Abstract
The iron (Fe) proteins of molybdenum (Mo)-, vanadium (V)- and iron (Fe)-only nitrogenases are encoded by nifH, vnfH and anfH, respectively. While the nifH-encoded Fe protein has been extensively studied over the past years, information regarding the properties of the vnfH- and anfH-encoded Fe proteins has remained scarce. Here we present a combined biochemical, electron paramagnetic resonance (EPR) and x-ray absorption spectroscopy (XAS) analysis of the [Fe4S4] clusters of NifH, VnfH and AnfH of Azotobacter vinelandii. Our data show that all three Fe proteins contain [Fe4S4] clusters of very similar spectroscopic and geometric structural properties, although NifH differs more from VnfH and AnfH with regard to the electronic structure. These observations have an interesting impact on the theory of the plausible sequence of evolution of nitrogenase Fe proteins. More importantly, the results presented herein provide a platform for future investigations of the differential activities of the three Fe proteins in nitrogenase biosynthesis and catalysis.
Keywords: Nitrogenase, Fe protein, NifH, VnfH, AnfH
1. Introduction
Nitrogenase is a complex metalloenzyme that catalyzes a key step in the global nitrogen cycle—the reduction of inert dinitrogen (N2) to bioavailable ammonia (NH3).1 Three homologous nitrogenases [i.e., the molybdenum (Mo), vanadium (V) and iron (Fe)-only nitrogenases] have been identified, which resemble one another in primary sequence and cluster type.2 All of them are two-component systems comprising a specific reductase (categorically termed Fe protein) and a catalytic component (designated MoFe, VFe and FeFe protein, respectively). The Fe protein of the Mo-nitrogenase is the best studied among the three homologous Fe proteins. It plays a key role in catalysis, serving as an obligate electron donor to the catalytic MoFe protein in an ATP hydrolysis-dependent manner.1,2 In addition, it is essential for the maturation of MoFe protein, functioning as an ATP-dependent reductase as well in this process.3
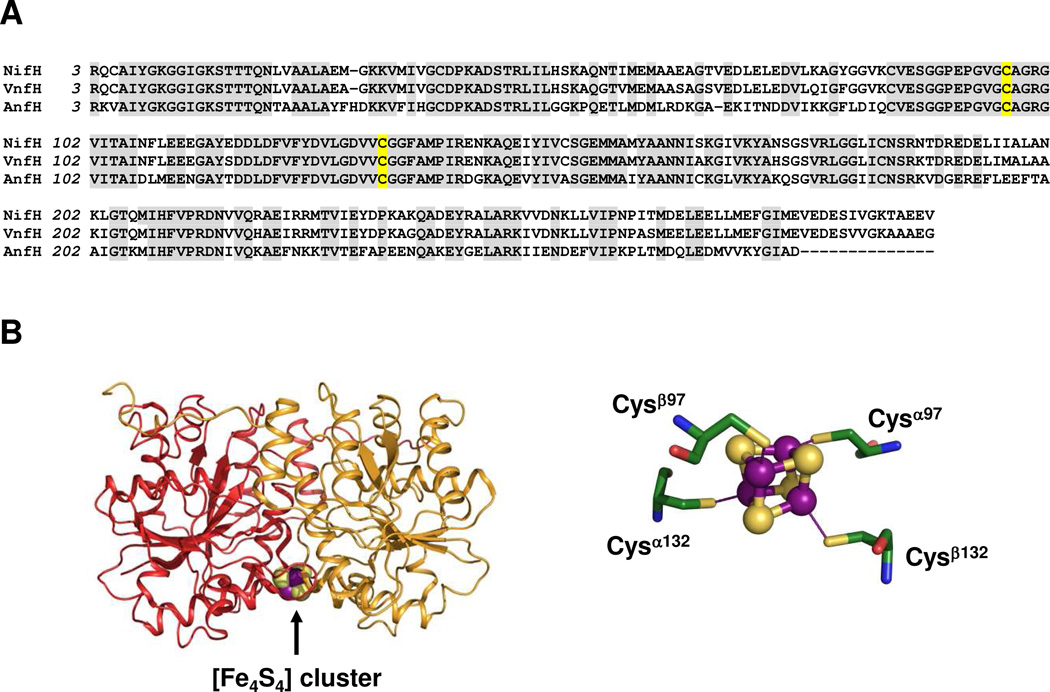
The Fe proteins of Mo-, V- and Fe-only nitrogenases are encoded by nifH, vnfH and anfH, respectively. The three Fe proteins (henceforth referred to as NifH, VnfH and AnfH) share a considerable degree of sequence homology with one another (Figure 1A), although NifH resembles VnfH (91% homology) much more closely than it does AnfH (61% homology) in primary sequence. X-ray crystallographic analysis reveals that NifH is a homodimer in which a surface-exposed [Fe4S4] cluster is ligated symmetrically through two cysteines (i.e., Cys97 and Cys132) from each subunit (Figure 1B).4 Additionally, NifH contains a nucleotide-binding site within each subunit and, in the case of MgADP, the two molecules are bound at locations near the dimer interface in a parallel orientation.5 The [Fe4S4] cluster of NifH can assume three oxidation states (i.e., 0, +1 and +2), although the [Fe4S4]2+/1+ couple is generally believed to be utilized under physiological conditions.6,7 Compared to what is known about the cluster in NifH, spectroscopic information on the clusters in VnfH and AnfH is rather limited. Nevertheless, sequence alignment reveals that the two Cys ligands in NifH are also conserved in VnfH and AnfH (Figure 1A), suggesting that VnfH and AnfH may contain [Fe4S4] clusters similar to that in NifH. Indeed, like NifH, VnfH contains a [Fe4S4]1+ cluster that exhibits analogous S = 1/2 and S = 3/2 ground-state electron paramagnetic resonance (EPR) signals in the reduced state.8 However, VnfH does differ from NifH with regard to the medium effects on the two spin mixtures in frozen solution, which is indicative of a subtle difference between the cluster species in these proteins.8 In the case of AnfH, so far, there has been no report on the properties of its associated cluster. Here, we present a combined biochemical, EPR and x-ray absorption spectroscopy (XAS) analysis of the [Fe4S4] clusters of NifH, VnfH and AnfH of Azotobacter vinelandii. Our results indicate that all three Fe proteins contain [Fe4S4] clusters of very similar spectroscopic and geometric structural properties; however, NifH differs more from VnfH and AnfH in terms of the electronic structure. A plausible sequence of evolution of nitrogenase Fe proteins can be proposed on the basis of these findings. Moreover, the results presented herein provide a framework for future investigations of the differential activities of the three Fe proteins in nitrogenase assembly and catalysis.
