Abstract
Background and Aims
The subgenus Ceratotropis in the genus Vigna is widely distributed from the Himalayan highlands to South, Southeast and East Asia. However, the interspecific and geographical relationships of its members are poorly understood. This study investigates the phylogeny and biogeography of the subgenus Ceratotropis using chloroplast DNA sequence data.
Methods
Sequence data from four intergenic spacer regions (petA-psbJ, psbD-trnT, trnT-trnE and trnT-trnL) of chloroplast DNA, alone and in combination, were analysed using Bayesian and parsimony methods. Divergence times for major clades were estimated with penalized likelihood. Character evolution was examined by means of parsimony optimization and MacClade.
Key Results
Parsimony and Bayesian phylogenetic analyses on the combined data demonstrated well-resolved species relationships in which 18 Vigna species were divided into two major geographical clades: the East Asia–Southeast Asian clade and the Indian subcontinent clade. Within these two clades, three well-supported eco-geographical groups, temperate and subtropical (the East Asia–Southeast Asian clade) and tropical (the Indian subcontinent clade), are recognized. The temperate group consists of V. minima, V. nepalensis and V. angularis. The subtropical group comprises the V. nakashimae–V. riukiuensis–V. minima subgroup and the V. hirtella–V. exilis–V. umbellata subgroup. The tropical group contains two subgroups: the V. trinervia–V. reflexo-pilosa–V. trilobata subgroup and the V. mungo–V. grandiflora subgroup. An evolutionary rate analysis estimated the divergence time between the East Asia–Southeast Asia clade and the Indian subcontinent clade as 3·62 ± 0·3 million years, and that between the temperate and subtropical groups as 2·0 ± 0·2 million years.
Conclusions
The findings provide an improved understanding of the interspecific relationships, and ecological and geographical phylogenetic structure of the subgenus Ceratotropis. The quaternary diversification of the subgenus Ceratotropis implicates its geographical dispersal in the south-eastern part of Asia involving adaptation to climatic condition after the collision of the Indian subcontinent with the Asian plate. The phylogenetic results indicate that the epigeal germination is plesiomorphic, and the germination type evolved independently multiple times in this subgenus, implying its limited taxonomic utility.
Keywords: Subgenus Ceratotropis, Vigna, Leguminosae, diversification, intergenic spacer, germination type
INTRODUCTION
The genus Vigna Savi (Leguminosae) comprises >80 species which are distributed throughout the Old World and New World. The genus is divided into six subgenera, Ceratotropis (Piper) Verdc., Haydonia (Wilczek) Verdc., Lasiospron (Benth.) Verdc., Plectotropis (Schum.) Baker, Sigmoidotropis (Piper) Verdc. and Vigna Savi (Verdcourt, 1970; Maréchal et al., 1978), and the subgenus Macrorhynchus Verdc. previously placed in genus Vigna, was transferred to genus Wajira Thulin (Thulin et al., 2004).
Subgenus Ceratotropis, one of the most economically important groups in the genus Vigna for food, forage and cover crops, contains five well-known domesticated species (Baudoin and Maréchal, 1988; Smartt, 1990; Lumpkin and McClary, 1994; Tomooka et al., 2002b). It is originally circumscribed as a group of the Asian Vigna (Verdcourt, 1970) and distinguished from the other subgenera by having peltate stipule, a pocket on the left keel petal, style extending beyond the stigma as a beak, keel petals curved to the left in the upper part, and pollen grains with a coarse reticulate sculpture (Verdcourt, 1970; Maréchal et al., 1978).
The species of subgenus Ceratotropis are widely distributed in South Asia, the Himalayan highlands, Southeast Asia, and East Asia (Maréchal et al., 1978; Tateishi, 1985, 1996; Tateishi and Ohashi, 1990; Tomooka et al., 2002b), while the members of other subgenera are endemic to Africa [Plectotropis (except V. vexillata (L.) A. Rich], Africa and Madagascar (Haydonia), or America (Lasiospron and Sigmoidotropis). The subgenus Vigna is found throughout sub-Saharan Africa, with representatives present in tropical Asia and the Americas (Maréchal et al., 1978).
The 21 known wild species in the subgenus Ceratotropis inhabit coastal sandy soil, limestone hills, forest margins and open fields (Tateishi, 1983, 1985; Tomooka et al., 2002a, b). East Asian and Southeast Asian species of the subgenus Ceratotropis occur naturally in temperate and subtropical regions [e.g. V. angularis (Willd.) Ohwi & H. Ohashi var. nipponensis (Ohwi) Ohwi & H. Ohashi and V. nepalensis Tateishi & Maxted in temperate regions and V. nakashimae (Ohwi) Ohwi & H. Ohashi, V. umbellata (Thunb.) Ohwi & H. Ohashi and V. tenuicaulis N. Tomooka & Maxted in subtropical regions], while Indian subcontinental species [i.e. V. mungo (L.) Hepper var. silvestris Lukoki, Maréchal & Otoul, V. radiata (L.) R. Wilczek var. sublobata (Roxb.) Verdc., V. trilobata (L.) Verdc., and V. aridicola N.Tomooka & Maxted] are mainly confined to tropical regions.
All of the species in the subgenus Ceratotropis are diploid (2n = 2x =22; Maréchal et al., 1978; Tateishi, 1985; Tomooka et al., 2002b) except tetraploid V. reflexo-pilosa Hayata (2n = 4x = 44; Swindell et al., 1973; Egawa et al., 1988). The ancestral donor of V. reflexo-pilosa has been assumed to be V. exilis Tateishi & Maxted or V. minima (Roxb.) Ohwi & H. Ohashi based on isozyme, interspecific hybridization (Tateishi, 1985; Egawa et al., 1996; Konarev et al., 2002) or V. trinervia (B. Heyne ex Wight & Arn.) Tateishi & Maxted as the maternal donor based on plastid DNA phylogeny (Yano et al., 2004; Ye Tun Tun and Yamaguchi, 2007).
In Leguminosae, seedling germination type [hypogeal (cotyledons may remain underground) vs. epigeal (cotyledons emerge above the soil surface following germination)] shows no variation in some tribes, while both types are found within the Trifolieae and Phaseoleae (Gates, 1951; Polhill, 1981; Endo and Ohashi, 1997) and papilionoid genera such as Phaseolus and Vigna (Gates, 1951; Polhill, 1981; Tomooka et al., 2002b). In some genera, epigeal and hypogeal germination are distinguished by different subgenera (Essig, 1992). In the subgenus Ceratotropis, seedling germination has been focused on as one of the diagnostic characters for inferring relationships among species of the subgenus (Maekawa, 1955; Baudoin and Maréchal, 1988; Tomooka et al., 1991; Tateishi, 1996). These studies recognized two morphological groups within the subgenus Ceratotropis, the azuki bean group (angularis–umbellata) with hypogeal germination having petiolate first and second leaves and the mung bean group (radiata–mungo) with epigeal germination having sessile first and second leaves, and ungrouped species showing epigeal germination with petiolate first and second leaves (Baudet, 1974) which later was considered as the intermediate group, aconitifoila–trilobata (Tomooka et al., 1991). Recently, three groups within the subgenus Ceratotropis were proposed as sections Angulares N.Tomooka & Maxted (azuki bean group), Ceratotropis N.Tomooka & Maxetd (mung bean group) and Aconitifoliae N.Tomooka & Maxted (Intermediate group), based on seedling characteristics, size of floral parts and growth habit (Tomooka et al., 2002a). In spite of the importance of morphological characters that have been used traditionally to define taxonomic relationships within the subgenus Ceratotropis, few works have evaluated them in a phylogenetic context. The only cladistic study in the subgenus Ceratotropis was conducted by Taeishi (1996) who considered hypogeal germination as the primitive state in the subgenus based on morphological data.
In recent phylogenetic studies on the subgenus Ceratotropis, relationships among the species remain in dispute. A phylogenetic tree based on DNA sequences from the ITS and atpB-rbcL regions showed a close relationship between V. reflexo-pilosa and the species of section Angulares such as V. umbellata, V. hirtella and V. exilis (Doi et al., 2002); however, the trnT-F sequence data indicated a closer relationship between V. reflexo-pilosa to V. trinervia, consisting of some species of section Ceratotropis (Yano et al., 2004; Ye Tun Tun and Yamaguchi, 2007). A molecular phylogeny by the cpDNA and nuclear ITS sequence data recognized three lineages corresponding to the three sections (Aconitifoliae, Angulares and Ceratotropis) in the subgenus Ceratotropis (Doi et al., 2002), but its clades are not well supported. In contrast, phylogenetic analyses based on DNA sequences of nuclear ribosomal ITS (Goel et al., 2002) and plastid DNA phylogenies (Yano et al., 2004; Ye Tun Tun and Yamaguchi, 2007) revealed two main groups in the subgenus Ceratotropis. The phylogenetic analysis using 5S IGS divided the ten Vigna species of subgenus Ceratotropis into two weakly supported clades: clade I which included the most species of sections Ceratotropis and Aconitifoliae and clade II consisting of some of the species in section Angulares (Saini and Jawali, 2009).
The biogeographic history of the subgenus Ceratotropis could be inferred from a phylogenetic analysis of the subgenus. However, previous studies have attempted to determine its molecular phylogenetic relationships with representative species from the limited geographical regions, e.g. samples mainly from Thailand based on AFLP marker (Seehalak et al., 2006) and those from Myanmar using trnT-F non-coding regions of chloroplast genome (Ye Tun Tun and Yamaguchi, 2007). These studies did not resolve the geographical relationships or interspecific relationships in the subgenus Ceratotropis. Furthermore, due to the limited molecular data, a low amount of variation and weak bootstrap support were found within each of the recognized taxonomic groups.
