Abstract
Objective
Most patients with erythropoietic protoporphyria (EPP) have deficient ferrochelatase (FECH) activity due to changes in FECH DNA. We evaluated seven patients with EPP phenotype in whom abnormalities of FECH DNA were not found by conventional analysis. The major focus was mitoferrin-1 (MFRN1), the mitochondrial transporter of Fe used for heme formation by FECH, and for 2Fe2S cluster synthesis that is critical to FECH activity/stability.
Patients and Methods
Four patients had a deletion in ALAS2 that causes enzyme gain-of-function, resulting in increased formation of protoporphyrin; one had a heterozygous major deletion in FECH DNA. All had an abnormal transcript of MFRN1 in mRNA extracted from blood leukocytes and/or liver tissue. The abnormal transcript contained an insert of intron 2 that had a stop codon. The consequences of abnormal MFRN1 expression were examined using zebrafish and yeast MFRN-deficient strains, and cultured lymphoblasts from the patients.
Results
Abnormal human MFRN1 cDNA showed loss-of-function in zebrafish and yeast mutants, whereas normal human MFRN1 cDNA rescued both. Using cultured lymphoblasts, qRT-PCR showed increased formation of abnormal transcript that was accompanied by decreased formation of normal transcript and reduced FECH activity in patients compared to normal lines. A positive correlation coefficient (0.75) was found between FECH activity and normal MFRN1 mRNA in lymphoblasts. However, no obvious cause for increased formation of abnormal transcript was identified in MFRN1 exons and splice junctions.
Conclusion
Abnormal MFRN1 expression may contribute to EPP phenotype in some patients, probably by causing a reduction in FECH activity.
Erythropoietic protoporphyria (EPP) is a genetic/metabolic disorder in which accumulation of protoporphyrin in skin causes photosensitivity, the principal clinical manifestation (1–3). Some patients have mild anemia with hypochromic microcytic indices (1). The most serious clinical feature is hepatobiliary disease (4, 5), which is caused by protoporphyrin-induced damage to liver cells and the formation of toxic bile (6–9), necessitating liver transplantation in some patients (10).
The enzyme defect in EPP is a deficiency of ferrochelatase (FECH, EC 4.99.1.1) activity (11,12), and bone marrow is the major tissue affected by the enzyme defect (10,13,14). FECH is located on the inner mitochondrial membrane, where it catalyzes the insertion of ferrous iron into protoporphyrin to form heme (15). The human FECH enzyme is a homodimer that contains two coordinated 2Fe2S clusters (15, 16), and FECH mRNA encodes a protein of 423 amino acid residues (17, 18). FECH DNA analysis in patients with symptomatic EPP has identified many mutations that alter structure/function of the enzyme, together with a polymorphism in intron 3 (IVS3-48c) that causes low expression of the nonmutant FECH allele (19–24).
However, not all patients with EPP have a FECH mutation detected by conventional sequencing of FECH cDNA and the 11 FECH exons. In some, gene dosage analysis has identified large deletions of the FECH gene (25). C-terminal deletions in the gene that encodes the erythroid-specific isoform of delta-aminolevulinic acid synthase (ALAS2), the regulatory enzyme in heme biosynthesis, also cause increased formation of protoporphyrin sufficient to produce EPP phenotype (26).
Proteins that may impact phenotypic expression in EPP include the protein that co-localizes with FECH in mitochondria (27), heme-regulated eIF2- kinase (28), and iron-regulatory protein 2 (IRP2) that post transcriptionally regulates expression of iron metabolism (29, 30). The protein responsible for mitochondrial iron transport in vertebrate erythroblasts is a member of the mitochondrial solute carrier family (Mitoferrin-1, MFRN1, SLC25A37) (31). An abnormality in MFRN1 synthesis or function could decrease transport of iron to FECH, and impair formation of the 2Fe2S cluster that is essential for human FECH activity (16). FECH protein is in the same oligomeric protein complex with MFRN1 and ABCB10, which directly integrates the importation of mitochondrial iron with heme synthesis (32).
In this manuscript, we report seven individuals with EPP phenotype, who had no abnormalities in FECH DNA detected by conventional molecular analysis. Additional studies examined for large deletions of FECH, C-terminal deletions in ALAS2, and abnormal MFRN1 expression in these patients, whom we have designated to have variant EPP. We here demonstrate abnormal expression of MFRN1 in variant EPP.
PATIENTS AND METHODS
Population
Seven individuals from five unrelated families had photosensitivity characteristic of EPP, with erythrocyte protoporphyrin levels ranging from 760 to 7,624 ug/dl (normal less than 100 in Bloomer Laboratory) (Table 1). Four patients underwent transplantation for advanced liver disease, and their livers had the black color and histological features described in classical EPP liver disease (4,10). Neither a FECH mutation nor the polymorphism IVS3-48c was found by FECH DNA analysis on at least three independent assays. The gender, age range, and erythrocyte protoporphyrin levels in the patients were similar to those in symptomatic patients with classical EPP (Table 1) (24). Studies were done with informed consent under the guidance of the Institutional Review Boards at the University of Alabama at Birmingham, Children’s Hospital Boston, and the University Hospital of Wales, Cardiff.
Table 1.
Clinical and Laboratory Parameters in Classical and Variant EPP Patients
| Classical EPP | Variant EPP | |
|---|---|---|
| Number | 32* | 7 |
| Gender | 15M / 17F | 4M/ 3F |
| Mean Age (Range) | 34 (10–68) | 31 (14–55) |
| Photosensitivity | 32 (100%) | 7 (100%) |
| Liver Transplantation | 10 (31%) | 4 (57%) |
| Mean RBC Protoporphyrin (range) (µg/dl) |
2274 (430–8240) |
4104 (760–7624) |
| Mean Hgb and Range (g/dl) | 10.8 (8.1–12.4) | 10.6 (8.8–14.1) |
| FECH Mutation | 32 (100%) | 0 |
| FECH Polymorphism IVS3-48c | 31 (97%) | 0 |
Patients previously reported by Risheg et al.24
Biochemical Studies
Routine biochemical parameters were assessed in clinical chemistry laboratories. Erythrocyte protoporphyrin levels were measured fluorometrically after solvent partitioning (4, 24). FECH enzyme activity was assayed in sonicates of EBV-transformed lymphoblasts from seven patients and five normal individuals, using an assay that measures the formation of zinc-deuteroporphyrin in nmol/mg protein/h (u) (33).
FECH DNA Analysis
Genomic DNA and total RNA were extracted from peripheral blood leukocytes, EBV-transformed lymphoblasts, and liver tissue as previously described (24, 33, 34). Specific PCR was used to amplify and sequence FECH DNA and the 11 exons and flanking intron regions of the FECH (24, 33, 34). Specific primers were used to amplify a 212-bp fragment of intron 3 containing the −48t/c base (24). Sequencing of purified products was done using the ABI 3730xl DNA Sequencer (Applied Biosystems).
Gene Dosage Analysis of FECH
Genomic DNA was extracted from liver tissue and blood leukocytes, (24, 33, 34), and gene dosage analysis was done as previously described (25), except that the FECH amplicons were analyzed singly and not as a multiplex.
ALAS2 DNA Analysis
Exon 11 of ALAS2, located on the X chromosome, was amplified and sequenced by specific PCR, using primer sequences and conditions previously described (26).
MFRN1 cDNA and Genomic DNA Analysis
Total RNA was isolated from blood leukocytes and liver tissue, and specific reverse transcriptase polymerase chain reactions (RT- PCR) were done to amplify and sequence MFRN1 cDNA (Table 2). The sequences for primers were taken from the sequence for Gene ID: 51312 (SLC25A37) in the NCBI website. SuperScript II Kit (Life Technologies) was used for first strand cDNA synthesis. Total RNA was denatured at 70°C for 10 min; cDNA was synthesized at 42°C for 50 min and terminated at 70°C for 15 min. RNA was removed using RNase H, and the first stand cDNA was amplified using GeneAmp PCR System 2400 (Perkin-Elmer) and Taq DNA Polymerase Kit (Life Technologies); 25–50 pmol of primer was used for 35 cycles of denaturation at 94°C for 10 sec, annealing of primers at 56 >58°C for 60 sec and elongation at 72°C for 120 sec.
