Abstract
The Krüppel-like family of transcription factors (KLFs) have been widely studied in proliferating cells, though very little is known about their role in post-mitotic cells, such as neurons. We have recently found that the KLFs play a role in regulating intrinsic axon growth ability in retinal ganglion cells (RGCs), a type of central nervous system (CNS) neuron. Previous KLF studies in other cell types suggest that there may be cell-type specific KLF expression patterns, and that their relative expression allows them to compete for binding sites, or to act redundantly to compensate for another’s function. With at least 15 of 17 KLF family members expressed in neurons, it will be important for us to determine how this complex family functions to regulate the intricate gene programs of axon growth and regeneration. By further characterizing the mechanisms of the KLF family in the nervous system, we may better understand how they regulate neurite growth and axon regeneration.
Keywords: axon, regeneration, KLF, Krüppel-like factor, CNS, growth, transcription factor, neuron
Introduction
Why do neurons in the central nervous system (CNS) fail to regenerate their axons after injury? This has remained a fundamental question in neuroscience, with obvious implications for human disease(Moore and Goldberg, 2010). In the CNS, embryonic or neonatal neurons can regenerate their axons after injury, whereas postnatal or adult neurons cannot (Bregman and Goldberger, 1982; Kunkel-Bagden et al., 1992). This has been partially attributed to the development of an inhibitory CNS environment. Both mature astrocytes and mature oligodendrocytes contribute to an inhibitory environment for axon regeneration in the injured adult mammalian CNS. Between the first and second postnatal week of development in most CNS tissues, oligodendrocytes begin to form myelin sheaths around axons to allow for increased conduction of electrical impulses (Foran and Peterson, 1992; Waxman, 1980). After injury, damaged axons are exposed to the myelin-associated lipids and proteins that are inhibitory to axon growth and regeneration (reviewed in (Yiu and He, 2006). Astrocytes respond to injury by secreting chondroitin sulfate proteoglycans (CSPGs) which are also inhibitory to growth (Becker and Becker, 2002; Jones et al., 2003; Jones et al., 2002; McKeon et al., 1999; Snow et al., 1990; Tang et al., 2003). Many strategies have been attempted to overcome glial-associated inhibitory cues and thus increase CNS regeneration. For example, many inhibitory proteins such as the myelin-derived axon growth inhibitor “Nogo”, myelin associated glycoprotein “MAG”, the oligodendrocyte myelin glycoprotein (OMgp) have been knocked out at the genetic level (Bartsch et al., 1995; Kim et al., 2003; Simonen et al., 2003; Su et al., 2008; Zheng et al., 2003), neutralized through antibody treatments (Bregman et al., 1995; Caroni and Schwab, 1988; Tang et al., 2001), or enzymatically digested (reviewed in (Crespo et al., 2007). These studies have resulted in modest regeneration, leading to alternative strategies targeting the downstream signaling of these inhibitory pathways (reviewed in (Schmandke and Strittmatter, 2007). The incomplete regeneration in all of these studies suggests that there may be additional inhibitory proteins that have yet to be discovered still acting to inhibit growth, and that there may be intrinsic changes within the neurons themselves that limit their regenerative ability.
An involvement of KLFs in the intrinsic control of axon regenerative ability
It has long been known that there is a developmental decrease in the ability of CNS axons to grow in vitro or regenerate in vivo(Blackmore and Letourneau, 2006; Chen et al., 1995; Dusart et al., 1997; Li et al., 1995; MacLaren and Taylor, 1995; Saunders et al., 1992; Saunders et al., 1995; Treherne et al., 1992). For example, in spinal cord injury experiments in whole CNS preparations from neonatal opossoms and embryonic rat, the injured neonatal CNS can regenerate, and this ability is lost postnatally(MacLaren and Taylor, 1995; Saunders et al., 1992; Saunders et al., 1995; Treherne et al., 1992). In purified in vitro cultures, where RGCs are removed from all other contaminating cell types, embryonic RGCs grow their axons ten-fold faster than postnatal RGCs, with this growth ability lost specifically around the time of birth(Goldberg et al., 2002b). These data suggest that CNS neurons lose their intrinsic capacity for rapid axon growth during development, and that this may play a role in the regenerative failure of CNS axons after injury.
What is the molecular basis for this loss? Prior work has pointed to possible roles for cyclic adenosine 3′,5′-monophosphate (cAMP; (Cai et al., 2001), cAMP response element-binding protein(CREB; (Gao et al., 2004), B-cell lymphoma/leukemia (Bcl-2; (Chen et al., 1997; Cho et al., 2005), anaphase promoting complex (APC) signaling pathways(Konishi et al., 2004; Lasorella et al., 2006)and phosphatase and tensin homology (PTEN; (Park et al., 2008)in this loss (Box 1). To identify new candidate genes that could contribute, we analyzed microarray-derived transcriptomes from different ages of RGCs to reveal developmentally regulated genes(Wang et al., 2007). These genes were screened in primary neurons for their effect on neurite outgrowth.
Box 1. Some of the intracellular signaling molecules with roles in CNS regeneration.
cAMP
Endogenous cAMP levels influences the developmental loss of regenerative capacity in retinal ganglion cells(Cai et al., 2001).
CREB
Downstream of cAMP, the activated transcription factor CREB is essential for spinal neurons to overcome the inhibitory injury environment(Gao et al., 2004).
Bcl-2
The proto-oncogene bcl-2 enhances retinal axon regeneration in certain assays (Chen et al., 1997).
Cdh1-APC
These and other cell cycle regulators also negatively influence axon growth in cerebellar granule neurons(Konishi et al., 2004).
PTEN
Deletion of this negative regulator of the mammalian mTOR pathway significantly promotes axon regeneration in retinal ganglion cells in vivo(Park et al., 2008).
KLFs
Kruppel-like factors regulate, positively and negatively, axon growth of CNS neurons in vitro and in vivo (Moore et al., 2009)..
Overexpression of the transcription factor Krüppel-like factor 4 (KLF4) resulted in a significant decrease in neurite outgrowth in hippocampal and cortical neurons, and RGCs(Moore et al., 2009). KLF4 knockout during early development resulted in increased neurite growth from RGCs in vitro, and increased axon regeneration in vivo after optic nerve injury which was unrelated to RGC differentiation(Moore et al., 2009). Interestingly, KLF4 expression increases postnatally in RGCs, specifically during the period around birth, which is when RGCs lose their intrinsic axon growth ability(Moore et al., 2009). These data support a model whereby the increase in KLF4 expression around birth, long after all RGCs have become post-mitotic, leads to a loss of regenerative ability of RGCs(Moore et al., 2009).
The KLF family of transcription factors and their effects on neurite growth
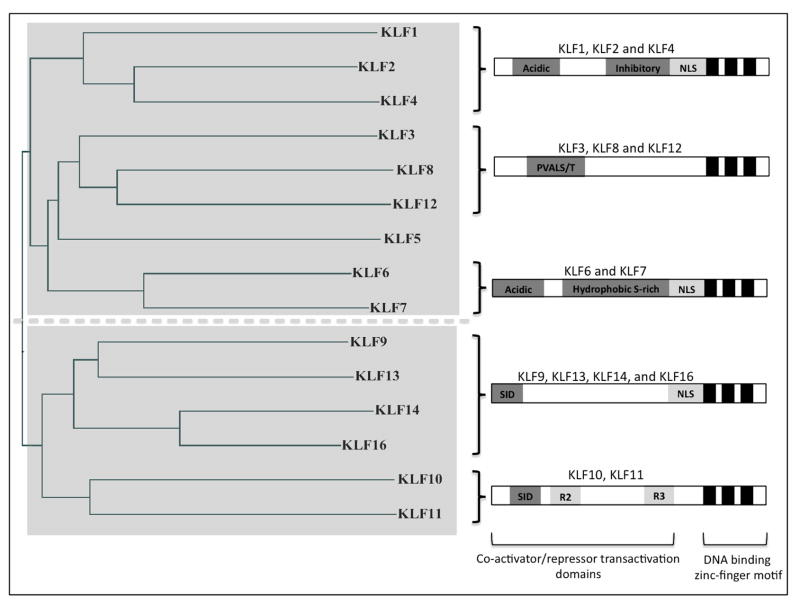
The enhanced regeneration seen by knocking out KLF4 was modest, raising the question of whether other KLF family members are also involved in regulating axon growth. KLF4 is one of 17 members of the KLF family of transcription factors (Fig. 1). Each family member contains 3 highly homologous Cys2/His2-type zinc fingers in their C termini with highly conserved DNA binding residues between them. KLFs bind DNA at CACCC/GC/GT boxes, which are highly represented throughout regulatory regions in the genome. They are often grouped with the Sp (specificity protein) family, though the KLF family is distinguished by the absence of the Sp family’s Buttonhead (BTD) box and Sp box (Suske et al., 2005). Little is known about the expression or function of the 17 KLFs in the mammalian nervous system.
Figure 1.
(Left) Phylogenetic tree based on KLF homology at the amino acid level, generated using Clustalw2 software. (Right) By phylogenetic cluster, diagrams detailing known human KLF family protein domains (after (Kaczynski et al., 2003; Moore et al., 2009).
We found that 15 of 17 KLF family members are expressed in RGCs (Moore et al., 2009), and a similar number are expressed in cortical neurons. We then asked if all KLF family members affect neurite outgrowth. We found that overexpression of KLF family members in cortical neurons resulted in differential effects on neurite growth which correlated with their structural subfamily groupings(Moore et al., 2009). Is the expression of all KLF family members developmentally regulated, and can this be correlated with their effect on neurite growth? The expression of many KLF family members was developmentally regulated, with direct correlation to the developmental loss of axon growth ability in RGCs: KLF6 and -7 enhanced growth when overexpressed but their expression decreased postnatally, and others including KLF4 and -9 suppressed neurite growth when overexpressed and increased in expression postnatally(Moore et al., 2009). Thus, the developmental regulation of at least several KLFs renders them additional candidates in the developmental loss of intrinsic axon growth ability. These studies looked only at levels of mRNA in the cell, however, and did not take into account protein levels. Further studies must be done to determine if these KLFs are regulated at the protein level, as well as the mRNA level.
