind, the Newton-Raphson algorithm and the forward-difference approximation to Jacobian are used.
4. Examples
In this section we consider the analyses of simulated and real-life data in order to illustrate the usage of DPpackage.
4.1. Bayesian density regression
We illustrate the DPcdensity and LDDPdensity functions by means of simulated data. We replicate the results reported by Dunson et al. (2007b), where a different approach is proposed. Following Dunson et al. (2007b), we simulate n = 500 observations from from a mixture of two normal linear regression models, with the mixture weights depending on the predictor, with different error variances and with a non-linear mean function for the second component,
where the predictor values xi are simulated from a uniform distribution, . The data was simulated using the following piece of code
################################################
# true conditional densities,
# mean function and
# simulation of the data.
################################################
dtrue <– function(grid,x)
{
exp(−2*x)*dnorm(grid,mean=x,sd=sqrt(0.01))+
(1–exp(−2 *x))*dnorm(grid,mean=x^4,sd=sqrt(0.04))
}
mtrue <– function(x)
{
exp(−2*x)*x+(1–exp(−2*x))*x^4
}
set.seed(0)
nrec <– 500
x <– runif(nrec)
y1 <– x + rnorm(nrec, 0, sqrt(0.01))
y2 <– x^4 + rnorm(nrec, 0, sqrt(0.04))
u <– runif(nrec)
prob <- exp(−2*x)
y <– ifelse(u<prob,y1,y2)
The extension of the DPM of normals approach of Müller et al. (1996) considered by the DPcdensity function, was fitted using the following hyper-parameters: a0 = 10, b0 = 1, ν1 = ν2 = 4, m2 = (ȳ,x̄)′, τ1 = 6.01, τ2 = 3.01, and , where S is the sample covariance matrix for the response and predictor. A total number of 25,000 scans of the Markov chain cycle implemented in the DPcdensity function were completed. A burn-in period of 5,000 samples was considered and the chain was subsampled every 4 iterates to get a final sample size of 5,000. The following commands were used to fit the model, where the conditional density estimates were evaluated on a grid of 100 points on the range of the response,
################################################
# prior information
################################################
w <– cbind(y,x)
wbar <– apply(w,2,mean)
wcov <– var(w)
prior <– list(a0=10,
b0=1,
nu1=4,
nu2=4,
s2=0.5*wcov,
m2=wbar,
psiinv2=2*solve(wcov),
tau1=6.01,
tau2=3.01)
################################################
# mcmc specification
################################################
mcmc <– list(nburn=5000,
nsave=5000,
nskip=3,
ndisplay=1000)
################################################
# covariate values where the density
# and mean function is evaluated
################################################
xpred <– seq(0,1,0.02)
################################################
# fitting the model
################################################
fitWDDP <– DPcdensity(y=y,x=x,
xpred=xpred,
ngrid=100,
prior=prior,
mcmc=mcmc,
state=NULL,
status=TRUE)
Using the same MCMC specification, the LDDP model was also fitted to the data. The LDDPdensity function was used to fit a a mixture of B-splines models with , where ψk(x) corresponds to the kth B-spline basis function evaluated at x as implemented in the bs function of the splines R package. The LDDP model was fitted using Zellner's g-prior (Zellner, 1983), with g = 103. The following values for the hyper-parameters were considered: a0 = 10, b0 = 1, m0 = (X′X)−1X′y, S0 = g(X′ X)−1, τ1 = 6.01, τs1 = 6.01, τs2 = 2.01, ν = 9, and Ψ−1 = S0. The following piece of code was used to fit the model:
################################################
# prior information
################################################
library(splines)
W <– cbind(rep(1,nrec),bs(x,df=6))
S0 <– 1000*solve(t(W)%*%W)
m0 <– solve(t(W)%*%W)%*%t(W)%*%y
prior<–list(a0=10,
b0=1,
m0=m0,
S0=S0,
tau1=6.01,
taus1=6.01,
taus2=2.01,
nu=9,
psiinv=solve(S0))
################################################
# covariate values where the density
# and mean function is evaluated
################################################
xpred <– seq(0,1,0.02)
Wpred <– cbind(rep(1,length(xpred)),bs(xpred,df=6))
################################################
# fitting the model
################################################
fitLDDP <– LDDPdensity(formula=y~W-1,zpred=Wpred,
ngrid=100,
prior=prior,
mcmc=mcmc,
state=NULL,
status=TRUE)
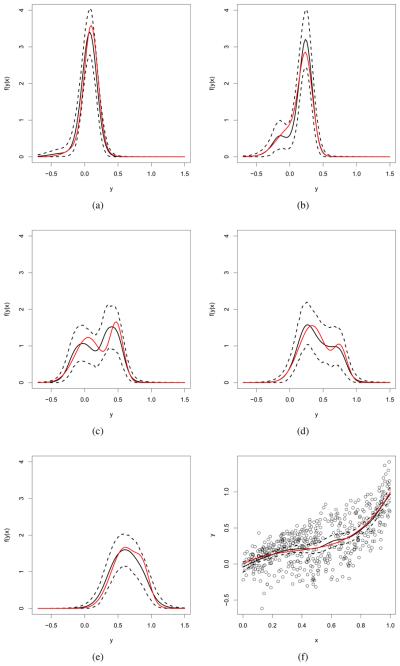
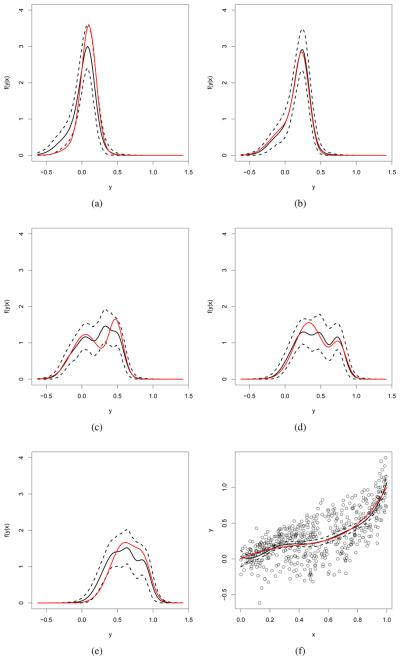
Figures 1 and 2 show the true density, the estimated density and point-wise 95% HPD intervals for a range of values of the predictor for the WDDP and LDDP model, respectively. The estimates correspond approximately to the true densities in each case. The figures also display the plot of the data along with the estimated mean function, which is very close to the true one under both models.