Figure 1.
Sequence alignment of NifH, VnfH and AnfH (A) and crystal structure of NifH and its associated [Fe4S4] cluster (B). The residues conserved among all three Fe proteins are highlighted in gray, with the Cys ligands highlighted in yellow (A). The atoms of the [Fe4S4] cluster are colored as follows: Fe, purple; S, lime (B). The presentation in B was generated in PYMOL using PDB coordinates 2NIP.4
2. Results and Discussion
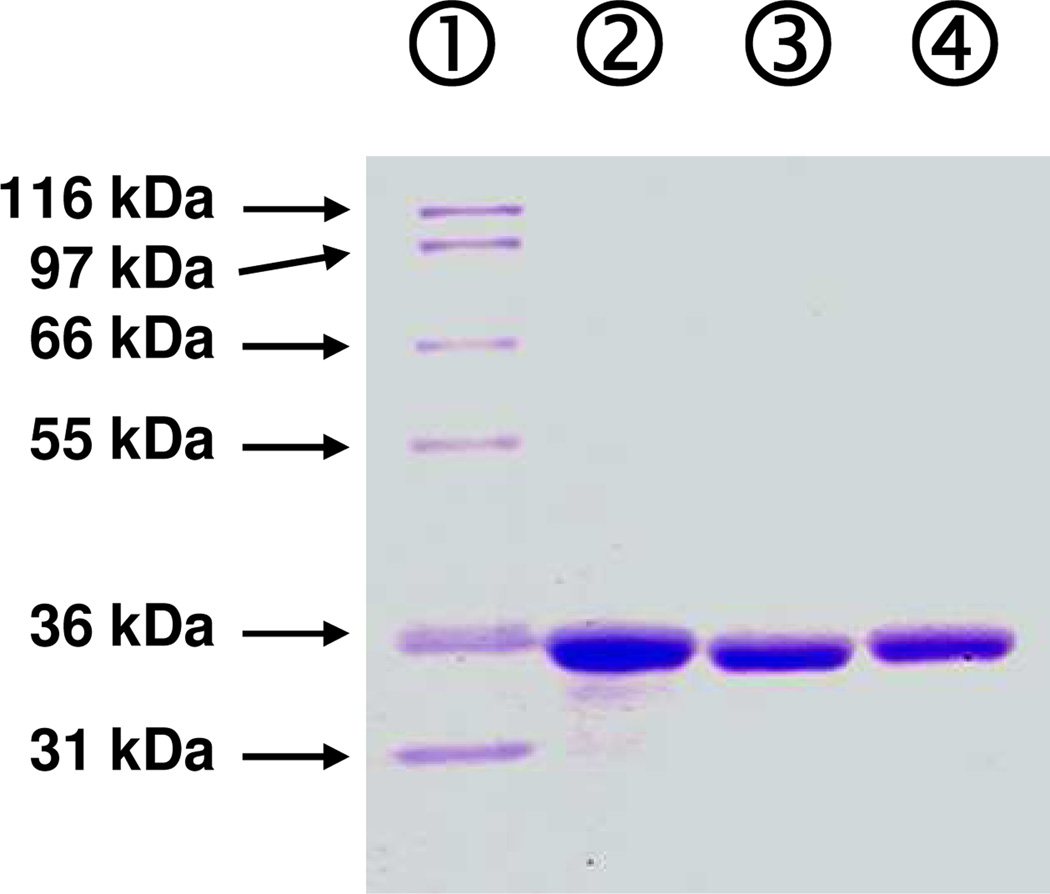
The identities of NifH, VnfH and AnfH are confirmed by N-terminus amino acid sequencing (data not shown). Each of them contains a subunit of ~30 kDa (Figure 2) and, based on gel filtration analysis, all three Fe proteins are homodimers of ~60 kDa (data not shown). Metal analysis reveals the presence of approximately 4 mol Fe atoms per mol protein in all three protein species (Table 1). Furthermore, in combination with the MoFe protein, both VnfH and AnfH exhibit activities that are comparable to those of NifH in the standard nitrogenase reactions (Table 1).
Figure 2.
SDS PAGE of NifH, VnfH and AnfH. Lane 1, Mark 12, 5 µg; lane 2, NifH, 3 µg; lane 3, VnfH, 3 µg; lane 4, AnfH, 3 µg.
Table 1.
Metal content (A) and substrate-reducing activities (B) of NifH, VnfH and AnfH.
