Abstract
Induction of NF-κB–dependent gene expression plays an important role in a number of biological processes including inflammation and ischemia-reperfusion injury. However, few attempts aimed at selective regulation of this transcription factor have been successful. We report here that a naturally occurring antibacterial peptide PR39 reversibly binds to the α7 subunit of the 26S proteasome and blocks degradation of NF-κB inhibitor IκBα by the ubiquitin-proteasome pathway without affecting overall proteasome activity. IκBα phosphorylation and ubiquitination occur normally after PR39 treatment, and binding of valosin-containing proteins is not impaired. The inhibition of IκBα degradation abolishes induction of NF-κB–dependent gene expression in cell culture and in mouse models of acute pancreatitis and myocardial infarction, including upregulation of endothelial adhesion proteins VCAM-1 and ICAM-1. In the latter model, sustained infusion of PR39 peptide resulted in significant reduction of myocardial infarct size. PR39 and related peptides may provide novel means to regulate cellular function and to control of NF-κB–dependent gene expression for therapeutic purposes.
Introduction
Degradation of proteins in mammalian cells proceeds via two distinct pathways: lysosome-dependent and proteasome-dependent. The 26S proteasome catalyses the hydrolysis of proteins marked for degradation by conjugation to ubiquitin, as well as certain nonubiquitinated proteins. This 2.4-MDa structure consists of the 20S proteasome “core” that contains multiple peptidase activities and two 19S regulatory complexes that bind ubiquitinated proteins and promote their transfer into the central 20S particle (1–3). The 20S proteasome is composed of four stacked rings, each containing seven distinct, but related, subunits. The outer rings, through which the protein substrates must enter, are composed of seven α subunits that play predominantly a structural role. The two central rings, composed of seven β subunits, of which three contain proteolytic sites, enclose a central chamber where proteins are hydrolyzed (4, 5).
Proteasome-mediated degradation is a principal factor controlling the intracellular levels of most cell proteins, including major regulators of gene expression such as NF-κB inhibitor IκBα (6); hypoxia-inducing factor-1α (HIF-1α) (7–9); protooncogenes c-Fos (10), c-Jun, and c-Mos (11); and various cyclins (11). In addition, partial degradation by the 26S proteasome converts the p105 NF-κB precursor to the active p50 subunit (12). Furthermore, some peptides generated by the proteasome during the course of protein breakdown are presented as antigens on the class I MHC (13). Selective inhibitors of the ubiquitin-proteasome (Ub-proteasome) pathway–mediated protein degradation can, therefore, serve as important tools for the study of a variety of biological processes and also may have potential as therapeutic agents (14).
In particular, NF-κB–dependent gene expression plays an important role in a number of biological processes of major medical importance, including immune, inflammatory, and antiapoptotic responses (15–17). NF-κB is a dimer composed of p50 and p65 (RelA) subunits. Binding of this complex to IκB inhibitor in the cytosol is the main cellular mechanism preventing NF-κB–dependent transcription under normal conditions. A number of extracellular stimuli including TNF-α, Il-1, and lipopolysaccharide trigger NF-κB activation by causing rapid degradation of this inhibitor protein by the Ub-proteasome pathway (6). Several steps necessary for IκBα degradation to occur have been identified, including phosphorylation of IκBα at two sites by a specific IκBα kinase (6, 18) of the SCF1 family, which then leads to ubiquitination of the phosphorylated IκBα by a specific E3 enzyme complex (19–23); this in turn allows binding to valosin-containing protein (VCP) (24), which appears to provide a physical link between the ubiquitinated IκBα and the 26S proteasome.
PR39 is a highly basic arginine/proline-rich peptide originally isolated from porcine intestine on the basis of its antibacterial activity (25). The peptide is secreted in a preproprotein form that includes a canonical leader sequence and rapidly undergoes cleavage of the NH2-terminal portion to generate the mature form composed of the 39 COOH-terminal amino acids (26). Whereas the sequence of the NH2-terminal part of the preproprotein is highly homologous to the cathelin gene family members, the sequence of the 39 COOH-terminal amino acids that make up the mature peptide has no homology to any other known protein (26). PR39 can rapidly cross cell membranes and, by virtue of its proline-rich composition, may interact with SH3 domains of p47phox (27) and p130Cas (28). The peptide, predominantly produced by blood-derived macrophages (29, 30), is found at the sites of active inflammation, including skin wounds and myocardial infarction, and may play an important role by inducing expression of heparan sulfate–carrying core proteins, syndecan 1 and 4 (30, 31), and inhibiting degradation of the hypoxia-inducible factor–1α (HIF-1α) protein (32). However, the molecular events involved in this peptide’s actions remain largely unknown.
The present study was undertaken to explore the molecular mechanisms of PR39 activity. We show that the peptide binds to the α7 subunit of the 20S proteasome and can block degradation of IκBα by the Ub-proteasome pathway without disrupting overall proteasome activity and suppress NF-κB–dependent gene expression both in cell culture and in two different models of acute injury in mice.
