Abstract
RNase P is a ribonucleoprotein (RNP) complex that utilizes a Mg2+-dependent RNA catalyst to cleave the 5′-leader of precursor tRNAs (pre-tRNAs) and generate mature tRNAs. The bacterial RNase P protein (RPP) aids RNase P RNA (RPR) catalysis by promoting substrate binding, Mg2+ coordination, and product release. Archaeal RNase P comprises an RPR and at least four RPPs, which have eukaryal homologs and function as two binary complexes (POP5•RPP30 and RPP21•RPP29). In this study, we employed a previously characterized substrate-enzyme conjugate [pre-tRNATyr-Methanocaldococcus jannaschii (Mja) RPR] to investigate the functional role of a universally conserved uridine in a bulge-helix structure in archaeal RPRs. Deletion of this bulged uridine resulted in an 80-fold decrease in the self-cleavage rate of pre-tRNATyr-MjaΔU RPR compared to the wildtype, and this defect was partially ameliorated upon addition of either RPP pair. The catalytic defect in the archaeal mutant RPR mirrors that reported in a bacterial RPR and highlights a parallel in their active sites. Furthermore, an N-terminal deletion mutant of Pyrococcus furiosus (Pfu) RPP29 that is defective in assembling with its binary partner RPP21, as assessed by isothermal titration calorimetry and NMR spectroscopy, is functional when reconstituted with the cognate Pfu RPR. Collectively, these results indicate that archaeal RPPs are able to compensate for structural defects in their cognate RPR and vice-versa, and provide striking examples of the cooperative subunit interactions critical for driving archaeal RNase P towards its functional conformation. (236 words)
Keywords: pre-tRNA processing, in vitro reconstitution, mutational rescue
INTRODUCTION
Key cellular processes such as RNA processing, splicing and translation are catalyzed by large ribonucleoprotein (RNP) complexes. Mapping the assembly pathways of these RNPs from multiple RNA and protein subunits and delineating the functional contributions of individual subunits remain challenging problems. RNase P, a catalytic RNP vital for tRNA biogenesis, presents tractable prospects for studying the structural and functional cooperation among the subunits of an RNP. In all domains of life, tRNAs are typically made as precursors with additional nucleotides at their 5′ termini. The endonucleolytic action of RNase P excises these 5′-leaders from precursor tRNAs (pre-tRNAs).1–4 With the exception of some organellar variants,5,6 RNase P typically functions as an RNP,1–4 albeit with differences in subunit make-up: while all these RNPs contain a catalytic RNase P RNA (RPR), there is one RNase P protein (RPP) subunit in bacteria, and at least four and nine subunits in archaea and eukarya (nuclear), respectively. Archaeal and eukaryal RPPs are related, but none of them shares any sequence homology with the bacterial RPP.7 Although RPRs (without RPPs) can cleave pre-tRNAs, their cleavage rates vary by ~106 fold (bacterial > archaeal > eukaryal RPR).8–10 The weaker activity of the archaeal/eukaryal RPRs, compared to their bacterial counterpart, together with the reverse trend in protein:RNA molecular mass ratios of RNase P (eukaryal > archaeal > bacterial), suggests a more acute dependence of the archaeal/eukaryal RPRs on their multiple protein cofactors.
Archaeal RNase P serves as an experimental alternative to its eukaryotic cousin,11 which has proven difficult to assemble in vitro despite the availability of constituent subunits in recombinant form, and as a paradigm to uncover the coordination among multiple proteins that aid an RNA catalyst. The latter objective has been assisted by recent advances in functional reconstitution of archaeal RNase P12–17 and elucidation of the high-resolution structures12,18–26 of the RPPs. Our reconstitution studies revealed that the four archaeal RPPs function as two binary RPP complexes (POP5•RPP30 and RPP21•RPP29), which have large effects on the RPR’s catalytic efficiency [e.g., a 4250-fold increase in kcat/KM in Pyrococcus furiosus (Pfu) RNase P].17 Subsequent kinetic studies demonstrated a principal role for POP5•RPP30 in enhancing the RPR’s rate of pre-tRNA cleavage and RPP21•RPP29 in increasing substrate affinity.13,16 The RPP structures were solved both individually and as binary complexes.12,18–26 These structures fall within established nucleic acid binding protein families: an RRM-like fold (POP5), a TIM barrel (RPP30), a zinc ribbon (RPP21) and an Sm-like fold (RPP29). The POP5•RPP30 and RPP21•RPP29 heterodimer structures reveal protein-protein binding interfaces and furnish clues as to possible RNA-binding sites. Collectively, these findings provide an ideal platform for uncovering structure-function relationships in a multi-subunit RNP, a focus of our ongoing work.
The functional importance of universally-conserved nucleotides in RPRs is borne out by their presence even in pared-down, active versions of bacterial and archaeal RPRs.13,16,17,27,28 To elucidate similarities in the RNA-mediated catalytic mechanism in bacterial and archaeal RNase P, we investigated whether a universally conserved, bulged uridine shown to participate in binding catalytically important Mg2+ ions in bacterial RPRs is also vital in the archaeal relative. Indeed, we found a severe catalytic defect in an archaeal RPR in which this bulged uridine was deleted; however, this defect was partially rescued upon the addition of RPPs. During our structural studies of archaeal RPP29, we also unexpectedly identified an N-terminal deletion mutant of RPP29 which fails to bind its partner RPP21 [as judged by isothermal titration calorimetry (ITC) and NMR spectroscopy] but is functional upon addition of RPR. These findings collectively illustrate the reciprocal subunit interactions vital for driving archaeal RNase P towards its functional structure.
RESULTS
Mutation in the P4 helix of an archaeal RPR decreases cleavage activity
A conserved feature in all RPRs is a bulge-helix structure in the P4 paired region (Fig. 1). The geometry of this P4 bulge-helix structure in bacterial RPRs is important for RNA structure and Mg2+ association, which in turn are critical for pre-tRNA binding and cleavage.29 Disrupting this bulge-helix structure either by eliminating the bulged uridine (ΔU) or inserting one additional uridine in the bulge (+U) decreases activity. For example, deleting this bulged uridine (U69) in Escherichia coli (Eco) RPR results in a 100-fold lower single-turnover reaction rate compared to the wild-type (WT) RPR even at saturating (300 mM) concentrations of Mg2+.29 Eliminating the bulge in the Eco RPR weakened the apparent affinity for Mg2+ and reduced the cooperativity for Mg2+ (the Hill coefficient, nH, decreased from 2.2 to 1.5).29 Replacement of non-bridging oxygen atoms with sulfur at positions proximal to U69 in Eco RPR (e.g., A67) lowered the cleavage rate by three to four orders of magnitude, with the deleterious effect in some instances largely rescued by thiophilic metal ions such as Mn2+;30,31 similar results were reported with Bacillus subtilis (Bsu) RPR.32 Moving the bulge away from the nearby sites of metal ion coordination also decreased activity by 70-fold indicating that the position of the bulge in P4 is important.29 A recent crystal structure of the bacterial RNase P holoenzyme-tRNA complex33 sheds light on the putative role(s) of the universally conserved bulged uridine. Some attributes of the RNase P catalytic site were inferred when a pre-tRNA 5′-leader was soaked into a crystal of the Thermotoga maritima RNase P-pre-tRNAPhe complex, with and without Sm3+ ions (used as Mg2+ mimics). Notably, a metal ion, likely to generate the attacking hydroxide nucleophile, is held in position by interactions with the O4 oxygen of the uridine and its proximal phosphate backbone.
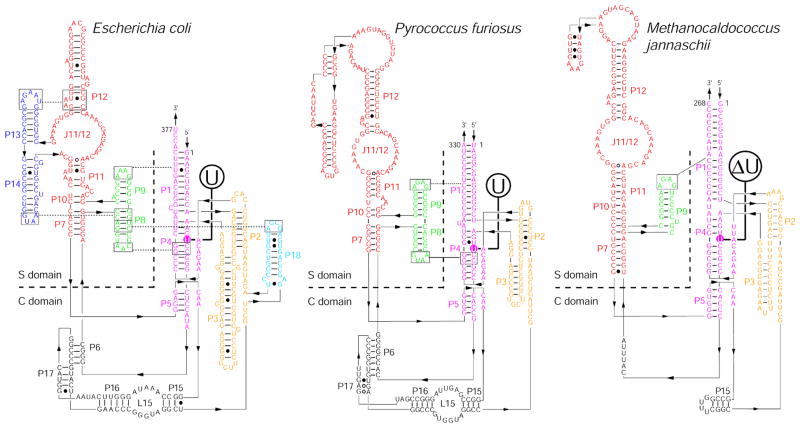
Figure 1.
Secondary structures of RPRs from Bacteria (e.g., Escherichia coli) and Archaea [e.g., Pyrococcus furiosus (Pfu; type A) and Methanocaldococcus jannaschii (Mja; type M)].27,75 The universally conserved bulged uridine in the P4 helix of all RPRs is enclosed in a colored circle. A deletion (ΔU) mutation made at this position in the Mja RPR is also indicated.
We tested the importance of the conserved bulge-helix structure in P4 of archaeal RPRs by deleting the bulge in the P4 helix (ΔU; Fig. 1). To focus solely on the chemical cleavage step, we introduced the ΔU mutation in a self-cleaving construct, pre-tRNATyr-S3-Mja RPR (hereafter referred as pre-tRNATyr-Mja RPR for simplicity). In this cis construct, the pre-tRNATyr substrate is tethered via a 3-nt spacer to Mja RPR to help overcome the type M RPR’s substrate-binding defects,10,16,34 which typically prevent pre-tRNA cleavage in trans. We previously showed this cis conjugate to be a good model for studying the chemical cleavage step [based on a slope of ~1 in plots of log(kobs) vs. pH].16 We found that the kobs for self-cleavage of pre-tRNATyr-MjaΔU RPR (at pH 6) is 0.0025 min−1, 80-fold lower than the wild type (WT) pre-tRNATyr-Mja RPR (Table 1 and Fig. 2; ref. 16).
