Abstract
CSX/NKX2.5 is an evolutionarily conserved homeodomain-containing (HD-containing) transcription factor that is essential for early cardiac development. Recently, ten different heterozygous CSX/NKX2.5 mutations were found in patients with congenital heart defects that are transmitted in an autosomal dominant fashion. To determine the consequence of these mutations, we analyzed nuclear localization, DNA binding, transcriptional activation, and dimerization of mutant CSX/NKX2.5 proteins. All mutant proteins were translated and located to the nucleus, except one splice-donor site mutant whose protein did not accumulate in the cell. All mutants that had truncation or missense mutations in the HD had severely reduced DNA binding activity and little or no transcriptional activation function. In contrast, mutants with intact HDs exhibit normal DNA binding to the monomeric binding site but had three- to ninefold reduction in DNA binding to the dimeric binding sites. HD missense mutations that preserved homodimerization ability inhibited the activation of atrial natriuretic factor by wild-type CSX/NKX2.5. Although our studies do not characterize the genotype-phenotype relationship of the ten human mutations, they identify specific abnormalities of CSX/NKX2.5 function essential for transactivation of target genes.
Introduction
Homeobox proteins comprise a large family of transcription factors that contain a highly conserved 60–amino acid DNA binding domain (homeodomain [HD]) (1, 2). Csx/Nkx2.5 belongs to the NK2 class of homeobox proteins characterized by a tyrosine residue at amino acid 54 of the HD (54Tyr in HD) and a conserved 23–amino acid NK2-specific domain (3–5). NK2 class HD proteins are expressed in a tissue-specific manner and are important for the determination of cell fate. Csx/Nkx2.5 is the earliest known marker of the heart field mesoderm in Drosophila, zebrafish, Xenopus, chick, and mouse and is shown to be essential for heart formation in Drosophila, Xenopus, and mouse (6–12). In mice, Csx/Nkx2.5 expression starts as early as 7.5 days postcoitum (dpc) in the precardiac mesoderm and its expression continues throughout adulthood (13–15). Csx/Nkx2.5-targeted homozygous mutant mice show normal heart tube formation, but die around 11 dpc before or just after completion of looping morphogenesis (11, 12). Analysis of Csx/Nkx2.5 homozygous mutant embryos showed downregulation of ventricular myosin light chain 2V, atrial natriuretic factor (ANF), and brain natriuretic factor, but most myofilament genes are normally expressed. Transcription factors including eHAND, MEF2C, N-Myc, CARP, Msx2, and Irx4 are downregulated or misexpressed in the mutant embryo (11, 12, 16–19).
In Xenopus and zebrafish, overexpression of wild-type Nkx2.5 causes cardiac enlargement due to an increase in the number of cardiac myocytes (8, 20, 21). On the other hand, dominant inhibitory mutants of Xenopus XNkx2.3 as well as XNkx2.5, created either by fusion to the engrailed repressor domain or by mutating a single amino acid within the HD to abolish DNA binding, causes small hearts or in the most severe case, a complete loss of heart in Xenopus embryos (9, 10).
Recently, heterozygous mutations of human CSX/NKX2.5 were identified in patients with congenital heart disease (22, 23). The most common phenotype was progressive atrioventricular conduction delays (AV block) and secundum atrial septal defect (ASD), but other anatomical abnormalities, such as ventricular septal defect (VSD), tetralogy of Fallot (TOF), or tricuspid valve abnormalities including Ebstein’s anomaly, and progressive left ventricular failure were also found (22, 23). These findings strongly suggest that CSX/NKX2.5 is important in the later stages of heart development and maturation in addition to its functions in early cardiac development.
To date, a number of mutations in HD proteins have been identified in a variety of congenital disorders; however, only CSX/NKX2.5 mutations have been identified in the NK2 class associated with congenital disease (24–26). Most of these mutations are point mutations in exons (nonsense or missense) or in RNA splicing signals. Others are small nucleotide insertions or deletions that cause intron-splicing abnormalities or produce frameshifts leading to truncated proteins. Several biochemical studies showed that the mutant homeoproteins may function as either loss of function or dominant inhibitory mutants (25–29). However, the mechanisms by which CSX/NKX2.5 mutations cause congenital heart disease have not been defined. Here, we report the initial biochemical characterization of ten different CSX/NKX2.5 mutations associated with human congenital heart disease.
Methods
Plasmid construct.
pBS SK(-)-CSX/NKX2.5 (ref. 30) digested with BglI-blunt–ended and EcoRI was ligated into SacI-blunt–ended and EcoRI-digested pMALC2 (New England Biolabs Inc., Beverly, Massachusetts, USA) to construct maltose binding protein–CSX/NKX2.5 (MBP-CSX/NKX2.5). BglI-blunt–ended and XhoI-digested pBS SK(-)-CSX/NKX2.5 was ligated into EcoRV-XhoI-digested pcDNA3 (Invitrogen Corp., San Diego, California, USA) to construct pcDNA3-CSX/NKX2.5. FLAG epitope tag (gac tac aaa gac gat gac gac aag) was inserted at the NH2-terminus of pcDNA3-CSX/NKX2.5. Site-directed mutations were introduced into both plasmids by using Quick Change Site-Directed Mutagenesis Kit (Stratagene, La Jolla, California, USA) with the following primers:
M25 (F, 5′-AACAGCAGCAGtGCAGCCTGGCT-3′; R, 5′-AGCCAGGCTGCaCTGCTGCTGTT-3′): M149 (F, 5′-TTCTCGCAGGCGtAGGTCTATGAGCT-3′; R, 5′-AGCTCATAGACCTaCGCCTGCGAGAA-3′): M170 (F, 5′-CGAACGCGACtAGCTGGCCAGC-3′; R, 5′-GCTGGCCAGCTaGTCGCGTTCG-3′): M178 (F, 5′-TGCTGAAACTCAtGTCCACGCAGG-3′; R, 5′-CCTGCGTGGACaTGAGTTTCAGCA-3′): M188 (F, 5′-TGGTTCCAGAAaCGGCGCTACAA-3′; R, 5′-TTGTAGCGCCGtTTCTGGAACCA-3′): M189 (F, 5′-GGTTCCAGAACgGGCGCTACAAG-3′; R, 5′-CTTGTAGCGCCcGTTCTGGAACC-3′): M191 (F, 5′-CAGAACCGGCGCTgCAAGTGCAAGCGG-3′; R, 5′-CCGCTTGCACTTGcAGCGCCGGTTCTG-3′): M198 (F, 5′-AAGCGGCAGCGGtAGGACCAGACT-3′; R, 5′-AGTCTGGTCCTaCCGCTGCCGCTT-3′): M259 (F, 5′-CTATCCGGGTTAaGGCGGCGCGGCCT-3′; R, 5′-AGGCCGCGCCGCCtTAACCCGGATAG-3′). pcDNA3-CSX/NKX2.5 was digested with pflMI-NotI, blunt ended and religated to construct pcDNA3-CSX/NKX2.5(1–200).