These findings were the first description of KLFs’ ability to regulate axon growth and regeneration. In addition, many KLFs’ function as a growth suppressive transcription factor is unique in the field of transcription factor modulation of neurite outgrowth, where most transcription factors (such as STAT3)act as growth enhancers. How KLFs limit regeneration and growth in neurons is not yet known. As KLF4 and other growth-suppressive family members can activate or repress gene transcription in other cell types, their function may be to turn off genes that promote neurite growth, and/or turn on genes that suppress neurite growth. Thus, during development, KLFs may play an integral role in slowing down axon growth as the axon nears or reaches its target, perhaps freezing the neuron in a morphologically stable state once the gross circuitry is established.
KLF family members in the nervous system
What else is known about KLF family members generally and in the nervous system? It is worth noting that all the KLF family members have highly conserved zinc-finger DNA binding domains, particularly in the region predicted to interact with DNA (nearly 100%) (Kaczynski et al., 2003). Therefore, all family members are presumed to be capable of binding the same downstream targets and differential effects seen by their overexpression in neurons may be due to structural and thus functional differences outside this region. However, functional differences in vivo may not only reflect structural differences outside the DNA-binding domain, but also differences in expression profiles. While the majority of research on KLF family members has been performed in other systems, some characterization has been performed in neurons and the nervous system as a whole. Here we briefly review some of the published data on KLF functions throughout the nervous system, focusing on neurons and organizing our discussion by subfamily groupings.
![]()
KLF1, 2, and 4.”AIN” Subfamily. (AIN=Acidic and Inhibitory N-terminal domain)
This subfamily is characterized by shared acidic activation and inhibitory domains in the KLF amino termini, a conserved nuclear localization signal sequence, as well as the common KLF Cys2His2 zinc-finger DNA-binding domain in their carboxy-termini. These N-terminal domains permit interaction with co-factor complexes to regulate downstream gene expression, epigenetic modifications and diverse functional phenotypes. Some known interacting binding partners include p300/CBP, SWI/SNF and mSin3A(Kaczynski et al., 2003), although none of these have been identified to interact with KLFs in the CNS. In cancer biology, where these interactions have been well studied, they often lead to complex functional outcomes that are heavily dependent on the cellular context. For example, in breast cancer tumor cells, KLF4 appears to promote cell growth while acting as a tumor suppressor in B-cell non-hodgkins lymphoma (Guan et al.). This raises the possibility that protein-protein interactions unique to cellular context may be mediating stable epigenetic modifications that result in different functional outcomes in different cell types. Interestingly, we find that all 3 subfamily members have a similar suppressive effect on neurite outgrowth in CNS regeneration, raising the possibility, given their common structural motifs, that they may all be acting through a common effector binding partner in neurons. If this proves to be accurate, it may be possible to efficiently disrupt all three subfamily members’ suppression of axon growth simultaneously by targeting the common binding partner, and thus promote an even more robust effect on neurite outgrowth. Much more is known about KLF4 than other members of this subfamily. We thus focus a more detailed discussion of this subfamily member and its known role in the nervous system below.
KLF4
What is known about KLF4?
Outside of the nervous system, KLF4has been most widely studied in stem cell reprogramming(Zhao and Daley, 2008), differentiation(Dai and Segre, 2004; Ghaleb et al., 2005), growth arrest(Chen et al., 2001; Chen et al., 2003; Shields et al., 1996; Yoon et al., 2003), and cancer progression(Black et al., 2001; Rowland and Peeper, 2006; Safe and Abdelrahim, 2005). It was first identified to inhibit proliferation(Shields et al., 1996), and as such is often mutated or de-regulated in tumors(McConnell et al., 2007). KLF4 recruits both co-activator and co-repressor complexes, with known protein-protein interactions with p300/CBP (CREB-binding protein)(Evans et al., 2007; Geiman et al., 2000), and histone deacetylase 3 (HDAC3)(Evans et al., 2007), and CtBP1(C-terminal-binding protein 1)(Liu et al., 2009), leading to powerful epigenetic consequences on target gene promoter occupancy.
KLF4 function in neurons had been reported only once previously, when it was shown to be upregulated by NMDA or AMPA treatment in cortical neuron cultures(Zhu et al., 2009). Overexpression of KLF4 in these neurons concurrent with NMDA treatment led to an increased activation of caspase-3, which was dependent on extracellular and intracellular calcium levels. Importantly, overexpression of KLF4 alone did not increase caspase-3 levels in these neurons. Therefore, KLF4 overexpression in cortical slices led to increased caspase-3 activation after NMDA insult. We have found that KLF4 overexpression or knockout had no effect on survival in any of the neuronal types tested(Moore et al., 2009). In addition, after optic nerve injury in vivo, KLF4 knockout had no effect on RGC survival(Moore et al., 2009). Thus, it is possible that KLF4 may affect survival in cultures in combination with other stressors, however, it does not appear to affect survival in primary neurons in vitro or in RGCs in vivo either during development or after injury. Thus, its seemingly confined effects on neurite outgrowth makes it an intriguing candidate as a potential developmental switch conferring limited intrinsic growth capacity in adult neurons.
What are the downstream mediators of KLF4’s effects on neurite growth and regeneration? Although little is known about KLF4’s targets in neurons, a number of KLF4 targets identified outside of the nervous system may be good candidates for mediating KLF4’s effect on axon growth, such as p53, urokinase plasminogen activator receptor (u-PAR), ornithine decarboxylase (ODC), three different laminin chains(Rowland and Peeper, 2006), activating transcription factor 3 (ATF3), small proline rich protein 1a (SPRR1a), and matrix metalloproteinases (MMPs). For example, p53 expression is required for neurite growth and regeneration (Di Giovanni et al., 2006; Tedeschi et al., 2009). u-PAR is the receptor for the plasminogen activator system which cleaves extracellular matrix molecules. Mice lacking any portion of this system show delayed regeneration in the peripheral nervous system following sciatic nerve crush(Siconolfi and Seeds, 2001). Upregulation of Arginase I (Arg I), which catalyzes arginine to ornithine, has been suggested to be the downstream modulator of elevated cAMP and CREB activation which allows for a decreased inhibition to myelin(Cai et al., 2002; Gao et al., 2004). ODC converts ornithine (the product of Arg I) to putrescine, which is subsequently catalyzed into the polyamines spermidine and spermine, found to promote regeneration of neurons in vitro and after optic nerve injury in vivo(Chu et al., 1995; Deng et al., 2009). The laminin gamma-1 chain is one of the laminin chains regulated by KLF4(Higaki et al., 2002). Removal of the gamma-1 chain in a mossy fiber CNS injury model resulted in decreased axon growth as well as an inability of regenerating fibers to cross the lesion site(Grimpe et al., 2002), whereas application of a portion of the laminin gamma-1 chain to a rat model of spinal cord injury resulted in enhanced regeneration of CNS axons(Wiksten et al., 2004). Laminins in general strongly promote RGC axon growth as well (Goldberg et al., 2002a; Goldberg et al., 2002b). Finally, in studies in which KLF4 was knocked out in cornea, multiple genes relevant to regeneration were found to be upregulated, such as ATF3(Swamynathan et al., 2008), a growth-enhancing transcription factor expressed in regenerating neurons(Campbell et al., 2005; Mason et al., 2003; Pearson et al., 2003; Seijffers et al., 2007). SPRR1a is upregulated after KLF4 knockout(Swamynathan et al., 2008), and also during peripheral nerve regeneration(Bonilla et al., 2002). In addition, multiple MMPs have been shown to be upregulated after KLF4 knockout in the cornea(Young et al., 2009), and these enzymes can enhance regeneration in multiple systems through degradation of inhibitory proteins(Ahmed et al., 2005). Interestingly, other previously identified gene targets, such as p21WAF1/Cip1 (p21), which has been implicated in neurite outgrowth through inhibition of Rho kinase (ROCK) (Tanaka et al., 2002), can also be regulated by other KLFs. For example, both KLF4 and KLF6 can induce p21 expression, yet overexpression of KLF6 in neurons enhances growth while KLF4 suppresses growth(Moore et al., 2009). Therefore, while there are similar targets between some family members, correlating growth phenotype with known gene regulation may further narrow down the list of downstream targets that may be mediating the effect of that particular KLF.
It is presently unknown whether KLF4 regulates the expression of these genes in neurons, or whether these or other target genes mediate the effect of KLF4 on CNS axon growth. To more fully understand the mechanism by which KLF4 exerts its influence on neurite growth, additional experiments to identify KLF4 gene targets in neurons will have to be completed.
![]()
KLF6, 7”AHN” Subfamily (AHN=Acidic and Hydrophobic N-terminal domain)
Shared acidic activation and hydrophobic S-rich domains in their amino termini, a conserved nuclear localization signal sequence, and the KLF zinc-finger binding domain characterize this subfamily. Although both are known to play roles as tumor suppressors in various cancers, including prostate carcinoma, colorectal tumors, glioblastoma, hepatocellular and lung–derived cancers, precisely how they exert their effect is less clear. No binding partners have been identified in the CNS or other systems, although regulation of many genes involved in survival and synaptogenesis in the nervous system has been observed. Some evidence (described below) indicates functional redundancy in positive enhancing effects seen in zebrafish models of axon regeneration between the two. This is consistent with our findings in RGCs, again suggesting the possibility of a structure-function relationship in this subfamily as seen in others. Below is a brief summary of both KLFs in the nervous system.
KLF6, KLF7
The growth enhancers KLF6 and -7 were both found to be highly expressed in the developing nervous system, consistent with our findings in RGCs(Moore et al., 2009). In particular, KLF7 is highly expressed in both the peripheral nervous system (PNS) and CNS throughout development (Laub et al., 2001a; Lei et al., 2001). KLF6is expressed in the developing nervous system(Laub et al., 2001b), and in the adult is present in neurons, endothelial cells and neuronal progenitors in the forebrain(Jeong et al., 2009). KLF6’s continued presence in adult neurons again suggests that this growth enhancer, though present, may no longer be able to enhance growth in the presence of increased growth suppressors.