Table 1.
Effect of Mja RPPs on the rate of cleavage of pre-tRNATyr-Mja (WT) and -Mja ΔU (MT) at 55°C
| Assayed under optimal conditions for each catalytic entity | [NH4+]; [Mg2+], M | kobs (WT), min−1* | Rel. kobs (WT) | kobs (MT), min−1** | Rel. kobs (MT) | kobs(WT)/kobs(MT) |
|---|---|---|---|---|---|---|
| pre-tRNATyr-Mja RPR (WT or ΔU MT) | 2.5; 0.5 | 0.2 ± 0.04 | 1 | 0.0025 ± 0.0002 | 1 | 80 |
| + RPP21•RPP29 | 0.8; 0.1 | 0.24 ± 0.04 | 1 | 0.017 ± 0.0014 | 7 | 14 |
| + POP5•RPP30 | 0.8; 0.1 | 20 ± 0.32 | 100 | 1.46 ± 0.12 | 584 | 14 |
| + Both binary RPP complexes | 0.8; 0.1 | 22 ± 0.16 | 105 | 1.84 ± 0.18* | 736 | 12 |
These rates at pH 6 were extrapolated from those determined at pH 5.4 and reported by us previously.16 We had also earlier established the direct relationship between log (kobs) versus pH.16
Experiments were performed at pH 6, except for pre-tRNATyr-Mja ΔU RPR reconstituted with both binary RPP complexes, which was assayed at pH 5.4. The rate at pH 6 for the reaction with four RPPs was obtained by extrapolation from 5.4. The standard errors of the curve fits shown in Figure 3 are indicated in the estimates of kobs.
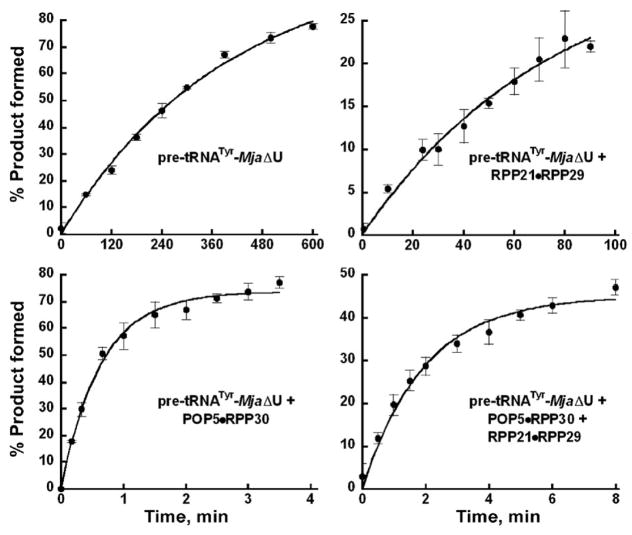
Figure 2.
Effects of Mja RPPs on the rate of pre-tRNATyr-MjaΔU RPR self-cleavage. By analyzing the time course for product formation, the kobs values for the reactions catalyzed by the RPR with and without RPPs were determined (see Table 1). The mean and standard deviation values were calculated from three independent experiments.
RPP21•RPP29 and POP5•RPP30 both partially rescue the decrease in cleavage rate caused by the ΔU mutation
We next proceeded to test the activity of the P4-mutated archaeal RPRs in the presence of their cognate RPPs since there is precedent for the bacterial RPP influencing substrate recognition, cleavage and metal ion coordination.32,35–42 For example, the absence or presence of the RPP caused differences in the ability of thiophilic metal ions to rescue the adverse effects caused by replacing different non-bridging oxygens with sulfur in the P4 helix of Bsu RPR.32 This observation taken together with subsequent studies37,41 supports the idea that the bacterial RPP increases the affinity of the RPR for catalytically relevant metal ions. Also, aberrant cleavage of select pre-tRNAs by the Eco ΔC92 RPR mutant was corrected by the Eco RPP, suggesting that the protein cofactor can alleviate RPR catalytic defects including altered substrate positioning.36
We had previously demonstrated that addition of Mja POP5•RPP30 enhanced kobs for self-cleavage of pre-tRNATyr-Mja RPR by ~100-fold while RPP21•RPP29 had no effect; however, both binary RPP complexes reduced the monovalent and divalent ionic requirement. It is important to note that these cis construct findings were mirrored in single-turnover, trans-cleavage studies with another archaeal (Methanothermobacter thermautotrophicus) RNase P.13 We sought to determine if Mja RPPs could rescue the MjaΔU RPR mutation. When RPP21•RPP29 was added to pre-tRNATyr-MjaΔU RPR (at pH 6), the rate increased by ~7-fold compared to the reaction in the absence of RPPs. Notably, the rate increased by 584-fold in the presence of POP5•RPP30 (Fig. 2; Table 1). When all four proteins were present, the rate is 736-fold higher compared to the RPR-alone reaction (Fig. 2; Table 1). However, the presence of all four RPPs was unable to raise the mutant RPR’s activity to that observed with the WT; the activity of the mutant holoenzyme is still 12-fold lower than the WT at 100 mM Mg2+ (Table 1). This difference is roughly halved at 500 mM Mg2+ (not shown).
Archaeal RPR-mediated rescue of structural defects in RPPs
The ability of archaeal RPPs to rescue an RPR mutant (Fig. 2) was mirrored in our adventitious finding that an N-terminal deletion mutant of Pyrococcus furiosus (Pfu) RPP29 which fails to bind RPP21 is nevertheless functional when assembled with the RPR. As described below, given the premise that the interaction between RPP29 and RPP21 is important for proper assembly of the holoenzyme, our observations suggest that the RPR rescues activity by overcoming the structural defect in the mutated protein.
We expect the findings in this report on Mja and Pfu RNase P to be broadly applicable to RNase P from various archaea. Due to technical reasons, we employed a different type of archaeal RNase P for the kinetic and biophysical studies. There are two broad classes of euryarchaeal RNase P: type A (Pfu) and M (Mja),34 and we have successfully reconstituted both holoenzymes.13,16 The type A RPRs are so called because of their resemblance to the bacterial type A (ancestral) RPRs; the type M RPRs (mostly from Methanococcales) resemble the eukaryal RPRs (Fig. 1). Due to the utility of the well-studied pre-tRNA-Mja RPR cis conjugate,16 we favored it for our kinetic analyses of the ΔU mutant, while the availability of structures of the Pfu (and not Mja) RPPs has led to use of Pfu RNase P as the preferred model for our ongoing biophysical studies.
NMR and ITC assays establish a role for the N-terminus of RPP29 in forming the RPP21•RPP29 binary complex
Limited trypsin proteolysis
To identify a deletion derivative of RPP29 that might be better suited than the full-length protein for structure determination of the RPP21•RPP29 complex, we first sought to identify a structured core in RPP29. Pfu RPP29 contains 127 residues, the first 35 of which are unique to the Pyrococcus family (Fig. 3). In the absence of RPP21, the first 47 residues of Pfu RPP29 gave no signals in the two-dimensional 1H-15N correlated NMR spectrum, suggesting that they might not be required for maintaining the core structure of Pfu RPP29. Limited trypsin proteolysis coupled with electrospray mass spectrometry (ESI-MS) revealed a trypsin-resistant core comprising residues 42–120 (data not shown) that was insensitive to further digestion after 40 min, while the remainder of RPP29 could be rapidly digested within 10 min. Since trypsin only cleaves after Arg/Lys residues and K52 is the next Arg/Lys residue after K41, K52 must be in a well-folded region, a premise confirmed by heteronuclear NMR spectra.25 The absence of putative trypsin cleavage sites between K41 and K52 prevented us from precisely defining the exact boundary of the unstructured N-terminus.
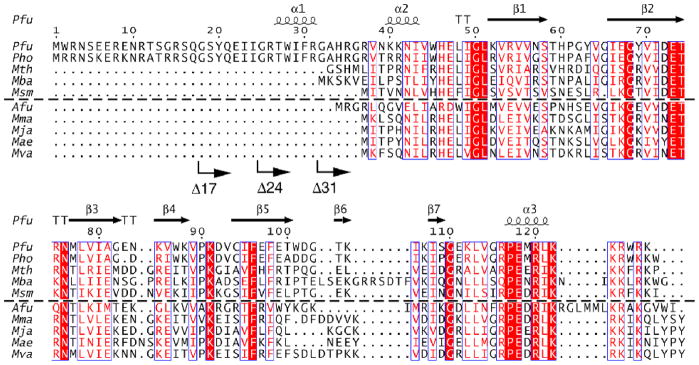
Figure 3.
Sequence alignment of select type A and M archaeal RPP29 homologs. The alignment was generated with CLUSTALW,76 and illustrated using ESPRIPT2.2 (http://espript.ibcp.fr/ESPript). Boxes indicate a global similarity score of 0.7, and block letters indicate invariant residues. Secondary structural elements represented in cartoon were observed in the NMR structure of the Pfu RPP21-RPP29 complex. Arrows indicate the sites of N-terminal deletions; for example, Δ17 has the first 17 residues deleted. Type A sequences are above the dashed line; type M below. Aligned sequences are from Pyrococcus furiosus (Pfu, NP_579545), Pyrococcus horikoshii (Pho, NP_143607), Methanothermobacter thermautotrophicus (Mth, 10QK_A), Methanosarcina barkeri (Mba, YP_303669), and Methanobrevibacter smithii (Msm, EFC93468), Archaeoglobus fulgidus (Afu, 1TSF_A), Methanococcus maripaludis (Mma, YP_001549331), Methanocaldococcus jannaschii (Mja, NP_247439), Methanococcus aeolicus (Mae, YP_001325580) and Methanococcus vanielli (Mva, YP_001323236).