The SphI-KpnI fragment of CSX/NKX2.5 genomic clone (hCsx10) was inserted into SphI-KpnI-digested MBP-CSX/NKX2.5 to construct MBP-hCsx10. A mutation was introduced by site-directed mutagenesis using 5′-CCGAAAAGAAAGtTGAGGAGGAA-3′; 5′-TTCCTCCTCAaCTTTCTTTTCGG-3′) to construct MBP-M112. PmlI-PflMI-digested fragments of MBP-hCsx10 and MBP-M112 were replaced with that of pcDNA3-CSX/NKX2.5 to construct pcDNA3-hCsx10 and pcDNA3-M112. All the PCR-amplified fragments were sequenced to rule out DNA misincorporations.
Electrophoretic mobility shift assay.
MBP fusion proteins were prepared as described previously (31). Briefly, cultured Escherichia coli BL21(DE3) (Novagen, Madison, Wisconsin, USA) induced with 0.3 mM of IPTG, were lysed by sonication in lysis buffer (20 mM HEPES [pH 7.5], 100 mM NaCl, 5 mM MgCl2, 1% Triton X-100, 2 μg/mL aprotinin, 0.7 μg/mL pepstatin, 0.1 mM PMSF, 1 mM DTT, and 10% glycerol), and lysates were incubated with amylose resin (New England Biolabs Inc.). Fusion proteins were eluted from the beads with lysis buffer (0.1% Triton X-100 instead of 1%) containing 10 mM maltose. The molecular weight of MBP fusion protein was estimated by the addition of the molecular weight of MBP protein (mol wt = 42 kDa) and full length (mol wt = 34.9 kDa) or M198 (mol wt = 22.1 kDa) or M259 (mol wt = 28.5 kDa) or M25 (mol wt = 34.9 kDa). Of note, MBP-fused CSX/NKX2.5 proteins do not remain completely intact during purification. Coomassie stained SDS-PAGE gel was scanned and the intensity of the top bands representing the intact proteins were compared with that of the BSA standard to estimate the protein concentration.
2 pM of end-labeled ANF Csx/Nkx2.5 binding site, 5′- –264TCACACCTTTGAAGTGGGGGCCTCTTGAGGCAAATC–227-3′ was annealed with 5′-GATTTGCCTCAAGAGGCCCCCACTTCAAAGGTGTGA-3′, and 5′- –264TCACACCTTTGAAGTGGGGGCCT-3′ was annealed with 5′-AGGCCCCCACTTCAAAGGTGTGA-3′ and used for electrophoretic mobility shift assay (EMSA). Threefold serial dilutions of 66 ng of bacterially expressed fusion proteins were incubated with 50,000 cpm of probe, 50 μg BSA, 0.5 μg poly (dG-dC) in 10 mM HEPES (pH 8.0), 50 mM KCl, 1 mM EGTA, 10% glycerol, 2.5 mM DTT, and 7 mM MgCl2 in 15 μL reaction volume for 20 minutes at room temperature, separated in 5% polyacrylamide gel with 0.5 × Tris-glycine buffer at 15 mA for approximately 20 minutes.
Reporter gene assays.
10T1/2 fibroblast cells cultured in six-well plates were transfected with 1.5 μg of ANF-luciferase reporter construct (–638, provided by K.R. Chien, University of California at San Diego, La Jolla, California, USA), 1 μg of pcDNA3-CSX/NKX2.5 expression vector and 0.5 μg of Rous sarcoma virus–β-galactosidase (RSV–β-galactosidase) (provided by B. Markham) using the calcium phosphate method. Total plasmid amount was adjusted to 3 μg with pcDNA3 vector plasmid. For cotransfection experiments, 1.2 μg of ANF-luciferase reporter construct, 0.3 μg of RSV–β-galactosidase construct, 0.7 μg of pcDNA3-CSX/NKX2.5 expression vector, 0.7 or 1.4 μg of pcDNA3 expression vector encoding mutant proteins and pcDNA3 empty vector to adjust the total plasmid amount 3.6 μg were used. After glycerol shock using 1X HEPES buffer containing 15% glycerol, cells were cultured for another 48 hours, lysed with 300 μL of reporter lysis buffer (Promega Corp., Madison, Wisconsin, USA) and assayed for luciferase activity (Promega Corp.) and β-galactosidase activity.
Protein-protein interaction.
Bacterially produced MBP-CSX/NKX2.5, MBP, glutathione-S-transferase-GATA4 (GST-GATA4; provided by D. Wilson) and GST protein were made as described previously (31). In vitro-transcribed and translated proteins were generated by using TNT-coupled reticulocyte lysate systems (Promega Corp.). A total of 1 μL of reticulocyte lysate containing [35S]-labeled wild-type or mutant CSX/NKX2.5 proteins was mixed with the fusion proteins bound on the beads in 400 μL of binding buffer (20 mM HEPES [pH 7.5], 100 mM NaCl, 5 mM MgCl2, 0.1% Triton X-100, aprotinin [2 μg/mL], pepstatin [0.7 μg/mL], 0.1 mM PMSF, 1 mM DTT, and 1% BSA) at 4°C for 2 hours. Beads were washed with binding buffer (without BSA) five times and the bound protein complexes were subjected to SDS-PAGE.
Splicing assays, RT-PCR and protein synthesis assay.
Transient transfection of pcDNA3-CSX/NKX2.5 wild-type and various mutants in COS 7 cells was performed by using Lipofectamine (GIBCO BRL, Gaithersburg, Maryland, USA) as described previously (15). Cells on 6 cm plates were harvested approximately 24 hours after transfection, spun down and directly lysed in 200 μl of SDS-PAGE sample buffer (62.5 mM Tris [pH 6.8], 2.3% SDS, 10% glycerol, 5% 2-mercaptoethanol), and 20 μL of protein (∼10 μg) was analyzed for protein expression by Western blotting using anti-FLAG mAb (Sigma Chemical Co., St. Louis, Missouri, USA), anti-Csx/Nkx2.5 mAb (15) and anti-GAPDH mAb (RDI Research Diagnostics Inc., Flanders, New Jersey, USA). Total RNA was extracted from transfected cells (RNeasy kit; Qiagen Inc., Valencia, California, USA), and 0.5 μg were used for RT-PCR (Perkin-Elmer Corp., Norwalk, Connecticut, USA) by the use of a sense primer (5′-CAAGGACCCTAGAGCCGAAAAG-3′) and an antisense primer (5′-TTGACCTGCGTGGACGTGAGTTTC-3′) for CSX/NKX2.5 that span the intron. In vitro transcription/translation was performed using the TNT-coupled reticulocyte lysate system (Promega Corp.) using 2 μg plasmid/ 50 μL reaction.
Results
Grouping of ten mutants according to predicted protein structure.