KLF6 and -7 have been studied previously for their effects on axon growth in zebrafish retinal explants, where KLF7 was found, together with KLF6, to be necessary for axonal outgrowth. Interestingly, KLF6 and -7 were able to compensate for each other in their ability to affect axon growth(Veldman et al., 2007), suggesting a possible redundancy in the genes that they regulate. Similarly, we have found that concurrent overexpression of KLF6 and -7 in cortical neurons was not synergistic, further supporting a redundant role in their function(Moore et al., 2009). A more comprehensive study on KLF7’s functions in the nervous system revealed that KLF7 knockout results in deficits in axon growth and path finding in the olfactory system, retina, and brain(Laub et al., 2006; Laub et al., 2005), again supporting a role for KLF7 in enhancing axon growth. Interestingly, KLF7 has been shown to positively regulate p21, p27, L1, GAP-43, TrkA, TrkB, and genes important for synaptogenesis and cytoskeleton dynamics in neurons(Kajimura et al., 2007; Kingsbury and Krueger, 2007; Laub et al., 2001a; Laub et al., 2005; Lei et al., 2006). KLF7’s ability to upregulate the Trk neurotrophin receptors may also play an important role in RGC survival after optic nerve injury. RGCs experience a loss of trophic responsiveness after injury, which may be due to the decreased expression of Trk receptors(Duan et al., 2009; Goldberg et al., 2002a; Meyer-Franke et al., 1998; Shen et al., 1999). Thus, therapeutically, overexpression of KLF7 may not only serve to enhance neurite growth, but also may increase trophic responsiveness in neurons, leading to increased neuroprotection after injury. In summary, these studies support our hypotheses that KLF6 and -7 act as growth-enhancers in neurons, and suggest possible downstream mechanisms for their effect.
![]()
KLF9, 13, 14, 16.BTEB-like Subfamily
A consensus Sin-interacting domain (SID) region and NLSsets apart exemplify this subfamily. This unique domain mediates their interaction with the known scaffold protein mSin3A as been shown in various systems (excluding the nervous system). Other known binding partners include p300/CBO and PCAF. Although the SID functions mainly to recruit the mSin3A co-repressor complex, it also overlaps with the rest of the amino-terminal region which can function as an activation domain in the right context. Thus, activating or repressive functional outcomes may be observed. Our findings again showed all members of this subfamily exerting a suppressive effect on axon growth in neurons, again suggesting a structure-function relationship. Here we review known roles of only members of this subfamily studied in the nervous system.
KLF9
KLF9 has also been studied in the nervous system and in neurite growth. It is a thyroid hormone (T3)-regulated transcription factor, with its expression in the brain developmentally regulated such that its expression is barely detectable from embryonic ages through P0, increases dramatically through one month, and then maintains this higher expression into adulthood(Denver et al., 1999; Martel et al., 2002; Morita et al., 2003), reflecting the pattern of plasma thyroid hormone levels. This pattern of expression is also consistent with KLF9’s developmentally increased mRNA levels in RGCs(Moore et al., 2009). KLF9’s expression is not only positively regulated by binding of T3 receptor-retinoid X receptor heterodimers to the T3 response element (T3RE) in the 5′ flanking region of the KLF9 gene(Denver and Williamson, 2009), but also by corticosterone(Bonett et al., 2009), and electrical activity(Lin et al., 2008; Scobie et al., 2009). KLF9 itself can repress transcription through recruitment of the co-repressor Sin3A(Imataka et al., 1992; Zhang et al., 2001). These findings make KLF9 an interesting candidate for a role in the loss of axon growth ability, due not only to its dramatic developmental regulation, but also to its ability to be modulated by electrical activity. As activity enhances trophic responsiveness and axon growth(Goldberg et al., 2002a), it will be interesting to explore the role of KLF9 in these processes.
Previous studies on the role of KLF9 in neurite outgrowth are opposite to our results. For example, in cell lines, overexpression of KLF9 led to an increase in the number of cells extending neurites as well as increasing the number and length of the neurites (Denver et al., 1999). However, this finding was obtained in a neuroblastoma (Neuro2a) cell line whose responsiveness to T3-regulated genes (such as KLF9) varies by the tumor subtype. Thus, its physiological relevance to primary neurons remains unknown. In agreement, additional studies in embryonic cortical neurons reported that knockdown of KLF9 decreased neurite branching to non-T3 treated levels, without affecting elongation. A similar effect was seen in the small subpopulation of small acetylcholinesterase (AChE) expressing cells, which are a target population for T3(Cayrou et al., 2002). Thus, previously reported data suggest that KLF9 functions to increase neurite growth, branching, and elongation. In contrast, we have found that in embryonic and postnatal RGCs, and young postnatal cortical neurons all supplemented with T3, KLF9 overexpression dramatically decreased neurite growth, suggesting possible differences for this transcription factor in different cell and neuronal types(Moore et al., 2009).
Despite multiple reports of effects of KLF9 on neurite growth, KLF9 knockout mice do not have defects in axon targeting and dendrite length(Scobie et al., 2009), although Purkinje cell dendrites may be slightly under-developed in the cerebellum(Morita et al., 2003). KLF9 knockout mice do display deficits in dentate granule (DG) neuron maturation, such that there is an increase in immature DG neurons in the hippocampus of developing knockout animals as defined by early and late maturation markers, spine maturation and electrophysiological properties(Scobie et al., 2009). This increase in the numbers of immature DG neurons is unrelated to proliferation and cell fate specification, suggesting that the early phase processes in neurogenesis in the KLF9 knockout hippocampus are normal. As newly born neurons are less able to functionally integrate into the adult hippocampus, there is reduced neurogenesis-dependent synaptic plasticity(Scobie et al., 2009). Behaviorally, KLF9 knockout mice have deficits in rotorod and contextual fear-conditioning tests(Morita et al., 2003; Scobie et al., 2009). Thus KLF9 is required for newly born DG neurons late-phase integration into the hippocampal circuitry(Scobie et al., 2009), but may have redundant factors that can compensate for its role in neurite growth.
Taken together, it appears that KLF9 may function differently between various neuronal types, and it is possible that it may act together with other KLFs in a redundant fashion to maintain its specific regulation of downstream targets.
KLF16
Three of the other KLFs we have found to suppress neurite growth, KLF5, -15, and -16, have been studied to a moderate degree in the nervous system. KLF16, a member of the basic transcription element binding protein (BTEB) subgroup of KLFs together with KLF9, -13, and -14, is expressed in the brain in embryos and adult animals (D’Souza et al., 2002; Hwang et al., 2001). It can bind to Sp1 sites in the promoters of at least 3 dopamine receptors to regulate their transcription, competing with Sp1 and Sp3 for binding site occupancy. KLF16 can either activate or repress transcription depending on the cell type studied(Hwang et al., 2001), suggesting that the endogenous population of other proteins available directly affects its function.
![]()
KLFs 3, 8 and 12.PVALS/T Subfamily
A PVALS/T N-terminal consensus sequence exemplifies this subfamily. The PVALS/T motif is known to interact with co-repressors of the C-terminal binding protein (CtBP) family and leads to transcriptional repression (Kaczynski et al., 2003). A putative nuclear localization sequence has been proposed immediately upstream of the zinc fingers for this group (van Vliet et al., 2000), however, lacking a consensus NLS sequence, it remains unclear precisely how it is trafficked into the nucleus. No KLFs from this subfamily affected neurite outgrowth in our experiments. Only KLF12 has been studied in the CNS, where it is expressed at its highest levels in the brain shortly after birth (Imhof et al., 1999).
![]()
KLFs 10, 11. SID-R2/3 Subfamily
Similar to the BTEB-like subfamily, these KLFs share an SID motif (although not as far upstream of the N-terminus) along with the TGF-β inducible repressor domains R2 and R3. Neither subfamily member affected neurite outgrowth in RGCs (Moore et al., 2009)nor have they been well studied in the nervous system. KLF11 exhibits post-translation modifications during epithelial cell malignancy (Ellenrieder, 2008) that prevent it from recruiting known binding partners (see below). Since it shares the same SID domain with the BTEB-like subfamily, this finding may prove instructive for identifying and characterizing potential binding partners in neurons for the BTEB-like subfamily.
Other KLFs
KLF’s5, 15, and 17 have not been formally classified into any subfamily as they lack any consensus motifs that might justify such grouping (Outside the zinc-finger binding domain). Only KLF15 was found to suppress neurite outgrowth in our experiments although KLF5 has also been studied elsewhere in the CNS (discussed below). KLF17 is known to be a negative regulator of epithelial-mesenchemal transition and metastasis in breast cancer (Gumireddy, et. al) but has not been studied in the CNS and thus, will be excluded from discussion.
KLF15
KLF15, a KLF which was not assigned a specific subfamily, is expressed in various parts of the brain and retina. In the retina, KLF15 is present in neurons of the inner nuclear layer and some neurons of the ganglion cell layer(Otteson et al., 2004). Our studies suggest that its expression is developmentally upregulated in purified RGCs(D.L. Moore, unpublished observations), further supporting a role for KLF15 as a growth suppressor in neurons. Interestingly, KLF15 can repress the rhodopsin promoter, although deletion of its N terminus results in a switch to a transcriptional activator(Otteson et al., 2004). Its function in neurons has not been described previously.