Methods
Cell lines, supplies, and reagents
ECV304 cells were cultured in M199 medium supplemented with 10% FBS. Human umbilical vein endothelial cells (HUVEC; Clonetics Inc., Walkersville, Maryland,USA) were cultured in DMEM supplemented with 20% FBS. U937 cells (courtesy of J. Chang, Beth Israel Deaconess Medical Center) were cultured in RPM-1640 medium supplemented with 10% FBS. A mixture of 100 μg/mL penicillin/streptomycin was added to all cultures. For stable transfection, a full-length porcine PR39 cDNA (containing the leader sequence) and a cDNA construct corresponding to the fourth exon of the porcine PR39 gene were cloned into the eukaryotic expression vector pGRE5-2 and used to stably transfect ECV304 as described previously (32).
Synthetic PR39 peptide was generated on the basis of porcine sequence (26) and purified by HPLC (Genemed Synthesis Inc., South San Francisco, California, USA). Lactacystin and MG132 were obtained from Calbiochem-Novabiochem Corp. (San Diego, California, USA) and hrTNF-α from Sigma Chemical Co. (St. Louis, Missouri, USA).
Coimmunoprecipitation
For coimmunoprecipitation, transfected cells, wild-type ECV, full-length PR39 (ECV-PR39), and exon 4 PR39 (ECV-E4), were lysed with the RIPA buffer, and equal amounts of total protein were precleared with nonimmune rabbit serum and protein G plus/protein A-agarose beads (Calbiochem-Novabiochem Corp.). The cleared samples were incubated at 4°C overnight with 20 μL of polyclonal anti-PR39 Ab (32) and 40 μL of protein G plus/protein A-agarose beads or with the beads alone. The beads were then washed three times with PBS, resuspended in Laemmli sample buffer (2% SDS, 10% glycerol, 0.5% β-mercaptoethanol, 0.004% bromphenol blue, 50 mM Tris-HCl, pH 6.8), resolved on 10% SDS-PAGE, transferred to PVDF membrane, and blotted with 1:1000 mouse monoclonal anti-α7 or anti-α2 Ab’s (Affiniti Research Products Ltd., Exeter, United Kingdom). As a control, the total cell lysate of ECV cells was subjected to SDS-PAGE and immunoblotting with anti-α7 and anti-α2 Ab’s.
Yeast two-hybrid screening
Two-hybrid screening was done using Clontech MATCHMAKER GAL4 System 2 (Clontech Laboratories, Palo Alto, California, USA). The cDNA of porcine PR39 peptide corresponding to the fourth exon (amino acids 131 to 169) was subcloned into pAS2-1 vector as bait. Mouse embryo 3T3 MATCHMAKER cDNA library (Clontech Laboratories) was screened in the yeast Y190 strain. Plasmids from HIS3/LacZ positive clones were sequenced and cotransformed with bait plasmid back into the Y190 strain to confirm the interaction.
IκBα ubiquitination studies
In vitro assay.
IκBα and its phosphorylation-site mutants 32S→32A and 36S→36A (S32A-IκBα and S36A-IκBα) in pBluescript plasmids were generously provided by T. Maniatis (Harvard University, Boston, Massachusetts, USA). Plasmid for the overproduction of GST-Ub (pGEX-2TK-Ub) was a gift of J. Huibregtse (Rutgers University, Piscataway, New Jersey, USA). GST-Ub was prepared as described (33).
Preparation of HeLa cell extract.
HeLa cells were harvested by centrifugation at 500 g. The packed cell volume was estimated and the cells osmotically lysed by the addition of 5 volumes of hypotonic buffer (10 mM Tris-Cl, pH 7.9, 1.5 mM MgCl2,10 mM KCl, 0.2 mM PMSF, 0.5 mM DTT). The mixture was incubated on ice for 10 minutes, then homogenized with 12 strokes of a Dounce homogenizer (B-type pestle). The crude lysate was centrifuged for 15 minutes at 500 g, followed by recentrifugation of the supernatant at 100,000 g for 30 minutes. This supernatant was concentrated by the addition of solid (NH4)2SO4 to 80% saturation, stirred at 4°C for 30 minutes, and centrifuged for 15 minutes at 21,000 g. The precipitate was resuspended in 1:5 volume of 20 mM Tris-Cl, pH 7.6, 20 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.5 mM PMSF, 50 μM chymostatin, and 20 μM E64, and dialyzed against more than 500 volumes of 20 mM Tris-Cl, pH 7.6, 20 mM KCl, 5 mM MgCl2, 1 mM DTT, 10% glycerol at 4°C overnight, and stored at –70°C until use.
Ubiquitination assay.
