Summary
Background
Enhancers are genomic cis-regulatory sequences that integrate spatio-temporal signals to control gene expression. Enhancer activity depends on the combination of bound transcription factors, as well as—in some cases—the arrangement and spacing of binding sites for these factors. Here, we examine evolutionary changes to the sequence and structure of sparkling, a Notch/EGFR/Runx-regulated enhancer which activates the dPax2 gene in cone cells of the developing Drosophila eye.
Results
Despite functional and structural constraints on its sequence, sparkling has undergone major reorganization in its recent evolutionary history. Our data suggest that the relative strengths of the various regulatory inputs into sparkling change rapidly over evolutionary time, such that reduced input from some factors is compensated by increased input from different regulators. These gains and losses are at least partly responsible for the changes in enhancer structure that we observe. Furthermore, stereotypical spatial relationships between certain binding sites (“grammar elements”) can be identified in all sparkling orthologs—although the sites themselves are often recently derived. We also find that low binding affinity for the Notch-regulated transcription factor Su(H), a conserved property of sparkling, is required to prevent ectopic responses to Notch in non-cone cells.
Conclusions
Rapid DNA sequence turnover does not imply either the absence of critical cis-regulatory information or the absence of structural rules. Our findings demonstrate that even a severely constrained cis-regulatory sequence can be significantly rewired over a short evolutionary timescale.
Introduction
Enhancers are genomic cis-regulatory sequences that integrate spatio-temporal signals to control the pattern, timing, and levels of gene expression [1, 2]. Enhancers often employ a complex combinatorial logic that allows precisely patterned developmental outputs to be generated from broadly patterned inputs. This regulatory complexity is necessary to restrict gene expression to a specific subset of cells, especially in multicellular organisms, which have many more developmental cell states than transcription factors (TFs). In some enhancers, the organization of TF binding sites is also critical for enhancer function [1–9]. Because cis-regulatory elements account for much of the patterning information encoded in the genome, they are also an important evolutionary engine of developmental change [10].
Despite regulatory and structural constraints, genome-scale evolutionary analyses reveal significant sequence turnover within enhancers and TF binding sites [11–15]. cis-regulatory sequence evolution can cause changes in gene expression, which can drive morphological differences among populations [e.g., 13, 16–20]. On the other hand, a number of enhancers have retained their function despite sequence divergence and binding site turnover [e.g., 8, 21, 22–28]. Proposed explanations for the latter phenomenon include binding site redundancy, compensatory mutations, and organizational flexibility [21, 29–32], but these hypotheses have rarely been tested experimentally. Consequently, both the prevalence and the significance of conserved binding site “grammars” are debated in the recent literature [2, 5, 6, 8, 21, 23, 29, 33–35].
Here we present a fine-scale evolutionary analysis of the structure and function of the sparkling (spa) eye enhancer within the genus Drosophila. In the developing fly retina, the spa enhancer drives expression of dPax2 in cone cells, where it is required for proper differentiation [36–40]. spa is directly bound and regulated by the Notch pathway effector Suppressor of Hairless [Su(H)], the Runx-family protein Lozenge (Lz),and two Ets-family EGFR/MAPK pathway effectors, PointedP2 (PntP2) and Yan/Aop, via motifs resembling Su(H)/Runx/Ets consensus binding sites (Figure 1a) [37]. Regions 1, 4, 5, and 6a of spa, which contain no binding sites for the above TFs, also harbor multiple essential regulatory sequences [8, 41]. The linear organization and spacing (“grammar”) of these regulatory sites is critically important for both robust transcriptional activation and correct cell-type specific expression [8].
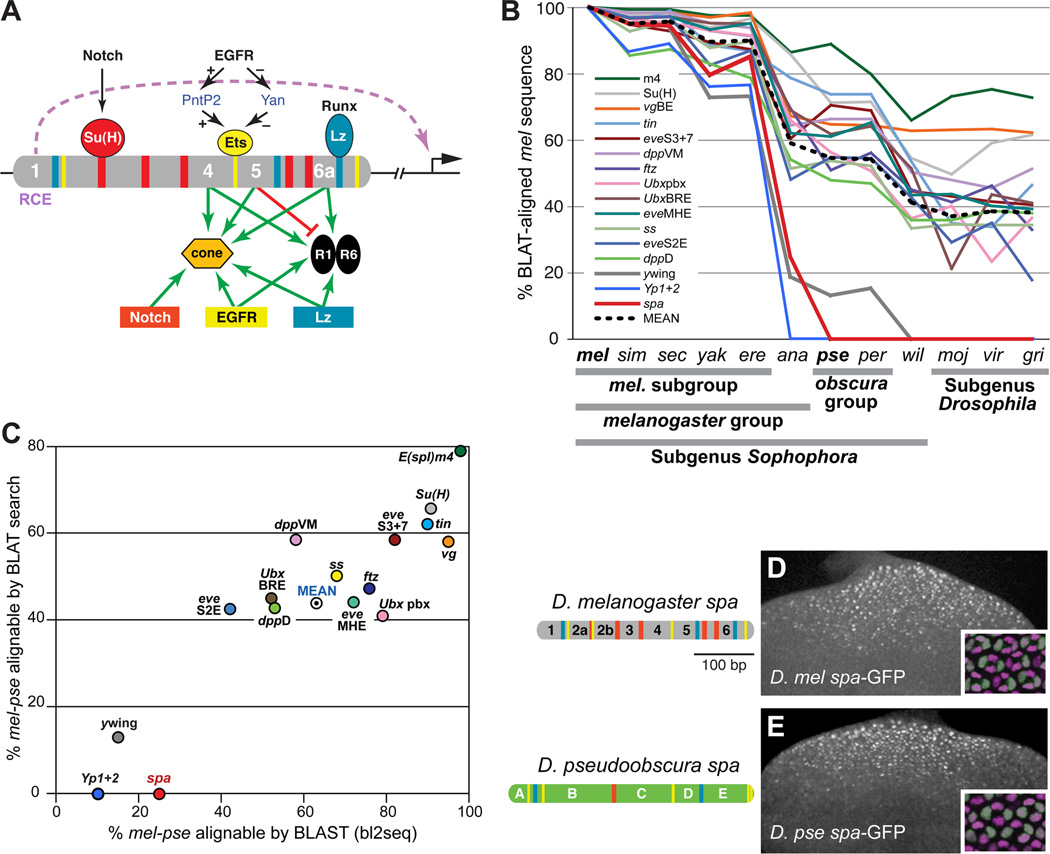
Figure 1. Despite Rapid Sequence Divergence, the Function and Cell-Type Specificity of the sparkling Enhancer Is Conserved.
(A) Summary of the regulatory logic of the sparkling (spa) cone cell enhancer of dPax2. Colored bars indicate known binding sites for Su(H), PntP2/Yan, and Lozenge (Lz); essential novel regulatory sequences 1, 4, 5, and 6a are numbered. RCE indicates the Remote Control Element in region 1 [8].
(B) Comparison of pairwise ortholog sequence similarity between D. melanogaster (mel) and 11 other sequenced Drosophila species, for 16 developmental enhancers. sim, D. simulans; sec, D. sechellia; yak, D. yakuba; ere, D. erecta; ana, D. ananassae; pse, D. pseudoobscura; per, D. persimilis; wil, D. willistoni; moj, D. mojavensis; vir, D. virilis; gri, D. grimshawi. Similarity is measured as the percentage of mel enhancer sequence that is aligned to the orthologous region of the comparison genome by BLAT. See Supplemental Experimental Procedures for detailed information on these reference enhancers.
(C) mel-pse pairwise sequence similarity for 16 developmental cis-regulatory sequences, measured as the percentage of mel enhancer sequence that is BLAST-alignable (x-axis) or BLAT-alignable (y-axis) to the pse ortholog.