Figure 1.
Simulated data - WDDP model: True conditional densities of y|x (in red), posterior mean estimates (black continuos line) and point-wise 95% HPD intervals (black dashed lines) for: (a) x = 0.1, (b) x = 0.25, (c) x = 0.48, (d) x = 0.76, and (e) x = 0.88. Panel (f) shows the data, along with the true and estimated mean regression curves.
Figure 2.
Simulated data - LDDP model: True conditional densities of y|x (in red), posterior mean estimates (black continuos line) and point-wise 95% HPD intervals (black dashed lines) for: (a) x = 0.1, (b) x = 0.25, (c) x = 0.48, (d) x = 0.76, and (e) x = 0.88. Panel (f) shows the data, along with the true and estimated mean regression curves.
In both functions, the posterior mean estimates and the limits of point-wise 95% HPD intervals for the conditional density for each value of the predictors are stored in the model objects densp.m, and densp.l and densp.h, respectively. The following piece of code illustrates how these objects can be used in order to get the posterior estimates for x = 0.1 in the LDDP model. This code was used to draw the plots displayed in Figures 1 and 2.
par(cex=1.5,mar=c(4.1, 4.1, 1, 1))
plot(fitLDDP$grid,fitLDDP$densp.h[6,],lwd=3,type=“l”,lty=2,
main=“”,xlab=“y”,ylab=“f(y|x)”,ylim=c(0,4))
lines(fitLDDP$grid,fitLDDP$densp.l[6,],lwd=3,type=“l”,lty=2)
lines(fitLDDP$grid,fitLDDP$densp.m[6,],lwd=3,type=“l”,lty=1)
lines(fitLDDP$grid,dtrue(fitLDDP$grid,xpred[6]),lwd=3,
type=“l”,lty=1,col=“red”)
Finally, both functions return the posterior mean estimates and the limits of point-wise 95% HPD intervals for the mean function in the model objects meanfp.m, and meanfp.l and meanfp.h, respectively. The following pice of code was used to obtain the estimated mean function under the LDDP model along with the true function.
par(cex=1.5,mar=c(4.1, 4.1, 1, 1)) plot(x,y,xlab=“x”,ylab=“y”,main=“”) lines(xpred,fitLDDP$meanfp.m,type=“l”,lwd=3,lty=1) lines(xpred,fitLDDP$meanfp.l,type=“l”,lwd=3,lty=2) lines(xpred,fitLDDP$meanfp.h,type=“l”,lwd=3,lty=2) lines(xpred,mtrue(xpred),col=“red”,lwd=3)
4.2. Dependent random effects distributions
We consider data from the Chilean system for educational quality measurement (Sistema de Medi-cición de la Calidad de la Educación, SIMCE). The Chilean education system is subject to several performance evaluations regularly at the school, teacher and student level. In the last case, SIMCE has developed mandatory census-type tests to regularly assess the educational progress at three stages: 4th and 8th grades in primary school (9 and 13 years old children, respectively), and 2nd grade in secondary school (16 years old children). The SIMCE instruments are designed to assess the achievement of fundamental goals and minimal contents of the curricular frame in different areas of knowledge, currently Spanish, mathematics and science. Here we focus on data from the math test applied in 2004 to 8 grader examinees in primary school. The test consists of 45 multiple choice items questions with 4 alternatives. The response yij ∈ {0,1} is a binary variable indicating whether the individual i answers item j correctly.
The main purpose of collecting these data is to monitor standards and progress of educational systems, focusing on characterizing the population (and its evolution) rather than individual examinees. It is of particular interest to understand the way in which some factors at individual and/or school level could explain systematic differences in the performance of students in order to establish policies to improve the education system. For instance, a significant characteristic of the Chilean elementary and secondary education system is a variety of different school types. These are grouped as Public I, financed by the state and administered by county governments; Public II, financed by the state and administered by county corporations; Private I, financed by the state and administered by the private sector; Private II, fee-paying schools that operate solely on payments from parents and administered by the private sector.
In order to evaluate the effect of the type of school and gender on the student performance we consider the LDDP mixture of normals prior for the ablities in a Rasch model as in Fariña et al. (2009). For illustration purposes, we consider a subset of 500 children. We refer to Fariña et al. (2009) for a full analysis of the complete data. The model is given by
Here, xi includes an intercept term, three dummy variables for the type of school and the gender indicator. The LDDP Rasch model was fitted using the LDDPrasch function and assuming β ~ N44(0,103 I44), a = 1, μ0 = 05, S0 = 100I5, τ1 = 6.01, τs1 = 6.01, τs2 = 2.01, ν = 8, Ψ = I5. A single Markov chain cycle of length 25, 000 was completed. The full chain was sub-sampled every 4 steps after a burn in period of 5,000 samples, to give a reduced chain of length 5,000. For each gender and type of school the density of the abilities distribution was evaluated on a grid of 100 equally spaced points in the range (−3, 8). The following commands were used to fit the model,