This study aimed to provide evidence for advancing our understanding of the phylogenetic relationships and historical biogeography of the subgenus Ceratotropis by using substantially increased molecular sequence data and improved species sampling in comparison to that of previous studies, and also to elucidate evolutional patterns of the seedling germination type on the molecular tree and to consider its taxonomic implication as well. To achieve the objectives, 18 species with four outgroups were selected and sequence data used from four plastid intergenic spacer regions, psbD-trnT, trnT-trnL, trnT-trnE and petA-psbJ, all of which are reported to include higher information content for phylogenetic analyses at lower taxonomic levels (Shaw et al., 2005, 2007).
MATERIALS AND METHODS
Plant materials
Out of 21 species of the subgenus Ceratotropis, 18 were included, representing the three taxonomical groups described by Tomooka et al. (2002a, b); material of the three exceptions, V. khandalensis (Santapau) Sundararagh. & Wadhwa, V. dalzelliana (Kuntze) Verdc. and V. subramaniana (Babu ex Raizada) M. Sharma, was not accessible at the time of the study. For those 18 species, accessions were obtained from varied sources, and included multiple accessions whenever possible (Table 1). Four outgroup taxa, V. unguiculata (L.) Walp. var. unguiculata and V. marina (Burm.) Merr. (subgenus Vigna), V. kirkii (Baker f.) J.B. Gillett (subgenus Plectotropis) and V. venulosa Baker (subgenus Haydonia), were selected (Table 1) based on previous molecular studies (Maréchal et al., 1978; Fatokun et al., 1993; Yasuda and Yamaguchi, 1996; Goel et al., 2002; Yano et al., 2004; Ye Tun Tun and Yamaguchi, 2007).
Table 1.
List of plant materials, inversion type in the psbD-trnT region, and seed germination
| Taxon | Accession number* (code) | Country | Inversion type† | Seed germination |
|---|---|---|---|---|
| Ingroup | ||||
| Subgenus Ceratotropis (Piper) Verdc. | ||||
| Section Angulares N.Tomooka & Maxted | ||||
| Vigna angularis (Willd.) Ohwi & H.Ohashi | Azn/90-J-41 (Van-2) | Japan | I | Hypogeal |
| V. angularis (Willd.) Ohwi & H.Ohashi var. angularis | Aza/88-J-26 (Van-1) | Japan | I | Hypogeal |
| V. angularis (Willd.) Ohwi & H.Ohashi var. nipponensis (Ohwi) Ohwi & H.Ohashi | Azn/91-J-14 (Van-3) | Japan | I | Hypogeal |
| I | Hypogeal | |||
| Azn/96-B-02 (Van-4) | Bhutan | |||
| V. aff. angularis (Willd.) Ohwi & H.Ohashi var. nipponensis (Ohwi) Ohwi & H.Ohashi | Azn/01-My-05 (Van-5) | Myanmar | I | Hypogeal |
| V. nepalensis Tateishi & Maxted | NI 971 (Vne-1) | India | I | Hypogeal |
| NI 1704 (Vne-2) | Nepal | I | Hypogeal | |
| V. tenuicaulis N. Tomooka & Maxted | Azte/01-My-8 (Vten-1)‡ | Myanmar | I | Hypogeal |
| Azte/01-My-9 (Vten-2)‡ | Myanmar | I | Hypogeal | |
| V. aff. minima (Roxb.) Ohwi & H.Ohashi | Azr/01-My-01 (Vmn-1) | Myanmar | I | Hypogeal |
| V. aff. minima (Roxb.) Ohwi & H.Ohashi | Azr/01-My-04 (Vmn-2) | Myanmar | I | Hypogeal |
| V. minima (Roxb.) Ohwi & H.Ohashi | NI 1363 (Vmn-3)‡ | Indonesia | I | Hypogeal |
| V. nakashimae (Ohwi) Ohwi & H.Ohashi | Azm/01-My-04 (Vnk-1) | Myanmar | I | Hypogeal |
| Azm/96-J-01 (Vnk-2) | Japan | I | Hypogeal | |
| V. riukiuensis (Ohwi) Ohwi & H.Ohashi | Azr/91-O-01 (Vri-1) | Japan | I | Hypogeal |
| NI 1635 (Vri-2) | Japan | I | Hypogeal | |
| V. hirtella Ridley | Azh/01-My-03 (Vhi-1) | Myanmar | I | Hypogeal |
| Azh/00-My-01 (Vhi-2) | Myanmar | I | Hypogeal | |
| NI 1394 (Vhi-3 )‡ | Thailand | I | Hypogeal | |
| Azh/06-C-01 (Vhi-4)‡ | China | I | Hypogeal | |
| Azh/06-C-04 (Vhi-5)‡ | China | I | Hypogeal | |
| V. umbellata (Thunb.) Ohwi & H.Ohashi | Azu/03-Th-01 (Vum-1) | Thailand | I | Hypogeal |
| Azu/01-My-04 (Vum-2) | Myanmar | I | Hypogeal | |
| V. exilis Tateishi & Maxted | Aze/01-My-01 (Vxi-1) | Myanmar | I | Hypogeal |
| Aze/01-My-02 (Vxi-2) | Myanmar | I | Hypogeal | |
| V. trinervia (B. Heyne ex Wight & Arn.) Tateishi & Maxted | Aztr/01-My-09 (Vtrn) | Myanmar | II | Hypogeal |
| V. reflexo-pilosa Hayata var. reflexo-pilosa | Azp/92-O-01 (Vrp-1) | Japan | II-i | Hypogeal |
| V. reflexo-pilosa Hayata var. glabra (Roxb.) N.Tomooka & Maxted | Azp/03-V-02 (Vrp-2) | Vietnam | II-i | Hypogeal |
| Section Ceratotropis N.Tomooka & Maxted | ||||
| V. radiata (L.) R. Wilczek var. radiata | Azd/My-004178 (Vrd-1) | Myanmar | II | Epigeal |
| V. radiata (L.) R. Wilczek var. sublobata (Roxb.) Verdc. | Azd/00-My-01 (Vrd-2) | Myanmar | II | Epigeal |
| NI634 (Vrd-3)‡ | India | II | Epigeal | |
| V. mungo (L.) Hepper var. mungo | Azg/My-003935 (Vmg-1) | Myanmar | II | Epigeal |
| V. mungo (L.) Hepper var. silvestris Lukoki, Maréchal & Otoul | NI 969 (Vmg-2)‡ | India | II | Epigeal |
| V. grandiflora (Prain) Tateishi & Maxted | NI 1721 (Vgrd-1)‡ | Thailand | II-ii | Epigeal |
| Azgr/06-Th-01 (Vgrd-2)‡ | Thailand | II-ii | Epigeal | |
| Azgr/06-Th-02 (Vgrd-3)‡ | Thailand | II-ii | Epigeal | |
| Section Aconitifoliae N.Tomooka & Maxted | ||||
| V. aridicola N.Tomooka & Maxted | 3227/Kew (Vari)‡ | Sri Lanka | II | Epigeal |
| V. aconitifolia (Jacq.) Maréchal | Azc/95-I-01 (Vac) | India | II-iii | Epigeal |
| V. stipulacea Kuntze | Azt/01-My-02 (Vst-1) | Myanmar | II | Hypogeal |
| Azt/01-My-04 (Vst-2) | Myanmar | II | Hypogeal | |
| V. trilobata (L.) Verdc. | Azt/96-I-01 (Vtb-1) | India | II | Epigeal |
| NI 453 (Vtb-2)‡ | Sri Lanka | I | Epigeal | |
| Outgroup | ||||
| Subgenus Vigna Savi | ||||
| V. unguiculata (L.) Walp var. unguiculata | Vuc/My-4208 (Out-1) | Myanmar | II-ii | Epigeal |
| V. marina (Burm.) Merr. | Vgm/90-O-01 (Out-2) | Japan | I | Epigeal |
| Subgenus Haydonia (Wilczek) Verdc. | ||||
| V. venulosa Baker | NI 548 (Out-3)‡ | Liberia | I | Epigeal |
| Subgenus Plectotropis (Schum.) Baker | ||||
| V. kirkii (Baker f.) J.B.Gillett | NI 448 (Out-4)‡ | Congo | I | Epigeal |
* NI, National Botanical Garden of Belgium; Kew, Royal Botanical Garden, Kew.
† Inversion type – see text. In the psbD-trnT region.
‡ Sequencing of the trnT-trnL region in this study.
DNA analysis
Total DNA was extracted from fresh leaves using the modified CTAB method (Doyle and Doyle, 1987) or obtained from herbarium samples from the Royal Botanic Gardens, Kew (Table 1). Primers used for amplification and sequencing of the psbD-trnT and trnT-trnE intergenic spacer regions were taken from a previous study (Ye Tun Tun and Yamaguchi, 2008). Those of the trnT-trnL region were taken from Taberlet et al. (1991), and those for the petA-psbJ intergenic spacer region were designed at the conservative region of alignment sequences of Meidcago truncatula L. (AC093544) and Lotus japonicus L. (AP002983) obtained from the GeneBank (Table 2). For the trnT-L spacer region, sequences were included from a previous study (Ye Tun Tun and Yamaguchi, 2007), and those for the 13 accessions of the ingroup and two outgroups were sequenced in this study (Table 1). PCR reactions were performed in a volume of 25 µL containing 2·5 µL of 10 × reaction buffer, 2·5 µL of 25 mm MgCl2, 2·5 µL of 1·25 mm dNTPs, 1·25 µL of each 10 µm primer, 0·1 µL of Takara rTaq (Takara Shuzo Co., Ltd, Japan), and 1·0 µL of 5–10 ng μL−1 genomic DNA. Amplifications were performed using a thermal cycler (GeneAmp® PCR System 2700; Applied Biosystems, Foster City, CA, USA) as follows: one cycle of 5 min at 95 °C, 35 cycles of 45 s at 95 °C, 45–60 s at 55 °C for trnT-trnE, 53 °C for petA-psbJ, 52 °C for psbD-trnT, 57 °C for trnT-trnL; 2 min at 68 °C; and finally one cycle of 5 min at 68 °C. Clean PCR products were sequenced with the BigDye Terminator v3·1 Cycle Sequencing Kit (Applied Biosystems), and cycle sequencing was performed according to the manufacturer's protocols. Sequencing was carried out on an ABI PRISM 3100 automated sequencer (Applied Biosystems).
