Abstract
Background and Aims
When rice seeds germinate under complete submergence, only the coleoptile elongates efficiently. It has been reported previously that coleoptile elongation is reduced in the rice alcohol dehydrogenase 1 (ADH1)-deficient mutant, reduced adh activity (rad). The aim of this study was to elucidate how expressions of genes responsible for coleoptile elongation are affected by the ADH1 deficiency in the rad mutant under submergence.
Methods
To identify genes whose expressions are changed in the rad coleoptile at an early stage in germination (i.e. 1 d after imbibition), coleoptiles of the rad mutant and its wild type (WT) were isolated by laser microdissection, and their mRNA levels were examined with a microarray.
Key Results
The microarray analysis identified 431 genes whose transcript levels were different between rad and WT. Interestingly, among the down-regulated genes in the rad coleoptile, 17·5 % were cell division-related genes and 5·1 % were cell elongation-related genes. It was found that cell division started at 1 d after imbibition and then gradually ceased, whereas in the WT coleoptile cell elongation started between 1 d and 2 d after imbibition and then continued. However, neither cell division nor cell elongation actively occurred in the rad coleoptile, in which the amounts of ATP were reduced.
Conclusions
These results indicate that cell division, as well as cell elongation, occur during coleoptile elongation in rice under complete submergence, and that the reduced ATP levels caused by the ADH1 deficiency repress both of them, thereby impairing coleoptile elongation in the rad mutant under submerged conditions.
Keywords: Cell division, cell elongation, rice, Oryza sativa, laser microdissection, coleoptile, microarray, alcohol dehydrogenase 1, ADH1, reduced adh activity, rad
INTRODUCTION
Rice (Oryza sativa) can germinate under complete submergence, whereas other cereal crops such as wheat, barley, sorghum and maize cannot. In anaerobically germinated rice seedlings, the coleoptile and mesocotyl rapidly elongate, whereas the root does not (Magneschi and Perata, 2009). When the elongated coleoptile reaches the water surface, aerenchyma is formed in the coleoptile, thereby supplying oxygen to leaves and root (Alpi and Beevers, 1983; Kawai and Uchimiya, 2000). Coleoptile elongation is thus a survival strategy under submergence.
When a rice seed germinates under water, sugars derived from starch hydrolysis in the endosperm move to the embryo where they are metabolized through glycolysis to produce energy (i.e. ATP) and other important metabolites (Perata and Alpi, 1993; Gibbs and Greenway, 2003; Magneschi and Perata, 2009). Anaerobic metabolic pathways such as glycolysis and fermentation are required for coleoptile elongation (Setter and Ella, 1994; Kato-Noguchi, 2001; Kato-Noguchi and Kugimiya, 2003). Recently, Lee et al. (2009) reported that rice CIPK15 (calcineurin B-like-interacting protein kinase 15) regulated carbohydrate metabolism and fermentation through a sugar signalling pathway during germination under submergence. Thus, coleoptile elongation was suppressed in a cipk15 knockout mutant under submergence, and was rescued by adding external sucrose (Lee et al., 2009).
Alcoholic fermentation [catalysed by pyruvate decarboxylase and alcohol dehydrogenase (ADH)] and lactate fermentation (catalysed by lactate dehydrogenase) support glycolysis and ATP synthesis by recycling NAD+ (Bailey-Serres and Voesenek, 2008). In a study of hypoxic rice coleoptiles, about 92 % of the carbon flux from pyruvate went to ethanol, 7 % went to alanine and 1 % went to lactate (Kato-Noguchi, 2006), suggesting that alcoholic fermentation is a more important source of NAD+ for glycolysis than lactate fermentation (Kato-Noguchi, 2006; Magneschi and Perata, 2009). Indeed, reduction of ADH activity in rice coleoptiles slows down their elongation under submergence (Matsumura et al., 1995, 1998; Rahman et al., 2001; Saika et al., 2006; see Fig. 1).
Coleoptile length is mainly increased by cell elongation (Wada, 1961; Opik, 1973; Atwell et al., 1982; Magneschi and Perata, 2009). Expansins probably have a major role in coleoptile cell elongation under anaerobic conditions (Huang et al., 2000; Choi et al., 2003; Magneschi et al., 2009). Expansins are classified into two groups, α-expansins and β-expansins, both of which are encoded by large gene families (Lee and Kende, 2001, 2002; Shin et al., 2005). Huang et al. (2000) reported that rice α-expansin genes, Os-EXP2 (EXPA2) and Os-EXP4 (EXPA4), were highly expressed in the submerged coleoptile, whereas their mRNAs were weakly detected in the coleoptile under aerobic or anoxic conditions. Additionally, coleoptile and mesocotyl lengths depended on the transcription level of Os-EXP4. Elongations of the coleoptile and mesocotyl were enhanced in Os-EXP4 over-expressing plants and repressed in Os-EXP4-antisense plants (Choi et al., 2003). Transcriptome analysis of an anoxic rice coleoptile recently revealed that EXPA7 and EXPB12 were up-regulated in the rice coleoptile under anoxic conditions (Lasanthi-Kudahettige et al., 2007). These results suggest that some expansins contribute to coleoptile elongation in rice under hypoxia or anoxia. Genes involved in carbohydrate and lipid metabolism and genes encoding transcription factors and heat-shock proteins have also been found to be up-regulated or down-regulated in rice embryos and coleoptiles under anaerobic conditions (Lasanthi-Kudahettige et al., 2007; Narsai et al., 2009), suggesting that their gene products are involved in anaerobic growth (i.e. coleoptile elongation) of rice seedlings. Coleoptile length also appears to also be increased partly by cell division because the number of cells in the coleoptile increased during germination under submerged conditions (Wada, 1961; Opik, 1973; Atwell et al., 1982). However, the results of these earlier studies have not yet been confirmed by modern molecular methods.
It has been reported previously that in the rice reduced adh activity (rad) mutant, the amount of ADH protein was greatly reduced because of a point mutation in the ADH1 gene (Matsumura et al., 1995, 1998; Saika et al., 2006). Coleoptile elongation in the rad mutant was reduced under submergence (Matsumura et al., 1995), but so far it remains unclear how the ADH1 deficiency affects coleoptile elongation in the rad mutant under submergence. Answering this question would help to elucidate the elongation mechanism in wild-type (WT) rice.