KLF5
KLF5, which also does not belong to a specific KLF subfamily, has been studied in the human prefrontal cortex where it is expressed, as well as in the granular and pyramidal cells of the hippocampus. Interestingly, KLF5 is downregulated in the prefrontal cortex in schizophrenia patients, and a polymorphism of KLF5 is associated with schizophrenia(Yanagi et al., 2008). Some targets of KLF5 identified in non-neuronal systems may prove to be good candidates for its effects on neurite growth in neurons. For example, KLF5 activates transcription of ILK (integrin-linked kinase), as well as the ILK targets Cdc42 and myosin light chain in keratinocytes(Yang et al., 2008). The signaling pathways and post-translational modifications of KLF5 have been well-characterized in other systems, and its function is highly regulated by these post-translational modifications (reviewed by (Dong and Chen, 2009), which may prove to be important to its function in neurite growth. Its expression pattern in RGCs is similar to that of KLF4, with a peak around the time of birth (D.L. Moore, unpublished observations). Whether it functions similarly or redundantly to KLF4 is unknown, but in embryonic stem cells, KLF5 has been shown to bind to the same sites as KLF4 when KLF4 is removed, compensating for its absence(Jiang et al., 2008). Despite these similarities and its comparable phenotype on neurite growth, in proliferating cellsKLF4 and -5 can have opposite effects on proliferation(Ghaleb et al., 2005). These studies further suggest a need for analysis of KLF function within the specific cell type of interest.
Brief summary of KLF subfamilies
Taken together, these data suggest that the KLF role in neuronal development may not be limited to effects on neurite growth. Less than half of the KLFs have had some previous characterization in the nervous system, leaving much to be revealed about their function, regulation, interactions, and downstream targets in neurons, and specifically asrelates to neurite growth. It appears that some structure-function relationship exists between the subfamilies and their effects on neurite outgrowth in neurons but this must be further explored and the players involved must be identified. A brief summary is shown on Table 1 below.
Table 1.
List of selected KLFs known to be expressed in primary neurons, summary of subfamily structure and functional domains and known binding partners. Data compiled from multiple sources (Kaczynski et al., 2003; Moore et al., 2009) and the National Center for Biotechnology Information (NCBI) database.
| Known structural/functional features of KLF Family members of the of Sp1-like/KLF family members
| |||
|---|---|---|---|
| KLFs | Species studied | Subfamily and Functional domains | Binding partners |
| KLF1 | Human/Mouse | AIN; Acidic, Activator. | p300/CBP, PCAF, SWI/SNF, mSin3A/TAF9/CKIIalpha/FLI- 1/p300/CBP |
| KLF2 | Human/Mouse | AIN; Acidic, Activator. | |
| KLF3 | Human/Mouse/Rat | PVALS/T;Activator/repressor. Context specific. | CtBP2/FHL3 |
| KLF4 | Human/Mouse/Rat/Zebrafish | AIN; Acidic, Activator or repressor. Context specific. | p300/CBP,HDAC5,ZF9,SP1 |
| KLF5 | Human/Mouse/Rat | Outlier. Activator | CBP,SET,GTF,NFκB,HDAC1, p300 |
| KLF6 | Human/Mouse | AHN; Activator | CyclinD1, HDAC3,SP1,p53/ |
| KLF7 | Human/Mouse | AHN; Activator | MoKA,CBP |
| KLF8 | Human | PVALS/T; Repressor. | CtBP2 |
| KLF9 | Human/Mouse | BTEB-like; Activator or repressor. Context specific. | mSin3A,PRB,SREBP-Sp1 |
| KLF10 | Human | SID-R2/3; Repressor. | mSin3A |
| KLF11 | Human | SID-R2/3; Activator or Repressor. Context specific. | mSin3A |
| KLF12 | Human/Mouse/Rat/Zebrafish | PVALS/T; Repressor. | CtBP1 |
| KLF13 | Human/Mouse/Rat | BTEB-like; Activator or repressor. Context specific. | mSin3A, p300/CBP, PRP4 and PCAF. |
| KLF14 | Human/Mouse | BTEB-like; Activator. | Unknown |
| KLF15 | Human/Mouse/Rat | Outlier; Repressor. | Unknown |
| KLF16 | Human/Mouse | BTEB-like; repressor. | mSin3A |
| KLF17 | Human/Chick | Unknown | Unknown |
KLF family members function as a “network”
As neurons express multiple KLFs, how do they interact to affect neurite growth? Eight growth suppressors are expressed during RGCs’ development, while only two growth enhancers are expressed(Moore et al., 2009). Knocking out a single growth suppressor, KLF4, increases axon growth and regeneration, suggesting that the sum of KLFs’ activity in postnatal neurons is suppressive. It is not known whether specific KLFs dominate in function over others, however. In overexpression experiments using combinations of growth suppressive (KLF4 and -9) and growth enhancing (KLF6 and -7) KLFs, we found that the suppressors dominated: KLF6 and -7 could never enhance growth in the presence of KLF4 or -9, and KLF4 could suppress neurite growth even when KLF6 or -7 were co-overexpressed (Moore et al., 2009). It is not yet known whether these factors are competing for similar binding sites or physically interacting with each other to inhibit function. These data imply that in the adult nervous system, the presence of many growth-suppressive KLFs is dominant, and suggest that to increase growth or regeneration, multiple suppressive KLFs may need to be removed.
Our discovery that multiple KLFs are expressed in RGCs suggests that this family acts as a “network”, as has been shown in other systems(Black et al., 2001; Eaton et al., 2008; Turner and Crossley, 1999). KLFs can function and interact differently in distinctcell types depending on the expression of other KLFs or other co-factors, or on differences in post-translational modifications of the transcription factors themselves. These features may explain how the KLF family of transcription factors creates a cell type-specific and perhaps developmental period-specific “code,” resulting in specific neurite growth and other phenotypes.
Cell-Type Specific Functions
The effects of KLFs differ depending on the cells in which they are studied. Outside of the nervous system, KLF4 can arrest the cell cycle through upregulation of p21 and inhibition of cyclins, resulting in a loss of proliferative capacity (reviewed in (Black et al., 2001). Similarly, it has been studied thoroughly in its role in differentiation of the skin cells of the epidermis(Segre et al., 1999). Recently, however, KLF4 has also been shown to be important in the induction of pluripotency in somatic cells(Takahashi and Yamanaka, 2006; Zhao and Daley, 2008). This role in maintaining a proliferative, undifferentiated state seems to be in direct opposition to KLF4’s role in arresting the cell cycle, inhibiting proliferation and promoting differentiation, suggesting that KLF function may be cell-specific.
The specific expression profile of different modulating co-factors in different neurons likely affects the function of these transcription factors. For example, KLF1 knockout mice display a reduced level of KLF3 in erythroid cells, whereas there is no effect of KLF1 knockout on KLF3 expression levels in the brain, demonstrating a differential regulation between tissue type(Crossley et al., 1996). Thus, there is a need to study each KLF in the specific neuron of interest to understand its function. In addition, such data suggest that research on single KLFs may not fully reveal their role in a specific phenotype, unless one also examines contributions of other KLFs in that specific cell and phenotype.
Relative Expression and Competition
KLF family members can compete for binding sites and regulation of the same target genes(Bieker, 2001; Black et al., 2001). KLF4 and KLF5, two highly related KLF family members, have opposing effects on shared target genes, such as on the transgelingene(Adam et al., 2000), or on the expression of KLF4 itself(Sun et al., 2001). In in vivo studies with KLF1, -3 and -8, the relative expression of the KLFs determines which KLF will out-compete the others at the binding site for specific genes. For example, KLF8’s two promoters contain multiple binding sites for KLFs. KLF1 can activate, and KLF3 can repress expression of KLF8. In vivo, KLF3 occupies both of the KLF8 promoters, repressing KLF8 expression, with little to no binding of KLF1 on these sites. In a KLF3 knockout animal, however, KLF1 binds these sites and activates the KLF8 promoter (Eaton et al., 2008). While the physiological function of KLF8 remains unknown, KLF3 is known to be widely expressed in erythroid tissues. Induction of KLF8 expression in cell lines is also known to promote proliferation, motility and invasiveness while suppressing KLF4 (a known tumor suppressor). This dynamic between KLFs 1,3,4, and 8 appears to be part of an elegant system primed for cell cycle control and any perturbation of this balance can dramatically alter the phenotype seen. All of this suggests that context is extremely critical to interpreting the differential effects of members of the KLF family.
KLF family members can also bind to different sites on a promoter to compete for functional transcriptional outcome. For example, KLF9 and -12 can both bind to a basic transcription element (BTE) site in the cis-regulated promoter elements of activating enhancer-binding protein 2 alpha (AP-2α) in separate locations. KLF9 activates and KLF12 represses AP-2α expression. Both bind overlapping nucleotides in an upstream regulatory sequence (position -336) of the promoter termed “A32” in a mutually exclusive manner. Hence, the relative amount of either KLF can tilt the balance in favor toward activation or repression. Indeed, after co-expression of KLF9 and -12 in equal amounts, the net expression of AP-2α transcription is unchanged(Imhof et al., 1999). Thus, all of these studies demonstrate that relative amounts of different KLFs present at a given time elicit differential regulation of target genes. This may be very important in studying the developmental loss of axon growth ability in neurons, as the balance of expression of different family members may shift at many specific developmental time periods.
Compensation and Redundancy
Upon removal of one KLF, other KLFs can bind to the same site and compensate for the loss of the first. For example, in embryonic stem cells, depletion of KLF2, -4 or -5 either singly or in pairs did not lead to differentiation; only with depletion of all three KLFs together did the stem cells finally differentiate (Jiang et al., 2008). Detailed chromatin immunoprecipitation (ChIP) analyses revealed that these three KLFs had many shared genome targets and upon depletion of one factor, the others were able to bind to that target, compensating for the loss of the removed KLF(Jiang et al., 2008).
Redundancy has also been shown in retinal explants from zebrafish, where KLF6a and KLF7a were identified as candidates for regeneration as they were upregulated after injury. Knockdown of either KLF6a or KLF7a singly had no effect on axon growth; however, concurrent knockdown of both factors resulted in a decrease in axon growth(Veldman et al., 2007). Similarly, in erythroid cells, double knockouts for both KLF1 and KLF2 yielded animals that died earlier, showed a greater reduction in embryonic Ey- and βh1-globin gene expression, and manifested cells with more morphological abnormalities when compared to single KLF1- or KLF2-single-knockout animals(Basu et al., 2007). Thus, members of the KLF family can compensate for the loss of one or more KLFs. Whether such redundancy limits the regenerative ability of KLF4-knockout RGCs(Moore et al., 2009) remains to be studied.