Table 2.
Primers used for amplification and sequencing of the four chloroplast intergenic spacer regions
| Region/primer | Sequence (5'-3') | Use | Reference |
|---|---|---|---|
| psbD-trnT | |||
| psbD-F | TCAACTACTTCAACCATTTCC | Amplification/sequencing | Ye Tun Tun and Yamaguchi (2008) |
| trnT-R | TGGTAAGGCGTAAGTCATCG | Amplification/sequencing | Ye Tun Tun and Yamaguchi (2008) |
| DT-F2 | TGGTGGAACTTGAAATTGGT | Sequencing | Ye Tun Tun and Yamaguchi (2008) |
| DT-R2 | ACCAATTTCAAGTTCCACCA | Sequencing | Ye Tun Tun and Yamaguchi (2008) |
| trnT-trnE | |||
| trnT-F | CGATGACTTACGCCTTACC | Amplification/sequencing | Ye Tun Tun and Yamaguchi (2008) |
| trnE-R | AGAGAGATGTCCTGAACCAC | Amplification/sequencing | Ye Tun Tun and Yamaguchi (2008) |
| petA-psbJ | |||
| petA-F | GTTACGTGTCCAAGGTCTC | Amplification/sequencing | This study |
| psbJ-R | CCGATACTACTGGAAGGATT | Amplification/sequencing | This study |
| trnT-trnL | |||
| trn-a | CATTACAAATGCGATGCTCT | Amplification/sequencing | Taberlet et al. (1991) |
| trn-b | TCTACCGATTTCGCCATATC | Amplification/sequencing | Taberlet et al. (1991) |
Phylogenetic analyses
Sequence alignment was initially performed using Clustal X v.1·83 (Thompson et al., 1997) and manually adjusted using MEGA v. 4 (Tamura et al., 2007). Phylogenetic analyses were performed using Bayesian and maximum-parsimony approaches. Maximum-parsimony analyses involved a heuristic search strategy with 1000 replicates of random addition of sequences, in combination with ACCTRAN character optimization, MULPARS + TBR branch swapping and STEEPEST DESCENT options off in PAUP* 4·0b10 (Swofford, 2002). All character states were treated as unordered and equally weighted. Informative insertions and deletions (indels) were coded as binary characters (0, 1) according to Graham et al. (2000). The strict consensus tree was constructed from the most-parsimonious trees. Support for individual branches or clades was estimated using resampling bootstrap analysis (Felsenstein, 1985). Bootstrap values were estimated with 1000 replicates, random addition sequence and TBR branch swapping.
Incongruence length difference (ILD) tests (Farris et al., 1995) as implemented under a partition homogeneity test in the PAUP* version 4·0b10 (Swofford, 2002), were conducted among pairwise combinations of the four datasets to determine whether the four regions were statistically incongruent. The ILD test was performed using 100 replications of heuristic searches with 100 random addition analyses and TBR branch swapping, holding ten trees per step, and using STEEPEST DESCENT off, with the MULTREES option enabled.
Bayesian inference of phylogeny using Monte Carlo Markov Chains (MCMC) was done with MrBayes 3·1·2 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003) under the optimal model of evolution. The dataset was partitioned into data partitions (trnT-trnE, petA-psbJ, psbD-trnT and trnT-trnL) and all non-partitioned combined datasets. Models of sequence evolution for each of the partitions and for all combined datasets were determined using the program ModelTest 3·7 (Posada and Crandall, 1998) based on the Akaike information criterion (AIC) (Posada and Buckley, 2004). The indels were treated as a ‘standard’ data type in MrBayes. For comparing the partitioned and non-partitioned datasets, Bayes factors were employed. Bayes factors measure the relative performance of two analyses as the ratio of their marginal likelihoods, i.e. the likelihood of the data under the model (Kass and Raftery, 1995; Nylander et al., 2004). The marginal likelihood may be approximated by the harmonic mean of likelihood values of evolutionary hypotheses sampled from the posterior distribution (Nylander et al., 2004), which is calculated by the sump command in MrBayes 3·1·2 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003). A Bayes factor of >10 provides strong evidence against the alternative hypothesis (Kass and Raftery, 1995; Brandley et al., 2005). For each analysis, four MCMC chains were run for 3 × 106 permutations of tree parameters and sampled every 3 × 104 permutation, such that the sampling yielded 100 Bayesian trees which excluded the burn-in and autocorrelated trees (Lavin et al., 2005; Delgado-Salinas et al., 2006). The trees were imported into PAUP* 4·0b10 to construct the 50 % majority rule consensus tree.
Molecular calibration and age estimation
A recent study of ages and evolutionary rates of Leguminosae (Lavin et al., 2005) provides estimated ages for many crown and stem clades which can serve as calibration points for legume groups lacking a fossil record. The hypothesis of rate constancy was evaluated with a likelihood ratio (LR) test comparing the likelihood scores from unconstrained and clock-constrained analyses (Felsenstein, 1981, 1988; Huelsenbeck and Crandall, 1997), where LR = 2 (lnLclock – lnLno clock) was assumed to be χ2 distributed with the degrees of freedom equal to n taxa minus two. The molecular clock was rejected because constrained and unconstrained analyses differed significantly [(–lnL = 2(7399·779 – 7359·102) = 81·534, d.f. = 44, P < 0·001]. Therefore, the penalized likelihood method with the truncated Newton algorithm (Sanderson, 2002) was used in the rate smoothing program r8s, version 1·70 (Sanderson, 2003) to estimate the ages of the groups, as described by Lavin et al. (2005), using multiple trees with branch lengths estimated by Bayesian analyses (Huelsenbeck and Ronquist, 2001; Huelsenbeck et al., 2001). The smoothing parameters for the penalized likelihood analyses were determined using cross-validation tests. The outgroups V. kirkii and V. marina were pruned prior to analysis using r8s (Sanderson, 2003; M. Lavin, Montana State University, USA, pers. comm.). Based on an analysis with multiple calibration points across the legumes (Lavin et al., 2005), the age of the root node on the trees was fixed at a maximum of 8·0 million years (Mya) which represents the root of the Old World Vigna clade (i.e. all of the endemic African Vigna lineages in which members of Ceratotropis are nested; node 68 in Lavin et al., 2005, which had a maximum age estimate of 10·4 Mya, minimum of 6·4, and a mean of 8·0; M. Lavin, pers. comm.). Ages were biased old by assigning the mean age estimate of 8·0 Mya rather than the youngest ages within the 95 % confidence interval. This was done for emphasis regardless of a bias in older estimates, and young ages were still estimated. Analyses using 8·0 Mya as the root age estimated 32 as the optimal smoothing value. Means and standard deviations of ages of specified clades were obtained from the input of 100 Bayesian trees (Lavin et al., 2005; Delgado-Salinas et al., 2006; Javadi et al., 2007).
Character state evolution was examined for seedling germination type (Table 1) by mapping character states onto the strict consensus tree of the most-parsimonious phylogenetic trees of the combined data with MacClade 4·06 (Maddison and Maddison, 2000). Both ACCTRAN (maximizing the proportion of the homoplasy accounted for parallelism) and DELTRAN (maximizing the proportion accounted for reversal) optimizations were considered and analysed.
RESULTS
Sequence characteristics of the four plastid regions
The length of the psbD-trnT region ranged from 1085 bp (V. mungo var. sylvestris) to 1122 bp (V. stipulacea) in the subgenus Ceratotropis. The total aligned length for psbD-trnT was 1189 bp in ingroup taxa, which included 93 variable and 72 (6·0 %) parsimony-informative characters (Table 3). Pairwise sequence divergence varied from 0·00 to 0·0517 within the ingroup. Three unique indels (insertion/deletion), 27 bp, 20 bp and 7 bp, were found in psbD-trnT: the 27-bp insertion in two accessions of V. stipulacea (Vst-1 and Vst-2), the 20-bp insertion in two accessions of V. tenuicaulis (Vten-1 and Vten-2), and the 7-bp deletion in two accessions of V. mungo (Vmg-1 and Vmg-2). There was a 42-bp inversion in this region. The inversion was bordered by a pair of inverted repeat sequences 14 bp long. Two types of 42-bp inversions were recognized: type I occurred in V. angularis, V. nepalensis, V. hirtella, V. exilis, V. umbellata, V. minima, V. riukiuensis, V. tenuicaulis, V. nakashimae and Sri Lankan accession of V. trilobata; and type II occurred in the remaining ingroup taxa, while single nucleotide substitutions were found in type II which further includes three subtypes, II-i, II-ii and II-iii (Fig. 1 and Table 1). The orientation of the loop in Type II was reverse complementary to that of type I (Fig. 1). Subtypes II-i, II-ii and II-iii differed from type II by one nucleotide substitution and from each other by two nucleotide substitutions (i.e. type II-i, V. reflexo-pilosa; type II-ii, V. grandiflora; type II-iii, V. aconitifolia) (Fig. 1 and Table 1).