In this study, coleoptiles were isolated from rad and WT embryos at an early stage in germination under submergence (before the start of coleoptile elongation) using laser microdissection (LM) (Nakazono et al., 2003; Nelson et al., 2006), and then their mRNA levels were examined with a microarray and semi-quantitative RT-PCR. As a result, many down-regulated genes related to cell division as well as cell elongation in the rad coleoptile were identified. To test the idea that cell division-related genes are at least partly responsible for the reduced elongation of rad coleoptile, the expressions of several of these genes, the mitotic index and 5′-bromo-2′-deoxyuridine (BrdU) incorporation in the WT and rad coleoptiles were investigated during germination and coleoptile elongation under submergence. The results show that cell division in the WT is active at the early stage of germination (i.e. 1–2 d after imbibition) and that during this period cell division in the rad coleoptile is much reduced.
MATERIALS AND METHODS
Plant materials and growth conditions
The rice (Oryza sativa L.) rad mutant and its WT (‘Kinmaze’) were used in this study. Dehulled caryopses (seeds) of rice were sterilized in a 0·6 % (v/v) sodium hypochlorite solution for 30 min. After washing with deionized water five times, 15 seeds were placed on the bottom of 1-L glass bottle filled with 1 L of deionized water. The seeds germinated and were grown under complete submerged or aerobic conditions in darkness at 28 °C. For the time-course experiments, seeds were placed in separate glass bottles for each time point and then harvested as seedlings. Dry seeds were used at 0 d after imbibition.
Extraction and quantification of ATP
ATP was extracted from freeze-dried rice coleoptiles using the luciferin–luciferase ATP assay system (Toyo Ink, Tokyo, Japan) according to the manufacturer's protocol. ATP amounts were determined by recording the relative light intensity for 10 s using a Junior LB9509 luminometer (Berthold Technologies, Bertech, Germany).
Preparation for frozen tissue sections
Frozen tissue sections were prepared following Nakazono et al. (2003) with minor modifications. Rice seeds or seedlings were fixed by a 5-min infiltration of 75 % (v/v) ethanol/25 % (v/v) acetic acid into the tissues under vacuum on ice. This fixation was repeated three times by replacing with new fixative solution and then the tissues were kept in fixative at 4 °C overnight. The fixed tissues were transferred to 10 % sucrose solution, which was prepared with PBS buffer (137 mm NaCl, 8·01 mm Na2HPO4, 2·68 mm KCl and 1·47 mm KH2PO4, pH 7·3). The sucrose solution infiltrated to the tissues under vacuum on ice for 5 min and the vials were kept at 4 °C overnight. The fixed tissues were embedded in RNase-free water, frozen in hexane cooled with dry ice, and stored at –80 °C. The tissues were sectioned into 8-μm-thick slices in a cryostat (Leica CM1850, Leica Microsystems, Wetzlar, Germany) by the method of Ishimaru et al. (2007). The frozen sections were dehydrated in 100 % ethanol for 10 s twice and dried at room temperature for 10 min.
Laser microdissection
The coleoptiles were cut from the frozen sections by UV laser (wavelength of 337 nm) using Leica AS LMD (Leica Microsystems), and then the cut coleoptile sections were dropped by gravity and were collected in a 0·5-mL tube cap containing mineral oil.
Extraction and quantification of total RNA and assessment of RNA quality
Total RNA was extracted from LM-isolated coleoptile sections (10–20 sections for each experiment) with a PicoPure™ RNA isolation kit (Molecular Devices, Toronto, Ontario, Canada) according to the manufacturer's protocol. The extracted total RNA was quantified with a Quant-iT™ RiboGreen RNA reagent and kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. The quality of total RNA extracted from LM-collected tissues was assessed using an RNA 6000 Pico kit on the Agilent 2100 bioanalyzer (Agilent Technologies Inc., Santa Clara, CA, USA). RNA integrity was judged by RNA Integrity Number (RIN), which was calculated with 2100 Expert Software (Agilent, version B.02·02, eukaryote total RNA pico mode), and total RNA, whose RIN is >6·0, was used for further analyses.
Microarray experiment
Aliquots of total RNAs (100 ng each) were labelled with a Quick Amp labelling kit (Agilent Technologies) according to the manufacturer's instructions. Aliquots of Cy5-labelled and Cy3-labelled cRNAs were used for hybridization in a rice 22K oligo-DNA microarray (Agilent Technologies). The array has 21 938 60-mer oligo probes to rice genes. Three biological replicates and a colour swap for each replicate were analysed. The hybridized slides were scanned using a DNA microarray scanner G2505B (Agilent Technologies), and signal intensities were extracted by Feature Extraction software (Version 8·5·1·1; Agilent Technologies). A complete set of microarray data was deposited with the Gene Expression Omnibus (GEO) repository under accession number GSE26632.
Microarray data analysis
Microarray signal intensities were digitized and log ratio or P-values were obtained by Feature Extraction software version 8·5·1·1 (Agilent Technologies). rad mutant genes that had >2·5- or <0·4-fold change in signal intensity and P-values <0·01 in all three replications and in each colour swap were identified. The fold change of each probe was calculated using the average of three replications and each colour swap. Gene descriptions in the 22K microarray were replaced with new descriptions based on RAP-DB (Build 5, http://rapdb.dna.affrc.go.jp/).
cDNA synthesis and amplification
A 1-ng aliquot of total RNA was used for cDNA synthesis and amplification. Reactions were performed by using a WT-Ovation™ RNA amplification system (NuGEN Technologies, San Carlos, CA, USA) according to the manufacturer's instructions.
Semi-quantitative RT-PCR
Semi-quantitative RT-PCR analysis was performed to examine the expression pattern of selected genes identified by the microarray analysis during germination. AmpliTaq® 360 (Applied Biosystems, Foster City, CA, USA) was used for subsequent PCR amplification with appropriate primers (Supplementary Data Table S3, available online): initial denaturation (95 °C for 10 min) and 22–37 cycles of denaturation (95 °C for 30 s), annealing (55 °C for 30 s), extension (72 °C for 30 s), and final extension (72 °C for 10 min).