Post-Translational Modifications Affect KLF Function
As has been shown with many other transcription factors, post-translational modifications of KLFs can affect transcriptional outcome. For example, sumoylation or acetylation of KLF5 in response to specific environmental stimuli can result in a complete switch of its function, likely due to changes in the recruitment of certain co-repressors or co-activators. These modifications can also affect the half-life of KLF5 protein (reviewed in (Dong and Chen, 2009). Similarly, while KLF1 is required for β-globin expression, it is also expressed in erythroid cell lines that do not yet express β-globin. Post-translational phosphorylation of KLF1 allows it to bind and activate the β-globin promoter, revealing a functional requirement for post-translational modification in gene regulation(Bieker et al., 1998). Another downstream effect of PTM involves regulation by co-factor recruitment. These interactions are well described in the case of KLF11 which has been shown to interact with the scaffold protein mSin3a, recruit histone deacetylases and produce lasting epigenetic changes that mediate repression of many downstream genes (Ellenrieder, 2008). Phosphorylation of KLF11 (by oncogenic Ras-ERK-MAPK pathways) at a specific residue in its N-terminal domain abolishes this interaction and promotes oncogenic growth. Given that KLFs seem to cluster into subfamilies based on common structural and functional features in their N-terminal domain (Fig. 1), these interactions may prove critical in explaining the redundant roles of KLF subfamiies in neurons. Indeed, targeting these interactions may prove useful in maximally enhancing neurite outgrowth by potentially suppressing all repressors and enhancing all enhancers simultaneously.
These examples emphasize the importance of the expression levels of not only the transcription factors themselves, but also of those proteins that modify these transcription factors in a specific cell type and at a specific stage of development. It is not yet known whether specific stimuli or cell-cell interactions lead to changes in KLF post-translational modifications in RGCs or other neurons.
Taken together, all of these studies suggest that the KLF family of transcription factors can function in unique ways that require an understanding of the simultaneous functions of the whole family as opposed to single factors. Thus, further analysis of KLF overexpression in neurons should consider the endogenous expression of (1) all KLFs in these neurons at the time of overexpression, (2) enzymes that can post-translationally modify KLFs, and (3) co-factors that interact with the KLFs to activate or repress transcription. KLF compensation or redundancy may also play an important role in axon regeneration of CNS neurons. Removing multiple axon growth suppressors that are acting redundantly may increase axon regeneration after injury above that seen with KLF4 knockout alone(Moore et al., 2009).
Other transcription factors in neurite growth and regeneration
A small number of other transcription factors have been studied for their ability to affect axon growth and regeneration (reviewed in (Zhou and Snider, 2006), suggesting that transcriptional regulation may be a fundamental mechanism for the loss of axon regenerative ability in neurons. The transcription factors p53, c-Jun, ATF3, CREB, STAT3, NFATs, NFκB, Sox11, and SnoN all affect neurite growth and axon regeneration in varied ways. Interestingly, many of these have been studied in proliferating cells, and can be involved in the cell cycle, suggesting that many cell cycle proteins may have important roles in post-mitotic cells.
In general, the expression of the majority of these transcription factors is developmentally regulated in neurons, such that they are highly present early in development, when CNS axons are able to regenerate, and decrease soon after birth through adulthood, when CNS axons lose axonal regenerative ability. The expression or activation of many of these factors increases after injury in regenerating neurons, but is absent in non-regenerating neurons. Interestingly, the majority of these other transcription factors enhance neurite growth, in contrast to our recent identification of multiple neurite growth-suppressing members of the KLF transcription factor family. Our studies suggest that in the presence of both growth-suppressors and growth-enhancers, the suppressors dominate the phenotypic outcome(Moore et al., 2009). Therefore, modulating only the levels of transcription factors which enhance axon growth may not be sufficient for functional regeneration without simultaneous repression or removal of growth-suppressing transcription factors.
Future Directions
How KLFs function in neurons is still unclear. Very little is known about the downstream targets of these factors in neurons, and as discussed above, this list of targets may be different in different cell types. Thus, one first step to better understand how these transcription factors affect neurite growth is to identify their downstream gene targets in CNS neurons. Revealing similar targets between growth-suppressors and separately between growth-enhancers may identify mechanisms of neurite growth regulation. Additionally, studying how these KLFs regulate and interact with each other in neurons will greatly contribute not only to the greater field of KLF function, but also to understanding their neuron-specific effects. Further characterization and analysis of the relevant co-factors and their regulation also may be necessary for understanding the mechanisms behind KLF effects. As many of these KLFs can recruit chromatin remodeling machinery, it will be important to determine the actual state of the chromatin at these different developmental ages, as KLFs may function to create more stable changes in gene expression. Finally, it will be interesting to more fully test the hypothesis that growth-suppressive KLFs are dominant over growth-enhancing KLFs.
Conclusions
The KLF family of transcription factors may limit the intrinsic axon growth capacity of RGCs and other CNS neurons. While much is known about the KLFs in proliferating cells, very little is known about the KLF family in neurons. As other studies suggest that their function may be cell-type specific, there is a need for specific knowledge about their function in neurons, and perhaps even in specific neuronal types. In other systems, KLFs have been shown to function in a “network,” such that the individual KLF contribution is only a part of the large interaction between family members which contributes to the phenotypic outcome. Any alteration in one KLF’s contribution could affect the final outcome. Therefore, it is clear that these unique functions in the KLF family will require a new way of analyzing and studying transcription factors in neurons, moving from an individual protein’s contribution to a more complete understanding of its place within the family interactions. A better understanding of the transcriptional control of axon growth may lead to new approaches to reverse the poor regenerative ability of adult mammalian CNS neurons to an embryonic growth potential, ultimately leading to therapeutic treatments for many types of CNS injuries.
Acknowledgments
This work was funded by the National Eye Institute (EY020913, J.L.G., and P30 EY014801 to Univ. of Miami), by the National Institute of Neurological Disorders and Stroke(NS061348, J.L.G.), and an unrestricted grant from Research to Prevent Blindness to the Univ. of Miami. D.L.M. was previously supported by NINDS training grants T32 NS07492 and T32NS007459, and A.A. by NINDS training grant T32 NS007492.
Abbreviations and acronyms
- CNS
central nervous system
- RGC
retinal ganglion cell
- KLF
Krüppel-like factor
- E
embryonic
- P
postnatal
- cAMP
cyclic adenosine 3′,5′-monophosphate
- CREB
cAMP response element-binding protein
- Bcl-2
B-cell lymphoma/leukemia
- Cdh1-APC
anaphase promoting complex
- PTEN
phosphatase and tensin homology
- CBP
CREB-binding protein
- HDAC3
histone deacetylase 3
- CtBP1
C-terminal-binding protein 1
- NMDA
N-methyl-D-aspartic acid
- AMPA
α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate
- u-PAR
urokinase plasminogen activator receptor
- ODC
ornithine decarboxylase
- ATF3
activating transcription factor 3
- SPRR1a
small proline rich protein 1a
- MMP
matrix metalloproteinases
- Arg I
Arginase I
- Sp
specificity protein
- BTD
Buttonhead
- PNS
peripheral nervous system
- GAP-43
growth-associated protein 43
- TrkA
neurotrophic tyrosine kinase, receptor, type 1
- TrkB
tropomyosin-related kinase B
- T3
thyroid hormone
- T3RE
T3 response element
- AChE
acetylcholinesterase
- DG
dentate granule
- BTEB
basic transcription element binding protein
- ILF
integrin-linked kinase
- Cdc42
Cell division control protein 42
- AP-2α
activating enhancer-binding protein 2α
- ChIP
chromatin immunoprecipitation
- ATF3
activating transcription factor 3
- STAT3
Signal transducer and activator of transcription 3
- NFAT
Nuclear factor of activated T-cells
- NFκB
Nuclear Factor-KappaB
- Sox11
SRY-box containing gene 11
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Darcie L. Moore, Email: dmoore@med.miami.edu.
Akintomide Apara, Email: aapara@med.miami.edu.
Jeffrey L. Goldberg, Email: jgoldberg@med.miami.edu.