Pfu RPP29 derivatives
In addition to the abovementioned findings from limited proteolysis of Pfu RPP29, the crystal structure of Pho RPP29 was solved by use of an N-terminal deletion mutant: RPP29Δ31, since crystals could not be obtained with the full-length protein.26 Based on these observations, two N-terminal deletion mutants of Pfu RPP29 were engineered, RPP29Δ42 and RPP29Δ36, in which the first 42 and 36 residues are removed, respectively. NMR spectra confirmed that the structured core is conserved in both mutants (data not shown), consistent with the solution structures of Mth and Afu RPP29.12,23 However, the addition of the unlabeled RPP21 to 15N-labeled Pfu RPP29 Δ36 and Δ42 variants did not induce significant spectral changes, indicating that deletion of the first 36 residues eliminated important binding determinants (data not shown). Thus, the interaction with Pfu RPP21 requires the N-terminal region of Pfu RPP29, despite the fact that these residues are missing in RPP29 from other archaea (Fig. 3). We then constructed five additional Pfu RPP29 mutants each with different length deletions at the N-terminus, namely RPP29Δ5, RPP29Δ13, RPP29Δ17, RPP29Δ24 and RPP29Δ31 (Fig. 3). RPP29Δ31 corresponds to the construct used for crystallization of Pho RPP29.26 This series of RPP29 derivatives also allowed us to explore more precisely the function of the N-terminus of RPP29. The ability of each mutant to bind RPP21 was examined by NMR spectroscopy and ITC, and the effect of these deletions on RNase P activity was tested in vitro.
NMR and ITC studies
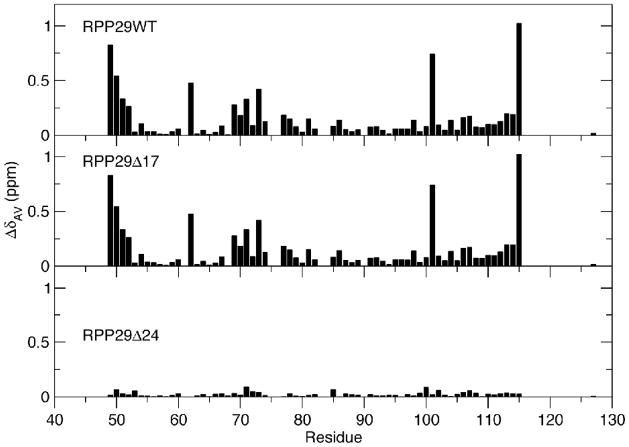
We used heteronuclear NMR chemical shift perturbations and ITC to examine the interactions between RPP21 and RPP29 deletion mutants. The extent and site of protein-protein interactions were mapped on the basis of changes in the positions of crosspeaks in two-dimensional 1H-15N-correlated spectra which result from changes in the environment sensed by individual backbone amides of one protein upon being bound by its partner.25 Using such chemical shift perturbations, we mapped the interactions when wild type RPP21 was bound to either RPP29 or one of its N-terminal mutant derivatives. As observed with RPP29Δ36 and RPP29Δ42, the five other RPP29 deletion mutants also adopt the same structured core, indicated by nearly identical 1H-15N NMR spectra (Figs. 4C and 4E, data shown for RPP29Δ17 and RPP29Δ24 only). Despite this conserved structure, among them only RPP29Δ5, RPP29Δ13 and RPP29Δ17 mutants resulted in dramatic spectral changes upon addition of unlabeled RPP21, while RPP29Δ24 and RPP29Δ31 did not (data shown for RPP29Δ17 in Figs. 4C and 4D, and for RPP29Δ24 in Figures 4E and 4F, as representatives of each group). The observation of small chemical shift perturbations in RPP29Δ24 suggests only weak binding to RPP21 (Fig. 4F and Fig. 5). The per-residue chemical shift perturbations are summarized in Figure 6 for RPP29 (WT), RPP29Δ17 or RPP29Δ24. Reverse titrations (i.e., unlabeled RPP29 deletion mutants titrated into U-[15N]-RPP21) were similarly consistent with tight RPP21 binding by RPP29 (WT) and RPP29Δ17 but not RPP29Δ24 (data not shown).
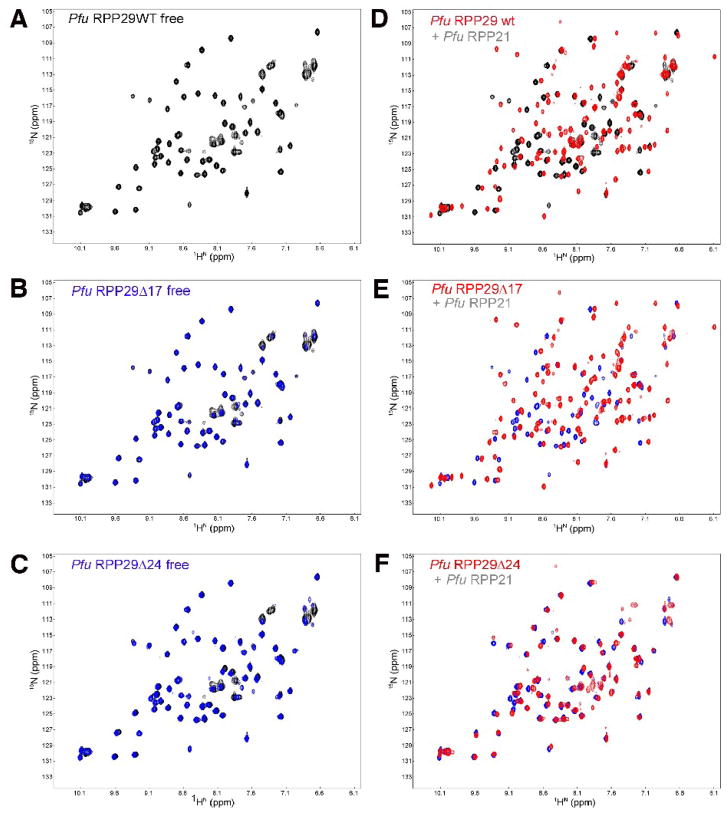
Figure 4.
NMR-based binding assay of Pfu RPP29 N-terminal deletion derivatives to RPP21. Both RPP29Δ17 (C) and RPP29Δ24 (E) adopt the same structured core as the wild type (A), indicated by the almost identical two-dimensional 1H-15N correlation spectra of the free 15N-labeled proteins. Titration of unlabeled RPP21 into 15N-labeled RPP29 induces dramatic spectral changes in the spectra of RPP29WT (B) and RPP29Δ17 (D), but not in the spectrum of RPP29Δ24 (F). In panels (C) and (E) the spectra of the free RPP29 derivative protein (blue) is overlaid on that of the free full-length protein (black). In panels (B), (D) and (F) the spectrum of each RPP29 variant is compared in the absence (back/blue) and presence (red) of equimolar RPP21.
Figure 5.
Chemical shift perturbations demonstrate equivalent binding by RPP21 to RPP29WT and RPP29Δ17 but not RPP29Δ24. The magnitude of the binding-induced chemical shift perturbations observed in two-dimensional 15N-1H correlated spectra (Fig. 4) are mapped to the sequence of RPP29. The similarity between the patterns for RPP29WT and RPP29Δ17 reflect similar affinities and binding interfaces. Very limited shift perturbations in RPP29Δ24 indicate severe RPP21-binding defects in that variant.
Figure 6.
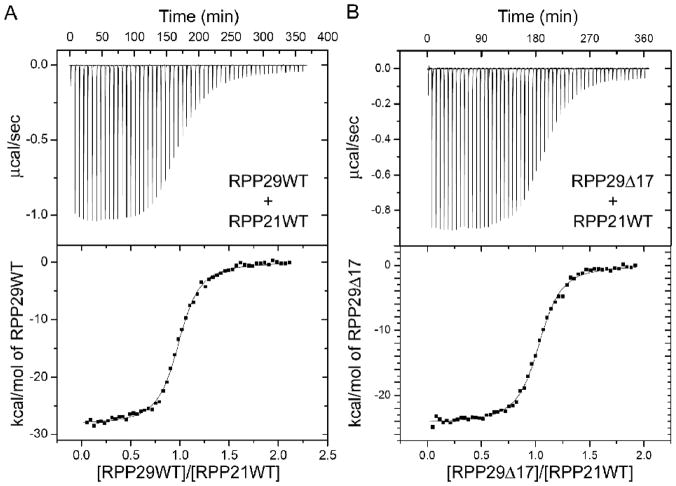
Calorimetric titration of Pfu RPP29WT and RPP29Δ17 into Pfu RPP21 at 55°C. Fitting of ITC thermograms to a single-site model reports similar binding enthalpies and dissociation constants KD of 0.25 ± 0.01 μM for RPP29WT (A) and 0.23 ± 0.01 μM for RPP29Δ17 (B). RPP29Δ24 shows no detectable binding to RPP21 under same conditions.
To assess the impact of the deletions in RPP29 quantitatively and inform subsequent NMR studies, we measured the binding affinity between RPP29 and RPP21 using ITC. Consistent with NMR data, RPP29Δ17 binds RPP21 similarly to RPP29WT, with comparable binding stoichiometry (N, 1:1), enthalpy (ΔH) and affinity (Table 2, Figure 6).43 On the other hand, no binding between RPP29Δ24 and RPP21 could be detected by ITC under the same conditions. The recently determined structure of the RPP21•RPP29 complex19,25 helps rationalize this large affinity difference for RPP21 binding to RPP29Δ17 versus RPP29Δ24 (see Discussion).