(D–E) The D. melanogaster (mel) and D. pseudoobscura (pse) orthologs of spa. Left, enhancer diagrams showing known or predicted Su(H), Ets, and Lz/Runx binding sites (red, yellow, and blue, respectively). Right, eye imaginal discs of transgenic mel larvae carrying mel spa-GFP (D) or pse spa-GFP (E). Insets show 24-hour transgenic pupal eyes stained with antibodies against GFP (green) and the cone cell nuclear marker Cut (magenta).
spa is a good candidate for evolutionary analysis for several reasons. First, its cis-regulatory circuitry, though complex, is well characterized; all essential regulatory sequences within a minimal 362-bp version of spa have been mapped [8, 37]. Second, unlike some other enhancers whose evolution has been examined, such as the well-studied even-skipped stripe 2 enhancer (eveS2E), spa is regulated by highly conserved cell signaling pathways and TFs [42]. Third, previous in vivo work has revealed strict functional constraints on the structure of spa; changing the spacing or arrangement of regulatory sites either kills the enhancer or changes its cell-type specificity [8]. Fourth, as will be discussed below, the sequence of spa is unusually rapidly evolving. We therefore investigated the evolutionary dynamics of spa, to determine the nature and extent of the evolutionary constraints on its sequence and structure.
Results
spa Has Undergone Rapid Sequence Divergence and Structural Rearrangement, While Maintaining Its Function and Cell-Type Specificity
Despite spa’s strict regulatory and structural constraints, described above, DNA sequence alignments reveal poor overall conservation within the genus Drosophila, and poor conservation of most critical regulatory sites (Figure 1B) [8]. Out of 11 mapped regulatory binding sites in spa—five Su(H) sites, three MGGAW PntP2/Ets sites, and three RACCRCA Lz/Runx sites [37, 43, 44], only two, a closely linked 5’ Lz-Ets pair, are unambiguously preserved throughout the genus (see Figure 5). spa sequence appears to be changing unusually rapidly among the 12 sequenced Drosophila species; unlike 13 other developmental enhancers used for comparison, no part of spa is BLAT-alignable between the melanogaster subgroup and the obscura group (Figures 1B and 1C). spa’s evolutionary dynamics differ from those of the 13 reference enhancers, including the eve stripe 2 enhancer (eveS2E), which is considered to be a rapidly evolving element [24, 45]. When the yellow wing enhancer (ywing), chosen because of its known functional adaptations within the genus Drosophila [46], was added to the analysis, it was comparable to spa in its rate of sequence divergence: both enhancers are mostly or entirely unalignable between D. melanogaster (mel) and D. pseudoobscura (pse), depending on the alignment method, while the original 13 reference enhancers, including eveS2E, are highly alignable over this distance (Figure 1C). We also examined the cis-regulatory region of the Yolk protein 1 and 2 genes (Yp1+2), which, like ywing, is “highly divergent” and thought to be subject to recent adaptive changes [25]. The conservation profile of Yp1+2 is similar to that of ywing and spa (Figures 1B and 1C). The poor alignability of ywing, Yp1+2, and spa is likely due to a lack of extended blocks of sequence conservation between D. mel and D. pse (Figure S1). spa, then, has undergone unusually rapid sequence divergence among Drosophila species, resembling that of an enhancer whose expression pattern has undergone significant adaptive change.
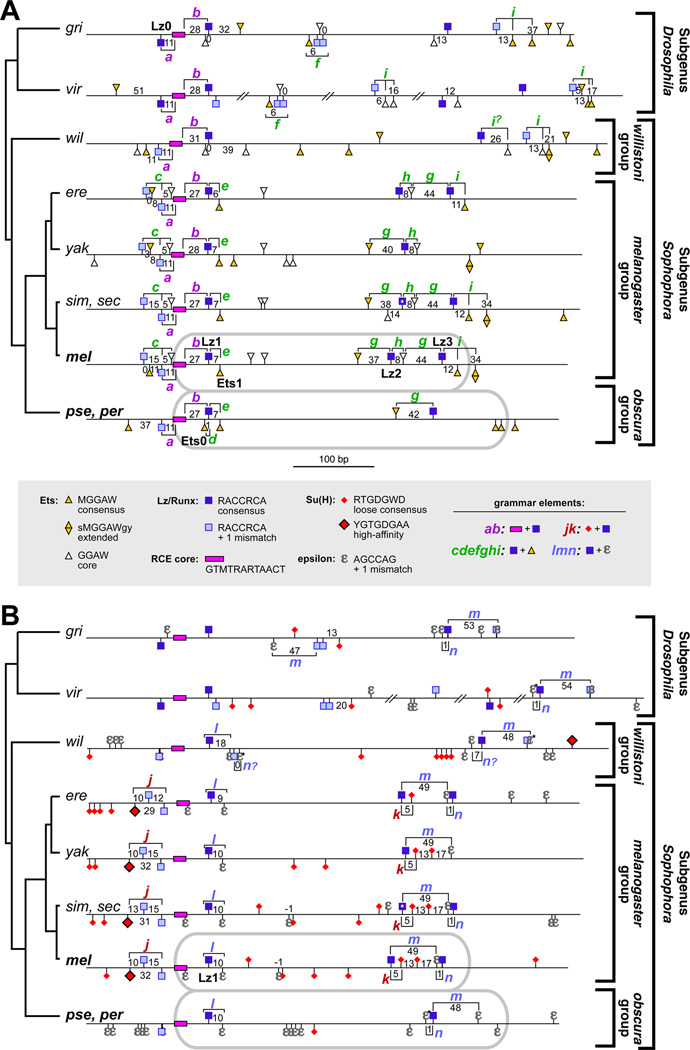
Figure 5. Evolutionary Dynamics of the Binding Site Grammar of sparkling.
Diagram of selected cis-regulatory motifs within spa orthologs of several Drosophila species. (A) Grammar elements involving Lz, Ets, and/or RCE motifs.
(B) Grammar elements involving Lz, Su(H), and/or ε motifs. Brackets, lettered a through n, show spatial relationships among binding sites (“grammar elements”) that are identifiable in multiple species; numbers indicate the base-pair spacing between two motifs. Gray ovals show the sequences included in the minimal mel and pse enhancer reporter constructs. Here, ε denotes a 5/6 or 6/6 match to the AGCCAG motif; selected sequences with a weaker match are designated ε*. The Lz site with a white asterisk in sim/sec indicates that that site is has a mismatch in sim only.
Despite sparkling’s sequence divergence and its poorly conserved regulatory binding sites, the D. mel and D. pse orthologs of spa drive indistinguishable, cone cell-specific patterns of gene expression in transgenic D. melanogaster (Figures 1D and 1E). Thus, spa could be an informative case study in enhancer evolution: a highly constrained cis-regulatory element that nevertheless undergoes rapid sequence change and binding site turnover, while maintaining its function and cell-type specificity. Because spa is finely mapped with respect to the in vivo function of its regulatory sequences [8, 37], it provides an opportunity to examine in detail how cis-regulatory sequences are rewired over evolutionary time.
