Abstract
Microarray-basedcomparative genomic hybridization (CGH) has become a powerful methodfor the genome-wide detection of chromosomal imbalances. Although BAC microarrays have been usedfor mouse CGH studies, the resolving power of these analyses was limitedbecause high-density whole-genome mouse BAC microarrays were not available. We therefore developed a mouse BAC microarray containing 2803 unique BAC clones from mouse genomic libraries at 1-Mb intervals. For the general amplification of BAC clone DNA prior to spotting, we designed a set of three novel degenerate oligonucleotide-primed (DOP) PCR primers that preferentially amplify mouse genomic sequences while minimizing unwantedamplification of contaminating Escherichia coli DNA. The resulting 3K mouse BAC microarrays reproducibly identified DNA copy number alterations in cell lines andprimary tumors, such as single-copy deletions, regional amplifications, and aneuploidy.
DNA copy number aberrations are commonly observed phenomena in most solid tumors (Knuutila et al. 1999; Ried et al. 1999). High-throughput simple sequence length polymorphism (SSLP) markers have been developed to screen for loss of heterozygosity (LOH) in tumors (Dietrich et al. 1994a,b). Unfortunately, tumors that are induced in inbred mouse strains are not amenable to this type of analysis. In addition, high-throughput analysis using large numbers of polymorphic markers across the whole genome is very time-consuming, and the resolution is limited by clustering of polymorphic markers. Comparative genomic hybridization (CGH) enables the evaluation of chromosomal imbalances in tumor tissues without preparation of metaphase chromosomes from tumor cells (Kallioniemi et al. 1992). In addition, this method permits genome-wide screening of DNA copy number changes in a single experiment. However, it is not possible to produce high-resolution data using conventional CGH analysis, for which the maximal resolution is around 10 Mb (Kallioniemi et al. 1992).
The recent development of CGH for arrays of mapped genomic DNA segments has greatly improved the resolution of the analysis (Solinas-Toldo et al. 1997; Pinkel et al. 1998; Hodgson et al. 2001; Snijders et al. 2001; Cai et al. 2002). It is therefore conceivable that array-based CGH strategies will replace more conventional cytogenetic CGH procedures. In most instances, BAC, PAC, or cosmid clones have been used as sources of mapped genomic DNA. One drawback of array-based CGH technology is that large-scale culture and DNA extraction of individual clones are required to obtain sufficient DNA to construct the microarrays. Such procedures become very costly and time-consuming when they are scaled up for the 3000 genomic clones that are needed to assemble a mouse array with 1-Mb resolution. To circumvent these difficulties, PCR strategies for random amplification of small amounts of genomic DNA have been employed, such as ligation-mediated PCR (Klein et al. 1999; Snijders et al. 2001) and degenerative oligonucleotide-primed (DOP) PCR (Telenius et al. 1992; Hodgson et al. 2001). The incorporation of amine residues into the PCR products during DOP-PCR resulted in a robust and covalent interaction of the spotted PCR products with amine-reactive glass surfaces (Hodgson et al. 2001). Fiegler et al. (2003) successfully employed a set of improved human-specific DOP-PCR primers to enhance the signal-to-noise ratio by reducingunwanted amplification of contaminating Escherichia coli (E. coli) DNA. In the present study, we designed a set of novel mouse-specific DOP-PCR primers and produced a high-density whole-genome mouse BAC microarray with 1-Mb spacing.
RESULTS
Production of a 3K Mouse BAC Array With 1-Mb Spacing
Selection and Validation of Mouse BAC Clones
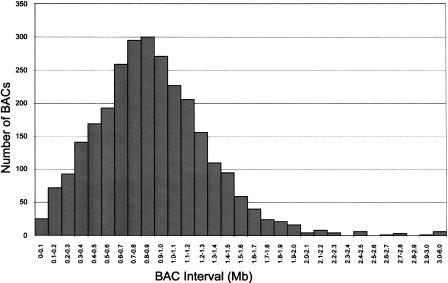
We selected a set of 3080 mouse BAC clones with an average spacingof 1 Mb across mouse chromosomes 1 to 19 and the X chromosome. Sequenced or end-sequenced BAC clones were preferentially selected from the mouse physical map (Zhao et al. 2001; Gregory et al. 2002). All clones were colony-purified, screened for T1 phage infection, and arrayed into 96-well plates. Fingerprinting revealed that 2825 of the 3080 clones had the expected HindIII restriction fragment patterns. The average spacing between the linear map positions of the 2825 BAC clones (calculated as the distance between the midpoint positions of two consecutive BACs) was 0.89 Mb. As shown in Figure 1, 93.1% of BAC intervals were smaller than 1.5 Mb. A total of 1532 clones (54.2 %) have been sequenced. A complete list of BAC clones, their linear positions, and other detailed information is available as supplemental research data. The complete 1-Mb clone set plus additional information on individual clones can also be viewed within the Ensembl mouse genome browser (Mouse CytoView, http://www.ensembl.org/mus_musculus/cytoview). Special buttons in Mouse CytoView allow downloadingof the 1-Mb clone set and the correspondingchromosomal locations. Upon request, glycerol stocks of the complete clone set will be provided.
Figure 1.
Distribution of spacing of the 2825 BACs used for the 3K mouse BAC microarray. The average spacing of the BACs was 0.89 Mb across the 19 autosomes and chromosome X. Linear map positions and other details of the 3K mouse BAC clone set are available online from the Ensembl database (http://www.ensembl.org/Mus_musculus/cytoview).
Design and Characterization of Mouse-Specific DOP-PCR Primers
The 3′ hexamer sequence of the previously published 6MW primer occurs at a frequency of 0.662 per Kb in mouse but also at the relatively high frequency of 0.396 per Kb in E. coli. Consequently, 6MW will amplify E. coli DNA almost as efficiently as mouse genomic DNA. In order to improve the discrimination of the primers, three mouse-specific DOP-PCR primers were designed (M-DOP1, M-DOP2, and M-DOP3), which specifically amplify mouse genomic sequences while minimizing the unwanted amplification of contaminating E. coli chromosomal DNA. The 3′ hexamer sequences of all three mouse DOP-PCR primers occur 200-300 times more frequently in mouse genomic sequences, compared to E. coli (Table 1).