Table 2.
Thermodynamic parameters determined by ITC for the interaction of Pfu RPP29WT and RPP29Δ17 to Pfu RPP21WT at 55°C§
| N | KA (1/106 M−1) | ΔG (kcal mol−1) | ΔH (kcal mol−1) | TΔS (kcal mol−1) | |
|---|---|---|---|---|---|
| RPP29WT | 1.001 ± 0.002 | 4.00 ± 0.15 | −9.90 ± 0.03 | −29.67 ± 0.11 | −19.77 ± 0.11 |
| RPP29Δ17 | 1.020 ± 0.002 | 4.42 ± 0.13 | −9.97 ± 0.02 | −24.66 ± 0.06 | −14.69 ± 0.06 |
The values for the stoichiometry (N), binding affinity (KA) and binding enthalpy (ΔH) were obtained from fit to a single site model. The binding free energy (ΔG) and binding entropy (ΔS) were deduced based on the relationships ΔG = −RTlnKA and ΔG = ΔH−TΔS, respectively. The reported errors are the standard deviation of the three replicas of each set of titrations.
In vitro reconstitution assays uncover the RPR’s ability to rescue a mutation in RPP29
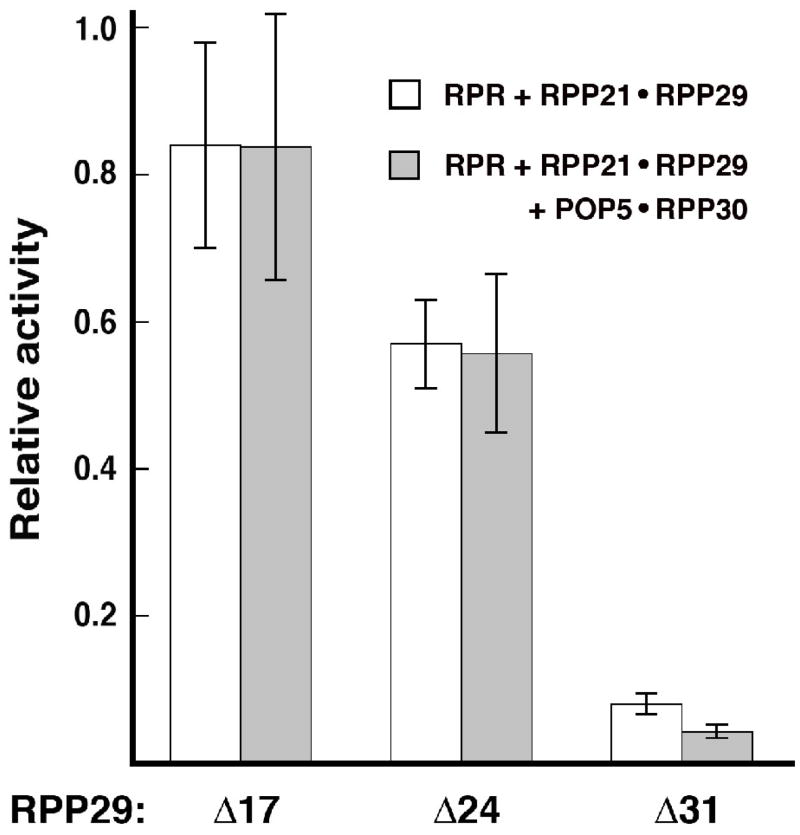
To examine the effect of Pfu RPP29 N-terminal deletions on RNase P activity, we performed steady-state kinetic assays using two different in vitro reconstituted holoenzymes: either RPR + RPP21•RPP29 or RPR + RPP21•RPP29 + POP5•RPP30. These complexes containing either two or four RPPs were assayed under their respective optimal assay condition17 and at substrate concentrations ~2.5 × KM. When Pfu RPR was assembled with RPP21 + RPP29Δ17 or RPP29Δ24 or RPP29Δ31, the partial holoenzymes displayed 84% or 57% or 8% of the activity observed with RPP29WT (Fig. 7). If POP5•RPP30 was also included, the reconstituted holoenzymes comprising RPP29Δ17 or RPP29Δ24 or RPP29Δ31 exhibited 84% or 56% or 4%, respectively, of the activity observed with RPP29WT (Fig. 7). Since deletion of the N-terminal 24 residues of RPP29 compromises its ability to bind RPP21, the nearly three-fifths of the wild-type activity observed with RPP21•RPP29Δ24 must reflect the RPR’s ability to rescue this binding defect in RPP29Δ24. Such a rescue, however, is absent with RPP29Δ31 (Fig. 7). The results obtained with RPR + RPP21•RPP29Δ31 in the absence or presence of POP5•RPP30 are nearly identical (Fig. 7), indicating that neither the RPR nor POP5•RPP30 could ameliorate the deleterious effects of the 31-amino acid N-terminal deletion in RPP29.
Figure 7.
Comparison of the relative pre-tRNATyr-processing activity of Pfu RNase P reconstituted in vitro using either wildtype (WT) or deletion mutant derivatives of RPP29. Activity of Pfu RPR + RPP21•RPP29 (WT or mutant) was determined without (white bars) or with (grey bars) Pfu POP5•RPP30. Relative activities of the mutant holoenzymes were calculated using as reference turnover numbers of 0.54 ± 0.03 min−1 for Pfu RPR + RPP21•RPP29 and 8.9 ± 1.6 min−1 for Pfu RPR + RPP21•RPP29 + POP5•RPP30. While Pfu RPR + RPP21•RPP29 was assayed at 55°C in 50 mM Tris-HCl (pH 7.1), 120 mM MgCl2 and 100 mM NH4OAc, the reaction with 4 RPPs contained 30 mM MgCl2 and 800 mM NH4OAc; these conditions were determined to be optimal in an earlier study.17 The mean and standard deviation values were calculated from three independent experiments.
DISCUSSION
Evolutionarily and functionally conserved motifs in both bacterial and archaeal RPRs
Despite the remarkable differences in the subunit composition of bacterial, archaeal and eukaryal RNase P, there is a striking conservation of various RPR features, notably the universally conserved bulge-helix P4 (Fig. 1). Altering the geometry of this bulge-helix structure in bacterial RPRs affects local RNA structure, substrate binding, catalysis and metal ion interactions.29–32 We have now demonstrated that alteration of this motif (ΔU mutation) in an archaeal RPR-substrate cis conjugate results in an 80-fold decrease in self-cleavage rate (Fig. 2; Table 1). These findings mirror the 100-fold decrease in cleavage rate reported for the bacterial (Eco) ΔU RPR and the complete loss of activity upon deletion of three nucleotides in the P4 helix of human RPR.9,29 Thus, the role of the conserved, bulged uridine in catalysis might be preserved in RNase P variants from all three domains of life.
Archaeal RPPs can mitigate catalytic defects in the cognate RPR
When the Mja RPR ΔU cis-cleaving construct was assembled with the cognate RPP21•RPP29 or POP5•RPP30 or all four RPPs, the differences in self-cleavage rates, relative to the WT RPR with the corresponding RPPs, was narrowed to an order of magnitude instead of the 80-fold observed in the RPR-alone reaction (Table 1). These observations shed light on the roles of RPPs in archaeal RNase P catalysis; however, before describing such inferences, it is useful to first consider a kinetic scheme (Fig. 8) for the RNase P-catalyzed pre-tRNA cleavage reaction.
Figure 8.
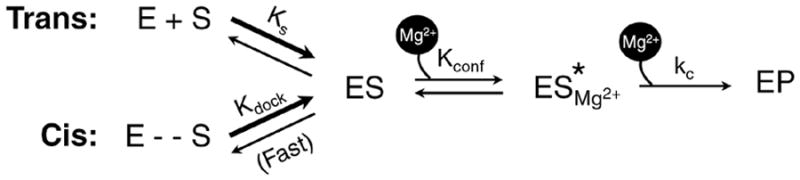
Schematic of the steps involved in RNase P-catalyzed pre-tRNA cleavage. E refers to either the RPR or the holoenzyme (RPR + RPPs). The self-cleaving enzyme-substrate construct pre-tRNATyr-MjaΔU is indicated by E -- S. The conformational transition (defined by Kconf) that precedes the cleavage step (with rate kc) involves the binding of an inner-sphere Mg2+; it is not known whether this divalent metal ion binds ES to form ES* or if it binds ES* and stabilizes it.48 If the RPPs and Mg2+ bind to the folded state of the RPR, their binding will be thermodynamically coupled (not shown).
Figure 8 depicts the individual steps during pre-tRNA processing and is based on a framework derived from investigations on bacterial RNase P. Kinetic and structural studies indicate that a two-step association between RNase P and pre-tRNA precedes the cleavage step.44–48 Stopped-flow kinetic studies revealed the rapid formation of an initial ES encounter complex (KS) before a slower uni-molecular conformational change converts ES to ES* (Kconf.).44 This conformational transition repositions the pre-tRNA cleavage site proximal to the RNA:protein interface in bacterial RNase P48 and appears to set the stage for favorable catalysis by the associated Mg2+ ions, whose roles in RNase P catalysis we discuss below.