################################################
# prediction's design matrix.
# columns: – intercept term.
# – 3 dummies for type of school.
# – gender indicator (1 = girl).
################################################
zpred <– matrix(c(1,0,0,0,0,
1,1,0,0,0,
1,0,1,0,0,
1,0,0,1,0,
1,0,0,0,1,
1,1,0,0,1,
1,0,1,0,1,
1,0,0,1,1),
nrow=8,ncol=5,byrow=T)
################################################
# prior information
################################################
prior <– list(alpha=1,
beta0=rep(0,44),
Sbeta0=diag(1000,44),
mu0=rep(0,5),
S0=diag(100,5),
tau1=6.01,
taus1=6.01,
taus2=2.01,
nu=8,
psiinv=diag(1,5))
################################################
# mcmc
################################################
mcmc <– list(nburn=5000,
nskip=3,
ndisplay=1000,
nsave=5000)
################################################
# fitting the model
################################################
fitLDDP <– LDDPrasch(formula=y ~ types+gender,
prior=prior,
mcmc=mcmc,
state=NULL,
status=TRUE,
zpred=zpred,
grid=seq(-3,8,len=100),
compute.band=TRUE)
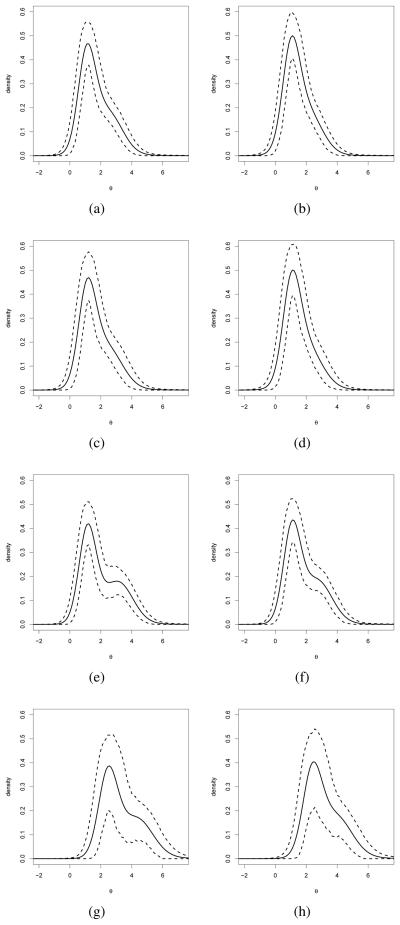
Different shapes in the resulting posterior densities were observed. Figure 3 displays the posterior mean and point wise 95%HPD interval for the random effects distribution for different combinations of the predictors. The density estimates show a clear departure from the commonly assumed normality of the random effects distributions. We found no important differences in the behavior of boys and girls. Children in Public I and II schools showed a similar skewed to the right random effects distribution. The estimated abilities distributions for children in private schools were shifted to the right in comparison with the distribution observed for children from public schools. This shift was more pronounced for children in fee-paying schools that operate solely on payments from parents and administered by the private sector (Private II) than those from schools financed by the state and administered by the private sector (Private I). A bimodal random effects distribution was observed in the abilities distributions from private schools.
Figure 3.
SIMCE data: Posterior estimates (mean and point-wise 95% HPD intervals) for the ability distribution for type of school and gender. The results for boys are shown in panels (a), (c), (e) and (g) for type of school Public I, Public II, Private I, and Private II, respectively. The results for girls are shown in panels (b), (d), (f) and (h) for type of school Public I, Public II, Private I, and Private II, respectively.
4.3. Proportional hazards regression with nonparametric frailties
Consider right censored survival data where failure times are repeatedly observed within a group or subject. Let i = 1,…, n denote the strata over which repeated times-to-event are recorded, and j = 1,…, ni denote the repeated observations within stratum i. The data are denoted {(wij, tij, δij) : i = 1,…, n; j = 1,…, ni} where tij is the recorded event time, δi = 1 if tij is an observed failure time and δij = 0 if the failure time is right censored at tij, and wij is a p-dimensional vector of covariates.
Functions fitting generalized linear mixed models (PTglmm, DPglmm, and DPMglmm) can be used to fit the Cox proportional hazards model (Cox, 1972) with nonparametric, multivariate frailties. Briefly, the baseline hazard function λ0 (t) corresponds to an individual with covariates w = 0 and survival time T0. Given that the baseline individual has made it up to t, T0 ≥ t, the baseline hazard is how the probability of expiring in the next instant is changing. In terms of the baseline survival function S0(t) = P(T0 > t) and density f0(t), this is given by
The conditionally proportional hazards assumption stipulates that
where θ = (θ1,…, θn)′ are random effects, termed frailties in the survival literature. Often the frailties θi, or exponentiated frailties eθi, are assumed to arise iid from some parametric distribution such as N(0, σ2), gamma, positive stable, etc. We consider a nonparametric MPT prior on the frailties below.
The specification is conditional because proportionality only holds for survival times within a given strata i, not across strata unless the distribution of θi is positive stable (see, e.g. Qiou, Ravishanker, and Dey, 1999). Precisely, for individuals j1 and j2 within strata i,
Often the baseline hazard is assumed to be piecewise constant on a partition of R+ comprised of K intervals, yielding the piecewise exponential model. References are too numerous to list; but see Walker and Mallick (1997), Aslanidou, Dey, and Sinha (1998), and Qiou et al. (1999). Assume
where a0 = 0 and aK = ∞, although in practice aK = max{tij} is sufficient. The prior hazard is specified by cutpoints and hazard values λ = (λ1,…, λK)′. If the prior on λ is taken to be independent gamma distributions, the model can approximate the gamma process on a fine mesh (Kalbfleisch, 1978). Regardless, the resulting model implies a Poisson likelihood for “data” yijk taking values yijk = 0 when tij ∉ (ak−1, ak] or δij = 0, and yijk = 1 when tij ∈ (ak−1, ak] and δij = 1, for k = 1,…, K(tij), where K(t) = max{k : ak ≤ t}. The likelihood for (β, λ, γ) is
where p(y|μ) is the probability mass function for a Poisson(μ) random variable, , and Δijk = min{ak,tij} − ak−1. Thus, the Cox model assuming a piecewise constant baseline hazard can be fitted in any software allowing for Poisson regression. Note that if covariates are time dependent as well, and change only at values included in , the likelihood is trivially extended to include wijk above for k = 1,…, K(tij) rather than wij.
We consider data on n = 38 kidney patients discussed by McGilchrist and Aisbett (1991). Each of the patients provides ni = 2 infection times, some of which are right censored. McGilchrist and Aisbett (1991) found that only gender was significant, and so we follow Aslanidou et al. (1998), Walker and Mallick (1997), Qiou et al. (1999), and Hemming and Shaw (2005) in considering only this covariate in what follows. We fitted the semiparametric proportional hazards regression model using a nonparametric prior for the frailties distribution. The following commands were used to prepare the data to fit the model. The original dataset, d[i, j], is a 38 by 6 matrix, which for each row (from left to right) contains the subject indicator, ti1, δi1, ti2, δi2, and the gender indicator. Ten intervals were considered with cutpoints {a1,…, a10} taken from the empirical distribution of the data.