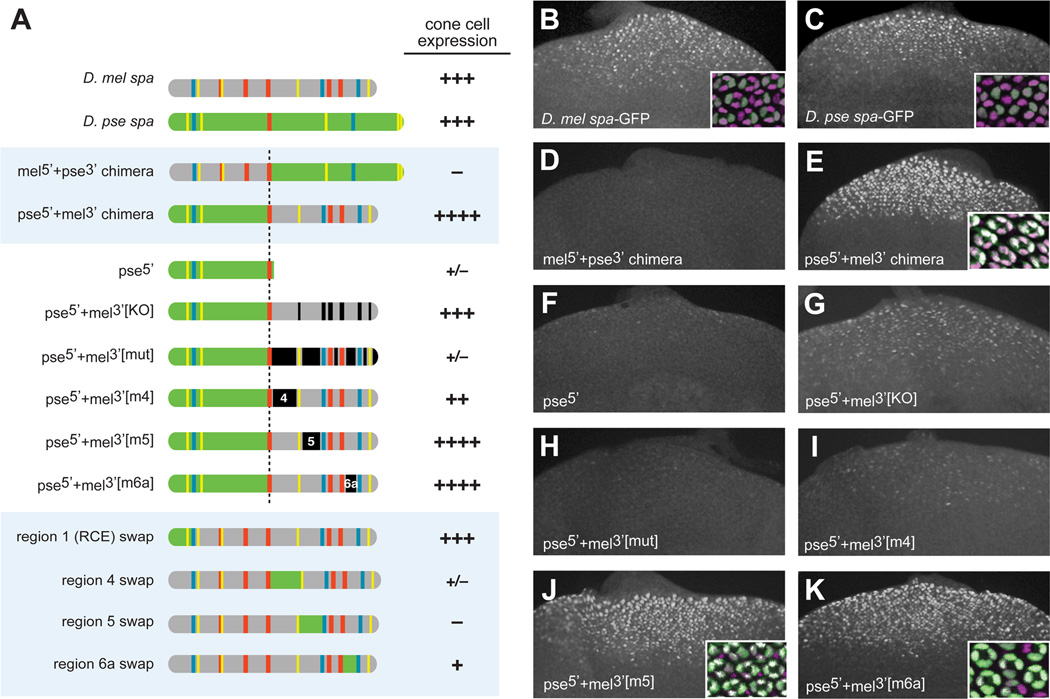
To understand how the patterning function of spa has been preserved despite extreme sequence divergence, we built chimeric constructs in which halves of the D. mel and D. pse orthologs of spa are spliced together (Figure 2A). When similar manipulations were performed with eveS2E in a classic set of experiments, both melpse chimeras were capable of driving a stripe of gene expression, albeit with shifted boundaries [45]. Our results from spa chimeras were very different: the mel5’+pse3’ construct was inactive in vivo (Figure 2D), while pse5’+mel3’ drives properly patterned cone cell-specific expression, at higher levels than either endogenous enhancer (Figure 2E). Thus, essential regulatory sites in the 3’ half of D. mel spa (mel3’) could not be replaced by the orthologous D. pse sequences, while, conversely, mel5’ failed to substitute for pse5’. This is consistent with a model in which essential activities are recruited to different regions of the orthologous enhancers.
Figure 2. Divergent cis-Regulatory Organization of sparkling Between D. melanogaster and D. pseudoobscura.
(A) Diagrams of mel-pse chimeric enhancer constructs, with GFP expression in cone cells of transgenic larvae summarized as follows: +++, wild-type levels of expression in cone cells; ++, moderately reduced; +, severely reduced; +/−, barely detectable or detectable in very few cells; –, no detectable expression; ++++, augmented levels of expression (greater than wild-type).
(B–K) GFP expression in eye discs of transgenic third-instar larvae carrying selected chimeric spa reporters depicted in panel A. Insets show 24-hour transgenic pupal eyes stained with antibodies against GFP (green) and Cut (magenta).
Fine-Scale Chimeric Analysis Tracks the Reorganization of Essential Regulatory Regions
We next undertook a detailed dissection of the hyper-active pse5’+mel3’ chimera. The 5’ half of D. pse spa alone (pse5’) alone was not active (Figure 2F), indicating that not all of the activities in mel3’ were duplicated in pse5’. In the context of the pse5’+mel3’ chimera, the Su(H)/Ets/Lz binding sites in mel3’ were not required for normal expression levels (construct pse5’+mel3’[KO], Figure 2G). Note that this construct, which drives expression comparable to wild-type spa, contains only one Su(H) site and one Lz/Runx site (Figure 2A). However, altering the mel3’ sequences surrounding these 3’ binding sites (regions 4, 5, and 6 of D. mel spa) abolished the function of the chimeric enhancer (Figure 2H). These mutations were sequence alterations, not deletions, preserving the native spacing of the remaining enhancer sequences.
Targeted mutations in regions 4, 5, and 6a, in the context of the pse5’+mel3’ construct, revealed the regulatory contributions of each region to the chimeric enhancer. Mutating only region 4 caused a reduction in expression levels (Figure 2I). By contrast, individually mutating regions 5 and 6a had no effect on the activity of the chimeric construct in third-instar larval eye discs (Figures 2J and 2K), although the loss of 6a caused a gradual loss of expression in cone cells, as well as ectopic expression in primary pigment cells, during pupal stages (not shown). Therefore, pse5’ contains sequences that can functionally replace region 5 and, to a lesser extent, 4 and 6a.
In finer-scale chimeric swaps, regions 4, 5, and 6a could not be functionally substituted by their D. pse orthologous sequences (Figure 2A). However, region 1, the Remote Control Element (RCE) [8]—the best-conserved region of spa—was fully substitutable (Figure 2A).
Taken together, these data show that the cis-regulatory organization of spa differs significantly between D. mel and D. pse. These structural differences are likely due to individual binding site turnover, rather than larger-scale DNA rearrangements, as dot-plot comparisons do not suggest sequence rearrangements within spa (Figure S2).
Novel Regulatory Motifs with Conserved Functions, But Divergent Locations
The above data suggest a model in which rapid losses and gains of regulatory sites have resulted in orthologous enhancers with conserved function, but distinct structures. 3’ regions of D. melanogaster spa are (wholly or in part) functionally equivalent to sequences in the 5’ half of D. pseudoobscura spa. These sequences could recruit the same TFs to different locations in both enhancers; if so, we might be able to identify common novel regulatory sites that have moved since the divergence of D. mel and D. pse, based on our new understanding of the organization of D. pse spa.
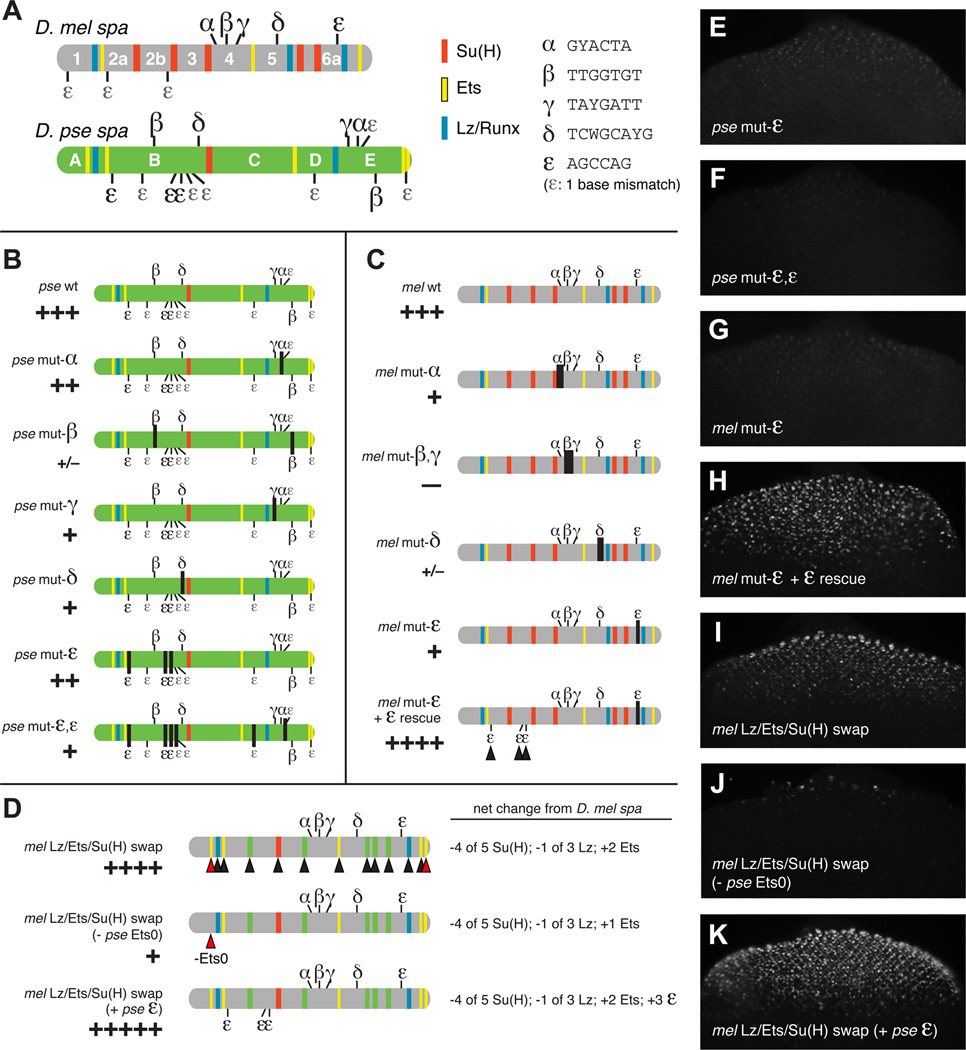
By MEME sequence comparisons among Drosophila spa orthologs, we identified five novel motifs (i.e., non-Su(H)/Ets/Lz sites) in D. mel regions 4, 5, and 6a, all of which are also found in D. pse spa—but not in corresponding positions. We named these motifs α through ε (Figure 3A). Since the proteins binding to these sequences (if any) are unknown, we were unable to define consensus sites based on binding properties. Instead, we chose potential regulatory motifs that fit the following criteria: (1) they reside in essential regulatory regions of mel spa; (2) they occur in spa-orthologous sequence in most sequenced Drosophila species; and (3) they are in non-corresponding positions in the mel and pse enhancers. Motif degeneracy was adjusted to optimally fit the above criteria.