Divalent metal ions play pleiotropic roles in RNA structure and function.49–51 These ions are broadly classified as diffusely- and site-bound, with the latter further categorized as outer- and inner-sphere interactions – the former involves water-mediated contacts with the RNA, while the latter the phosphate backbone and nucleobases. Both types screen electrostatic repulsion and promote RNA tertiary structure, but site-specific metal ions additionally participate directly in RNA catalysis. In fact, during RNase P RNA catalysis, a hydrated, active site-bound Mg2+ is believed to generate the nucleophile to attack the scissile phosphodiester linkage in the pre-tRNA.52,53 Fluorescently-labeled pre-tRNAs were employed in binding and cleavage assays, performed in the presence of Ca2+, Mg2+ or Co(NH3)63+, to dissect the types of divalent ions participating in the different steps of the RNase P reaction mechanism.48 While even Co(NH3)63+, an exchange-inert mimic of the Mg(H 2O)62+, permits formation of the initial encounter complex (ES), a high-affinity Mg2+ that supports inner-sphere contacts is absolutely essential for the ES → ES* transition; this metal ion stabilizes the ES* conformer but it is unclear if it does so before or after the conformational change.48 A second, lower-affinity Mg2+ then activates cleavage of ES* → EP. The recently solved crystal structure of the T. maritima RNase P-pre-tRNAPhe complex33 is also consistent with a two metal-ion mechanism.
We can interpret our findings on the ΔU mutant using the abovementioned kinetic scheme (Fig. 8). The 80-fold decrease in the self-cleavage rate of pre-tRNATyr-MjaΔU RPR (relative to the WT) likely results from an altered local structure and/or weaker coordination of Mg2+ ions critical for catalysis post-ES complex formation. We have drawn a parallel with bacterial RNase P where deletion of the bulged uridine (in P4) caused only a four-fold weaker pre-tRNA binding while decreasing the cleavage rate by 100-fold, largely due to defects in Mg2+ coordination,29 a premise bolstered by the recent structure of the bacterial RNase P holoenzyme33 which places the O4 oxygen of the bulged uridine proximal to a putative catalytic metal ion.
Our previous kinetic studies of trans cleavage of a pre-tRNA by an archaeal (type A) RNase P and cis cleavage of pre-tRNATyr-Mja (type M) RPR revealed that POP5•RPP30 is solely responsible for increasing the rate of chemical cleavage by ~100-fold (through effects on Kconf.) while RPP21•RPP29 enhances substrate binding by 16-fold (by influencing KS).13,16 Thus, the ability of POP5•RPP30 to increase the cleavage of pre-tRNATyr-MjaΔU RPR by 584-fold (Table 1) was not unanticipated. However, it was surprising that Mja RPP21•RPP29, which does not increase the self-cleavage of pre-tRNATyr-Mja (WT) RPR, does so by 7-fold for the ΔU mutant. In the latter instance, RPP21•RPP29 must somehow help position the substrate optimally for cleavage to occur, and this effect manifests only when the conserved uridine in P4 is absent. We outline how RPP21•RPP29 and the bulged uridine might be functionally redundant in terms of binding substrate/metal ions vital for RNase P catalysis.
Bacterial RPRs can be divided into two independently-folded domains: a specificity (S) domain with conserved nucleotides making up a T-stem-loop binding site (TBS) that recognizes the T stem-loop (TSL) of the pre-tRNA, and a catalytic (C) domain that cleaves the pre-tRNA while binding to the 5′-leader sequence, acceptor stem and the 3′-RCCA sequence.54–62 Results from various studies indicate that a similar demarcation is likely in archaeal RPRs.13,16,17,63 Moreover, footprinting studies indicate that POP5•RPP30 binds to the C domain and RPP21•RPP29 binds to the S domain;17,25 such a delineation is consistent with the idea that binding of POP5•RPP30 in the vicinity of the active site contributes directly to cleavage, and binding of RPP21•RPP29 to the S domain contributes to substrate binding presumably by facilitating the TSL (pre-tRNA)-TBS (RPR) interaction.54,57 Given that P4 is in the C domain (Fig. 1) and RPP21•RPP29 footprints on the S domain, this binary complex may affect Mg2+ coordination in the C domain by enabling long-distance interactions between the S and C domains. Integrating two earlier observations provides support for this notion. First, in bacterial RPRs, the TSL-S domain interaction is believed to trigger a conformational change that aids catalysis by positioning the chemical groups and catalytically important Mg2+ near the cleavage site in the C domain.54,57 Second, cleavage-site selection in model substrates revealed that when archaeal RPRs are bound to RPP21•RPP29, their substrate-recognition properties coincide with those of bacterial RPRs.64 Such inter-domain crosstalk mediated by RPP21•RPP29 and its direct relevance to catalytic metal ion interactions remain to be deciphered.
Both POP5•RPP30 and RPP21•RPP29 permit pre-tRNA cleavage at lower Mg2+ concentrations (100 mM) than the reaction catalyzed by the RPR alone (500 mM). Rescue of the ΔU mutant by POP5•RPP30 and RPP21•RPP29 (likely through different mechanisms) coupled with the possibility that the bulged uridine coordinates a catalytic-site Mg 2+ suggests a role for RPPs in binding active-site metal ions, a premise that needs to be experimentally validated.
Archaeal RPR can rescue the deleterious effects of a large deletion in a cognate RPP
We have demonstrated that a protein-protein interaction defect in a Pfu RPP binary complex is rescued by the Pfu RPR. Removal of the first 24 residues in Pfu RPP29 largely abolishes its ability to bind RPP21, yet the holoenzyme assembled in vitro using RPP29Δ24 retains three-fifths of the activity of wild-type Pfu RNase P (Fig. 8). This near-normal functional behavior, together with the previous finding that neither RPP21 nor RPP29 can individually activate the RPR, likely reflects the ability of the RPR to restore the interactions between RPP29Δ24 and RPP21, an observation which we elaborate below.
High-resolution structures of the Pho and Pfu RPP21•RPP29 complexes highlight how different structural elements in RPP21 and RPP29 form a tight interface mediated by several polar, ionic and hydrophobic interactions.19,25 An examination of the N-termini of the two proteins in this binary complex reveals that residues I23 (α1) and I29 (α2) of Pfu RPP29 make close contacts with the side-chains of V28 and L35 (α1) of Pfu RPP21. Thus, it is not surprising that Pfu RPP29Δ24 (lacking part of α1, residues 19–24) and RPP29Δ31 (missing parts of α1 and α2, residues 26–31) would fail to bind Pfu RPP21 with high affinity (Figs. 5 and 6).
In our solution structure of Pfu RPP21•RPP29,25 we noted the presence of a large electropositive surface patch (site 1) which spans both proteins, and a smaller electropositive patch on the opposite side of RPP21 (site 2). We proposed that site 1 may be used for RPR docking and site 2 for substrate binding, as suggested for human RPP21.25,65 Results from our RNase T1/V1-based footprinting studies25 indicated that archaeal RPP21•RPP29 footprints the RPR’s S-domain, although the exact regions in these proteins responsible for these RPR interactions remain to be mapped. It is reasonable to infer that in the absence of the RPR, the N-terminus (residues 25–35) of Pfu RPP29Δ24 cannot adopt a stable conformation needed for RPP21 binding; in the presence of Pfu RPR, however, the large RNA may serve as a scaffold to recruit both proteins and then induce the appropriate conformation of each protein required for their interaction. While the formation of heterodimers (e.g., RPP21•RPP29) might be obligatory for generating a suitable RNA-binding platform, and even enhance the affinity and specificity for the intended RNA target, our data suggest that the RNA reciprocates by providing a platform that promotes proper protein-protein interactions.
Since RPP29 from different Thermococcaceae possess the long N-terminal addition not seen in other archaeal relatives (Fig. 4 and data not shown), it is conceivable that deleting its N-terminal 24 residues, although lacking a severe phenotype under in vitro assay conditions (pre-tRNA cleavage at 55°C; Fig. 7), might prove deleterious in vivo. With Pfu’s optimal growth at ~95 to 100°C, the entire suite of protein-protein interactions (including those involving α1 in Pfu RPP29) is likely needed for forming the RPP21•RPP29 complex under such harsh habitats.
Concluding remarks
Our observations of mutual rescue between archaeal RPR and RPPs reveal an intimate, inter-dependent relationship among the subunits in RNase P, and extends earlier findings from NMR studies on the Pfu RPP21•RPP29 complex25 in which we uncovered binding-coupled folding of structural elements in both proteins. These results are consistent with induced-fit mechanisms, a recurring theme in large dynamic RNPs.66,67 The structural flexibility afforded by co-folding of different interacting pairs might offer alternative, equally productive paths to the final assemblage in large multi-subunit complexes (e.g., RNase P, RNase MRP,68,69 ribosome70,71).
MATERIALS & METHODS
Construction of mutant derivatives of archaeal RPRs
The gene encoding the Mja RPR ΔU mutant was generated using PCR-based mutagenesis. To generate pBT7-ptRNATyr-MjaΔU RPR, pBT7-ptRNATyr-Mja RPR16,72 was utilized as the template. T4 polynucleotide kinase was used to phosphorylate the primers pTyr-S3-Mja RPR ΔU-F (5′-TCCGCCCACCCCATTTAT-3′) and pTyr-S3-Mja RPR-R (5′-CTTCCTCCCCTCTTAAAG-3′); these primers flank the nucleotide to be deleted and are oriented outward to ensure amplification of the entire pBT7-ptRNATyr-S3-Mja RPR plasmid (except the single position to be deleted). The resulting PCR product was circularized by ligation with T4 DNA ligase and transformed into E. coli DH5α. Transformants were then screened to identify those harboring pBT7-ptRNATyr-MjaΔU. The sequence of the RPR mutant was confirmed by automated DNA sequencing.
In vitro transcription of RPRs used in this study
The Pfu RPR was generated as described previously.17 For the cis-cleaving Mja ΔU RPR, we generated the RNA using a PCR product as the template for transcription. A high-fidelity PCR [using Phusion DNA polymerase (New England Biolabs)] was performed with pBT7-ptRNATyr-MjaΔU RPR as the template, and 5′-TAATACGACTCACTATAGGGAGCAGGCCAGTAAA-3′ (forward) and 5′-CTATTTCGGCTTGCACCCC-3′ (reverse) as primers. These primers generate a PCR product containing the coding sequence of ptRNATyr-Mja ΔU RPR under the control of a T7 RNA polymerase promoter (whose sequence is shown in italics in the forward primer).