Table 2.
Primers for Sequencing MFRN1 cDNA and Genomic DNA, and Quantitative Real-Time PCR Measurement of MFRN1 RNA
| MFRN1 cDNA | |||
|---|---|---|---|
| Primer | Sequence | Product Size (bp) | |
| Set 1 | Forward ___________ Reverse |
GGAGGATGGATGGGGACA ______________________________ TGGCCATACTCCCAGCTATC |
414 |
| Set 2 | Forward ___________ Reverse |
CAACGGGATAGCTGGGAGTA ______________________________ GTATGGAGCTCGATTTTCCAG |
586 |
| Set 3 | Forward ___________ Reverse |
TCTACGGAGCCCTCAAGAAA ______________________________ TGGCCATACTCCCAGCTATC |
180 |
| MFRN1 exons and flanking intron regions | |||
|---|---|---|---|
| Exon | Forward Primer | Reverse Primer | Product size (bp) |
| 1 | GGTGGCTCCCACTTTAAGAA | CGCTCGTTGCGTCCCCGAAG | 330 |
| 2 | TTTTTCTCTCTCTCCTTCTC | GGGTCACAGGGAGGAACA | 404 |
| 3 | AAGTCCTGGGTTCCGTCTGA | CTCTCCACTCTCCCACAAC | 225 |
| 4 | CTCTTTATGGCTTGGCTT | CGGTGGAGGGCATCTGGTAG | 543 |
| qRT-PCR measurement of MFRN1 RNA | |||
|---|---|---|---|
| Normal RNA | |||
| Oligo | Length (bases) |
Position | Sequence |
| Forward primer | 20 | Exon 2 | ATGAAAACATGAAAAGGACT |
| Reverse primer | 18 | Exon 2 – Exon 3 | GCCATACTCCCAGCTATC |
| Quantiprobe | 17 | Exon 2 | CACCAAGGAAACAGCCA |
| Abnormal RNA | |||
|---|---|---|---|
| Oligo | Length (bases) |
Position | Sequence |
| Forward primer | 19 | Intron 2 | ACAAACAGGTCTCTACAAC |
| Reverse primer | 18 | Exon 3– Exon 4 | CGCTGCTTCACCACTTCT |
| Quantiprobe | 17 | Exon 3 | ACCCTGCTCCACGATGC |
Amplified PCR products were analyzed using electrophoresis on 1.5–1.8% agarose gel. Products of expected size and different size were excised and purified using QIA quick Gel Extraction Kit (Qiagen). Direct sequencing was done, and the sequence analyzed using DNASIS-Mac v2.0 software.
Extraction of genomic DNA used Puregene DNA Isolation Kit (Gentra Systems). Specific primers for amplification of the exons and their flanking intron regions were used (Table 2), and subcloning and sequencing were performed.
Lymphoblast Culture
EBV-transformed lymphoblasts were cultured in RPMI 1640 containing penicillin/streptomycin sulfate (Cellgro) and 20% heat-inactivated fetal bovine serum (Biomeda) until cells grew well. They were then cultured in the same medium containing 15% fetal bovine serum for three to four days to reach log phase of growth, and harvested for measurement of FECH activity and MFRN1 mRNA.
Quantitative Real-Time PCR Measurement of Normal and Abnormal MFRN1 mRNA
Levels of MFRN1 mRNA were measured by qRT-PCR in lymphoblasts from seven individuals with variant EPP and five normal individuals, using the QuantiTect™ Custom Assay (Qiagen). RNA was extracted from human liver, and amplicons for normal and abnormal mRNA were generated by RT-PCR, purified, and cloned into pCR2.1 vector (Invitrogen). Ligated fragments were transformed into DH5-competent cells (Life Technologies). Plasmid DNA was prepared and cloned amplicons sequenced; cDNA plasmid concentrations were measured by optical density spectrophotometry (Spectronic Genesys 5). Serial dilutions of the cDNA plasmid were used as standard curve. Primers and probes were designed by Qiagen Quantiprobe Design Software (Qiagen) (Table 2). Real-time PCR was performed on ABI prism 7700 sequence detection system (Applied Biosystems). Standard wells contained 10 to 4000 copies of plasmid MFRN1 cDNA.
Zebrafish Strains and Complementation by cRNA Microinjection
The MFRN1-deficient strain, frascati (frstq223) was maintained in compliance with institutional standards. Normal and abnormal human MFRN1 cDNA were subcloned into the pCS2+ vector, and 5’-capped cRNA was prepared using the SP6 mMessage mMachine kit (Ambion) (31). Fertilized eggs from frs heterozygote pairs were injected with cRNA, and embryos at three days post-fertilization were assayed for hemoglobinization with o-dianisidine, and genotyped for the frstq223 allele (31).
Yeast Complementation and Analysis
Human MFRN1 cDNA was subcloned into pRS426-ADH vector and transformed into yeast Δmrs3/4 and wild type strains for complementation analysis on iron-depleted media (31, 35).
Transient Transfection of Mammalian Cells, Immunohistochemistry, and 55Fe-Flux Assay
Human MFRN1 cDNA clones were transiently transfected into Cos7 cells using Lipofectamine (Invitrogen). After 48 h post-transfection, the cells were harvested and mitochondrial and cytosolic fractions prepared using the Mitochondrial Isolation Kit (Pierce) (36). Transfected cells were processed for confocal immunofluoresence microscopy (31) at the Harvard Digestive Disease Center Imaging Facility (Children’s Hospital Boston).
Human K562 cells were cultured in supplemented DMEM media and transfected, using the Amaxa nucleofection device with Solution V and Program T16. Fe-saturated transferrin preparation, using 55FeCl3 (Perkin Elmer), metabolic labeling of Fe-starved K562 cells (~1 × 106 ), and quantification of 55Fe-complexed as heme in a liquid scintillation counter were done (31). The concentration of diferric Tf as 55Fe-Tf was 100 ug/ml media, and the time of incubation with 55Fe-Tf was 5–6 hrs at 37°C.
Minigene Expression for RNA Processing
Genomic DNA from lymphoblasts of control and EPP patients were amplified by PCR, encompassing exon 2, intron 2 and exon 3. This DNA fragment was subcloned into the pcDNA3.1 vector to generate MFRN “minigene” constructs, which were transfected into Friend mouse erythroleukemia (MEL) cells. Stable MEL cells were selected for zeocin-resistance. Total RNA was processed by RT-PCR, and the PCR products were analyzed (37).
RESULTS
Clinical and Biochemical Features in Variant EPP
Clinical and biochemical features of patients with variant EPP are listed in Tables 1 and 3. All had photosensitivity; four had liver transplantation because of advanced EPP liver disease. However, the high frequency reflects ascertainment bias, as they were specifically referred to one of the authors (JRB) because of liver disease. Five remain alive, while two (A.1 and E.1.) have died from complications of recurrent EPP liver disease in the graft.
Table 3.