Table 1.
Comparison of the Frequencies of the 3′ Hexanucleotide Anchors of DOP-PCR Primers in Mouse and E. coli Genomic Sequences
|
Number of matches per kilobase
|
||||
|---|---|---|---|---|
| Primer | Sequencea | M. musculus | E. coli | RatiobMm/Ec |
| M-DOP1 | CCGACTCGAGNNNNNNCTAGAG | 0.523 | 0.014 | 331.7 |
| M-DOP2 | CCGACTCGAGNNNNNNCTAGAA | 0.579 | 0.020 | 254.7 |
| M-DOP3 | CCGACTCGAGNNNNNNCTAGGT | 0.338 | 0.014 | 217.8 |
| 6MW | CCGACTCGAGNNNNNNATGTGG | 0.662 | 0.396 | 1.67 |
Bold characters indicate 3′ end hexamer sequence
Mm, M. musculus; Ec, E. coli
To verify whether two rounds of random amplification with the three DOP-PCR primers preserve a proper representation of mouse genomic sequences, we amplified C57BL/6J genomic DNA with primers M-DOP1, M-DOP2, and M-DOP3 and tested the resultingproducts for representation of 38 MIT markers from all autosomes as well as the X chromosome (Table 2). In our control study, all MIT markers were amplified using25 ngof mouse genomic DNA but failed to amplify to a detectable level using1:1000 dilution of the same genomic DNA under identical PCR conditions (data not shown). A positive MIT marker PCR usinga 1:100 dilution of DOP-PCR product as template DNA (correspondingto a 1:1000 dilution of the genomic template DNA) would demonstrate that the MIT marker-containing genomic region was efficiently amplified by our DOP-PCR strategy. Therefore, all MIT marker PCR reactions were performed on a 1:100 dilution of the DOP-PCR product. The PCR products from individual DOP-PCR primers M-DOP1, M-DOP2, and M-DOP3 were positive for 80%-95 % of the MIT markers. However, marker dropout differed amongthe different DOP-PCR products, and when the results from DOP-PCR primers 1, 2, and 3 results were combined, all of the 38 MIT markers tested positive. In the case of the 6MW DOP-PCR product, 34/38 of the MIT markers tested positive (Table 2).
Table 2.
Representation of MIT Loci in DOP-PCR Products
|
MIT marker PCR
|
|||||||
|---|---|---|---|---|---|---|---|
| Chr | MIT markers | Linear location (Mb) | M-DOP1 | M-DOP2 | M-DOP3 | 6MW | G-DNA |
| 1 | D1Mit319 | 32.8 | + | + | + | + | + |
| D1Mit249 | 61.4 | + | + | + | + | + | |
| 2 | D2Mit239 | 34.3 | + | + | + | + | + |
| D2Mit474 | 81.2 | + | + | + | + | + | |
| 3 | D3Mit329 | 22.6 | + | + | + | + | + |
| D3Mit189 | 101.2 | + | + | + | + | + | |
| 4 | D4Mit259 | 142.4 | + | + | + | + | + |
| 5 | D5Mit225 | 15.1 | + | + | + | + | + |
| D5Mit357 | 76.2 | + | + | + | + | + | |
| 6 | D6Mit122 | 59.4 | + | + | 0 | + | + |
| D6Mit9 | 87.9 | 0 | + | + | + | + | |
| 7 | D7Mit121 | 47.0 | + | + | 0 | + | + |
| D7Mit301 | 80.0 | + | + | + | + | + | |
| 8 | D8Mit64 | 31.6 | + | + | 0 | + | + |
| D8Mit304 | 73.9 | + | + | + | + | + | |
| 9 | D9Mit234 | 62.0 | + | + | + | + | + |
| D9Mit135 | 87.0 | + | + | + | + | + | |
| 10 | D10Mit111 | 65.0 | + | + | + | + | + |
| D10Mit71 | 112.1 | + | + | + | + | + | |
| 11 | D11Mit100 | 111.1 | + | + | + | + | + |
| 12 | D12Mit269 | 16.8 | + | + | + | + | + |
| D12Mit247 | 52.9 | + | + | + | + | + | |
| 13 | D13Mit173 | 12.3 | + | + | + | + | + |
| D13Mit118 | 40.9 | + | + | + | + | + | |
| 14 | D14Mit153 | 47.6 | + | + | + | + | + |
| D14Mit197 | 96.3 | + | + | + | + | + | |
| 15 | D15Mit191 | 9.1 | + | + | + | + | + |
| D15Mit211 | 66.9 | + | + | + | + | + | |
| 16 | D16Mit61 | 48.3 | + | + | + | + | + |
| D16Mit86 | 93.7 | + | + | + | + | + | |
| 17 | D17Mit84 | 47.8 | + | + | + | + | + |
| D17Mit190 | 84.3 | + | 0 | 0 | + | + | |
| 18 | D18Mit131 | 87.4 | 0 | 0 | + | 0 | + |
| D18Mit73 | 48.4 | + | 0 | 0 | + | + | |
| 19 | D19Mit34 | 54.7 | + | 0 | 0 | 0 | + |
| D19Mit118 | 37.8 | + | + | 0 | 0 | + | |
| X | DXMit82 | 34.9 | + | + | 0 | 0 | + |
| DXMit93 | 71.7 | + | + | + | + | + | |
| % of MIT marker positive | 94.7 | 89.5 | 78.9 | 89.5 | |||
| Sum (%) | 100 | ||||||
Chr, chromosome; G-DNA, genomic DNA.