Following in vitro transcription, all RNAs were subjected to extensive dialysis to remove unincorporated rNTPs and their concentrations determined from their respective extinction coefficients at Abs260.
Construction of Pfu RPP29 deletion mutant derivatives
Seven Pfu RPP29 mutants were constructed, with different N-terminal deletions, named RPP29Δ5, RPP29Δ13, RPP29Δ17, RPP29Δ24, RPP29Δ31, RPP29Δ36, and RPP29Δ42, based on the number of N-terminal residues removed. Each open reading frame encoding the specific RPP29 N-terminal deletion mutant was amplified by PCR using the full-length Pfu pET-33b/RPP29 plasmid as the template and gene-specific DNA primers (Table 3); the same reverse primer was used in all cases. These PCR products were individually digested with NcoI and XhoI, whose recognition sites were included in the forward and reverse primers, respectively, and then cloned into pET-33b (Novagen) also digested with NcoI and XhoI. The sequences of all these RPP29 mutant derivatives were confirmed by automated DNA sequencing.
Table 3.
Oligonucleotide primers used to construct Pfu RPP29 derivatives
| PfuP29Δ5-F | 5′-cctagtccATGGAAGAACGTGAGAATAGAACTTCAG-3′ |
| PfuP29Δ13-F | 5′-gctattccATGGGGAGATCACAGGGATCG-3′ |
| PfuP29Δ17-F | 5′-cctcgaccATGGGATCGTATCAAGAAATTATTGG-3′ |
| PfuP29Δ24-F | 5′-gcatttccATGGGTAGAACTTGGATTTTCAGAGG-3′ |
| PfuP29Δ31-F | 5′-cctaaaccATGGGAGCTCATAGAGGTAGAGTAAAC-3′ |
| PfuP29Δ36-F | 5′-cgtggaccATGGGTAGAGTAAACAAGAAAAATATAGTATGG-3′ |
| PfuP29Δ42-F | 5′-cgaattccATGGGCAATATAGTATGGCACGAACTAATC-3′ |
| PfuP29C127-R | 5′-cctaactcgagTCATTTACGCCAACGC-3′ |
Start and stop codons are highlighted in bold, and NcoI (in Forward primers, denoted as F) and XhoI (in Reverse primer, denoted as R) restriction sites are underlined. Uppercase letters correspond to the open reading frame.
Expression and purification of the protein subunits of Mja and Pfu RNase P
The preparation of Mja and Pfu RPPs followed established methods.13,16,18,24 The unlabeled and U-[15N]-Pfu RPP29ΔX proteins (as well as Pfu RPP21) were overexpressed and purified as previously described.18,25
RNase P assays
All assays were performed in a thermal cycler. Regardless of how the reactions were set up, they were always terminated using a stop dye [10 M urea, 1 mM EDTA, 0.05% (w/v) xylene cyanol, 0.05% (w/v) bromophenol blue, 20% (v/v) phenol]. The reaction products were then subjected to 8% (w/v) polyacrylamide/7 M urea gel electrophoresis and cleavage rates analyzed as described in the next section.
Multiple-turnover reactions
Multiple-turnover trans-cleavage reactions with Pfu RNase P were performed essentially as described elsewhere (see also Fig. 7 for additional details).13,17
Single-turnover reactions
The self-cleavage rates of pre-tRNATyr-MjaΔU RPR were determined in the absence and presence of Mja RPPs. In vitro transcribed pre-tRNATyr-MjaΔU RPR was first folded as follows: incubation at 50°C for 50 min in water followed by 37°C for 30 min in 50 mM Tris-acetate (pH 8), 800 mM NH4OAc and 10 mM Mg(OAc)2. For all cis cleavage reactions, we mixed 25,000 dpm of folded 5′-[P32]-labeled pre-tRNATyr-MjaΔU RPR with 50 nM of the same unlabeled, folded transcript. Assays were performed in two phases: a pre-incubation followed by a cleavage time-course. We optimized conditions to minimize self-cleavage during pre-incubation at 55°C, which was required for temperature equilibration and for reconstitution with RPPs (see description below). In all cases, once cleavage was initiated, aliquots were removed at defined time intervals and the reactions were terminated by adding an equal volume of stop dye.
RPR-alone
The folded RPR [50 μM in 20 μl of 50 mM 2-(N-morpholino) ethanesulfonic acid (MES) pH 6, 2.5 M NH4OAc, 10 mM Mg(OAc)2] was incubated at 55°C for 5 min. Cleavage at 55°C was then initiated by addition of an equal volume of identical buffer containing 1 M Mg(OAc)2, which had been pre-warmed to 55°C.
RPR + RPPs
The reconstitutions with different combinations of RPPs involved variations in pH and divalent ions used since care had to be taken to minimize self-cleavage while maximizing RNP assembly and product formation. We empirically determined these optimal conditions, although they largely mirrored those in our earlier study when we examined self-cleavage of pre-tRNATyr-Mja (WT) RPR.16 For determining the self-cleavage rates of pre-tRNATyr-MjaΔU RPR in the presence of RPPs, we first mixed the RPR with RPPs (final concentration of 50 and 500 nM, respectively) at 37°C for 10 min followed by 55°C for 10 min. With pre-tRNATyr-Mja ΔU RPR + RPP21•RPP29, the RNP was first reconstituted in 20 μl of 50 mM MES (pH 6), 800 mM NH4OAc and 10 mM Mg(OAc)2; at the end of this pre-incubation, cleavage was initiated by addition of 20 μl of 50 mM MES pH 6, 800 mM NH4OAc and 200 mM Mg(OAc)2. In the case of RPR + POP5•RPP30, the reconstitution was in 20 μl of 50 mM MES (pH 6), 800 mM NH4OAc and 25 mM Ca(OAc)2 and cleavage initiated by addition of 20 μl of 50 mM MES (pH 6), 800 mM NH4OAc and 200 mM Mg(OAc)2. When all four RPPs were present, the reconstitution was in 20 μl of 50 mM MES (pH 5.4), 800 mM NH4OAc and 1 mM Mg(OAc)2, and cleavage initiated by addition of 20 μl of 50 mM MES (pH 5.4), 800 mM NH4OAc and 200 mM Mg(OAc)2.
RNase P activity data analysis
Regardless of trans or cis cleavage reactions, the reaction products separated by denaturing PAGE were visualized by phosphorimager analysis (Typhoon, GE Healthcare) and quantitated with ImageQuant (GE Healthcare) to assess the extent of substrate cleaved. For the trans cleavage reactions, we calculated the initial velocities by determining the product formed during a time course; typically, the times of incubation were chosen to restrict the amount of substrate cleaved to < 30%. To obtain the rate of product formation (kobs) in cis reactions, the percent of product formed at time t (Pt) was fit to Pt = P∞(1 − e−kt) using Kaleidagraph software (Synergy). The standard errors for the best-fit values of kobs did not exceed 20%. The reported kinetic parameters are the mean and standard deviation values calculated from at least three independent experiments.
In the cis cleavage reactions, the ES complexes formed using MjaΔU RPR are largely productive in the case of the RPR-alone reaction and the RPR + POP5•RPP30 as indicated by their ≥ 80% amplitudes (Fig. 2, left panels). However, this is not the case in the reactions that have RPP21•RPP29 (either 25 or 50%, Fig. 2, right panels). Since we use a 10-fold stoichiometric excess of RPPs:RPR to facilitate assembly of the respective RNP, it is possible that any RPP21•RPP29 uncomplexed with the RPR engages in RPR-independent interactions with the covalently-tethered pre-tRNA13,65 and thereby lowers the amplitude; although the presence of POP5•RPP30 ameliorates this problem, it is not entirely overcome (Fig. 2).
Limited proteolysis to identify the folded core of RPP29
One hundred μg of purified Pfu RPP29WT was subjected to limited proteolysis by 1% (w/w) trypsin at room temperature for 1 h, and the proteolytic reaction was quenched by adding 5% (w/v) phenylmethanesulphonylfluoride and stored on ice. The digested proteins were analyzed by electrospray mass spectroscopy (Q-TOF-II, Micromass).
NMR spectroscopy
NMR spectra were recorded at 55°C on a 600 MHz spectrometer equipped with a cryogenically-cooled triple-resonance single-axis gradient probe. For each U-[15N]-Pfu RPP29ΔX mutant, a two-dimensional 1H-15N correlation spectrum was recorded on the free protein (~0.3 mM), then another spectrum was recorded in the presence of a small molar excess of unlabeled Pfu RPP21. We also performed the NMR assay with the reversed labeling pattern, that is, with U[15N]-Pfu RPP21 and unlabeled Pfu RPP29ΔX (not shown). NMR spectra were processed and analyzed using NMRPipe and NMRView.73,74
Backbone resonance assignments of the Pfu RPP29 derivatives in the absence and presence of Pfu RPP21 were inferred from the assignments of the free and bound Pfu RPP29WT, respectively.25 Weighted average chemical shift perturbations (CSPs) of amide resonances between the free and the bound spectra were calculated using the relation , where f = 8 for Gly, 6 otherwise.
