Abstract
Evolutionary sequence conservation of non-coding DNA can be indicative of functional conservation and has enabled major insights into the regulatory architecture of mammalian genomes. However, a new study in this issue cautions against a generalization of this notion by demonstrating notable differences between human and mouse stem cell regulatory networks, in part due to binding sites derived from lineage-specific transposable elements.
Deciphering the gene regulatory architecture embedded in mammalian genomes remains a pressing problem since it is an essential prerequisite for understanding the role of regulatory sequences in human biology and disease. Cross-species sequence comparisons have proven successful for identifying core sets of gene regulatory elements by means of their evolutionary conservation. In contrast, conservation-based approaches have severe limitations in the exploration of species-specific changes in gene regulatory architecture. In this issue of Nature Genetics, Kunarso et al. 1 set out to overcome this challenge using an elegant series of experiments through which they compared the functional, rather than the sequence conservation of gene regulatory sites between the human and the mouse genome in embryonic stem (ES) cells. Remarkably, they find that the genomic locations of binding sites for some key regulatory proteins are poorly conserved across species, despite their widely perceived fundamental importance in mammalian ES cell biology.
Functional Divergence Between Species
To date, the vast majority of studies exploring the question of functional conservation across species have focused on experimental data sets from only one species, followed by their post-hoc comparative genomic analysis to infer degrees of DNA conservation across species 2-5. These indirect studies have shown that some molecular marks associated with regulatory sequences tend to be found at sites whose sequence is highly conserved across species 5 whereas others tend to be found at sites with little or no sequence conservation 4. It is therefore a particular strength of the new study by Kunarso et al. to tackle this problem by obtaining genome-wide experimental data from both human and mouse by identical methodology.
To compare the genome-wide binding profiles of regulatory proteins between species, the authors performed ChIP-seq for three well-studied regulatory proteins (OCT4, NANOG, and CTCF) from human and mouse ES cells. OCT4 (also known as POU5F1) and NANOG are transcription factors that play major roles in maintenance of ES cell pluripotency, whereas the CTCF protein is associated with genomic insulator elements that prevent enhancer-promoter interactions. Unexpectedly, only ~5% of binding sites for the two transcription factors, OCT4 and NANOG, were found in orthologous positions in human and mouse ES cells, supporting major differences in genome-wide binding between species. While subsets of these differences may be due to technical limitations of the approach, analysis of CTCF binding sites by identical methods revealed that, depending on statistical stringency, up to 50% of binding sites are functionally conserved between mouse and human. It is therefore reasonable to assume that the genome-wide binding profiles of OCT4 and NANOG in ES cells have substantially changed during the 75 million years of evolution that separate the two species from their last common ancestor.
Transposon-Mediated Rewiring
The marked changes in the genome-wide binding profiles of OCT4 and NANOG in human compared to mouse ES cells raise two important questions: 1. What molecular mechanisms have brought forth these changes and 2. do these observed changes functionally affect the transcriptional landscape of human and mouse ES cells?
To answer the first question, the authors examined the evolutionary origins of the sequences in which experimentally identified binding sites were located. Consistent with previous observations of regulatory sequences that arose through exaptation from transposable elements (TEs) 6-8, between 10% and 30% of binding sites overlapped repeat elements (RABS, repeat-associated binding sites). However, many of these RABS were found in lineage-specific repeat elements that are absent in the respective other species, raising the intriguing possibility that large numbers of binding sites arose more recently in evolution and may have rewired the regulatory architecture in ES cells on a substantial scale.
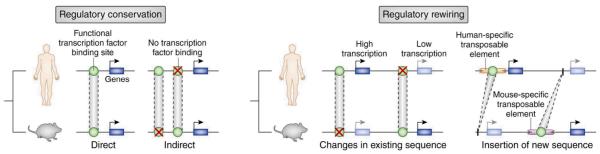
To examine the second question, the authors quantified the impact and relative contributions of different modes of regulatory conservation and rewiring (Figure 1). For this purpose, they obtained transcriptome-wide expression data from normal human ES cells, as well as from ES cells that had been depleted of OCT4 by RNAi and compared these results to equivalent data from mouse ES cells. Overall, the genomic location of OCT4 binding sites correlated with the location of genes that were down-regulated upon OCT4 depletion. However, among genes whose OCT4-dependence was conserved between human and mouse, the majority of identified OCT4 binding sites was not directly, but rather indirectly conserved, i.e. disappearance of a binding site was compensated for by nearby emergence of a new binding site for the same transcription factor (Figure 1, left). Moreover, the authors identified 50 cases in which human-specific OCT4-regulation could be directly linked to RABS, i.e. cases of regulatory repeat-associated rewiring in human compared to mouse ES cells (Figure 1, right).
Figure 1. Modes of regulatory conservation and rewiring.
Emerging and disappearing binding sites for a given transcription factor can have different effects on the transcriptional output of individual genes, as shown schematically for some possible modes of conservation and rewiring. Transcriptional output is preserved through direct sequence conservation between species (left panel, left), but indirect conservation, where one binding site is created while another is destroyed, is also possible (left panel, right). In contrast, transcriptional output is altered when binding sites are destroyed or created without compensatory nearby changes (right panel, left) or when new binding sites are created by insertion and exaptation of transposable elements (right panel, right).
Taken together, the study by Kunarso et al. 1 provides evidence that significant differences in the transcriptome of ES cells between human and mouse are caused by a considerable divergence in genome-wide binding profiles of major ES cell transcription factors. The work also provides direct insights into the unexpectedly large role that local binding site turnover, as well as RABS play in the conservation and rewiring of mammalian regulatory networks. Sequence conservation has been useful as a predictor of functional regulatory elements in the genome 2,9 but the observations of Kunarso et al. 1 serve as another important reminder that it is not justified to assume in turn that all functional regulatory elements show evidence of sequence constraint. The functional relevance of the new human-specific OCT4 target genes identified through this work will remain to be determined, but provides important leads for future studies. It is noteworthy that for OCT4 and NANOG relatively strong differences were observed between human and mouse ES cells, whereas binding of CTCF was highly conserved. Thus, it is expected that additional DNA-binding proteins and chromatin marks not examined in the present study will fall into a spectrum from strong to weak similarity between these two species. Indeed, a recent work comparing binding sites in liver tissue across five vertebrate species supports the notion that genome-wide occupancy profiles of some transcription factors have undergone substantial turnover in evolution 10. The insights from these studies highlight the need to assess such inter-species similarities and differences carefully by experimental approaches in order to evaluate the potential of data derived from the mouse or any other model organism for the functional annotation of the human genome.
References
- 1.Kunarso G, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010 doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 2.Cooper GM, Brown CD. Qualifying the relationship between sequence conservation and molecular function. Genome Res. 2008;18:201–5. doi: 10.1101/gr.7205808. [DOI] [PubMed] [Google Scholar]
- 3.King DC, et al. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res. 2005;15:1051–60. doi: 10.1101/gr.3642605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.ENCODE Project Consortium et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–8. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourque G, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–62. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang T, et al. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci U S A. 2007;104:18613–8. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bejerano G, et al. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 9.Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt D, et al. Five-Vertebrate ChIP-seq Reveals the Evolutionary Dynamics of Transcription Factor Binding. Science. 2010 doi: 10.1126/science.1186176. E-pub April 8, 2010, DOI: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]