Validation of the 3K Mouse BAC Array
Detection of DNA Copy Number Differences Between Male and Female DNA
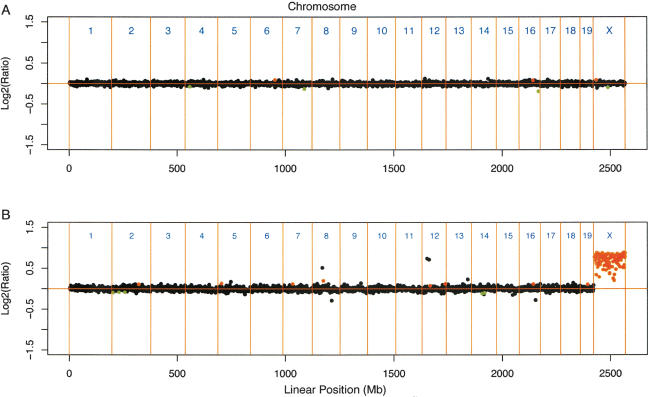
In total, 2819 of the 2825 BAC clones were successfully amplified and spotted to produce a 3K mouse BAC microarray. As a first step, we performed sex-matched normal versus normal hybridizations to determine the hybridization variation in normal genomic DNA (Fig. 2A). We performed all CGH experiments in fluorochrome-reversed pairs of two-color hybridizations. For all data points, we obtained quadruple measurements derived from duplicate spots on arrays from both hybridizations. In order to weigh gains and losses equally, measurements were converted to log2 values. We used the Rosetta Error Model (Hughes et al. 2000) to calculate weighted averages and confidence levels. Statistical analysis of the sex-matched normal versus normal hybridization yielded stable and reproducible ratios for all of the data points (mean log2 ratio = -0.0066; s.d. of the log2 ratios = 0.064). Use of the Rosetta Error Model with a significance threshold set at P < 0.01 yielded seven false-positive data points, which is well below the expected number of 28. The use of lower P-values resulted in a further reduction of false-positives, but also in a concomitant increase in false-negative data points in certain experiments. Therefore, all subsequent analyses were performed with a significance cut-off of P < 0.01.
Figure 2.
Detection of single-copy differences. (A) Normal vs. normal hybridization of spleen DNA from a C57BL/6J female. (B) Comparative genomic hybridization of normal C57BL/6J female DNA vs. normal C57BL/6J male DNA. Plotted are log2-transformed hybridization ratios against the linear map position of the BACs (in Mb). Confidence levels were calculated according to the Rosetta Error Model. Red indicates significant gain in DNA copy number, green indicates significant loss, and black indicates no significant change. The CGH profile of the female vs. male hybridization shows identical DNA copy numbers for all autosomes (mean log2 ratio -0.0066, s.d. of the log2 ratios = 0.064); the copy numbers of male and female chromosome X sequences were clearly different (mean log2 ratio of 0.63).
To test the utility of this mouse array CGH for the detection of single-copy changes, we labeled male and female C57BL/6J mouse genomic DNA with different Cy dyes and hybridized the probe mixture to the BAC microarray (Fig. 2B). Analysis of three independent paired-slide hybridizations identified seven autosomal BACs that showed significantly divergent ratios (gains or losses) in all three female versus male hybridizations. These seven BACs might either contain inserts derived from the sex chromosomes, or autosomal sequences with stronghomology to sex-chromosomal sequences; therefore, they were flagged and excluded from further analyses. Conversely, two X-chromosomal BAC clones consistently showed nonsignificant variations in log2 ratios, suggesting an autosomal BAC insert, an X-chromosomal insert with stronghomology to autosomal sequences, or the presence of repeats that are refractory to blocking with C0t-1 DNA. These BAC clones were permanently excluded from further analysis. The mean log2 ratio of the remaining131 chromosome X clones in the female-versus-male hybridization was 0.63, with log2 ratios ranging from 0.11-0.83.
Detection of Single-Copy Deletions
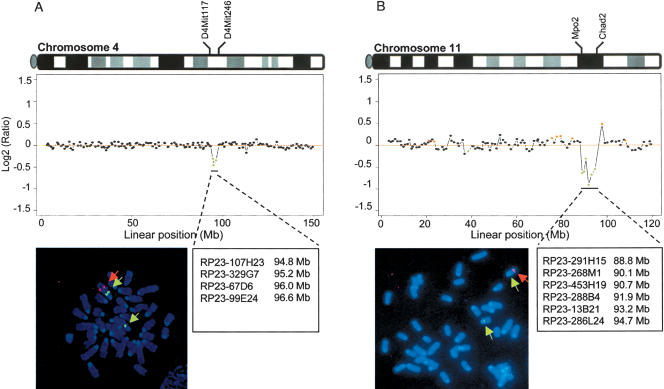
We tested the utility of the 3K mouse BAC array for CGH analysis of single-copy deletions, one of the most frequent chromosomal alterations in malignancies. For this purpose, we performed array CGH with genomic DNA isolated from two genetically engineered mouse embryonic stem (ES) cell lines harboring regional single-copy deletions on chromosome 4 and chromosome 11. The 7A9 ES cell line contains a 2-Mb heterozygous deletion between D4Mit117 (94.6 Mb) and D4Mit246 (96.6 Mb) on chromosome 4, and the 3D5 ES cell line contains a 6.9-Mb heterozygous deletion from Mpo2 (88.5 Mb) to Chad2 (95.4 Mb). As shown in Figure 3, mouse array CGH analysis was able to precisely measure both single-copy deletions. In the case of 7A9 (Fig. 3A), four BAC clones, located between 94.6 and 96.6 Mb on chromosome 4, revealed a single-copy deletion (green points). In the 3D5 clone (Fig. 3B), six BAC clones between 88.5 and 95.4 Mb on chromosome 11 demonstrated a single-copy deletion (green points).
Figure 3.
Detection of regional single-copy deletions. (A) Comparative genomic hybridization of normal AB2.2 ES cell DNA vs. DNA from 7A9 ES cells, harboring a regional single-copy deletion between D4Mit117 (94.6 Mb) and D4Mit246 (96.6 Mb) on chromosome 4. (B) Comparative genomic hybridization of normal AB2.2 ES cell DNA vs. DNA from 3D5 ES cells, harboring a 6.9-Mb single-copy deletion from Mpo2 (88.5 Mb) to Chad2 (95.4 Mb) on chromosome 11. The BAC clones spanning the deletion regions (green dots) are denoted. Both deletions were confirmed by FISH analysis. On prometaphase chromosomes from 7A9 and 3D5 ES cells, only one hybridization signal was observed, with probes corresponding to the deleted region (red signals, indicated by red arrow), whereas two hybridization signals, derived from both alleles, were detected with probes located outside of the deletion regions (green signals, indicated by green arrows).