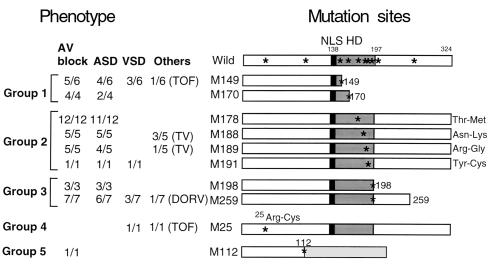
Human CSX/NKX2.5 is a 324–amino acid protein, that includes an HD between amino acids 138 and 197 (30, 32) and an Arg-Lys cluster of a nuclear localization signal at the NH2-terminus of the HD (Figure 1) (31). Ten mutation sites identified in patients (22, 23) (Figure 1, asterisks) are distributed throughout the CSX/NKX2.5 molecule. Two mutations are located at the NH2-terminus to the HD, six are within the HD, and two are located COOH-terminus to the HD. As shown in Figure 1, these mutation sites were divided into five groups based on the predicted protein structure: group 1: two nonsense mutations in the HD (M149 and M170); group 2: four missense mutations within the HD [178Thr-Met (M178), 188Asn-Lys (M188), 189Arg-Gly (M189), and 191Tyr-Cys (M191)]; group 3: two truncation mutants occurring COOH-terminal to the HD [Δ198-324 (M198) and Δ259-324 (M259)]; group 4: one missense mutation NH2-terminal to the HD (25Arg-Cys, M25); and group 5: one point mutation at the exon-intron splicing site (M112). The corresponding phenotype associated with each of these mutations is indicated in Figure 1 (23).
Figure 1.
Diagram of CSX/NKX2.5 cDNA with the location of ten mutation sites identified in congenital heart disease. Ten mutation sites (asterisks in Wild) were divided into five groups based on the predicted protein structure: nonsense mutation in the HD (group 1: M149 and M170); missense mutation in the HD (group 2: M178, M188, M189, and M191); premature termination after HD (group 3: M198 and M259); 25Arg-Cys missense mutation (group 4: M25); and mutation at the intron-splicing donor site (group 5: M112). Phenotypes observed in patients are listed on the left. For example, “11/12” indicates that 11 patients show the phenotype among 12 patients examined. These mutation sites were mapped on CSX/NKX2.5 cDNA, which encodes 324 amino acids including 60 amino acids of HD (shaded box). Nuclear localization signal at the NH2-terminus of the HD is indicated (black box). Predicted translated product of M112 mutation in splicing donor site is indicated with a light gray box. AV block, atrioventricular conduction block; ASD, atrial septal defect; VSD, ventricular septal defect; TOF, tetralogy of Fallot; TV, tricuspid valve abnormality; DORV, double outlet right ventricle; NLS, nuclear localization signal; HD, homeodomain.
All mutant proteins were translated and localized to the nucleus, except one splice-donor site mutant protein (M112) that did not accumulate in the cell.
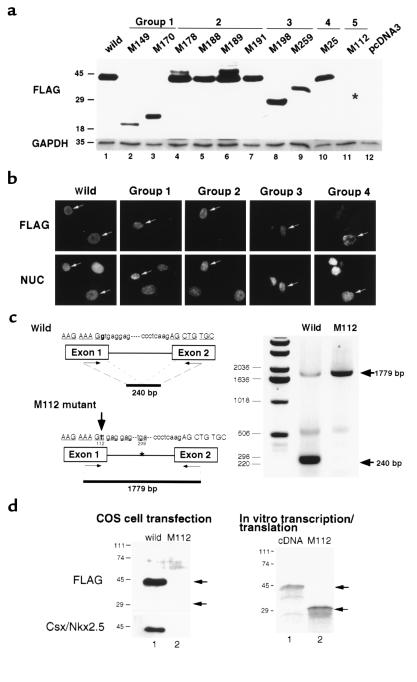
To determine whether mutant proteins accumulate in the cell, FLAG epitope tagged mutant cDNAs were subcloned into the mammalian expression vector pcDNA3 and transfected into COS cells. By Western blotting using anti-FLAG mAb (Figure 2a), proteins with expected molecular weight were detected in group 1, 2, 3 and 4 mutants, whereas group 5 (M112) mutant protein was not detected. As shown in Figure 2b, all accumulated proteins were localized in the nucleus.
Figure 2.
Expression of translated products in cells: intron-splicing defect in M112 mutant resulted in poor protein accumulation. (a) Wild-type and mutant CSX/NKX2.5 expression vectors were transfected into COS 7 cells, and the protein expression was examined by Western blotting using anti-FLAG Ab approximately 24 hours after transfection (FLAG, top). All mutant proteins except M112 (lane 11, asterisk indicates the expected molecular weight of M112 protein) were detected at the expected molecular weight. GAPDH expression in each lane was also shown (GAPDH, bottom). (b) Wild-type and mutant CSX/NKX2.5 proteins accumulated in the nucleus colocalizing with Hoechst nuclear staining (NUC, lower panels). The results presented are wild-type, group 1 (M170), group 2 (M191), group 3 (M198), and group 4 (M25). (c) G→T substitution (large arrow) identified in M112 mutant on the CSX/NKX2.5 splicing donor site was examined by RT-PCR. RNA-purified form COS 7 cells transfected with the wild-type and M112 mutant of CSX/NKX2.5 were amplified with two primers spanning the intron. In the wild-type transfectant, the intron was spliced out, resulting in the generation of 240-bp product, whereas in the mutant transfectant, G→T substitution of the first codon of the intron splicing site abolished the normal splicing, resulting in generation of 1,779-bp product. CSX/NKX2.5 protein was encoded by the two exons represented. (d) Translated products were examined approximately 24 hours after transfection by Western blotting using anti-FLAG mAb (top) and anti-Csx/Nkx2.5 mAb (bottom). Wild-type CSX/NKX2.5 gene was translated into approximately 42-kDa protein and was detected with anti-FLAG and anti-Csx/Nkx2.5 mAb (lane 1). In contrast, M112 mutant protein that is expected to migrate approximately 29 kDa was not detected in the cell lysate (lane 2). In in vitro transcription and translation system, cDNA produced approximately 42-kDa protein (lane 1), and M112 genomic construct produced about 29-kDa protein (lane 2).
The M112 expression construct contained a mutated intron (G→T transversion) involving the first nucleotide of the splice-donor site and results in a change in sequence from GTGAGG to TTGAGG (Figure 2c). To examine whether the failure of protein accumulation in cells is due to transcriptional or translational regulation, we compared mRNA and protein expression in transfected cells. As a control, we inserted the wild-type intron (1,539 bp) in the wild-type cDNA construct used in Figure 2a. RNA isolated from transfected cells was amplified by RT-PCR with two primers spanning the intron. The wild-type construct amplified a major spliced product (240 bp) with a low amount of nonspliced product (1,779 bp). In contrast, RT-PCR of RNA isolated from cells transfected with the M112 construct produced only the nonspliced product (1,779 bp). In wild-type transfectants, CSX/NKX2.5 protein was detected as a 42-kDa band by both anti-FLAG mAb and the anti-Csx/Nkx2.5 mAb (Figure 2d, left, lane 1). However, M112 protein was not detected (Figure 2d, left, lane 2), even though the corresponding mRNA was as easily detected by RT-PCR as that of wild-type (Figure 2c). In vitro transcription and translation of M112 produced a protein with an approximate molecular eight of 29 kDa (Figure 2d, right, lane 2), but in the cell, the translation product did not accumulate. Thus, it is likely that the M112 allele functions as a heterozygous null allele in vivo.