| Fe content | Specific Activities | ||||
|---|---|---|---|---|---|
| Fe protein | mol Fe/ mol protein |
C2H4-formation under C2H2/Ar |
H2-formation under Ar |
H2-formation under N2 |
NH3-formation under N2 |
| NifH | 4.0 ± 0.1 | 1780 ± 27 | 2170 ± 28 | 291 ± 39 | 750 ± 66 |
| VnfH | 4.1 ± 0.2 | 1848 ± 34 | 2321 ± 30 | 314 ± 33 | 737 ± 33 |
| AnfH | 4.0 ± 0.1 | 1916 ± 37 | 2640 ± 40 | 380 ± 30 | 813 ± 19 |
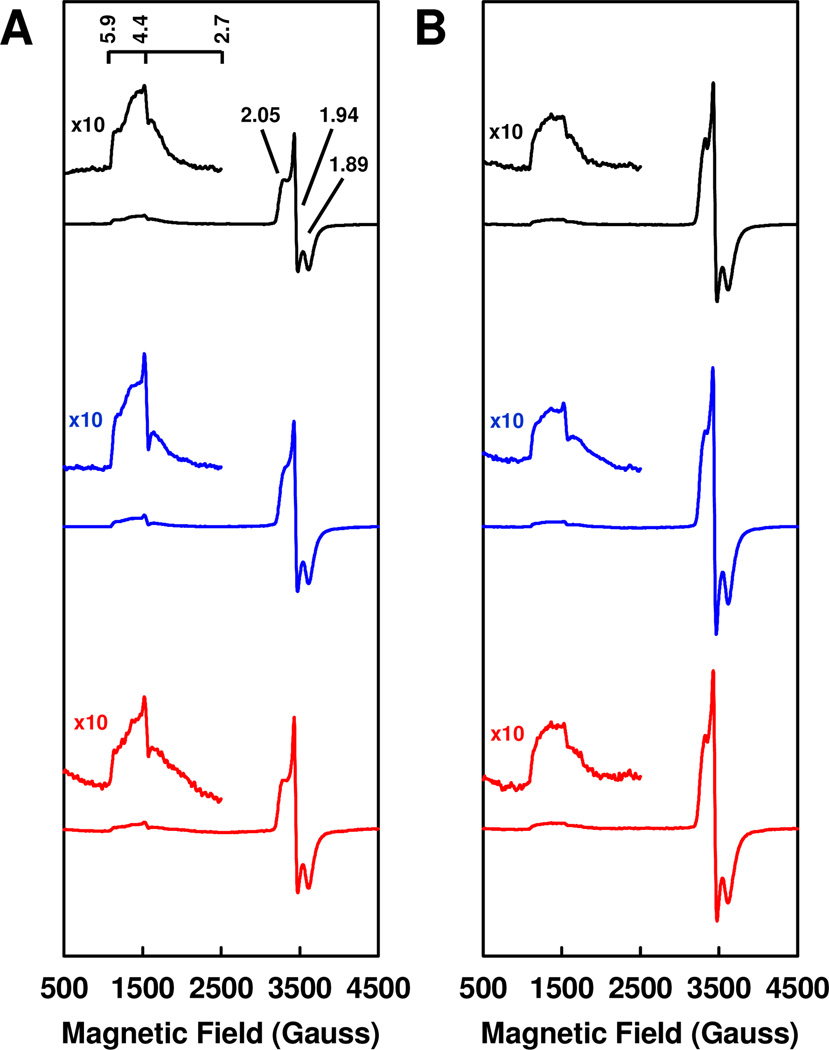
The similarity among NifH, VnfH and AnfH is further demonstrated by EPR analysis. The dithionite-reduced NifH, VnfH and AnfH exhibit a highly similar mixture of S = 1/2 and S = 3/2 signals, which originate from the [Fe4S4]+ clusters in these proteins (Figure 3). In the absence of glycerol, the ratios between the S = 1/2 and S = 3/2 signals of NifH, VnfH and AnfH are very similar to one another, with those of VnfH and AnfH slightly higher than that of NifH (Figure 3A). However, the intensities of the two signals in NifH are consistently smaller than those of their counterparts in VnfH and AnfH, which may reflect a slightly different oxidation state of the [Fe4S4] cluster in the former protein; additionally, there are small differences in the line-shapes of the S = 3/2 signals, particularly between the signal of NifH and those of the other two Fe proteins (Figure 3A). In the presence of 50% glycerol, the intensities of the S = 1/2 signals of NifH, VnfH and AnfH increase by 1.6-, 1.5- and 1.5-fold, respectively; whereas the intensities of the S = 3/2 signals of all three proteins decrease by 30% (Figure 3B). As a result, the ratios between the S = 1/2 and S = 3/2 signals of NifH, VnfH and AnfH become nearly indistinguishable from one another (Figure 3A). Interestingly, a similar, albeit more pronounced, spin-redistribution effect was previously documented for NifH and VnfH in the presence of 50% ethylene glycol.8
Figure 3.
EPR spectra of dithionite-reduced NifH (black), VnfH (blue) and AnfH (red) in the absence (A) and presence (B) of 50% glycerol. Spectra were measured at 10 K. The g values are indicated. In the absence of glycerol, the intensities of the S = 1/2 and S = 3/2 signals of NifH are 79% and 91%, respectively, of those of VnfH and 80% and 90%, respectively, of those of AnfH. Nevertheless, the ratios between the S = 1/2 and S = 3/2 signals of all Fe proteins are very similar to one another, with those of VnfH and AnfH being 116% and 112% of that of NifH, respectively. In the presence of 50% glycerol, the intensities of the S = 1/2 signals of NifH, VnfH and AnfH increase by 1.6-, 1.5- and 1.5-fold, respectively; whereas the intensities of the S = 3/2 signals of all three proteins decrease by 30%. Consequently, the ratios between the S = 1/2 and S = 3/2 signals of all three Fe proteins become nearly indistinguishable, with those of VnfH and AnfH being 103% and 94% of that of NifH.