Clinical Parameters and Molecular Defects in Patients with Variant EPP
| Patient # |
Age (years) |
Race* | Gender | Hgb† | RBC PP‡ |
Liver Transplant |
ALAS2 DNA | Large FECH Deletion |
|---|---|---|---|---|---|---|---|---|
| A.1 | 15 | C | M | 9.1 | 6470 | Yes | WT | None |
| B.1 | 55 | C | M | 8.8 | 3136 | Yes | delAGTG§ | Not done |
| B.2 | 45 | C | M | - | 1994 | No | delAGTG | Not done |
| B.3 | 27 | C | F | - | 760 | No | delAGTG / WT | Not done |
| C.1 | 47 | C | F | 14.1 | 3079 | No | delAGTG/WT | Not done |
| D.1 | 17 | AA | F | 9.0 | 7624 | Yes | WT/WT | None |
| E.1 | 14 | C | M | 12.0 | 5665 | Yes | WT | Heterozygous¶ |
C (Caucasian), AA (African-American);
Blood Hemoglobin level (g/dL);
Red Blood Cell Protoporphyrin level (µg/dL);
c.1706-1709 deletion in exon 11;
c.1−7887c.67+2425del10379insTTCA
Three patients (B.1, 2, 3) were in the same family; B.1 and B.2 were brothers, and B.3 was the daughter of B.1. Patient A.1 was an orphan, with no known family members. Patient C.1 has three sons, none of whom have photosensitivity or elevated erythrocyte protoporphyrin levels; two half-sisters, a full sister, and her father also do not have photosensitivity. Her mother has avoided sunlight all her life because of photosensitivity; she refused biochemical studies. Neither parent nor siblings of patient D.1 have photosensitivity, and refused biochemical or genetic studies. Neither parent nor the siblings of Patient E.1 have photosensitivity, and have normal erythrocyte protoporphyrin levels.
ALAS2 DNA and FECH Gene Dosage Analysis
ALAS2 analysis was done to search for deletions in exon 11 that cause replacement or deletion of C-terminal residues of the enzyme, resulting in gain of function that leads to an increase in erythrocyte protoporphyrin production (26). Four patients had c.1706-1709 delAGTG (Table 3), as described by Whatley et al (26). Two female patients had deletions in ALAS2, which is located on X chromosome. Patient B.3 was heterozygous for the deletion and had erythrocyte protoporphyrin level of 760 ug/dl. Patient C.1 was also heterozygous for the deletion but had a significantly higher level of erythrocyte protoporphyrin (3079 µg/dl, of which 30% was zinc protoporphyrin).
FECH gene dosage analysis on the remaining three patients showed no abnormality in patient A.1 or D.1, but patient E.1 had heterozygous deletion of the 5´-untranslated region and exon 1 (c.17887c.67+2425del10379insTTCA) as previously described (Table 3) (25).
MFRN1 cDNA Analysis
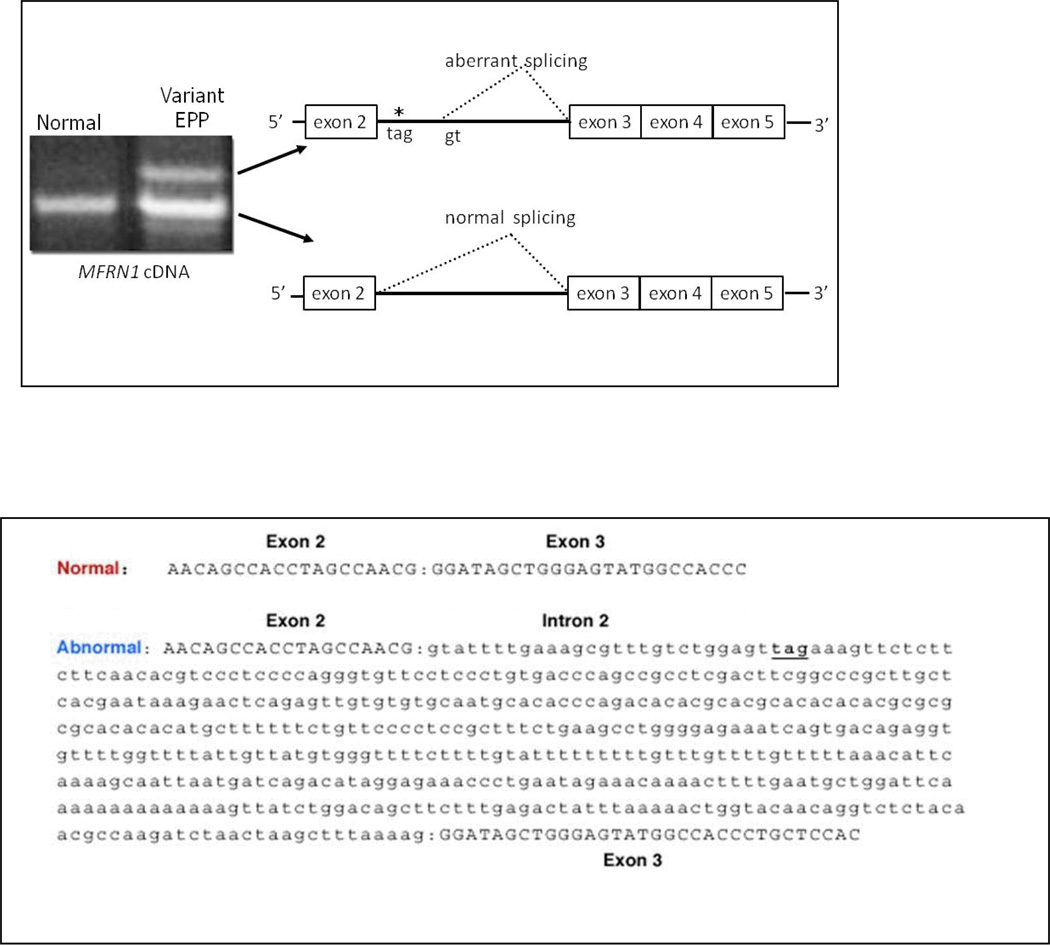
MFRN1 cDNA was examined using RNA extracted from peripheral blood leukocytes and/or liver tissue. In addition to the species of normal size, an abnormal species of MFRN1 cDNA was present in all variant EPP patients due to insertion of a 477 base segment of intron 2 between exons 2 and 3 (Fig. 1). The dinucleotide sequence in intron 2 that immediately follows the 477 base segment is gt, indicating that the abnormal species may arise due to activation of a cryptic donor splice site in intron 2 which causes a stop codon at amino acid position 156.
Figure 1. Aberrant Mitoferrin-1 (MFRN1) splicing in variant EPP.
Variant EPP patients exhibited two distinct MFRN1 transcripts. In addition to the transcript of normal size, they had a longer transcript due to the insertion of 477 bases of intron 2, which causes a nonsense codon (tag) at amino acid position 156 as shown by the asterisk. The aberrant splicing may result from activation of a cryptic donor splice site (gt) in intron 2. The cDNA sequences for the normally processed and aberrantly processed mRNA transcripts are shown. A faint product shorter than the normal product was also analyzed and found to have a nonspecific DNA sequence. When the stringency of the PCR reaction was enhanced by increasing the annealing temperature to 62°C and decreasing the primer concentration, the short product disappeared, indicating that it was a PCR artifact.
MFRN1 Expression and Level of FECH activity in Cultured Lymphoblasts
MFRN1 expression was quantatively examined in cultured EBV-transformed lymphoblasts from the seven patients with variant EPP and compared to that in five normal EBV-transformed lymphoblast lines. Levels of FECH enzyme activity were also measured.
