Abstract
Fragile X syndrome (FXS), caused by loss of fragile X mental retardation 1 (FMR1) gene function, is the most common heritable cause of intellectual disability and autism spectrum disorders. The FMR1 product (FMRP) is an RNA-binding protein best established to function in activity-dependent modulation of synaptic connections. In the Drosophila FXS disease model, loss of functionally-conserved dFMRP causes synaptic overgrowth and overelaboration in pigment dispersing factor (PDF) peptidergic neurons in the adult brain. Here, we identify a very different component of PDF neuron misregulation in dfmr1 mutants: the aberrant retention of normally developmentally-transient PDF tritocerebral (PDF-TRI) neurons. In wild-type animals, PDF-TRI neurons in the central brain undergo programmed cell death and complete, processive clearance within days of eclosion. In the absence of dFMRP, a defective apoptotic program leads to constitutive maintenance of these peptidergic neurons. We tested whether this apoptotic defect is circuit-specific by examining crustacean cardioactive peptide (CCAP) and bursicon circuits, which are similarly developmentally-transient and normally eliminated immediately post-eclosion. In dfmr1 null mutants, CCAP/bursicon neurons also exhibit significantly delayed clearance dynamics, but are subsequently eliminated from the nervous system, in contrast to the fully persistent PDF-TRI neurons. Thus, the requirement of dFMRP for the retention of transitory peptidergic neurons shows evident circuit specificity. The novel defect of impaired apoptosis and aberrant neuron persistence in the Drosophila FXS model suggests an entirely new level of “pruning” dysfunction may contribute to the FXS disease state.
Keywords: pigment dispersing factor, crustacean cardioactive peptide, bursicon, synapse, calcium, apoptosis, Fragile X syndrome, Drosophila
Introduction
Fragile X syndrome (FXS) is an X-linked neurodevelopmental disorder caused by CGG-trinucleotide repeat expansion in the 5' UTR of the fragile X mental retardation 1 (FMR1) gene (reviewed in Bassell and Warren, 2008; Garber et al., 2008; Gatto and Broadie, 2009b; Penagarikano et al., 2007; Reiss and Hall, 2007). The FMR1 repeat expansion causes hypermethylation and transcriptional silencing, preventing expression of the FMR1 gene product (FMRP) (Sutcliffe et al., 1992; Verkerk et al., 1991), an RNA-binding, polysome-associated protein that functions in mRNA translational regulation (Gross et al., 2010; Laggerbauer et al., 2001; Li et al., 2001; Lu et al., 2004; Qin et al., 2005; Sung et al., 2003; Tessier and Broadie, 2008; Zalfa et al., 2003), stability (Zalfa et al., 2005; Zhang et al., 2007) and trafficking (Dictenberg et al., 2008; Estes et al., 2008; Kao et al., 2010). Full FMR1 mutation (>200 CGG repeats) prevalence has recently been estimated at 1:2,500 (Hagerman, 2008), making FXS the leading heritable monogenic cause of intellectual disability and autism spectrum disorders.
FXS patients harbor the hallmark cytological presentation of morphologically aberrant synaptic connections, i.e., supernumerary, elongated, immature dendritic spines (Hinton et al., 1991; Irwin et al., 2001; Rudelli et al., 1985; Wisniewski et al., 1991). Murine and Drosophila FXS models allow determination of mechanisms that underlie the disease state (Bakker et al., 1994; Mientjes et al., 2006; Zhang et al., 2001). Both animal models recapitulate synaptic connectivity defects, with aberrant synaptogenesis, impaired synaptic refinement, synaptic overgrowth and abnormal activity-dependent synaptic modulation, which are coupled to behavioral changes including defective learning and memory, social interaction abilities and circadian rhythms (reviewed in Bassell and Warren, 2008; Bhogal and Jongens, 2010; Gatto and Broadie, 2009b; Mercaldo et al., 2009; Pfeiffer and Huber, 2009; Tessier and Broadie, 2009). Importantly, in addition to these synaptic functions, FMRP in both murine and Drosophila models has recently been implicated in controlling cellular proliferation and fate specification, both in germline (Epstein et al., 2009; Yang et al., 2007) and neural stem cells (Callan et al., 2010; Castren et al., 2005; Luo et al., 2010; Tervonen et al., 2009). These functions may correlate with hyperplasia and associated increased organ size in FXS patients, both in the testes (Lachiewicz and Dawson, 1994; Nielsen et al., 1982; Rocchi et al., 1987) and certain brain regions including the hippocampus, amygdala, caudate nucleus and thalamus (for review see Hessl et al., 2004). This suggests that FMRP may regulate neuron number in circuits underlying behavioral manifestations of the disease condition.
In light of the recently reported role of FMRP in neurogenesis, we hypothesized that FXS neural circuit defects may be influenced by aberrant connectivity due to superfluous, supernumerary neuronal integration. In testing this prediction, we observed that Drosophila pigment dispersing factor (PDF) peptidergic neurons showed just such a defect; in the absence of dFMRP, there is an overabundance of these neurons in the adult brain. The PDF neuropeptide is expressed in a small subset of neurons, including clock neurons driving locomotor rhythmicity (Blanchardon et al., 2001; Grima et al., 2004; Helfrich-Forster, 1995, 2005; Lin et al., 2004; Renn et al., 1999; Shafer and Taghert, 2009; Stoleru et al., 2004). Luciferase reporter studies reveal that PDF facilitates circadian oscillations in individual neurons via adjustments of amplitude, period and phase, while mediating synchrony within neuronal subgroups (Sheeba et al., 2008; Yoshii et al., 2009). Though its receptor has been identified (Hyun et al., 2005; Lear et al., 2005; Mertens et al., 2005), only recent signaling studies employing a genetically encoded cAMP fluorescence resonance energy transfer (FRET) sensor have demonstrated widespread receptivity to PDF throughout the circadian circuit, conclusively assigning PDF a central network modulatory function (Shafer et al., 2008).
Our further developmental studies revealed that the excess PDF neurons in the dfmr1 null were restricted to the tritocerebral (TRI) brain region. PDF-TRI neurons are characterized as a developmentally-transient population in wild-type animals (Helfrich-Forster, 1997). Consistently, we show that PDF-TRI neuron retention in the dfmr1 null brain is due to defective initiation of programmed cell death and not a result of over-proliferation. We further show that aberrantly persistent PDF-TRI neurons contribute inappropriate synaptic connection in the mature brain and remain functionally competent. Other peptidergic neuron classes are similarly developmentally-transient (Choi et al., 2006; Draizen et al., 1999), apparently performing temporally restricted roles before being eliminated. For example, crustacean cardioactive peptide (CCAP) neurons have a transient role initiating the ecdysis motor program and are then programmed to die immediately following eclosion (Draizen et al., 1999; Park et al., 2003). Likewise, peptidergic bursicon neurons have a transient role in epidermal maturation and wing inflation and then undergo immediate post-eclosion apoptosis (Dewey et al., 2004; Honegger et al., 2008; Loveall and Deitcher, 2010; Luan et al., 2006; Peabody et al., 2008). We show here that these other neuromodulatory circuits also display a significant delay in their developmental elimination in the dfmr1 null, although the defect is mild compared to the long-term persistence of PDF-TRI neurons. Thus, the dfmr1 defect is widespread but manifests circuit specificity. Taken together, these results demonstrate a novel role for dFMRP in regulating neuronal survival and suggest that FXS disease symptoms may arise from inappropriate synaptic incorporation of abnormally persistent neuron populations causing excessive neuromodulatory signaling.
Materials and Methods
Drosophila Genetics
Drosophila stocks were maintained on standard cornmeal/agar/molasses medium supplemented with yeast at 25°C in humidity-controlled incubators with a 12 hour light:dark cycle. The w1118 line served as the genetic background control for the dfmr1 null allele (dfmr150M) typically used throughout, unless otherwise specified (Gatto and Broadie, 2008, 2009a; Tessier and Broadie, 2008, 2011; Zhang et al., 2001). Two other independently isolated dfmr1 null alleles were also examined, dfmr12 (Dockendorff et al., 2002) and dfmr1B55 (Inoue et al., 2002). Additionally, a dfmr150M line carrying a genomic rescue construct (Dockendorff et al., 2002) was generated (p(dfmr1)w+/+ ; dfmr150M/TM6). Drosophila Stock Center (Bloomington, IN) lines carrying Pdf-Gal4 and UAS-mCD8::GFP were introduced into the dfmr150M null background using standard genetic techniques, as previously described (Gatto and Broadie, 2009a). The UAS-synaptotagmin-GFP line was used to label synaptic connections (Gatto and Broadie, 2009a; Zhang et al., 2002). The UAS-GCaMP1.3 line (Nakai et al., 2001) was used as a Ca2+ reporter (Tessier and Broadie, 2011). Both lines were crossed together with the Pdf-Gal4 driver into genetic control and dfmr150M null backgrounds.
Immunocytochemistry
Antibodies employed included: anti-GFP (1:2000; ab290, rabbit, Abcam Inc., Cambridge, MA), anti-PDF (1:5; C7, mouse, Developmental Studies Hybridoma Bank (DSHB), University of Iowa), anti-period (PER, 1:50; d-300, rabbit, Santa Cruz Biotechnology, Inc., Santa Cruz, CA), anti-CCAP (1:5000; rabbit (Woodruff et al., 2008)), anti-bursicon α subunit (burs, 1:5000; rabbit (Luan et al., 2006)), and anti-bursicon β subunit, partner-of-bursicon (pburs, 1:500; mouse (Luo et al., 2005)). For GFP and PDF labeling, brains from staged animals were dissected in standard saline (128 mM NaCl, 2 mM KCl, 4 mM MgCl2, 35.5 mM sucrose, 5mM HEPES, 2 mM CaCl2, pH 7.2) and fixed for 40 minutes with 4% paraformaldehyde/4% sucrose in phosphate buffered saline (PBS), pH 7.4 at room temperature. Preparations were rinsed with PBS, then blocked and permeablized with 0.2% triton X-100 in PBS (PBST) supplemented with 1% bovine serum albumin (BSA) and 0.5% normal goat serum (NGS) for 1 hour at room temperature. Primary and secondary antibodies were diluted in PBST with 0.2% BSA and 0.1% NGS and incubated overnight at 4°C and 2 hours at room temperature, respectively. Alexa-Fluor secondaries (1:250; Invitrogen-Molecular Probes) were used. For PER labeling, dissection was carried out in Ca2+-free PEM (0.1M PIPES, 5mM EGTA, 2mM MgCl2, pH 6.9) and fixation with 4% paraformaldehyde/4% sucrose in PEM. These dissections were performed at zeitgeber time 0–2 hours (ZT 0–2). For CCAP and bursicon labeling, the brain alone or whole nervous system including the brain and thoracic/abdominal ganglia was dissected from staged animals and fixed for 2 hours at 4°C with Bouin's fixative. Tissue was then transferred into isopropanol until processing completion. Preparations were rinsed with PBS, incubated with 0.02% collagenase for 10 minutes, and rinsed again. Permeablization was done with 0.3% triton X-100 in PBS followed by preincubation with 10% NGS in 0.3% triton X-100 in PBS for 2 hours at room temperature. Primary antibodies were diluted in PBS with 0.3% triton X-100 and incubated for 72 hours at 4°C. Preparations were sequentially washed with PBS plus 0.3% and 0.1% triton X-100. Alexa-Fluor secondaries (1:250–500; Invitrogen-Molecular Probes) were incubated overnight at 4°C. All specimens were mounted in Fluoromount-G (Electron Microscopy Sciences, Hatfield, PA). Fluorescent images were collected using a ZEISS LSM 510 META laser scanning confocal microscope and displayed as maximal Z-stack projections, unless otherwise indicated.
Western Blot Analyses
Western blot analyses were carried out as previously described (Gatto and Broadie, 2008, 2009a). Flies were decapitated at 1 day post-eclosion with heads further homogenized and boiled in 1× NuPage sample buffer (Invitrogen, Carlsbad, CA) supplemented with 40 mM DTT. The total protein from 2 heads per sample was loaded onto 4–12% Bis-Tris gels and electrophoresed in NuPage MES Buffer (Invitrogen) for 1 hour at 200V. Transfer to nitrocellulose was carried out for 1 hour at 100V in NuPage transfer buffer (Invitrogen) with 10% MeOH. Processing was completed using the Odyssey near infrared fluorescence detection system (Li-COR, Lincoln, NE). Antibodies used included: anti-dFMRP (1:3,000; 6A15, mouse monoclonal, Sigma, St. Louis, MO), anti-α-tubulin (1:100,000; B512, mouse monoclonal, Sigma), and Alexa-goat-anti-mouse-680 (1:10,000; Invitrogen–Molecular Probes).
Calcium Imaging and Quantification
The UAS-GCaMP1.3 transgenic calcium reporter was used to measure depolarization-induced calcium transients (Tessier and Broadie, 2011). To examine this in PDF-TRI neuronal somas, brains from staged animals were dissected in physiological saline containing 128 mM NaCl, 2 mM KCl, 4 mM MgCl2, 35.5 mM sucrose, 5 mM Hepes, and 1.5 mM CaCl2, pH 7.2. Isolated brains were immediately imaged under a 63× water-immersion objective with maximal pinhole aperture on a ZEISS LSM 510 META laser scanning confocal microscope. Preparations were scanned for 1 minute to ascertain baseline fluorescence and then acutely depolarized using bolus application of 3M KCl to a final concentration of 70 mM KCl (Tessier and Broadie, 2011). Calcium transient development and recovery were then typically monitored for at least 1 additional minute. ImageJ (NIH, Bethesda, MD, http://rsb.info.nih.gov/ij/) was used for image registration and determination of fluorescence intensity values for regions of interest. The baseline fluorescence level was defined as the average value during the initial period from 40–50 seconds of acquisition and used to normalize the data set. For induced calcium transient measurements, the absolute value of the difference between the lowest and highest values post-stimulation was taken.