35S-IκBα and 35S-S32A/S36A IκBα were prepared by coupled in vitro transcription/translation in wheat germ extract (Promega, Madison, Wisconsin, USA) using Tran35S-Label (ICN Radiochemicals, Costa Mesa, California, USA) according to the manufacturer’s instructions. 35S-IκBα and 35S-S32A/S36A IκBα were removed from unincorporated radioactivity by gel filtration using a NICK-Spin column (Pharmacia Biotech, Piscataway, New Jersey, USA). 35S-IκBα or 35S-S32A/S36A IκBα (∼40,000 cpm) were added to a 20-μL reaction containing 2 mM ATP, 10 mM creatine phosphate, 0.2 mg/mL creatine kinase, 70 μM GST-ubiquitin, 3.3 μM okadaic acid (Sigma Chemical Co.), 30 μM MG132, 2 μM ubiquitin aldehyde, 200 μM bestatin, 10 μM E64, 0.5 mM PMSF, and 80 μg HeLa cell extract in 20 mM Tris-Cl, pH 7.6, 20 mM KCl, 10 mM MgCl2, 1 mM DTT, 10% glycerol. PR39 or control peptides were also added to individual reactions. The samples were incubated at 37°C for 90 minutes followed by the addition of Laemmli sample buffer to stop the conjugation reactions. The samples were then heated for 5 minutes at 95°C and analyzed by SDS-PAGE on 4–15% acrylamide gradient gels (Bio-Rad, Hercules, California, USA). After electrophoresis, the gels were incubated with gentle stirring in 30% methanol, 10% acetic acid, dried, and analyzed using a Fuji PhosphoImager (Fuji, Tokyo, Japan).
Cell-culture assay.
ECV304 cells were stably transfected with a HA-tagged ubiquitin plasmid MT123 (kind gift of G. Walz, Beth Israel Deaconess Medical Center). The clone with the highest expression of HA-ubiquitin was treated with 10 μM PR39 for 45 minutes followed by treatment with 1 ng/mL TNF-α for 20 minutes. At that point, the cells were washed and lysed and IκBα was immunoprecipitated. After SDS-PAGE and membrane transfer of the immunoprecipitated material, the HA-ubiquitinated IκBα was visualized by Western blotting with an anti-HA Ab (Santa Cruz Biotechnology, Santa Cruz, California, USA).
Transcription assays
Stable cell lines expressing luciferase reporter gene under control of a tandem of four NF-κB–binding sites in front of a minimal TK promoter (NF-κB-TK-Luc, courtesy of G. Walz) or a cytomegalovirus (CMV) promoter (CMV-Luc, pRLCMV; Promega) were generated by electroporation of ECV304 cells followed by selection in 1 mg/mL G418-supplemented media (GIBCO-BRL Life Technologies, Gaithersburg, Maryland, USA)as described previously (34). Twenty to 30 clones expressing each construct were pooled for subsequent assay. Following cell lysis, luciferase activity was determined using Luciferase Assay System (Promega) (34).
Protein-degradation assay
Exponentially proliferating U937 cells were grown in 10% FBS–RPMI-1640 methionine-free medium supplemented with 200 μCi 35S methionine for 16 hours. Cells were then washed with complete RPMI-1640 medium three times and cultured in 10% FBS-RPMI (chase) in the presence of 10 μM of either PR39 or MG132 or an equal volume of PBS. Chloroquine (45 μM) was added to all cultures to prevent lysosomal protein degradation. One hour later the cells were lysed, and TCA-soluble 35S counts were measured in a liquid scintillation counter (Beckman Instruments Inc., Fullerton, California, USA). All experiments were carried out in triplicate and repeated three times.
Electrophoretic mobility shift assay
Nuclear extracts were prepared as described (35). In brief, pancreatic tissue was homogenized in ice-cold 0.3 M sucrose, cells were subjected to iso-osmolar lysis in buffer containing NP-40, and the nuclei were isolated. Nuclear protein was extracted from intact nuclei in a buffer containing 0.4 M KCl and stored at –70°C before analysis.
Aliquots of 7.5–10 μg of nuclear protein were mixed in 25-μL reactions containing 5 mM Tris, pH 7.5, 100 mM NaCl, 1 mM DDT, 1 mM EDTA, 4% (vol/vol) glycerol, 0.08 mg/mL salmon sperm DNA, and H2O. The oligonucleotide probe (5′-AGT TGA GGG GAC TTT CCC AGG C-3′; Promega) containing the κB–binding motif was end-labeled with [γ-32P]ATP using T4 polynucleotide kinase. Then, 106 cpm of the probe was added to the mixture, and the binding reaction was allowed to proceed for 20 minutes at room temperature. The unlabeled oligonucleotide was used in the specific competition assay. DNA-protein complexes were resolved in a 6% nondenaturing polyacrylamide gel in a Tris-borate-EDTA buffer at 150 V. Gels were dried and exposed to Kodak Bio Max MR film (Eastman Kodak Co., Rochester, New York, USA) at –70°C.
Mouse acute injury models
Acute pancreatitis.
Male mice weighing 20–25 g (ICR; Charles River Laboratories, Wilmington, Massachusetts, USA) were pretreated with PR39 (10 mg/kg, intravenously) or physiological saline 1 hour before injecting 50 μg/kg caerulein intraperitoneally (Research Plus, Bayonne, New Jersey, USA) to induce acute pancreatitis (36). Thirty minutes later the animals were sacrificed in a CO2 chamber and a 75–100 mg piece of pancreas was removed for Western blotting and NF-κB gel-shift assays.