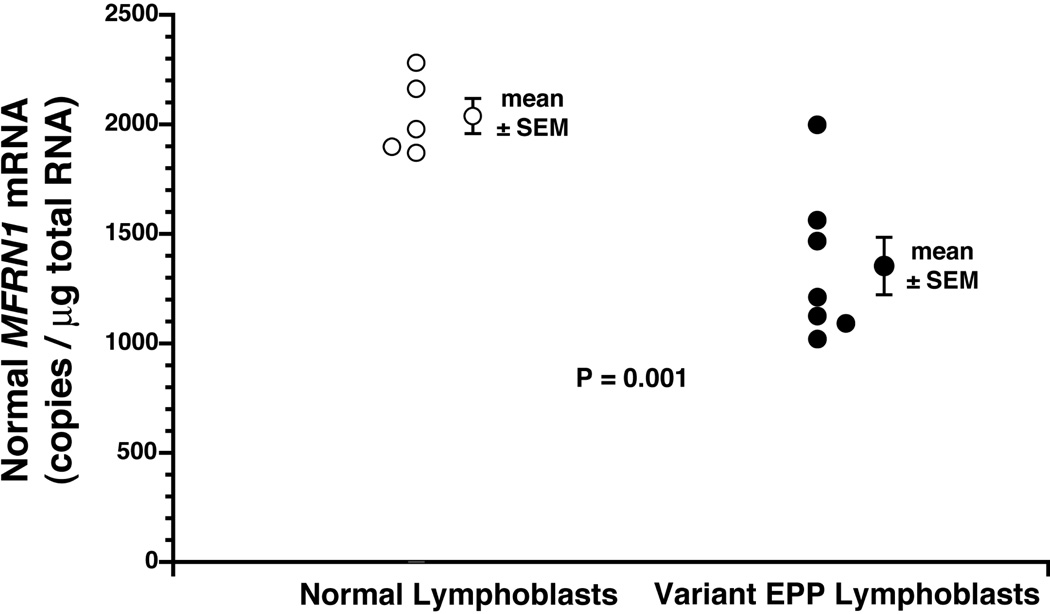
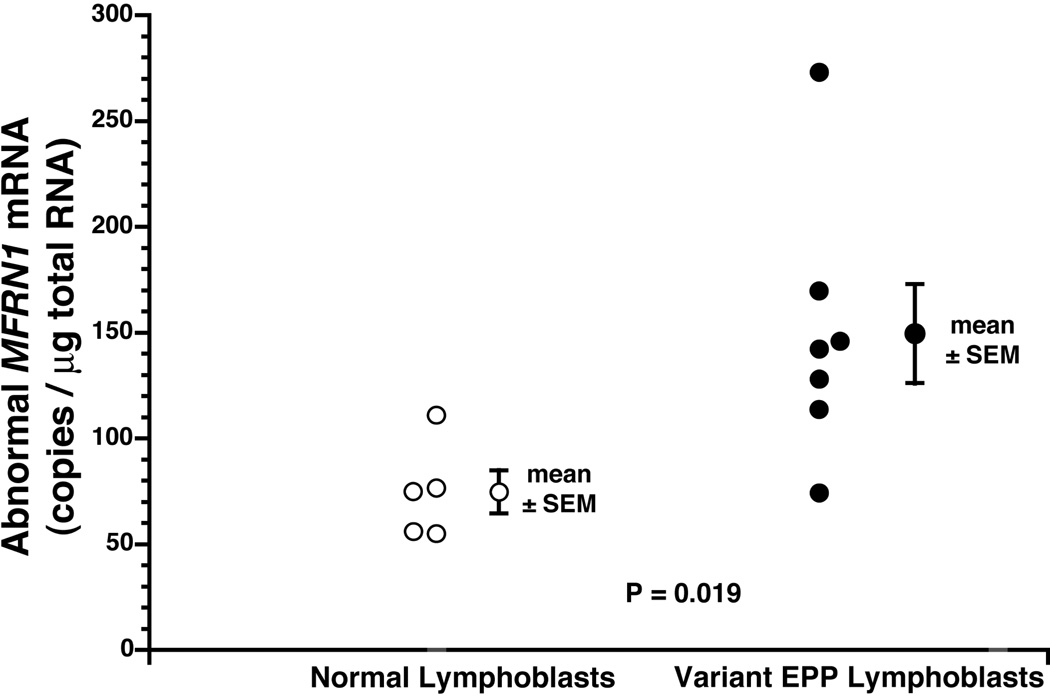
One patient with variant EPP (D.1) had a normal level of normal MFRN1 mRNA, whereas the other six all had levels below the range (1871 to 2282 copies/µg total RNA) in the normal lymphoblast lines (p=0.001) (Fig. 2). The mean level in the six patients was reduced to 61% of that in the normal lines. In contrast, the mean level of abnormal MFRN1 mRNA was significantly increased in the patients with variant EPP (150 ± 24 copies per µg total RNA) compared to that in the normal lines (75 ± 10 copies) (p=0.019) (Fig.3).
Figure 2. Normal MFRN1 mRNA levels in lymphoblasts.
Figure 3. Abnormal MFRN1 mRNA levels in lymphoblasts.
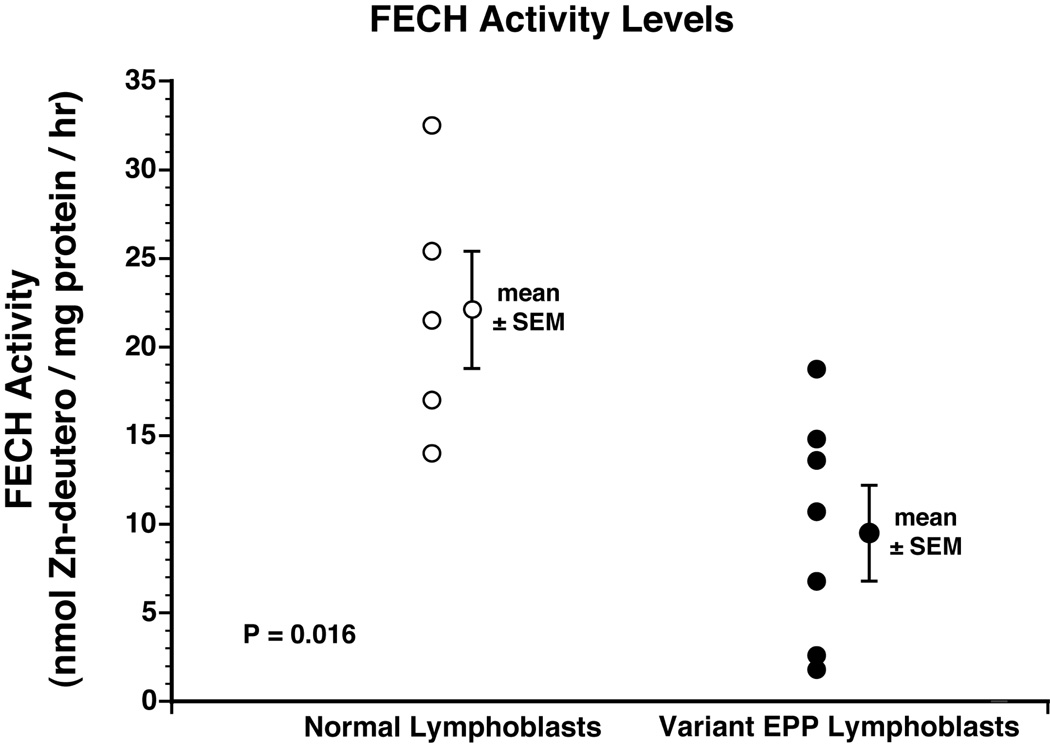
FECH enzyme activity in sonicated lymphoblasts of the seven variant EPP patients (9.9 ± 2.4 u) was significantly lower than that in the normal lymphoblast lines (22.1 ± 3.2 u) (p=0.011) (Fig. 4). The patient with normal level of normal MFRN1 mRNA also had a normal level of FECH enzyme activity (18.6 u).
Figure 4. FECH activity levels in lymphoblasts.