Measurement of longitudinal cell length and counting cells in mitotic phase
Rice seeds or seedlings were fixed by a 5-min infiltration of 4 % (w/v) paraformaldehyde in PBS buffer (pH 7·4) into the tissues under vacuum on ice. This fixation was repeated three times by replacing with new fixative solution and then the tissues were kept in fixative at 4 °C overnight. After fixation, embryos were separated from seeds and were embedded in paraffin by the microwave method reported by Takahashi et al. (2010)
Serial longitudinal sections of the paraffin-embedded embryos, 6 µm thick, were cut, floated on water on glass slides, and incubated at 42 °C for 10 min. Subsequently, the solution was removed with a micropipette, and then sections were dried at 42 °C overnight. To remove paraffin, slides were gently immersed in xylene for 15 min twice, and then immersed in 50 % xylene/50 % ethanol for 15 min. The deparaffinized slides were rinsed with 100 % ethanol for 1 min, followed by acclimation steps of 1 min each in 90 % ethanol, 80 % ethanol, 70 % ethanol, 50 % ethanol and water for 1 min. Finally, slides were washed with PBS buffer three times. DNA in the nuclei on the sections were stained with 2·5 µg mL−1 DAPI (4',6-diamidino-2-phenylindole) solution [containing 1 % (v/v) Triton X-100 in PBS buffer] for 20 min. After staining, slides were washed with PBS buffer for 5 min.
Longitudinal cell length was measured and the numbers of cells in the mitotic phase were counted under a microscope. The results were expressed as the fraction of cells in the mitotic phase (the mitotic index).
Incorporation of BrdU and immunostaining
Labelling and immunostaining of BrdU were performed using a 5′-bromo-2′-deoxyuridine labelling and detection kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's protocol with minor modifications. WT and rad mutant seeds germinated in 10 µm BrdU and 1 µm 5-fluorodeoxyuridine solution under darkness at 28 °C for 1 d, and then were fixed with 4 % (v/v) paraformaldehyde solution. After fixation, embryos were separated from seeds. The fixed embryos were embedded in paraffin and sections were prepared by the microwave method reported by Takahashi et al. (2010).
The immunostained sections were further stained with DAPI solution [containing 1 % (v/v) Triton X-100 in PBS buffer] for 20 min. After staining, slides were washed with PBS buffer for 5 min. BrdU and DAPI signals were observed under a confocal laser scanning microscope (Nikon, Tokyo, Japan).
RESULTS
When seeds of the rad mutant and its WT (‘Kinmaze’) germinated completely submerged, the WT coleoptiles began to elongate between 1 d and 2 d after imbibition and reached 51·2–67·4 mm at 5–7 d after imbibition (Fig. 1A, B). On the other hand, coleoptiles of the rad mutant elongated more slowly, reaching only 15·4 ± 8·9 mm at 7 d after imbibition (Fig. 1B). Under aerobic conditions, the elongation patterns of WT and rad coleoptiles were comparable (Fig. 1C). Under submerged conditions, the amount of ATP in the rad coleoptiles at 1 d after imbibition was only about 37 % of that in the WT (Fig. 1D), which is probably the reason for its slow elongation.
Fig. 1.
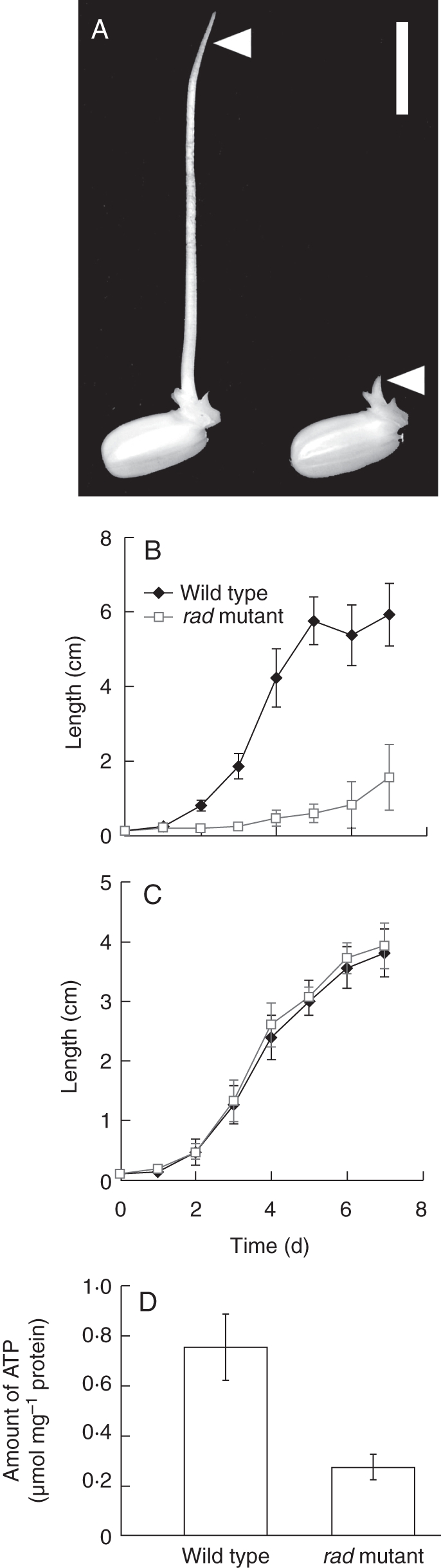
Comparison of WT and rad mutant rice coleoptiles. (A) WT and rad mutant seeds were germinated for 5 d under complete submergence. Left: WT; right: rad mutant. Arrowheads indicate the coleoptiles. Scale bar = 1 cm. (B, C) Growth of coleoptiles under submerged (B) and aerobic (C) conditions. Data are means of 15 replicates ± s.d. (D) ATP concentrations in coleoptiles under submerged conditions at 1 d after imbibition. Data are means of three replicates.
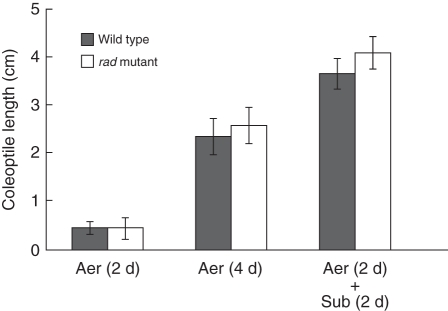
Next, we examined whether rad coleoptiles can actively elongate under submergence once coleoptile elongation, which occurs by both cell division and cell elongation, starts under aerobic conditions. Seeds were germinated and seedlings grown under aerobic conditions for 2 d. Then the seedlings, whose coleoptiles started elongation (Fig. 1C), were completely submerged for 2 d. Interestingly, the WT and rad coleoptiles elongated equally (Fig. 2).