Acute myocardial infarction.
Transgenic mice stably expressing PR39 in cardiac myocytes (αMHC-PR39 mice) and littermate controls (both op/op strain background), or C57BL/6 mice were subjected to acute coronary artery ligation as described (30). C57BL/6 mice were randomized to intraperitoneal implantation of Alzet Minipumps (Alza Corp., Palo Alto, California, USA) delivering 1 μg/kg/24 h of PR39 or buffer. Myocardial tissues collected 1, 3, or 7 days later from αMHC-PR39 and littermate control mice were subjected to Western blot analyses for VCAM-1 gene expression, which was normalized to expression levels before induction of infarction. Infarct size was determined from serial cross-sections of myocardial tissues after sacrifice at day 7 using digital camera and quantitative image analysis software (Optimas 6.0, Media Cybernetics, Silver Spring, Maryland, USA).
Results
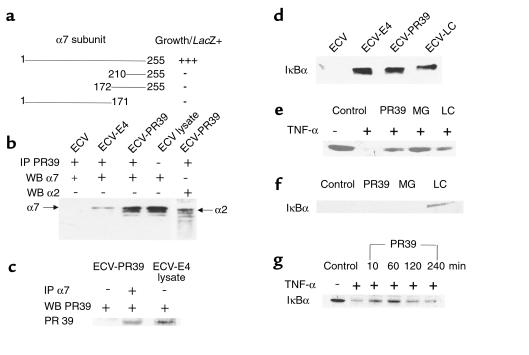
To identify possible intracellular targets of PR39 action, we carried out a yeast two-hybrid screen of a mouse cDNA library using the unique fourth-exon DNA sequence of porcine PR39 gene as bait. Four clones growing on selective media and demonstrating lacZ staining were purified and sequenced. All four encoded overlapping, identical cDNA sequences, which are highly homologous to the sequence of the human α7 (HC8) subunit of the 20S proteasome (GeneBank accession no. AF055983). Deletion analysis showed that the presence of both the COOH-terminal as well as the NH2-terminal sequences of α7 are required for PR39 binding (Figure 1a). To confirm that PR39 and the α7 subunit interact in cells, a polyclonal anti-PR39 Ab was used to immunoprecipitate PR39 protein from ECV304 cells transfected with cDNA constructs corresponding either to the full length (ECV-PR39) or exon 4 (39 amino acid COOH-terminal domain) (ECV-E4) of the porcine PR39 gene. Western blot analysis of the material immunoprecipitated from the whole cell lysate with the anti-α7 subunit mAb demonstrated the presence of a 29-kD band corresponding to the known size of the α7 proteasome subunit in the ECV-PR39 and ECV-E4, but not wild-type ECV304 cells (compare lanes 2 and 3 and lane 1 in Figure 1b). Western blotting of the ECV cell lysate with or without an anti-α7 subunit Ab was essentially identical in appearance (Figure 1b, lane 4). The appearance of additional bands below the α7-subunit band in Western blots of anti-PR39 Ab-precipitated material (Figure 1b, lane 3) suggests potential presence of other 20S proteasome subunits. To confirm this directly, we subjected the anti-PR39 antibody immunoprecipitate from ECV-PR39 cells to Western blotting with a mAb directed against the α2 subunit of the 20S proteasome and demonstrated the presence of this subunit in the immunoprecipitate (Figure 1b). Western blot of the whole-cell lysate with an anti-α2 Ab confirmed the identity of the α2 band in the PR39 Ab immunoprecipitate (not shown).
Figure 1.