Table 3.
Sequence characteristics of four plastid regions in the subgenus Ceratotropis
| Character | trnT-trnL | psbD-trnT | petA-psbJ | trnT-trnE |
|---|---|---|---|---|
| Length range (bp) | ||||
| Ingroup | 796–849 | 1085–1122 | 527–594 | 754–795 |
| Outgroup included | 754–789 | 1044–1089 | 509–553 | 733–800 |
| Aligned length (bp) | ||||
| Ingroup | 903 | 1189 | 615 | 801 |
| Outgroup included | 957 | 1193 | 655 | 830 |
| Number of variable characters (%) | ||||
| Ingroup | 90 (9·9) | 93 (7·8) | 44 (7·2) | 84 (10·5) |
| Outgroup included | 156 (16·3) | 167 (13·9) | 96 (14·6) | 131 (15·7) |
| Parsimony-informative characters (%) | ||||
| Ingroup | 60 (6·6) | 72 (6·0) | 28 (4·5) | 54 (6·7) |
| Outgroup included | 75 (7·8) | 91 (7·6) | 35 (5·3) | 61 (7·3) |
Fig. 1.
Inversion types in the psbD-trnT intergenic spacer region in the Vigna subgenus Ceratotropis. Arrows below the sequence indicate inverted repeats. Nucleotides bordered by inverted repeats have undergone inversions. The bold letters on a grey background show single nucleotide substitution. The different types of each accession are shown in Table 1 and Fig. 2I.
The length of the trnT-trnL region ranged from 796 bp (V. trinervia) to 849 bp (V. grandiflora) within the ingroup taxa. The total aligned length for trnT-trnL was 903 bp in ingroup taxa, which included 90 variable and 60 (6·6 %) parsimony-informative characters (Table 3). Pairwise sequence divergence ranged from 0·00 to 0·0323 among taxa in the subgenus Ceratotropis. Two unique insertions (19 bp and 17 bp) were observed in V. grandiflora in this region.
Within the subgenus Ceratotropis, the length of the trnT-trnE region ranged from 754 bp (V. radiata) to 795 bp (V. hirtella). The total aligned length for trnT-trnE was 801 bp in ingroup taxa, which included 84 variable and 54 (6·7 %) parsimony-informative characters (Table 3). Pairwise sequence divergence ranged from 0·00 to 0·0295 within the ingroup. Two unique gaps were found in this region, a 13-bp insertion shared by four out of five accessions of V. hirtella (except for accession Vhi-5, Table 1) and a 28-bp deletion shared by three accessions of V. radiata.
The length of petA-psbJ ranged from 527 bp to 594 bp within the ingroup. The total aligned length for petA-psbJ was 615 bp in ingroup taxa, which included 44 variable and 28 (4·5 %) parsimony-informative characters (Table 3). Pairwise sequence divergence varied from 0·00 to 0·029 among taxa in this subgenus. Within the petA-psbJ data, a 67-bp insertion was found in one accession of V. exilis (Vxi-1; Table 1).
Phylogenetic analysis
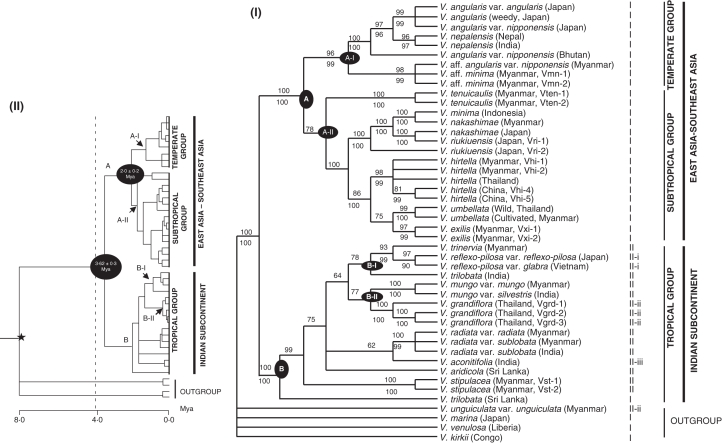
The suitability of combined phylogenetic analysis was conducted using ILD tests. Pairwise ILD tests indicated that there were no conflicts among any of the four chloroplast regions (Table 4). Therefore, phylogenetic analyses based on the combined dataset were conducted. For the phylogenetic analysis, the inversion was excluded, and potentially informative indels were added to the data matrix. The combined dataset comprised 2832 bp, of which 465 were variable and 224 were parsimony-informative. Maximum-parsimony analysis resulted in 15 most-parsimonious trees (TL = 613, CI = 0·809, RI = 0·901), and the strict consensus tree of the 15 most-parsimonious trees is shown in Fig. 2I.
Table 4.
Results of the ILD tests for pairwise comparisons between the data partitions
| Partitions | petA-psbJ | trnT-trnL | trnT-trnE | psbD-trnT |
|---|---|---|---|---|
| petA-psbJ | – | |||
| trnT-trnL | 0·70 | – | ||
| trnT-trnE | 0·82 | 0·99 | – | |
| psbD-trnT | 0·47 | 0·11 | 0·11 | – |
Fig. 2.
(I) Strict consensus tree of 15 parsimonious trees inferred from petA-psbJ, psbD-trnT, trnT-trnE, and trnT-trnL spacer sequences. Numbers above branches refer to bootstrap support in parsimony analysis, and those below branches are Bayesian posterior probability. CI = 0·809, RI = 0·901. Roman numerals following species names represent the inversion type in the psbD-trnT region. (II) A penalized likelihood chronogram of the Bayesian consensus tree of the combined data. The star indicates the calibration point used (Lavin et al., 2005, for details, see Materials and methods), The time scale is in million years ago (Mya).
Bayesian analyses were also performed for individual DNA regions; the best fitting models and parameter values are shown in Table 5. The optimal models identified were TIM (four rate classes) for the petA-psbJ, TIM + G (four rate classes following a gamma distribution) for the trnT-trnL, TVM + G (five rate classes following a gamma distribution) for the trnT-trnE, K81uf + I + G (three rate classes following a gamma distribution and invariant sites) for the psbD-trnT, and TVM + I + G (five rate classes following a gamma distribution and invariant sites) for the combined datasets. Since MrBayes only allows the choices of 1, 2 or 6 rate categories, all of the 3+ rate category submodels of the general time reversible model were placed in the six rate classes. Bayesian analysis using a single evolutionary model across all DNA regions and the partitioned analysis of each cpDNA region produced trees with –ln L = 17 111·61 and –ln L = 18 949·80, respectively. The Bayes factor between the single and partitioned models was therefore 1838·19, indicating that application of a single evolutionary model was more appropriate than partitioned analysis. The results presented here are therefore based on homogeneous Bayesian inference.
Table 5.
Best-fitting models and parameter values for separate (petA-psbJ, trnT-trnL, trnT-trnE, psbD-trnT) and combined datasets in this study
| Base frequencies |
Substitution model (rate matrix) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Region | AIC selected model | A | C | G | T | A–C | A–G | A–T | C–G | C–T | G–T | I* | G* |
| petA-psbJ | TIM | 0·3623 | 0·1419 | 0·1185 | 0·3773 | 1·0000 | 0·8642 | 0·2735 | 0·2735 | 0·5067 | 1·0000 | 0 | Equal |
| trnT-trnL | TIM + G | 0·4190 | 0·0884 | 0·1238 | 0·3688 | 1·0000 | 0·0865 | 0·0596 | 0·0596 | 0·2375 | 1·0000 | 0 | 0·3479 |
| trnT-trnE | TVM + G | 0·3709 | 0·1337 | 0·1260 | 0·3695 | 1·0000 | 0·5281 | 0·2382 | 0·8342 | 0·5281 | 1·0000 | 0 | 0·6412 |
| psbD-trnT | K81uf + I + G | 0·3510 | 0·1391 | 0·1247 | 0·3853 | 1·0000 | 0·3110 | 0·1832 | 0·1832 | 0·3110 | 1·0000 | 0·5654 | 0·9177 |
| Combined analyses | TVM + I + G | 0·3731 | 0·1240 | 0·1254 | 0·3775 | 1·1519 | 0·3672 | 0·1489 | 0·4059 | 0·3672 | 1·0000 | 0·3310 | 0·8910 |
* I, Proportion of invariable sites; G, gamma distribution.
The strict consensus tree and Bayesian tree were identical in the topology and revealed two clades with robust support, clade A and clade B (Fig. 2I). Nine taxa of the subgenus Ceratotropis formed clade A with high support (BS = 100 %; Fig. 2I) having inversion type I, and they fell into two subclades, A-I and A-II (Fig. 2I). Clade A-I constitutes V. angularis (wild, weedy and cultivated forms), two Myanmar accessions of V. aff. minima (Vmn-1 and Vmn-2; Table 1) and V. nepalensis (Fig. 2I) which were collected from the temperate regions of East Asia and the Himalayan highlands. Clade A-II included V. tenuicaulis, V. hirtella, V. exilis, an Indonesian accession of V. minima (Vmn-3; Table 1), V. umbellata, V. riukiuensis and V. nakashimae (Fig. 2I) which were mainly found in a subtropical climate. The V. hirtella, V. umbellata and V. exilis clade was highly supported (BS = 86 %) in parsimony but not in Bayesian analysis (Fig. 2I). Twelve taxa of the subgenus Ceratotropis form clade B with high support (BS = 100 %; Fig. 2I), including four domesticated and eight wild species of the subgenus Ceratotropis which are mainly distributed in the tropical region of the Indian subcontinent and Southeast Asia. This clade B included taxa belonging to three sections of Ceratotropis and mainly has inversion type II and its three subtypes, except Sri Lankan accession of V. trilobata (Table 1 and Fig. 2I). The species relationships in this clade were not clearly resolved; however, there were two distinct subclades: subclade B-I, including V. trinervia, V. reflexo-pilosa and an Indian accession of V. trilobata (Fig. 2I), and subclade B-II, consisting of V. mungo and V. grandiflora (Fig. 2I).