TUNEL Assay
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) was performed as previously described (Phillips et al., 2008), according to the manufacturer's instructions using the In Situ Cell Death Detection Kit, TMR Red (Roche, Indianapolis, IN). Co-immunostaining for PDF-TRI neurons was done with anti-PDF (DSHB) in staged brains employing an assay-compatible Alexa-Fluor-488 secondary antibody (Invitrogen-Molecular Probes). Preparations were then incubated for 1 hour in a humidified chamber at 37°C with a 1:9 mixture of the detection kit, containing calf thymus terminal deoxynucleotidyl transferase and TMR red labeled nucleotides. A positive control was generated by treatment with 250 U/mL DNaseI (BioRad, Hercules, CA) in 10 mM TrisCl, pH 7.5 for 10 minutes at room temperature. A negative control was omission of enzyme. After labeling, samples were washed with PBS and water before mounting in Fluoromount-G (Electron Microscopy Sciences). Fluorescent images were collected using a ZEISS LSM 510 META laser scanning confocal microscope.
Statistics
Statistical analyses were performed using GraphPad InStat 3 (GraphPad Software, Inc., San Diego, CA). Unpaired, two-tailed t-tests with Welch correction were applied, allowing for populations with variable standard deviations. Where appropriate, i.e., in the dfmr1 null allelic examinations and CCAP/pburs phenotypic classifications, a one-way analysis of variance (ANOVA) was performed with subsequent Tukey-Kramer multiple comparison post-tests. Significance levels in figures are represented as p<0.05 (*); p<0.01 (**); p<0.001 (***). All error bars represent standard error of the mean (s.e.m.).
Results
Null dfmr1 mutants retain developmentally-transient PDF-TRI neurons
Null dfmr1 mutants display a range of well-defined defects in brain synaptic connectivity in PDF-expressing neural circuitry (Coffee et al., 2010; Dockendorff et al., 2002; Gatto and Broadie, 2009a; Morales et al., 2002; Reeve et al., 2005; Reeve et al., 2008; Sekine et al., 2008). PDF is expressed in small (sLNv) and large (lLNv) ventrolateral neurons, and in mutants these neurons exhibit mild architectural defects including misdirected collateral branching and defasiculation resulting in posterior optic tract splitting (Morales et al., 2002; Reeve et al., 2005). More robustly, in the dorsal protocerebrum, sLNvs show overgrowth and overelaboration of their PDF-expressing synaptic bouton arborizations (Coffee et al., 2010; Dockendorff et al., 2002; Gatto and Broadie, 2009a; Reeve et al., 2005; Reeve et al., 2008). Visualization of these peptidergic neurons, their processes and synaptic connections can be accomplished with anti-PDF immunocytochemistry, Pdf-Gal4 driven expression of the UAS-mCD8∷GFP membrane marker or both labels in parallel (Fig. 1).
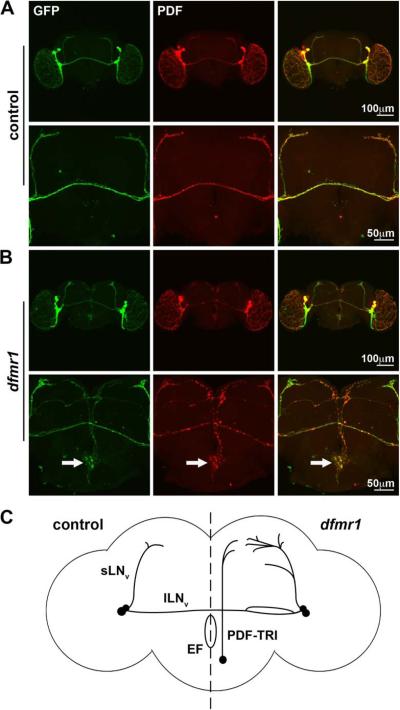
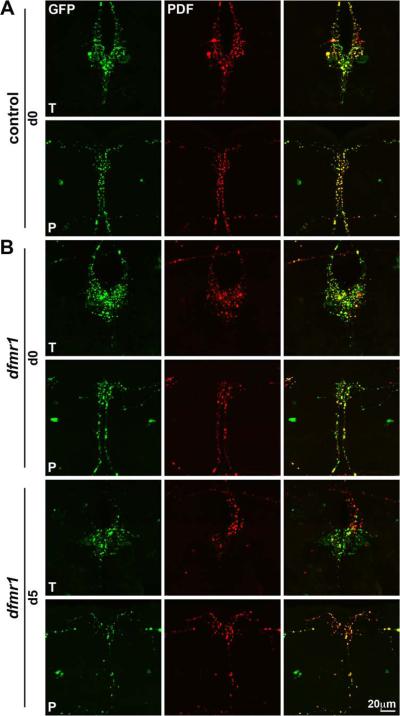
Figure 1. Null dfmr1 mutant brains exhibit aberrant PDF-TRI neurons.
Immunocytochemistry in the adult (day 5) brain is shown to visualize Pdf-Gal4 driven UAS-mCD8∷GFP (anti-GFP, green) and endogenous PDF immunolabeling (anti-PDF, red). A) A representative control brain demonstrates the relative positioning of PDF neurons at low magnification (top row) and at higher magnification focuses on the central brain region (bottom row). B) A representative dfmr150M null brain is shown with arrows to indicate the tritocerebral PDF neurons present only in the mutant. C) Model summarizing PDF neuron abnormalities in wild-type (control, left) compared to dfmr1 null brain (mutant, right). Posterior optic tract splitting, collateral branching and synaptic overgrowth have been described in dfmr1 mutants. We report here novel presentation of PDF-neurons near the midline of the mutant brain. sLNv: small ventrolateral neuron, lLNv: large ventrolateral neuron, PDF-TRI: PDF tritocerebral neuron, EF: esophageal foramen.
A particularly prominent defect in PDF neurons has gone entirely unreported in previous studies of the FXS disease model: the aberrant appearance of another class of PDF-positive neurons in the central brain of dfmr1 null mutants (compare Fig. 1A to 1B). In adult brains (day 5 post-eclosion; d5) isolated from control and dfmr1 nulls, mutants consistently present with a midline/median bundle of PDF immunoreactive projections from between 2–4 neurons originating from the ventral aspect of the brain (Fig. 1B). These neurons were not observed in the age-matched adult wild-type brain (Fig. 1A). In mutants, these neurons were often present as a bilaterally oriented pair with cell body positioning in the tritocerebrum, near the esophageal foramen (Fig. 1B, arrows at higher magnification). Mutant PDF neurons project their axons upward beyond the posterior optic tract and elaborate a synaptic field in the protocerebral area adjacent to the sLNv synaptic arbors. Thus, in addition to connectivity defects within the s- and lLNv neurons, the PDF circuit may be compromised by the encroaching aberrant contributions of the midline PDF tritocerebral (PDF-TRI) neurons present only in the dfmr1 null mutant condition (Fig. 1C).
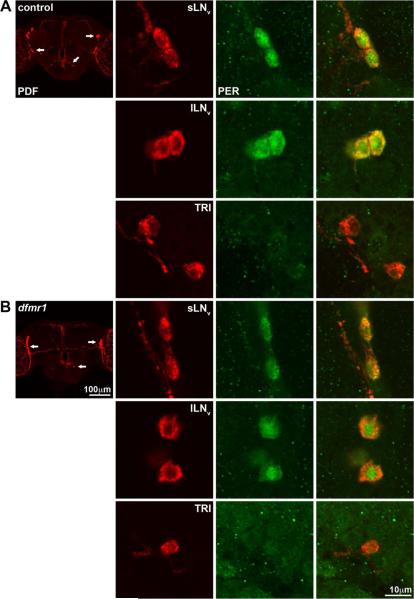
A developmentally-transient population of PDF-TRI neurons has been previously described (Helfrich-Forster, 1997). These peptidergic neurons initially appear during mid-pupal development (P+50%) as one or two highly branched, bilateral neurons forming a network surrounding the ventral and lateral esophageal foramen. They project into the median bundle and often extend to fasiculate with the terminals of the sLNv neurons, as we describe above. These PDF-TRI neurons are suggested to have a developmentally-transient, eclosion-related role within the clock circuit (Helfrich-Forster, 1997). However, it has been unclear whether PDF-TRI neurons express oscillating circadian clock proteins and thus serve as an additional class of clock neurons regulating circadian rhythms. Previous studies suggest that the PDF-TRI neurons do not mediate this function, as inducing their aberrant retention did not alter the circadian rhythmicity of free-running locomotor output (Renn et al., 1999). To test this possibility further, we labeled brains for the clock protein period (PER) at zeitgeber time 0–2 hours (ZT 0–2) in control (w1118) and dfmr1 null mutants (Fig. 2). Assays were done at late-stage pupal day 4 (P4), when PDF-TRI neurons are equally present in both genotypes. We readily detected PER within sLNv and lLNv PDF neurons (Fig. 2), which therefore served as an internal control. We note that the PER expression was consistently more robust in control (Fig. 2A) compared to the dfmr1 null (Fig. 2B), suggesting perturbation of the circadian clock mechanism. Importantly, we never detected PER within the PDF-TRI neurons in either control (Fig. 2A) or dfmr1 nulls (Fig. 2B). Additional experiments at ZT 4–6 (data not shown) also revealed no PER expression in the PDF-TRI neurons. Thus, we have seen no evidence that PDF-TRI neurons oscillate out of phase as compared to the s- and lLNvs, at least within this temporal window. Consistent with Renn et al. (1999), these data show that PDF-TRI neurons do not function as clock neurons in the circadian circuit.
Figure 2. PDF-TRI neurons do not express the clock protein PER.
Immunocytochemistry at late-stage pupal day 4 (P4) brain is shown to visualize endogenous PDF (anti-PDF, red) and PER (anti-PER, green) in A) control (w1118) and B) dfmr150M null. The lower magnification images (left) indicate the position of sLNv, lLNv and PDF-TRI cell bodies (arrows) in the context of the whole brain. The higher magnification images (right) show single confocal optical sections through the different classes of PDF neuron cell bodies. PER expression is evident in both the sLNvs and lLNvs, while undetectable in the PDF-TRI neurons in both genotypes.
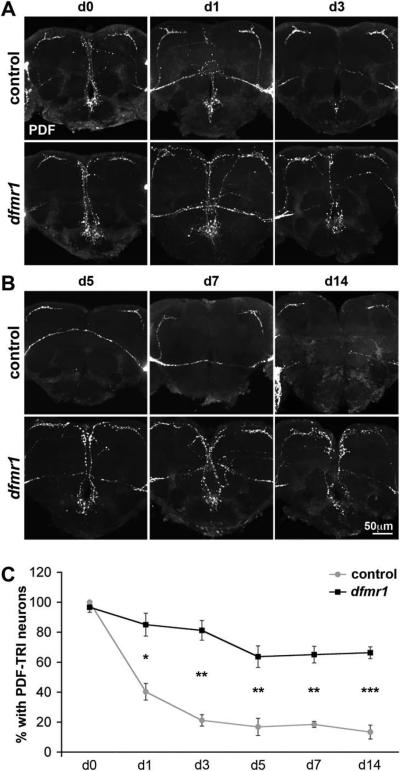
In the wild-type brain, PDF immunoreactivity within PDF-TRI neuronal soma is lost within 24 hours of eclosion, and cell processes are no longer discernible within 2–4 days (Fernandez et al., 2008; Helfrich-Forster, 1997; Renn et al., 1999). Based on this description, we examined the time course of PDF labeling in control (w1118) and dfmr1 null animals (Fig. 3). Upon eclosion (d0), control and mutant brains showed an indistinguishable array of PDF-TRI neurons (Fig. 3A). However, in the proceeding days, these cells were rapidly lost in control brains, while retained in the dfmr1 null. By day 1 post-eclosion, PDF-TRI neurons were manifest in only 40±6% of control brains versus 85±8% in dfmr1 nulls (p=0.017, n=3; n=number of independent trials comparing ≥10 brains of each genotype in each trial; Fig. 3C). At later time periods, the great majority of control brains lacked any detectable PDF-TRI neurons, whereas most dfmr1 null brains retained these cells (Fig. 3B). At day 5 post-eclosion, PDF-TRI neurons were evident in only 17±6% of control brains compared to 64±7% of dfmr1 null brains (p=0.003, n=4; Fig. 3C). At maturity (2 weeks), a similar difference was maintained; PDF-TRI neurons were observed in 13±5% of control versus 66±4% of mutant brains (p<0.0001, n=6; Fig. 3B,C). Thus, PDF-TRI neurons are present at a 5-fold higher incidence in the dfmr1 null mutant central brain compared to controls.
Figure 3. Null dfmr1 mutants retain developmentally-transient PDF-TRI neurons.