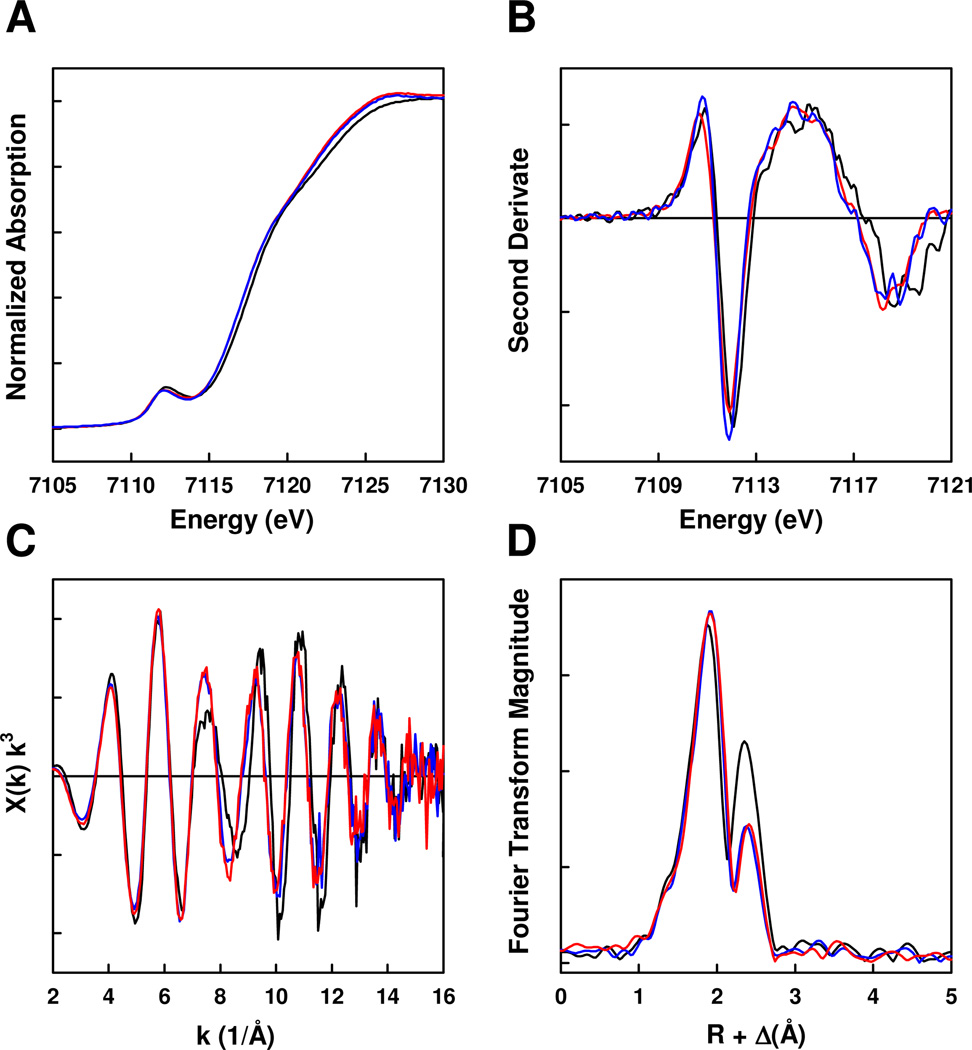
Consistent with the outcome of the EPR analysis, Fe K-edge XAS data reveal similarities and differences among NifH, VnfH and AnfH. All three Fe proteins (prepared in 50% glycerol) display pre-edge features of nearly identical intensity at ~7112.2 eV in their dithionite-reduced forms (Figure 4A), suggesting that the clusters in these proteins are similar in the coordination geometry of Fe atoms.9 However, in the case of NifH, this feature is shifted by 0.2 eV toward higher energy (Figure 4A). Likewise, the inflection point of the rising edge of NifH is 0.2 eV higher in energy than those of the rising edges of VnfH and AnfH (Figure 4A). The same difference among the three Fe proteins is more clearly displayed in the second derivatives of their Fe K-edges, with the derivative of NifH spectrum showing a somewhat different shape and a higher energy than those of VnfH and AnfH (Figure 4B). The shift of both the pre-edge feature and the rising edge position to higher energy suggests that the Fe atoms of the [Fe4S4] cluster in NifH are at a greater average Zeff than those of the [Fe4S4] clusters in VnfH and AnfH.10 Although the difference in Zeff is a subtle one, it points to a more ferric nature of the Fe atoms in the cluster of NifH than those in the clusters of VnfH and AnfH.
Figure 4.
Fe K-edge XAS (A) and second derivatives (B) of NifH (black), VnfH (blue) and AnfH (red); Fe K-edge EXAFS (C) and Fourier transforms (D) of data for NifH (black), VnfH (blue) and AnfH (red) in their dithionite-reduced states.
The k3 weighted Fe K-edge extended x-ray fine structure (EXAFS) spectra (Figure 4C) are characteristic of those originating from both biological [FeS] clusters (e.g., the P-cluster precursor) and synthetic compounds (e.g., (Et4N)2[Fe4S4(SPh)4]).11,12 The EXAFS signals of all three Fe proteins are of nearly the same amplitude (Figure 4C), suggesting the presence of intact, well-ordered clusters in these proteins.13 On the other hand, NifH again exhibits features that are noticeably different than those of VnfH and AnfH in the EXAFS spectra. While VnfH and AnfH show almost identical frequency and intensity throughout the EXAFS region of k = 2–16 Å−1, NifH diverges markedly from them in the beat region near k = 8 Å−1, which arises from the intersection of the Fe-S and Fe-Fe waves (Figure 4C). The third maximum in the NifH EXAFS spectrum (~7.5 Å−1) is broader in shape, yet lower in height than the corresponding peaks in the VnfH and AnfH EXAFS spectra (Figure 4C). Moreover, at higher k (i.e., k > 8 Å−1), the EXAFS waveform of NifH maintains roughly the same frequency as, but shifts slightly out of phase from, those of VnfH and AnfH until k = ~14 Å−1 (Figure 4C).
As is expected from the EXAFS data, the intensity of the first peak of the Fourier transform (FT), which results from the S shell backscattering, is roughly the same for all three Fe proteins; however, in the case of NifH, the maximum of this peak, as well as its left shoulder, is shifted to a shorter distance (Figure 4D). In contrast, the intensity of the second peak of the Fourier transform, which originates mainly from the Fe backscattering, varies substantially between NifH and the other two Fe proteins; additionally, the position of the maximum of this peak is very similar, but not identical, for all three proteins (Figure 4D). The differences in the second FT peak would reflect a significant degree the difference in the EXAFS data beyond k = 8 Å−1 (Figure 4C), which suggest a variation in the Fe coordination shell of NifH from those of the other two Fe proteins.