################################################
# function to make a row with ‘1’ at ind
################################################
onv <– function(ind,len)
{
onv <– rep(0,len)
onv[ind] <– 1
return(onv)
}
################################################
# Create data to fit Cox model using
# Poisson likelihood for piecewise
# exponential model.
################################################
newdat <– matrix(1:(38*2*2),nrow=38*2,ncol=2)
tt <– rep(0,38*2)
delta <– tt
for(i in 1:38)
{
newdat[i*2-1,1] <– d[i,1]
newdat[i*2-1,2] <– d[i,6]
newdat[i*2 ,1] <– d[i,1]
newdat[i*2 ,2] <– d[i,6]
tt[i*2-1] <– d[i,2]
delta[i*2-1] <– d[i,3]
tt[i*2] <– d[i,4]
delta[i*2] <– d[i,5]
}
y <– NULL
mat <– NULL
tot <– 0
p <– ncol(newdat)
off <– NULL
n <– length(tt)
intervals <– 10
cutpoint <– quantile(tt,(1:intervals)/intervals,names=FALSE)
for(i in 1:n)
{
tot <– tot+1
mat <– matrix(append(mat,c(newdat[i,1:p],onv(1,intervals))),
c(p+intervals,tot))
off <– append(off,min(cutpoint[1],tt[i]))
if(tt[i]<=cutpoint[1] && delta[i]==1)
{
y <– append(y,1)
}
else
{
y <– append(y,0)
}
for(j in 1:(intervals-1))
{
if(tt[i]>cutpoint[j])
{
off <– append(off,min(cutpoint[j+1],
tt[i])-cutpoint[j])
tot <– tot+1
mat <– matrix(append(mat,c(newdat[i,1:p],
onv(j+1,intervals))),
c(p+intervals,tot))
if(tt[i] <= cutpoint[j+1] && delta[i]==1)
{
y <– append(y,1)
}
else
{
y <– append(y,0)
}
}
}
}
mat <– t(mat)
id <– mat[,1]
gender <– mat[,2]
loghazard <– mat[,3:12]
We performed the analysis using the PTglmm function to the responses
and where xij is a 11-dimensional design vector containing the gender indicator and the indicator for the interval associated to the corresponding response. Finally, we set β = (γ′, λ′)′, and assume
and
We consider a M = 5 finite PT prior which was centered around a N(0, σ2) distribution and constrained to have median-0 (frstlprob=TRUE in the prior object below). The values for the hyper-parameters β0 and Sβ0 were obtained from a penalized quasi-likelihood (PQL) fit using the glmmPQL function available from the MASS pakage (Venables and Ripley, 2002). The matrix Sβ0 was inflated by a factor of 100. The remaining hyper-parameters were a0 = b0 = 1, ν0 = 3, and T = I1. Starting values for the model parameters were obtained from the PQL fit. A single Markov chain cycle of length 25,000 was completed. The full chain was sub-sampled every 4 steps after a burn in period of 5,000 samples, to give a reduced chain of length 5,000. The code for fitting the model using PTglmm was
################################################
# PQL estimation
################################################
library(MASS)
fit0 <– glmmPQL(fixed=y~gender+loghazard−1+
offset(log(off)),
random=~1|id,family=poisson(log))
################################################
# prior
################################################
beta0 <– fit0$coefficients$fixed
Sbeta0 <– vcov(fit0)
prior <– list(M=5,
a0=1,
b0=1,
nu0=3,
tinv=diag(1,1),
mu=rep(0,1),
beta0=beta0,
Sbeta0=Sbeta0,
frstlprob=TRUE)
################################################
# starting values from PQL estimation
################################################
beta <– fit0$coefficients$fixed
b <– as.vector(fit0$coefficients$random$id)
mu <– rep(0,1)
sigma <– getVarCov(fit0)[1,1]
state <– list(alpha=1,
beta=beta,
b=b,
mu=mu,
sigma=sigma)
################################################
# mcmc
################################################
mcmc <– list(nburn=5000,
nsave=5000,
nskip=19,
ndisplay=1000,
tune3=1.5)
################################################
# fitting the model
################################################
fitPT <– PTglmm(fixed=y~gender+loghazard,
offset=log(off),
random=~1|id,
family=poisson(log),
prior=prior,
mcmc=mcmc,
state=state,
status=FALSE)
################################################
# posterior inferences
################################################
summary(fitPT)
################################################
# frailties density estimate
################################################
predPT <– PTrandom(fitPT,predictive=TRUE,
gridl=c(-2.3,2.3))
plot(predPT)
The abridged output is given below. The output lists the estimated effect for gender β̂1 = −1.13 followed by K = 10 estimated log-hazard values. Notice that the intercept term in the posterior information for the “fixed” effects (regression coefficients in the output), corresponds to the mean of the frailties distribution G. The posterior median estimate of the centering variance was σ̂2 = 0.35 and close to the posterior median of the frailties variance (0.33). Further, the posterior median (95% credible interval) for α was 0.75 (0.04; 3.77). The trace plots of the parameters (not shown) indicate a good mixing of the chain. The acceptance rates for the MH steps associated to the regression coefficients, frailties, centering variance and precision parameter was 36, 61, 43 and 0.46%, respectively. Notice that the 0 values for the acceptance rates in the output corresponds to the centering mean, which is sampled, and the decomposition of the centering covariance matrix. The latter is only sampled for dimensions greater than or equal to 2.
Walker and Mallick (1997) analyzed these data with piecewise exponential model and frailties following a Polya tree with fixed centering variance, PT8(Π100, A0.1) and find β̂1 = −1.0. McGilchrist and Aisbett (1991) obtain β̂1 = −1.8, but with other nonsignificant covariates included. Aslanidou et al. (1998) also reportes β̂ = −1.0. Hemming and Shaw (2005) obtain β̂ = −1.7 and Qiou et al. (1999) obtain β̂ = −1.1 under the positive stable and β̂ = −1.6 under gamma frailties, respectively. The the deviance information criterion (DIC), as presented by Spiegelhalter, Best, Carlin, and Van der Linde (2002), was 398 for either PT or normal model (not shown), so the normal model does about the same from a predictive standpoint based on the DIC.