Multiple accessions of 13 species were sampled, and most species formed monophyletic clades. Three accessions of V. minima, however, fell into two independent clades. The two accessions of V. trilobata were paraphyletic (clade B; Fig. 2I). One accession of V. trilobata (India, Vtb-1) was sister to V. reflexo-pilosa and V. trinervia in subclade B-I (Fig. 2I; Table 1). The other accession of V. trilobata (Sri Lanka, Vtb-2) was placed in the basal part of clade B (Fig. 2I and Table 1).
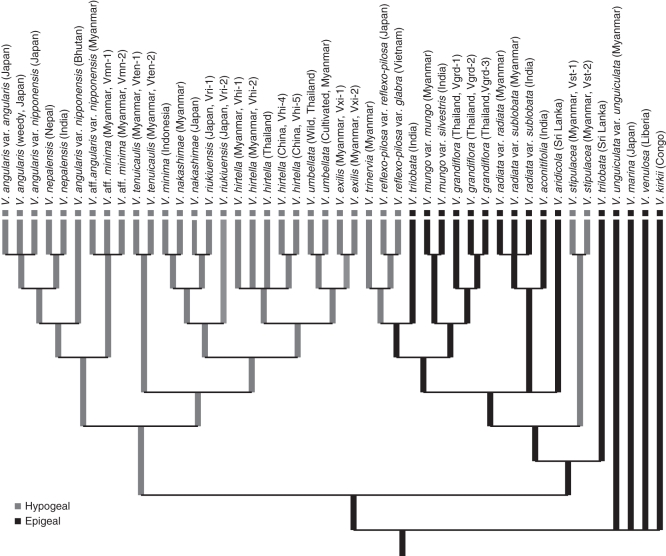
Character state mapping for seedling germination
The character changes and ancestral-state reconstruction on the tree were explored using the total-evidence analysis of plastid sequences. ACCTRAN and DELTRAN optimizations showed similar results, and only results using ACCTRAN are presented. Epigeal seedling germination is polyphyletic according to the trees (Fig. 3). In clade B, Vigna mungo, V. grandiflora, V. radiata and V. aridicola are plesiomorphically characterized as having epigeal germination. Two separate gains in hypogeal germination are also inferred in clade B: once in the evolution of V. stipulacea and again in the clade containing V. trinervia and V. reflexo-pilosa (Fig. 3). Hypogeal germination is also a synapomorphy for clade A (Figs 2I and 3). A parsimony interpretation of character evolution reveals that hypogeal germination was derived independently on three occasions from epigeal germination.
Fig. 3.
Mapping of seedling germination type on the strict consensus tree of Vigna subgenus Ceratotropis generated from the combined DNA dataset using MacClade 4·0 (Maddison and Maddison, 2000).
Age estimation
By setting the root node on the trees at a maximum of 8·0 Mya, the estimated divergence time for clade A and clade B was 3·62 ± 0·3, and that of two subclades, A-I and A-II, within clade A (Fig. 2I) was 2·0 ± 0·2 Mya. The chronogram showing the range of estimated divergence times is shown in Fig. 2II.
DISCUSSION
Phylogenetic utility of the four plastid regions
The psbD-trnT, trnT-trnE and trnT-trnL intergenic spacer sequences showed a significant amount of variation in Vigna, in accordance with that of Shaw et al. (2005, 2007). The psbD-trnT region has a higher substitution rate than those of the three other regions (Table 3). The trnT-trnE and the trnT-trnL regions, although evolving more slowly than the psbD-trnT region, have a higher percentage of phylogenetically informative sites among the variable sites (6·6 % vs. 6·7 %; Table 3). The indels in the psbD-trnT, trnT-trnE and trnT-trnL regions provided phylogenetically informative and species-specific characters, and a few indels showed intraspecific variation. These DNA regions have good potential for phylogenetic analyses at the interspecific level.
The inversion found in the psbD-trnT region was bordered by inverted repeat sequences (Fig. 1), suggesting formation of stem–loop structures and recombination in the stems at inversion process (Sang et al., 1997). In contrast to some large inversions in cpDNA which provided reliable phylogenetic information at the higher taxonomic levels (i.e. Doyle, 1992; Raubeson and Jansen, 1992), short inversions in the intergenic spacer easily yielded homoplasious information even at the interspecific level (Sang et al., 1997). Thus, sequence data of inversion were not included in the present phylogenetic analyses, following the recommendation of Sang et al. (1997). Nevertheless, the inversion might provide additional information on certain levels in the present study, e.g. both clades differed in their inversion type (Fig. 2I).
The tree derived from the combined sequence data of the four chloroplast intergenic spacers strongly supported the interspecific relationships of the subgenus Ceratotropis (Fig. 2I). This phylogenetic view is in general agreement with that of previously published subgenus Ceratotropis phylogenies (Goel et al., 2002; Yano et al., 2004; Ye Tun Tun and Yamaguchi, 2007), although the results of the present study show that the inclusion of more molecular data under an increasing taxon-sampling scheme was crucial to increase the resolution and support of the phylogenetic tree.
Phylogenetic and biogeographical relationships
The topology of the cpDNA phylogeny clearly indicates geographical and ecological relationships within the subgenus Ceratotropis (Fig. 2I). According to known geographical distribution patterns of the subgenus Ceratotropis (Tomooka et al., 2002b; authors' own field surveys), two major clades in the phylogenetic tree show geographic structure: East Asia–Southeast Asia clade (clade A) and the Indian subcontinent clade (clade B), and the two ecogeographic substructures within clade A, temperate group (subclade A-I) and subtropical group (subclade A-II) (Fig. 2I).
East Asia–Southeast Asia clade (temperate group and subtropical group)
The monophyletic temperate group contains domesticated azuki bean (V. angularis var. angularis), wild azuki bean (V. angularis var. nipponensis), weedy azuki (V. angularis), V. nepalensis and V. aff. minima (Myanmar accessions) (Fig. 2I), which are distributed throughout the temperate East Asia regions and the cooler parts of the Himalayan foothills. The well-supported monophyly of wild, weedy and cultivated V. angularis (Fig. 2I) is congruent with that of previous findings in terms of morphology (Yamaguchi, 1992), cross compatibility (Sawa, 1983), isozyme features (Yasuda and Yamaguchi, 1996), RAPD (Mimura et al., 2000), AFLP (Zong et al., 2003) and cpDNA sequences (Yano et al., 2004; Ye Tun Tun and Yamaguchi, 2007, 2008). Despite the affinity of V. nepalensis and V. tenuicaulis in AFLP analysis (Tomooka et al., 2002c), present and previous studies (Doi et al., 2002; Zong et al., 2003; Ye Tun Tun and Yamaguchi, 2007, 2008) reveal a close relationship between V. angularis and V. nepalensis (Fig. 2I). These two species have a number of morphological similarities such as flower colour, shape of calyx, hypogeal germination (Tomooka et al., 2002b) and share a unique 8-bp insertion in the trnT-L spacer region. They differ in hairiness of primary and secondary bracts and bracteole (pubescent in V. angularis vs. glabrous in V. nepalensis) and length of bracteole/calyx (bracteole much longer than calyx in V. angularis vs. as long as calyx in V. nepalensis) (Tomooka et al., 2002b). In this monophyletic temperate group, V. angularis aff. var. nipponensis (Van-5; Table 1) from the hilly region of northern Myanmar and two accessions of V. aff. minima from Myanmar are grouped together with high support (BS = 98 %; Fig. 2I) and share three synapomorphic substitutions. This Myanmar accession (Van-5, Table 1) is shorter and has smaller floral parts than the other accessions of V. angularis var. nipponensis, and they share a 11-bp deletion of the trnT-L spacer region with accessions from Bhutan and China (Ye Tun Tun and Yamaguchi, 2007). Since the Myanmar accessions, V. aff. minima (Vmn-1 and Vmn-2; Table 1) resemble V. minima in many key characters, including size of stipule, bract and bracteole, glabrous pod, and well-developed aril (Tomooka et al., 2002b), differ from V. angularis aff. var. nipponensis in a 51-bp insertion of the trnL-F spacer region (Ye Tun Tun and Yamaguchi, 2007), and its flower is morphologically similar to that of V. tenuicaulis, it is considered to be a distinct taxon in temperate Ceratotropis. The relationship between V. aff. minima and V. angularis aff. var. nipponensis in the tree reveals that the ecological and geographical aspects of species ranges reflect the phylogenetic relationships and further support the finding that ecological characteristics are an obvious determinant of geographical distribution in Vigna and relatives (Lavin and Beyra-Matos, 2008).