Fig. 2.
Effect of switching from aerobic to submerged conditions on coleoptile elongation in WT and rad mutant. Aerobic growth for 2 d followed by submerged growth for 2 d. Control seedlings were grown under aerobic conditions for 4 d. Data are means of eight replicates ± s.d.
To identify which genes are involved in coleoptile elongation during submergence, RNA samples were prepared on day 1 (1 d after imbibition) when the coleoptile lengths of the rad mutant (1·7 ± 0·3 mm) and the WT (2·3 ± 0·3 mm) were just beginning to diverge (Fig. 1B). The seeds which imbibed under submergence for 1 d were fixed, and frozen tissue sections were prepared for LM. Coleoptile cross-sections were collected from the tissue sections via LM (Fig. 3). The RNA samples extracted from the LM-isolated coleoptiles were labelled with Cy3 or Cy5 dye, and the labelled cDNA from each of three biological replications was hybridized to rice oligo-microarrays. Genes were selected whose intensities differed by a factor of at least 2·5 in the rad and WT coleoptiles. After submergence, 294 genes were up-regulated in the rad coleoptile compared with the WT (Supplementary Data Table S1, available online) and 137 genes were down-regulated (Supplementary Data Table S2).
Fig. 3.
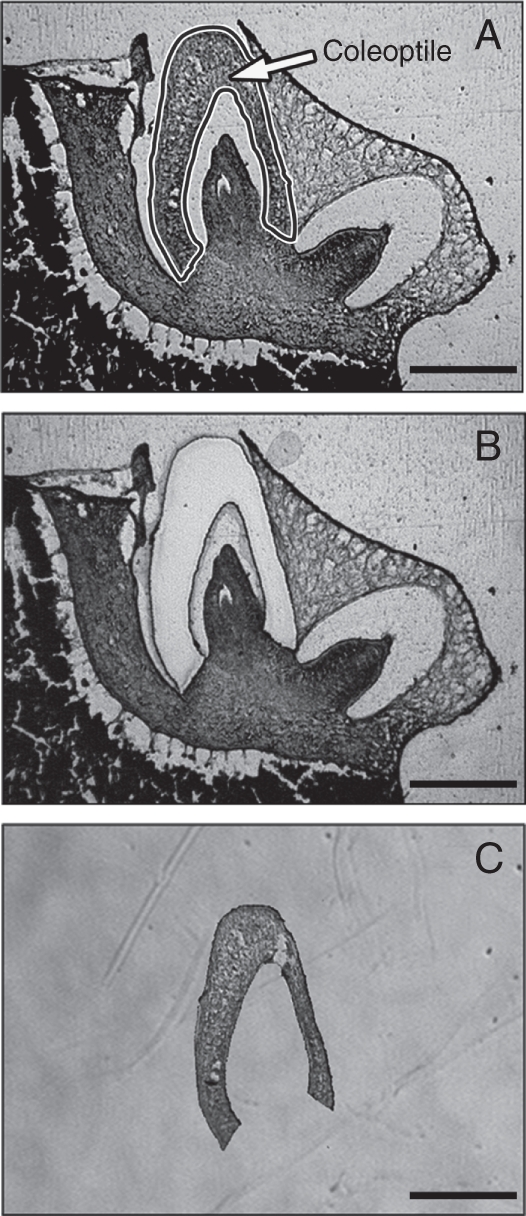
Collection of a vertical coleoptile section from a rice embryo using LM. (A) An 8-μm-thick frozen section of a rice embryo before LM. The area enclosed with a black line (i.e. the coleoptile) was cut by LM and the cut coleoptile was collected. (B) The same frozen section after LM. (C) LM-isolated coleoptile. Scale bars = 500 µm.
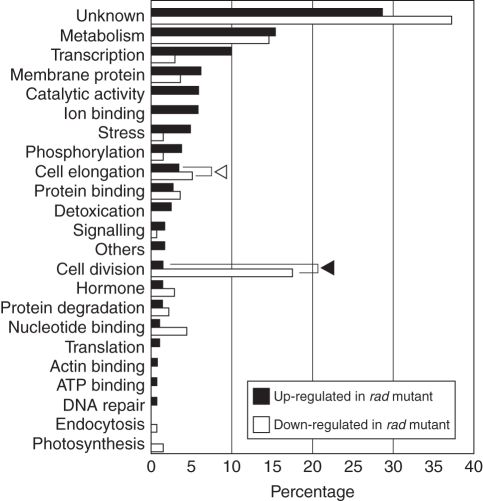
These genes were classified by function in Fig. 4. What stands out in this figure is that most of the genes involved in cell division and some of the genes involved in cell elongation (arrowheads in Fig. 4) were less expressed in the rad mutant, which suggests that the down-regulation of these genes is responsible for the slow elongation of the rad coleoptile under submergence. Thus, we examined whether cell division, as well as cell elongation, occurred during coleoptile growth in rice when it is submerged.
Fig. 4.
Percentages of up-regulated and down-regulated genes in the rad mutant that are involved in different cellular functions. Up-regulated genes were those whose expressions were >2·5-fold greater in the rad mutant than in the WT; down-regulated genes were those whose expressions were >2·5 fold greater in the WT than in the rad mutant (P < 0·01). The white arrowhead indicates cell elongation-related genes, the black arrowhead cell division-related genes.
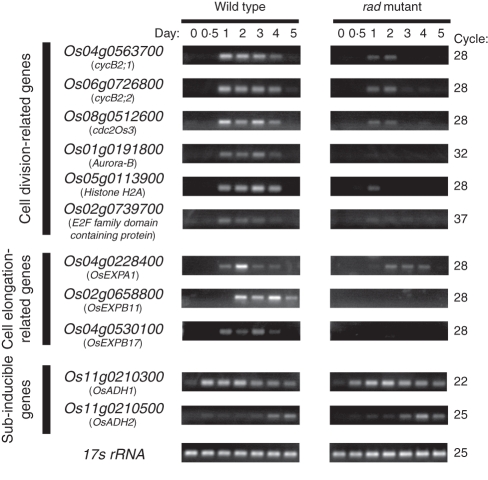
To confirm that the cell division-related genes are less expressed in the rad coleoptile than in the WT coleoptile, transcript levels of some cell division-related genes in the WT and rad coleoptiles were measured during germination and early growth of seedlings under submergence by semi-quantitative RT-PCR. The cell-division- and cell-elongation-related genes making up the bars in Fig. 4 and their rad/WT expression ratios are listed in Table 1. Six of these genes were selected for the semi-quantitative RT-PCR analyses: three G2- and M-phase-expressed genes (cycB2;1, cycB2;2 and cdc2Os3) (Umeda et al., 1999a, b; Lee et al., 2003), one M-phase-expressed gene (Aurora-B), one S-phase-expressed gene (Histone H2A) and one G1- and S-phase-expressed gene (E2F family domain-containing protein). In the WT coleoptile, the transcript levels dramatically increased at 1 d after imbibition and then gradually decreased, whereas in the rad coleoptile, the induction levels were much lower (Fig. 5).