PR39-proteasome interaction and the effect on IκBα degradation and NF-κB activity. (a) Binding of PR39 to α7 and its fragments in the yeast two-hybrid assay. The extent of interaction of deletion mutants of the mouse α7 subunit and PR39 was analyzed using the yeast two-hybrid system. Only the full-length α7 construct was able to bind to PR39. (b and c) Coimmunoprecipitation of PR39 and 20S subunits in ECV cells. (b) Western blotting of anti-PR39 Ab immunoprecipitate from ECV, ECV-E4, or ECV-PR39 cells was carried out with anti-α7 (lanes 1–3) or anti-α2 (lane 5) Ab’s. Note the presence of bands corresponding to the α7 or α2 subunits in the immunoprecipitate of ECV-E4 or ECV-PR39; but not wild-type ECV cells, as well as the presence of additional bands, that likely correspond to other 20S proteasome subunits. Western blotting with anti-α7 Ab of the whole ECV cell lysate was used as a control (lane 4). (c) Western blotting with anti-PR39 Ab of anti-α7 Ab (or nonimmune IgG) immunoprecipitate of ECV-PR39 cells. A Western blot of ECV-E4 cell lysate (third lane) is used as a control for PR39 band. Note the presence of the PR39 band only in the anti-α7 Ab immunoprecipitate (lane 2 vs. 1). (d) Stable expression of PR39 increases IκBα expression in ECV304 cells. Western blot analysis of IκBα protein levels in normally proliferating wild-type ECV304 cells (ECV) or ECV-PR39 and ECV-E4 cells (lanes 1–3). ECV304 cells treated for 2 hours with 10 μM lactacystin (LC) were used as control. Note increased IκBα levels in ECV-E4 and ECV-PR39 cells as well as in lactacystin-treated cells. (e) PR39 prevents TNF-α–induced degradation of IκBα. U937 cells cultured in the absence (–) or presence of TNF-α (1 ng/mL). Ten minutes after TNF-α addition, the cells were washed, and IκBα protein levels were assessed by Western blotting. Note complete disappearance of IκBα in TNF-α–treated cells. Forty-five minutes of pretreatment with 100 nM of PR39 or 10 μM of MG132 (MG) or lactacystin (LC) before the addition of TNF-α blocked cytokine-induced degradation of IκBα. (f) Reversibility of PR39 inhibition of IκBα degradation. U937 cells were pretreated with 100 nM of PR39, 10 μM of either MG132 or lactacystin (LC), or buffer (control) for 45 minutes. The cells were then washed extensively with fresh medium. Then, after 45 minutes, TNF-α (1 ng/mL) was added to the medium, and the extent of IκBα degradation was determined 10 minutes later by Western blotting. Note preservation of IκBα by lactacystin (an irreversible proteasome inhibitor) but not by MG132 or PR39. (g) Time course of PR39-mediated inhibition of TNF-α–induced IκBα degradation. HUVEC cells were challenged with TNF-α 10, 60, 120, or 240 minutes after addition of PR39. Ten minutes later, the cells were lysed and IκBα levels were determined by Western blotting (lanes 3–6). Note peak protection 60 minutes after PR39 exposure and wearing off of the effect by 240 minutes. TNF-α induced full degradation of IκBα in the absence of PR39 (compare lanes 1 and 2).
To further confirm PR39-α7 interaction, we performed Western blotting of ECV-PR39–cell immunoprecipitate obtained with an anti-α7 Ab or a nonimmune IgG with an anti-PR39 Ab. In this experiment, the band corresponding to the PR39 peptide was present only in the anti-α7, but not control, immunoprecipitate (Figure 1c). Taken together, these results suggest that PR39 can bind to the 20S proteasome particle through an interaction with the α7 subunit.
Since such an interaction with a proteasome subunit may result in inhibition of proteasome-mediated protein degradation, we assessed the effect of PR39 expression on the degradation of IκBα, an important regulator of gene transcription. Stable expression of PR39 construct in ECV304 cells resulted in increased levels of IκBα (Figure 1d, compare lanes 1 and 2). The same increase in IκBα protein levels was seen after exposure of ECV304 cells to proteasome-specific inhibitor lactacystin or synthetic PR39 peptide (Figure 1d, lanes 3 and 4).
TNF-α induces rapid degradation of IκBα by the Ub-proteasome pathway (16). We therefore tested the ability of PR39 to inhibit TNF-α–induced degradation of IκBα in U937 cells that normally exhibit significant baseline levels of IκBα (37). TNF-α caused a rapid decline of the level of IκBα in these cells, and this effect was markedly reduced by pretreatment with PR39 or proteasome inhibitors lactacystin or MG132 (Figure 1e). To test whether treatment with PR39 irreversibly blocked IκBα degradation, U937 cells were pretreated with the peptide and then were extensively washed, and 45 minutes later were exposed to TNF-α. In parallel, U937 cells were exposed to MG132, a rapidly reversible competitive inhibitor of the proteasome, or with lactacystin, which covalently and irreversibly modifies the active-site threonine residues. Western blot analysis demonstrated the complete disappearance of IκBα protein in PR39 and MG132-treated cells, whereas pretreatment with lactacystin irreversibly blocked proteasome-mediated degradation of IκBα (Figure 1f), suggesting that PR39 effect, like that of MG132, is reversible. To further study the time course of PR39-mediated inhibition of TNF-α–induced degradation of IκBα, primary HUVEC cells were pretreated with PR39 for a variable period of time before TNF-α administration. Whereas as little as 10 minutes of pretreatment was sufficient to partially block TNF-α–induced IκBα degradation, full protection was seen after 60 minutes of pretreatment, and the effect was lost if TNF-α administration was delayed for more than 2 hours after exposure to PR39 (Figure 1g). These results further support the notion that PR39 has a transient and reversible effect on TNF-α–induced IκBα degradation.