Assessment of mutant DNA binding using EMSA.
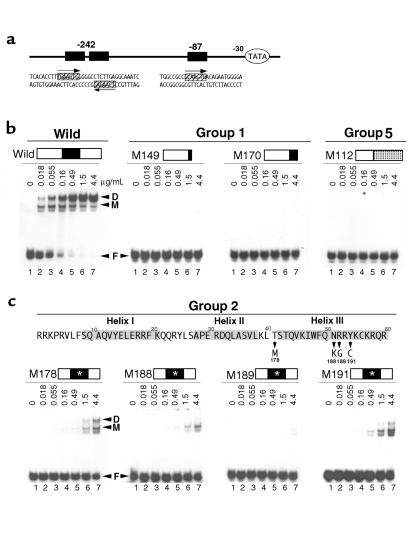
We examined DNA binding affinity by using the electrophoretic mobility shift assay (EMSA). The ANF promoter, an in vivo Csx/Nkx2.5 target, contains three specific Csx/Nkx2.5 binding sites (TNAAGTG) (3, 33) that are located upstream of the transcription start site at approximately –408, –242, and –87 bp (34, 35). We used the –242 bp binding site as a probe to examine CSX/NKX2.5 binding affinity, since this site contains two binding sites spaced by 5 nucleotides for CSX/NKX2.5 (Figure 3a) and had more than 80-fold higher affinity than the -87 bp site (data not shown). The [32P]-labeled 32 bp oligonucleotide of the ANF promoter (–248 to –217, ANF –242) was mixed with wild-type or a mutant protein purified from E. coli as an MBP fusion protein (Figure 3b). In each panel, lane 1 shows free DNA probe without protein (F), and lanes 2–7 show threefold serial increase in protein concentration (0.018–4.4 μg/mL) mixed with the same amount of DNA probe. The wild-type CSX/NKX2.5 formed two specifically shifted bands in each lane (Figure 3b, wild). The faster migrating band most likely corresponds to the occupation of one of the two specific DNA binding sites of the ANF –242 (M, monomer), and the slower migrating band, to the simultaneous occupation of the two DNA binding sites (D, dimer). The intensity of these bands was dependent on the protein concentration of CSX/NKX2.5 in the reaction (Figure 3b, wild).
Figure 3.
DNA binding affinity of group 1, 2, and 5 mutant proteins versus wild-type CSX/NKX2.5. (a) Sequence of two consensus CSX/NKX2.5 binding sites (–242 bp and –87 bp sites) in rat ANF promoter; the paired binding sites in –242 bp site, and a single binding site in –87 bp site. (b) –242 bp site was used for the DNA binding assay (lane 1) mixed with threefold serially increased CSX/NKX2.5 fusion proteins (0.018–4.4 μg/mL of MBP-CSX/NKX2.5 fusion protein) (lanes 2–7). Wild-type CSX/NKX2.5 bound as a monomer (M) as well as a dimer (D) depending on the protein concentration. No shifted bands were observed in groups 1 and 5 (M149, M170, and M112). (c) The EMSA of the group 2 mutant proteins that have a single missense mutation in the HD. 178Thr-Met (M178) mutation was located just before the third helix, and 188Asn-Lys (M188), 189Arg-Gly (M189), and 191Tyr-Cys (M191) were located in the third helix. Two conserved amino acid mutations were identified: 188Asn (51Asn in HD) is conserved in all the HD protein that is directly bound to the major groove of DNA, and 191Tyr (54Tyr in HD) is conserved in all NK2 class homeoprotein. All four mutant proteins show dramatically reduced DNA binding affinity compared with the wild-type CSX/NKX2.5. D, dimer; M, monomer; F, free probe. Mutation sites of group 2 are indicated with white asterisks.
Group 1 and group 5 do not bind DNA due to the lack of the third helix of HD.
As expected, group 1 mutant proteins (M149 and M170) with truncated HD did not bind DNA (Figure 3b, group 1). Similarly, the mutant MBP fusion protein M112 (group 5), which does not encode HD, did not bind DNA (Figure 3b, group 5).
Group 2 mutants have markedly reduced DNA binding affinity.
Four mutants that have a single missense mutation in the HD comprise group 2 (Figure 3c); all group 2 mutants had markedly reduced DNA binding affinity. 178Thr-Met (M178) mutation is located just before the third helix of the HD. 178Thr-Met (M178) mutation required approximately 34-fold (81-fold) higher protein concentration to shift the DNA probe to the same degree as wild-type. 188Asn (amino acid 51 in HD) that is conserved among all the HD proteins and is critical for DNA binding (1, 2). 188Asn-Gly (M188) mutant had more than 35-fold (273)-fold lower DNA binding affinity than wild-type. Also, 189Arg-Gly (M189) mutation showed barely detectable DNA binding. The 191Tyr residue (54Tyr in HD) is unique for the NK2 class HD proteins (3–5, 8). 191Tyr-Cys (M191) mutant protein also showed a markedly reduced DNA binding affinity by approximately 34-fold (81-fold) (Figure 3c).
Group 3 and group 4 mutants showed similar DNA binding affinity as a monomer but reduced DNA binding affinity as a dimer.
In three mutants, the HD was unaffected; two COOH-terminus deletion mutants (M198 and M259) comprised group 3, and an NH2-terminus missense mutation (M25) comprised group 4. All group 3 and 4 mutant proteins bound to DNA (Figure 4), but there were subtle but significant differences in DNA binding characteristics. CSX/NKX2.5 bound to the dimeric NK2 specific binding sites predominantly as a monomer at a low protein concentration and transited into a dimer at higher protein concentration. In wild-type CSX/NKX2.5 protein, the monomer-dimer transition was observed in lane 3 (Figure 4a, asterisk in wild-type) at an approximate protein concentration of 0.055 μg/mL or 7.1 × 10–10 M). In contrast, M198 required approximately 9 (32) times higher protein concentration to yield a monomer/dimer ratio similar to that of wild-type (Figure 4b, asterisk in M198; approximate protein concentration 7.7 × 10–9 M). A 31–32 times higher protein concentration was necessary for equimolar monomer-dimer formation in M259 (Figure 4c, asterisk in M259; approximate protein concentration 2.3 × 10–9 to 7.0 × 10–9 M). We also detected a subtle defect of dimer formation in the M25 mutant, which required approximately three times higher protein concentration to yield a monomer/dimer ratio similar to that of wild-type (Figure 4d, asterisk in M25; approximate protein concentration 2.1 × 10–9 M).
Figure 4.
DNA binding affinity of the group 3 and 4 mutant proteins versus wild-type CSX/NKX2.5. Sequence of the native –242 bp site (top) and a mutated –242 bp binding site (bottom) in the ANF promoter. EMSA of group 3 (M198 and M259) and group 4 (M25, the mutation site is marked with an asterisk) protein compared with that of the wild-type CSX/NKX2.5. Proteins were mixed with probes containing either tandemly repeated binding sites (a–d) or single binding site (e–h). Lanes showing similar monomer/dimer ratios are indicated with asterisks in the top panels (a–d). In all three mutant proteins, binding affinity as a dimer is reduced approximately 3- to 32 fold (b–d versus a), whereas they show similar DNA binding affinity as wild-type to the single binding site (f–h versus e). D, dimer; M, monomer; F, free probe.