Detection of Genomic Amplifications
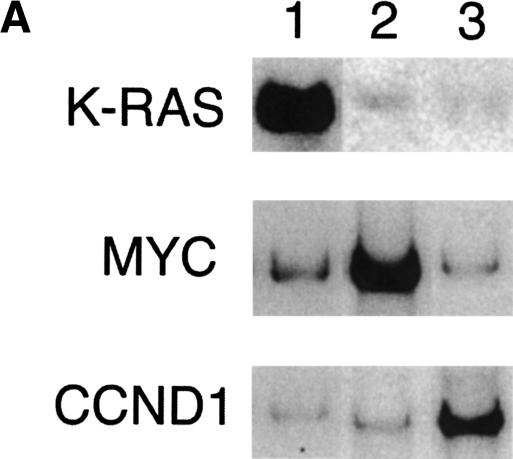
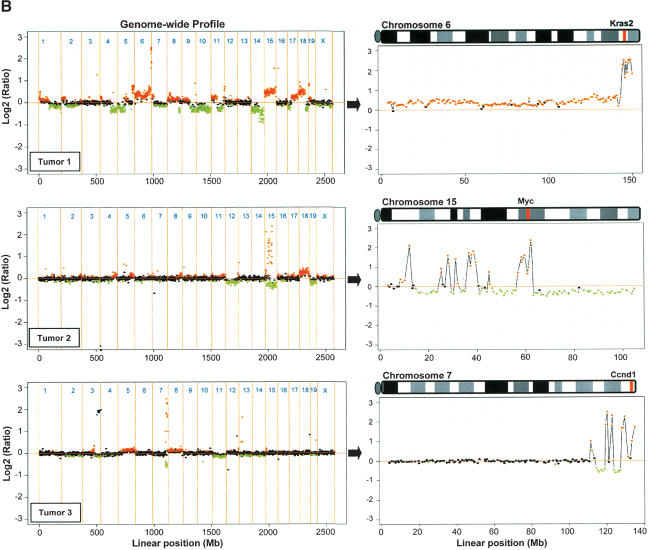
Usingconventional Southern analysis, we previously detected amplification of several oncogenes, including Kras2, Myc, and Ccnd1, in mouse mammary tumors induced by conditional mutation of Brca2 and Trp53 (Fig. 4A). To test whether the 3K mouse BAC microarray can be effectively used to detect regional amplifications, we performed CGH analysis on the individual mouse mammary tumors, usingsplenic DNA from the same animal as a control (Fig. 4B). In mammary tumor 1, amplification of the telomeric end of chromosome 6 (144.7 Mb - end) was detected by array CGH. The Kras2 gene (145.6 Mb) is located within this region. Mammary tumor 2 showed amplification of a chromosome 15 region between 56 and 63 Mb that includes Myc (map position 62.2 Mb). In tumor 3, amplification of Ccnd1 (134 Mb) was clearly detected by array CGH. Interestingly, the chromosomal regions outside the amplification on chromosome 6 were silent in tumor 1, whereas extensive chromosomal rearrangements were observed around the Myc and Ccnd1 amplification in mammary tumors 2 and 3, respectively.
Figure 4.
Array CGH analysis of oncogene amplifications in mouse mammary tumors. (A) Southern analysis of tumor DNA using Kras2, Myc, and Ccnd1 specific probes. Tumors 1, 2, and 3 showed amplification of Kras2, Myc, and Ccnd1, respectively. (B) Whole-genome array CGH profiles (left panels) and CGH profiles of the relevant chromosomes (right panels) from mouse mammary tumors 1, 2, and 3. Plotted are log2-transformed hybridization ratios of tumor DNA vs. control DNA. Vertical red bars on each chromosome indicate the positions of Kras2, Myc, and Ccnd1, respectively.
Detection of Chromosome Instability
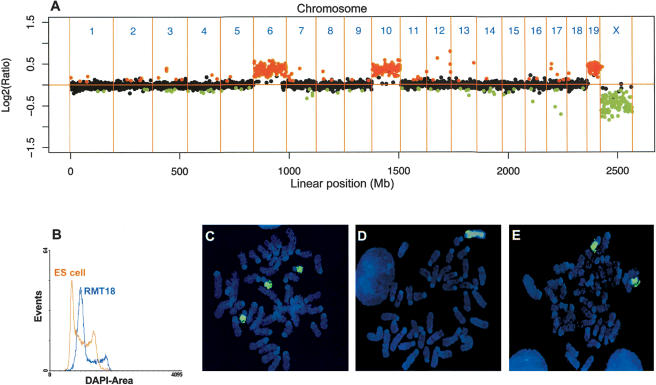
Aneuploidy is another frequently observed phenomenon in tumorigenesis. To test the utility of array CGH for measuring aneuploidy, we performed an array CGH analysis on a female mouse embryonic fibroblast (MEF) cell line that was transformed in vitro by Tbx2, activated K-Ras, and γ-irradiation (RMT18). The resulting CGH profile revealed single-copy changes for chromosomes 6, 10, 19, and X (Fig. 5A). The mean log2 ratios of the trisomic chromosomal regions corresponding to chromosomes 6, 10, and 19 were 0.35, 0.40, and 0.42, respectively, compared to the ideal value of 0.58 for a 3/2 ratio. The mean log2 ratios of the chromosome X monosomy were -0.48, compared to the ideal value of -1.0 for a 1/2 ratio. Subsequent ploidy analysis of the same MEF cell line showed that the RMT18 cells were aneuploid (DNA index = 1.37, Fig. 5B). This finding suggests that most of the chromosomes are diploid and that there is only limited gain of chromosomes in RMT18 cells. We performed chromosome paintingfor chromosomes 19 and X to confirm that the copy number gains and losses detected by array CGH on RMT18 corresponded to chromosomal trisomy and monosomy, respectively (Fig. 5C,D). We also carried out chromosome painting for chromosome 18 to verify the overall diploid status of RMT18 cells (Fig. 5E).
Figure 5.
Array CGH analysis of in vitro transformed RMT18 MEFs derived from a female embryo. (A) CGH profiles of all autosomes and the X-chromosome, showing copy number gains for chromosomes 6, 10, and 19, and copy number loss for chromosome X. (B) Ploidy analysis of RMT18 cells. Normal mouse ES cells were used as a normal diploid control. DNA index is 1.37. (C-E) Chromosome painting of RMT18 metaphase spreads for chromosome 19, X, and 18, respectively.