The monophyletic subtropical group contains Vigna minima (from Indonesia), V. nakashimae, V. riukiuensis, V. hirtella, V. exillis and V. umbellata (Fig. 2I; clade A-II). Almost all the species of this group are found in the plant communities that are marginal to the subtropical rain forests of Southeast Asia except natural populations of V. nakashimae in the temperate Far East. The three morphologically close species, V. nakashimae, V. minima (Indonesia) and V. riukiuensis, are included in a well-supported subgroup (Fig. 2I), and they are distinguished by a large golden flower in V. minima, small glossy leaflets in Vigna riukiuensis and a protruding hilum and a small, pale yellow flower in V. nakashimae (Tomooka et al. 2002b). Vigna nakashimae was first described in Japan and has only been reported in East Asia (Tateishi, 1985; Tateishi and Ohashi, 1990; Tomooka et al., 2002b); however, it was recently found in the area from the Far East through southern China to the southern part of Myanmar (Mon state; Ye Tun Tun and Yamaguchi, 2007). Accessions of V. nakashimae in this study show close similar plastic sequences despite their intraspecific differentiation in AFLP data (Seehalak et al., 2006). Since V. riukiuensis is endemic to the southern part of the Ryukyu archipelago (Miyako and Sakishima Islands, Okinawa) and these islands constitute an extended peninsula from Taiwan and the Chinese mainland during the last glacial age, the present results support the hypothesis that the island species V. riukiuensis might have evolved from a continental species, V. nakashimae (Ye Tun Tun and Yamaguchi, 2007); we further propose that V. nakashimae, V. riukiuensis and V. minima evolved from a common ancestor (Fig. 2I). As V. nakashimae and V. riukiuensis were treated as distinct taxa (Tomooka et al., 2002b) or considered as subspecies/varieties of V. minima (Tateishi, 1985), their taxonomic delimitation remains crucial due to a high variation of Vigna minima. The Indonesian accession (Vmn-3; Table 1) has the same morphological features of the plants recognized as V. minima, representing small stipule, small primary and secondary bracts, small bracteoles, glabrous pod and hilum with well-developed aril. Since the lack of monophyly of even morphologically well-defined species can result from incomplete lineage sorting of shared ancestral polymorphism or contemporary gene flow (Funk and Omland, 2003), the polyphyly of V. minima and V. aff. minima is probably due to the ancestral polymorphism, and thus there is a need for further examination of this group.
The close relationship between the three species, V. hirtella, V. exillis and V. umbellata, is well-resolved and highly supported (BS = 86 %; Fig. 2I) as is also the case in the AFLP study (Seehalak et al., 2006). They share a unique single base-pair deletion in the trnT-E region. Vigna umbellata and V. exillis share some characters such as protruding hilum and well-developed aril in their seed morphology, although these characters are homoplasious in the subgenus Ceratotropis. Despite the AFLP results (Seehalak et al., 2006), in V. hirtella, five accessions from a wide area (Myanmar, Thailand and China) are monophyletic (BS = 98 %, BI = 99 %; Fig. 2I) and share a 13-bp insertion in the trnT-trnE region, except for one accession from China (Vhi-5). In this monophyletic of V. hirtella, two accessions from China (Vhi-4 and Vhi-5; Table 1 and Fig. 2I) share one unique 1-bp substitution and a single base-pair deletion. The present combined cpDNA tree suggests that AFLP differentiation among V. hirtella accessions (Seehalak et al., 2006) might be due to geographical or population differentiation within species but not to a distant relationship.
The wild and cultivated accessions of V. umbellata (rice bean) show only two nucleotide substitution differences in the four plastid regions which form a monophyletic clade (Fig. 2I). This result agrees with the findings of the AFLP analysis (Seehalak et al., 2006) and the molecular tree based on the trnT-F region (Ye Tun Tun and Yamaguchi, 2007). Vigna tenuicaulis has the early branching position within the subtropical group with moderate clade support (BS = 78 %; Fig. 2I), although this species was considered to be close to V. angularis and V. nepalensis (Tomooka et al., 2002b).
Indian subcontinent clade (tropical group)
Vigna species of clade B inhabit hot-dry or sub-humid tropical lowlands mainly on the Indian subcontinent, and this species is considered as the tropical group (Fig. 2I). Although the phylogenetic relationships among members of this group were unresolved in the previous studies (Doi et al., 2002; Konarev et al., 2002; Tomooka et al., 2002c; Ye Tun Tun and Yamaguchi, 2007), two well-supported subgroups of the tropical group were recognized in the molecular tree, B-I and B-II (Fig. 2I). The close relationship between V. trinervia and V. reflexo-pilosa (BS = 93 %, BI = 99 %) and their inclusion in the tropical group is clear as clade B-I which was also the case with the AFLP results (Seehalak et al., 2006), although the two species have been placed as members of the azuki bean group (Maekawa, 1955; Tateishi and Ohashi, 1990) or in section Angulares (Tomooka et al., 2002a, b). Vigna trinervia has a distribution pattern in Africa, Asia and the Pacific Islands and shares characters such as rectangular seed and hypogeal germination with V. reflexo-pilosa (Tomooka et al., 2002b). The placement of V. reflexo-pilosa within the Indian subcontinent clade (Fig. 2I) conflicts with the result of the nrDNA ITS tree (Doi et al., 2002). The present results are in agreement with the assumption that V. trinervia is a diploid donor (Egawa et al., 1996; Konarev et al., 2002) or a maternal donor (Yano et al., 2004; Ye Tun Tun and Yamaguchi, 2007) of allotetraploid V. reflexo-pilosa. Its sister relationship of an Indian accession of V. trilobata to V. reflexo-pilosa and V. trinervia with moderate support (BS = 78 %, BI = 81 %; Fig. 2I) is worth noting, as seen in an earlier analysis based on trnT-F data (Ye Tun Tun and Yamaguchi, 2007).
The grouping of V. grandiflora and V. mungo received moderate support as clade B-II (BS = 77 %, BI = 84 %; Fig. 2I) sharing one synapomorphic substitution and a unique single base-pair deletion in the trnT-trnL and petA-psbJ regions, respectively. Although V. mungo differs morphologically from V. grandiflora by the brightness of its yellow flower, protruding ovate hilum and well-developed aril (Tomooka et al., 2002b), a close relationship between V. grandiflora and V. mungo in the present tree is congruent with that of the nrDNA ITS phylogenetic tree (Doi et al., 2002). Vigna mungo var. silvestris is widely distributed throughout India, Myanmar and Thailand, while V. grandiflora is narrowly distributed in Thailand and Cambodia (Baudoin and Maréchal, 1988; Tateishi, 1996; Tomooka et al., 2002b). This phylogeographical pattern leads to the assumption that V. grandiflora may have gained its present distribution area after differentiation from V. mungo var. silvestris. Vigna aconitifolia is sister to V. radiata in the present plastid tree with low support (BS = 62 %; Fig. 2I), though both of them have one shared synapomorphic character in the petA-psbJ spacer region. However, these two species differ in numerous morphological characters such as shape of leaflet (deeply five-lobed terminal and deeply four-lobed lateral leaflets in V. aconitifolia and ovate terminal and obliquely lateral leaflets in V. radiata), flower size and colour (small bright yellow flower in V. aconitifolia and large pale greyish yellow flower in V. radiata) and seed shape (elliptic in V. aconitifolia and rectangular in V. radiata), despite their molecular relatedness.
Although the phylogenetic relationship is not so well resolved, the domesticated and wild types of V. radiata are monophyletic (Fig. 2I) and their relationship is supported by a 28-bp deletion in the trnT-L region. Vigna radiata var. sublobata is widely distributed in Africa, Asia and the Pacific islands (Tomooka et al., 2002b) by its weedy habit (Chandel et al., 1984), and it occupies mostly disturbed habitat such as roadsides (Tomooka et al., 2002b) which may contribute to its rapid dispersal. A possible explanation for its low molecular divergence could be that divergence within V. radiata occurred recently, such that neutral cpDNA intergenic spacers variation would not have had sufficient time to be fixed in the different taxa. The basal position of V. stipulacea in the tropical group is supported by a unique 27-bp insertion in psbD-trnT and the sister-species relationships of V. stipulacea and Sri Lankan V. trilobata to the tropical group by 3-bp synapomorphic insertion in the petA-psbJ region.
The two accessions of V. trilobata sampled from Sri Lanka and India are resolved as paraphyletic. Vigna trilobata is distributed in Sri Lanka, India and Myanmar, and is characterized by its orbicular to ovate stipule, golden yellow flower, glabrous mature pod, seed with protruding orbicular, and hilum having well-developed aril (Tomooka et al., 2002b). The Indian accession of V. trilobata has longer hairs on stems and leaves and less protruding hilum in comparison with that of the Sri Lankan accession, and it shares a rectangular seed shape with V. reflexo-pilosa. At the molecular level, Sri Lankan and Indian accessions are differentiated by several substitutions and indels, and also type II of the 42-bp inversion in the petA-psbD region in Indian accession of V. trilobata (Table 1 and Fig. 2I). Because of the apparent frequency of rapidly radiating clades that create conditions favourable for incomplete lineage sorting, plant species paraphyly is a common finding (i.e. Wood and Nakazato, 2009; Fishbein et al., 2010). We suppose that placement of Indian and Sri Lankan accessions of V. trilobata in different phylogenetic positions is probably due to the ancient differentiation between the island population (Sri Lanka) and the mainland population (India) or due to delimitation status of V. trilobata.