Table 1.
List of genes related to cell division, whose expression was up-regulated or down-regulated in the rad coleoptiles
| Fold change (rad/WT) * | Locus ID† | Full-length cDNA accession no.† | Description† | Functional classification |
|---|---|---|---|---|
| 4·77 | Os03g0107700 | AK063743 | Similar to EL2 protein | Cell cycle |
| 3·14 | Os12g0147800 | AK059860 | Similar to phytosulfokines 5 precursor (secretory protein SH27A) | Cell proliferation |
| 3·04 | Os07g0596600 | AK067238 | Similar to Cdc2MsC protein | Cell cycle |
| 2·66 | Os03g0285800 | AK067339 | MAP kinase. (OsMAPK2) | Phosphorylation |
| 0·18 | Os01g0191800 | AK102360 | Similar to serine/threonine-protein kinase 12 (EC 2·7·1·37) (Aurora-B) (fragment) | Cell cycle |
| 0·20 | Os04g0563700 | AK070211 | Cyclin (cycB2;1) | Cell cycle |
| 0·22 | Os06g0726800 | AK070518 | G2/mitotic-specific cyclin 2 (B-like cyclin) (CycOs2) (cycB2;2) | Cell cycle |
| 0·24 | Os04g0375900 | AK101769 | Kinesin, motor region domain-containing protein | Motor protein |
| 0·25 | Os02g0699700 | AK072471 | Similar to DNA topoisomerase II | DNA replication |
| 0·26 | Os08g0421800 | AK103087 | Similar to mitogen-activated protein kinase kinase kinase 1 (EC 2·7·1.–) (arabidospsis NPK1-related protein kinase 1); splice isoform 1S | Cell cycle |
| 0·26 | Os03g0114000 | AK063381 | Similar to kinesin | Motor protein |
| 0·27 | Os03g0712100 | AK106258 | Cell division cycle-associated protein domain-containing protein | Cell cycle |
| 0·29 | Os01g0685900 | AK064024 | Similar to 65-kD microtubule-associated protein | Cell cycle |
| 0·29 | Os02g0739700 | AK111537 | E2F Family domain-containing protein | Cell cycle |
| 0·31 | Os03g0162200 | AK061605 | Similar to histone H2A | DNA replication |
| 0·32 | Os07g0659500 | AK073537 | Non-SMC condensin subunit, XCAP-D2/Cnd1 family protein | Chromosome condensation |
| 0·32 | Os05g0497100 | AK064293 | Similar to SMC4 protein | Chromosome segregation |
| 0·33 | Os08g0512600 | AK059682 | Protein cdc2 kinase (cdc2Os3) | Cell cycle |
| 0·33 | Os02g0193600 | AK060499 | Mitotic checkpoint serine/threonine protein kinase, Bub1 domain-containing protein | Cell cycle |
| 0·34 | Os07g0507200 | AK102722 | Targeting for Xklp2 family protein | Spindle assembly |
| 0·35 | Os03g0119900 | AK058741 | Similar to histone H4 | DNA replication |
| 0·35 | Os02g0699700 | AK063335 | Similar to DNA topoisomerase II | DNA replication |
| 0·37 | Os04g0583600 | AK059019 | Similar to histone H4 | DNA replication |
| 0·37 | Os05g0459400 | AK073413 | Kinesin, motor region domain-containing protein | Motor protein |
| 0·38 | Os05g0113900 | AK074018 | Similar to histone H2A | DNA replication |
| 0·39 | Os01g0748000 | AK103720 | Similar to dynamin family protein | GTPase activity |
| 0·39 | Os01g0243100 | AK064212 | Similar to kinesin | Motor protein |
| 0·40 | Os03g0214100 | AK060582 | Replication protein A1 | DNA replication |
* Fold change shows average expression ratio of three biological replicates and a colour swap for each replicate.
† Locus ID, full-length cDNA accession number and description in Rice Annotation Project Database (RAP-DB; http://rapdb.dna.affrc.go.jp/).
Fig. 5.
Changes in the expressions of cell division- and cell elongation-related genes. Coleoptiles of seedlings grown under complete submergence for the indicated number of days after imbibition were isolated by LM. Semi-quantitative RT-PCR analysis of the selected genes was performed with appropriate primers (Supplementary Data Table S3, available online). Six cell division-related genes and three cell elongation-related genes were selected from the microarray result. Submergence-inducible OsADH1 and OsADH2 genes and 17s rRNA were used as controls.
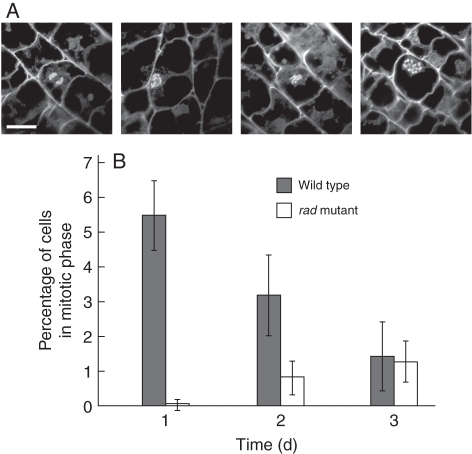
In the WT coleoptile, the mitotic index was highest (5·5 ± 1·0 %) at 1 d after imbibition and then gradually decreased to 3·2 ± 1·2 % at 2 d and to 1·4 ± 1·1 % at 3 d (Fig. 6). In contrast, in the rad coleoptile, the mitotic index was zero at 1 d, 0·8 ± 0·5 % at 2 d and 1·2 ± 0·6 % at 3 d (Fig. 6), indicating a much lower rate of cell division.