Cited Literature
- Adam PJ, Regan CP0, Hautmann MB, Owens GK. Positive- and negative-acting Kruppel-like transcription factors bind a transforming growth factor beta control element required for expression of the smooth muscle cell differentiation marker SM22alpha in vivo. J Biol Chem. 2000;275:37798–37806. doi: 10.1074/jbc.M006323200. [DOI] [PubMed] [Google Scholar]
- Ahmed Z, Dent RG, Leadbeater WE, Smith C, Berry M, Logan A. Matrix metalloproteases: degradation of the inhibitory environment of the transected optic nerve and the scar by regenerating axons. Mol Cell Neurosci. 2005;28:64–78. doi: 10.1016/j.mcn.2004.08.013. [DOI] [PubMed] [Google Scholar]
- Bartsch U, Bandtlow CE, Schnell L, Bartsch S, Spillmann AA, Rubin BP, Hillenbrand R, Montag D, Schwab ME, Schachner M. Lack of evidence that myelin-associated glycoprotein is a major inhibitor of axonal regeneration in the CNS. Neuron. 1995;15:1375–1381. doi: 10.1016/0896-6273(95)90015-2. [DOI] [PubMed] [Google Scholar]
- Basu P, Lung TK, Lemsaddek W, Sargent TG, Williams DC, Jr, Basu M, Redmond LC, Lingrel JB, Haar JL, Lloyd JA. EKLF and KLF2 have compensatory roles in embryonic beta-globin gene expression and primitive erythropoiesis. Blood. 2007;110:3417–3425. doi: 10.1182/blood-2006-11-057307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker CG, Becker T. Repellent guidance of regenerating optic axons by chondroitin sulfate glycosaminoglycans in zebrafish. J Neurosci. 2002;22:842–853. doi: 10.1523/JNEUROSCI.22-03-00842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieker JJ. Kruppel-like factors: three fingers in many pies. J Biol Chem. 2001;276:34355–34358. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- Bieker JJ, Ouyang L, Chen X. Transcriptional factors for specific globin genes. Ann N Y Acad Sci. 1998;850:64–69. doi: 10.1111/j.1749-6632.1998.tb10463.x. [DOI] [PubMed] [Google Scholar]
- Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- Blackmore M, Letourneau PC. Changes within maturing neurons limit axonal regeneration in the developing spinal cord. J Neurobiol. 2006;66:348–360. doi: 10.1002/neu.20224. [DOI] [PubMed] [Google Scholar]
- Bonett RM, Hu F, Bagamasbad P, Denver RJ. Stressor and glucocorticoid-dependent induction of the immediate early gene kruppel-like factor 9: implications for neural development and plasticity. Endocrinology. 2009;150:1757–1765. doi: 10.1210/en.2008-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla IE, Tanabe K, Strittmatter SM. Small proline-rich repeat protein 1A is expressed by axotomized neurons and promotes axonal outgrowth. J Neurosci. 2002;22:1303–1315. doi: 10.1523/JNEUROSCI.22-04-01303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman BS, Goldberger ME. Anatomical plasticity and sparing of function after spinal cord damage in neonatal cats. Science. 1982;217:553–555. doi: 10.1126/science.7089581. [DOI] [PubMed] [Google Scholar]
- Bregman BS, Kunkel-Bagden E, Schnell L, Dai HN, Gao D, Schwab ME. Recovery from spinal cord injury mediated by antibodies to neurite growth inhibitors. Nature. 1995;378:498–501. doi: 10.1038/378498a0. [DOI] [PubMed] [Google Scholar]
- Cai D, Deng K, Mellado W, Lee J, Ratan RR, Filbin MT. Arginase I and polyamines act downstream from cyclic AMP in overcoming inhibition of axonal growth MAG and myelin in vitro. Neuron. 2002;35:711–719. doi: 10.1016/s0896-6273(02)00826-7. [DOI] [PubMed] [Google Scholar]
- Cai D, Qiu J, Cao Z, McAtee M, Bregman BS, Filbin MT. Neuronal cyclic AMP controls the developmental loss in ability of axons to regenerate. J Neurosci. 2001;21:4731–4739. doi: 10.1523/JNEUROSCI.21-13-04731.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell G, Hutchins K, Winterbottom J, Grenningloh G, Lieberman AR, Anderson PN. Upregulation of activating transcription factor 3 (ATF3) by intrinsic CNS neurons regenerating axons into peripheral nerve grafts. Exp Neurol. 2005;192:340–347. doi: 10.1016/j.expneurol.2004.11.026. [DOI] [PubMed] [Google Scholar]
- Caroni P, Schwab ME. Antibody against myelin-associated inhibitor of neurite growth neutralizes nonpermissive substrate properties of CNS white matter. Neuron. 1988;1:85–96. doi: 10.1016/0896-6273(88)90212-7. [DOI] [PubMed] [Google Scholar]
- Cayrou C, Denver RJ, Puymirat J. Suppression of the basic transcription element-binding protein in brain neuronal cultures inhibits thyroid hormone-induced neurite branching. Endocrinology. 2002;143:2242–2249. doi: 10.1210/endo.143.6.8856. [DOI] [PubMed] [Google Scholar]
- Chen DF, Jhaveri S, Schneider GE. Intrinsic changes in developing retinal neurons result in regenerative failure of their axons. Proc Natl Acad Sci U S A. 1995;92:7287–7291. doi: 10.1073/pnas.92.16.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DF, Schneider GE, Martinou JC, Tonegawa S. Bcl-2 promotes regeneration of severed axons in mammalian CNS. Nature. 1997;385:434–439. doi: 10.1038/385434a0. [DOI] [PubMed] [Google Scholar]
- Chen X, Johns DC, Geiman DE, Marban E, Dang DT, Hamlin G, Sun R, Yang VW. Kruppel-like factor 4 (gut-enriched Kruppel-like factor) inhibits cell proliferation by blocking G1/S progression of the cell cycle. J Biol Chem. 2001;276:30423–30428. doi: 10.1074/jbc.M101194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Whitney EM, Gao SY, Yang VW. Transcriptional profiling of Kruppel-like factor 4 reveals a function in cell cycle regulation and epithelial differentiation. J Mol Biol. 2003;326:665–677. doi: 10.1016/S0022-2836(02)01449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KS, Yang L, Lu B, Feng Ma H, Huang X, Pekny M, Chen DF. Re-establishing the regenerative potential of central nervous system axons in postnatal mice. J Cell Sci. 2005;118:863–872. doi: 10.1242/jcs.01658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu PJ, Saito H, Abe K. Polyamines promote regeneration of injured axons of cultured rat hippocampal neurons. Brain Res. 1995;673:233–241. doi: 10.1016/0006-8993(94)01419-i. [DOI] [PubMed] [Google Scholar]
- Crespo D, Asher RA, Lin R, Rhodes KE, Fawcett JW. How does chondroitinase promote functional recovery in the damaged CNS? Exp Neurol. 2007;206:159–171. doi: 10.1016/j.expneurol.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Crossley M, Whitelaw E, Perkins A, Williams G, Fujiwara Y, Orkin SH. Isolation and characterization of the cDNA encoding BKLF/TEF-2, a major CACCC-box-binding protein in erythroid cells and selected other cells. Mol Cell Biol. 1996;16:1695–1705. doi: 10.1128/mcb.16.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza UM, Lammers CH, Hwang CK, Yajima S, Mouradian MM. Developmental expression of the zinc finger transcription factor DRRF (dopamine receptor regulating factor) Mech Dev. 2002;110:197–201. doi: 10.1016/s0925-4773(01)00564-0. [DOI] [PubMed] [Google Scholar]
- Dai X, Segre JA. Transcriptional control of epidermal specification and differentiation. Curr Opin Genet Dev. 2004;14:485–491. doi: 10.1016/j.gde.2004.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng K, He H, Qiu J, Lorber B, Bryson JB, Filbin MT. Increased synthesis of spermidine as a result of upregulation of arginase I promotes axonal regeneration in culture and in vivo. J Neurosci. 2009;29:9545–9552. doi: 10.1523/JNEUROSCI.1175-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver RJ, Ouellet L, Furling D, Kobayashi A, Fujii-Kuriyama Y, Puymirat J. Basic transcription element-binding protein (BTEB) is a thyroid hormone-regulated gene in the developing central nervous system. Evidence for a role in neurite outgrowth. J Biol Chem. 1999;274:23128–23134. doi: 10.1074/jbc.274.33.23128. [DOI] [PubMed] [Google Scholar]
- Denver RJ, Williamson KE. Identification of a thyroid hormone response element in the mouse Kruppel-like factor 9 gene to explain its postnatal expression in the brain. Endocrinology. 2009;150:3935–3943. doi: 10.1210/en.2009-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giovanni S, Knights CD, Rao M, Yakovlev A, Beers J, Catania J, Avantaggiati ML, Faden AI. The tumor suppressor protein p53 is required for neurite outgrowth and axon regeneration. Embo J. 2006;25:4084–4096. doi: 10.1038/sj.emboj.7601292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong JT, Chen C. Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cell Mol Life Sci. 2009;66:2691–2706. doi: 10.1007/s00018-009-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Kong W, Benny Klimek M, Goldberg JL. Loss of Retinal Ganglion Cell Trophic Responsiveness Is Correlated With Reduced Electrical Activity. ARVO; Fort Lauderdale, FL: 2009. p. 127.p. A171. [Google Scholar]
- Dusart I, Airaksinen MS, Sotelo C. Purkinje cell survival and axonal regeneration are age dependent: an in vitro study. J Neurosci. 1997;17:3710–3726. doi: 10.1523/JNEUROSCI.17-10-03710.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton SA, Funnell AP, Sue N, Nicholas H, Pearson RC, Crossley M. A network of Kruppel-like Factors (Klfs). Klf8 is repressed by Klf3 and activated by Klf1 in vivo. J Biol Chem. 2008;283:26937–26947. doi: 10.1074/jbc.M804831200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenrieder V. TGFbeta regulated gene expression by Smads and Sp1/KLF-like transcription factors in cancer. Anticancer Res. 2008;28:1531–1539. [PubMed] [Google Scholar]
- Evans PM, Zhang W, Chen X, Yang J, Bhakat KK, Liu C. Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J Biol Chem. 2007;282:33994–34002. doi: 10.1074/jbc.M701847200. [DOI] [PubMed] [Google Scholar]