Patterns of character evolution
The well-resolved phylogeny in this study provides an opportunity for an analysis of character evolution that has been used to define groups at various taxonomic levels. Hypogeal and epigeal germination was used as one of the key morphological characters in sectional classification of Vigna subgenus Ceratotropis (Sects. Angulares, Ceratotropis and Aconitifoliae; Table 1) (Tomooka et al., 2002a, b). The cpDNA phylogeny present here indicates that the germination type evolved independently multiple times (Fig. 3), suggesting the taxonomic limit of the utility of seedling germination. Based on morphological cladistic analysis, the hypogeal germination was suggested as the primitive state in the subgenus Ceratotropis (Tateishi, 1985, 1996); however, the present results assume its derived condition and that it evolved in three independent lineages (Fig. 3). Furthermore, seedling traits are evolutionarily conservative, reflecting phylogenetic niche conservatism (Ibarra-Manrìquez et al., 2001). Dispersal of Sophora tomentosa in the tropics appears to be correlated with its hypogeal germination (Heenan et al., 2004). In the genus Manihot, a phylogenetically based survey of seedling functional morphology is associated with habitat and life form (Pujol et al., 2005), although it is difficult to identify the relationship between habitat and germination type within some families such as Restionaceae (Linder and Caddick, 2001). The phylogeny generated in this study suggests that seedling germination type is probably associated with a shift in habitat conditions since species with epigeal germination are mainly distributed in dry and tropical lowland habitats, while the subhumid and warm temperate regions are mainly occupied by species with hypogeal germination except for V. stipulaceae, V. trinervia and V. reflexo-pilosa (Figs 2I and 3).
Diversification time in the subgenus Ceratotropis
The combined cpDNA phylogenetic tree showed two major geographical groups in the subgenus Ceratotropis; the Indian subcontinent group and East Asia–Southeast Asia group, suggesting the process of interspecific diversification to different climatic and ecological habitats, e.g. temperate, subtropical and tropical groups (Fig. 2I). The estimated divergence time (3·62 ± 0·3 Mya) between the Indian subcontinent group and East Asia–Southeast Asia group was less than that of the Tibetan uplift (8 Mya or 15 Mya; Harrison et al., 1992; Spicer et al., 2003), suggesting the diversification within the subgenus Ceratotropis may have occurred after the collision of the Indian subcontinent and the Asian plate. The recent diversification of the subgenus Ceratotropis was congruent with the findings of Lavin et al. (2005) who proposed that the age of the African crown clade including African and Asian Vigna (V. mungo and V. umbellata) was about 5·1 Mya. Moreover, molecular dating indicates late Pliocene diversification of the temperate and subtropical groups (2·0 ± 0·2 Mya) in the subgenus Ceratotropis. However, diversification spread through late Pliocene and into the Pleistocene. The north-east Indian passageway (Cordaux et al., 2004), a narrow land bridge between the Indian subcontinent and East Asia which was created by the collision of Indian subcontinent with Asia, might be an important geographic barrier for species migration, together with the formation of high mountain ranges between the two regions. Therefore, formation and persistence of seasonal climate after the collision of the Indian subcontinent with the Asian plate and long-distance dispersal could have subsequently facilitated the Pliocene–Pleistocene diversification of the subgenus Ceratotropis into different eco-geographic groups.
Conclusions
The evidence presented here clearly demonstrates the geography and ecological habitat as important factors in shaping phylogenetic relationships of the subgenus Ceratotropis. Indels in the four cpDNA intergenic spacers provide useful information to infer the phylogenetic relationships at the interspecific levels in the subgenus Ceratotropis. It is important to note that DNA-based identification in the subgenus Ceratotropis would be much more challenging for a future study. Mapping seedling germination used taxonomically in the subgenus Ceratotropis into the phylogeny indicates the multiple origins, thus limiting its taxonomic utility, and a reassessment of diagnostic characters in the current classification of subgenus is thus necessary. Ultimately, this study provides a framework for future studies that deal taxonomically with Vigna species.
ACKNOWLEDGEMENTS
We are particularly grateful to Matthew Lavin for correcting the English and providing extremely useful suggestions for improving the manuscript. We appreciate the attention of Pat Heslop-Harrison, Jeannette Whitton and two anonymous reviewers, whose detailed comments greatly improved the manuscript. We also thank Yuichiro Nakayama and Kyoko Yamane for technical advice during this research. The study was funded by Grant-in Aid for Scientific Research (B) nos 15310161, 1831054 and 23310168 from the Japan Society for the Promotion of Science.
LITERATURE CITED
- Baudet JC. Signification taxonomique des caractéres blastogéniques dans la tribu des Papilionaceae-Phaseoleae. Bulletin du Jardin Botanique National de Belgiquè. 1974;44:259–293. [Google Scholar]
- Baudoin JP, Maréchal R. Taxonomy and evolution of the genus Vigna. In: Shanmugasundaram S, McLean BT, editors. Mungbean: Proceedings of the Second International Symposium. Taiwan: Asian Vegetable Research and Development Center; 1988. pp. 2–12. [Google Scholar]
- Brandley MC, Schmitz A, Reeder TW. Partitioned Bayesian analyses, partition choice, and the phylogenetic relationships of scincid lizards. Systematic Biology. 2005;54:373–390. doi: 10.1080/10635150590946808. [DOI] [PubMed] [Google Scholar]
- Chandel KPS, Lester RN, Starling RJ. The wild ancestors of urid and mung beans [V. mungo (L.) Hepper and V. radiata (L.) Wilcz.] Botanical Journal of the Linnean Society. 1984;89:85–96. [Google Scholar]
- Cordaux R, Weiss G, Saha N, Stoneking M. The northeast Indian passageway: a barrier or corridor for human migrations? Molecular Biology and Evolution. 2004;21:1525–1533. doi: 10.1093/molbev/msh151. [DOI] [PubMed] [Google Scholar]
- Delgado-Salinas A, Bibler R, Lavin M. Phylogeny of the genus Phaseolus (Leguminosae): a recent diversification in an ancient landscape. Systematic Botany. 2006;31:779–791. [Google Scholar]
- Doi K, Kaga A, Tomooka N, Vaughan DA. Molecular phylogeny of genus Vigna subgenus Ceratotropis based on rDNA ITS and atpB-rbcL intergenic spacer region of cpDNA sequences. Genetica. 2002;114:129–145. doi: 10.1023/a:1015158408227. [DOI] [PubMed] [Google Scholar]
- Doyle JJ. Chloroplast DNA inversions and the origin of the grass family (Poaceae) Proceeding of the National Academy of Sciences of the United States of America. 1992;89:7722–7726. doi: 10.1073/pnas.89.16.7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin of the Botanical Society of America. 1987;19:11–15. [Google Scholar]
- Egawa Y, Nakagahra M, Fernandez GCJ. Cytogenetical analysis of tetraploid Vigna glabrescens by interspecific hybridization involving diploid Asian Vigna species. In: Shanmugasundaram S, McLean BT, editors. Mungbean: Proceedings of the Second International Symposium. Taiwan: Asian Vegetable Research and Development Center; 1988. pp. 200–204. [Google Scholar]
- Egawa Y, Bujang IB, Chotechuen S, Tomooka N, Tateishi Y. Phyloegenetic differentiation of tetraploid Vigna species, V. glabrescens and V. reflexo-pilosa. JIRCAS Journal. 1996;3:49–58. [Google Scholar]
- Endo Y, Ohashi H. Cladistic analysis of phylogenetics relationships among tribes Cicereae, Trifolieae, and Vicieae (Leguminosae) American Journal of Botany. 1997;84:523–529. [PubMed] [Google Scholar]
- Essig FB. Seedling morphology in Clematis (Ranunculaceae) and its taxonomic implications. Sida. 1992;15:377–390. [Google Scholar]
- Farris JS, Källersjö M, Kluge AG, Bult C. Testing significance of incongruence. Cladistics. 1995;10:315–320. [Google Scholar]
- Fatokun CA, Danesh D, Young ND, Stewart EL. Molecular taxonomic relationships in the genus Vigna based on RFLP analysis. Theoretical and Applied Genetics. 1993;86:97–104. doi: 10.1007/BF00223813. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. Journal of Molecular Evolution. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annual Review of Genetics. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- Fishbein M, Kephart SR, Wilder M, Halpin KM, Datwyler SL. Phylogeny of Camassia (Agavaceae) inferred from plastid rpl16 intron and trnD-trnY-trnE-trnT intergenic spacer DNA sequences: implications for species delimitation. Systematic Botany. 2010;35:77–85. [Google Scholar]
- Funk DJ, Omland KE. Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annual Review of Ecology, Evolution, and Systematics. 2003;34:397–423. [Google Scholar]
- Gates RR. Epigeal germination in the Leguminosae. Botanical Gazette. 1951;113:151–157. [Google Scholar]
- Goel S, Raina SN, Ogihara Y. Molecular evolution and phylogenetic implications of internal transcribed spacer sequences of nuclear ribosomal DNA in the Phaseolus–Vigna complex. Molecular Phylogenetics and Evolution. 2002;22:1–19. doi: 10.1006/mpev.2001.1037. [DOI] [PubMed] [Google Scholar]
- Graham SW, Reeves PA, Burns ACE, Olmstead RG. Microstructural changes in non-coding chloroplast DNA: interpretation, evolution, and utility of indels and inversions in basal angiosperm phylogenetic inference. International Journal of Plant Sciences. 2000;161:S83–S96. [Google Scholar]