Despite the differences in the EXAFS data of the three Fe proteins, a common, overall structural model can be generated for the clusters of all three proteins. A total of four S ligands at a distance of ~2.30 Å—which provide the vast majority of the intensity of the first peak in the Fourier transform—represent one of the key components of the final fit (Table 2). These S atoms are assigned to the four Cys residues—two from each subunit—of the homodimeric Fe protein (Figure 1B).4 Consistent with the shift of the first FT peak and its left shoulder to shorter distances (Figure 4D), the Fe-S distance in NifH is the shortest among the three Fe proteins (Table 2). Nevertheless, within the resolution limit of XAS technique (i.e., 0.02 Å), the Fe-S backscattering paths in all three Fe proteins are approximately equal in length, which is anticipated for three highly similar [Fe4S4] clusters with equivalent Cys ligands. In addition to the Fe-S path, two distinct Fe-Fe paths—one short path of ~2.50 Å and two longer ones of ~2.70 Å—represent the other key component of the final fit (Table 2). These Fe-Fe paths describe the unique shape and intensity of the second peak in the Fourier transform and are common elements of EXAFS fits to all three Fe proteins. Although the longer path is not present in the crystal structure of NifH, it can be seen in the previous EXAFS fits to both Ti(III) citrate- and dithionite-reduced NifH, as well as those to the P-cluster precursor comprising paired [Fe4S4]-like clusters.14 Crystallographic and Fe K-edge XAS studies of both synthetic [Fe4S4]+ clusters and the [Fe4S4]+ clusters of reduced bacterial ferredoxins have shown that an Fe-Fe distance of approximately 2.70 Å is a conserved feature of this type of [FeS] cubanes;15–19 whereas an Fe-Fe distance of approximately 2.50 Å is typical of the [Fe4S4] cluster of reduced NifH, the P-cluster precursor and the fused [Fe8S8] clusters.7,12–14,20,21 The ratios between these two distances, however, vary substantially depending on the structure and electronic state of the cluster.
Table 2.
Two-Fe path fits for the Fe K-edge EXAFS data of NifH, VnfH and AnfH over the k-range of 2–16 Å−1. Coordination number (N), interatomic distance (R, Å), mean-square thermal and static disorder in distance (σ2, Å2), and EXAFS threshold energy adjustment from 7030 eV (ΔE0, eV) were varied in the fits. Estimated errors are ± 0.02 Å in R, ± 0.0001 Å2 in σ2, and ± 20% in N. The goodness of fit, F, is defined as F = [Σk6(χexper. − χcalc.)2/Σk6(χexper.)2]0.5.
| NifH | VnfH | AnfH | |||||||
|---|---|---|---|---|---|---|---|---|---|
| N | R (Å) | σ2 (Å2) | N | R (Å) | σ2 (Å2) | N | R (Å) | σ2 (Å2) | |
| Scatterer | |||||||||
| Fe-S | 4 | 2.30 | 0.0050 | 4 | 2.32 | 0.0052 | 4 | 2.31 | 0.0054 |
| Fe-Fe | 1 | 2.50 | 0.0128 | 1 | 2.51 | 0.0065 | 1 | 2.52 | 0.0038 |
| Fe-Fe | 2 | 2.72 | 0.0041 | 2 | 2.72 | 0.0061 | 2 | 2.71 | 0.0056 |
| ΔE0 (eV) | −12.0 | −12.8 | −13.2 | ||||||
| Weighted F | 0.190 | 0.165 | 0.183 | ||||||
Interestingly, while there is an excellent agreement on the distance (R) and mean static and thermal disorder (σ2) of the longer path among all three Fe proteins, the short Fe-Fe path in NifH is the shortest in distance (2.50 Å) with a mean displacement factor (0.0128) approximately twice and three times as large as those of the corresponding paths in VnfH and AnfH, respectively (Table 2). Such a combination of a shorter distance with a higher static disorder is largely responsible for the unique EXAFS features of NifH that are distinct from those of the other two Fe proteins. In support of this argument, the EXAFS data of NifH can be alternatively fit by including three (instead of two) Fe-Fe paths: 2.46 Å, 2.62 Å and 2.74 Å with coordination numbers of 0.5, 1 and 1.5, respectively (Table 3). Both of the two shorter Fe-Fe distances have been reported earlier: a Fe-Fe path of 2.62 Å was observed in the crystallographic models of bacterial ferredoxin, whereas a even shorter Fe-Fe path of 2.46 Å was seen in the EXAFS fits to the oxidized P-cluster of MoFe protein.22,23 In contrast, no reasonable fits that include three Fe-Fe paths can be generated for either VnfH or AnfH consistently, resulting in a very high σ2 value on the 2.68 Å path for VnfH (σ 2=0.0540) and the 2.72 Å path for AnfH (σ 2=0.0456) (Table 3), thereby effectively reducing the contribution of this path to a very minor degree. It should be noted that, although the inclusion of an extra Fe-Fe path improves the fit to the NifH EXAFS data, it does not improve the fit appreciably enough to merit its selection over the two-path fit. Nevertheless, the fact that only the NifH data can be fit with such a three Fe-path model once again suggests that the cluster in NifH possesses some features that distinguish it from the clusters in the other two Fe proteins.
Table 3.
Three-Fe path fits for the Fe K-edge EXAFS data of NifH, VnfH and AnfH over the k-range of 2–16 Å−1. Coordination number (N), interatomic distance (R, Å), mean-square thermal and static disorder in distance (σ2, Å2), and EXAFS threshold energy adjustment from 7030 eV (ΔE0, eV) were varied in the fits. Estimated errors are ± 0.02 Å in R, ± 0.0001 Å2 in σ2, and ± 20% in N. The goodness of fit, F, is defined as F = [Σk6(χexper. − χcalc.)2/Σk6(χexper.)2]0.5.