- Foran DR, Peterson AC. Myelin acquisition in the central nervous system of the mouse revealed by an MBP-Lac Z transgene. J Neurosci. 1992;12:4890–4897. doi: 10.1523/JNEUROSCI.12-12-04890.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Deng K, Hou J, Bryson JB, Barco A, Nikulina E, Spencer T, Mellado W, Kandel ER, Filbin MT. Activated CREB is sufficient to overcome inhibitors in myelin and promote spinal axon regeneration in vivo. Neuron. 2004;44:609–621. doi: 10.1016/j.neuron.2004.10.030. [DOI] [PubMed] [Google Scholar]
- Geiman DE, Ton-That H, Johnson JM, Yang VW. Transactivation and growth suppression by the gut-enriched Kruppel-like factor (Kruppel-like factor 4) are dependent on acidic amino acid residues and protein-protein interaction. Nucleic Acids Res. 2000;28:1106–1113. doi: 10.1093/nar/28.5.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Yang VW. Kruppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res. 2005;15:92–96. doi: 10.1038/sj.cr.7290271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg JL, Espinosa JS, Xu Y, Davidson N, Kovacs GT, Barres BA. Retinal ganglion cells do not extend axons by default: promotion by neurotrophic signaling and electrical activity. Neuron. 2002a;33:689–702. doi: 10.1016/s0896-6273(02)00602-5. [DOI] [PubMed] [Google Scholar]
- Goldberg JL, Klassen MP, Hua Y, Barres BA. Amacrine-signaled loss of intrinsic axon growth ability by retinal ganglion cells. Science. 2002b;296:1860–1864. doi: 10.1126/science.1068428. [DOI] [PubMed] [Google Scholar]
- Grimpe B, Dong S, Doller C, Temple K, Malouf AT, Silver J. The critical role of basement membrane-independent laminin gamma 1 chain during axon regeneration in the CNS. J Neurosci. 2002;22:3144–3160. doi: 10.1523/JNEUROSCI.22-08-03144.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan H, Xie L, Leithauser F, Flossbach L, Moller P, Wirth T, Ushmorov A. KLF4 is a tumor suppressor in B-cell non-Hodgkin lymphoma and in classic Hodgkin lymphoma. Blood. 116:1469–1478. doi: 10.1182/blood-2009-12-256446. [DOI] [PubMed] [Google Scholar]
- Higaki Y, Schullery D, Kawata Y, Shnyreva M, Abrass C, Bomsztyk K. Synergistic activation of the rat laminin gamma1 chain promoter by the gut-enriched Kruppel-like factor (GKLF/KLF4) and Sp1. Nucleic Acids Res. 2002;30:2270–2279. doi: 10.1093/nar/30.11.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CK, D’Souza UM, Eisch AJ, Yajima S, Lammers CH, Yang Y, Lee SH, Kim YM, Nestler EJ, Mouradian MM. Dopamine receptor regulating factor, DRRF: a zinc finger transcription factor. Proc Natl Acad Sci U S A. 2001;98:7558–7563. doi: 10.1073/pnas.121635798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H, Sogawa K, Yasumoto K, Kikuchi Y, Sasano K, Kobayashi A, Hayami M, Fujii-Kuriyama Y. Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene. EMBO J. 1992;11:3663–3671. doi: 10.1002/j.1460-2075.1992.tb05451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhof A, Schuierer M, Werner O, Moser M, Roth C, Bauer R, Buettner R. Transcriptional regulation of the AP-2alpha promoter by BTEB-1 and AP-2rep, a novel wt-1/egr-related zinc finger repressor. Mol Cell Biol. 1999;19:194–204. doi: 10.1128/mcb.19.1.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong KH, Kim SK, Kim SY, Cho KO. Immunohistochemical localization of Kruppel-like factor 6 in the mouse forebrain. Neurosci Lett. 2009;453:16–20. doi: 10.1016/j.neulet.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- Jones LL, Margolis RU, Tuszynski MH. The chondroitin sulfate proteoglycans neurocan, brevican, phosphacan, and versican are differentially regulated following spinal cord injury. Exp Neurol. 2003;182:399–411. doi: 10.1016/s0014-4886(03)00087-6. [DOI] [PubMed] [Google Scholar]
- Jones LL, Yamaguchi Y, Stallcup WB, Tuszynski MH. NG2 is a major chondroitin sulfate proteoglycan produced after spinal cord injury and is expressed by macrophages and oligodendrocyte progenitors. J Neurosci. 2002;22:2792–2803. doi: 10.1523/JNEUROSCI.22-07-02792.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura D, Dragomir C, Ramirez F, Laub F. Identification of genes regulated by transcription factor KLF7 in differentiating olfactory sensory neurons. Gene. 2007;388:34–42. doi: 10.1016/j.gene.2006.09.027. [DOI] [PubMed] [Google Scholar]
- Kim JE, Li S, GrandPre T, Qiu D, Strittmatter SM. Axon regeneration in young adult mice lacking Nogo-A/B. Neuron. 2003;38:187–199. doi: 10.1016/s0896-6273(03)00147-8. [DOI] [PubMed] [Google Scholar]
- Kingsbury TJ, Krueger BK. Ca2+, CREB and kruppel: a novel KLF7-binding element conserved in mouse and human TRKB promoters is required for CREB-dependent transcription. Mol Cell Neurosci. 2007;35:447–455. doi: 10.1016/j.mcn.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi Y, Stegmuller J, Matsuda T, Bonni S, Bonni A. Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science. 2004;303:1026–1030. doi: 10.1126/science.1093712. [DOI] [PubMed] [Google Scholar]
- Kunkel-Bagden E, Dai HN, Bregman BS. Recovery of function after spinal cord hemisection in newborn and adult rats: differential effects on reflex and locomotor function. Exp Neurol. 1992;116:40–51. doi: 10.1016/0014-4886(92)90174-o. [DOI] [PubMed] [Google Scholar]
- Lasorella A, Stegmuller J, Guardavaccaro D, Liu G, Carro MS, Rothschild G, de la Torre-Ubieta L, Pagano M, Bonni A, Iavarone A. Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature. 2006;442:471–474. doi: 10.1038/nature04895. [DOI] [PubMed] [Google Scholar]
- Laub F, Aldabe R, Friedrich V, Jr, Ohnishi S, Yoshida T, Ramirez F. Developmental expression of mouse Kruppel-like transcription factor KLF7 suggests a potential role in neurogenesis. Dev Biol. 2001a;233:305–318. doi: 10.1006/dbio.2001.0243. [DOI] [PubMed] [Google Scholar]
- Laub F, Aldabe R, Ramirez F, Friedman S. Embryonic expression of Kruppel-like factor 6 in neural and non-neural tissues. Mech Dev. 2001b;106:167–170. doi: 10.1016/s0925-4773(01)00419-1. [DOI] [PubMed] [Google Scholar]
- Laub F, Dragomir C, Ramirez F. Mice without transcription factor KLF7 provide new insight into olfactory bulb development. Brain Res. 2006;1103:108–113. doi: 10.1016/j.brainres.2006.05.065. [DOI] [PubMed] [Google Scholar]
- Laub F, Lei L, Sumiyoshi H, Kajimura D, Dragomir C, Smaldone S, Puche AC, Petros TJ, Mason C, Parada LF, Ramirez F. Transcription factor KLF7 is important for neuronal morphogenesis in selected regions of the nervous system. Mol Cell Biol. 2005;25:5699–5711. doi: 10.1128/MCB.25.13.5699-5711.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Ma L, Nef S, Thai T, Parada LF. mKlf7, a potential transcriptional regulator of TrkA nerve growth factor receptor expression in sensory and sympathetic neurons. Development. 2001;128:1147–1158. doi: 10.1242/dev.128.7.1147. [DOI] [PubMed] [Google Scholar]
- Lei L, Zhou J, Lin L, Parada LF. Brn3a and Klf7 cooperate to control TrkA expression in sensory neurons. Dev Biol. 2006;300:758–769. doi: 10.1016/j.ydbio.2006.08.062. [DOI] [PubMed] [Google Scholar]
- Li D, Field PM, Raisman G. Failure of axon regeneration in postnatal rat entorhinohippocampal slice coculture is due to maturation of the axon, not that of the pathway or target. Eur J Neurosci. 1995;7:1164–1171. doi: 10.1111/j.1460-9568.1995.tb01106.x. [DOI] [PubMed] [Google Scholar]
- Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, Kim TK, Hu LS, Malik AN, Greenberg ME. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455:1198–1204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Zheng H, Ai W. C-terminal binding proteins (CtBPs) attenuate KLF4-mediated transcriptional activation. FEBS Lett. 2009 doi: 10.1016/j.febslet.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLaren RE, Taylor JS. A critical period for axon regrowth through a lesion in the developing mammalian retina. Eur J Neurosci. 1995;7:2111–2118. doi: 10.1111/j.1460-9568.1995.tb00633.x. [DOI] [PubMed] [Google Scholar]
- Martel J, Cayrou C, Puymirat J. Identification of new thyroid hormone-regulated genes in rat brain neuronal cultures. Neuroreport. 2002;13:1849–1851. doi: 10.1097/00001756-200210280-00003. [DOI] [PubMed] [Google Scholar]
- Mason MR, Lieberman AR, Anderson PN. Corticospinal neurons up-regulate a range of growth-associated genes following intracortical, but not spinal, axotomy. Eur J Neurosci. 2003;18:789–802. doi: 10.1046/j.1460-9568.2003.02809.x. [DOI] [PubMed] [Google Scholar]
- McConnell BB, Ghaleb AM, Nandan MO, Yang VW. The diverse functions of Kruppel-like factors 4 and 5 in epithelial biology and pathobiology. Bioessays. 2007;29:549–557. doi: 10.1002/bies.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon RJ, Jurynec MJ, Buck CR. The chondroitin sulfate proteoglycans neurocan and phosphacan are expressed by reactive astrocytes in the chronic CNS glial scar. J Neurosci. 1999;19:10778–10788. doi: 10.1523/JNEUROSCI.19-24-10778.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Franke A, Wilkinson GA, Kruttgen A, Hu M, Munro E, Hanson MG, Jr, Reichardt LF, Barres BA. Depolarization and cAMP elevation rapidly recruit TrkB to the plasma membrane of CNS neurons. Neuron. 1998;21:681–693. doi: 10.1016/s0896-6273(00)80586-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DL, Blackmore MG, Hu Y, Kaestner KH, Bixby JL, Lemmon VP, Goldberg JL. KLF family members regulate intrinsic axon regeneration ability. Science. 2009;326:298–301. doi: 10.1126/science.1175737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DL, Goldberg JL. Four steps to optic nerve regeneration. J Neuroophthalmol. 2010;30:347–360. doi: 10.1097/WNO.0b013e3181e755af. [DOI] [PubMed] [Google Scholar]