DISCUSSION
The physical map and the draft sequence of the mouse genome have provided a large collection of mapped and sequenced BAC clones for the construction of DNA microarrays (Gregory et al. 2002; Mouse Genome Sequencing Consortium 2002). Here, we utilized this resource to select and validate a set of 2803 BAC clones with an average spacing of 1 Mb across all mouse autosomes and chromosome X. This BAC clone set was used as a template for the production of a high-density mouse BAC microarray for genome-wide detection of chromosomal imbalances by array CGH.
For the amplification of the individual BAC clones prior to printing, we designed three new DOP-PCR primers that show improved amplification properties compared to the widely used DOP-PCR primer 6MW. DOP-PCR has been designed for the uniform amplification of any target sequence. Nevertheless, the efficiency of the reaction is greatly dependent on the frequency of the 3′ hexanucleotide anchor of the DOP-PCR primer in the template DNA. DOP-PCR amplification will be inefficient if the target sequences are widely separated. Fiegler et al. (2003) utilized this aspect of DOP-PCR to bias amplification specificity, in order to minimize any unwanted co-amplification of E. coli DNA during the DOP-PCR amplification of BAC and PAC clones for human DNA microarrays. We used the same approach to select the sequences for the 3′ hexanucleotide anchors of the primers M-DOP1, M-DOP2, and M-DOP3 so that they were frequently represented in mouse DNA but were rare in E. coli DNA. Accordingly, these new DOP-PCR primers are ineffective in the amplification of contaminating E. coli DNA, but retain their capacity to produce unbiased representations of mouse genomic DNA.
Because the different primers will amplify a different subset of sequences, a combination of the products from different DOP-PCR primers should result in a more universal representation of the target DNA than the DOP-PCR products from individual primers. In keepingwith this notion, in sum all MIT markers we tested were represented in at least one of the DOP-PCR products. Hence, combiningthe three primer products can be expected to normalize sequence amplification biases.
With the 3K mouse BAC arrays, we obtained highly reproducible CGH results over a wide dynamic range. In sex-matched normal versus normal hybridizations, we obtained very reproducible ratios for all clones (s.d. of the log2 ratios = 0.064). In female versus male hybridizations on the 3K mouse BAC array, we found a mean ratio for the chromosome X clones of 0.63, compared to the expected value of 1.0. The underestimation of the chromosome X ratios was reported previously in human array CGH experiments with normal female versus male DNA, and is thought to result from incomplete suppression of repetitive sequences in the X-chromosomal BACs, or from cross-hybridization of autosomal or Y-chromosomal sequences with strong homology to chromosome X (Snijders et al. 2001; Fiegler et al. 2003). Female versus male hybridizations consistently showed aberrant hybridization ratios for seven autosomal and two X-chromosomal BAC clones (0.3%). Extrapolatingautosomal cross-hybridization can be calculated from the ratio of the total length of the euchromatic mouse genome (2.5Gb) and X chromosome (0.15Gb), and we estimate that a maximum of 150 BAC clones might behave incorrectly. These might be incorrectly mapped, chimeric, or display strongcross-hybridization. This number corresponds to 5.3% of the total number of BAC clones, indicatingthat the vast majority of data points on the 3K mouse BAC array are informative.
To test the utility of our BAC arrays for detectingsingle-copy changes, which are commonly observed in most tumors, we used the 3K mouse BAC arrays to obtain CGH measurements in cell lines and primary tumors. We readily detected single-copy losses in ES cells with engineered regional heterozygous deletions. Here, the advantage of array CGH with 1-Mb resolution becomes apparent, as even a small heterozygous deletion of 2 Mb in the 7A9 ES cells was covered by four different BACs on the array.
Usingthe 3K mouse BAC array, we were also able to detect previously identified oncogene amplifications in primary mouse mammary tumors. For the detection of regional amplifications, array-based CGH has unique advantages over conventional CGH or Southern analysis in that it combines high throughput with high resolution. Consequently, complex amplifications as exemplified in mouse mammary tumors 2 and 3 may only be faithfully analyzed by array CGH, because conventional CGH would produce single signals from regions that contain multiple independent amplicons.
The utility of our 3K mouse BAC array for measuringpartial or whole-chromosome aneuploidies was demonstrated by the successful detection of trisomies and monosomy in the RMT18 fibroblast cell line. The copy number changes detected by array CGH were clearly confirmed by chromosome painting. For most autosomal BAC clones, we observed a comparable response of the hybridization ratios to DNA copy number changes, as the standard deviations of the log2 ratios for autosomal clones were 0.082 and 0.102 for two and three copies, respectively. In contrast, the standard deviation of the X-chromosomal log2 ratios increased from 0.045 in sex-matched normal versus normal hybridizations to 0.173 in the case of the RMT18 chromosome X monosomy or in female versus male hybridizations. In line with previous results from human array CGH studies (Snijders et al. 2001), we found the ratio differences between individual chromosome X clones to be quite reproducible (Fig 2B; Y.J. Chung, J. Jonkers, and A. Bradley, unpubl.), suggesting that these differences are related to the sequence characteristics of individual clones, such as repeat content or the presence of sequences with stronghomology to regions outside chromosome X. In this respect it is noteworthy that LINE repeats in both human and mouse show a preference for accumulatingon sex chromosomes, resultingin two- to threefold higher densities of L1 copies in human and mouse chromosomes X compared to the autosomes (Mouse Genome Sequencing Consortium 2002).
We have described the generation of a 3K mouse BAC array for genome-wide analysis of DNA copy number changes with high resolution and high sensitivity. Although mouse BAC arrays have been described previously, they have variable (2-20 Mb) and moderate (3 Mb) resolution (Hodgson et al. 2001; Cai et al. 2002). The design of improved mouse DOP-PCR primers and the use of three admixed DOP-PCR amplification products for the production of the DNA microarrays has led to a significant improvement of both quality and reproducibility of the array CGH data. The high sensitivity of the resulting microarrays allows single-copy gains and losses in cell lines and primary tumors to be detected unambiguously and mapped with 1-Mb resolution. The use of the Rosetta Error Model for computation of weighted averages and confidence levels results in an unprejudiced identification of significant copy number alterations, thus providing extremely reliable CGH data, even in heterogeneous or aneuploid tumor samples. Application of the described methodology to expand the current 1-Mb mouse BAC array towards a full-coverage tile path array will no doubt result in an ultimate resource for diverse biological studies.