| NifH | VnfH | AnfH | |||||||
|---|---|---|---|---|---|---|---|---|---|
| N | R(Å) | σ2 (Å2) | N | R(Å) | σ2 (Å2) | N | R(Å) | σ2 (Å2) | |
| Scatterer | |||||||||
| Fe-S | 4 | 2.29 | 0.0058 | 4 | 2.32 | 0.0049 | 4 | 2.32 | 0.0049 |
| Fe-Fe | 0.5 | 2.46 | 0.0010 | 0.5 | 2.52 | 0.0050 | 0.5 | 2.53 | 0.0018 |
| Fe-Fe | 1 | 2.62 | 0.0020 | 1 | 2.68 | 0.0540 | 1 | 2.72 | 0.0456 |
| Fe-Fe | 1.5 | 2.74 | 0.0017 | 1. 5 | 2.72 | 0.0051 | 1. 5 | 2.72 | 0.0048 |
| ΔE0 (eV) | −13.3 | −12.2 | −12.3 | ||||||
| Weighted F | 0.188 | 0.165 | 0.183 | ||||||
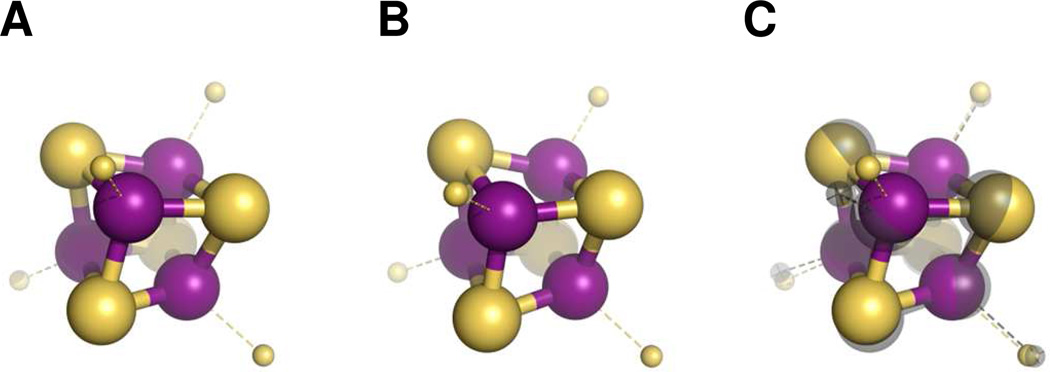
On the basis of the EXAFS analysis above, two structural models can be proposed for the [Fe4S4] clusters in the three Fe proteins (Figure 5). Both models have the four Fe-S paths in common, yet differ in the number of Fe-Fe paths. The first model contains two Fe-Fe paths and, thus, represents the fit to the clusters in all three Fe proteins (Figure 5A). It consists of two stacked [Fe2S2] rhomboids that are offset by 90 degrees, and every Fe atom in this cluster roughly assumes a tetrahedral geometry with one Fe neighbor at ~2.50 Å and two more at ~2.70 Å. The second model contains three Fe-Fe paths and, therefore, is unique to the cluster in NifH (Figure 5B). Compared to the first model, it consists of two unequal [Fe2S2] rhomboids that are bent to a greater degree out-of-plane relative to each other. The Fe-S distances are uniformly decreased in this model, providing further credence to the observed increase in the average Zeff of NifH. Additionally, the Fe sites in this cluster are not equivalent, with each Fe atom neighboring 0.5 Fe at 2.46 Å, 1 Fe at 2.62 Å and 1.5 Fe at 2.74 Å. It needs to be pointed out that, while the second model highlights the subtle difference between the cluster in NifH and those in VnfH and AnfH, it still bears a considerable resemblance to the first model that suitably describes the clusters in all three Fe proteins (Figure 5C).
Figure 5.
Two-Fe path model of the [Fe4S4] clusters in NifH, VnfH and AnfH (A); three-Fe path model of the [Fe4S4] cluster in NifH (B); and overlay of the two-Fe path (colored) and the three-Fe path (gray) models (C). The structural models were adapted from the crystallographic coordinates of NifH4 but modified for distances on the basis of the EXAFS fits. The atoms are colored as follows: Fe, purple; S, lime. The S atoms of the Cys ligands are represented by small lime spheres.
The structural similarity among the clusters of NifH, VnfH and AnfH comes as little surprise, as it has been demonstrated that the structure of Fe protein (and its associated [Fe4S4] cluster) is generally insensitive to the sequence variations among different Fe proteins in the absence of nucleotide.4 Moreover, the residues at or near the cluster-binding sites are highly conserved in all three Fe proteins. Apart from the two Cys ligands, all residues within 10 Å of the cluster of NifH are conserved in NifH and VnfH (Figure 1A).4,24 In particular, Phe135 and Val130, which form an interlocking screen to trap the cluster in place in NifH, are present in the sequences of the other two Fe proteins (Figure 1A).4,24 On the other hand, given the lesser degree of sequence homology between AnfH and VnfH (61%) than that between VnfH and NifH (91%), it is somewhat surprising that the clusters in AnfH and VnfH are very similar to each other, yet more distinct from their counterpart in NifH. From the sole perspective of the Fe protein, it can be theorized that AnfH preceded the other two Fe proteins during the course of evolution and that, upon the appearance of a heterometal-dependent nitrogenase (likely in the order of first the V-nitrogenase and then the Mo-nitrogenase), the conversion from VnfH to NifH required a substantially lesser degree of changes in the amino acid sequence, but rather a fine tuning of the structure of the [Fe4S4] cluster within the protein. Given the specific pairing of Fe proteins with other proteins during the cofactor assembly (i.e., AnfH/VnfEN, VnfH/VnfEN and NifH/NifEN) and substrate-reducing (i.e., AnfH/FeFe protein, VnfH/VFe protein and NifH/MoFe protein) events, it is plausible that the variation of the primary sequence and the concomitant adjustment of the cluster properties are the determining factors for the high selectivity of Fe proteins for their respective “partners” in these processes, an interesting evolutionary relic that may provide useful insights into the mechanism of nitrogenase.