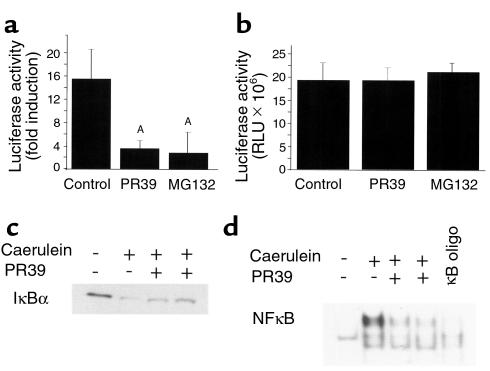
To show that the PR39 not only prevented inhibition of IκBα degradation but also inhibited NF-κB–dependent transcription (i.e., to show that the IκBα protein is fully functional), we employed pooled stable ECV304 clones expressing a luciferase reporter construct under control of either NF-κB-TK or CMV promoters. Stimulation with TNF-α of NF-κB-TK-Luc–expressing cells induced a 15.8-fold increase in the luciferase activity that was almost completely inhibited by pretreatment with either PR39 or MG132 (Figure 2, a and b). At the same time, PR39 or MG132 had no effect on the luciferase activity in cells expressing CMV-Luc construct.
Figure 2.
PR39 inhibits NF-κB–dependent gene expression in cell culture and in mice. (a and b) PR39 administration blocks NF-κB–dependent gene expression in cell culture. (a) Luciferase activity (fold increase), after exposure to 1 ng/mL TNF-α of ECV cells transfected with (NF-κB)4-TK-Luc construct, was measured in the absence (control) or presence of 100 nM PR39 or 10 μM MG132. Note significant inhibition of induction of NF-κB activity by TNF-α in PR39-treated cells. AP < 0.01 versus control. (b) Luciferase activity in ECV cells expressing CMV-Luc construct in the absence (control) or presence of 100 nM of PR39 or 10 μM of MG132. Note the lack of any effect of PR39 on CMV-driven luciferase activity. (c and d) PR39 blocks IκBα degradation and NF-κB–dependent transcription in the mouse pancreas after induction of acute pancreatitis. (c) Western blot analysis of IκBα in the mouse pancreas after exposure to caerulein in the absence (–) or presence (+) of pretreatment with 10 mg/kg intravenously injected PR39. (d) NF-κB activation in mouse pancreatic tissues in the setting of caerulein-induced pancreatitis examined using electromobility-shift assay. Note almost complete disappearance of NF-κB gel shift in PR39-treated animals. Excess of oligonucleotide competitor of NF-κB binding site (100×) was used to demonstrate band specificity (κB oligo).
To study whether PR39 can also inhibit NF-κB–dependent transcription in animals, we examined IκBα protein levels and the presence of NF-κB transcriptional activity in a model of acute pancreatitis. In this model, induction of acute pancreatitis by treatment with the cholecystokinin analogue caerulein (36) leads to the prompt disappearance of IκBα (Figure 2c) and activation of NF-κB–dependent transcription in the pancreas (Figure 2d). Intravenous injection of 10 mg/kg of PR39 1 hour before caerulein administration largely prevented IκBα degradation and, consequently, NF-κB–dependent gene expression as assessed by DNA gel shifts (Figure 2, c and d).
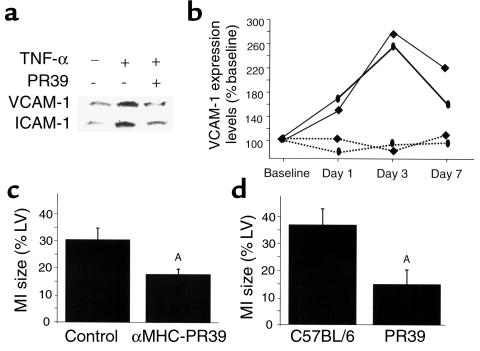
To examine the functional consequences of PR39-mediated inhibition of NF-κB–dependent gene expression, we evaluated the effect of peptide administration on expression of NF-κB–dependent adhesion proteins VCAM-1 and ICAM-1 in cell culture and in the setting of acute myocardial infarction in mice. Exposure of HUVEC to TNF-α in culture (38) or induction of ischemia in the heart (39) leads to increased VCAM-1 and ICAM-1 gene expression, which is thought to play a major role in mediation of postischemic myocardial injury (40). However, pretreatment with PR39 prevented TNF-α–induced induction of expression of both adhesion proteins in HUVEC cells (Figure 3a).
Figure 3.
Effect of PR39 on NF-kB–dependent gene expression and infarct size in mice. (a) PR39 blocks TNF-α–induced activation of VCAM-1 and ICAM-1 expression in cell culture. The expression levels of adhesion protein levels in HUVEC cells were measured by Western blotting 2 hours (VCAM-1) and 8 hours (ICAM-1) after exposure to TNF-α in absence (–) or presence (+) of PR39. The addition of PR39 completely blocked TNF-α–dependent induction of both VCAM-1 and ICAM-1 expression. (b) Time course of VCAM-1 and ICAM-1 expression after induction of myocardial infarction. Western blot analysis of VCAM-1 (oval) and ICAM-1 (square) expression in the hearts of control (solid lines) and αMHC-PR39 (dotted lines) mice 1, 3, or 7 days after induction of myocardial infarction. Note increased expression of VCAM-1 and ICAM-1 in the control, but not αMHC-PR39 mice (n = 2 for each time point). (c) Effect of transgenic PR39 expression on myocardial infarct size. Histologically determined extent of myocardial necrosis (expressed as percentage of cross-sectional left ventricle [LV] area) in αMHC-PR39 (n = 6) and littermate control (n = 8) mice. Note significantly smaller infarct size in PR39 transgenic mice (mean ± SD). (d) Effect of PR39 treatment on myocardial infarction size. Histologically determined extent of myocardial necrosis (expressed as percentage of cross-sectional LV area) in C57BL/6 mice randomized to intraperitoneal infusion of PR39 (n = 6) or saline (n = 13). Note significantly smaller infarct size in PR39-treated mice (mean ± SD). AP < 0.05.