To confirm that these mutant proteins bound to the monomeric binding site with a similar affinity, we performed EMSA using oligonucleotides in which one of the DNA binding sites was deleted from the ANF -242 site. Group 3 and group 4 mutants bound to the mutated monomeric site with identical affinity as wild-type CSX/NKX2.5 (Figure 4, e–h). These findings suggest that group 3 and group 4 mutants bound to monomeric DNA binding sites with similar affinity as wild-type CSX/NKX2.5, but the mutations decreased the ability to form dimers on the dimeric site.
Transcriptional activation function of CSX/NKX2.5 mutants.
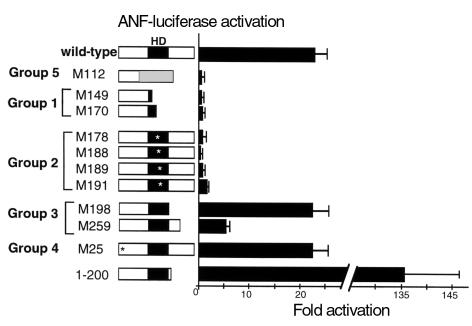
We examined the transcriptional activation function using the ANF-luciferase reporter construct. The mutants subcloned into pcDNA3 were transfected into 10T1/2 cells with ANF luciferase reporter plasmid (Figure 5). Wild-type CSX/NKX2.5 activated the ANF-luciferase reporter gene 23.0 ± 2.2 fold compared with the pcDNA3 parental vector, indicating that human CSX/NKX2.5 is a transcriptional activator as is the mouse Csx/Nkx2.5 (31, 33–35), which shows 87% overall amino acid homology (30).
Figure 5.
Effect of ten mutations on transcriptional activation. 10T1/2 cells were transfected with pcDNA3 expression vectors encoding wild-type or each of ten mutations with the reporter gene ANF-luciferase. When the wild-type CSX/NKX2.5 was transfected with the ANF reporter gene, luciferase activity was increased 23-fold compared with cells transfected with the empty expression vector pcDNA3. M112, group 1, and group 2 expression vectors did not activate the ANF promoter. M25 and M198 transactivated the reporter gene similarly to the wild-type CSX/NKX2.5; however, M259 transactivated only 5.2-fold. Another COOH-terminus deletion mutant, CSX/NKX2.5(1–200), transactivated the reporter construct approximately 136-fold. Bars represent means ± SEM of at least three separate transfection assays done in duplicate.
As expected from the EMSA data, group 1 and group 5 (M112) with truncated HD (Figure 3b) did not activate the ANF-luciferase reporter gene (Figure 5). Group 2 mutants, which have markedly reduced DNA binding, also failed to activate the ANF reporter. M198 in group 3 and M25 in group 4 mutants transactivated the reporter construct as effectively as wild-type, whereas M259 in group 3 had a reduced transcriptional activation function (5.2 ± 0.9 fold). Because the COOH-terminus deletions of the mouse Csx/Nkx2.5 were shown to cause markedly increased transcriptional activity (31, 33), the finding of unchanged or lower transcriptional activities of group 3 mutants was unexpected. Accordingly, we constructed another COOH-terminus deletion mutant, CSX/NKX2.5(1–200) that includes an additional three amino acid COOH-terminus to the HD and corresponds exactly to the murine Csx/Nkx2.5(1–199) mutant and was previously shown to have very high transcriptional activity (240-fold) (31). Similar to the mouse mutant, CSX/NKX2.5(1–200) had markedly increased transcriptional activity (136 ± 10 fold; Figure 5). These results demonstrate that mutants that do not encode the complete HD (group 1 and group 5) or with single missense mutation in the HD did not transactivate the ANF promoter. Although CSX/NKX2.5(1–200) showed “gain of function,” neither M198 nor M259 COOH-terminus deletion mutants were gain-of-function mutations.
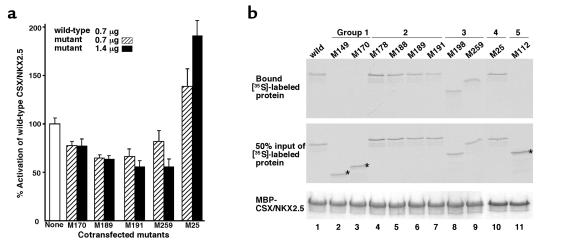
Effects of group 1, 2, 3, and 4 mutants on wild-type CSX/NKX2.5.
To examine whether these mutations affect transcriptional activity of wild-type CSX/NKX2.5, we cotransfected the expression plasmid encoding M170 (group 1), M189 (group 2), M191 (group 2), M259 (group 3), or M25 (group 4) with the plasmid encoding the wild-type CSX/NKX2.5 and examined reporter gene activity compared with that of wild-type cotransfected with pcDNA3 empty plasmid (Figure 6a, open bar, none). Luciferase activation after cotransfection of wild-type with pcDNA3 empty plasmid was defined as 100%. When the M170 protein expression plasmid was cotransfected with wild-type CSX/NKX2.5 expression plasmid, we noted an approximately 23% decrease in activation of ANF-luciferase reporter gene compared with wild-type alone at a plasmid ratio of 1:1 (0.7 μg) (Figure 6a, hatched bar, M170) and 2:1 (1.4 μg) (Figure 6a, black bar, M170). Reduction of luciferase activity was observed in M189; 34% reduction at 1:1 ratio, and 35% reduction at 2:1 ratio, as well as in M191 (34% and 44% reduction, respectively) and M259 (18% and 44% reduction, respectively). In contrast, when the plasmid encoding the M25 mutant was cotransfected, we noted 39% increase in luciferase activity at 1:1 ratio, and 91% increase with 2:1 ratio. These data demonstrate that cotransfection of the plasmid-encoding groups 2 and 3 inhibited transactivation function of wild-type CSX/NKX2.5 moderately, and further reduction of luciferase activity was observed with cotransfection of plasmid encoding M189, M191 (group 2), and M259 (group 3) mutants, suggesting that these mutants act in a dominant inhibitory manner on wild-type CSX/NKX2.5 in transient transfection assays. However, these mutants did not act in a typical dominant inhibitory manner (see Discussion). Interestingly, the M25 mutant did not exhibit an inhibitory effect, rather it transactivated the ANF promoter.
Figure 6.