3. Experimental Procedures
Cell Growth and Protein Purification
NifH, VnfH and AnfH were purified from A. vinelandii strains YM13A,25 YM68A25 and YM10A, respectively. Using a previously described method,26 YM10A was constructed by deleting both nifH and vnfH genes in the chromosomal DNA of YM68A. All strains were grown in 180-L batches in a 200-L New Brunswick fermentor (New Brunswick Scientific, Edison, NJ) in Burke’s minimal medium supplemented with 2 mM ammonium acetate, except that Mo in Burke’s medium was replaced by an equal amount of V (for the growth of YM68A) or completely left out (for the growth of YM10A). The growth rate was measured by cell density at 436 nm using a Spectronic 20 Genesys (Thermo scientific, Waltham, MA). Cells were harvested in the late exponential phase by a flow-through centrifugal harvester (Cepa, Lahr, Schwarzwald, Germany). The cell paste was washed with a buffer containing 50 mM Tris-HCl (pH 8.0). A published method was used for the purification of non-tagged NifH, VnfH and AnfH proteins.27
Protein Characterization and Metal Analysis
An equal amount (3 µg) of NifH, VnfH and AnfH was loaded on a 10% Tris-glycine-SDS gel and the gel was run at 60 V for 2.5 h. The native molecular weight of each Fe protein was determined by gel filtration chromatography on a Sephacryl S-300 HR column (GE Healthcare, Piscataway, NJ) as described previously.28 The N-terminal amino acid sequences of the three Fe proteins were analyzed by the Molecular Structure Facility at University of California-Davis. Iron content was determined using a published method.29 The products of activity assays, H2 and C2H4, were analyzed as described elsewhere;30 whereas NH4+ was determined by a high performance liquid chromatography fluorescence method.31
EPR Spectroscopy
All EPR samples were prepared in a Vacuum Atmospheres dry box at an oxygen level of less than 4 ppm. The dithionite-reduced samples contained 25 mM Tris-HCl (pH 8.0), 0.5 M NaCl and 2 mM Na2S2O4. All EPR spectra were recorded in perpendicular mode by a Bruker ESP 300 Ez spectrometer (Bruker, Madison, WI) interfaced with an Oxford Instruments ESR-9002 liquid helium continuous-flow cryostat. Unless noted otherwise, all EPR spectra were recorded at a microwave power of 50 mW, a microwave frequency of 9.62, a gain of 5 × 104, a modulation frequency of 100 kHz, and a modulation amplitude of 5 G. Ten scans were collected for each sample at 10 K. The S = 1/2: S = 3/2 ratios of the three Fe proteins were calculated based on previously published methods.8,32
XAS Data Acquisition
The XAS data of VnfH and AnfH were measured at XAS stations BL9-3 (focused) and BL7-3 (unfocused) at the Stanford Synchrotron Radiation Lightsource (SSRL) under 3 GeV and 80–100 mA storage ring operating conditions. Energy selection was attained using a Si(220) double-crystal monochromator tuned at ~8 keV and, for both beam lines, a pre-monochromator collimating, harmonic rejection mirror was used. Protein samples were loaded into Kapton sheathed Lucite cells and flash frozen and preserved in liquid N2. An Oxford Instruments CF1208 liquid He continuous-flow cryostat held samples at a constant temperature of 10 K during XAS measurement. Fe Kα fluorescence data were recorded to k = 16 Å−1 (for VnfH and AnfH) and to k = 17 Å−1 (for NifH) using a Canberra 30-element Ge detector array. The concentrations of all Fe protein samples were approximately 100 mg/mL and contained 25 mM Tris-HCl buffer (pH 8.0), 50% glycerol, 500 mM NaCl, 200 mM imidazole and 2 mM sodium dithionite. In all cases, internal energy calibration was performed by the concurrent measurement of the absorption of an Fe foil placed between two ionization chambers filled with N2 located after the sample. The first inflection point of the foil XAS edge was assigned to 7111.2 eV. No signs of photoreduction of the metal sites, as observed by shifts in edge energy with time, were observed. A total of 30, 29 and 14 scans were measured for VnfH, AnfH and NifH, respectively.
XAS Data Analysis
Following the inspection of raw data and the removal of specious detector channels and scans, average files of all scans for each sample were generated. The program PYSPLINE was used to normalize the average data files for each sample by selecting control points and fitting a first- or second-order polynomial to the pre-edge region prior to the subtraction of this function from the entire data range.33 All data were normalized to an edge jump of 1.0 at 7130 eV. A four-region spline function of orders 2, 3 and 3 over the post-edge region was employed to yield the EXAFS spectra. Rising edge comparisons were made by first truncating the data to k = 13.5 Å−1, then performing a similar background subtraction, and finally fitting the data with a three/four-region spline. The smoothed second-derivative of the rising edge was produced by using a third-order polynomial with five-point data smoothing. By means of the least-squares fitting program OPT, a component of the EXAFSPAK suite of software, EXAFS data over the k range of 2–16 Å−1 (truncated from 16 Å−1 onward in the case of NifH) were fit using initial ab initio theoretical phase and amplitude functions calculated from FEFF 7.0 on the basis of the 2NIP crystallographic starting model.4,34,35 Atomic coordinates from the crystal model were modulated iteratively as fitting progressed to generate improved, chemically plausible models. During the process of fit optimization, the interatomic distance between the absorbing and backscattering atom (R) and the mean-square thermal and static deviation in R (σ2) were varied for all components. The threshold energy (ΔE0) was allowed to vary for each fit but constrained to the same value for all components. The amplitude reduction factor (S02) was maintained at a value of 1.0 throughout fitting. Coordination numbers (N) were systematically adjusted from crystallographic values to provide the best chemically viable agreement to the EXAFS data and their Fourier transforms. Inclusion or exclusion of various scattering paths was methodically tested to fully explore the average atomic environment at the Fe atoms.36
Acknowledgments
This work was supported by Herman Frasch Foundation Grant 617-HF07 (M.W.R.), NIH Grant GM 67626 (M.W.R.) and NIH Grant RR 001209 (K.O.H.). SSRL operations are funded by the U.S. Department of Energy, Office of basic Energy Sciences, and the SSRL Structural Molecular Biology Program by National Institutes of Health, National Center for Research Resources’ Biomedical Technology Program and the DOE Office of Biological and Environmental Research. This publication was made possible by Grant Number RR 001209 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH).