To study this in vivo, we assessed VCAM-1 and ICAM-1 protein levels after induction of myocardial infarction in transgenic mice expressing PR39 cDNA in cardiac myocytes (αMHC-PR39) (32) or age- and strain-matched control mice. The αMHC-PR39 mice express PR39 peptide selectively in cardiac myocytes, and the peptide expression is not associated with any detectable changes in any baseline heart morphometric or physiologic parameters (weight, wall thickness, left-ventricular function), except for increased vascularity (32).
Coronary ligation in control (wild-type C57BL/6) mice led to a significant rise in the VCAM-1 and ICAM-1 expression by 24 hours, reached a peak by 72 hours, and then gradually declined. In contrast, induction of myocardial infarction had no effect on VCAM-1 or ICAM-1 expression in α-MHC-PR39 mice (Figure 3b). Morphometric evaluation of the infarct size in these mice 7 days after coronary ligation showed a significantly smaller area of necrosis in αMHC-PR39 (n = 6) compared with control animals (n = 8; Figure 3c). To test whether exogenously administered peptide would have a similar effect on myocardial infarct size, C57BL/6 mice were randomized to sustained 7-day intraperitoneal infusion of PR39 peptide (1 μg/kg/day; n = 6) or saline control (n = 13) via an implantable osmotic pump immediately after coronary ligation. Morphometric assessment of the infarct size at day 7 demonstrated significantly smaller infarcts in PR39-treated, compared with saline-treated, mice (Figure 3d).
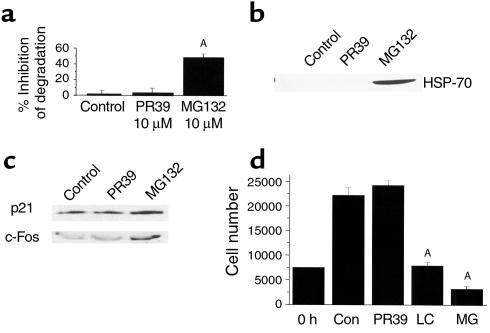
One possible explanation of the PR39-dependent inhibition of IκBα degradation would be that the peptide, like MG132, reversibly inhibits all proteasome function, thus generally suppresses intracellular protein degradation. Therefore, we compared the extent of degradation of long-lived cell proteins before or after exposure to either PR39 or MG132. After 16-hour metabolic labeling of total cell protein in ECV304 cells, MG132 treatment resulted in approximately 50% inhibition of total cellular protein degradation, in accordance with results reported previously (41). In contrast, administration of PR39 had little or no effect on this process (Figure 4a). To confirm that PR39 does not cause a general inhibition of protein degradation, we studied the expression level of HSP-70, a major heat shock protein whose expression is stimulated by proteasome inhibitors (37, 41, 42). Western blot analysis demonstrated a striking increase in HSP-70 levels in cultured ECV304 cells after treatment with 10 μM of MG132. However, exposure to a similar concentration of PR39 failed to stimulate HSP-70 accumulation (Figure 4b). In addition, studies in HUVEC and ECV304 cells with specific antibodies failed to demonstrate any effect of PR39 treatment on the levels of two cell-cycle proteins principally controlled by proteasome-dependent degradation — the cell-cycle inhibitor p21Cip1/Waf1 and the transcription factor c-Fos (Figure 4c).
Figure 4.
Selective nature of PR39 effect. (a) PR39 does not substantially affect proteasome-dependent degradation of total cellular protein. Pulse-chase analysis of total cellular protein degradation in the U937 cells in the absence (control) or presence of 1 μM PR39 or 1 μM MG132. Note significantly lower inhibition of total cell protein degradation by PR39 than by MG132 (mean ± SD). (b) Lack of induction of HSP-70 expression in PR39-treated cells. Western blotting of cell extract from untreated (control) and PR39-treated (1 μM), or MG132-treated (1 μM) U937 cells demonstrated the appearance of HSP-70 expression after 3 hours of exposure to MG132 but not PR39. (c) Effect of PR39 administration on proteasome-dependent protein levels in cells. The level of proteasome-regulated cell-cycle repressor p21, and c-Fos transcription factor were determined by Western blotting in HUVEC cells pretreated with 100 nM of PR39 or 10 μM of MG132. Note the lack of the effect of PR39 treatment on the expression level of these proteins whereas the treatment with MG132 increased expression of both by inhibiting their degradation by the proteasome. (d) PR39 does not affect cell growth in culture. The effect of PR39 or proteasome inhibitors MG132 (MG) and lactacystin (LC) on cell growth in culture was tested using ECV304 cells. Cell counts 48 hours after stimulation of synchronized ECV304 cells with 10% serum in the presence of buffer (control) or 1 μM of PR39, lactacystin, or MG132, demonstrated significant inhibition of cell growth by both proteasome inhibitors, but not by PR39 (mean ± SD). AP < 0.01 versus control. Con, control.