METHODS
BAC Clone Selection, BAC DNA Isolation, and Fingerprinting
First, 3080 mouse BAC clones were selected from the RPCI-23 mouse genomic library (Osoegawa et al. 2000) to be spaced at approximately 1-Mb intervals across the whole genome. All clones were screened for T1 phage contamination and characterized by fingerprinting (Marra et al. 1997). In brief, nonphage-contaminated BAC clones were colony-isolated, grown in 96-well deep-well plates (1.5 mL) and 250 μL of culture were pelleted for DNA isolation usingan alkaline lysis method.
Aliquots of the isolated BAC DNA were digested with Hind-III, separated on agarose gels, and stained with Vistra Green. Fingerprint gel images were collected using a Typhoon 8600 Scanner (Molecular Dynamics) and processed using Image software (http://www.sanger.ac.uk/Software/Image). The restriction fragment patterns of the isolated BAC clones were compared with reference fingerprints generated at the University of British Columbia, Vancouver, Canada (http://www.bcgsc.ca/lab/mapping/mouse) using FPC software (Soderlund et al. 2000). The complete 1-Mb BAC clone set and detailed information are available through Ensembl Mouse CytoView (http://www.ensembl.org/Mus_musculus/cytoview).
Design of a Mouse-Specific DOP-PCR Primer Set
We used a custom Perl script to count the occurrence of all possible hexamers in mouse genomic sequence contigs and in the E. coli genome. We selected three hexamers that showed the lowest matchingfrequency in E. coli DNA (<0.02 matches per kilobase) and a high frequency in mouse genomic sequences (>0.33 matches per kilobase). These three hexamers were used as the 3′ anchors in the respective mouse DOP-PCR primers. The 5′ anchor sequences and six nucleotides of random sequences were identical to the previously published 6MW primer (Telenius et al. 1992). The sequences of 6MW and the three mouse-specific DOP-PCR primers are shown in Table 1.
DOP-PCR Amplification of Mouse BAC Clones
Two rounds of PCR were performed to amplify genomic segments and to link amine residues to the amplified DNA. A first-round DOP-PCR was performed with each of the mouse DOP-PCR primers in a 50-μL reaction volume. PCR reactions were performed as described (Fiegler et al. 2003). After checking that DOP-PCR amplification products ranged from 200 bp to 2 Kb, a second-round amine-linking PCR was performed by using5′-aminolinked universal primer (Amine-GGAAACAGCCCGACTC GAG). The 3′ end of this primer matches the conserved 5′ anchor sequences in M-DOP1, 2, and 3. Forty μL of each amine-linked PCR product (M-DOP1, 2, and 3) were combined, and 40 μL of 4X spottingbuffer (1M sodium phosphate buffer, pH 8.5, 0.001% sarcosyl) were added prior to spotting. The final concentration of the PCR products was ∼200 ng/μL.
BAC Array Fabrication
Amine-linked PCR products were spotted in duplicate onto amine-bindingslides (Amersham) usinga MicroGrid II arrayer (BioRobotics). Post-spottingprocessingof the slides was performed accordingto published protocols (http://www.sanger.ac.uk/Projects/Microarrays/arraylab/protocol4).
Genomic DNA Isolation, Labeling, and BAC Array Hybridization
Genomic DNA was extracted and purified by standard methods (Sambrook and Russel 2001). Test DNA and control DNA were digested with HaeIII for 2 h. Digested genomic DNA was labeled with Cy3- or Cy5-dCTP by random priming(Bioprime Labeling Kit, Invitrogen). Briefly, 1 μg of DNA was denatured in random primer solution. To this mixture was added 15 μL 10X dNTP mix (0.5 mM dCTP, 2 mM dATP, 2 mM dTTP, and 2 mM dGTP), 6 μL Cy3- or Cy5-dCTP (1 mM, NEN), 3 μL Klenow enzyme and distilled water to a final volume of 150 μL. After 37°C overnight incubation, labeled genomic DNA was purified through Sephadex G-50 columns and precipitated together with 135 μg of mouse C0t-1 DNA (Invitrogen). The DNA pellet was redissolved with 60 μL of hybridization buffer (50% formamide, 10% dextran sulfate, 0.1% Tween 20, 2x SSC, and 10 mM Tris-HCl, pH 7.4) and 600 μgyeast t-RNA (Invitrogen). Probe DNA was denatured for 10 min at 70°C and incubated for 1 h at 37°C prior to application on the prehybridized BAC microarray slides. Array slides were prehybridized for 1 h at 37°C with 90 μL of denatured hybridization buffer containing540 μgherringsperm DNA and 90 μgof mouse C0t-1 DNA. Hybridization was performed without cover slips, in a humid chamber. In brief, a rubber cement ringwas applied around the microarray. After application of the hybridization solution on the array, slides were incubated in a light tight humid chamber on an orbital shaker (400 rpm) for 48 h at 37°C. Slides were washed serially in solution 1 (PBS containing 0.05% Tween 20) for 10 min at room temperature, in solution 2 (50% formamide, 2X SSC) at 42°C for 30 min, and in solution 1 at room temperature for 10 min. Finally, slides were spin-dried for 3 min at 1000 rpm.
Image Scanning and Data Processing
Hybridized microarrays were scanned using a ScanArray 5000XL scanner (Packard BioChip Technology) and processed using Quantarray software (Packard BioChip Technology) according to the manufacturer's instructions. Fluorescent intensities were normalized per block to the median raw Cy5:Cy3 ratio. All experiments were performed in fluor-reversed pairs of two-color hybridizations. All values were converted to log2 values to weight gains and losses equally. Weighted averages, errors, and confidence levels were computed from quadruple measurements obtained from duplicate spots on arrays from both hybridizations, accordingto the Rosetta Error Model (Hughes et al. 2000, http://download.cell.com/supplementarydata/cell/102/1/109/DC1/ErModlv2.htm). In this model, errors in the measurements of Cy5:Cy3 ratio are estimated usingcontrol experiments where two identical DNA samples are independently labeled with Cy3 and Cy5, respectively. The resultingdistribution of the log2-transformed ratios provides a model for the error distribution. This distribution widens with decreasingintensity, and the error model is designed to fit this intensity dependence closely.