REFERENCES
- 1.Burgess BK, Lowe DJ. Chem. Rev. 1996;96:2983. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- 2.Eady RR. Chem. Rev. 1996;96:3013. doi: 10.1021/cr950057h. [DOI] [PubMed] [Google Scholar]
- 3.Hu Y, Fay AW, Lee CC, Yoshizawa J, Ribbe MW. Biochemistry. 2008;47:3973. doi: 10.1021/bi7025003. [DOI] [PubMed] [Google Scholar]
- 4.Schlessman JL, Woo D, Joshua-Tor L, Howard JB, Rees DC. J. Mol. Biol. 1998;280:669. doi: 10.1006/jmbi.1998.1898. [DOI] [PubMed] [Google Scholar]
- 5.Jang SB, Seefeldt LC, Peters JW. Biochemistry. 2000;48:14745. doi: 10.1021/bi001705g. [DOI] [PubMed] [Google Scholar]
- 6.Lawson DM, Smith BE. Met. Ions. Biol. Syst. 2002;39:75. [PubMed] [Google Scholar]
- 7.Strop P, Takahara PM, Chiu H, Angove HC, Burgess BK, Rees DC. Biochemistry. 2001;3:651. doi: 10.1021/bi0016467. [DOI] [PubMed] [Google Scholar]
- 8.Onate YA, Finnegan MG, Hales BJ, Johnson MK. Biochim. Biophys. Acta. 1993;1164:113. doi: 10.1016/0167-4838(93)90237-l. [DOI] [PubMed] [Google Scholar]
- 9.Westre TE, Kennepohl P, DeWitt JG, Hedman B, Hodgson KO, Solomon EI. J. Am. Chem. Soc. 1997;119:6297. [Google Scholar]
- 10.Sarangi R, Aboelella N, Fujisawa K, Tolman WB, Hedman B, Hodgson KO, Solomon EI. J. Am. Chem. Soc. 2006;128:8286. doi: 10.1021/ja0615223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee CC, Blank MA, Fay AW, Yoshizawa JM, Hu Y, Hodgson KO, Hedman B, Ribbe MW. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18474. doi: 10.1073/pnas.0909149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corbett MC, Hu Y, Naderi F, Ribbe MW, Hedman B, Hodgson KO. J. Biol. Chem. 2004;279:28276. doi: 10.1074/jbc.M403156200. [DOI] [PubMed] [Google Scholar]
- 13.Lindahl PA, Teo B-K, Orme-Johnson WH. Inorg. Chem. 1987;26:3912. [Google Scholar]
- 14.Musgrave KB, Angove HC, Burgess BK, Hedman B, Hodgson KO. J. Am. Chem. Soc. 1998;120:5325. [Google Scholar]
- 15.Rao PV, Holm RH. Chem. Rev. 2004;104:527. doi: 10.1021/cr020615+. [DOI] [PubMed] [Google Scholar]
- 16.Segal BM, Hoveyda HR, Holm RH. Inorg. Chem. 1998;37:3440. [Google Scholar]
- 17.Carney MJ, Papaefthymiou GC, Whitener MA, Spartalian K, Frankel RB, Holm RH. Inorg. Chem. 1988;27:346. doi: 10.1021/ja00226a025. [DOI] [PubMed] [Google Scholar]
- 18.Dauter Z, Wilson KS, Sieker LC, Meyer J, Moulis JM. Biochemistry. 1997;36:16065. doi: 10.1021/bi972155y. [DOI] [PubMed] [Google Scholar]
- 19.Stephens PJ, Morgan TV, Devlin F, Penner-Hahn JE, Hodgson KO, Scott RA, Stout CD, Burgess BK. Proc. Natl. Acad. Sci. U. S. A. 1985;82:5661. doi: 10.1073/pnas.82.17.5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohki Y, Sunada Y, Honda M, Katada M, Tatsumi K. J. Am. Chem. Soc. 2003;125:4052. doi: 10.1021/ja029383m. [DOI] [PubMed] [Google Scholar]
- 21.Zhou HC, Holm RH. Inorg. Chem. 2003;42:11. doi: 10.1021/ic020464t. [DOI] [PubMed] [Google Scholar]
- 22.Saridakis E, Giastas P, Efthymiou G, Thoma V, Moulis JM, Kyritsis P, Mavridis IM. J. Biol. Inorg. Chem. 2009;14:783. doi: 10.1007/s00775-009-0492-x. [DOI] [PubMed] [Google Scholar]
- 23.Peters JW, Stowell MH, Soltis SM, Finnegan MG, Johnson MK, Rees DC. Biochemistry. 1997;36:1181. doi: 10.1021/bi9626665. [DOI] [PubMed] [Google Scholar]
- 24.Setubal JC, et al. J. Bacteriol. 2009;191:4534. doi: 10.1128/JB.00504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee CC, Hu Y, Ribbe MW. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9209. doi: 10.1073/pnas.0904408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christiansen J, Goodwin PJ, Lanzilotta WN, Seefeldt LC, Dean DR. Biochemistry. 1998;36:12611. doi: 10.1021/bi981165b. [DOI] [PubMed] [Google Scholar]
- 27.Burgess BK, Jacobs DB, Stiefel EI. Biochim. Biophys. Acta. 1980;614:196. doi: 10.1016/0005-2744(80)90180-1. [DOI] [PubMed] [Google Scholar]
- 28.Eady RR, Richardson TH, Miller RW, Hawkins M, Lowe DJ. Biochem. J. 1988;256:189. doi: 10.1042/bj2560189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van de Bogart M, Beinert H. Anal. Biochem. 1967;20:325. doi: 10.1016/0003-2697(67)90038-3. [DOI] [PubMed] [Google Scholar]
- 30.Gavini N, Burgess BK. J. Biol. Chem. 1992;267:21179. [PubMed] [Google Scholar]
- 31.Corbin JL. Microbiol. 1984;47:1027. doi: 10.1128/aem.47.5.1027-1030.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Y, Fay AW, Dos Santos PC, Naderi F, Ribbe MW. J. Biol. Chem. 2004;279:54963. doi: 10.1074/jbc.M408983200. [DOI] [PubMed] [Google Scholar]
- 33.Tenderholt A. PYSPLINE. Stanford, CA: Stanford Synchrotron Radiation Laboratory; 2006. [Google Scholar]
- 34.George GN. EXAFSPAK. Stanford, CA: Stanford Synchrotron Radiation Laboratory; 1990. [Google Scholar]
- 35.Rehr JJ, Albers RC. Rev. Mod. Phys. 2000;72:621. [Google Scholar]
- 36.Pickering IJ, George GN, Dameron CT, Kurz B, Winge DR, Dance IG. J. Am. Chem. Soc. 1993;115:9498. [Google Scholar]