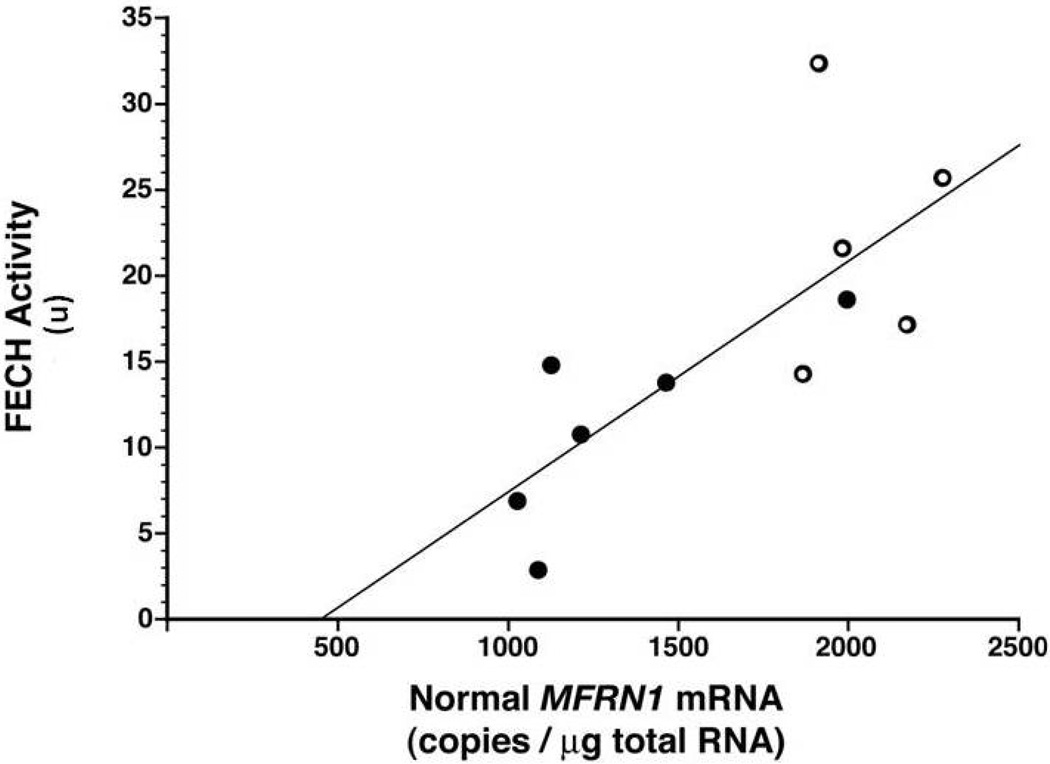
The relationship between FECH activity and normal MFRN1 mRNA was examined in the combined group of normal lymphoblast lines and six variant EPP lines. Patient E.1 was excluded because he had a major deletion in one of his FECH alleles. Using Microsoft Excel, there was a positive correlation coefficient of 0.75, and the relationship was defined by the formula: FECH activity = (0.0134 × normal MFRN1 mRNA level) – 5.9 (Fig 5).
Figure 5. Relationship between FECH activity and normal MFRN1 mRNA in lymphoblasts.
A positive correlation coefficient of 0.75 was found between these two measurements in the combined normal (open circles) and variant EPP (closed circles) lymphoblast lines.
Functional Studies of the Abnormal MFRN1 Transcript
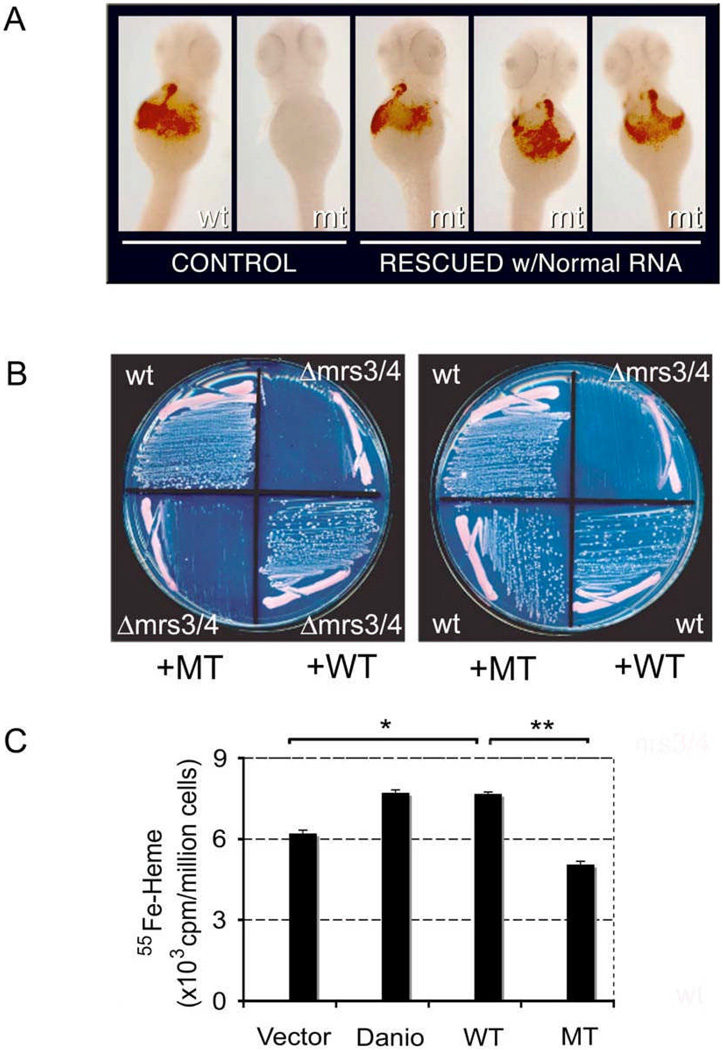
In order to examine the biological consequences of the abnormal MFRN1 transcript, normal and abnormal human MFRN1 cDNA were subcloned into pCR2.1TA vector. Normal and abnormal MFRN1 cDNA programmed in vitro translation of polypeptides 37 kDa and 17 kDa, respectively, as predicted from cDNA sequences. Normal and abnormal MFRN1 cDNA were subcloned into expression vectors pCS2+ and pRS426-ADH for functional interrogation in zebrafish and yeast. By microinjection of cRNA into fertilized eggs from a MFRN - deficient zebrafish strain, frascati (frs), normal human MFRN1 cDNA was able to fully complement the anemia in mutant embryos (Fig. 6A). The number of hemoglobinized cells in frs anemic embryos (80% rescued, n=210) was comparable to the endogenous zebrafish and mouse cDNA clones (31). In contrast, the abnormal MFRN1 cDNA failed to complement anemia in frs mutants (0%, n=135), which indicates that abnormal MFRN1 cDNA has loss-of-function activity.
Figure 6. Functional analysis of the normally and abnormally processed MFRN1 cDNA.
(A) Complementation of anemia in frascati zebrafish embryos with normal MFRN1 cRNA. Wild type (wt) and frascati (mt) embryos are shown in first two panels. Genotypic mt embryos exhibit restored hemoglobinized cells after injection with the normal MFRN1 cRNA. (B) Complementation of Δmrs3/4 growth defect on Fe-depleted media by normal (WT) MFRN1 cDNA, but not abnormal (MT) MFRN1 (left panel). Abnormal (MT) MFRN1 is not a gain-of-function, dominant mutation when expressed in a wild type (wt) yeast strain (right panel). (C) 55Fe-heme synthesized in K562 cells nucleofected with empty vector, zebrafish (Danio) mfrn1, human normal (WT) and abnormal (MT) MFRN1 cDNA. cDNA from Danio and normal human MFRN1 programmed increased 55Fe-heme production above control vector, * p<0.05 (t-test). Abnormal (MT) MFRN1 cDNA generated significantly less 55Fe-heme compared to WT, ** p=0.004 (t-test). Experiments were done in triplicates.
Similarly, studies in the Δmrs3/4 yeast mutant, which is deficient in the yeast ortholog for MFRN, showed that normal human MFRN1 cDNA rescued the iron auxotrophy when cultured on iron-depleted media, whereas the abnormal MFRN1 cDNA could not (Fig. 6B). The truncated MFRN1 polypeptide could potentially act as a dominant-negative protein since members of the SLC25 carriers normally exist as homodimers (38). To examine this possibility, mutant MFRN1 cDNA was transformed into wild type yeast, and there was no inhibition of growth on iron-depleted media (Fig. 6B).