Fig. 6.
Frequency of occurrence of mitotic-phase cells. Sections of rice embryos at 1–3 d after imbibition were embedded in paraffin. (A) Nuclei of cells of the WT coleoptiles in mitotic phase. Nuclei were stained with DAPI. Scale bar = 20 µm. (B) Percentage of cells in the mitotic phase. Data are means of three replicates ± s.d.
Table 2.
List of genes related to cell elongation, whose expression was up-regulated or down-regulated in the rad coleoptiles
| Fold change (rad/WT)* | Locus ID† | Full-length cDNA accession no.† | Description† | Functional classification |
|---|---|---|---|---|
| 10·05 | Os02g0275200 | AK071664 | Xyloglucan fucosyltransferase family protein (FUT) | Cell wall biogenesis |
| 7·38 | Os05g0542800 | AK106049 | Pectin lyase fold/virulence factor domain-containing protein | Cell wall loosening |
| 6·45 | Os05g0542800 | AK105858 | Similar to polygalacturonase | Cell wall loosening |
| 6·05 | Os04g0604300 | AK070734 | Similar to xyloglucan endotransglucosylase/hydrolase protein 24 precursor (EC 2·4·1·207) (At-XTH24) (XTH-24) (meristem protein 5) (MERI-5 protein) (MERI5 protein) (endo-xyloglucan transferase) (xyloglucan endo-1,4-beta-d-glucanas e) (OsXTH6) | Cell wall loosening |
| 5·73 | Os10g0450900 | AK064310 | Similar to glycine-rich cell wall structural protein 2 precursor | Cell wall component |
| 4·82 | Os10g0521100 | AK064063 | Similar to actin-depolymerizing factor 6 (ADF-6) (AtADF6) | Actin binding |
| 4·59 | Os10g0450900 | AK064065 | Similar to glycine-rich cell wall structural protein 2 precursor | Cell wall component |
| 4·09 | Os03g0133400 | AK073032 | Peptidoglycan-binding LysM domain-containing protein | Cell wall catabolic process |
| 4·08 | Os06g0621900 | AK101940 | Similar to alpha-expansin OsEXPA 16 (fragment) | Cell wall loosening |
| 2·71 | Os10g0407000 | AK101494 | Pectin lyase fold/virulence factor domain-containing protein | Cell wall loosening |
| 0·14 | Os06g0199000 | AK106381 | Similar to glycine-rich cell wall structural protein 2 precursor | Cell wall component |
| 0·24 | Os10g0498900 | AK108732 | Similar to microtubule-associated protein EB1-like protein | Actin binding |
| 0·32 | Os02g0658800 | AK059638 | Beta-expansin (OsEXPB11) | Cell wall loosening |
| 0·34 | Os04g0228400 | AK069548 | Expansin precursor (alpha-expansin OsEXPA1) | Cell wall loosening |
| 0·35 | Os04g0530100 | AK107184 | Similar to beta-expansin 1 precursor (AtEXPB1) (At-EXPB1) (Ath-ExpBeta-1·5) (OsEXPB17) | Cell wall loosening |
| 0·39 | Os07g0551700 | AK067424 | Similar to cellulose synthase (fragment) | Cell wall synthesis |
| 0·39 | Os04g0602500 | AK058634 | Similar to pectin acetylesterase | Cell wall loosening |
* Fold change shows average expression ratio of three biological replicates and a colour swap for each replicate.
† Locus ID, full-length cDNA accession number and description in Rice Annotation Project Database (RAP-DB; http://rapdb.dna.affrc.go.jp/).
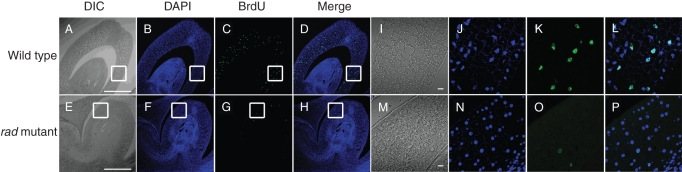
Proliferating cells were identified by incorporation of BrdU into newly synthesized DNA of replicating cells. At 1 d after imbibition, many BrdU-labelled nuclei were observed in the WT coleoptiles, but only a few were observed in the rad coleoptiles (Fig. 7). These results suggest that cell division occurs in the WT coleoptile at the early stages (i.e. 1–2 d) of seedling growth under submergence and does not actively occur in the rad coleoptile.
Fig. 7.
Immunostaining of BrdU in rice coleoptiles. All photographs were obtained with a confocal laser scanning microscopy: (A–H) low magnification: (I–P) high magnification of square areas in A–H; (A, E, I, M) differential interference contrast images; (B, F, J, N) nuclei stained with DAPI; (C, G, K, O) fluorescent immunostaining of BrdU-incorporated nuclei; (D, H, L, P) merged images of the DAPI and BrdU signals. Scale bars = 300 µm. (A and E), 10 µm (I and M).
The expressions of three cell elongation-related genes identified by the LM-microarray analysis (the expansins OsEXPA1, OsEXPB11 and OsEXPB17) were measured by semi-quantitative RT-PCR. Their mRNA levels dramatically increased in the WT coleoptile during germination and early growth, peaking at 2–4 d after imbibition, but were much lower in the rad coleoptile (Fig. 5). However, no significant differences were observed between the WT and rad coleoptiles in the transcript levels of other expansins (OsEXPA2, OsEXPA4, OsEXPA7 and OsEXPB12) (data not shown), which were previously suggested to be involved in coleoptile elongation (Huang et al., 2000; Choi et al., 2003; Lasanthi-Kudahettige et al., 2007). This suggests that the expressions of these genes are not affected by the ADH1 deficiency in the rad mutant at 1 d after imbibition.
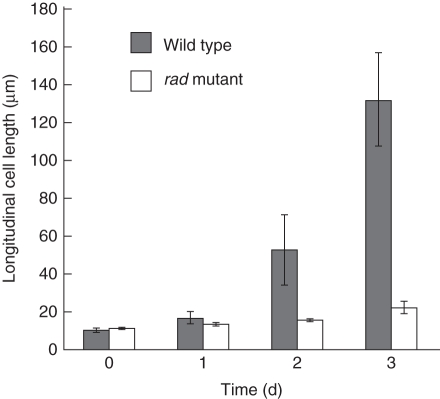
Next, longitudinal cell lengths were measured in the WT and rad coleoptiles during germination and early growth under submergence. The longitudinal cell lengths at 0 d and 1 d after imbibition were comparable between the WT and the rad mutant (Fig. 8). Cell elongation started at between 1 d and 2 d and was enhanced at 3 d in the WT coleoptile, but not in the rad coleoptile.