Figure 3. Cross-Species Sequence Comparisons Identify Novel cis-Regulatory Motifs at Rapidly Changing Positions.
(A) Distribution of Su(H), Ets, and Lz/Runx binding sites, along with putative novel regulatory motifs α, β, γ, δ, and ε, in D. mel and D. pse orthologs of spa. Sequence motifs are listed to the right.
(B,C) In vivo mutational analysis of novel regulatory motifs in D. pse spa (B) and D. mel spa (C).
(D) Experiments in which the Lz/Ets/Su(H) sites in mel are replaced with their orthologous pse sequences. Black arrowheads show D. mel Lz, Ets, and Su(H) sites (blue, yellow, and red bands) that have been replaced by orthologous D. pse sequences; green indicates that the orthologous D. pse sequence is not a predicted site. Red arrowheads show D. pse-specific Ets and ε sites added to D. mel spa, including the 5’ site Ets0.
(E–K) GFP expression in eye discs of transgenic third-instar larvae carrying mutated or chimeric spa reporters depicted in panels B–D.
The α, β, and γ motifs reside in mel region 4; in pse, α and γ are found near the 3’ end of spa, in region E, while β motifs are present in both regions B and E (Figure 3A). The δ motif is present in mel region 5 and pse region B. The ε motif, AGCCAG, is present in mel region 6a and in three copies in pse region B, with similar sequences (containing a one-base mismatch) in pse regions B, D, and E (Figure 3A). The relocation of any of these novel motifs, if they are functionally significant, could help to explain the evolving cis-regulatory structure of spa.
Regions 4, 5, and 6a of mel spa, which contain motifs α-ε, are all critical for enhancer function in vivo [8]. Finer-scale mutations in mel spa, which alter the putative novel regulatory motifs described above, all caused enhancer failure in vivo [Figures 3C and 3G; see also ref. 8]. Targeted mutation of the α motif in pse spa weakened its activity, while mutation of β, γ, or δ resulted in a severe loss of enhancer function (Figure 3B). These motifs may therefore represent novel, labile regulatory sites present in both spa orthologs.
The ε motif was of particular interest to us because it is present in three identical copies in pse5’ (plus six more sites with a one-base mismatch), but only one AGCCAG motif is present in D. mel, which suggested a possible compensatory mechanism that might account for the reduced numbers of Su(H) and Lz sites in D. pse spa, relative to D. mel. Mutation of the three ε sites in pse region B reduced reporter gene expression, while additionally mutating three sites with a one-base mismatch to the AGCCAG motif resulted in a severe loss of enhancer function (Figures 3B, 3E and 3F). The latter result suggests that some of the sites containing a “mismatch” to our ε motif may be functional regulatory sites in vivo. Targeted mutation of the sole ε motif in D. mel spa causes a severe loss of reporter gene expression (Figures 3B and 3G). This loss was fully rescued—to a level of expression greater than that of wild-type spa—by altering three “mismatched” ε -like motifs in region 2 to create three “perfect” AGCCAG ε sites (mel mut-ε + ε rescue; Figures 3C and 3H). These results are consistent with the possibility that ε is a binding site for an important activator of spa in both D. mel and D. pse—though via different numbers of sites, in different regions of spa, in the two species.
Tracking Compensatory Changes to the Regulatory Inputs of spa
The identification of novel motifs regulating both D. melanogaster spa and D. pseudoobscura spa, and the importance of Lz, Ets, and Su(H) sites in both orthologs for proper enhancer function [8, 37; this study; additional data not shown], suggests that both spa orthologs may recruit largely the same set of regulatory factors. However, as the divergent numbers of Su(H), Lz/Runx, Ets, and ε motifs suggest, the relative contributions of individual regulatory factors may have diverged in the two species. D. mel spa contains more RACCRCA Lz/Runx motifs than does D. pse spa (3 vs. 2), and many more Su(H) sites (5 vs. 1), but has fewer MGGAW Ets motifs (3 vs. 5) and fewer AGCCAG ε motifs (1 vs. 3; allowing one mismatch, 4 vs. 9).
In order to compare the relative contributions of Lz/Ets/Su(H) to mel and pse spa, we designed a chimera in which mel spa’s Lz/Ets/Su(H) sites were replaced with orthologous sequences from pse (Figure 3D), based on a pairwise BLASTZ alignment (Figure S3A). The result of this substitution was the loss of four non-conserved Su(H) sites and one Lz site, along with the gain of two Ets motifs unique to pse spa, which were added, at their orthologous positions, to mel spa (red arrowheads). The Lz+Ets+Su(H) input into mel spa was thus replaced with that of pse spa. Despite a reduced number of Lz and Su(H) sites (relative to mel spa), this construct drives higher levels of cone cell expression than wild-type spa from either mel or pse (Figure 3I). We also observed occasional ectopic photoreceptor expression in these lines (Table S1).
Note that the pse-specific “Ets0” site is immediately adjacent to the conserved Lz1 site (Figures S3A and S3B). Lz (a Runx factor) and PntP2 (an Ets factor) physically interact and synergistically activate gene expression through neighboring DNA binding sites, as do their mammalian Runx and Ets orthologs [47–51]. When the Ets0 site was excluded from the binding-site chimera described above, in vivo gene expression was drastically reduced (Figure 3J), suggesting that this site (a unique innovation of the obscura group lineage; see Figure 5 and Figure S3B) helps to compensate for the relative reduction of Lz and Su(H) sites in D. pse spa.
We next created a version of mel spa that not only contains the Lz/Ets/Su(H) input from D. pse, but also includes three ε motifs (AGCCAG) taken from the 5’ half of pse spa. The resulting Frankensteinian sequence drives augmented cone cell expression, and also exhibits some ectopic activity in photoreceptors and primary pigment cells (Figure 3D and 3K; Table S1; additional data not shown). Important inter-site spatial relationships may have been disrupted in this chimeric context, resulting in ectopic enhancer activity (see below and [8] for further discussion of site arrangement). The above data suggest the 5’ Ets and ε motifs that are unique to the obscura group are functionally significant compensatory adaptations.