MFRN1 carriers import ferrous iron into mitochondria for biogenesis of heme and Fe/S clusters (31, 36, 39). 55Fe-heme synthesis was examined in hematopoietic K562 cells nucleofected with normal and abnormal human MFRN1 cDNA. This showed that normal human MFRN1 cDNA significantly increased synthesis of 55Fe-heme in K562 cells compared to an empty vector control (p<0.05) (Fig. 6C). In contrast, the abnormal MRFN1 cDNA did not result in 55Fe-heme synthesis compared to the normal cDNA (p=0.004). Zebrafish cDNA was comparable to the human clone in the amount of 55Fe incorporation into heme. Some basal level of 55Fe-heme synthesis occurs because endogenous MFRN1 and MFRN2 transporters are functional.
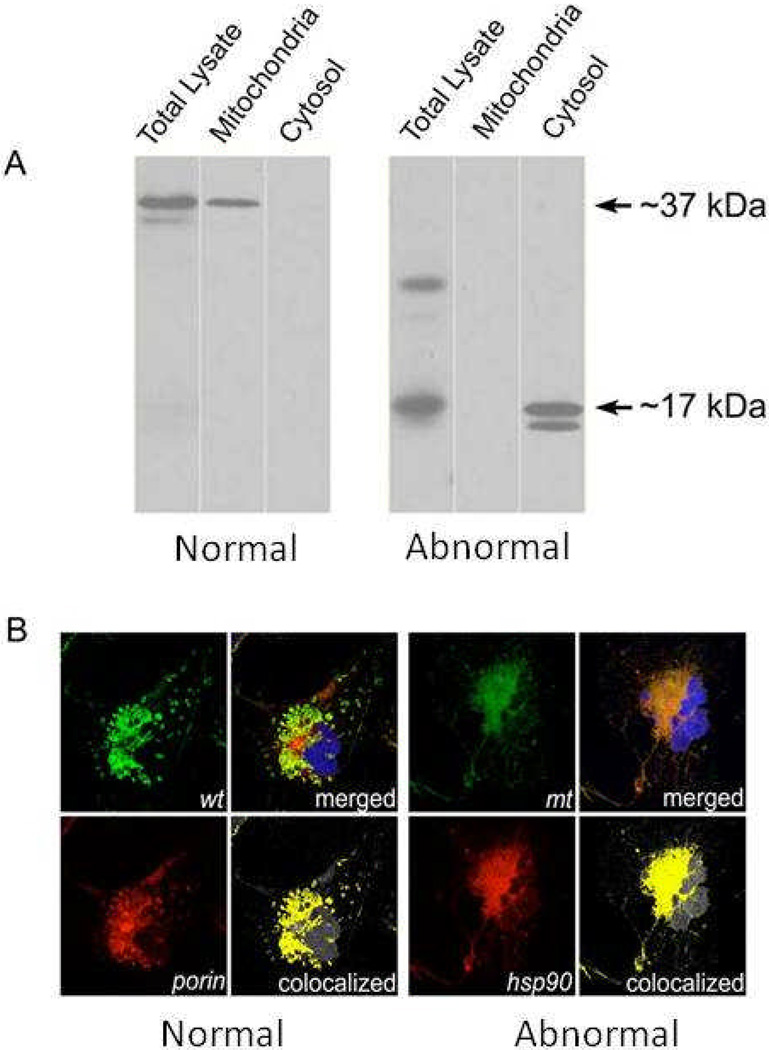
To evaluate the subcellular localization of abnormal MFRN1 protein, as protein mistargeting could explain its null activity, normal and abnormal MFRN1 cDNA were tagged with FLAG-epitope and transfected into Cos7 cells. Western blot analysis of total lysate, as well as the mitochondrial and cytosol fractions, showed that normal human MFRN1 protein targeted properly to the mitochondria, whereas abnormal human MFRN1 was retained in cytosol (Fig 7A). Immunofluorescence confocal microscopy validated this finding, showing colocalization of normal MFRN1 with mitochondria resident protein porin; whereas, abnormal MFRN protein colocalized with cytosolic marker hsp90 (Fig. 7B).
Figure 7. Mis-targeting of abnormal MFRN1 protein in mammalian cells.
(A) FLAG-tagged normal and abnormal MFRN1 cDNA were transfected into Cos7 cells. Western analysis shows proper mitochondrial targeting of the normal human MFRN1 protein, whereas the abnormal human MFRN1 protein is prematurely truncated and retained in the cytosol. (B) Confocal immunofluorescence microscopy analysis of subcellular localization of MFRN1 proteins. Normal (wt) MFRN1 protein colocalizes with porin in the mitochondria (left panels), whereas, abnormal (mt) MFRN1 protein is retained in the cytosol by its colocalization with heat-shock hsp90 (right panels). DAPI dye stains the cell nucleus (blue).
MFRN Genomic DNA Analysis
Using genomic DNA isolated from EBV-transformed lymphoblasts of the variant EPP patients, the exons and their flanking intron regions in MFRN1 were sequenced to search for mutations that might alter the function of the protein. None were identified.
A search was also made for single nucleotide polymorphism in MFRN1 that might be associated with aberrant splicing. Three sites of polymorphism were identified in exon 2, and three in intron 2 (+289 t/g, +293 t/g, +298 t/g). The relative frequency of the polymorphism at these sites in the variant EPP patients was not significantly different from that in 27 normal/control individuals.
“Minigene” cassettes of genomic DNA fragments from normal and EPP patients, encompassing exon 2, intron 2, and exon 3, were expressed in murine MEL and human HEK293 cells. Stable clones were selected for analysis of RNA splicing of the MFRN1 transcript. No obvious difference was noted in the MFRN1 transcripts from these “minigene” constructs.
DISCUSSION
Patients with EPP phenotype usually are heterozygous for a mutation in FECH, the gene coding for the mitochondrial enzyme that inserts ferrous iron into protoporphyrin to form heme, accompanied by a polymorphism in the non-mutant FECH allele (IVS3-48c) that is associated with low expression (19–24, 40). The combination reduces FECH activity to less than 30% of normal (12, 22–24), sufficient to cause a level of excess protoporphyrin accumulation that leads to clinical manifestations.
However, not all patients with EPP phenotype have a mutation in FECH when conventional DNA analysis is done (25, 26). In this report, we describe seven North American patients with EPP phenotype, in whom neither a mutation nor the IVS3-48c polymorphism was found in FECH. Four patients had liver transplantation for EPP liver disease, indicating severe phenotype. Analysis of ALAS2 showed that four patients, three in the same family, had c.1706-1709 delAGTG in exon 11 (Table 3). A fifth patient had a large hemizygous deletion in FECH previously described (25). The other two patients (A.1 and D.1) had no obvious defects in either ALAS2 or FECH.
Studies in human erythroleukemia K562 cells and Cos7 cells expressing human FECH indicated that the level of FECH activity is regulated by intracellular iron via the 2Fe2S cluster bound to the C-terminal region of the enzyme protein (41). In the presence of low intracellular iron, there was rapid degradation of FECH protein (41). Post-translational stability of FECH is dependent on iron availability and intact 2Fe2S cluster assembly machinery (42), providing a link between Fe-S biogenesis and completion of heme synthesis (42, 43).
If iron is an important regulator of the final step in heme biosynthesis, the protein that transports iron in mitochondria should also be a factor, since it could be a determinant of the concentration and availability of mitochondrial iron. MFRN1 has been identified as the principal mitochondrial iron transporter in vertebrate erythroblasts through studies in a zebrafish mutant with profound anemia and erythroid maturation arrest due to defective mitochondrial iron uptake (31). MFRN1 is a member of the SLC25 solute carrier family that shuttles a diverse group of compounds across the inner mitochondrial membrane (38). FECH is part of an oligomeric complex with MFRN1 and ABCB10, implicating the synergistic integration of mitochondrial iron import with utilization for heme biosynthesis (32).