Fig. 8.
Changes in longitudinal length of coleoptile epidermal cells. Measurements were made with paraffin-embedded sections of rice embryos under a microscope. Lengths of 50–80 epidermal cells were measured for one replicate. Data are means of three replicates ± s.d.
DISCUSSION
It was found that the rad coleoptiles, unlike the WT coleoptiles, slowly elongated under submergence (Fig. 1B), whereas the rad and WT coleoptiles were equally elongated under aerobic conditions (Fig. 1C). The slow elongation of the rad coleoptile may be due to the reduced ATP level in the coleoptile under submerged conditions (Fig. 1D). The finding that rad coleoptiles grown under aerobic conditions for 2 d elongated at a normal rate following submergence treatment (Fig. 2) may be due to aerobic ATP production, which compensates for the reduction of ATP levels in the rad coleoptile and allows the rad coleoptile active cell division and cell elongation under submergence.
The LM-microarray analysis identified 294 genes whose mRNA levels were higher in rad than WT (Supplementary Data Table S1) and 137 genes whose mRNA levels were lower in rad than WT (Supplementary Data Table S2) under submerged conditions. Whereas we compared the expressions of genes in the WT and rad mutant after submergence, Narsai et al. (2009) compared the expressions of rice genes in the WT under aerobic and anaerobic conditions. Most of the genes that were expressed more strongly in the rad coleoptiles than in the WT coleoptiles during submergence were found by Narsai et al. (2009) to be up-regulated in the WT under anaerobic conditions (Supplementary Data Fig. S1). On the other hand, among genes that were expressed more strongly in the WT coleoptiles than in the rad coleoptiles during submergence, most were down-regulated in the WT under anaerobic conditions (Supplementary Data Fig. S1). Expressions of these genes may rather be regulated by ATP levels because more ATP can be produced under aerobic conditions than under anaerobic conditions (Bailey-Serres and Voesenek, 2008) and ATP levels in the coleoptile were lower in the rad mutant than in WT under submerged conditions (Fig. 1D). It is noteworthy that most of cell division-related genes that are less expressed in the rad mutant (Table 1) were down-regulated in the WT under anaerobic conditions (Supplementary Data Fig. S1 and Table S2). This result suggests that the expressions of these cell division-related genes are regulated by ATP levels.
In rice, coleoptile elongation during submergence is thought to mainly occur by cell elongation (Wada, 1961; Opik, 1973; Magneschi and Perata, 2009), although cell division may contribute to coleoptile elongation at an early stage in germination (Wada, 1961; Opik, 1973; Atwell et al., 1982). In this study, the occurrence of cell division in WT coleoptiles under submergence was confirmed by the expressions of cell division-related genes (Fig. 5), a relatively high mitotic index (Fig. 6) and BrdU incorporation (Fig. 7). Also a shift from cell division to cell elongation was found in the coleoptile. This shift may be important for anaerobic coleoptile growth and survival, because it is likely that cell elongation requires less energy (i.e. ATP) than does cell division (Atwell et al., 1982; Magneschi and Perata, 2009).
In the natural environment, coleoptile elongation during flooding is needed to obtain oxygen from the air. Flooding (e.g. 5–10 cm water depth) would thus be expected to select for rice cultivars that grow longer coleoptiles under submergence. The final length of a coleoptile is determined by both the number of cells and the length of each cell. Thus, some flood-tolerant rice cultivars may increase both cell length and the number of cells in the coleoptile under submergence. However, this strategy may be costly and thus the flooding water depth may determine whether the strategy is useful. In submerged rice, coleoptile lengths vary widely among different cultivars (Setter and Ella, 1994; Magneschi et al., 2009). It would be interesting to investigate the cell numbers and cell lengths of coleoptiles of these cultivars and to examine how cell division contributes to the difference in coleoptile lengths.
In conclusion, the present results indicate that cell division starts within 1 d after imbibition and then gradually ceases, and subsequently cell elongation starts between 1 d and 2 d after imbibition and continues at the later stages of the coleoptile growth under submergence. Deficiency of the ADH1 gene in the rad mutant, which results in reduction of ATP synthesis in the submerged coleoptile (Fig. 1D; Saika et al., 2006), represses both cell division and cell elongation, thereby impairing efficient coleoptile elongation under submergence. Some transcription-factor genes identified by the LM-microarray analysis may also have a role in coleoptile elongation and are currently being examined.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Dr Mafumi Abiko for assistance in preparing the cryosections, Ms Ritsuko Motoyama for assistance in the microarray analysis, Dr Yoshiaki Inukai for assisting with the BrdU experiment, and Drs Tim Colmer, Hank Greenway, Kimiharu Ishizawa, Pierdomenico Perata, Jim Raymond and Shin-ichi Arimura for stimulating discussions. This work was supported by Research Fellowships of the Japan Society for the promotion of Science for Young Scientists to H.T.