Evidence for Selective Pressure Maintaining Low Binding Affinity for Su(H)
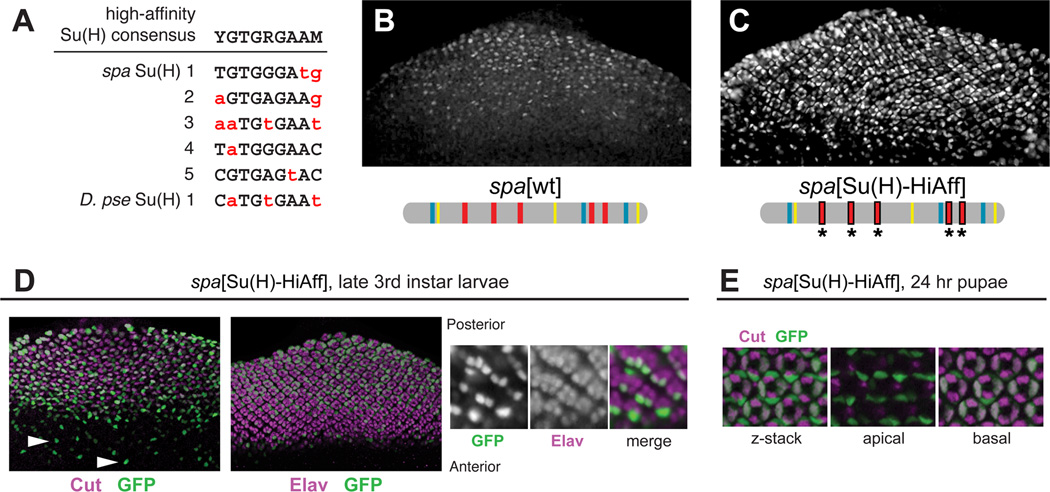
The predicted Su(H) binding sites in all twelve spa orthologs are almost exclusively non-consensus, low-affinity sites (see Figure 5B). All five of the confirmed Su(H) sites in D. mel spa, as well as the lone predicted site in D. pse spa, deviate from the well-established high-affinity consensus YGTGRGAAM by one to four bases (Figure 4A), and four of these six sites also deviate from the lower-affinity binding consensus RTGRGAR [21, 37, 43]. These sub-optimal Su(H) sites are essential in both spa orthologs for normal expression in cone cells, which respond to Notch [37, 42; additional data not shown]. To determine whether spa’s low affinity for Su(H) might be a significant functional adaptation, we made targeted nucleotide substitutions to increase the affinity of the five Su(H) sites in D. mel spa, based on previous binding data [37, 43]. The resulting enhancer, spa[Su(H)-HiAff], drives increased levels of GFP expression in cone cells (Figures 4B and 4C). Perhaps more significantly, spa[Su(H)-HiAff] is ectopically active in multiple non-cone cell types of the eye. Expression was observed in a variable subset of larval photoreceptors (Figure 4D); Notch plays complex, sequential roles in photoreceptor specification [42, 52]. We also observed robust reporter gene expression in the primary pigment cells of 24-hour pupal eyes (Figure 4E). Primary pigment cells depend on high levels of Notch signaling [42, 53; see Supplemental Discussion]. We take these findings to be strong evidence supporting the idea that the Notch/Su(H) input into spa must be balanced at a relatively weak level: some input is required for activation in Notch-responsive cone cells, but strong input enables the ectopic activation of dPax2 elsewhere in the eye.
Figure 4. Low Binding Affinity for Su(H) Is Essential for Proper Cell Type Specificity.
(A) The five previously identified Su(H) binding sites in D. mel spa [37], and the single identifiable putative Su(H) site in D. pse spa, deviate from the high-affinity consensus binding motif (non-matching bases are in red lowercase).
(B,C) Levels of eye disc GFP expression driven by wild-type spa (B) and spa[Su(H)-HiAff] (C), in which the Su(H) sites were altered to the high-affinity sequence CGTGGGAA.
(D) spa[Su(H)-HiAff]-GFP is ectopically expressed in a subset of photoreceptors. Left, GFP expression (green) precedes cone cell specification, as marked by Cut expression (magenta). Middle, GFP expression overlaps temporally and spatially with Elav, a photoreceptor marker (magenta). Right, GFP labels a varying subset of early-specified photoreceptors.
(E) Unlike spa[wt], spa[Su(H)-HiAff]-GFP (green) is strongly active in Cut-negative, apically located, Notch-responsive primary pigment cells in the 24-hour pupal eye.
Evolutionary Dynamics of the Binding Site Grammar of sparkling
Given that the arrangement of cis-regulatory sites within spa plays a critical role in enhancer function and proper patterning [8], we were surprised to see little obvious conservation of binding site “structure” (Figure 5 and Figure S3). Very few TF binding sites are indisputably conserved across the genus, or even between mel and pse. How can an enhancer obey structural rules without highly conserved binding sites?
Even if regulatory sites are rapidly turned over, they may still be preferentially found in structurally optimal positions, relative to other sites. For example, although Lz and PntP2 (Runx and Ets) sites may be rapidly lost and gained, the fact that they physically interact could bias the location of newly acquired sites [30]. This could result in the preservation of characteristic arrangements of sites, even if the sites themselves are recently derived. In principle, such configurations could move around the enhancer as individual sites are gained and lost [29, 33].
In order to identify any such “grammar elements,” as we will refer to them here, we inspected the sequence of spa orthologs, including 5' and 3' flanking DNA, in the 12 sequenced Drosophila species. Since no preferred arrangements or spacings are known for any of the regulators of spa (excepting the Su(H) “paired site” [43], which does not appear in spa), all possible configurations were considered. The results of this analysis are depicted in Figure 5.
Spacing between the well-conserved core of the RCE and the Lz1 site is maintained at 27–31 bp across the genus (grammar element b; Figure 5A and Figure S3B). We identified an additional Lz/Runx motif, Lz0, 5’ of the RCE (just outside of the minimal 362-bp spa element); Lz0-RCE spacing is exactly 11 bp in all species (grammar element a), suggesting that Lz0 may be a significant regulatory site. However, it is not very surprising that, within the only well-conserved region of spa, binding site spacing is also conserved [24, 29, 54]; grammar elements in highly divergent sequences, described below, are potentially more informative.
Of the potential grammar elements that we were able to identify, a large proportion consisted of Lz and Ets sites (labeled c-i in Figure 5A). Only one such element, i, occurs in both Drosophila subgenera, and no single Lz-Ets element is identifiable in all species. Most of these elements are fairly recently derived: for example, c and h are unique to the melanogaster group, while d is a novel feature of the obscura group, and f is found only in the subgenus Drosophila (Figure 5A; additional data not shown). Nevertheless, all spa orthologs contain at least one recognizable Lz-Ets grammar element that is shared with other orthologs.
Three potential Lz-ε grammar elements older than the melanogaster subgroup were identified: l is restricted to the subgenus Sophophora, while m and n could be identified in both subgenera (Figure 5B and Figure S3C). Elements m and n appear to have switched their relative positions in mel and pse. Because of poor sequence conservation in this region, it is difficult to trace the most likely scenario of site gain and loss explaining the rearrangement of m and n. However, based on outgroup species comparisons (Figure 5B), the nm arrangement found in pse seems to be the ancestral state.
We could only identify two grammar elements involving Su(H) sites in the vicinity of sparkling (j and k), both of which are exclusive to the closely related species of the melanogaster subgroup (Figure 5B). Further, we could detect no conservation of individual Su(H) sites beyond the melanogaster subgroup. In this respect, dPax2 differs from other Notch targets such as numb, Su(H), and genes of the Enhancer of split complex, whose enhancers contain many conserved Su(H) binding sites [55–57]. Interestingly, the highly conserved Su(H) sites in those enhancers are generally of much higher predicted affinity than the poorly conserved, but functionally significant, Su(H) sites in spa.
Discussion
Because of spa's rapid structural evolution and binding site turnover, multi-species sequence alignments do not reveal many conserved features. Only the extreme 5' end of spa is unequivocally alignable across 12 Drosophila genomes [8] (Figure 5A and Figure S3B). Given spa’s complex regulatory circuitry and structure, its unusually rapid sequence divergence between D. mel and D. pse was surprising, especially since both orthologs of spa have identical cell type specificities. Here we demonstrate that even an enhancer that is subject to structural constraints can be evolutionarily flexible; therefore, an apparent lack of conserved cis-regulatory structure does not imply an absence of organizational rules within an enhancer.