Isothermal titration calorimetry (ITC)
Purified Pfu RPPs were dialyzed into ITC buffer [20 mM cacodylate (pH 6.7), 10 mM KCl, 0.3 mM ZnCl2, and 0.02% (w/v) NaN3] twice, each for at least 6 h. The protein solutions were thoroughly degassed under vacuum immediately before use at an experimental temperature of 55°C. All ITC experiments were performed on a VP-ITC calorimeter (MicroCal, Inc., Northhampton, MA), with the sample cell containing ~20 μM of Pfu RPP21 and the syringe containing ~200 μM of Pfu RPP29 or its mutant derivatives. Titration of RPP29 into RPP21 started with a 3-μl injection, followed by a series of 5-μl injections, with a spacing of 400 s. ITC data were analyzed with Origin V.7 SR4 (Microcal, Inc.) as follows. The heat pulse from each injection (μcal s−1) was integrated (yielding enthalpy change ΔH, kcal mol−1), corrected for heat of dilution obtained form the average of last ten injections post saturation, and plotted as a function of molar ratio. Nonlinear least squares fitting of the data to a single-site binding model yielded stoichiometry (N) and the association constant of the binding reaction (KA).
Acknowledgments
We are grateful to Lien Lai (OSU) for comments on the manuscript, and the Foster and Gopalan laboratory members for reagents and discussions. This research was supported by grants from the National Institutes of Health (GM067807 to MPF and VG) and National Science Foundation (MCB0843543, to VG). NMR and mass spectrometry data were recorded at the OSU Campus Chemical Instrument Center, with generous assistance from Center staff.
ABBREVIATIONS USED
- Mja
Methanocaldococcus jannaschii
- Mth
Methanothermobacter thermautotrophicus
- pre-tRNA
precursor tRNA
- Pfu
Pyrococcus furiosus
- Pho
Pyrococcus horikoshii
- RNase P
ribonuclease P
- RNP
ribonucleoprotein
- RPP
RNase P protein subunit
- RPR
RNase P RNA
- tRNA
transfer RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Evans D, Marquez SM, Pace NR. RNase P: interface of the RNA and protein worlds. Trends Biochem Sci. 2006;31:333–41. doi: 10.1016/j.tibs.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Lai LB, Vioque A, Kirsebom LA, Gopalan V. Unexpected diversity of RNase P, an ancient tRNA processing enzyme: challenges and prospects. FEBS Lett. 2010;584:287–96. doi: 10.1016/j.febslet.2009.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu F, Altman S. Ribonuclease P Protein Reviews Series. Springer-Verlag; New York: 2010. [Google Scholar]
- 4.Walker SC, Engelke DR. Ribonuclease P: the evolution of an ancient RNA enzyme. Crit Rev Biochem Mol Biol. 2006;41:77–102. doi: 10.1080/10409230600602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gobert A, Gutmann B, Taschner A, Gossringer M, Holzmann J, Hartmann RK, Rossmanith W, Giege P. A single Arabidopsis organellar protein has RNase P activity. Nat Struct Mol Biol. 2010;17:740–744. doi: 10.1038/nsmb.1812. [DOI] [PubMed] [Google Scholar]
- 6.Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–74. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 7.Hall TA, Brown JW. Archaeal RNase P has multiple protein subunits homologous to eukaryotic nuclear RNase P proteins. RNA. 2002;8:296–306. doi: 10.1017/s1355838202028492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–57. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 9.Kikovska E, Svard SG, Kirsebom LA. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc Natl Acad Sci USA. 2007;104:2062–7. doi: 10.1073/pnas.0607326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pannucci JA, Haas ES, Hall TA, Harris JK, Brown JW. RNase P RNAs from some Archaea are catalytically active. Proc Natl Acad Sci USA. 1999;96:7803–8. doi: 10.1073/pnas.96.14.7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai LB, Cho I-M, Chen W-Y, Gopalan V. Archael RNase P: A mosaic of its bacterial and eukaryal relatives. In: Liu F, Altman S, editors. Ribonuclease P. Springer Verlag; New York: 2010. pp. 153–172. [Google Scholar]
- 12.Boomershine WP, McElroy CA, Tsai HY, Wilson RC, Gopalan V, Foster MP. Structure of Mth11/Mth Rpp29, an essential protein subunit of archaeal and eukaryotic RNase P. Proc Natl Acad Sci U S A. 2003;100:15398–403. doi: 10.1073/pnas.2535887100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen WY, Pulukkunat DK, Cho IM, Tsai HY, Gopalan V. Dissecting functional cooperation among protein subunits in archaeal RNase P, a catalytic ribonucleoprotein complex. Nucleic Acids Res. 2010;38:8316–27. doi: 10.1093/nar/gkq668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho IM, Lai LB, Susanti D, Mukhopadhyay B, Gopalan V. Ribosomal protein L7Ae is a subunit of archaeal RNase P. Proc Natl Acad Sci USA. 2010;107:14573–8. doi: 10.1073/pnas.1005556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kouzuma Y, Mizoguchi M, Takagi H, Fukuhara H, Tsukamoto M, Numata T, Kimura M. Reconstitution of archaeal ribonuclease P from RNA and four protein components. Biochem Biophys Res Commun. 2003;306:666–73. doi: 10.1016/s0006-291x(03)01034-9. [DOI] [PubMed] [Google Scholar]
- 16.Pulukkunat DK, Gopalan V. Studies on Methanocaldococcus jannaschii RNase P reveal insights into the roles of RNA and protein cofactors in RNase P catalysis. Nucleic Acids Res. 2008;36:4172–80. doi: 10.1093/nar/gkn360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai HY, Pulukkunat DK, Woznick WK, Gopalan V. Functional reconstitution and characterization of Pyrococcus furiosus RNase P. Proc Natl Acad Sci USA. 2006;103:16147–52. doi: 10.1073/pnas.0608000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amero CD, Boomershine WP, Xu Y, Foster M. Solution structure of Pyrococcus furiosus RPP21, a component of the archaeal RNase P holoenzyme, and interactions with its RPP29 protein partner. Biochemistry. 2008;47:11704–10. doi: 10.1021/bi8015982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honda T, Kakuta Y, Kimura K, Saho J, Kimura M. Structure of an archaeal homolog of the human protein complex Rpp21-Rpp29 that is a key core component for the assembly of active ribonuclease P. J Mol Biol. 2008;384:652–62. doi: 10.1016/j.jmb.2008.09.056. [DOI] [PubMed] [Google Scholar]
- 20.Kakuta Y, Ishimatsu I, Numata T, Kimura K, Yao M, Tanaka I, Kimura M. Crystal structure of a ribonuclease P protein Ph1601p from Pyrococcus horikoshii OT3: an archaeal homologue of human nuclear ribonuclease P protein Rpp21. Biochemistry. 2005;44:12086–93. doi: 10.1021/bi050738z. [DOI] [PubMed] [Google Scholar]
- 21.Kawano S, Nakashima T, Kakuta Y, Tanaka I, Kimura M. Crystal structure of protein Ph1481p in complex with protein Ph1877p of archaeal RNase P from Pyrococcus horikoshii OT3: implication of dimer formation of the holoenzyme. J Mol Biol. 2006;357:583–91. doi: 10.1016/j.jmb.2005.12.086. [DOI] [PubMed] [Google Scholar]
- 22.Sidote DJ, Heideker J, Hoffman DW. Crystal structure of archaeal ribonuclease P protein aRpp29 from Archaeoglobus fulgidus. Biochemistry. 2004;43:14128–38. doi: 10.1021/bi048578z. [DOI] [PubMed] [Google Scholar]
- 23.Sidote DJ, Hoffman DW. NMR structure of an archaeal homologue of ribonuclease P protein Rpp29. Biochemistry. 2003;42:13541–50. doi: 10.1021/bi030170z. [DOI] [PubMed] [Google Scholar]
- 24.Wilson RC, Bohlen CJ, Foster MP, Bell CE. Structure of Pfu Pop5, an archaeal RNase P protein. Proc Natl Acad Sci U S A. 2006;103:873–8. doi: 10.1073/pnas.0508004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Y, Amero CD, Pulukkunat DK, Gopalan V, Foster MP. Solution structure of an archaeal RNase P binary protein complex: formation of the 30-kDa complex between Pyrococcus furiosus RPP21 and RPP29 is accompanied by coupled protein folding and highlights critical features for protein-protein and protein-RNA interactions. J Mol Biol. 2009;393:1043–55. doi: 10.1016/j.jmb.2009.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Numata T, Ishimatsu I, Kakuta Y, Tanaka I, Kimura M. Crystal structure of archaeal ribonuclease P protein Ph1771p from Pyrococcus horikoshii OT3: an archaeal homolog of eukaryotic ribonuclease P protein Rpp29. RNA. 2004;10:1423–32. doi: 10.1261/rna.7560904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown JW. The Ribonuclease P Database. Nucleic Acids Res. 1999;27:314. doi: 10.1093/nar/27.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gopalan V. Uniformity amid diversity in RNase P. Proc Natl Acad Sci U S A. 2007;104:2031–2. doi: 10.1073/pnas.0611193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaye NM, Zahler NH, Christian EL, Harris ME. Conservation of helical structure contributes to functional metal ion interactions in the catalytic domain of ribonuclease P RNA. J Mol Biol. 2002;324:429–42. doi: 10.1016/s0022-2836(02)01094-x. [DOI] [PubMed] [Google Scholar]