- Morita M, Kobayashi A, Yamashita T, Shimanuki T, Nakajima O, Takahashi S, Ikegami S, Inokuchi K, Yamashita K, Yamamoto M, Fujii-Kuriyama Y. Functional analysis of basic transcription element binding protein by gene targeting technology. Mol Cell Biol. 2003;23:2489–2500. doi: 10.1128/MCB.23.7.2489-2500.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otteson DC, Liu Y, Lai H, Wang C, Gray S, Jain MK, Zack DJ. Kruppel-like factor 15, a zinc-finger transcriptional regulator, represses the rhodopsin and interphotoreceptor retinoid-binding protein promoters. Invest Ophthalmol Vis Sci. 2004;45:2522–2530. doi: 10.1167/iovs.04-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, Xu B, Connolly L, Kramvis I, Sahin M, He Z. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson AG, Gray CW, Pearson JF, Greenwood JM, During MJ, Dragunow M. ATF3 enhances c-Jun-mediated neurite sprouting. Brain Res Mol Brain Res. 2003;120:38–45. doi: 10.1016/j.molbrainres.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Rowland BD, Peeper DS. KLF4, p21 and context-dependent opposing forces in cancer. Nat Rev Cancer. 2006;6:11–23. doi: 10.1038/nrc1780. [DOI] [PubMed] [Google Scholar]
- Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur J Cancer. 2005;41:2438–2448. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Saunders NR, Balkwill P, Knott G, Habgood MD, Mollgard K, Treherne JM, Nicholls JG. Growth of axons through a lesion in the intact CNS of fetal rat maintained in long-term culture. Proc Biol Sci. 1992;250:171–180. doi: 10.1098/rspb.1992.0146. [DOI] [PubMed] [Google Scholar]
- Saunders NR, Deal A, Knott GW, Varga ZM, Nicholls JG. Repair and recovery following spinal cord injury in a neonatal marsupial (Monodelphis domestica) Clin Exp Pharmacol Physiol. 1995;22:518–526. doi: 10.1111/j.1440-1681.1995.tb02060.x. [DOI] [PubMed] [Google Scholar]
- Schmandke A, Strittmatter SM. ROCK and Rho: biochemistry and neuronal functions of Rho-associated protein kinases. Neuroscientist. 2007;13:454–469. doi: 10.1177/1073858407303611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobie KN, Hall BJ, Wilke SA, Klemenhagen KC, Fujii-Kuriyama Y, Ghosh A, Hen R, Sahay A. Kruppel-like factor 9 is necessary for late-phase neuronal maturation in the developing dentate gyrus and during adult hippocampal neurogenesis. J Neurosci. 2009;29:9875–9887. doi: 10.1523/JNEUROSCI.2260-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Seijffers R, Mills CD, Woolf CJ. ATF3 increases the intrinsic growth state of DRG neurons to enhance peripheral nerve regeneration. J Neurosci. 2007;27:7911–7920. doi: 10.1523/JNEUROSCI.5313-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Wiemelt AP, McMorris FA, Barres BA. Retinal ganglion cells lose trophic responsiveness after axotomy. Neuron. 1999;23:285–295. doi: 10.1016/s0896-6273(00)80780-1. [DOI] [PubMed] [Google Scholar]
- Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siconolfi LB, Seeds NW. Mice lacking tPA, uPA, or plasminogen genes showed delayed functional recovery after sciatic nerve crush. J Neurosci. 2001;21:4348–4355. doi: 10.1523/JNEUROSCI.21-12-04348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonen M, Pedersen V, Weinmann O, Schnell L, Buss A, Ledermann B, Christ F, Sansig G, van der Putten H, Schwab ME. Systemic deletion of the myelin-associated outgrowth inhibitor Nogo-A improves regenerative and plastic responses after spinal cord injury. Neuron. 2003;38:201–211. doi: 10.1016/s0896-6273(03)00226-5. [DOI] [PubMed] [Google Scholar]
- Snow DM, Lemmon V, Carrino DA, Caplan AI, Silver J. Sulfated proteoglycans in astroglial barriers inhibit neurite outgrowth in vitro. Exp Neurol. 1990;109:111–130. doi: 10.1016/s0014-4886(05)80013-5. [DOI] [PubMed] [Google Scholar]
- Su Y, Wang F, Zhao SG, Pan SH, Liu P, Teng Y, Cui H. Axonal regeneration after optic nerve crush in Nogo-A/B/C knockout mice. Mol Vis. 2008;14:268–273. [PMC free article] [PubMed] [Google Scholar]
- Sun R, Chen X, Yang VW. Intestinal-enriched Kruppel-like factor (Kruppel-like factor 5) is a positive regulator of cellular proliferation. J Biol Chem. 2001;276:6897–6900. doi: 10.1074/jbc.C000870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics. 2005;85:551–556. doi: 10.1016/j.ygeno.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Swamynathan SK, Davis J, Piatigorsky J. Identification of candidate Klf4 target genes reveals the molecular basis of the diverse regulatory roles of Klf4 in the mouse cornea. Invest Ophthalmol Vis Sci. 2008;49:3360–3370. doi: 10.1167/iovs.08-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Yamashita T, Asada M, Mizutani S, Yoshikawa H, Tohyama M. Cytoplasmic p21(Cip1/WAF1) regulates neurite remodeling by inhibiting Rho-kinase activity. J Cell Biol. 2002;158:321–329. doi: 10.1083/jcb.200202071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Qiu J, Nikulina E, Filbin MT. Soluble myelin-associated glycoprotein released from damaged white matter inhibits axonal regeneration. Mol Cell Neurosci. 2001;18:259–269. doi: 10.1006/mcne.2001.1020. [DOI] [PubMed] [Google Scholar]
- Tang X, Davies JE, Davies SJ. Changes in distribution, cell associations, and protein expression levels of NG2, neurocan, phosphacan, brevican, versican V2, and tenascin-C during acute to chronic maturation of spinal cord scar tissue. J Neurosci Res. 2003;71:427–444. doi: 10.1002/jnr.10523. [DOI] [PubMed] [Google Scholar]
- Tedeschi A, Nguyen T, Puttagunta R, Gaub P, Di Giovanni S. A p53-CBP/p300 transcription module is required for GAP-43 expression, axon outgrowth, and regeneration. Cell Death Differ. 2009;16:543–554. doi: 10.1038/cdd.2008.175. [DOI] [PubMed] [Google Scholar]
- Treherne JM, Woodward SK, Varga ZM, Ritchie JM, Nicholls JG. Restoration of conduction and growth of axons through injured spinal cord of neonatal opossum in culture. Proc Natl Acad Sci U S A. 1992;89:431–434. doi: 10.1073/pnas.89.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J, Crossley M. Basic Kruppel-like factor functions within a network of interacting haematopoietic transcription factors. Int J Biochem Cell Biol. 1999;31:1169–1174. doi: 10.1016/s1357-2725(99)00067-9. [DOI] [PubMed] [Google Scholar]
- van Vliet J, Turner J, Crossley M. Human Kruppel-like factor 8: a CACCC-box binding protein that associates with CtBP and represses transcription. Nucleic Acids Res. 2000;28:1955–1962. doi: 10.1093/nar/28.9.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman MB, Bemben MA, Thompson RC, Goldman D. Gene expression analysis of zebrafish retinal ganglion cells during optic nerve regeneration identifies KLF6a and KLF7a as important regulators of axon regeneration. Dev Biol. 2007;312:596–612. doi: 10.1016/j.ydbio.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Wang JT, Kunzevitzky NJ, Dugas JC, Cameron M, Barres BA, Goldberg JL. Disease gene candidates revealed by expression profiling of retinal ganglion cell development. J Neurosci. 2007;27:8593–8603. doi: 10.1523/JNEUROSCI.4488-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman SG. Determinants of conduction velocity in myelinated nerve fibers. Muscle Nerve. 1980;3:141–150. doi: 10.1002/mus.880030207. [DOI] [PubMed] [Google Scholar]
- Wiksten M, Vaananen AJ, Liebkind R, Liesi P. Regeneration of adult rat spinal cord is promoted by the soluble KDI domain of gamma1 laminin. J Neurosci Res. 2004;78:403–410. doi: 10.1002/jnr.20159. [DOI] [PubMed] [Google Scholar]
- Yanagi M, Hashimoto T, Kitamura N, Fukutake M, Komure O, Nishiguchi N, Kawamata T, Maeda K, Shirakawa O. Expression of Kruppel-like factor 5 gene in human brain and association of the gene with the susceptibility to schizophrenia. Schizophr Res. 2008;100:291–301. doi: 10.1016/j.schres.2007.11.042. [DOI] [PubMed] [Google Scholar]
- Yang Y, Tetreault MP, Yermolina YA, Goldstein BG, Katz JP. Kruppel-like factor 5 controls keratinocyte migration via the integrin-linked kinase. J Biol Chem. 2008;283:18812–18820. doi: 10.1074/jbc.M801384200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiu G, He Z. Glial inhibition of CNS axon regeneration. Nat Rev Neurosci. 2006;7:617–627. doi: 10.1038/nrn1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Chen X, Yang VW. Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J Biol Chem. 2003;278:2101–2105. doi: 10.1074/jbc.M211027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RD, Swamynathan SK, Boote C, Mann M, Quantock AJ, Piatigorsky J, Funderburgh JL, Meek KM. Stromal edema in klf4 conditional null mouse cornea is associated with altered collagen fibril organization and reduced proteoglycans. Invest Ophthalmol Vis Sci. 2009;50:4155–4161. doi: 10.1167/iovs.09-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JS, Moncrieffe MC, Kaczynski J, Ellenrieder V, Prendergast FG, Urrutia R. A conserved alpha-helical motif mediates the interaction of Sp1-like transcriptional repressors with the corepressor mSin3A. Mol Cell Biol. 2001;21:5041–5049. doi: 10.1128/MCB.21.15.5041-5049.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Daley GQ. From fibroblasts to iPS cells: Induced pluripotency by defined factors. J Cell Biochem. 2008;105:949–955. doi: 10.1002/jcb.21871. [DOI] [PubMed] [Google Scholar]
- Zheng B, Ho C, Li S, Keirstead H, Steward O, Tessier-Lavigne M. Lack of enhanced spinal regeneration in Nogo-deficient mice. Neuron. 2003;38:213–224. doi: 10.1016/s0896-6273(03)00225-3. [DOI] [PubMed] [Google Scholar]
- Zhou FQ, Snider WD. Intracellular control of developmental and regenerative axon growth. Philos Trans R Soc Lond B Biol Sci. 2006;361:1575–1592. doi: 10.1098/rstb.2006.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Tai C, MacVicar BA, Jia W, Cynader MS. Glutamatergic stimulation triggers rapid Krupple-like factor 4 expression in neurons and the overexpression of KLF4 sensitizes neurons to NMDA-induced caspase-3 activity. Brain Res. 2009;1250:49–62. doi: 10.1016/j.brainres.2008.11.013. [DOI] [PubMed] [Google Scholar]