The developmental time course of PDF neuron remodeling is shown with representative images of anti-PDF immunocytochemistry in the central brain region of age-matched controls (w1118) and dfmr150M null mutants compared. A) Days 0–3 (d0–3) post-eclosion show PDF-TRI neurons progressively lost in the control but maintained in mutant. B) Days 5–14 (d5–14) in mature adults show no PDF-TRI neurons in control, but perseverance of these cells in the dfmr1 null. C) Developmental time course quantification of PDF-TRI neuron prevalence in control (gray) and dfmr1 null (black) animals. Data are shown as the percentage of brains maintaining identifiable PDF-TRI neuronal soma and midline process reactivity originating below the esophageal foramen extending beyond the posterior optic tract. Significance levels are represented throughout as p<0.05 (*); p<0.01 (**); p<0.001 (***). All error bars represent s.e.m.
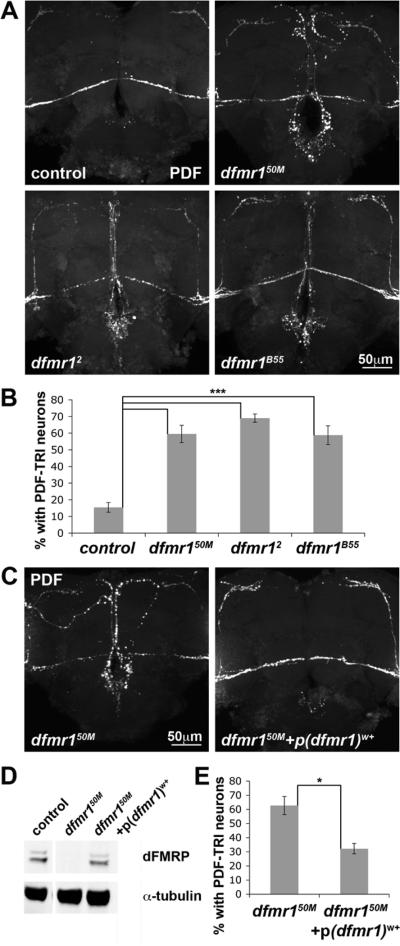
To test the specificity of this mutant phenotype, we confirmed PDF-TRI neuron retention using two additional, independently isolated dfmr1 null alleles; dfmr12 (Dockendorff et al., 2002) and dfmr1B55 (Inoue et al., 2002) (Fig. 4A). PDF labeling at day 5 revealed equivalent PDF-TRI neuron maintenance in all three alleles (dfmr150M: 60±5%, n=4; dfmr12: 69±2%, n=3; dfmr1B55: 59±6%, n=3; n=number of independent trials comparing ≥10 brains of each genotype in each trial; p>0.05 for each pair-wise comparison; Fig. 4B), which were all very significantly elevated as compared to control (w1118: 15±3%, n=4; p<0.001 for each pair-wise comparison; Fig 4B). To also verify that PDF-TRI neuron retention was due to dfmr1 gene disruption, a genomic rescue construct (p(dfmr1)w+) (Dockendorff et al., 2002) was introduced into the dfmr150M null background (Fig. 4C). One copy of the rescue construct was introduced into an otherwise dfmr1 null animal to drive dFMRP expression confirmed by Western blot (Fig. 4D). The genomic construct strongly diminished PDF-TRI neuron retention (Fig. 4C,E). Quantification at day 5 showed significant (p=0.026; n=3) rescue of PDF-TRI retention by introduction of wild-type dFMRP (Fig. 4E). We conclude that PDF-TRI neurons are abnormally retained throughout life within the tritocerebral brain region due specifically to the loss of dFMRP function.
Figure 4. Specificity of PDF-TRI neuron retention to loss of dFMRP.
A) PDF immunocytochemistry in the central brain region of day 5 (d5) age-matched animals compared between control (w1118) and three independent dfmr1 null alleles (dfmr150M, dfmr12, and dfmr1B55). B) Quantification of PDF-TRI neuron prevalence in all four genotypes at d5. Data are shown as the percentage of brains maintaining identifiable PDF-TRI cells. Significance levels are represented as p<0.001 (***). All error bars represent s.e.m. C) PDF immunocytochemistry in the central brain region of d5 age-matched dfmr150M null with and without a genomic rescue construct (p(dfmr1)w+). D) Quantification of PDF-TRI neuron prevalence in dfmr150M null mutants with and without p(dfmr1)w+. Significance level (p=0.026) represented as p<0.05 (*). All error bars represent s.e.m.
PDF-TRI neurons maintain synaptic connectivity at maturity
During late pupal development, the PDF-TRI neurons emanate from the tritocerebral brain region and project dorsally to infiltrate the protocerebral regions, including the pars intercerebralis and pars lateralis (Helfrich-Forster, 1997). We sought to characterize the synaptic organization of these neurons and their complex processes in both the trito- and protocerebral brain regions (Supplemental Fig. 1). As we demonstrated previously in studies of the sLNv synaptic arbors (Gatto and Broadie, 2009a), Pdf-Gal4 driven expression of the UAS-synaptotagmin-GFP transgene, an integral synaptic vesicle protein marker, specifically labels synaptic boutons overlapping and largely coincident with the anti-PDF neuropeptide distribution (Supplemental Fig. 1). At eclosion (d0), the PDF-TRI neurons project processes within the tritocerebral region, which localize both PDF transmitter and synaptic vesicles within boutons near to their somas, as do their dorsally projecting axons. These PDF-TRI synaptic arbors extend past the posterior optic tract to elaborate an extensive synaptic bouton array in the protocerebrum, closely approaching the sLNv synaptic arbors (Supplemental Fig. 1).
At eclosion, control and dfmr1 null brains exhibit indistinguishable patterns of PDF/synaptotagmin colocalization and spatial synaptic bouton arrangements in both the tritocerebral and protocerebral brain regions (compare Figs. 5A and B, d0). Considerable synaptic establishment is present in both regions, suggesting a high degree of interconnectivity with surrounding neurons. PDF-TRI synaptic connectivity is absent in the wild-type brain at maturity (Supplemental Fig. 1; d5 inset). In sharp contrast, the synaptic array from the aberrant PDF-TRI neurons is maintained in the mature dfmr1 null brain. Pdf-Gal4 driven UAS-synaptotagmin-GFP showed the continued presence of synaptic boutons bearing the PDF neuropeptide in both tritocerebral and protocerebral brain regions (Fig. 5B, d5). As expected, this synaptic array is progressively refined and remodeled over developmental time (compare Figs. 5B d0 vs. d5). Nevertheless, it is clear that the inappropriately retained PDF-TRI neurons persist in the dfmr1 nulls and contribute synaptic connections in close proximity to both the clock circuit and surrounding cells.
Figure 5. Null dfmr1 PDF-TRI neurons maintain synaptic architecture.
Representative images of Pdf-Gal4 driven synaptic marker UAS-synaptotagmin-GFP in PDF-TRI neurons (GFP, green) double-labeled for the PDF neuropeptide (red). As oriented in Supplemental Fig. 1, trito- (T) and protocerebral (P) regions are shown. A) Control brains immediately after eclosion (day 0; d0) show developmentally-transient PDF-TRI neurons with an extensive array of differentiated synaptic boutons. B) Null dfmr150M mutants at d0 (top) and d5 (bottom). Similar synaptic arrays are present in control and mutant at the early time point, immediately following eclosion. PDF-TRI synaptic boutons are maintained at maturity (d5) only in the dfmr1 null brain, with an extensive array of remodeled synaptic boutons in both trito- (T) and protocerebral (P) brain regions.
PDF-TRI neurons maintain depolarization-dependent Ca2+ transients at maturity
In the dfmr1 null brain, PDF-TRI neurons likely contribute to neural dysfunction via aberrant synaptic connectivity maintained throughout adult life. However, it was critical to assess whether the function of the normally transient PDF-TRI neurons was altered in wildtype versus dfmr1 nulls and to examine the potentially maintained PDF-TRI function in the dfmr1 null mature brain. As one test of function, we assayed calcium dynamics incurred upon depolarizing stimulation using the transgenic GCaMP calcium reporter (reviewed in Mao et al., 2008; Pologruto et al., 2004; Tian et al., 2009). The Pdf-Gal4 driver was used to express UASGCaMP1.3, a calcium responsive probe incorporating a calmodulin sensor tethered to GFP, which upon calcium influx alters GFP conformation to increase fluorescence output (Nakai et al., 2001; Tessier and Broadie, 2011). Whole mount brain preparations were first imaged to establish baseline reporter fluorescence within PDF-TRI neuron somas, then acutely depolarized with 70 mM KCl application while continuously monitoring calcium transient development and dissolution (Fig. 6 and Supplemental Movies 1–3).
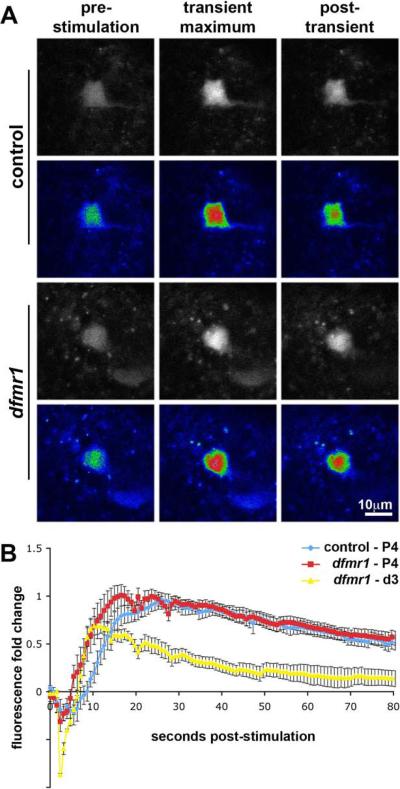
Figure 6. Developmentally-transient PDF-TRI neurons are responsive to depolarization and maintain this functionality when persistent in the dfmr1 null brain.
Examples of the transgenic fluorescent calcium reporter UAS-GCaMP driven by Pdf-Gal4 in the PDF-TRI neurons are depicted. Neurons were acutely depolarized with 70 mM KCl while continuously monitoring fluorescence intensity. A) Still frames of PDF-TRI neuron cell somas from time-lapse imaging (see Supplemental Movies for full imaging sequence) at pupal day 4 (P4) in control and dfmr150M null brains are shown. The raw data are presented in black and white (top rows) and pseudocolored for intensity (bottom rows). The baseline pre-stimulation, transient KCl-induced maximum and post-transient recovery fluorescence are shown for each genotype. B) The quantification of fluorescence intensity as fold change from baseline during KCl depolarizing stimulation. Control and dfmr1 nulls at pupal day 4 (P4) in blue and red, respectively, when PDF-TRI neurons are present in both genotypes. Calcium transients are similar in magnitude, but with a more rapid rise time in the dfmr1 nulls. Null dfmr1 persistent PDF-TRI neurons in the adult (day 3 post-eclosion (d3); in yellow) remain functional as assayed. All error bars represent s.e.m.
Initial studies were performed at late-stage pupal day 4 (P4), a period in which PDF-TRI neurons are similarly present and arrayed in control and dfmr1 null brains. Both genotypes exhibited similar reporter basal fluorescence, and generally indistinguishable depolarization-induced calcium transients were elicited (Fig. 6A). Following an initial depression caused by osmotic shock (Fiala and Spall, 2003; Tessier and Broadie, 2011), reporter fluorescence intensity in the PDF-TRI cell bodies rose rapidly to a maximal level and then declined gradually toward the initial baseline (Fig. 6B and Supplemental Movies 1–2). By measuring the absolute value of the relative transient amplitudes from trough to peak, accounting for the entirety of the depolarization-induced calcium influx (Tessier and Broadie, 2011), we quantified any differential function between control and dfmr1 null PDF-TRI neurons. The amplitude of peak response was indistinguishable between genotypes (control: 1.78±0.09, n=11; n=number of individual PDF-TRI neurons examined; dfmr1 null: 1.82±0.12, n=13; p=0.78; Fig. 6B). At each individual time point, the genotypes were not statistically different, except at 5.5, 8.7, 9.5 and 10.3 seconds post-stimulation (p<0.05; Fig. 6B). The average fluorescence rise time to achieve maximal intensity in dfmr1 null neurons was more rapid (control: 19.14±1.96 sec, n=11 vs. dfmr1 null: 13.1±1.42 sec, n=13; p=0.02; Fig. 6B). This phenotype suggests equivalent signaling capacity with an increased excitability in the dfmr1 null neurons, possibly due to a lower activation threshold.
We next assayed the persistent PDF-TRI neurons present only in the dfmr1 null at maturity. The mutant cells maintained a robust calcium influx response to depolarizing stimulation (Fig. 6B and Supplemental Movie 3). Due to a more substantial fluorescence depression with KCl stimulation (2.7–5.5 seconds post-KCl stimulation dfmr1 null P4 vs. d3; minimally p<0.05), the relative calcium transient amplitude was similar in the adult compared to pupal day 4 (1.59±0.09, n=5; P4 vs. d3 adult, p=0.15; Fig. 6B). However, the fluorescence response rate was modified over this developmental time period, with a significantly more rapid rise time at maturity (8.83±0.58 sec, n=5; P4 vs. d3 adult, p=0.01; Fig. 6B). We have recently reported similar developmental changes in calcium dynamics in the Mushroom Body brain region (Tessier and Broadie, 2011). We conclude that PDF-TRI neurons are capable of eliciting depolarization-induced calcium transients during development in both control and dfmr1 null brains, and retain this competence in the null mutant into adult life.