- 30.Christian EL, Kaye NM, Harris ME. Helix P4 is a divalent metal ion binding site in the conserved core of the ribonuclease P ribozyme. RNA. 2000;6:511–9. doi: 10.1017/s1355838200000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christian EL, Kaye NM, Harris ME. Evidence for a polynuclear metal ion binding site in the catalytic domain of ribonuclease P RNA. EMBO J. 2002;21:2253–62. doi: 10.1093/emboj/21.9.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crary SM, Kurz JC, Fierke CA. Specific phosphorothioate substitutions probe the active site of Bacillus subtilis ribonuclease P. RNA. 2002;8:933–47. doi: 10.1017/s1355838202025025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragon A. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010;468:784–789. doi: 10.1038/nature09516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris JK, Haas ES, Williams D, Frank DN, Brown JW. New insight into RNase P RNA structure from comparative analysis of the archaeal RNA. RNA. 2001;7:220–32. doi: 10.1017/s1355838201001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crary SM, Niranjanakumari S, Fierke CA. The protein component of Bacillus subtilis ribonuclease P increases catalytic efficiency by enhancing interactions with the 5′ leader sequence of pre-tRNAAsp. Biochemistry. 1998;37:9409–16. doi: 10.1021/bi980613c. [DOI] [PubMed] [Google Scholar]
- 36.Guerrier-Takada C, Lumelsky N, Altman S. Specific interactions in RNA enzyme-substrate complexes. Science. 1989;246:1578–84. doi: 10.1126/science.2480641. [DOI] [PubMed] [Google Scholar]
- 37.Kurz JC, Fierke CA. The affinity of magnesium binding sites in the Bacillus subtilis RNase P•pre-tRNA complex is enhanced by the protein subunit. Biochemistry. 2002;41:9545–58. doi: 10.1021/bi025553w. [DOI] [PubMed] [Google Scholar]
- 38.Kurz JC, Niranjanakumari S, Fierke CA. Protein component of Bacillus subtilis RNase P specifically enhances the affinity for precursor-tRNAAsp. Biochemistry. 1998;37:2393–400. doi: 10.1021/bi972530m. [DOI] [PubMed] [Google Scholar]
- 39.Peck-Miller KA, Altman S. Kinetics of the processing of the precursor to 4.5 S RNA, a naturally occurring substrate for RNase P from Escherichia coli. J Mol Biol. 1991;221:1–5. doi: 10.1016/0022-2836(91)80194-y. [DOI] [PubMed] [Google Scholar]
- 40.Sun L, Campbell FE, Yandek LE, Harris ME. Binding of C5 protein to P RNA enhances the rate constant for catalysis for P RNA processing of pre-tRNAs lacking a consensus (+ 1)/C(+ 72) pair. J Mol Biol. 2010;395:1019–37. doi: 10.1016/j.jmb.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun L, Harris ME. Evidence that binding of C5 protein to P RNA enhances ribozyme catalysis by influencing active site metal ion affinity. RNA. 2007;13:1505–15. doi: 10.1261/rna.571007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai HY, Masquida B, Biswas R, Westhof E, Gopalan V. Molecular modeling of the three-dimensional structure of the bacterial RNase P holoenzyme. J Mol Biol. 2003;325:661–75. doi: 10.1016/s0022-2836(02)01267-6. [DOI] [PubMed] [Google Scholar]
- 43.Xu Y. PhD thesis. The Ohio State University; Columbus, OH: 2009. [Google Scholar]
- 44.Hsieh J, Fierke CA. Conformational change in the Bacillus subtilis RNase P holoenzyme--pre-tRNA complex enhances substrate affinity and limits cleavage rate. RNA. 2009;15:1565–77. doi: 10.1261/rna.1639409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loria A, Pan T. Recognition of the 5′ leader and the acceptor stem of a pre-tRNA substrate by the ribozyme from Bacillus subtilis RNase P. Biochemistry. 1998;37:10126–33. doi: 10.1021/bi980220d. [DOI] [PubMed] [Google Scholar]
- 46.Pomeranz Krummel DA, Altman S. Multiple binding modes of substrate to the catalytic RNA subunit of RNase P from Escherichia coli. RNA. 1999;5:1021–33. doi: 10.1017/s1355838299990416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun L, Campbell FE, Zahler NH, Harris ME. Evidence that substrate-specific effects of C5 protein lead to uniformity in binding and catalysis by RNase P. EMBO J. 2006;25:3998–4007. doi: 10.1038/sj.emboj.7601290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsieh J, Koutmou KS, Rueda D, Koutmos M, Walter NG, Fierke CA. A divalent cation stabilizes the active conformation of the B. subtilis RNase P x pre-tRNA complex: a role for an inner-sphere metal ion in RNase P. J Mol Biol. 2010;400:38–51. doi: 10.1016/j.jmb.2010.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pyle AM. Metal ions in the structure and function of RNA. J Biol Inorg Chem. 2002;7:679–90. doi: 10.1007/s00775-002-0387-6. [DOI] [PubMed] [Google Scholar]
- 50.Draper DE, Grilley D, Soto AM. Ions and RNA folding. Annu Rev Biophys Biomol Struct. 2005;34:221–43. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 51.Draper DE. RNA folding: thermodynamic and molecular descriptions of the roles of ions. Biophys J. 2008;95:5489–95. doi: 10.1529/biophysj.108.131813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cassano AG, Anderson VE, Harris ME. Analysis of solvent nucleophile isotope effects: evidence for concerted mechanisms and nucleophilic activation by metal coordination in nonenzymatic and ribozyme-catalyzed phosphodiester hydrolysis. Biochemistry. 2004;43:10547–59. doi: 10.1021/bi049188f. [DOI] [PubMed] [Google Scholar]
- 53.Smith D, Pace NR. Multiple magnesium ions in the ribonuclease P reaction mechanism. Biochemistry. 1993;32:5273–81. doi: 10.1021/bi00071a001. [DOI] [PubMed] [Google Scholar]
- 54.Brannvall M, Kikovska E, Wu S, Kirsebom LA. Evidence for induced fit in bacterial RNase P RNA-mediated cleavage. J Mol Biol. 2007;372:1149–64. doi: 10.1016/j.jmb.2007.07.030. [DOI] [PubMed] [Google Scholar]
- 55.Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR. Crystal structure of a bacterial ribonuclease P RNA. Proc Natl Acad Sci U S A. 2005;102:13392–7. doi: 10.1073/pnas.0506662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kirsebom LA, Svard SG. Base pairing between Escherichia coli RNase P RNA and its substrate. EMBO J. 1994;13:4870–6. doi: 10.1002/j.1460-2075.1994.tb06814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kirsebom LA, Trobro S. RNase P RNA-mediated cleavage. IUBMB Life. 2009;61:189–200. doi: 10.1002/iub.160. [DOI] [PubMed] [Google Scholar]
- 58.Loria A, Pan T. Domain structure of the ribozyme from eubacterial ribonuclease P. RNA. 1996;2:551–63. [PMC free article] [PubMed] [Google Scholar]
- 59.Pan T, Loria A, Zhong K. Probing of tertiary interactions in RNA: 2′-hydroxyl-base contacts between the RNase P RNA and pre-tRNA. Proc Natl Acad Sci U S A. 1995;92:12510–4. doi: 10.1073/pnas.92.26.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature. 2005;437:584–7. doi: 10.1038/nature04074. [DOI] [PubMed] [Google Scholar]
- 61.Zahler NH, Christian EL, Harris ME. Recognition of the 5′ leader of pre-tRNA substrates by the active site of ribonuclease P. RNA. 2003;9:734–45. doi: 10.1261/rna.5220703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oh BK, Pace NR. Interaction of the 3′-end of tRNA with ribonuclease P RNA. Nucleic Acids Res. 1994;22:4087–94. doi: 10.1093/nar/22.20.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Terada A, Yoshida T, Kimura M. Identification of nucleotide residues essential for RNase P activity from the hyperthermophilic archaeon Pyrococcus horikoshii OT3. Biosci Biotechnol Biochem. 2007;71:1940–5. doi: 10.1271/bbb.70145. [DOI] [PubMed] [Google Scholar]
- 64.Sinapah S, Wu S, Chen Y, Pettersson BMF, Gopalan V, Kirsebom LA. Cleavage of model substrated by archaeal RNase P: role of protein cofactors in cleavage-site selection. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq732. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jarrous N, Reiner R, Wesolowski D, Mann H, Guerrier-Takada C, Altman S. Function and subnuclear distribution of Rpp21, a protein subunit of the human ribonucleoprotein ribonuclease P. RNA. 2001;7:1153–64. doi: 10.1017/s1355838201010469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leulliot N, Varani G. Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–56. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 67.Williamson JR. Cooperativity in macromolecular assembly. Nat Chem Biol. 2008;4:458–65. doi: 10.1038/nchembio.102. [DOI] [PubMed] [Google Scholar]
- 68.Hands-Taylor KL, Martino L, Tata R, Babon JJ, Bui TT, Drake AF, Beavil RL, Pruijn GJ, Brown PR, Conte MR. Heterodimerization of the human RNase P/MRP subunits Rpp20 and Rpp25 is a prerequisite for interaction with the P3 arm of RNase MRP RNA. Nucleic Acids Res. 2010;38:4052–66. doi: 10.1093/nar/gkq141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perederina A, Esakova O, Quan C, Khanova E, Krasilnikov AS. Eukaryotic ribonucleases P/MRP: the crystal structure of the P3 domain. EMBO J. 2010;29:761–9. doi: 10.1038/emboj.2009.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Adilakshmi T, Bellur DL, Woodson SA. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature. 2008;455:1268–72. doi: 10.1038/nature07298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mulder AM, Yoshioka C, Beck AH, Bunner AE, Milligan RA, Potter CS, Carragher B, Williamson JR. Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science. 2010;330:673–7. doi: 10.1126/science.1193220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsai HY, Lai LB, Gopalan V. A Modified pBluescript-based vector for facile cloning and transcription of RNAs. Anal Biochem. 2002;303:214–7. doi: 10.1006/abio.2001.5567. [DOI] [PubMed] [Google Scholar]
- 73.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 74.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–52. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 75.Massire C, Jaeger L, Westhof E. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J Mol Biol. 1998;279:773–93. doi: 10.1006/jmbi.1998.1797. [DOI] [PubMed] [Google Scholar]
- 76.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]