Bayesian semiparametric generalized linear mixed effect model
Call:
PTglmm.default(fixed = y ~ gender + loghazard, random = ~1 |
id, family = poisson(log), offset = log(off), prior = prior,
mcmc = mcmc, state = state, status = FALSE)
Posterior Predictive Distributions (log):
Min. 1st Qu. Median Mean 3rd Qu. Max.
−5.99200 −0.22250 −0.10970 −0.48500 −0.05714 −0.01381
Model's performance:
Dbar Dhat pD DIC LPML
379.21 360.63 18.58 397.79 −200.29
Regression coefficients:
Mean Median Std. Dev. Naive Std.Error 95%CI-Low 95%CI-Upp
(Intercept) −0.0004443 0.0015210 0.0960076 0.0013578 −0.2066125 0.2021371
gender −1.1321281 −1.1296717 0.3219508 0.0045531 −1.7762785 −0.5117994
loghazard1 −4.2608268 −4.2375512 0.4412274 0.0062399 −5.1598904 −3.4611046
loghazard2 −3.7898628 −3.7638395 0.5018976 0.0070979 −4.8383288 −2.8794989
loghazard3 −3.9792281 −3.9691425 0.4556631 0.0064440 −4.9028932 −3.1213276
loghazard4 −3.0627136 −3.0526713 0.4526581 0.0064016 −4.0124879 −2.2353213
loghazard5 −3.2581084 −3.2477986 0.4219626 0.0059675 −4.1039312 −2.4603991
loghazard6 −3.9951390 −3.9805448 0.4544001 0.0064262 −4.9103962 −3.1403702
loghazard7 −4.9343777 −4.9183270 0.5365962 0.0075886 −6.0496817 −3.9150135
loghazard8 −3.6883152 −3.6845014 0.4479935 0.0063356 −4.5692123 −2.8232222
loghazard9 −3.6723423 −3.6673231 0.4810002 0.0068024 −4.6112294 −2.7315973
loghazard10 −4.1246955 −4.1272752 0.4966618 0.0070239 −5.0749243 −3.1886274
Baseline distribution:
Mean Median Std. Dev. Naive Std.Error 95%CI-Low 95%CI-Upp
mu-(Intercept) 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000
sigma-(Intercept) 0.430385 0.354618 0.294752 0.004168 0.119319 1.212674
Precision parameter:
Mean Median Std. Dev. Naive Std.Error 95%CI-Low 95%CI-Upp
alpha 1.05875 0.75117 1.02204 0.01445 0.04448 3.76967
Random effects variance:
Mean Median Std. Dev. Naive Std.Error 95%CI-Low 95%CI-Upp
R.E.Cov-(Intercept) 0.378637 0.331281 0.222121 0.003141 0.096121 0.948495
Acceptance Rate for Metropolis Steps = 0.3570935 0.6072718 0 0.428972 0.463486 0
Number of Observations: 413
Number of Groups: 38
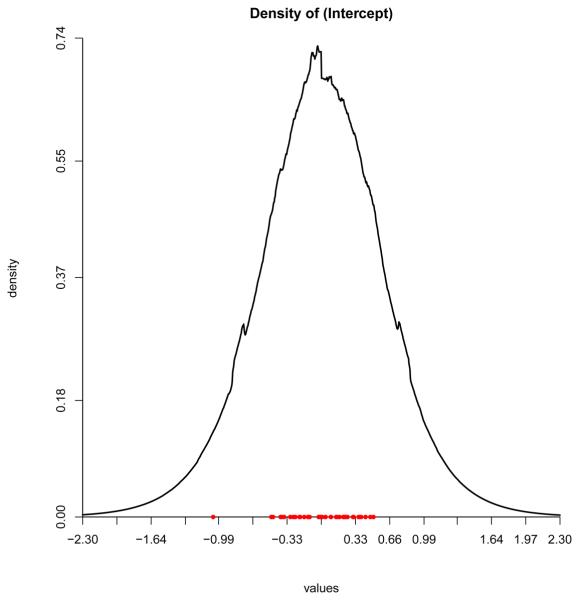
Figure 4 shows the estimated frailty distribution from these data along with the posterior mean of the frailty term for each patient. The distribution is remarkably Gaussian-shaped, in contrast to the analysis presented in Walker and Mallick (1997), which showed two well defined density modes corresponding to men and women. We were unable to duplicate this result across several sets of hyper-prior values, including the consideration of PT8 (Π100, A0.1). In retrospect, this is not surprising. Two well separated modes would typically indicate an omitted covariate, yet gender was included as a risk factor in the model.
Figure 4.
Kidney data: Posterior mean of the frailty distribution. The density is overlaid on a plot of the posterior mean of the individuals frailty terms.
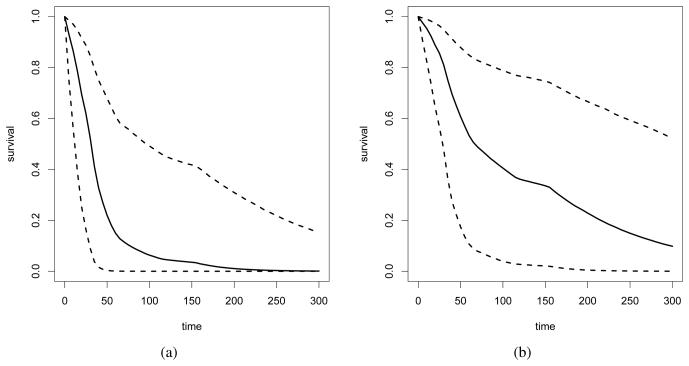
Finally, Figure 5 show the posterior median and 95% credible interval for survival curves for males and females, taking the individual-level heterogeneity modeled through the frailty distribution into account.
Figure 5.
Kidney data: Posterior estimates (median and point-wise 95% credible intervals) for the survival function for time to infection. The results for males and females are shown in panels (a) and (b), respectively.