Bioinformatic data analysis was performed usingthe Ensembl mouse genome server (http://www.ensembl.org/Mus_musculus/) and NCBI mouse genome resources (http://www.ncbi.nlm.nih.gov/genome/guide/mouse/).
Cell Lines and Tumor Tissues
Two genetically engineered mouse ES cell lines harboring single-copy deletions on chromosome 4, clone 7A9 (I. Nishijima and A. Bradley, unpubl.), and chromosome 11, clone 3D5 (Y. Yu and A. Bradley, unpubl.) were used to test the utility of the 3K mouse BAC array for the detection of single-copy deletions. A mouse embryonic fibroblast (MEF) cell line that was transformed in vitro by Tbx2, activated K-Ras, and γ-irradiation (RMT18, Y.J. Chungand A. Bradley, unpubl.) was used for simulatingchromosome aneuploidy. Three primary mouse mammary tumors from K14cre;Brca2F11/+;Trp53F2-10/F2-10 females (Jonkers et al. 2001) were used to test the utility of the 3K mouse BAC array for the detection of regional amplifications. Amplification of the Kras2, Myc, and Ccnd1 oncogenes was confirmed by Southern blot analysis with gene-specific probes.
FISH, Chromosome Painting, and DNA Ploidy Analysis
Metaphase spreads for fluorescence in situ hybridization (FISH) analysis were prepared as described (Robertson 1987). RP23-67D6 and RP23-86E8 BAC clones were labeled with Texas red and FITC, respectively, and hybridized to metaphase spreads from ES cell line 7A9. For FISH analysis of 3D5 cells, DNA from BAC clones RP23-257C13 and RP23-276G11 was labeled with Texas red and FITC, respectively. Probe labeling, DNA hybridization, and signal detection were carried out usingstandard methods (Trask 1999). Chromosome paintingwas performed as described previously with minor modifications (Carter et al. 1992). FITC-labeled probes, specific for mouse chromosome 18, 19, and X (Cambio), were hybridized onto metaphase spreads from RMT18 cells. Hybridizations were performed accordingto the manufacturer's protocol. DNA ploidy analysis was performed accordingto published methods (Darzynkiewicz and Juan 1997). In brief, RMT18 cells were trypsinized and fixed with 70% ethanol. Following stainingwith DAPI, ploidy analysis was performed usinga fluorescence-activated flow cytometer (Mo-Flo, DakoCytomation). Normal mouse ES cells were used as a normal diploid control.
MIT Marker PCR
C57BL/6J mouse genomic DNA was amplified with DOP-PCR primers usingthe PCR conditions described above. The PCR product was diluted to 1:100. MIT marker PCR was performed in a final volume of 20 μL containing10 picomole MIT marker pair, 2 μL 10x PCR buffer, 1 unit Taq polymerase (Promega), 0.25 mM dNTPs, and 1 μL diluted PCR product. Reaction conditions were as follows: one cycle at 94°C for 3 min for initial denaturation, 35 cycles at 94°C for 30 sec, 55°C for 1 min, and 72°C for 1 min, and one cycle at 72°C for 2 min. The MIT markers used are listed in Table 2.
Acknowledgments
We thank the Wellcome Trust Sanger Institute Chromosome Mapping Core for providingclones; Seon-Hee Yim for help with the BAC DNA extractions; Wei-Wen Cai and Philippa Carr for help and suggestions with setting up the DOP-PCR amplifications and BAC array hybridizations; Ruby Banerjee and Bee Ling Ngfor FISH, chromosome painting, and ploidy analysis; David Vetrie, Cordelia Langford, and Oliver Dovey for preparation of the microarray slides; Nurcan Yagci for help with the image analyses; Xiaoling Liu for providingthe Southern blottingdata for the Kras2, Myc, and Ccnd1 amplifications; and David Adams and Patrick Biggs for comments on the manuscript. This work was supported by grants from the Wellcome Trust. J.J. is a recipient of an NWO Genomics Fellowship from the Netherlands Organization for Scientific Research.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1878804.
References
- Cai, W.W., Mao, J.H., Chow, C.W., Damani, S., Balmain, A., and Bradley, A. 2002. Genome-wide detection of chromosomal imbalances in tumors using BAC microarrays. Nat. Biotech. 20: 393-396. [DOI] [PubMed] [Google Scholar]
- Carter, N.P., Ferguson-Smith, M.A., Perryman, M.T., Telenius, H., Pelmear, A.H., Leversha, M.A., Glancy, M.T., Wood, S.L., Cook, K., Dyson, H.M., et al. 1992. Reverse chromosome painting: A method for the rapid analysis of aberrant chromosomes in clinical cytogenetics. J. Med. Genet. 29: 299-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz, Z. and Juan, G. 1997. DNA content analysis of fixed cells with DAPI. In Current protocols in cytometry (ed. J.P. Robinson), pp. 7.5.3. J. Wiley, New York.