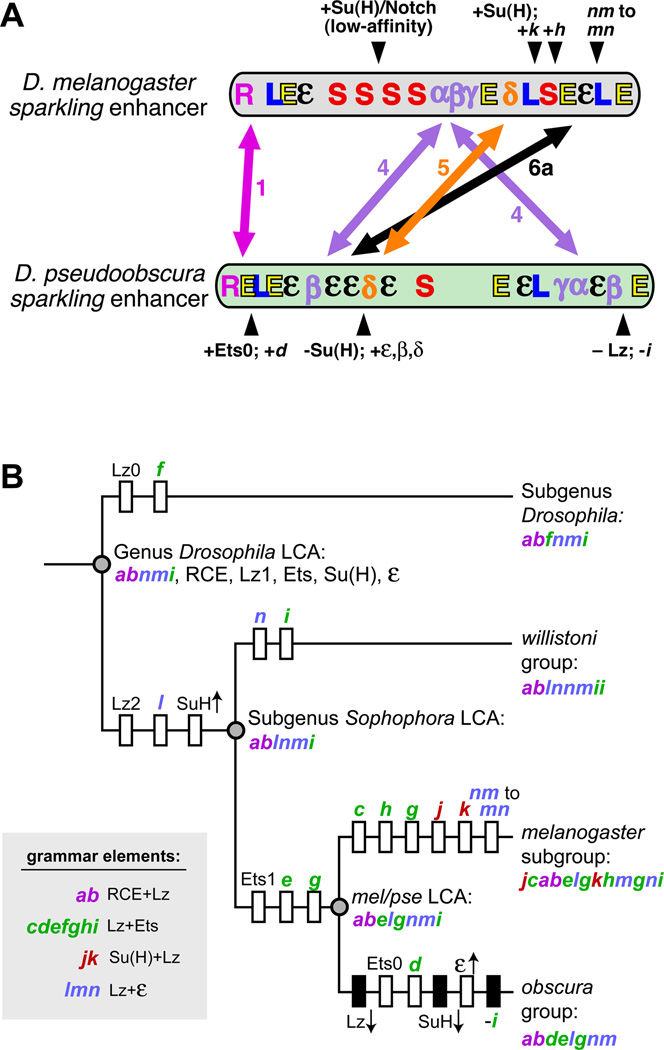
We propose a model for the structural divergence of spa between the melanogaster and obscura groups (Figure 6A), based on our sequence analyses and experimental data. While the RCE and its flanking Lz1-Ets1 pair are relatively stable, many other essential regulatory sites have been relocated. Within regions 4, 5, and 6a, we have identified putative novel regulatory motifs, essential for full-strength activation of both spa orthologs, whose movements are consistent with our experimental data on spa's evolutionary restructuring (Figure 6A).
Figure 6. Inferred Evolutionary History of the Vocabulary and Grammar of the sparkling Enhancer.
(A) Summary of conserved, divergent, and relocated cis-regulatory features of sparkling in D. melanogaster (top) and D. pseudoobscura (bottom). Symbols represent binding sites for known regulators (L, Lz/Runx; E, Ets; S, Su(H)), the Remote Control Element (R), and novel regulatory motifs α, β, γ, δ, and ε. Double-headed arrows show changes in position of the regulatory motifs comprising essential enhancer regions 1, 4, 5, and 6a (arrows). Selected lineage-specific innovations are indicated with black arrowheads. Letters in italics refer to grammar elements described in Figure 5.
(B) A maximum parsimony tree describing the cis-regulatory features of the sparkling enhancer in the inferred last common ancestors (LCAs) of four Drosophila subtaxa, based on sequence analysis and functional assays. Acquired novel features—binding sites or grammar elements—are shown as white boxes crossing a particular lineage, while lost ancestral features are shown as black boxes. Arrows pointing up or down, next to the name of a transcription factor, indicate increased or decreased input of that regulator. Grammar elements are listed in 5’-3’ order.
Important changes to the Lz/Ets/Su(H) inputs have also occurred: D. pse has fewer Su(H) and Lz sites, relative to the melanogaster group—which can be compensated by newly acquired, functionally significant 5' Ets and ε sites. Meanwhile, the melanogaster group has gained a new Lz site, and also has a relative abundance of Su(H) sites, which may compensate for relatively few ε and Ets sites (Figures 6A and 6B).
By tracking the reorganization of Su(H), Lz, Ets, and ε motifs across multiple species, we can propose a speculative phylogeny of the spa enhancer within the genus Drosophila, and predict the cis-regulatory content of the last common ancestors (LCAs) of several species groups, by reconstructing the gain and loss of sites, and the changing strengths of trans-regulatory inputs, in specific lineages (Figure 6B). The main conclusions to be drawn from this evolutionary view of spa, informed by our functional experiments, are: (1) significant enhancer rewiring has occurred since the divergence of the mel and pse lineages; (2) this rewiring involves the loss and gain of individual regulatory motifs, as well as compensatory changes in the overall strength of several trans-regulatory inputs through changes in binding site number, position, and possibly affinity; (3) despite very rapid site turnover, characteristic configurations of sites (“grammar elements”) can be identified; (4) these grammar elements can be relocated within the enhancer, suggesting that a specific arrangement of sites can be more ancient than the individual sites that comprise it. These last two points, taken together, may explain how spa can continue to obey structural rules while being significantly reconfigured.
A large proportion of the grammar elements we have identified involve Lz/Runx and Ets motifs. Unlike the case of linked sites for Dorsal, Twist, and other factors in insect neurogenic enhancers [3, 21, 58], there is no single, clearly preferred arrangement of Lz and Ets sites within spa: we identified seven distinct types of Lz/Ets grammar element that are at least as ancient as the LCA of the melanogaster group (Figure 5). Perhaps Runx and Ets factors, which are known to directly interact and to cooperatively activate transcription in flies and vertebrates [47–51], can synergize productively in several different spatial configurations. This is consistent with mapped Runx and Ets sites in vertebrate genomes, which are frequently associated with one another in target enhancers, but not with a single rigid arrangement or spacing [59–61].
We have also discovered a non-structural constraint on the sequence of spa: a requirement for non-consensus, low-affinity Su(H) sites for proper cone-specific patterning. Since ectopic dPax2 expression in photoreceptor precursors causes faulty cell fate specification and differentiation, resulting in defective eye morphology [40], it is reasonable to suppose that the expression pattern of spa[Su(H)-HiAff] would have negative fitness consequences for the fly. Taken together with previous work, the data presented here suggest that spa requires input from Notch/Su(H), but also requires that input to be attenuated at the cis-regulatory level, in order to generate the proper levels and cell-type specificity of dPax2 expression in a tissue with widespread Notch signaling.
Like Notch/Su(H), EGFR/Ets signaling and Lz are also used to specify multiple cell types in the retina, which presents a challenge for combinatorial gene regulation: enhancers must be able to make fine qualitative distinctions in regulatory inputs, and often must translate this information into relatively sharp on/off decisions [2, 6, 40–43, 53, 62, 63]. These pressures could result in a cis-regulatory logic for genes like dPax2 in which many weak inputs are independently tuned—and spatially arranged—to maximize activation in the proper cell type, while minimizing ectopic activation. Our previous studies of spa present a picture of an enhancer operating just above a functional threshold, such that the loss of a single regulatory site, or a loss of proper grammar, can result in transcriptional failure in cone cells. One of our main conclusions from this study is that, over a relatively short evolutionary timescale, a cis-regulatory module can find multiple solutions to this complex computational problem.
The presence of weak, non-consensus binding sites for signal-regulated TFs is a common, but little remarked upon, feature of developmental enhancers [64]. Low-affinity TF binding sites have well-documented functions in shaping a stripe of gene expression across a morphogen gradient, and in determining temporal responses to developmental regulators [58, 65–68]. Here, we provide direct evidence supporting a role for weak signal response elements in preventing ectopic transcriptional responses to highly pleiotropic signaling pathways such as Notch.