5. Concluding remarks
Because the main obstacle for the practical use of BSP and BNP methods has been the lack of estimation tools, we presented an R package for fitting some frequently used models. Until the release of DPpackage, the two options for researchers who wished to fit a BSP or BNP model were to write their own code or to rely heavily on particular parametric approximations to some specific processes using the BUGS code given in Peter Congdon's books (see e.g., Congdon, 2001). DPpackage is geared primarily towards users who are not willing to bear the costs associated with both of these options.
Chambers (2000) conceptualized statistical software as a set of tools to organize, analyze and visualize data. Data organization and visualization of results is based on R capabilities. Chambers (2000) also proposed requirements and guidelines for developing and assessing statistical softwares. These requirements may be discussed with respect to DPpackage:
Easy specification of simple tasks: The documentation contains examples, and similar problems can be analyzed by moderate modifications of the model description files. The examples have been chosen so that they demonstrate the functionality of DPpackage with well-known data sets.
Gradual refinement of the tasks: The user can enhance a nonparametric model by adding covariates, and by fixing part of the baseline distributions and the precision parameters.
Arbitrarily extensive programming: DPpackage has a programming environment for implementing sophisticated proposal distributions, if the default proposals are not sufficient.
Implementing high-quality computations: Also, because the source code in a compiled language is available, new procedures can be added and the old ones modified to improve performance and flexibility.
Embedding the results of items 2–4 as new simple tools: DPpackage has the capability of continuing a Markov chain from the last value of the parameters of a previous analysis. As the MCMC samples are saved in matrix objects, both parts of the Markov chain can be easily merged.
Many improvements to the current status of the package can be made. For example, all DPpackage modeling functions compute CPOs for model comparison. However, only some of them compute the effective number of parameters pD and DIC, as presented by Spiegelhalter et al. (2002). These and other model comparison criterion will be included for all functions in future versions of DPpackage.
The implementation of more models, the development of general-purpose sampling functions, realtime visualization of simulation progress, and the ability to handle large dataset problems, through the use of sparse matrix techniques (George and Liu, 1981), are the topic of further improvements.
6. Acknowledgments
The first author is supported by Fondecyt grant 3095003. Partial support from the KUL-PUC bilateral (Belgium-Chile) grant BIL05/03 and of the IAP research network grant Nr P6/03 of the Belgian government (Belgian Science Policy) for previous versions of DPpackage is also acknowledged. The work of the second author was supported in part by NIH grant 2-R01-CA95955-05. The third author was partially supported by grant Fondecyt 1060729. The last two authors were partially supported by grant NIH/NCI R01CA75981. The SIMCE Office from the Chilean Government kindly allowed us access to the databases used in this paper.
References
- Albert JH, Chib S. Bayesian analysis of binary and polychotomous response data. Journal of the American Statistical Association. 1993;88:669–679. [Google Scholar]
- Antoniak CE. Mixtures of Dirichlet processes with applications to Bayesian nonparametric problems. The Annals of Statistics. 1974;2:1152–1174. [Google Scholar]
- Aslanidou H, Dey DK, Sinha D. Bayesian analysis of multivariate survival data using Monte Carlo methods. Canadian Journal of Statistics. 1998;26:33–48. [Google Scholar]
- Besag J, Green P, Higdon D, Mengersen K. Bayesian computation and stochastic systems (with Discussion) Statistical Science. 1995;10:3–66. [Google Scholar]
- Branscum A, Hanson T. Bayesian nonparametric meta-analysis using Polya tree mixture models. Biometrics. 2008;64:825–833. doi: 10.1111/j.1541-0420.2007.00946.x. [DOI] [PubMed] [Google Scholar]
- Bush CA, MacEachern SN. A semiparametric Bayesian model for randomised block designs. Biometrika. 1996;83:275–285. [Google Scholar]
- Carlin BP, Louis TA. Bayesian methods for data analysis. 3rd Ed. Chapman and Hall/CRC; New York, USA: 2008. [Google Scholar]
- Chambers JM. Users, programmers, and statistical software. Journal of Computational and Graphical Statistics. 2000;9(3):402–422. [Google Scholar]
- Chen MH, Shao QM. Monte Carlo estimation of Bayesian credible and HPD intervals. Journal of Computational and Graphical Statistics. 1999;8:69–92. [Google Scholar]
- Christensen R, Hanson T, Jara A. Parametric nonparametric statistics: An introduction to mixtures of finite Polya trees. The American Statistician. 2008;62:296–306. [Google Scholar]
- Congdon P. Bayesian statistical modelling. John Wiley and Sons; New York, USA: 2001. [Google Scholar]
- Cox DR. Regression models and life-tables (with Discussion) Journal of the Royal Statistical Society, Series B. 1972;34:187–220. [Google Scholar]
- De Boeck P, Wilson M. Explanatory item response models. A generalized linear and nonlinear approach. Springer; New York, USA: 2004. [Google Scholar]
- De Iorio M, Johnson WO, Müller P, Rosner GL. Bayesian nonparametric non-proportional hazards survival modelling. Biometrics. 2009;65:762–771. doi: 10.1111/j.1541-0420.2008.01166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iorio M, Müller P, Rosner GL, MacEachern SN. An ANOVA model for dependent random measures. Journal of the American Statistical Association. 2004;99:205–215. [Google Scholar]
- Dey D, Müller P, Sinha D. Practical nonparametric and semiparametric Bayesian statistics. Springer; New York, USA: 1998. [Google Scholar]
- Doss H. Bayesian nonparametric estimation for incomplete data via successive substitution sampling. The Annals of Statistics. 1994;22:1763–1786. [Google Scholar]