- Dietrich, W.F., Miller, J.C., Steen, R.G., Merchant, M., Damron, D., Nahf, R., Gross, A., Joyce, D.C., Wessel, M., Dredge, R.D., et al. 1994a. A genetic map of the mouse with 4,006 simple sequence length polymorphisms. Nat. Genet. 7: 220-245. [DOI] [PubMed] [Google Scholar]
- Dietrich, W.F., Radany, E.H., Smith, J.S., Bishop, J.M., Hanahan, D., and Lander, E.S. 1994b. Genome-wide search for loss of heterozygosity in transgenic mouse tumors reveals candidate tumor suppressor genes on chromosomes 9 and 16. Proc. Natl. Acad. Sci. 91: 9451-9455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiegler, H., Carr, P., Douglas, E.J., Burford, D.C., Hunt, S., Scott, C.E., Smith, J., Vetrie, D., Gorman, P., Tomlinson, I.P., et al. 2003. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer 36: 361-374. [DOI] [PubMed] [Google Scholar]
- Gregory, S.G., Sekhon, M., Schein, J., Zhao, S., Osoegawa, K., Scott, C.E., Evans, R.S., Burridge, P.W., Cox, T.V., Fox, C.A., et al. 2002. A physical map of the mouse genome. Nature 418: 743-750. [DOI] [PubMed] [Google Scholar]
- Hodgson, G., Hager, J.H., Volik, S., Hariono, S., Wernick, M., Moore, D., Nowak, N., Albertson, D.G., Pinkel, D., Collins, C., et al. 2001. Genome scanningwith array CGH delineates regional alterations in mouse islet carcinomas. Nat. Genet. 29: 459-464. [DOI] [PubMed] [Google Scholar]
- Hughes, T.R., Marton, M.J., Jones, A.R., Roberts, C.J., Stoughton, R., Armour, C.D., Bennett, H.A., Coffey, E., Dai, H., He, Y.D., et al. 2000. Functional discovery via a compendium of expression profiles. Cell 102: 109-126. [DOI] [PubMed] [Google Scholar]
- Jonkers, J., Meuwissen, R., van der Gulden, H., Peterse, H., van der Valk, M., and Berns, A. 2001. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29: 418-425. [DOI] [PubMed] [Google Scholar]
- Kallioniemi, A., Kallioniemi, O.P., Sudar, D., Rutovitz, D., Gray, J.W., Waldman, F., and Pinkel, D. 1992. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 258: 818-821. [DOI] [PubMed] [Google Scholar]
- Klein, C.A., Schmidt-Kittler, O., Schardt, J.A., Pantel, K., Speicher, M.R., and Riethmuller, G. 1999. Comparative genomic hybridization, loss of heterozygosity, and DNA sequence analysis of single cells. Proc. Natl. Acad. Sci. 96: 4494-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuutila, S., Aalto, Y., Autio, K., Bjorkqvist, A.M., El-Rifai, W., Hemmer, S., Huhta, T., Kettunen, E., Kiuru-Kuhlefelt, S., Larramendy, M.L., et al. 1999. DNA copy number losses in human neoplasms. Am. J. Pathol. 155: 683-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra, M.A., Kucaba, T.A., Dietrich, N.L., Green, E.D., Brownstein, B., Wilson, R.K., McDonald, K.M., Hillier, L.W., McPherson, J.D., and Waterston, R.H. 1997. High throughput fingerprint analysis of large-insert clones. Genome Res. 7: 1072-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. 2002. Initial sequencingand comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Osoegawa, K., Tateno, M., Woon, P.Y., Frengen, E., Mammoser, A.G., Catanese, J.J., Hayashizaki, Y., and de Jong, P.J. 2000. Bacterial artificial chromosome libraries for mouse sequencingand functional analysis. Genome Res. 10: 116-128. [PMC free article] [PubMed] [Google Scholar]
- Pinkel, D., Segraves, R., Sudar, D., Clark, S., Poole, I., Kowbel, D., Collins, C., Kuo, W.L., Chen, C., Zhai, Y., et al. 1998. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 20: 207-211. [DOI] [PubMed] [Google Scholar]
- Ried, T., Heselmeyer-Haddad, K., Blegen, H., Schrock, E., and Auer, G. 1999. Genomic changes defining the genesis, progression, and malignancy potential in solid human tumors: A phenotype/genotype correlation. Genes Chromosomes Cancer 25: 195-204. [DOI] [PubMed] [Google Scholar]
- Robertson, E.J. 1987. In Teratocarcinomas and embryonic stem cells: A practical approach (ed. E.J. Robertson), pp. 71-112. IRL, Oxford, UK.
- Sambrook, J. and Russel, D.W. 2001. Molecular cloning: A laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Snijders, A.M., Nowak, N., Segraves, R., Blackwood, S., Brown, N., Conroy, J., Hamilton, G., Hindle, A.K., Huey, B., Kimura, K., et al. 2001. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat. Genet. 29: 263-264. [DOI] [PubMed] [Google Scholar]
- Soderlund, C., Humphray, S., Dunham, A., and French, L. 2000. Contigs built with fingerprints, markers, and FPC V4.7. Genome Res. 10: 1772-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinas-Toldo, S., Lampel, S., Stilgenbauer, S., Nickolenko, J., Benner, A., Dohner, H., Cremer, T., and Lichter, P. 1997. Matrix-based comparative genomic hybridization: Biochips to screen for genomic imbalances. Genes Chromosomes Cancer 20: 399-407. [PubMed] [Google Scholar]
- Telenius, H., Carter, N.P., Bebb, C.E., Nordenskjold, M., Ponder, B.A., and Tunnacliffe, A. 1992. Degenerate oligonucleotide-primed PCR: General amplification of target DNA by a single degenerate primer. Genomics 13: 718-725. [DOI] [PubMed] [Google Scholar]
- Trask, B.J. 1999. In Genome analysis: A laboratory manual, Vol. 4: Mapping genomes (eds. B. Birren et al.), pp. 303-413. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Zhao, S., Shatsman, S., Ayodeji, B., Geer, K., Tsegaye, G., Krol, M., Gebregeorgis, E., Shvartsbeyn, A., Russell, D., Overton, L., et al. 2001. Mouse BAC ends quality assessment and sequence analyses. Genome Res. 11: 1736-1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.ensembl.org/mus_musculus/cytoview; map positions of 1-Mb mouse BAC clone set.
- http://www.sanger.ac.uk/Software/Image; fingerprint gel image analysis software.
- http://www.bcgsc.ca/lab/mapping/mouse; mouse BAC fingerprints reference database.
- http://www.sanger.ac.uk/Projects/Microarrays/arraylab/protocol4; microarray spotting and post-spotting processing protocols.
- http://download.cell.com/supplementarydata/cell/102/1/109/DC1/ErModlv2.htm; Rosetta error model.
- http://www.ensembl.org/Mus_musculus/; Ensembl mouse genome resources.
- http://www.ncbi.nlm.nih.gov/genome/guide/mouse/; NCBI mouse genome resources.