Inhibition of ANF-luciferase transcriptional activity of wild-type CSX/NKX2.5 by group 1, 2, and 3 mutants but not by group 4 mutant. (a) Inhibition of transactivation of the ANF-luciferase reporter gene in the presence of both 1:1 and 2:1 ratio of M170, M189, M191, and M259 expression plasmid to wild-type expression plasmid. 10T1/2 cells were transiently transacted with 1.2 μg of ANF-luciferase(-638), 0.3 μg of RSV–β-galactosidase, 0.7 μg of wild-type CSX/NKX2.5 expression plasmid, and 0.7 μg (hatched bars) or 1.4 μg (filled bars) of mutant expression plasmid or empty pcDNA3 plasmid to adjust the total amount of plasmid. A moderate reduction of luciferase activity was observed in M170 mutants, and a further decrease of luciferase activity was detected in M189, M191, and M250 mutants. In contrast, M25 mutant increased luciferase activity. Results are presented as a percent of the ANF reporter activity when cotransfected with wild-type and mutant plasmid compared with that of the wild type alone (open bar). Bars represent means ± SEM of the means of at least three separate transfection assays done in duplicate. (b) Protein-protein interaction of wild-type CSX/NKX2.5 with mutant proteins. MBP-fused wild-type CSX/NKX2.5 protein was mixed with [35S]-labeled wild-type (lane 1) or ten mutants (lanes 2–11). After washing five times with binding buffer, the protein complexes were resolved on SDS-PAGE and autoradiographed (top panel). Group 1 (lanes 2 and 3) and group 5 (lane 11) mutants did not associate with MBP-CSX/NKX2.5, whereas group 2, 3, and 4 mutants associated with MBP-CSX/NKX2.5 (lanes 4–10). Fifty percent input of [35S]-labeled proteins are shown in the middle panel, and Coomassie blue–stained MBP-fused wild-type CSX/NKX2.5 proteins are shown in the bottom panel.
Protein-protein interaction of mutants with wild-type CSX/NKX2.5.
As shown in Figure 3b and Figure 4, we found that CSX/NKX2.5 bound to the ANF -242 site as a monomer as well as a dimer. Our recent studies demonstrated that the mouse Csx/Nkx2.5 physically interact with each other in vitro as well as in the cell in the absence of DNA (Kasahara et al., unpublished data). Because DNA binding of group 2 mutants was totally abolished or markedly reduced (Figure 3, b and c), and these mutants themselves did not activate the ANF promoter (Figure 5), it is of interest to examine whether the inhibitory effect (Figure 6a) is due to dimer formation of the mutant proteins with wild-type CSX/NKX2.5. Accordingly, MBP-fused CSX/NKX2.5 or MBP alone were mixed with in vitro translated [35S]-labeled mutant proteins. After extensive washing, the protein complexes were resolved on SDS-PAGE and autoradiographed (Figure 6b). [35S]-labeled wild-type CSX/NKX2.5 bound to MBP-CSX/NKX2.5 fusion protein was detected in lane 1 (Figure 6b, top), but not with MBP alone (data not shown). In contrast, group 1 (M149 and M170) and group 5 (M112) mutants did not interact with wild-type CSX/NKX2.5 (asterisks in lanes 2, 3, and 11), whereas group 2 (M178, M188, M189, and M191), group 3 (M198 and M259), and group 4 (M25) mutants interacted with CSX/NKX2.5 (lanes 4–10). Therefore, group 1 and 5 mutants, which completely or partially lack the HD, did not associate with wild MBP-CSX/NKX2.5, whereas mutants with the HD associated with MBP-CSX/NKX2.5. These data demonstrate that the HD region is necessary for dimerization with wild-type CSX/NKX2.5, but the amino acid residues mutated in group 2 mutants (amino acids 178, 188, 189, and 191) did not significantly affect the dimer formation with wild-type CSX/NKX2.5.
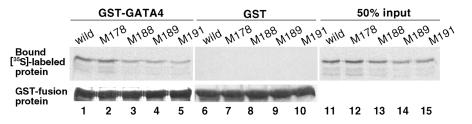
Group 2 mutants can associate with GATA4 protein.
Previous studies demonstrated that Csx/Nkx2.5 transactivates the ANF promoter synergistically with the zinc-finger transcription factor GATA4 (34–37). We also demonstrated that the second zinc finger of GATA4 is necessary and sufficient in the specific interaction with Csx/Nkx2.5, and the third helix is required for direct interaction with GATA4 (35). Because three of group 2 mutation sites are within in the third helix (Figure 3c), we examined whether these group 2 mutation sites affect the association with GATA4. As demonstrated in Figure 7, lane 1, [35S]-labeled wild-type CSX/NKX2.5 associated with GATA4, but not with GST alone (lane 6). Similar protein-protein interactions with GATA4 were observed in all four group 2 mutants (lanes 2–5), but none of these proteins associated with GST protein alone (lanes 7–10). These results demonstrate that the missense mutations found in group 2 patients do not significantly affect the interaction with GATA4. Therefore, they potentially sequester GATA4 from wild-type CSX/NKX2.5.
Figure 7.
Interaction of group 2 mutants with GATA4 protein. [35S]-labeled wild-type CSX/NKX2.5 and four group 2 mutant proteins were mixed with GST-GATA4 protein (lanes 1–5) or GST alone (lanes 6–10). Bound labeled proteins were resolved on SDS-PAGE and autoradiographed (top panel). Fifty percent input of [35S]-labeled proteins is also shown. Coomassie blue–stained GST-GATA4 (lanes 1–5) or GST (lanes 6–10) fusion proteins are shown (bottom panel).
Discussion
Heterozygous mutations of the homeoprotein CSX/NKX2.5 are associated with progressive AV block with secundum ASD as well as several other cardiac malformations that are transmitted as an autosomal dominant trait (22, 23) In this study, we examined the function of ten known mutations of CSX/NKX2.5 found in patients in order to gain insights into the nature and mechanism of the molecular defects responsible for the clinical abnormalities. We were able to categorize these mutation sites into five groups (Figure 1).
In spite of the extensive in vitro characterization of these CSX/NKX2.5 mutations presented here, it is not yet possible to define clear genotype-phenotype associations. However, some generalizations are possible. For example, the M112 mutation (group 5) abolished intron splicing (Figure 2c) and is likely to function as loss of function allele in vivo, as the mutant protein did not accumulate in transfected cells (Figure 2d). Patients with M112 mutation show second-degree AV block without morphological defects, which is similar to the phenotype of hemizygous Csx/Nkx2.5–null mice, who showed a high penetrance of AV block and low incidence of ASD (20%) without other morphological defects (M. Tanaka et al., unpublished data). These findings suggest that a loss of function of one allele may cause AV conduction delay but additional dominant effects of “hypomorphic” mutant proteins (groups 2 and 3) may cause various anatomical anomalies such as ASD, VSD, TOF, and left ventricular failure. (Note that group 2 and 3 mutants have altered DNA binding but preserved protein-protein interactions). In contrast, the patient with the M25 mutation (group 4, 25Arg-Cys missense mutation in the NH2-terminus), exhibited VSD with TOF without AV block (Figure 1). In EMSA as well as in transient transfection assays, M25 appeared to function similarly to the wild-type CSX/NKX2.5 (Figures 4 and 5). Cotransfection of plasmid encoding M25 and wild-type CSX/NKX2.5 increased the ANF luciferase activity, which is different from other mutants examined (Figure 6a). Although, M25 is genetically a dominant mutation vivo, our in vitro assays of CSX/NKX2.5 function is limited in explaining the nature of the molecular defect of the M25 mutation.