There is one striking question not addressed by this study: why is this enhancer evolving at an unusually high rate, given that its expression pattern is stable? We can think of two plausible explanations for which supporting data exist. First, dPax2 is on chromosome 4, the “dot” chromosome of Drosophila, which has a severely reduced recombination rate, resulting in inefficient selection and relaxed sequence constraint [69]. No other cis-regulatory module on the fourth chromosome has been subjected to an extensive evolutionary analysis, nor is any as well-mapped as sparkling, but enhancers of the fourth-chromosome genes eyeless and toy contain fairly large blocks of sequence conservation, compared to spa [70]. An alternative explanation for the rapid turnover observed within spa involves the presence of non-consensus, predicted low-affinity sites for Su(H) and, in some cases, Lz and PntP2 (Figure 4 and Figure 5). For a typical TF, there are many more possible low-affinity binding sites than high-affinity sites: for example, the highest-affinity Su(H) consensus YGTGDGAAM encompasses only 12 variants (TGTGGGAAA, etc.), while the lower-affinity consensus of the same length nRTGDGWDn, which accommodates all of the known Su(H) sites within spa, contains 576 possible sequences. Accordingly, it is much more likely that an enhancer will acquire a low-affinity binding site via a single mutational event than a high-affinity site. Thus, an enhancer that does not require high-affinity binding sites for given trans-regulator may rapidly sample a variety of configurations of weak sites, and may thereby undergo considerable sequence turnover without losing the input from that regulator. In other words, an enhancer such as spa, which must maintain a weak regulatory linkage with Notch/Su(H), may be less constrained than a high-affinity target with respect to the sequence, number, and position of its Su(H) binding sites. Whatever the reason for the rapid sequence divergence of spa, it provides an opportunity to examine in detail the evolutionary mechanisms by which a complex cis-regulatory module can be significantly reorganized, while still conforming to specific constraints of combinatorial logic and grammar.
Supplementary Material
Acknowledgments
This research was supported by a Center for Organogenesis Predoctoral Fellowship (5T32HD007505) to C.I.S. and by NIH grant GM076509 and ARRA supplement GM07650903S1 to S.B. We thank Aditi Ravindrananth, Lisa Johnson, Andy Vo, and Autumn Holmes for research support, and Ben Novitch and Ken Cadigan for generously sharing reagents. We are grateful to Niki Evans and other members of the Barolo lab, members of the UM developmental genetics group meeting, and the UMfly Drosophila community, especially Trisha Wittkopp, for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental Information includes Supplemental Discussion, Experimental Procedures, three figures, and one table.
References
- 1.Bulger M, Groudine M. Enhancers: the abundance and function of regulatory sequences beyond promoters. Dev Biol. 2010;339:250–257. doi: 10.1016/j.ydbio.2009.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine M. Transcriptional enhancers in animal development and evolution. Curr Biol. 2010;20:R754–R763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erives A, Levine M. Coordinate enhancers share common organizational features in the Drosophila genome. Proc. Natl. Acad. Sci. USA. 2004;101:3851–3856. doi: 10.1073/pnas.0400611101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giese K, Kingsley C, Kirshner JR, Grosschedl R. Assembly and function of a TCR enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 5.Papatsenko D, Goltsev Y, Levine M. Organization of developmental enhancers in the Drosophila embryo. Nucleic Acids Res. 2009;37:5665–5677. doi: 10.1093/nar/gkp619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papatsenko D, Levine M. A rationale for the enhanceosome and other evolutionarily constrained enhancers. Curr Biol. 2007;17:R955–R957. doi: 10.1016/j.cub.2007.09.035. [DOI] [PubMed] [Google Scholar]
- 7.Senger K, Armstrong GW, Rowell WJ, Kwan JM, Markstein M, Levine M. Immunity regulatory DNAs share common organizational features in Drosophila. Mol Cell. 2004;13:19–32. doi: 10.1016/s1097-2765(03)00500-8. [DOI] [PubMed] [Google Scholar]
- 8.Swanson CI, Evans NC, Barolo S. Structural rules and complex regulatory circuitry constrain expression of a Notch- and EGFR-regulated eye enhancer. Dev Cell. 2010;18:359–370. doi: 10.1016/j.devcel.2009.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thanos D, Maniatis T. Virus induction of human IFN- gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 10.Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 11.Richards S, Liu Y, Bettencourt BR, Hradecky P, Letovsky S, Nielsen R, Thornton K, Hubisz MJ, Chen R, Meisel RP, et al. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moses AM, Pollard DA, Nix DA, Iyer VN, Li XY, Biggin MD, Eisen MB. Large-scale turnover of functional transcription factor binding sites in Drosophila. PLoS Comput Biol. 2006;2:e130. doi: 10.1371/journal.pcbi.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wittkopp PJ. Evolution of cis-regulatory sequence and function in Diptera. Heredity. 2006;97:139–147. doi: 10.1038/sj.hdy.6800869. [DOI] [PubMed] [Google Scholar]
- 14.Li L, Zhu Q, He X, Sinha S, Halfon MS. Large-scale analysis of transcriptional cis-regulatory modules reveals both common features and distinct subclasses. Genome Biol. 2007;8:R101. doi: 10.1186/gb-2007-8-6-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balhoff JP, Wray GA. Evolutionary analysis of the well characterized endo16 promoter reveals substantial variation within functional sites. Proc Natl Acad Sci U S A. 2005;102:8591–8596. doi: 10.1073/pnas.0409638102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcellini S, Simpson P. Two or four bristles: functional evolution of an enhancer of scute in Drosophilidae. PLoS Biol. 2006;4:e386. doi: 10.1371/journal.pbio.0040386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong S, Rebeiz M, Andolfatto P, Werner T, True J, Carroll SB. The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell. 2008;132:783–793. doi: 10.1016/j.cell.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 18.Williams TM, Selegue JE, Werner T, Gompel N, Kopp A, Carroll SB. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell. 2008;134:610–623. doi: 10.1016/j.cell.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rebeiz M, Pool JE, Kassner VA, Aquadro CF, Carroll SB. Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science. 2009;326:1663–1667. doi: 10.1126/science.1178357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan YF, Marks ME, Jones FC, Villarreal G, Jr, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crocker J, Potter N, Erives A. Dynamic evolution of precise regulatory encodings creates the clustered site signature of enhancers. Nat Commun. 2010;1:99. doi: 10.1038/ncomms1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006;312:276–279. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- 23.Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet. 2008;4:e1000106. doi: 10.1371/journal.pgen.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ludwig MZ, Palsson A, Alekseeva E, Bergman CM, Nathan J, Kreitman M. Functional evolution of a cis-regulatory module. PLoS Biol. 2005;3:e93. doi: 10.1371/journal.pbio.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piano F, Parisi MJ, Karess R, Kambysellis MP. Evidence for redundancy but not trans factor-cis element coevolution in the regulation of Drosophila Yp genes. Genetics. 1999;152:605–616. doi: 10.1093/genetics/152.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weirauch MT, Hughes TR. Conserved expression without conserved regulatory sequence: the more things change, the more they stay the same. Trends Genet. 2010;26:66–74. doi: 10.1016/j.tig.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Kalay G, Wittkopp PJ. Nomadic enhancers: tissue-specific cis-regulatory elements of yellow have divergent genomic positions among Drosophila species. PLoS Genet. 2010;6:e1001222. doi: 10.1371/journal.pgen.1001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crocker J, Erives A. A closer look at the eve stripe 2 enhancers of Drosophila and Themira. PLoS Genet. 2008;4:e1000276. doi: 10.1371/journal.pgen.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Johnson AD. Evolution of transcription networks--lessons from yeasts. Curr Biol. 2010;20:R746–R753. doi: 10.1016/j.cub.2010.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludwig MZ. Functional evolution of noncoding DNA. Curr Opin Genet Dev. 2002;12:634–639. doi: 10.1016/s0959-437x(02)00355-6. [DOI] [PubMed] [Google Scholar]