Null dfmr1 PDF-TRI neurons exhibit impaired apoptotic initiation
How are PDF-TRI neurons eliminated from the wild-type brain? What is the developmental mechanism facilitating their persistence in the dfmr1 null compared to the trigger for their rapid removal following eclosion in wildtype? In previous cell ablation studies using Pdf-Gal4 mediated expression of the cell death promoting gene, reaper (UAS-rpr), it was shown that the caspase inhibitor, p35, can rescue rpr-mediated cell death of the PDF-TRI neurons (Renn et al., 1999). In this study, the neurons persisted into maturity, at least to day 10 post-eclosion. This work suggests that PDF-TRI neurons normally die via a developmental apoptotic program. However, recent studies have clearly shown in contrast that FMRP serves as a negative regulator of cellular proliferation. Increased cell numbers have been observed in mutant animals as the loss of FMRP increases neural stem cell proliferative capacity in both Drosophila and murine FXS models (Callan et al., 2010; Luo et al., 2010). Thus, two distinct possibilities could contribute to the PDF-TRI neuron survival in the dfmr1 nulls, i.e., a failure to remove extraneous cells from the circuit via a defective apoptotic program or an initial overabundance of these cells owing to an over-proliferative neurogenesis defect.
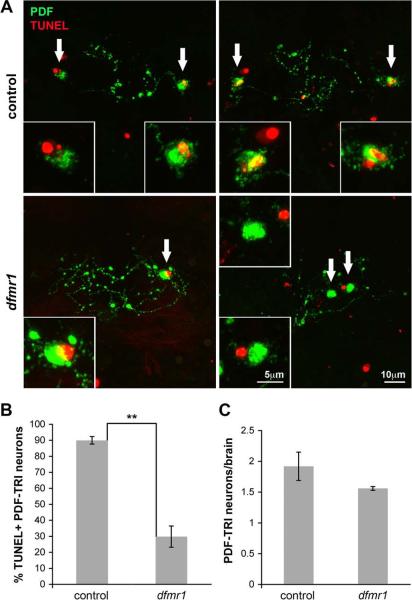
To formally distinguish between these mechanisms, we first examined an apoptotic indicator of DNA fragmentation, TUNEL labeling, in the PDF-TRI neuron cell bodies (Fig. 7). We focused on the first day post-eclosion (day 0; <24 hours), the period immediately preceding the disappearance of these cells in the wild-type brain. A TMR-red coupled assay to reveal compromised DNA integrity was employed in conjunction with anti-PDF labeling to visualize the PDF-TRI cell bodies and associated tritocerebral processes (Fig. 7A). With this imaging approach, the vast majority of PDF-TRI neurons in control (w1118) brains were TUNEL-positive at day 0 post-eclosion, whereas many fewer PDF-TRI neurons were undergoing apoptosis in the dfmr1 nulls. In control brains, 90±2% of PDF-TRI neurons demonstrated TUNEL labeling as compared to 30±7% in the dfmr1 mutants, a highly significant difference (p=0.003, n=4; n=number of independent trials with ≥10 brains of each genotype examined per trial; Fig. 7B). In sharp contrast, at this early time point (d0), there was no detectable difference in the number of PDF-TRI neurons in the control versus the mutant (control: 1.9±0.2 vs. dfmr1: 1.6±0.03, n=4; p=0.22; Fig. 7C). Therefore, there is no evidence of increased proliferation for this neuron class, but rather clear indication of impaired apoptosis in the dfmr1 null brain. We conclude that although FMRP is implicated in contributing to enhanced neurogenesis in other contexts, the prolonged presentation of PDF-TRI neurons in the dfmr1 null is due to an inability to initiate normal programmed cell death.
Figure 7. PDF-TRI neurons undergo programmed cell death immediately post-eclosion, which is depressed in the dfmr1 null brain.
TUNEL labeling reveals programmed cell death in the first day after eclosion. A) PDF-TRI neuron cell bodies and local tritocerebral processes are labeled with anti-PDF immunocytochemistry (PDF, green) in conjunction with TUNEL labeling (red) in control (w1118; top) and dfmr150M null (bottom) brains at day 0. Left and right panels show 2 independent examples of TUNEL/PDF staining per genotype. Arrows indicate PDF-TRI cell bodies shown at higher magnification (insets). Nuclear TUNEL staining enveloped by anti-PDF is indicative of an apoptotic TRI neuron. Notably, other non-PDF associated TUNEL staining is observed in some panels, highlighting the efficacy of the assay and the relative positioning of other apoptotic cells within this brain region. B) Quantification of TUNEL-positive PDF-TRI neurons shown as a percentage of total identifiable PDF-TRI neurons. Significance level is represented as p<0.01 (**). C) Quantification of the total number of PDF-TRI neurons present upon eclosion is presented. There is no statistical difference between control and mutant. All error bars represent s.e.m.
dFMRP influences clearance of other transient peptidergic neurons
To determine whether the anti-apoptotic effect of the dfmr1 mutation is a generalized phenomenon among developmentally transient peptidergic neurons, we next examined other neuronal populations known to be similarly targeted for developmental programmed cell death (Draizen et al., 1999; Ewer et al., 1998; Peabody et al., 2008). Neurons of this class include neuropeptidergic cells that express CCAP, which are thought to trigger the ecdysis motor program (Baker et al., 1999; Park et al., 2003), and a CCAP neuron subset that also expresses bursicon, mediating cuticle tanning and wing inflation (Dewey et al., 2004; Luan et al., 2006; Peabody et al., 2008). As reported previously in the pharate adult brain (Luan et al., 2006), upon eclosion we detected four bilaterally symmetric pairs of CCAP neurons within the subesophageal ganglion (SEG), one pair of which coordinately expresses bursicon (Supplemental Fig. 2). In addition, bilateral pairs of CCAP- and bursicon immunoreactive neurons were found in the third thoracic (T3) and abdominal (Ab) neuromeres (Supplemental Fig. 2) (Dewey et al., 2004; Luan et al., 2006). These T3/Ab neurons display clear signs of apoptosis within 6–8 hours post-eclosion, extensive degeneration by 10–12 hours, and almost complete clearance by 14 hours, with only 2% of brains displaying detectable cells by 24 hours (Draizen et al., 1999; Peabody et al., 2008).
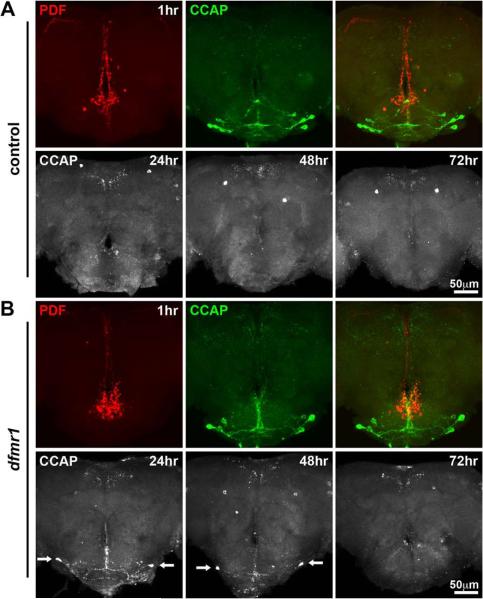
We first examined the CCAP peptidergic neurons in the SEG (Fig. 8). At 1 hour following eclosion, CCAP neurons were readily visualized and, as previously reported (Park et al., 2003), some overlap of the CCAP and PDF-TRI neuron processes was detected (Fig. 8A, top row). In the dfmr1 null brain, equivalent profiles of CCAP and PDF neurons were present immediately following eclosion (Fig. 8B, top row), indicating no evidence of over-proliferation. At later time points, these neurons were rapidly eliminated from the control brain, and at 24, 48 and 72 hours post-eclosion, the CCAP immunoreactivity was absent in the SEG (Fig. 8A, bottom row). Over this developmental time course, an obvious delay in the clearance of these neurons and their processes was observed in the dfmr1 mutants, as visualized at 24 and 48 hours post-eclosion (Fig. 8B, bottom row, arrows). Although their removal was delayed, the neurons were largely undetectable in both control and dfmr1 null brains by 72 hours post-eclosion. Thus, it appears that dFMRP facilitates the apoptosis and cell clearance of CCAP neurons but is not required at the same level as for the PDF-TRI neurons.
Figure 8. Clearance of CCAP neurons is delayed in the dfmr1 null brain.
Representative images depict staged brains double-labeled with anti-PDF (red) and anti-CCAP (green) in control (w1118) and dfmr150M null mutants. A) Control brain at 1 hour post-elcosion (top row) shows CCAP neurons within the SEG. PDF-TRI neurons cluster more dorsally near the esophageal foramen. The bottom row shows anti-CCAP immunocytochemistry alone at 24, 48 and 72 hours post-eclosion. All SEG CCAP neurons are absent at these later time points in wildtype. B) Comparable images from the dfmr1 null brain at 1 hour post-elcosion (top row) show a neuron array indistinguishable from control. The time course for CCAP neuron elimination over the next 72 hours reveals prolonged presence of cells in the dfmr1 null (arrows at 24 and 48 hours post-eclosion), although almost all SEG CCAP neurons have been lost by 72 hours post-eclosion.
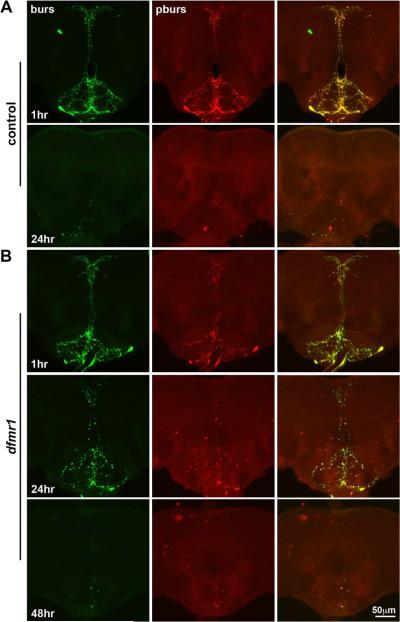
We next examined peptidergic bursicon neurons by labeling for bursicon and its functionally heterodimerizing β-subunit, pburs (Fig. 9) (Luo et al., 2005). Immediately following eclosion, two bursicon cells ramify widely within the SEG region of the brain (Fig. 9A). Cell numbers were indistinguishable between control and dfmr1 nulls at this early time point, again indicating no change in the proliferation for this neuron type (compare Figs. 9A and B, top rows). In wildtype, these cells were completely eliminated by 24 hours post-eclosion, whereas extensive bursicon immunoreactivity and process elaboration persisted in the dfmr1 null brain (compare Figs. 9A and B, second rows). This indicates a delay in the initiation of the apoptosis program and a defect in the cellular mechanisms required to eliminate these cells in the absence of dFMRP. Nevertheless, by 72 hours post-eclosion, bursicon peptidergic neurons were principally absent from the dfmr1 null brain (Fig. 9B, bottow row). Examination of the bursicon neurons in the thoracic T3 and abdominal ganglia revealed very similar cellular dynamics (Supplemental Fig. 3). Thus, once again dFMRP facilitates apoptosis and removal of bursicon neurons, although the cells are eliminated in a delayed fashion in dfmr1 null mutants.
Figure 9. Clearance of bursicon neurons is likewise delayed in the dfmr1 null brain.
Representative images of the ventral brain SEG double-labeled for bursicon (green) and pburs (red) in control (w1118) and dfmr150M null mutants. A) A control brain at 1 hour post-elcosion (top row) shows the two bursicon peptidergic neurons' somal positioning and process elaboration. The bottom row reveals that the neurons have been eliminated by 24 hours post-eclosion in the wild-type brain. B) Comparable images from the dfmr1 null brain at 1 hour post-elcosion (top row) show a complex neuronal array indistinguishable from control. Bursicon processes are aberrantly retained at 24 hours in the dfmr1 null mutant (middle row), although largely lost from the brain SEG by 48 hours (bottom row).
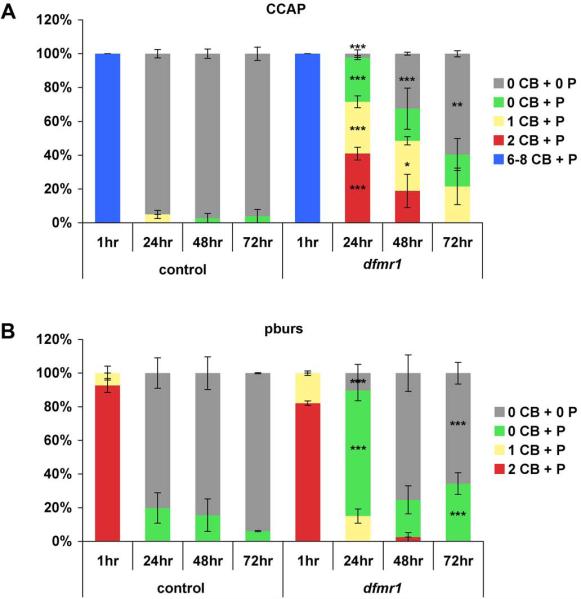
We conclude that different populations of developmentally-transient peptidergic neurons are influenced by dFMRP in aspects of programmed cell death mediation, from initiation to terminal process clearance. Quantitative examination of the central CCAP and pburs SEG profiles, assessing the presence or absence of associated cell bodies and/or processes observed in control and dfmr1 null animals, clearly demonstrated this influence (Fig. 10). These experiments were performed in triplicate, examining 13±1 brains per genotype per time point. Immediately after eclosion (1 hour), the number and organization of CCAP and pburs neurons were indistinguishable between controls and dfmr1 nulls, with the full complement of 8 CCAP and 2 pburs cells seen in both. Thereafter, a delayed clearance of these populations in the dfmr1 nulls was evident and more protracted in the CCAP neurons. At 24 hours, 95±2% of control brains had no SEG CCAP-reactive cell bodies (CB) or processes (P) detectable compared to 2±2% in dfmr1 nulls (p<0.001) (Fig. 10A). By contrast in the dfmr1 nulls at 24 hours, 40±4% of brains maintained 2 CB+P (p<0.001), 30±4% 1CB+P (p<0.001), and 24±3% 0CB+P (p<0.001). By 48 hours, 30±2% of dfmr1 brains still harbored 1 CCAP CB+P (p<0.05) and only 32±1% were fully cleared (p<0.001) (Fig. 10A). By 72 hours, 60±2% of the dfmr1 null brains showed no CCAP cells (p<0.01) with another 19±9% showing only residual processes. The pattern was similar for pburs. At 24 hours, controls had 80±9% of brains devoid of pburs SEG cells with the remainder (20±10%) showing discernable processes (Fig. 10B). In contrast, only 16±2% were fully cleared in the dfmr1 nulls (p<0.001) and 70±6% retained processes (p<0.001). By 48 hours, the genotypes were similar with 85±10% of brains lacking pburs in control vs. 75±11% in dfmr1 (Fig. 10B). As the pburs cells are a subset of the CCAP neurons, it appears that the CCAP/non-pburs neurons are more persistent in the dfmr1 nulls. Despite this generalized role of dFMRP in regulating apoptosis and clearance of normally developmentally-transient peptidergic neurons from the brain, we conclude that the PDF-TRI neurons show the greatest degree of dependence on dFMRP. In the dfmr1 null brain, only PDF-TRI neurons persist into adult maturity and, thereby, continue to contribute inappropriate synaptic connections within the tritocerebral and protocerebral brain regions throughout life.