Patients with the other eight mutations showed both AV block and cardiac malformations (Figure 1). Six mutations were found in the HD either as a nonsense (M149 and M170 in group 2) or a missense (M178, M188, M189, M191 in group 3) mutation. All six HD mutant proteins had markedly reduced DNA binding affinity (Figures 3 and 4). Even in the presence of two repeated CSX/NKX2.5 binding sites, M149 and M170 (group 1) and M189 (group 2) did not show any shifted bands (Figure 3, b and c). M178, 178Thr-Met mutation in between the second and third helix, is likely to change the angle of the helix III, resulting in the reduction of contact to the major groove of DNA (10, 38, 39). Other group 2 mutations were mapped in the third helix. Interestingly, two conserved amino acids in the HD were mutated in patients. 188Asn (51Asn in HD), which is conserved in all members of the homeoprotein family and is known to directly contact adenine in the major groove of DNA (1), was mutated into Lys in M188 mutant, resulting in markedly reduced DNA binding affinity by more than 243-fold. Also, 191Tyr (54Tyr in HD), which is conserved in all NK2 class homeoproteins and is speculated to specify the DNA binding (4), was mutated into Cys in M191, which reduced its DNA binding affinity by 81-fold. Consistent with absent or markedly reduced DNA binding, group 1 and 2 mutants had markedly reduced transcriptional activity on the ANF-luciferase reporter (Figure 5).
Previous studies in Xenopus demonstrated that non–DNA-binding missense mutants act in a dominant inhibitory manner on wild-type homeoproteins, Mix1, XVent2, XNkx2.3, and XNkx2.5 (10, 38, 39). In Mix1 HD proteins, the dominant inhibitory effect of mutant proteins is likely due to their ability to homo- or heterodimerize with other HD proteins, and these mutant–wild-type homeoprotein complexes may change the transcriptional activity (38). Accordingly, we examined the homodimerization of CSX/NKX2.5 and found that wild-type CSX/NKX2.5 homodimerize on DNA that contains two binding sites (Figure 3b). We also demonstrated that CSX/NKX2.5 can homodimerize without DNA by in vitro pulldown assay (Figure 6b). All the group 2 mutations with a single missense mutations in the HD had markedly reduced DNA binding (Figure 3c) but preserved dimerization ability (Figure 6b). The mutants themselves did not transactivate or suppress the ANF promoter (Figure 5) but inhibited the transcriptional activity of wild-type CSX/NKX2.5 (Figure 6a). However, the degree of inhibition increased little by the increase in the wild-type versus mutant plasmid from 1:1 to 1:2. This suggests that the mutant may not simply act as a dominant inhibitor of wild-type CSX/NKX2.5, but may inhibit CSX/NKX2.5 function in a more complex manner.
The COOH-terminus deletion mutant of CSX/NKX2.5(1–200) had a significantly increased transcriptional activity on ANF promoter (136-fold) compared to wild-type (23-fold). In contrast, two COOH-terminus deletion mutants found in patients (group 3, M198 and M259) exhibited similar (22-fold in M198) or reduced (fivefold in M259) transcriptional activity. This suggests that the region COOH-terminus to the HD may contain several transcriptional activation and repression domains, or the deletion mutations may change the conformation of CSX/NKX2.5 to increase or decrease transcriptional activity. Considering that no mutations seen in patients showed higher transcriptional activity than wild-type, it is possible that constitutively active mutations may cause a different phenotype or may even cause an embryonic lethal phenotype, therefore escaping clinical detection.
In summary, although nuclear localization was intact in all mutants except M112, the mutations in the HD (groups 1 and 2) results in the loss or severely impaired DNA binding with the concomitant loss of their transactivation function. Mutations located outside of the HD had normal DNA binding to the monomeric target site but had reduced DNA binding to the dimeric target site compared with wild-type CSX/NKX2.5. Group 2 mutations had preserved protein dimerization function and exhibited an inhibitory function on wild-type CSX/NKX2.5. Group 4 mutant with a single missense mutations NH2-terminal to the HD acted very similarly as the wild-type CSX/NKX2.5 except for subtle DNA binding defect to the dimeric binding site. CSX/NKX2.5 is likely to form multifactor complexes to transactivate target genes (34–37, 40), and these factors in each complex could be different depending on the context of the target sites. Through a complex mechanism, CSX/NKX2.5 seems to regulate precisely target gene activation and repression at each developmental stage. Further analysis of target genes of CSX/NKX2.5 may aid in the understanding of the role of the CSX/NKX2.5 in cardiac development as well as the genotype-phenotype associations resulting from its mutations.
Acknowledgments
We thank E.O. Weinberg for critical reading of the manuscript; I. Lee and D. Turbay for CSX/NKX2.5 genomic DNA and cDNA; and A. Usheva and N. Horikoshi for valuable suggestions. This work was supported by Charles Hood Foundation and AHA Massachusetts Affiliate Fellowship (H. Kasahara), NIH R01-HL51253 (S. Izumo), and SCOR in Congenital Heart Disease grant from NIH P50-HL61036 (S. Izumo, C.E. Seidman, and J.G. Seidman).
Footnotes
Jean-Jacques Schott’s present address is: Genetic Department, Institut National de la Santé et de la Recherche Médicale Unité 533, Nantes, France.
Seigo Izumo and Hideko Kasahara contributed equally to this work.
References
- 1.Gehring WJ, Affolter M, Burglin T. Homeodomain proteins. Annu Rev Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- 2.Duboule, D. 1994. Guidebook to the homeobox genes. Oxford University Press. New York, New York, USA. 1–284.
- 3.Damante G, et al. Sequence-specific DNA recognition by the thyroid transcription factor-1 homeodomain. Nucleic Acids Res. 1994;22:3075–3083. doi: 10.1093/nar/22.15.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harvey RP. NK-2 homeobox genes and heart development. Dev Biol. 1996;178:203–216. doi: 10.1006/dbio.1996.0212. [DOI] [PubMed] [Google Scholar]
- 5.Gruschus JM, Tsao DH, Wang LH, Nirenberg M, Ferretti JA. Interactions of the vnd/NK-2 homeodomain with DNA by nuclear magnetic resonance spectroscopy: basis of binding specificity. Biochemistry. 1997;36:5372–5380. doi: 10.1021/bi9620060. [DOI] [PubMed] [Google Scholar]
- 6.Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- 7.Azpiazu N, Frasch M. Tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 1993;7:1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]
- 8.Chen JN, Fishman MC. Zebrafish tinman homolog demarcates the heart field and initiates myocardial differentiation. Development. 1996;122:3809–3816. doi: 10.1242/dev.122.12.3809. [DOI] [PubMed] [Google Scholar]
- 9.Fu Y, Yan W, Mohun TJ, Evans SM. Vertebrate tinman homologues XNkx2-3 and XNkx2-5 are required for heart formation in a functionally redundant manner. Development. 1998;125:4439–4449. doi: 10.1242/dev.125.22.4439. [DOI] [PubMed] [Google Scholar]
- 10.Grow MW, Krieg PA. Tinman function is essential for vertebrate heart development: elimination of cardiac differentiation by dominant inhibitory mutants of the tinman-related genes, XNkx2-3 and XNkx2-5. Dev Biol. 1998;204:187–196. doi: 10.1006/dbio.1998.9080. [DOI] [PubMed] [Google Scholar]
- 11.Lyons I, et al. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- 13.Lints TJ, Parsons LM, Hartley L, Lyons I, Harvey RP. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993;119:419–431. doi: 10.1242/dev.119.2.419. [DOI] [PubMed] [Google Scholar]
- 14.Komuro I, Izumo S. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proc Natl Acad Sci USA. 1993;90:8145–8149. doi: 10.1073/pnas.90.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasahara H, Bartunkova S, Schinke M, Tanaka M, Izumo S. Cardiac and extra-cardiac expression of Csx/Nkx2.5 homeodomain protein. Circ Res. 1998;82:936–946. doi: 10.1161/01.res.82.9.936. [DOI] [PubMed] [Google Scholar]
- 16.Zou Y, et al. CARP, a cardiac ankyrin repeat protein, is downstream in the Nkx2-5 homeobox gene pathway. Development. 1997;124:793–804. doi: 10.1242/dev.124.4.793. [DOI] [PubMed] [Google Scholar]
- 17.Biben C, Harvey RP. Homeodomain factor Nkx2-5 controls left/right asymmetric expression of bHLH gene eHand during murine heart development. Genes Dev. 1997;11:1357–1369. doi: 10.1101/gad.11.11.1357. [DOI] [PubMed] [Google Scholar]
- 18.Harvey, R.P., and Rosental, N. 1999. Heart development. Academic Press. San Diego, California, USA. 122–123.