- Duan JA, Guindani M, Gelfand AE. Generalized spatial Dirichlet process models. Biometrika. 2007;94:809–825. [Google Scholar]
- Dunson D, Yang M, Baird D. Technical report. Department of Statistical Science, Duke University; 2007a. Semiparametric Bayes hierarchical models with mean and variance constraints. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunson DB, Park JH. Kernel stick-breaking processes. Biometrika. 2008;95:307–323. doi: 10.1093/biomet/asn012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunson DB, Pillai N, Park JH. Bayesian density regression. Journal of the Royal Statistical Society, Series B. 2007b;69:163–183. [Google Scholar]
- Eilers PHC, Marx BD. Flexible smoothing with B-splines and penalties. Statistical Science. 1996;11(2):89–121. [Google Scholar]
- Escobar MD. Estimating normal means with a Dirichlet process prior. Journal of the American Statistical Association. 1994;89:268–277. [Google Scholar]
- Escobar MD, West M. Bayesian density estimation and inference using mixtures. Journal of the American Statistical Association. 1995;90:577–588. [Google Scholar]
- Fariña P, Quintana FA, San Martín E, Jara A. Technical report. Department of Statistics, Pontificia Universidad Católica de Chile; 2009. A dependent semiparametric Rasch model for the analysis of Chilean educational data. [Google Scholar]
- Ferguson TS. A Bayesian analysis of some nonparametric problems. Annals of Statistics. 1973;1:209–230. [Google Scholar]
- Ferguson TS. Prior distribution on the spaces of probability measures. Annals of Statistics. 1974;2:615–629. [Google Scholar]
- Gamerman D. Sampling from the posterior distribution in generalized linear mixed models. Statistics and Computing. 1997;7:57–68. [Google Scholar]
- Gelfand AE, Kottas A. A computational approach for full nonparametric Bayesian inference under Dirichlet Process Mixture models. Journal of Computational and Graphical Statistics. 2002;11:289–304. [Google Scholar]
- George A, Liu JW. Computer solution of large sparse positive definite systems. Prentice-Hall; New York, USA: 1981. [Google Scholar]
- Ghosh JK, Ramamoorthi RV. Bayesian nonparametrics. Springer; New York, USA: 2003. [Google Scholar]
- Gilks WR, Thomas A, Spiegelhalter DJ. A language and program for complex Bayesian modelling. The Statistician. 1994;43:169–178. [Google Scholar]
- Green PJ. Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika. 1995;82:711–732. [Google Scholar]
- Griffin JE, Steel MFJ. Order-based dependent Dirichlet processes. Journal of the American Statistical Association. 2006;101:179–194. [Google Scholar]
- Hanson T. Inference for mixtures of finite Polya tree models. Journal of the American Statistical Association. 2006;101:1548–1565. [Google Scholar]
- Hanson T, Branscum A, Johnson W. Bayesian nonparametric modeling and data analysis: an introduction. In: Dey DK, Rao CR, editors. Bayesian Thinking: Modeling and Computation (Handbook of Statistics, volume 25) Elsevier; Amsterdam, The Netherlands: 2005. pp. 245–278. [Google Scholar]
- Hanson T, Johnson WO. Modeling regression error with a mixture of Polya trees. Journal of the American Statistical Association. 2002;97:1020–1033. [Google Scholar]
- Hanson T, Johnson WO. A Bayesian semiparametric AFT model for interval-censored data. Journal of Computational and Graphical Statistics. 2004;13:341–361. [Google Scholar]
- Hastie T, Tibshirani R. Generalized additive models. Chapman and Hall; New York, USA: 1990. [DOI] [PubMed] [Google Scholar]
- Hemming K, Shaw JEH. A class of parametric dynamic survival models. Lifetime Data Analysis. 2005;11:81–98. doi: 10.1007/s10985-004-5641-5. [DOI] [PubMed] [Google Scholar]
- Ishwaran H, James LF. Approximate Dirichlet process computing in finite normal mixtures: smoothing and prior information. Journal of Computational and Graphical Statistics. 2002;11:508–532. [Google Scholar]
- Jara A. Applied Bayesian non- and semi-parametric inference using DPpackage. Rnews. 2007;7:17–26. [Google Scholar]
- Jara A, García-Zattera MJ, Lesaffre E. A Dirichlet process mixture model for the analysis of correlated binary responses. Computational Statistics and Data Analysis. 2007;51:5402–5415. [Google Scholar]
- Jara A, Hanson T, Lesaffre E. Robustifying generalized linear mixed models using a new class of mixture of multivariate Polya trees. Journal of Computational and Graphical Statistics. 2009 To appear. [Google Scholar]
- Kalbfleisch JD. Nonparametric Bayesian analysis of survival time data. Journal of the Royal Statistical Society, Series B. 1978;40:214–221. [Google Scholar]
- Kleinman KP, Ibrahim JG. A semi-parametric Bayesian approach to generalized linear mixed models. Statistics in Medicine. 1998a;17:2579–2596. doi: 10.1002/(sici)1097-0258(19981130)17:22<2579::aid-sim948>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Kleinman KP, Ibrahim JG. A semiparametric Bayesian approach to the random effects model. Biometrics. 1998b;54:921–938. [PubMed] [Google Scholar]
- Kottas A, Müller P, Quintana F. Nonparametric Bayesian modeling for multivariate ordinal data. Journal of Computational and Graphical Statistics. 2005;14:610–625. [Google Scholar]
- Kraemer HC. Evaluating medical tests. Sage Publications; New York, USA: 1992. [Google Scholar]
- Lang S, Brezger A. Bayesian P-splines. Journal of Computational and Graphical Statistics. 2004;13:183–212. [Google Scholar]
- Lavine M. Some aspects of Polya tree distributions for statistical modeling. The Annals of Statistics. 1992;20:1222–1235. [Google Scholar]
- Lavine M. More aspects of Polya tree distributions for statistical modeling. The Annals of Statistics. 1994;22:1161–1176. [Google Scholar]
- Li Y, Müller P, Lin X. Technical report. Department of Biostatistics, The MD Anderson Cancer Center; 2007. Center-adjusted inference for a nonparametric Bayesian random effect distribution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JS. Nonparametric hierarchical Bayes via sequential imputations. The Annals of Statistics. 1996;24:911–930. [Google Scholar]
- Lo AY. On a class of Bayesian nonparametric estimates I: Density estimates. The Annals of Statistics. 1984;12:351–357. [Google Scholar]