- Harrison TM, Copeland P, Kidd WSF, Yin A. Raising Tibet. Science. 1992;255:1663–1670. doi: 10.1126/science.255.5052.1663. [DOI] [PubMed] [Google Scholar]
- Heenan PB, Dawson MI, Wagstaff SJ. The relationship of Sophora sect. Edwardsia (Fabaceae) to Sophora tomentosa, the type species of the genus Sophora, observed from DNA sequence data and morphological characters. Botanical Journal of the Linnean Society. 2004;146:439–446. [Google Scholar]
- Huelsenbeck JP, Crandall KA. Phylogeny estimation and hypothesis testing using maximum likelihood. Annual Review of Ecology and Systematics. 1997;28:437–466. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielson R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- Ibarra-Manrìquez G, Martinez Ramos M, Oyama K. Seedling functional types in a lowland rain forest in Mexico. American Journal of Botany. 2001;88:1801–1812. [PubMed] [Google Scholar]
- Javadi F, Wojciechowski MF, Yamaguchi H. Geographical diversification of the genus Cicer (Leguminosae: Papilionoideae) inferred from molecular phylogenetic analyses of chloroplast and nuclear DNA sequences. Botanical Journal of the Linnean Society. 2007;154:175–186. [Google Scholar]
- Kass RE, Raftery AE. Bayes factors. Journal of the American Statistical Association. 1995;90:773–795. [Google Scholar]
- Konarev AV, Tomooka N, Vaughan DA. Proteinase inhibitor polymorphism in the genus Vigna Savi subgenus Ceratotropis and its biosystematic implications. Euphytica. 2002;123:165–177. [Google Scholar]
- Lavin M, Beyra-Matos A. The impact of ecology and biogeography on legume diversity, endemism, and phylogeny in the Caribbean region: a new direction in historical biogeography. The Botanical Review. 2008;74:178–196. [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the Tertiary. Systematic Biology. 2005;54:530–549. doi: 10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
- Linder HP, Caddick LR. Restionaceae seedlings: morphology, anatomy and systematic implications. Feddes Repertorium. 2001;112:59–80. [Google Scholar]
- Lumpkin TA, McClary DC. Azuki bean: botany, production and uses. Wallingford: CAB International; 1994. [Google Scholar]
- Maddison DR, Maddison WP. MacClade 4·06: analysis of phylogeny and character evolution. Sunderland, MA: Sinauer; 2000. [DOI] [PubMed] [Google Scholar]
- Maekawa F. Topo-morphological and taxonomical studies in Phaseoleae, Leguminosae. The Journal of Japanese Botany. 1955;15:103–116. [Google Scholar]
- Maréchal R, Mascherpa JM, Stainer F. Etude taxonomique d'un groupe complexe d'espèces des genres Phaseolus et Vigna (Papilionaceae) sur la base de données morphologiques et polliniques, traitées par l'analyse informatique. Boissiera. 1978;28:1–273. [Google Scholar]
- Mimura M, Yasuda K, Yamaguchi H. RAPD variation in wild, weedy and cultivated azuki beans in Asia. Genetic Resources and Crop Evolution. 2000;47:603–610. [Google Scholar]
- Nylander JAA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey JL. Bayesian phylogenetic analysis of combined data. Systematic Biology. 2004;53:47–67. doi: 10.1080/10635150490264699. [DOI] [PubMed] [Google Scholar]
- Polhill RM. Papilionoideae. In: Polhill RM, Raven PH, editors. Advances in legume systematics. Vol. 1. Richmond: Royal Botanic Gardens, Kew; 1981. pp. 191–208. [Google Scholar]
- Posada D, Buckley TR. Model selection and model averaging in phylognetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Systematic Biology. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Pujol B, Mühlen G, Garwood N, Horoszowski Y, Douzery EJP, McKey D. Evolution under domestication: contrasting functional morphology of seedlings in domesticated cassava and its closest wild relatives. New Phytologist. 2005;166:305–318. doi: 10.1111/j.1469-8137.2004.01295.x. [DOI] [PubMed] [Google Scholar]
- Raubeson LA, Jansen RK. Chloroplast DNA evidence on the ancient evolutionary split in vascular land plants. Science. 1992;255:1697–1699. doi: 10.1126/science.255.5052.1697. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Saini A, Jawali N. Molecular evolution of 5S rDNA region in Vigna subgenus Ceratotropis and its phylogenetic implications. Plant Systematics and Evolution. 2009;280:187–206. [Google Scholar]
- Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Molecular Biology and Evolution. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- Sang T, Crawford DJ, Stuessy TF. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae) American Journal of Botany. 1997;84:1120–1136. [PubMed] [Google Scholar]
- Sawa M. Study on the breeding of azuki bean (V. angularis) by interspecific crossing. Journal of Michurin Biology. 1983;18:3–26. [Google Scholar]
- Seehalak W, Tomooka N, Waranyuwat A, et al. Genetic diversity of the Vigna germplasm from Thailand and neighboring regions revealed by AFLP analysis. Genetic Resources and Crop Evolution. 2006;53:1043–1059. [Google Scholar]
- Shaw J, Lickey EB, Beck JT, et al. The tortoise and the hare. II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. American Journal of Botany. 2005;92:142–166. doi: 10.3732/ajb.92.1.142. [DOI] [PubMed] [Google Scholar]
- Shaw J, Lickey EB, Schilling EE, Small RL. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. American Journal of Botany. 2007;94:275–288. doi: 10.3732/ajb.94.3.275. [DOI] [PubMed] [Google Scholar]
- Smartt J. Grain legumes: evolution and genetic resources. Cambridge: Cambridge University Press; 1990. [Google Scholar]
- Spicer RA, Harris NBW, Widdowson M, et al. Constant elevation of southern Tibet over the past 15 million years. Nature. 2003;421:622–624. doi: 10.1038/nature01356. [DOI] [PubMed] [Google Scholar]
- Swindell RE, Watt EE, Evans GM. A natural tetraploid mung bean of suspected amphidiploid origin. Journal of Heredity. 1973;64:107. [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods). Ver. 4·0b10. Sunderland, MA: Sinauer Associates; 2002. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4·0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tateishi Y. Leguminosae collected in the Arun valley, East Nepal. In: Numata M, editor. Structure and dynamics of vegetation in eastern Nepal. Japan: Chiba University; 1983. pp. 131–146. [Google Scholar]
- Tateishi Y. A revision of the azuki bean group, the subgenus Ceratotropis of the genus Vigna (Leguminosae) 1985 PhD Thesis, Tohoku University, Japan. [Google Scholar]
- Tateishi Y. Systematics of the species of Vigna subgenus Ceratotropis. In: Srinives P, Kitabamroong C, Miyazaki S, editors. Mungbean germplasm: collection, evaluation and utilization for breeding program. Japan: JIRCAS; 1996. pp. 9–24. [Google Scholar]
- Tateishi Y, Ohashi H. Systematics of the Azuki bean group in the genus Vigna. In: Fujii K, Gatehouse AMR, Johnson CD, Mitchel R, Yoshida T, editors. Bruchids and legumes: economics, ecology and coevolution. Dordrecht: Kluwer Academic Publishers; 1990. pp. 189–199. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thulin M, Lavin M, Pasquet R, Delgado-Salinas A. Phylogeny and biogeography of Wajira (Leguminosae): a monophyletic segregate of Vigna centered in the Horn of Africa region. Systematic Botany. 2004;29:903–920. [Google Scholar]
- Tomooka N, Lairungreang C, Nakeeraks P, Egawa Y, Thavarasook C. Mungbean and the genetic resources, the subgenus Ceratotropis. 1991. Tsukuba: Tropical Agriculture Research Center. [Google Scholar]
- Tomooka N, Maxted N, Thavarasook C, Jayasuriya AHM. Two new species, new species combinations and sectional designations in Vigna subgenus Ceratotropis (Piper) Verdcourt (Leguminosae, Phaseoleae) Kew Bulletin. 2002a;57:613–624. [Google Scholar]
- Tomooka N, Vaughan DA, Moss H, Maxted N. The Asian Vigna: genus Vigna subgenus Ceratotropis genetic resources. Dordrecht: Kluwer; 2002b. [Google Scholar]
- Tomooka N, Yoon MS, Doi K, Kaga A, Vaughan DA. AFLP analysis of diploid species in the genus Vigna subgenus Ceratotropis. Genetic Resources and Crop Evolution. 2002c;49:521–530. [Google Scholar]
- Verdcourt B. Studies in the Leguminosae–Papilionoideae for the flora of tropical East Africa, IV. Kew Bulletin. 1970;24:507–569. [Google Scholar]
- Wood TE, Nakazato T. Investigating species boundaries in the Giliopsis group of Ipomopsis (Polemoniaceae): strong discordance among molecular and morphological markers. American Journal of Botany. 2009;96:853–861. doi: 10.3732/ajb.0800153. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H. Wild and weed azuki beans in Japan. Economic Botany. 1992;46:384–394. [Google Scholar]
- Yano A, Yasuda K, Yamaguchi H. A test for molecular identification of Japanese archaeological beans and phylogenetic relationship of wild and cultivated species of subgenus Ceratotropis (Genus Vigna, Papilionaceae) using sequence variation in two non-coding regions of the trnL and trnF genes. Economic Botany. 2004;58:S135–S146. [Google Scholar]
- Yasuda K, Yamaguchi H. Phylogenetic analysis of the subgenus Ceratotropis (genus Vigna) and an assumption of the progenitor of azuki bean using isozyme variation. Breeding Science. 1996;46:337–342. [Google Scholar]
- Ye Tun Tun, Yamaguchi H. Phylogenetic relationship of wild and cultivated Vigna (Subgenus Ceratotropis, Fabaceae) from Myanmar based on sequence variations in non-coding regions of trnT-F. Breeding Science. 2007;57:271–280. [Google Scholar]
- Ye Tun Tun, Yamaguchi H. Sequence variation of four chloroplast non-coding regions among wild, weedy and cultivated Vigna angularis accessions. Breeding Science. 2008;58:325–330. [Google Scholar]
- Zong XX, Kaga A, Tomooka N, Wang XW, Han OK, Vaughan D. The genetic diversity of the Vigna angularis complex in Asia. Genome. 2003;46:647–658. doi: 10.1139/g03-041. [DOI] [PubMed] [Google Scholar]