In our studies, six of the seven patients with variant EPP had a significant decrease in the level of normal MFRN1 mRNA in cultured lymphoblasts. The decrease in normal MFRN1 mRNA may be caused by aberrant splicing, in which there is increased formation of abnormal MFRN1 mRNA that is rapidly degraded by nonsense mediated decay (40). The abnormal steady-state MFRN1 mRNA remaining codes for a defective protein that does not target properly to mitochondria. The net result is a significant decrease in the amount of the functional mitochondrial iron transporter. This in turn may cause a decrease in FECH activity due to reduced formation of the 2Fe2S complex that is critical to FECH stability and activity.
The mechanism responsible for aberrant splicing of MFRN1 mRNA was not determined by our studies, however. In particular, polymorphisms in intron 2 or exon 2 that could be associated with aberrant splicing were not identified. Given the complexity of RNA splicing because of defective splicing due to mutations acting through long distances (44), splicing polymorphisms in distal regions of large introns of MFRN1, such as intron 1 (36,895 bp) and intron 3 (2,956 bp), are possible. The mini-gene constructs that we used to assay RNA processing may not mimick the in vivo splicing pattern without the full genomic context of adjacent exons (45). Polymorphic changes in distal genomic DNA could potentially alter the affinity of positive and negative splicing regulatory factors, which regulate the efficiency of splice site selection. A precedence for the complexity of interplay between positive and negative splicing regulators in exon inclusion/skipping and splice site selection is well documented (46, 47). Finally, splicing factors such as those associated with retinitis pigmentosa and spinal muscular atrophy have been shown to alter the repertoire of small nuclear ribonucleoprotein particles (snRNPs) and perturb pre-mRNA processing, leading to differential splice site usage and splicing defects (48–50).
Acknowledgments
ACKNOWLEDGMENT AND SUPPORT
We are grateful to Dr. Sharon Whatley in the Department of Medical Biochemistry and Immunology at University Hospital of Wales, Cardiff, U.K., for gene dosage analysis of FECH. Confocal immunofluorescence microscopy was performed at the Harvard Digestive Disease Center Imaging Facility (Children’s Hospital Boston). The work was supported by grants from American Society of Hematology (G.C.S.), March of Dimes Foundation (B.H.P.), America Porphyria Foundation (J.R.B.), and National Institutes of Health grants R01 DK026466 and 5U54DK038909 (J.R.B.), R01 DK052380 (J.K.), R01 DK070838 (B.H.P.), P01 HL032262 (B.H.P.), and P30 DK072437 (J.K., B.H.P.).
Footnotes
The authors declare no conflicts of interest.
LITERATURE CITED
- 1.Lecha M, Puy H, Deybach JC. Erythropoietic protoporphyria. Orphanet Journal of Rare Diseases. 2009;4:19–29. doi: 10.1186/1750-1172-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poh-Fitzpatrick MB. Molecular and cellular mechanisms of porphyrin photosensitization. Photodermatology. 1986;3:148–157. [PubMed] [Google Scholar]
- 3.Timonen K, Kariniem A, Niemi K, Teppo A, Tenlvunen R, Kauppinen R. Vascular changes in erythropoietic protoporphyria: Histopathologic and immunohistochemical study. J Am Acad Dermatol. 2000;43:489–497. doi: 10.1067/mjd.2000.107498. [DOI] [PubMed] [Google Scholar]
- 4.Bloomer JR, Phillips MJ, Davidson DL, Klatskin G. Hepatic disease in erythropoietic protoporphyria. Am J Med. 1975;58:869–882. doi: 10.1016/0002-9343(75)90644-0. [DOI] [PubMed] [Google Scholar]
- 5.Doss MO, Frank M. Hepatobiliary implications and complications in protoporphyria, a 20 year study. Clin Biochem. 1989;22:223–229. doi: 10.1016/s0009-9120(89)80081-5. [DOI] [PubMed] [Google Scholar]
- 6.Avner DL, Lee RG, Berenson MM. Protoporphyrin-induced cholestasis in the isolated in situ perfused rat liver. J Clin Invest. 1981;67:385–394. doi: 10.1172/JCI110046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rademakers LH, Cleton MI, Kooijman C, baart de la Faille H, van Hattum J. Early involvement of hepatic parenchymal cells in erythrohepatic protoporphyria? An ultrastructural study of patients with and without overt liver disease and the effect of chenodeoxycholic acid treatment. Hepatology. 1990;11:449–457. doi: 10.1002/hep.1840110316. [DOI] [PubMed] [Google Scholar]
- 8.Meerman L, Koopen NR, Bloks B, et al. Biliary fibrosis associated with altered bile composition in a mouse model of erythropoietic protoporphyria. Gastroenterology. 1999;117:696–705. doi: 10.1016/s0016-5085(99)70464-6. [DOI] [PubMed] [Google Scholar]
- 9.Komatsu H, Sajima Y, Imamura K, et al. An ultrastructural study of the liver in erythropoietic protoporphyria. Med Electron Microsc. 2000;33:32–38. doi: 10.1007/s007950000005. [DOI] [PubMed] [Google Scholar]
- 10.McGuire BM, Bonkovsky HL, Carithers RL, et al. Liver transplantation for erythropoietic protoporphyria liver disease. Liver Transplantation. 2005;11:1590–1596. doi: 10.1002/lt.20620. [DOI] [PubMed] [Google Scholar]
- 11.Bottomley SS, Tanaka M, Everet MA. Diminished erythroid ferrochelatase activity in protoporphyria. J Lab Clin Med. 1975;86:126–131. [PubMed] [Google Scholar]
- 12.Bonkovsky HL, Bloomer JR, Ebert PS, Mahoney MJ. Heme synthetase deficiency in human protoporphyria. Demonstration of the defect in liver and cultured skin fibroblasts. J Clin Invest. 1975;56:1139–1148. doi: 10.1172/JCI108189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scholnick P, Marver HS, Schmid R. Erythropoietic protoporphyria: evidence for multiple sites of excess protoporphyrin formation. J Clin Invest. 1971;50:203–207. doi: 10.1172/JCI106474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poh-Fitzpatrick MB. Protoporphyrin metabolic balance in human protoporphyria. Gastroenterology. 1985;88:1239–1242. doi: 10.1016/s0016-5085(85)80085-8. [DOI] [PubMed] [Google Scholar]
- 15.Dailey HA, Dailey TA, Wu CK, Medlock AE, Wang KF. Ferrochelatase at the millennium: structures, mechanisms and [2Fe-2S] clusters. Cell Mol Life Sci. 2000;57:1909–1926. doi: 10.1007/PL00000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu CK, Dailey HA, Rese JP, Burden AM, Sellers VA, Wang BC. The 2A structure of human ferrochelatase, the terminal enzyme of heme biosynthesis. Nat Struct Bio. 2001;8:156–160. doi: 10.1038/84152. [DOI] [PubMed] [Google Scholar]
- 17.Nakahashi Y, Taketani S, Okuda M, Inoue K, Tokunaga R. Molecular cloning and sequence analysis of cDNA endoding human ferrochelatase. Biochem Biophys Res Comm. 1990;173:748–755. doi: 10.1016/s0006-291x(05)80099-3. [DOI] [PubMed] [Google Scholar]