- MacEachern SN. Computational methods for mixture of Dirichlet process models. In: Dey D, Müller P, Sinha D, editors. Practical Nonparametric and Semiparametric Bayesian Statistics. Springer; 1998. pp. 1–22. [Google Scholar]
- MacEachern SN. ASA Proceedings of the Section on Bayesian Statistical Science. American Statistical Association; Alexandria, VA: 1999. Dependent nonparametric processes. [Google Scholar]
- MacEachern SN. Technical report. Department of Statistics, The Ohio State University; 2000. Dependent Dirichlet processes. [Google Scholar]
- MacEachern SN, Müller P. Estimating mixture of Dirichlet Process models. Journal of Computational and Graphical Statistics. 1998;7(7(2)):223–338. [Google Scholar]
- Mauldin RD, Sudderth WD, Williams SC. Polya trees and random distributions. Annals of Statistics. 1992;20:1203–1221. [Google Scholar]
- McGilchrist CA, Aisbett CW. Regression with frailty in survival analysis. Biometrics. 1991;47:461–466. [PubMed] [Google Scholar]
- Mukhopadhyay S, Gelfand AE. Dirichlet process mixed generalized linear models. Journal of the American Statistical Association. 1997;92:633–647. [Google Scholar]
- Muliere P, Tardella L. Approximating distributions of random functionals of Ferguson-Dirichlet priors. The Canadian Journal of Statistics. 1998;26:283–297. [Google Scholar]
- Müller P, Erkanli A, West M. Bayesian curve fitting using multivariate normal mixtures. Biometrika. 1996;83:67–79. [Google Scholar]
- Müller P, Quintana FA. Nonparametric Bayesian data analysis. Statistical Science. 2004;19:95–110. [Google Scholar]
- Müller P, Quintana FA, Rosner G. A method for combining inference across related nonpara-metric Bayesian models. Journal of the Royal Statistical Society, Series B. 2004;66:735–749. [Google Scholar]
- Müller P, Rosner GL. A Bayesian population model with hierarchical mixture priors applied to blood count data. Journal of the American Statistical Association. 1997;92:1279–1292. [Google Scholar]
- Neal R. Markov chain sampling methods for Dirichlet process mixture models. Journal of Computational and Graphical Statistics. 2000;9:249–265. [Google Scholar]
- Neal R. Slice sampling. The Annals of Statistics. 2003;31:705–767. [Google Scholar]
- Newton MA. Technical report, N. 905. University of Wisconsin-Madison, Department of Statistics; 1994. Computing with priors that support identifiable semiparametric models. [Google Scholar]
- Newton MA, Czado C, Chapell R. Bayesian inference for semiparametric binary regression. Journal of the American Statistical Association. 1996;91:142–153. [Google Scholar]
- Perron F, Mengersen K. Bayesian nonparametric modeling using mixtures of triangular distributions. Biometrics. 2001;57:518–528. doi: 10.1111/j.0006-341x.2001.00518.x. [DOI] [PubMed] [Google Scholar]
- Petrone S. Bayesian density estimation using Bernstein polynomials. The Canadian Journal of Statistics. 1999a;27:105–126. [Google Scholar]
- Petrone S. Random Bernstein polynomials. Scandinavian Journal of Statistics. 1999b;26:373–393. [Google Scholar]
- Petrone S, Wasserman L. Consistency of Bernstein polynomial posterior. Journal of the Royal Statistical Society, Series B. 2002;64:79–100. [Google Scholar]
- Plummer M, Best N, Cowles K, Vines K. CODA: Output analysis and diagnostics for MCMC. 2006 R package version 0.10-7. [Google Scholar]
- Qiou Z, Ravishanker N, Dey DK. Multivariate survival analysis with positive stable frailties. Biometrics. 1999;55:81–88. doi: 10.1111/j.0006-341x.1999.00637.x. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2009. ISBN 3-900051-07-0, URL http://www.R-project.org. [Google Scholar]
- Rasch G. Probabilistic models for some intelligence and attainment tests. The Danish Institute for Educational Research (Expanded Edition, 1980, The University Chicago Press); Chicago, USA: 1960. [Google Scholar]
- Rossi P, Allenby G, McCulloch R. Bayesian statistics and marketing. John Wiley and Sons; New York, USA: 2005. [Google Scholar]
- Rossi P, McCulloch R. bayesm: Bayesian inference for marketing/micro-econometrics. 2008 R package version 2.2-2, URL http://faculty.chicagogsb.edu/peter.rossi/research/bsm.html.
- San Martín E, Jara A, Rolin JM, Mouchart M. On the analysis of Bayesian semiparametric IRT-type models. 2007 p. (Submitted) [Google Scholar]
- Sethuraman J. A constructive definition of Dirichlet prior. Statistica Sinica. 1994;2:639–650. [Google Scholar]
- Smith BJ. BOA: An R package for MCMC output convergence assessment and posterior inference. Journal of Statistical Software. 2007;21:1–37. [Google Scholar]
- Spiegelhalter SD, Best NG, Carlin BP, Van der Linde A. Bayesian measures of model complexity and fit. Journal of the Royal Statistical Society: Series B. 2002;64:583–639. [Google Scholar]
- Strurtz S, Ligges U, Gelman A. R2WinBUGS: A package for running WinBUGS from R. Journal of Statistical Software. 2005;12:1–16. [Google Scholar]
- Thomas A, O'Hara B, Ligges U, Sibylle S. Making BUGS open. Rnews. 2006;6:12–17. [Google Scholar]
- Tierney L. Markov chains for exploring posterior distributions. The Annals of Statistics. 1994;22:1701–1762. [Google Scholar]
- Venables WN, Ripley BD. Modern applied statistics with S. fourth edition. Springer; New York: 2002. ISBN 0-387-95457-0, URL http://www.stats.ox.ac.uk/pub/MASS4. [Google Scholar]
- Walker SG, Damien P, Laud PW, Smith AFM. Bayesian nonparametric inference for random distributions and related functions (with discussion) Journal of the Royal Statistical Society, Series B. 1999;61:485–527. [Google Scholar]
- Walker SG, Mallick BK. Hierarchical generalized linear models and frailty models with Bayesian nonparametric mixing. Journal of the Royal Statistical Society, Series B. 1997;59:845–860. [Google Scholar]
- West M. Generalized linear models: outlier accomodation, scale parameter and prior distributions. In: Bernardo JM, DeGroot MH, Lindley DV, Smith AFM, editors. Proceedings of the Second Valencia International Meeting. North Holland, Amsterdam: 1985. [Google Scholar]
- Zellner A. Applications of Bayesian analysis in econometrics. The Statistician. 1983;32:23–34. [Google Scholar]