- 19.Bruneau BG, et al. Cardiac expression of the ventricle-specific homeobox gene irx4 is modulated by Nkx2-5 and dHand. Dev Biol. 2000;217:266–277. doi: 10.1006/dbio.1999.9548. [DOI] [PubMed] [Google Scholar]
- 20.Fu Y, Izumo S. Cardiac myogenesis:overexpression of XCsx2 or XMEF2 in whole Xenopus embryos induces the precocious expression of XMHC gene. Roux Arch Dev Biol. 1995;205:198–202. doi: 10.1007/BF00357766. [DOI] [PubMed] [Google Scholar]
- 21.Cleaver OB, Patterson KD, Krieg PA. Overexpression of the tinman-related genes XNkx-2.5 and XNkx-2.3 in Xenopus embryos results in myocardial hyperplasia. Development. 1996;122:3549–3556. doi: 10.1242/dev.122.11.3549. [DOI] [PubMed] [Google Scholar]
- 22.Schott JJ, et al. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- 23.Benson W, et al. Mutations in the cardiac transcription factor Nkx2-5 affect diverse cardiac developmental pathways. J Clin Invest. 1999;104:1567–1573. doi: 10.1172/JCI8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boncinelli E. Homeobox genes and disease. Curr Opin Genet Dev. 1997;7:331–337. doi: 10.1016/s0959-437x(97)80146-3. [DOI] [PubMed] [Google Scholar]
- 25.Swain PK, et al. Mutations in the cone-rod homeobox gene are associated with the cone-rod dystrophy photoreceptor degeneration. Neuron. 1997;19:1329–1336. doi: 10.1016/s0896-6273(00)80423-7. [DOI] [PubMed] [Google Scholar]
- 26.Duquesnoy P, et al. Human Prop-1: cloning, mapping, genomic structure. Mutations in familial combined pituitary hormone deficiency. FEBS Lett. 1998;437:216–220. doi: 10.1016/s0014-5793(98)01234-4. [DOI] [PubMed] [Google Scholar]
- 27.Hu G, et al. Haploinsufficiency of MSX1: a mechanism for selective tooth agenesis. Mol Cell Biol. 1998;18:6044–6051. doi: 10.1128/mcb.18.10.6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomura H, et al. Loss-of-function and dominant-negative mechanisms associated with hepatocyte nuclear factor-1beta mutations in familial type 2 diabetes mellitus. J Biol Chem. 1999;274:12975–12978. doi: 10.1074/jbc.274.19.12975. [DOI] [PubMed] [Google Scholar]
- 29.Liu YH, et al. Premature suture closure and ectopic cranial bone in mice expressing Msx2 transgenes in the developing skull. Proc Natl Acad Sci USA. 1995;92:6137–6141. doi: 10.1073/pnas.92.13.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turbay D, Wechsler SB, Blanchard KM, Izumo S. Molecular cloning, chromosomal mapping, and characterization of the human cardiac-specific homeobox gene hCsx. Mol Med. 1996;2:86–96. [PMC free article] [PubMed] [Google Scholar]
- 31.Kasahara H, Izumo S. Identification of the in vivo casein kinase II phosphorylation site within the homeodomain of the cardiac tissue-specifying homeobox gene product Csx/Nkx2.5. Mol Cell Biol. 1999;19:526–536. doi: 10.1128/mcb.19.1.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiojima I, et al. Molecular cloning and characterization of human cardiac homeobox gene CSX1. Circ Res. 1996;79:920–929. doi: 10.1161/01.res.79.5.920. [DOI] [PubMed] [Google Scholar]
- 33.Chen CY, Schwartz RJ. Identification of novel DNA binding targets and regulatory domains of a murine tinman homeodomain factor, nkx-2.5. J Biol Chem. 1995;270:15628–15633. doi: 10.1074/jbc.270.26.15628. [DOI] [PubMed] [Google Scholar]
- 34.Durocher D, Charron F, Warren R, Schwartz RJ, Nemer M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee Y, et al. The cardiac-restricted homeobox protein Csx/Nkx2.5 physically associates with the zinc finger protein GATA4 and cooperatively activates atrial natriuretic factor gene expression. Mol Cell Biol. 1998;18:3120–3129. doi: 10.1128/mcb.18.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sepulveda JL, et al. GATA-4 and Nkx-2.5 coactivate Nkx-2 DNA binding targets: role for regulating early cardiac gene expression. Mol Cell Biol. 1998;18:3405–3415. doi: 10.1128/mcb.18.6.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shiojima I, et al. Context-dependent transcriptional cooperation mediated by cardiac transcription factors Csx/Nkx-2.5 and GATA-4. J Biol Chem. 1999;274:8231–8239. doi: 10.1074/jbc.274.12.8231. [DOI] [PubMed] [Google Scholar]
- 38.Mead PE, Brivanlou IH, Kelley CM, Zon LI. BMP-4-responsive regulation of dorsal-ventral patterning by the homeobox protein Mix.1. Nature. 1996;382:357–360. doi: 10.1038/382357a0. [DOI] [PubMed] [Google Scholar]
- 39.Onichtchouk D, Glinka A, Niehrs C. Requirement for Xvent-1 and Xvent-2 gene function in dorsoventral patterning of Xenopus mesoderm. Development. 1998;125:1447–1456. doi: 10.1242/dev.125.8.1447. [DOI] [PubMed] [Google Scholar]
- 40.Chen CY, et al. Activation of the cardiac alpha-actin promoter depends upon serum response factor, Tinman homologue, Nkx-2.5, and intact serum response elements. Dev Genet. 1996;19:119–130. doi: 10.1002/(SICI)1520-6408(1996)19:2<119::AID-DVG3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]