- 32.Wittkopp PJ. Evolutionary genetics: how flies get naked. Curr Biol. 2007;17:R881–R883. doi: 10.1016/j.cub.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 33.Hare EE, Peterson BK, Eisen MB. A careful look at binding site reorganization in the even-skipped enhancers of Drosophila and sepsids. PLoS Genet. 2008;4:e1000268. doi: 10.1371/journal.pgen.1000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kulkarni MM, Arnosti DN. cis-regulatory logic of short-range transcriptional repression in Drosophila melanogaster. Mol Cell Biol. 2005;25:3411–3420. doi: 10.1128/MCB.25.9.3411-3420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rastegar S, Hess I, Dickmeis T, Nicod JC, Ertzer R, Hadzhiev Y, Thies WG, Scherer G, Strahle U. The words of the regulatory code are arranged in a variable manner in highly conserved enhancers. Dev Biol. 2008;318:366–377. doi: 10.1016/j.ydbio.2008.03.034. [DOI] [PubMed] [Google Scholar]
- 36.Dziedzic K, Heaphy J, Prescott H, Kavaler J. The transcription factor D-Pax2 regulates crystallin production during eye development in Drosophila melanogaster. Dev Dyn. 2009;238:2530–2539. doi: 10.1002/dvdy.22082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flores GV, Duan H, Yan H, Nagaraj R, Fu W, Zou Y, Noll M, Banerjee U. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75–85. doi: 10.1016/s0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- 38.Fu W, Duan H, Frei E, Noll M. shaven and sparkling are mutations in separate enhancers of the Drosophila Pax2 homolog. Development. 1998;125:2943–2950. doi: 10.1242/dev.125.15.2943. [DOI] [PubMed] [Google Scholar]
- 39.Fu W, Noll M. The Pax2 homolog sparkling is required for development of cone and pigment cells in the Drosophila eye. Genes Dev. 1997;11:2066–2078. doi: 10.1101/gad.11.16.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi Y, Noll M. Determination of cell fates in the R7 equivalence group of the Drosophila eye by the concerted regulation of D-Pax2 and TTK88. Dev Biol. 2009;331:68–77. doi: 10.1016/j.ydbio.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 41.Johnson LA, Zhao Y, Golden K, Barolo S. Reverse-engineering a transcriptional enhancer: a case study in Drosophila. Tissue Eng Part A. 2008;14:1549–1559. doi: 10.1089/ten.tea.2008.0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voas MG, Rebay I. Signal integration during development: insights from the Drosophila eye. Dev Dyn. 2004;229:162–175. doi: 10.1002/dvdy.10449. [DOI] [PubMed] [Google Scholar]
- 43.Nellesen DT, Lai EC, Posakony JW. Discrete enhancer elements mediate selective responsiveness of Enhancer of split Complex genes to common transcriptional activators. Dev. Biol. 1999;213:33–53. doi: 10.1006/dbio.1999.9324. [DOI] [PubMed] [Google Scholar]
- 44.Wasylyk B, Hahn SL, Giovane A. The Ets family of transcription factors. Eur J Biochem. 1993;211:7–18. doi: 10.1007/978-3-642-78757-7_2. [DOI] [PubMed] [Google Scholar]
- 45.Ludwig MZ, Bergman C, Patel NH, Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- 46.Gompel N, Prud'homme B, Wittkopp PJ, Kassner VA, Carroll SB. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- 47.Dittmer J. The biology of the Ets1 proto-oncogene. Mol Cancer. 2003;2:29. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goetz TL, Gu TL, Speck NA, Graves BJ. Auto-inhibition of Ets-1 is counteracted by DNA binding cooperativity with core-binding factor alpha2. Mol Cell Biol. 2000;20:81–90. doi: 10.1128/mcb.20.1.81-90.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jackson Behan K, Fair J, Singh S, Bogwitz M, Perry T, Grubor V, Cunningham F, Nichols CD, Cheung TL, Batterham P, et al. Alternative splicing removes an Ets interaction domain from Lozenge during Drosophila eye development. Dev Genes Evol. 2005;215:423–435. doi: 10.1007/s00427-005-0490-0. [DOI] [PubMed] [Google Scholar]
- 50.Kim WY, Sieweke M, Ogawa E, Wee HJ, Englmeier U, Graf T, Ito Y. Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. Embo J. 1999;18:1609–1620. doi: 10.1093/emboj/18.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu H, Holm M, Xie XQ, Wolf-Watz M, Grundstrom T. AML1/Runx1 recruits calcineurin to regulate granulocyte macrophage colony-stimulating factor by Ets1 activation. J Biol Chem. 2004;279:29398–29408. doi: 10.1074/jbc.M403173200. [DOI] [PubMed] [Google Scholar]
- 52.Brennan CA, Moses K. Determination of Drosophila photoreceptors: timing is everything. Cell Mol Life Sci. 2000;57:195–214. doi: 10.1007/PL00000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagaraj R, Banerjee U. Combinatorial signaling in the specification of primary pigment cells in the Drosophila eye. Development. 2007;134:825–831. doi: 10.1242/dev.02788. [DOI] [PubMed] [Google Scholar]
- 54.Lusk RW, Eisen MB. Evolutionary mirages: selection on binding site composition creates the illusion of conserved grammars in Drosophila enhancers. PLoS Genet. 2010;6:e1000829. doi: 10.1371/journal.pgen.1000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barolo S, Walker RG, Polyanovsky AD, Freschi G, Keil T, Posakony JW. A Notch-independent activity of Suppressor of Hairless is required for normal mechanoreceptor physiology. Cell. 2000;103:957–969. doi: 10.1016/s0092-8674(00)00198-7. [DOI] [PubMed] [Google Scholar]
- 56.Macdonald SJ, Long AD. Identifying signatures of selection at the Enhancer of split neurogenic gene complex in Drosophila. Mol Biol Evol. 2005;22:607–619. doi: 10.1093/molbev/msi046. [DOI] [PubMed] [Google Scholar]
- 57.Rebeiz M, Miller SW, Posakony JW. Notch regulates numb: integration of conditional and autonomous cell fate specification. Development. 2011;138:215–225. doi: 10.1242/dev.050161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Papatsenko D, Levine M. Quantitative analysis of binding motifs mediating diverse spatial readouts of the Dorsal gradient in the Drosophila embryo. Proc Natl Acad Sci U S A. 2005;102:4966–4971. doi: 10.1073/pnas.0409414102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Del Blanco B, Roberts JL, Zamarreno N, Balmelle-Devaux N, Hernandez-Munain C. Flexible stereospecific interactions and composition within nucleoprotein complexes assembled on the TCR gene enhancer. J Immunol. 2009;183:1871–1883. doi: 10.4049/jimmunol.0803351. [DOI] [PubMed] [Google Scholar]
- 60.Hollenhorst PC, Chandler KJ, Poulsen RL, Johnson WE, Speck NA, Graves BJ. DNA specificity determinants associate with distinct transcription factor functions. PLoS Genet. 2009;5:e1000778. doi: 10.1371/journal.pgen.1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pencovich N, Jaschek R, Tanay A, Groner Y. Dynamic combinatorial interactions of RUNX1 and cooperating partners regulates megakaryocytic differentiation in cell line models. Blood. 2011;117:e1–e14. doi: 10.1182/blood-2010-07-295113. [DOI] [PubMed] [Google Scholar]
- 62.Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- 63.Hong JW, Hendrix DA, Papatsenko D, Levine MS. How the Dorsal gradient works: insights from postgenome technologies. Proc Natl Acad Sci U S A. 2008;105:20072–20076. doi: 10.1073/pnas.0806476105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barolo S, Posakony JW. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 2002;16:1167–1181. doi: 10.1101/gad.976502. [DOI] [PubMed] [Google Scholar]
- 65.Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- 66.Jiang J, Levine M. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell. 1993;72:741–752. doi: 10.1016/0092-8674(93)90402-c. [DOI] [PubMed] [Google Scholar]
- 67.Rowan S, Siggers T, Lachke SA, Yue Y, Bulyk ML, Maas RL. Precise temporal control of the eye regulatory gene Pax6 via enhancer-binding site affinity. Genes Dev. 2010;24:980–985. doi: 10.1101/gad.1890410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parker DS, White MA, Ramos AI, Cohen BA, Barolo S. The cis-regulatory logic of Hedgehog gradient responses: Key roles for Gli binding affinity, competition, and cooperativity. Sci Signal. 2011;4:ra38. doi: 10.1126/scisignal.2002077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arguello JR, Zhang Y, Kado T, Fan C, Zhao R, Innan H, Wang W, Long M. Recombination yet inefficient selection along the Drosophila melanogaster subgroup's fourth chromosome. Mol Biol Evol. 2010;27:848–861. doi: 10.1093/molbev/msp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Adachi Y, Hauck B, Clements J, Kawauchi H, Kurusu M, Totani Y, Kang YY, Eggert T, Walldorf U, Furukubo-Tokunaga K, et al. Conserved cis-regulatory modules mediate complex neural expression patterns of the eyeless gene in the Drosophila brain. Mech Dev. 2003;120:1113–1126. doi: 10.1016/j.mod.2003.08.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.