Figure 10. Quantification of CCAP/pburs neuron clearance after eclosion.
A) Control (w1118) and dfmr150M null brains were examined by CCAP immunoreactivity at 1, 24, 48 and 72 hours post-eclosion. Classifications were assigned based on the presence or absence of identifiable CCAP cell bodies (CB) and processes (P). Blue: 6–8 CB + P, red: 2 CB + P, yellow: 1 CB + P, green: 0 CB + P, and gray: 0 CB + 0 P. An average of 13±1 brains were examined per time point per genotype in triplicate. Significance levels are represented throughout as p<0.05 (*); p<0.01 (**); p<0.001 (***). All error bars represent s.e.m. B) The same control and dfmr1 null brains were examined by pburs immunoreactivity at 1, 24, 48 and 72 hours post-eclosion. Classifications were assigned based on the presence or absence of identifiable bursicon cell bodies (CB) and processes (P). Red: 2 CB + P, yellow: 1 CB + P, green: 0 CB + P, and gray: 0 CB + 0 P. Significance levels are represented throughout as p<0.001 (***). All error bars represent s.e.m.
Discussion
It has long been known that loss of dFMRP compromises the architecture of the small and large ventrolateral PDF neuron circuitry in Drosophila brain (Coffee et al., 2010; Dockendorff et al., 2002; Gatto and Broadie, 2009a; Morales et al., 2002; Reeve et al., 2005; Reeve et al., 2008; Sekine et al., 2008). Here, we have surprisingly identified an additional, novel defect harbored within the PDF neuron population; the dfmr1 null mutation enables inappropriate life-long survival of PDF-TRI neurons normally destined for death via an apoptosis program immediately after eclosion. Although we show that PDF-TRI neurons are not circadian pacemakers, the retention of these cells into adulthood introduces functional neurons with elaborate synaptic contacts that may alter information flow and processing by contributing an aberrant source of PDF. The role of dFMRP in regulating the initiation of apoptotic events appears to be somewhat neuron-type specific and selective to PDF-TRI neurons. Nevertheless, the dfmr1 null mutants do present a generalized delay in peptidergic neuronal process removal upon cell death, including the delayed elimination of CCAP and bursicon cells. As these studies were conducted in ubiquitously dfmr1 null animals, determination of the cell autonomy assigned to these novel dFMRP functions, as compared to potentially coordinate and impinging function(s) of dFMRP in neighboring cells, will require future experimentation.
The key finding of this study is a novel role for dFMRP in mediating neuronal survival by influencing apoptotic initiation. In contrast, the recent focus of cell population regulation has centered on the role of dFMRP in controlling proliferation and differentiation. For example, dFMRP has been implicated in germline stem cell maintenance and proliferative capacity in the Drosophila ovary (Yang et al., 2007) in association with the micro-RNA bantam (Yang et al., 2009) and the E3 ubiquitin ligase cbl (Casitas B-lineage lymphoma) (Epstein et al., 2009). Earlier mammalian embryonic studies demonstrated altered cellular differentiation from both Fmr1 deficient mouse and human stem cells in vitro (Castren et al., 2005). Fmr1-deficient neurospheres yielded 3–5 fold increases in cells of the TuJ1-positive neuronal lineage and marked decreases in their GFAP-positive glial counterparts, likely due to increased apoptosis (Castren et al., 2005). In the mouse Fmr1 knockout, there is also over-proliferation in the subventricular zone (Castren, 2006). However, human neural progenitor cells isolated from FMR1-deficient fetal cortex were later shown to have proliferative and specification indices comparable to control, albeit with altered gene expression profiles (Bhattacharyya et al., 2008). More recent studies have demonstrated that FMRP plays a role in regulating the differentiation and proliferative capacity of adult neural stem cells. In the mouse Fmr1 knockout, this was examined in relation to hippocampal neurogenesis, thought to be involved in the plasticity required for learning and memory (Luo et al., 2010). FMRP insufficiency yielded increased proliferation, with a bias toward decreased neuronal and increased glial differentiation, both in vitro and in vivo. In studies of the Drosophila FXS model, dfmr1 deficiency resulted in increased mitotic activity in neuroblasts during larval development, from which increased numbers of adult neurons were derived (Callan et al., 2010). The dfmr1 null neuroblasts exit quiescence prematurely to execute excess proliferative activities, demonstrating cell cycle misregulation.
Despite this evidence that FMRP plays a role in neural stem cell proliferation, we found that normal peptidergic neuron numbers were present in the PDF, CCAP and bursicon populations in the dfmr1 null mutant. Thus, there is no evidence for a proliferative defect in these neuronal types. In contrast, we observed adult retention of the PDF-TRI neuron subpopulation that is usually present only transiently during development, owing to a requirement for dFMRP in apoptotic initiation following eclosion. The novel role of dFMRP in this mechanism is presently unknown. As an RNA-binding protein known to either repress or activate the translation of particular target mRNAs (reviewed in Bassell and Warren, 2008; De Rubeis and Bagni, 2010), does dFMRP serve as a negative regulator of a survival or anti-apoptotic factor, like Drosophila inhibitor of apoptosis 1 (DIAP1) (Orme and Meier, 2009; Wang et al., 1999)? Does dFMRP itself promote the expression of a pro-apoptotic factor, such as Reaper, Head involution defective, or Grim, which could antagonize DIAP1 function (Goyal et al., 2000; Steller, 2008; Yoo et al., 2002)? While no obvious candidates in these RNA target classifications have yet been uncovered for dFMRP in extensive screening efforts (Bittel et al., 2007; Brown et al., 2001; Chen et al., 2003; Darnell et al., 2001; Liao et al., 2008; Miyashiro et al., 2003; Zarnescu et al., 2005; Zhang et al., 2005; Zou et al., 2008), it is of potential interest that down-regulation of the ENA/VASP actin regulatory proteins has been recently implicated in neuronal degeneration via an apoptosis program (Franco et al., 2010; Rezaval et al., 2008).
The ENA/VASP family is crucial for appropriate filopodia formation, serving to block F-actin filament capping and encourage elongation (reviewed in Bear and Gertler, 2009; Drees and Gertler, 2008; Krause et al., 2003). The sole representative of this family in Drosophila, enabled (ena), has been shown to promote dendritic branching and actin-rich spine-like protrusions in dendritic arborization (DA) sensory neurons (Gao et al., 1999; Li et al., 2005). In dfmr1 null mutants, DA neurons in the peripheral nervous system display an increased number of higher-order branches, whereas dFMRP over-expression reciprocally yields dramatically decreased branching (Lee et al., 2003). Mechanistically, these defects have been linked to the small GTPase Rac1 (Lee et al., 2003); however, the structural phenocopy suggests that ena could likewise be upregulated in the Drosophila FXS model. Precedent for this type of misregulation has been demonstrated by the observations that dFMRP negatively regulates other cytoskeleton organizing proteins, including Futsch, the mictrobule-binding MAP1B homolog, and Chickadee, the actin-binding Profilin homolog (Reeve et al., 2005; Tessier and Broadie, 2008; Zhang et al., 2001). Thus, if the PDF-TRI neurons are sensitive to ena-mediated cell death, the dfmr1 mutation may be neuroprotective via this mechanism.
An outstanding question centers on the relative specificity of dFMRP-mediated influence on the commencement of programmed cell death. While serving to largely prevent developmental eradication of the PDF-TRI neurons, the dfmr1 mutation conferred no such long-term maintenance to neurons expressing the neuropeptides CCAP and/or bursicon. What segregating factors might differentiate these classes of peptidergic neurons and the relative requirement of dFMRP for their survival? Mechanistically, apoptotic initiating factors may differ between these neuron classes. The apoptosis of CCAP neurons has been linked to declining levels of the steroid hormone 20-hydroxyecdysone, which allow the pro-apoptotic accumulation of reaper and grim mRNA (Draizen et al., 1999). No such requirement has yet been defined for the PDF-TRI neurons. Perhaps within the subset of CCAP neurons expressing bursicon (Dewey et al., 2004; Luan et al., 2006; Peabody et al., 2008; Zhao et al., 2008), the temporal paracrine role of these neurons requires more stringent ablational regulation, and their effective removal is therefore less susceptible to reprogramming. Moreover, it is plausible that these other neuron classes may be more reliant upon other RNA-binding proteins, rather than dFMRP. For example, LARK is an RNA-binding protein that interacts with and stabilizes dFMRP (Sofola et al., 2008) and also notably influences the development of PDF LNv neurons (Huang et al., 2009). Although reportedly pan-neuronal, LARK is known to be enriched within the cytoplasm of CCAP neurons where it oscillates in a circadian manner (McNeil et al., 1998; Zhang et al., 2000). If key opportunistic translational regulation of an apoptotic factor underlies initiation of programmed cell death, and if LARK is the predominant translational regulator in these cells, it would be of interest to examine the cell death profiles of the CCAP/bursicon neurons in LARK-deficient animals to assess this mechanistic possibility.
Although dfmr1 mutation did not result in long-term survival of CCAP/bursicon neurons, the dfmr1 nulls did display a significant delay in the clearance of these cells, suggesting a delay in the initiation or progression of the apoptotic process. This is similar to observations in the bursicon receptor mutant rickets (rk4) (Peabody et al., 2008). In rk4 mutants, one-third of the abdominal bursicon neurons were still evident at 24 hours post-eclosion, which was then reduced to approximately 8% in 2-day old mutants. Clearly then, normal bursicon signaling is required for the appropriate initiation and regulation of death in these neurons. This suggests that the dfmr1 mutation may alter bursicon signaling or its downstream consequences. As immunocytochemistry failed to reveal any obvious differences in the observable levels of bursicon in dfmr1 nulls as compared to controls, dFMRP may more likely influence bursicon release, the stability or function of the bursicon receptor, or the downstream signaling cascade of this neurotransmission. Importantly, post-ecdysial wing epidermis cell death mediated via bursicon requires activation of the cAMP/PKA pathway (Kimura et al., 2004), and it is therefore interesting to note that significant impairment in cAMP production occurs both in dfmr1 mutants and human FXS patients (Berry-Kravis and Ciurlionis, 1998; Kelley et al., 2007). Thus, defective cAMP pathway signaling may underlie the neuronal clearance defect in the FXS condition.
Another factor that could contribute to the delayed or defective removal of these peptidergic neurons and their processes is aberrant glial activation. Drosophila glia have been implicated as active participants in the phagocytosis of apoptotic neurons (Kurant et al., 2008), developmental pruning of excessive axonal tracts (Awasaki and Ito, 2004; Awasaki et al., 2006; Watts et al., 2004), and clearance of degenerating axonal processes (Doherty et al., 2009; MacDonald et al., 2006). In addition, neuronal programmed cell death induces glial cell division during a critical period in the first week post-eclosion in adult Drosophila (Kato et al., 2009), perhaps facilitating cell corpse removal. Though dFMRP has not been readily detected in CNS repo-expressing glial cells (Morales et al., 2002), hFMRP has been detected in the early development of both astrocytic (Pacey and Doering, 2007) and oligodendrocyte (Wang et al., 2004) lineages. If in this specialized role glial responsiveness is altered in the dfmr1 null, this could effect the impaired elimination of the transient peptidergic neurons.
Future studies of PDF-TRI neuron retention in the dfmr1 null brain will focus on the potential role of synaptic activity in the critical survival versus apoptosis decision. Other PDF-expressing neurons, including the s- and lLNvs, appear to function downstream of GABAergic synaptic inputs (Hamasaka et al., 2005) and express the Rdl (resistance to dieldrin) GABAAreceptor subunit (Chung et al., 2009; Parisky et al., 2008). In addition, the LNvs also express the DmGluRA metabotropic glutamate receptor although, confoundingly, glutamate may serve an inhibitory role on the LNvs, as its dosage dependent application decreases sLNv intracellular calcium levels (Hamasaka et al., 2007). By determining how the PDF-TRI neurons may integrate such signals, we hope to gain further insight as to their contribution to neural dysfunction in the Drosophila FXS model. Such investigations will also allow us to assess the circuit-specific applicability of the `mGluR Theory of FXS', implicating FMRP function downstream of mGluR activation (Bear, 2005; Bear et al., 2004), compared to the `GABAAR Theory of FXS' (D'Hulst and Kooy, 2007, 2009), which suggests disruption of inhibitory GABAergic signaling is a key to the progression of the disease state. Although these theories are by no means mutually exclusive, each disruption may have a differential relative contribution to neuronal defects in the disease state, including the new discovery here of altered developmental pruning of neurons from specific modulatory neural circuits.