- 18.Taketani S, Inazawa J, Nakahashi Y, Abe T, Tokunaga R. Structure of the human ferrochelatase gene. Eur J Biochem. 1992;205:217–222. doi: 10.1111/j.1432-1033.1992.tb16771.x. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Poh-Fitzpatrick M, Taketani S, Chen T, Piomelli S. Screening for ferrochelatase mutations: molecular heterogeneity of erythropoietic protoporphyria. Biochem Biophys Acta. 1994;1225:187–190. doi: 10.1016/0925-4439(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 20.Rufenacht UB, Gouya L, Schneider-Yin X, et al. Systematic analysis of molecular defects in the ferrochelatase gene from patients with erythropoietic protoporphyria. Am J. Hum Genet. 1998;62:1341–1352. doi: 10.1086/301870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sellers VM, Dailey TA, Dailey HA. Examination of ferrochelatase mutations that cause erythropoietic protoporphyria. Blood. 1998;91:3980–3985. [PubMed] [Google Scholar]
- 22.Gouya L, Puy H, Lamoril J, et al. Inheritance in erythropoietic protoporphyria: a common wild-type ferrochelatase allelic variant with low expression accounts for clinical manifestation. Blood. 1999;93:2105–2110. [PubMed] [Google Scholar]
- 23.Gouya L, Puy H, Robreau A, et al. The penetrance of dominant erythropoietic protoporphyria is modulated by expression of wildtype FECH. Nat Genet. 2002;30:27–28. doi: 10.1038/ng809. [DOI] [PubMed] [Google Scholar]
- 24.Risheg H, Chen FP, Bloomer JR. Genotypic determinants of phenotype in North American patients with erythropoietic protoporphyria. Mol Genet Metab. 2003;80:196–206. doi: 10.1016/j.ymgme.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Whatley SD, Mason NG, Holme SA, Anstey AV, Elder GH, Badminton MN. Gene dosage analysis identifies large deletions of FECH gene in 10% of families with erythropoietic protoporphyria. J Invest Dermatol. 2007;127:2790–2794. doi: 10.1038/sj.jid.5700924. [DOI] [PubMed] [Google Scholar]
- 26.Whatley SD, Ducamp S, Gouya L, et al. C-terminal deletions in the ALAS2 gene lead to gain of function and cause x-linked dominant protoporphyria without anemia or iron overload. Am J Hum Genet. 2008;83:408–414. doi: 10.1016/j.ajhg.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taketani S, Kakimoto K, Veta H, Masaki R, Furukawa T. Involvement of ABC7 in the biosynthesis of heme in erythroid cells: interaction of ABC7 with ferrochelatase. Blood. 2003;101:3274–3280. doi: 10.1182/blood-2002-04-1212. [DOI] [PubMed] [Google Scholar]
- 28.Han AP, Fleming MD, Chen JJ. Heme-regulated eIF2-kinase modifies the phenotypic severity of murine models of erythropoietic protoporphyria and B-thalassemia. J Clin Invest. 2005;115:1562–1570. doi: 10.1172/JCI24141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooperman SS, Meyroen-Holtz EG, Olivierre-Wilson H, Ghosh MC, McConnell JP, Rouault TA. Microcytic anemia, erythropoietic protoporphyria and neurodegeration in mice with targeted deletion of iron- regulatory protein 2. Blood. 2005;106:1084–1091. doi: 10.1182/blood-2004-12-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galy B, Ferring D, Minana B, et al. Altered body iron distribution and microcytosis in mice deficient in iron regulatory protein 2 (IRP2) Blood. 2005 Oct 1;106(7):2580–2589. doi: 10.1182/blood-2005-04-1365. Epub 2005 Jun 14. [DOI] [PubMed] [Google Scholar]
- 31.Shaw GC, Cope JJ, Li L, et al. Mitoferrin is essential for erythroid iron assimilation. Nature. 2006;440:96–100. doi: 10.1038/nature04512. [DOI] [PubMed] [Google Scholar]
- 32.Chen W, Dailey HA, Paw BH. Ferrochelatase forms an oligomeric complex with mitoferrin1 and Abcb10 for erythroid heme biosynthesis. Blood. 2010;116:628–630. doi: 10.1182/blood-2009-12-259614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bloomer J, Bruzzone C, Zhu L, Scarlett Y, Magness S, Brenner D. Molecular defects in ferrochelatase in patients with protoporphyria requiring liver transplantation. J Clin Invest. 1998;102:107–114. doi: 10.1172/JCI1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen FP, Risheg H, Liu Y, Bloomer JR. Ferrochelatase gene mutations in erythropoietic protoporphyria: focus on liver disease. Cell Mol Biol. 2002;48:83–89. [PubMed] [Google Scholar]
- 35.Li L, Kaplan J. A mitochondrial-vacuolar signaling pathway in yeast that affects iron and copper metabolism. J Biol Chem. 2004;279:33653–33661. doi: 10.1074/jbc.M403146200. [DOI] [PubMed] [Google Scholar]
- 36.Chen W, Paradkar PN, Li L, et al. Abcb10 physically interacts with mitoferrin1 (Slc25a37) to enhance its stability and function in the erythroid mitochondria. Proc Natl Acad Sci USA. 2009;106:16263–16268. doi: 10.1073/pnas.0904519106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang G, Huang SC, Wu JY, Benz EJ. Regulated Fox-2 isoform expression mediates protein 4.1R splicing during erythroid differentiation. Blood. 2008;111:392–401. doi: 10.1182/blood-2007-01-068940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wohlrab H. Transport proteins (carriers) of mitochondria. IUBMB Life. 2009;61:40–46. doi: 10.1002/iub.139. [DOI] [PubMed] [Google Scholar]
- 39.Paradkar PN, Zumbrennen KB, Paw BH, Ward DM, Kaplan J. Regulation of mitochondrial iron import through differential turnover of mitoferrin 1 and mitoferrin 2. Mol Cell Biol. 2009;29:1007–1016. doi: 10.1128/MCB.01685-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik A. Nonsense-mediated decay approaches the clinic. Nature Genetics. 2004;36:801–808. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- 41.Taketani S, Adachi Y, Nakahashi Y. Regulation of the expression of human ferrochelatase by intracellular iron levels. Eur J Biochem. 2000;267:4685–4692. doi: 10.1046/j.1432-1327.2000.01519.x. [DOI] [PubMed] [Google Scholar]
- 42.Crooks DR, Ghosh MC, Haller RG, Tong WH, Rouault TA. Post-translational stability of the heme biosynthetic enzyme ferrochelatase is dependent on iron availability and intact iron-sulfur cluster assembly machinery. Blood. 2010;115:860–869. doi: 10.1182/blood-2009-09-243105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wingert RA, et al. Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature. 2005;436:1035–1039. doi: 10.1038/nature03887. [DOI] [PubMed] [Google Scholar]
- 44.Buratti E, Baralle M, Bralle FE. Defective splicing, disease and therapy: searching for master checkpoints in exon definition. Nucl Acid Res. 2006;34:3494–3501. doi: 10.1093/nar/gkl498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baralle M, et al. NF1 mRNA biogenesis: effects of the genomic milieu in splicing regulation of the NF1 exon 37. FEBS Lett. 2006;580:4449–4456. doi: 10.1016/j.febslet.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 46.Skoko N, Baralle M, Buratti E, Baralle FE. The pathological splicing mutation c.6792C>G in NF1 exon 37 causes a change in tenancy between antagonistic splicing factors. FEBS Lett. 2008;582:2231–2236. doi: 10.1016/j.febslet.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 47.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chakarova CF, Hims MM, Bolz H, et al. Mutations in HPRP3, a third member of pre-mRNA splicing factor genes, implicated in autosomal dominant retinitis pigmentosa. Hum Mol Genet. 2002;11(1):87–92. doi: 10.1093/hmg/11.1.87. [DOI] [PubMed] [Google Scholar]
- 49.Mordes D, Lou X, Kar A, et al. Pre-mRNA splicing and retinitis pigmentosa. Mol Vis. 2006;12:1259–1271. [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Z, Lotti F, Dittmar K, et al. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133(4):585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]