LITERATURE CITED
- Alpi A, Beevers H. Effects of O2 concentration on rice seedlings. Plant Physiology. 1983;71:30–34. doi: 10.1104/pp.71.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell BJ, Waters I, Greenway H. The effect of oxygen and turbulence on elongation of coleoptiles of submergence-tolerant and -intolerant rice cultivars. Journal of Experimental Botany. 1982;33:1030–1044. [Google Scholar]
- Bailey-Serres J, Voesenek LACJ. Flooding stress: acclimations and genetic diversity. Annual Review of Plant Biology. 2008;59:313–339. doi: 10.1146/annurev.arplant.59.032607.092752. [DOI] [PubMed] [Google Scholar]
- Choi D, Lee Y, Cho HT, Kende H. Regulation of expansin gene expression affects growth and development in transgenic rice plants. The Plant Cell. 2003;15:1386–1398. doi: 10.1105/tpc.011965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J, Greenway H. Mechanisms of anoxia tolerance in plants. I. Growth, survival and anaerobic catabolism. Functional Plant Biology. 2003;30:353–399. doi: 10.1071/PP98095_ER. [DOI] [PubMed] [Google Scholar]
- Huang J, Takano T, Akita S. Expression of alpha-expansin genes in young seedlings of rice (Oryza sativa L.) Planta. 2000;211:467–473. doi: 10.1007/s004250000311. [DOI] [PubMed] [Google Scholar]
- Ishimaru T, Nakazono M, Masumura T, et al. A method for obtaining high integrity RNA from developing aleurone cells and starchy endosperm in rice (Oryza sativa L.) by laser microdissection. Plant Science. 2007;173:321–326. [Google Scholar]
- Kato-Noguchi H. Submergence tolerance and ethanolic fermentation in rice coleoptiles. Plant Production Science. 2001;4:62–65. [Google Scholar]
- Kato-Noguchi H. Pyruvate metabolism in rice coleoptiles under anaerobiosis. Plant Growth Regulation. 2006;50:41–46. [Google Scholar]
- Kato-Noguchi H, Kugimiya T. Preferential induction of alcohol dehydrogenase in coleoptiles of rice seedlings germinated in submergence condition. Biologia Plantarum. 2003;46:153–155. [Google Scholar]
- Kawai M, Uchimiya H. Coleoptile senescence in rice (Oryza sativa L.) Annals of Botany. 2000;86:405–414. [Google Scholar]
- Lasanthi-Kudahettige R, Magneschi L, Loreti E, et al. Transcript profiling of the anoxic rice coleoptile. Plant Physiology. 2007;144:218–231. doi: 10.1104/pp.106.093997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Das A, Yamaguchi M, et al. Cell cycle function of a rice B2-type cyclin interacting with a B-type cyclin-dependent kinase. The Plant Journal. 2003;34:417–425. doi: 10.1046/j.1365-313x.2003.01736.x. [DOI] [PubMed] [Google Scholar]
- Lee KW, Chen PW, Lu CA, Chen S, Ho TH, Yu SM. Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Science Signaling. 2009;2 doi: 10.1126/scisignal.2000333. ra61. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kende H. Expression of beta-expansins is correlated with internodal elongation in deepwater rice. Plant Physiology. 2001;127:645–654. [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kende H. Expression of alpha-expansin and expansin-like genes in deepwater rice. Plant Physiology. 2002;130:1396–1405. doi: 10.1104/pp.008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magneschi L, Perata P. Rice germination and seedling growth in the absence of oxygen. Annals of Botany. 2009;103:181–196. doi: 10.1093/aob/mcn121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magneschi L, Kudahettige RL, Alpi A, Perata P. Expansin gene expression and anoxic coleoptile elongation in rice cultivars. Journal of Plant Physiology. 2009;166:1576–1580. doi: 10.1016/j.jplph.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Matsumura H, Takano T, Yoshida KT, Takeda G. A rice mutant lacking alcohol dehydorogenase. Breading Science. 1995;45:365–367. [Google Scholar]
- Matsumura H, Takano T, Takeda G, Uchimiya H. Adh1 is transcriptionally active but its translational product is reduced in a rad mutant of rice (Oryza sativa L.), which is vulnerable to submergence stress. Theoretical and Applied Genetics. 1998;97:1197–1203. [Google Scholar]
- Nakazono M, Qiu F, Borsuk LA, Schnable PS. Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: identification of genes expressed differentially in epidermal cells or vascular tissues of maize. The Plant Cell. 2003;15:583–596. doi: 10.1105/tpc.008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R, Howell KA, Carroll A, Ivanova A, Millar AH, Whelan J. Defining core metabolic and transcriptomic responses to oxygen availability in rice embryo and young seedlings. Plant Physiology. 2009;151:306–322. doi: 10.1104/pp.109.142026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T, Tausta SL, Gandotra N, Liu T. Laser microdissection of plant tissue: what you see is what you get. Annual Review of Plant Biology. 2006;57:181–201. doi: 10.1146/annurev.arplant.56.032604.144138. [DOI] [PubMed] [Google Scholar]
- Opik H. Effect of anaerobiosis on respiratory rate, cytochrome-oxidase activity and mitochondrial structures in coleoptiles of rice (Oryza sativa L) Journal of Cell Science. 1973;12:725–739. doi: 10.1242/jcs.12.3.725. [DOI] [PubMed] [Google Scholar]
- Perata P, Alpi A. Plant-responses to anaerobiosis. Plant Science. 1993;93:1–17. [Google Scholar]
- Rahman M, Grover A, Peacock WJ, Dennis ES, Ellis MH. Effects of manipulation of pyruvate decarboxylase and alcohol dehydrogenase levels on the submergence tolerance of rice. Australian Journal of Plant Physiology. 2001;28:1231–1241. [Google Scholar]
- Saika H, Matsumura H, Takano T, Tsutsumi N, Nakazono M. A point mutation of Adh1 gene is involved in the repression of coleoptile elongation under submergence in rice. Breeding Science. 2006;56:69–74. [Google Scholar]
- Setter TL, Ella ES. Relationship between coleoptile elongation and alcoholic fermentation in rice exposed to anoxia. I. Importance of treatment conditions and different tissues. Annals of Botany. 1994;74:265–271. [Google Scholar]
- Shin JH, Jeong DH, Park MC, An G. Characterization and transcriptional expression of the alpha-expansin gene family in rice. Molecules and Cells. 2005;20:210–218. [PubMed] [Google Scholar]
- Takahashi H, Kamakura H, Sato Y, et al. A method for obtaining high quality RNA from paraffin sections of plant tissues by laser microdissection. Journal of Plant Reserch. 2010;123:807–813. doi: 10.1007/s10265-010-0319-4. [DOI] [PubMed] [Google Scholar]
- Umeda M, Iwamoto N, Umeda-Hara C, Yamaguchi M, Hashimoto J, Uchimiya H. Molecular characterization of mitotic cyclins in rice plants. Molecular and General Genetics. 1999a;262:230–238. doi: 10.1007/s004380051079. [DOI] [PubMed] [Google Scholar]
- Umeda M, Umeda-Hara C, Yamaguchi M, Hashimoto J, Uchimiya H. Differential expression of genes for cyclin-dependent protein kinases in rice plants. Plant Physiology. 1999b;119:31–40. doi: 10.1104/pp.119.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S. Growth patterns of rice coleoptiles grown on water and under water. Science Reports of the Tohoku University. Fourth series (Biology) 1961;27:199–207. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.