Supplementary Material
Representative images are presented of PDF-TRI neurons in the central brain of a newly eclosed wild-type animal double-labeled with Pdf-Gal4 driven UAS-synaptotagmin-GFP (GFP, green; left panel) and immunocytochemistry against the PDF neuropeptide (red; right panel). Output synapses are shown in the ventral brain tritocerebral region (trito), where the PDF-TRI neurons originate, and the dorsal brain protocerebral region (proto), to which the PDF-TRI neurons project. Relative positioning of the sLNv and PDF-TRI axonal arbors are indicated. Comparison of the panels at day 0 (d0) post-eclosion to the d5 mature adult brain (inset) demonstrates the absence of PDF-TRI synapses in the wild-type brain at maturity.
The entire adult nervous system (brain, thoracic ganglia and abdominal ganglion) of a newlyeclosed wildtype (w1118) animal double-labeled for CCAP (green) and bursicon β-subunit (pburs; red). The locations of developmentally transient CCAP and bursicon peptidergic neurons are shown. OL: brain optic lobe, SEG: brain subesophageal ganglion, T1–T3: three thoracic ganglia, Ab: abdominal ganglion.
Representative images highlight the terminal thoracic (T3) and abdominal ganglia double-labeled with anti-bursicon (green) and anti-pburs (red) in controls (w1118) and dfmr150M null mutants. A) A control at 1 hour post-elcosion (top row) demonstrates the positioning and process elaboration of bursicon peptidergic neurons. The bottom row shows all bursicon neurons have been eliminated by 24 hours post-eclosion in the wild-type animal. B) Comparable images from the dfmr1 null brain at 1 hour post-elcosion (top row) show a bursicon neuron array indistinguishable from control. Processes from bursicon neurons are aberrantly retained at 24 hours in the dfmr1 null mutant (middle row), although fully eliminated by 48 hours (bottom row).
The transgenic fluorescent calcium reporter UAS-GCaMP was driven by Pdf-Gal4 and imaged in the PDF-TRI neurons to assay functionality. After an initial baseline fluorescence measurement for 1 minute, neurons were acutely depolarized with 70 mM KCl while continuously monitoring fluorescence intensity. A control from late pupal day 4 (P4) is shown over 140 frames spanning 109.6 seconds, with a frame rate of 15 frames per second. The video is pseudocolored for fluorescence intensity.
The transgenic fluorescent calcium reporter UAS-GCaMP was driven by Pdf-Gal4 and imaged in the PDF-TRI neurons to assay functionality. After initial baseline measurement for 60 seconds, neurons were acutely depolarized with 70 mM KCl while continuously monitoring fluorescence intensity. A dfmr150M null PDF-TRI neuron at late P4 is shown over 182 frames spanning 142.7 seconds, with a frame rate of 15 frames per second. The video is pseudocolored for fluorescence intensity.
The transgenic fluorescent calcium reporter UAS-GCaMP was driven by Pdf-Gal4 and imaged in the PDF-TRI neurons to assay functionality. After initial baseline measurement for 1 minute, neurons were acutely depolarized with 70 mM KCl while continuously monitoring fluorescence intensity. A null dfmr150M PDF-TRI neuron at post-eclosion day 3 (d3) is shown over 184 frames spanning 144.3 seconds, with a frame rate of 15 frames per second. The video is pseudocolored for fluorescence intensity.
Research Highlights.
Null dfmr1 mutants aberrantly retain normally transient PDF-TRI neurons.
Null dfmr1 PDF-TRI neurons maintain synaptic contacts and remain functional.
Null dfmr1 TRI-neurons persevere due to compromised apoptotic initiation.
CCAP/bursicon neuron elimination is significantly delayed in dfmr1 nulls.
Acknowledgements
We gratefully acknowledge Dr. Hans-Willi Honegger for providing invaluable experimental suggestions, guidance and essential commentary during the preparation of this manuscript. We also most sincerely thank donors of key reagents: the kind gifts of anti-CCAP from Dr. Hans-Jürgen Agricola (University of Jena, Germany), anti-bursicon α from Dr. Benjamin White (National Institute of Mental Health, NIH, Bethesda, Maryland), anti-bursicon β from Dr. Aaron Hsueh (Stanford University School of Medicine, Stanford, California) and dfmr1 stocks (dfmr12/TM6 and p(dfmr1)w+/+;dfmr13/TM6) from Thomas Dockendorff (University of Tennessee, Knoxville, Tennessee). We would also like to thank members of the Broadie Lab, especially Dr. Charles Tessier, R. Lane Coffee and Saul Siller for insightful discussion during this study and critical feedback during manuscript preparation and Allison Gordon for continued interest and technical assistance at the project inception. This work was supported by a Postdoctoral Fellowship from the FRAXA Research Foundation to C.G. and by National Institutes of Health R01 grant MH084989 to K.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest The authors declare that the research presented was completed in the absence of any commercial or financial relationships that could be possibly construed as a potential conflict of interest.
References
- Awasaki T, Ito K. Engulfing action of glial cells is required for programmed axon pruning during Drosophila metamorphosis. Curr Biol. 2004;14:668–677. doi: 10.1016/j.cub.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Awasaki T, Tatsumi R, Takahashi K, Arai K, Nakanishi Y, Ueda R, Ito K. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron. 2006;50:855–867. doi: 10.1016/j.neuron.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Baker JD, McNabb SL, Truman JW. The hormonal coordination of behavior and physiology at adult ecdysis in Drosophila melanogaster. J Exp Biol. 1999;202:3037–3048. doi: 10.1242/jeb.202.21.3037. [DOI] [PubMed] [Google Scholar]
- Bakker C, Verheij C, Willemsen R, van der Helm R. Fmr1 knockout mice: a model to study fragile X mental retardation. The Dutch-Belgian Fragile X Consortium. Cell. 1994;78:23–33. [PubMed] [Google Scholar]
- Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Gertler FB. Ena/VASP: towards resolving a pointed controversy at the barbed end. Journal of cell science. 2009;122:1947–1953. doi: 10.1242/jcs.038125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear MF. Therapeutic implications of the mGluR theory of fragile X mental retardation. Genes, brain, and behavior. 2005;4:393–398. doi: 10.1111/j.1601-183X.2005.00135.x. [DOI] [PubMed] [Google Scholar]
- Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends in neurosciences. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Berry-Kravis E, Ciurlionis R. Overexpression of fragile X gene (FMR-1) transcripts increases cAMP production in neural cells. Journal of neuroscience research. 1998;51:41–48. doi: 10.1002/(SICI)1097-4547(19980101)51:1<41::AID-JNR4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A, McMillan E, Wallace K, Tubon TC, Jr., Capowski EE, Svendsen CN. Normal Neurogenesis but Abnormal Gene Expression in Human Fragile X Cortical Progenitor Cells. Stem cells and development. 2008;17:107–117. doi: 10.1089/scd.2007.0073. [DOI] [PubMed] [Google Scholar]
- Bhogal B, Jongens TA. Fragile X syndrome and model organisms: identifying potential routes of therapeutic intervention. Disease models & mechanisms. 2010;3:693–700. doi: 10.1242/dmm.002006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittel DC, Kibiryeva N, Butler MG. Whole genome microarray analysis of gene expression in subjects with fragile X syndrome. Genet Med. 2007;9:464–472. doi: 10.1097/gim.0b013e3180ca9a9a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchardon E, Grima B, Klarsfeld A, Chelot E, Hardin PE, Preat T, Rouyer F. Defining the role of Drosophila lateral neurons in the control of circadian rhythms in motor activity and eclosion by targeted genetic ablation and PERIOD protein overexpression. The European journal of neuroscience. 2001;13:871–888. doi: 10.1046/j.0953-816x.2000.01450.x. [DOI] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Callan MA, Cabernard C, Heck J, Luois S, Doe CQ, Zarnescu DC. Fragile X protein controls neural stem cell proliferation in the Drosophila brain. Human molecular genetics. 2010;19:3068–3079. doi: 10.1093/hmg/ddq213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castren M. Differentiation of neuronal cells in fragile X syndrome. Cell cycle (Georgetown, Tex) 2006;5:1528–1530. doi: 10.4161/cc.5.14.3096. [DOI] [PubMed] [Google Scholar]
- Castren M, Tervonen T, Karkkainen V, Heinonen S, Castren E, Larsson K, Bakker CE, Oostra BA, Akerman K. Altered differentiation of neural stem cells in fragile X syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17834–17839. doi: 10.1073/pnas.0508995102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Yun SW, Seto J, Liu W, Toth M. The fragile X mental retardation protein binds and regulates a novel class of mRNAs containing U rich target sequences. Neuroscience. 2003;120:1005–1017. doi: 10.1016/s0306-4522(03)00406-8. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Lee G, Park JH. Programmed cell death mechanisms of identifiable peptidergic neurons in Drosophila melanogaster. Development (Cambridge, England) 2006;133:2223–2232. doi: 10.1242/dev.02376. [DOI] [PubMed] [Google Scholar]
- Chung BY, Kilman VL, Keath JR, Pitman JL, Allada R. The GABA(A) receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr Biol. 2009;19:386–390. doi: 10.1016/j.cub.2009.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffee RL, Jr., Tessier CR, Woodruff EA, 3rd, Broadie K. Fragile X mental retardation protein has a unique, evolutionarily conserved neuronal function not shared with FXR1P or FXR2P. Disease models & mechanisms. 2010;3:471–485. doi: 10.1242/dmm.004598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Hulst C, Kooy RF. The GABAA receptor: a novel target for treatment of fragile X? Trends in neurosciences. 2007;30:425–431. doi: 10.1016/j.tins.2007.06.003. [DOI] [PubMed] [Google Scholar]
- D'Hulst C, Kooy RF. Fragile X syndrome: from molecular genetics to therapy. Journal of medical genetics. 2009;46:577–584. doi: 10.1136/jmg.2008.064667. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- De Rubeis S, Bagni C. Fragile X mental retardation protein control of neuronal mRNA metabolism: Insights into mRNA stability. Molecular and cellular neurosciences. 2010;43:43–50. doi: 10.1016/j.mcn.2009.09.013. [DOI] [PubMed] [Google Scholar]
- Dewey EM, McNabb SL, Ewer J, Kuo GR, Takanishi CL, Truman JW, Honegger HW. Identification of the gene encoding bursicon, an insect neuropeptide responsible for cuticle sclerotization and wing spreading. Curr Biol. 2004;14:1208–1213. doi: 10.1016/j.cub.2004.06.051. [DOI] [PubMed] [Google Scholar]
- Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Developmental cell. 2008;14:926–939. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockendorff TC, Su HS, McBride SM, Yang Z, Choi CH, Siwicki KK, Sehgal A, Jongens TA. Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron. 2002;34:973–984. doi: 10.1016/s0896-6273(02)00724-9. [DOI] [PubMed] [Google Scholar]
- Doherty J, Logan MA, Tasdemir OE, Freeman MR. Ensheathing glia function as phagocytes in the adult Drosophila brain. J Neurosci. 2009;29:4768–4781. doi: 10.1523/JNEUROSCI.5951-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draizen TA, Ewer J, Robinow S. Genetic and hormonal regulation of the death of peptidergic neurons in the Drosophila central nervous system. J Neurobiol. 1999;38:455–465. doi: 10.1002/(sici)1097-4695(199903)38:4<455::aid-neu2>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Drees F, Gertler FB. Ena/VASP: proteins at the tip of the nervous system. Current opinion in neurobiology. 2008;18:53–59. doi: 10.1016/j.conb.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein AM, Bauer CR, Ho A, Bosco G, Zarnescu DC. Drosophila Fragile X protein controls cellular proliferation by regulating cbl levels in the ovary. Developmental biology. 2009;330:83–92. doi: 10.1016/j.ydbio.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Estes PS, O'Shea M, Clasen S, Zarnescu DC. Fragile X protein controls the efficacy of mRNA transport in Drosophila neurons. Molecular and cellular neurosciences. 2008;39:170–179. doi: 10.1016/j.mcn.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Ewer J, Wang CM, Klukas KA, Mesce KA, Truman JW, Fahrbach SE. Programmed cell death of identified peptidergic neurons involved in ecdysis behavior in the Moth, Manduca sexta. J Neurobiol. 1998;37:265–280. doi: 10.1002/(sici)1097-4695(19981105)37:2<265::aid-neu6>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Fernandez MP, Berni J, Ceriani MF. Circadian remodeling of neuronal circuits involved in rhythmic behavior. PLoS biology. 2008;6:e69. doi: 10.1371/journal.pbio.0060069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala A, Spall T. In vivo calcium imaging of brain activity in Drosophila by transgenic cameleon expression. Sci STKE. 2003;2003:PL6. doi: 10.1126/stke.2003.174.pl6. [DOI] [PubMed] [Google Scholar]
- Franco DL, Rezaval C, Caceres A, Schinder AF, Ceriani MF. ENA/VASP downregulation triggers cell death by impairing axonal maintenance in hippocampal neurons. Molecular and cellular neurosciences. 2010;44:154–164. doi: 10.1016/j.mcn.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Gao FB, Brenman JE, Jan LY, Jan YN. Genes regulating dendritic outgrowth, branching, and routing in Drosophila. Genes & development. 1999;13:2549–2561. doi: 10.1101/gad.13.19.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber KB, Visootsak J, Warren ST. Fragile X syndrome. Eur J Hum Genet. 2008;16:666–672. doi: 10.1038/ejhg.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto CL, Broadie K. Temporal requirements of the fragile X mental retardation protein in the regulation of synaptic structure. Development (Cambridge, England) 2008;135:2637–2648. doi: 10.1242/dev.022244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto CL, Broadie K. Temporal requirements of the fragile x mental retardation protein in modulating circadian clock circuit synaptic architecture. Frontiers in neural circuits. 2009a;3:8. doi: 10.3389/neuro.04.008.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto CL, Broadie K. The fragile x mental retardation protein in circadian rhythmicity and memory consolidation. Molecular neurobiology. 2009b;39:107–129. doi: 10.1007/s12035-009-8057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal L, McCall K, Agapite J, Hartwieg E, Steller H. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. The EMBO journal. 2000;19:589–597. doi: 10.1093/emboj/19.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B, Chelot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Gross C, Nakamoto M, Yao X, Chan CB, Yim SY, Ye K, Warren ST, Bassell GJ. Excess phosphoinositide 3-kinase subunit synthesis and activity as a novel therapeutic target in fragile X syndrome. J Neurosci. 2010;30:10624–10638. doi: 10.1523/JNEUROSCI.0402-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman PJ. The fragile X prevalence paradox. Journal of medical genetics. 2008;45:498–499. doi: 10.1136/jmg.2008.059055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaka Y, Rieger D, Parmentier ML, Grau Y, Helfrich-Forster C, Nassel DR. Glutamate and its metabotropic receptor in Drosophila clock neuron circuits. The Journal of comparative neurology. 2007;505:32–45. doi: 10.1002/cne.21471. [DOI] [PubMed] [Google Scholar]
- Hamasaka Y, Wegener C, Nassel DR. GABA modulates Drosophila circadian clock neurons via GABAB receptors and decreases in calcium. J Neurobiol. 2005;65:225–240. doi: 10.1002/neu.20184. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C. The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:612–616. doi: 10.1073/pnas.92.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster C. Development of pigment-dispersing hormone-immunoreactive neurons in the nervous system of Drosophila melanogaster. The Journal of comparative neurology. 1997;380:335–354. doi: 10.1002/(sici)1096-9861(19970414)380:3<335::aid-cne4>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C. Neurobiology of the fruit fly's circadian clock. Genes, brain, and behavior. 2005;4:65–76. doi: 10.1111/j.1601-183X.2004.00092.x. [DOI] [PubMed] [Google Scholar]
- Hessl D, Rivera SM, Reiss AL. The neuroanatomy and neuroendocrinology of fragile X syndrome. Mental retardation and developmental disabilities research reviews. 2004;10:17–24. doi: 10.1002/mrdd.20004. [DOI] [PubMed] [Google Scholar]
- Hinton VJ, Brown WT, Wisniewski K, Rudelli RD. Analysis of neocortex in three males with the fragile X syndrome. Am J Med Genet. 1991;41:289–294. doi: 10.1002/ajmg.1320410306. [DOI] [PubMed] [Google Scholar]
- Honegger HW, Dewey EM, Ewer J. Bursicon, the tanning hormone of insects: recent advances following the discovery of its molecular identity. Journal of comparative physiology. 2008;194:989–1005. doi: 10.1007/s00359-008-0386-3. [DOI] [PubMed] [Google Scholar]
- Huang Y, Howlett E, Stern M, Jackson FR. Altered LARK expression perturbs development and physiology of the Drosophila PDF clock neurons. Molecular and cellular neurosciences. 2009;41:196–205. doi: 10.1016/j.mcn.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun S, Lee Y, Hong ST, Bang S, Paik D, Kang J, Shin J, Lee J, Jeon K, Hwang S, Bae E, Kim J. Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron. 2005;48:267–278. doi: 10.1016/j.neuron.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Inoue S, Shimoda M, Nishinokubi I, Siomi MC, Okamura M, Nakamura A, Kobayashi S, Ishida N, Siomi H. A role for the Drosophila fragile X-related gene in circadian output. Curr Biol. 2002;12:1331–1335. doi: 10.1016/s0960-9822(02)01036-9. [DOI] [PubMed] [Google Scholar]
- Irwin SA, Patel B, Idupulapati M, Harris JB, Crisostomo RA, Larsen BP, Kooy F, Willems PJ, Cras P, Kozlowski PB, Swain RA, Weiler IJ, Greenough WT. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am J Med Genet. 2001;98:161–167. doi: 10.1002/1096-8628(20010115)98:2<161::aid-ajmg1025>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Kao DI, Aldridge GM, Weiler IJ, Greenough WT. Altered mRNA transport, docking, and protein translation in neurons lacking fragile X mental retardation protein. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15601–15606. doi: 10.1073/pnas.1010564107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Awasaki T, Ito K. Neuronal programmed cell death induces glial cell division in the adult Drosophila brain. Development (Cambridge, England) 2009;136:51–59. doi: 10.1242/dev.023366. [DOI] [PubMed] [Google Scholar]
- Kelley DJ, Davidson RJ, Elliott JL, Lahvis GP, Yin JC, Bhattacharyya A. The cyclic AMP cascade is altered in the fragile X nervous system. PLoS ONE. 2007;2:e931. doi: 10.1371/journal.pone.0000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Kodama A, Hayasaka Y, Ohta T. Activation of the cAMP/PKA signaling pathway is required for post-ecdysial cell death in wing epidermal cells of Drosophila melanogaster. Development (Cambridge, England) 2004;131:1597–1606. doi: 10.1242/dev.01049. [DOI] [PubMed] [Google Scholar]
- Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annual review of cell and developmental biology. 2003;19:541–564. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- Kurant E, Axelrod S, Leaman D, Gaul U. Six-microns-under acts upstream of Draper in the glial phagocytosis of apoptotic neurons. Cell. 2008;133:498–509. doi: 10.1016/j.cell.2008.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachiewicz AM, Dawson DV. Do young boys with fragile X syndrome have macroorchidism? Pediatrics. 1994;93:992–995. [PubMed] [Google Scholar]
- Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Human molecular genetics. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- Lear BC, Merrill CE, Lin JM, Schroeder A, Zhang L, Allada R. A G protein-coupled receptor, groom-of-PDF, is required for PDF neuron action in circadian behavior. Neuron. 2005;48:221–227. doi: 10.1016/j.neuron.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Lee A, Li W, Xu K, Bogert BA, Su K, Gao FB. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development (Cambridge, England) 2003;130:5543–5552. doi: 10.1242/dev.00792. [DOI] [PubMed] [Google Scholar]
- Li W, Li Y, Gao FB. Abelson, enabled, and p120 catenin exert distinct effects on dendritic morphogenesis in Drosophila. Dev Dyn. 2005;234:512–522. doi: 10.1002/dvdy.20496. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Ku L, Wilkinson KD, Warren ST, Feng Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic acids research. 2001;29:2276–2283. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L, Park SK, Xu T, Vanderklish P, Yates JR., 3rd Quantitative proteomic analysis of primary neurons reveals diverse changes in synaptic protein content in fmr1 knockout mice. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15281–15286. doi: 10.1073/pnas.0804678105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Stormo GD, Taghert PH. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J Neurosci. 2004;24:7951–7957. doi: 10.1523/JNEUROSCI.2370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveall BJ, Deitcher DL. The essential role of bursicon during Drosophila development. BMC developmental biology. 2010;10:92. doi: 10.1186/1471-213X-10-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Wang H, Liang Z, Ku L, O'Donnell W T, Li W, Warren ST, Feng Y. The fragile X protein controls microtubule-associated protein 1B translation and microtubule stability in brain neuron development. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15201–15206. doi: 10.1073/pnas.0404995101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H, Lemon WC, Peabody NC, Pohl JB, Zelensky PK, Wang D, Nitabach MN, Holmes TC, White BH. Functional dissection of a neuronal network required for cuticle tanning and wing expansion in Drosophila. J Neurosci. 2006;26:573–584. doi: 10.1523/JNEUROSCI.3916-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo CW, Dewey EM, Sudo S, Ewer J, Hsu SY, Honegger HW, Hsueh AJ. Bursicon, the insect cuticle-hardening hormone, is a heterodimeric cystine knot protein that activates G protein-coupled receptor LGR2. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2820–2825. doi: 10.1073/pnas.0409916102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Shan G, Guo W, Smrt RD, Johnson EB, Li X, Pfeiffer RL, Szulwach KE, Duan R, Barkho BZ, Li W, Liu C, Jin P, Zhao X. Fragile x mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS genetics. 2010;6:e1000898. doi: 10.1371/journal.pgen.1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald JM, Beach MG, Porpiglia E, Sheehan AE, Watts RJ, Freeman MR. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron. 2006;50:869–881. doi: 10.1016/j.neuron.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Mao T, O'Connor DH, Scheuss V, Nakai J, Svoboda K. Characterization and subcellular targeting of GCaMP-type genetically-encoded calcium indicators. PLoS ONE. 2008;3:e1796. doi: 10.1371/journal.pone.0001796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil GP, Zhang X, Genova G, Jackson FR. A molecular rhythm mediating circadian clock output in Drosophila. Neuron. 1998;20:297–303. doi: 10.1016/s0896-6273(00)80457-2. [DOI] [PubMed] [Google Scholar]
- Mercaldo V, Descalzi G, Zhuo M. Fragile X mental retardation protein in learning-related synaptic plasticity. Molecules and cells. 2009;28:501–507. doi: 10.1007/s10059-009-0193-x. [DOI] [PubMed] [Google Scholar]
- Mertens I, Vandingenen A, Johnson EC, Shafer OT, Li W, Trigg JS, De Loof A, Schoofs L, Taghert PH. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron. 2005;48:213–219. doi: 10.1016/j.neuron.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Mientjes EJ, Nieuwenhuizen I, Kirkpatrick L, Zu T, Hoogeveen-Westerveld M, Severijnen L, Rife M, Willemsen R, Nelson DL, Oostra BA. The generation of a conditional Fmr1 knock out mouse model to study Fmrp function in vivo. Neurobiology of disease. 2006;21:549–555. doi: 10.1016/j.nbd.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Miyashiro KY, Beckel-Mitchener A, Purk TP, Becker KG, Barret T, Liu L, Carbonetto S, Weiler IJ, Greenough WT, Eberwine J. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Morales J, Hiesinger PR, Schroeder AJ, Kume K, Verstreken P, Jackson FR, Nelson DL, Hassan BA. Drosophila fragile X protein, DFXR, regulates neuronal morphology and function in the brain. Neuron. 2002;34:961–972. doi: 10.1016/s0896-6273(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Nakai J, Ohkura M, Imoto K. A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nature biotechnology. 2001;19:137–141. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- Nielsen KB, Tommerup N, Dyggve HV, Schou C. Macroorchidism and fragile X in mentally retarded males. Clinical, cytogenetic, and some hormonal investigations in mentally retarded males, including two with the fragile site at Xq28, fra(X)(q28) Human genetics. 1982;61:113–117. doi: 10.1007/BF00274199. [DOI] [PubMed] [Google Scholar]
- Orme M, Meier P. Inhibitor of apoptosis proteins in Drosophila: gatekeepers of death. Apoptosis. 2009;14:950–960. doi: 10.1007/s10495-009-0358-2. [DOI] [PubMed] [Google Scholar]
- Pacey LK, Doering LC. Developmental expression of FMRP in the astrocyte lineage: implications for fragile X syndrome. Glia. 2007;55:1601–1609. doi: 10.1002/glia.20573. [DOI] [PubMed] [Google Scholar]
- Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJ, Kang K, Liu X, Garrity PA, Rosbash M, Griffith LC. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron. 2008;60:672–682. doi: 10.1016/j.neuron.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Schroeder AJ, Helfrich-Forster C, Jackson FR, Ewer J. Targeted ablation of CCAP neuropeptide-containing neurons of Drosophila causes specific defects in execution and circadian timing of ecdysis behavior. Development (Cambridge, England) 2003;130:2645–2656. doi: 10.1242/dev.00503. [DOI] [PubMed] [Google Scholar]
- Peabody NC, Diao F, Luan H, Wang H, Dewey EM, Honegger HW, White BH. Bursicon functions within the Drosophila CNS to modulate wing expansion behavior, hormone secretion, and cell death. J Neurosci. 2008;28:14379–14391. doi: 10.1523/JNEUROSCI.2842-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penagarikano O, Mulle JG, Warren ST. The pathophysiology of fragile x syndrome. Annual review of genomics and human genetics. 2007;8:109–129. doi: 10.1146/annurev.genom.8.080706.092249. [DOI] [PubMed] [Google Scholar]
- Pfeiffer BE, Huber KM. The state of synapses in fragile X syndrome. Neuroscientist. 2009;15:549–567. doi: 10.1177/1073858409333075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips SE, Woodruff EA, 3rd, Liang P, Patten M, Broadie K. Neuronal loss of Drosophila NPC1a causes cholesterol aggregation and age-progressive neurodegeneration. J Neurosci. 2008;28:6569–6582. doi: 10.1523/JNEUROSCI.5529-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pologruto TA, Yasuda R, Svoboda K. Monitoring neural activity and [Ca2+] with genetically encoded Ca2+ indicators. J Neurosci. 2004;24:9572–9579. doi: 10.1523/JNEUROSCI.2854-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin M, Kang J, Burlin TV, Jiang C, Smith CB. Postadolescent changes in regional cerebral protein synthesis: an in vivo study in the FMR1 null mouse. J Neurosci. 2005;25:5087–5095. doi: 10.1523/JNEUROSCI.0093-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve SP, Bassetto L, Genova GK, Kleyner Y, Leyssen M, Jackson FR, Hassan BA. The Drosophila fragile X mental retardation protein controls actin dynamics by directly regulating profilin in the brain. Curr Biol. 2005;15:1156–1163. doi: 10.1016/j.cub.2005.05.050. [DOI] [PubMed] [Google Scholar]
- Reeve SP, Lin X, Sahin BH, Jiang F, Yao A, Liu Z, Zhi H, Broadie K, Li W, Giangrande A, Hassan BA, Zhang YQ. Mutational analysis establishes a critical role for the N terminus of fragile X mental retardation protein FMRP. J Neurosci. 2008;28:3221–3226. doi: 10.1523/JNEUROSCI.5528-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss AL, Hall SS. Fragile X syndrome: assessment and treatment implications. Child and adolescent psychiatric clinics of North America. 2007;16:663–675. doi: 10.1016/j.chc.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Rezaval C, Berni J, Gorostiza EA, Werbajh S, Fagilde MM, Fernandez MP, Beckwith EJ, Aranovich EJ, Sabio y Garcia CA, Ceriani MF. A functional misexpression screen uncovers a role for enabled in progressive neurodegeneration. PLoS ONE. 2008;3:e3332. doi: 10.1371/journal.pone.0003332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchi M, Archidiacono N, Filippi G. X-linked mental retardation. I. Martin-Bell syndrome (report of 18 families) Journal de genetique humaine. 1987;35:351–379. [PubMed] [Google Scholar]
- Rudelli RD, Brown WT, Wisniewski K, Jenkins EC, Laure-Kamionowska M, Connell F, Wisniewski HM. Adult fragile X syndrome. Clinico-neuropathologic findings. Acta Neuropathol. 1985;67:289–295. doi: 10.1007/BF00687814. [DOI] [PubMed] [Google Scholar]
- Sekine T, Yamaguchi T, Hamano K, Siomi H, Saez L, Ishida N, Shimoda M. Circadian phenotypes of Drosophila fragile x mutants in alternative genetic backgrounds. Zoological science. 2008;25:561–571. doi: 10.2108/zsj.25.561. [DOI] [PubMed] [Google Scholar]
- Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, Taghert PH. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron. 2008;58:223–237. doi: 10.1016/j.neuron.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Taghert PH. RNA-interference knockdown of Drosophila pigment dispersing factor in neuronal subsets: the anatomical basis of a neuropeptide's circadian functions. PLoS ONE. 2009;4:e8298. doi: 10.1371/journal.pone.0008298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Sharma VK, Gu H, Chou YT, O'Dowd DK, Holmes TC. Pigment dispersing factor-dependent and -independent circadian locomotor behavioral rhythms. J Neurosci. 2008;28:217–227. doi: 10.1523/JNEUROSCI.4087-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofola O, Sundram V, Ng F, Kleyner Y, Morales J, Botas J, Jackson FR, Nelson DL. The Drosophila FMRP and LARK RNA-binding proteins function together to regulate eye development and circadian behavior. J Neurosci. 2008;28:10200–10205. doi: 10.1523/JNEUROSCI.2786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steller H. Regulation of apoptosis in Drosophila. Cell death and differentiation. 2008;15:1132–1138. doi: 10.1038/cdd.2008.50. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- Sung YJ, Dolzhanskaya N, Nolin SL, Brown T, Currie JR, Denman RB. The fragile X mental retardation protein FMRP binds elongation factor 1A mRNA and negatively regulates its translation in vivo. The Journal of biological chemistry. 2003;278:15669–15678. doi: 10.1074/jbc.M211117200. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, Warren ST. DNA methylation represses FMR-1 transcription in fragile X syndrome. Human molecular genetics. 1992;1:397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- Tervonen TA, Louhivuori V, Sun X, Hokkanen ME, Kratochwil CF, Zebryk P, Castren E, Castren ML. Aberrant differentiation of glutamatergic cells in neocortex of mouse model for fragile X syndrome. Neurobiology of disease. 2009;33:250–259. doi: 10.1016/j.nbd.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Tessier CR, Broadie K. Drosophila fragile X mental retardation protein developmentally regulates activity-dependent axon pruning. Development (Cambridge, England) 2008;135:1547–1557. doi: 10.1242/dev.015867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier CR, Broadie K. Activity-dependent modulation of neural circuit synaptic connectivity. Frontiers in molecular neuroscience. 2009;2:8. doi: 10.3389/neuro.02.008.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier CR, Broadie K. The fragile X mental retardation protein developmentally regulates the strength and fidelity of calcium signaling in Drosophila mushroom body neurons. Neurobiology of disease. 2011;41:147–159. doi: 10.1016/j.nbd.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, Bargmann CI, Jayaraman V, Svoboda K, Looger LL. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nature methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, Eussen BE, van Ommen G-JB, Blonden LAJ, Riggins GJ, Chastain JL, Kunst CB, Galjaard H, Caskey CT, Nelson DL, Oostra BA, Warren ST. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Wang H, Ku L, Osterhout DJ, Li W, Ahmadian A, Liang Z, Feng Y. Developmentally-programmed FMRP expression in oligodendrocytes: a potential role of FMRP in regulating translation in oligodendroglia progenitors. Human molecular genetics. 2004;13:79–89. doi: 10.1093/hmg/ddh009. [DOI] [PubMed] [Google Scholar]
- Wang SL, Hawkins CJ, Yoo SJ, Muller HA, Hay BA. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- Watts RJ, Schuldiner O, Perrino J, Larsen C, Luo L. Glia engulf degenerating axons during developmental axon pruning. Curr Biol. 2004;14:678–684. doi: 10.1016/j.cub.2004.03.035. [DOI] [PubMed] [Google Scholar]
- Wisniewski KE, Segan SM, Miezejeski CM, Sersen EA, Rudelli RD. The Fra(X) syndrome: neurological, electrophysiological, and neuropathological abnormalities. Am J Med Genet. 1991;38:476–480. doi: 10.1002/ajmg.1320380267. [DOI] [PubMed] [Google Scholar]
- Woodruff EA, 3rd, Broadie K, Honegger HW. Two peptide transmitters co-packaged in a single neurosecretory vesicle. Peptides. 2008;29:2276–2280. doi: 10.1016/j.peptides.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Duan R, Chen D, Wang J, Chen D, Jin P. Fragile X mental retardation protein modulates the fate of germline stem cells in Drosophila. Human molecular genetics. 2007;16:1814–1820. doi: 10.1093/hmg/ddm129. [DOI] [PubMed] [Google Scholar]
- Yang Y, Xu S, Xia L, Wang J, Wen S, Jin P, Chen D. The bantam microRNA is associated with drosophila fragile X mental retardation protein and regulates the fate of germline stem cells. PLoS genetics. 2009;5:e1000444. doi: 10.1371/journal.pgen.1000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SJ, Huh JR, Muro I, Yu H, Wang L, Wang SL, Feldman RM, Clem RJ, Muller HA, Hay BA. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nature cell biology. 2002;4:416–424. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Wulbeck C, Sehadova H, Veleri S, Bichler D, Stanewsky R, Helfrich-Forster C. The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila's clock. J Neurosci. 2009;29:2597–2610. doi: 10.1523/JNEUROSCI.5439-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa F, Adinolfi S, Napoli I, Kuhn-Holsken E, Urlaub H, Achsel T, Pastore A, Bagni C. Fragile X mental retardation protein (FMRP) binds specifically to the brain cytoplasmic RNAs BC1/BC200 via a novel RNA-binding motif. The Journal of biological chemistry. 2005;280:33403–33410. doi: 10.1074/jbc.M504286200. [DOI] [PubMed] [Google Scholar]
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- Zarnescu DC, Jin P, Betschinger J, Nakamoto M, Wang Y, Dockendorff TC, Feng Y, Jongens TA, Sisson JC, Knoblich JA, Warren ST, Moses K. Fragile X protein functions with lgl and the par complex in flies and mice. Developmental cell. 2005;8:43–52. doi: 10.1016/j.devcel.2004.10.020. [DOI] [PubMed] [Google Scholar]
- Zhang M, Wang Q, Huang Y. Fragile X mental retardation protein FMRP and the RNA export factor NXF2 associate with and destabilize Nxf1 mRNA in neuronal cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:10057–10062. doi: 10.1073/pnas.0700169104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, McNeil GP, Hilderbrand-Chae MJ, Franklin TM, Schroeder AJ, Jackson FR. Circadian regulation of the lark RNA-binding protein within identifiable neurosecretory cells. J Neurobiol. 2000;45:14–29. doi: 10.1002/1097-4695(200010)45:1<14::aid-neu2>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD, Rubin GM, Broadie K. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Friedman DB, Wang Z, Woodruff E, 3rd, Pan L, O'Donnell J, Broadie K. Protein expression profiling of the drosophila fragile X mutant brain reveals up-regulation of monoamine synthesis. Mol Cell Proteomics. 2005;4:278–290. doi: 10.1074/mcp.M400174-MCP200. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Rodesch CK, Broadie K. Living synaptic vesicle marker: synaptotagmin-GFP. Genesis. 2002;34:142–145. doi: 10.1002/gene.10144. [DOI] [PubMed] [Google Scholar]
- Zhao T, Gu T, Rice HC, McAdams KL, Roark KM, Lawson K, Gauthier SA, Reagan KL, Hewes RS. A Drosophila gain-of-function screen for candidate genes involved in steroid-dependent neuroendocrine cell remodeling. Genetics. 2008;178:883–901. doi: 10.1534/genetics.107.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou K, Liu J, Zhu N, Lin J, Liang Q, Brown WT, Shen Y, Zhong N. Identification of FMRP-associated mRNAs using yeast three-hybrid system. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:769–777. doi: 10.1002/ajmg.b.30678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative images are presented of PDF-TRI neurons in the central brain of a newly eclosed wild-type animal double-labeled with Pdf-Gal4 driven UAS-synaptotagmin-GFP (GFP, green; left panel) and immunocytochemistry against the PDF neuropeptide (red; right panel). Output synapses are shown in the ventral brain tritocerebral region (trito), where the PDF-TRI neurons originate, and the dorsal brain protocerebral region (proto), to which the PDF-TRI neurons project. Relative positioning of the sLNv and PDF-TRI axonal arbors are indicated. Comparison of the panels at day 0 (d0) post-eclosion to the d5 mature adult brain (inset) demonstrates the absence of PDF-TRI synapses in the wild-type brain at maturity.
The entire adult nervous system (brain, thoracic ganglia and abdominal ganglion) of a newlyeclosed wildtype (w1118) animal double-labeled for CCAP (green) and bursicon β-subunit (pburs; red). The locations of developmentally transient CCAP and bursicon peptidergic neurons are shown. OL: brain optic lobe, SEG: brain subesophageal ganglion, T1–T3: three thoracic ganglia, Ab: abdominal ganglion.
Representative images highlight the terminal thoracic (T3) and abdominal ganglia double-labeled with anti-bursicon (green) and anti-pburs (red) in controls (w1118) and dfmr150M null mutants. A) A control at 1 hour post-elcosion (top row) demonstrates the positioning and process elaboration of bursicon peptidergic neurons. The bottom row shows all bursicon neurons have been eliminated by 24 hours post-eclosion in the wild-type animal. B) Comparable images from the dfmr1 null brain at 1 hour post-elcosion (top row) show a bursicon neuron array indistinguishable from control. Processes from bursicon neurons are aberrantly retained at 24 hours in the dfmr1 null mutant (middle row), although fully eliminated by 48 hours (bottom row).
The transgenic fluorescent calcium reporter UAS-GCaMP was driven by Pdf-Gal4 and imaged in the PDF-TRI neurons to assay functionality. After an initial baseline fluorescence measurement for 1 minute, neurons were acutely depolarized with 70 mM KCl while continuously monitoring fluorescence intensity. A control from late pupal day 4 (P4) is shown over 140 frames spanning 109.6 seconds, with a frame rate of 15 frames per second. The video is pseudocolored for fluorescence intensity.
The transgenic fluorescent calcium reporter UAS-GCaMP was driven by Pdf-Gal4 and imaged in the PDF-TRI neurons to assay functionality. After initial baseline measurement for 60 seconds, neurons were acutely depolarized with 70 mM KCl while continuously monitoring fluorescence intensity. A dfmr150M null PDF-TRI neuron at late P4 is shown over 182 frames spanning 142.7 seconds, with a frame rate of 15 frames per second. The video is pseudocolored for fluorescence intensity.
The transgenic fluorescent calcium reporter UAS-GCaMP was driven by Pdf-Gal4 and imaged in the PDF-TRI neurons to assay functionality. After initial baseline measurement for 1 minute, neurons were acutely depolarized with 70 mM KCl while continuously monitoring fluorescence intensity. A null dfmr150M PDF-TRI neuron at post-eclosion day 3 (d3) is shown over 184 frames spanning 144.3 seconds, with a frame rate of 15 frames per second. The video is pseudocolored for fluorescence intensity.