Prolonged exposures to proteasome inhibitors tend to induce apoptosis in cell culture (43). To assess the potential cytotoxicity of PR39, ECV304 cells were cultured in the presence of 10 μM of PR39, MG132, or lactacystin. Cell counts 48 hours later documented that, while addition of PR39 did not affect the cells’ ability to proliferate, exposure to either MG132 or lactacystin led to a significant decline in cell counts (Figure 4d). These results demonstrate that PR39 is able to inhibit IκBα degradation without significantly affecting overall protein degradation in cells.
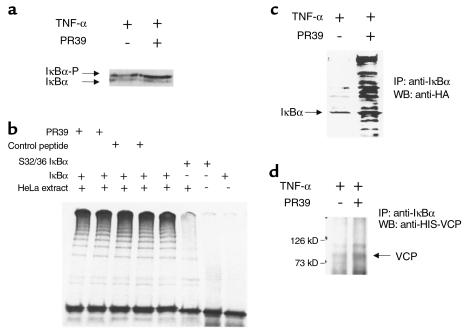
The preceding experiments suggest that PR39 inhibits IκBα degradation in a relatively selective manner. To explore potential mechanisms of this inhibition, we studied the effect of peptide administration on IκBα phosphorylation and ubiquitination. TNF-α treatment leads to phosphorylation of IκBα, which is required for its subsequent ubiquitination. Western blotting of U937 cell lysates after in vivo 32P labeling demonstrated the appearance of the phosphorylated form of IκBα after TNF-α exposure. Pretreatment with PR39 increased the amount of the phosphorylated form of IκBα, presumably due to inhibition of its degradation (Figure 5a). To test the effect of PR39 on IκBα ubiquitination in a cell-free system, HeLa cell extract was used to ubiquitinate in vitro transcribed, translated, and phosphorylated IκBα. In the absence of PR39, incubation of phosphorylated IκBα protein with a HeLa extract led to the appearance of multiple high-molecular-weight ubiquitinated forms of IκBα. The addition of PR39 in an amount sufficient to inhibit IκBα degradation in cell culture had no effect on the extent of IκBα ubiquitination in this assay (Figure 5b).
Figure 5.
PR39 does not affect known steps in IκBα processing. (a) The effect of PR39 on IκBα phosphorylation in cell culture. U937 cells cultured in the absence (–) or presence (+) of pretreatment with 10 μM PR39 were exposed to 1 ng/mL TNF-α; 45 minutes later the cells were lysed and subjected to SDS-PAGE and Western blotting with anti-IκBα Ab. Note increased amount of the typical-appearing phosphorylated IκBα band in PR39-treated cells. (b) The effect of PR39 on IκBα ubiquitination in vitro. In vitro ubiquitination of phosphorylated IκBα protein was carried out using crude HeLa-cell extract in the presence of control peptides (lanes 3 and 4), PR39 (lanes 1 and 2), or in the absence of both (lane 5). Note that the addition of either PR39 at two different concentrations or of a control (random) peptide failed to affect IκBα ubiquitination. At the same time, no ubiquitination of phosphorylation site IκBα mutants (lanes 6 and 7) or unphosphorylated IκBα (weight IκBα, lane 8) was detected. (c) The effect of PR39 on IκBα ubiquitination in cell culture. HA-Ub–expressing ECV304 cells were exposed to 1 ng/mL TNF-α in the presence (+) or absence (–) of 10 μM PR39 pretreatment; 45 minutes later the cells were lysed and subjected to IκBα immunoprecipitation followed by Western blotting of the pellet material with anti-HA antibody. Note multiple ubiquitinated IκBα intermediaries in PR39-treated, but not control cells. (d) PR39 does not inhibit IκBα-VCP binding. Wild-type ECV304 cells treated as described in c were subjected to immunoprecipitation with anti-IκBα antibody (IκBα-IP) followed by Western blotting with the anti–VCP-3 Ab. Note the presence of 90-kD VCP band in the presence or absence of PR39 treatment.
To assess the effect of PR39 treatment on IκBα ubiquitination in intact cells, we generated a stable ECV304-derived cell line expressing HA-tagged ubiquitin. Immunoprecipitation of IκBα followed by Western blotting with anti-HA Ab demonstrated the accumulation of more ubiquitinated IκBα complexes in cells pretreated with 10 μM of PR39 than in untreated cells (Figure 5c). Together these findings argue that PR39 does not block the ubiquitination process, but does inhibit the degradation of ubiquitinated IκBα by the 26S proteasome.
A recent study suggested that binding of the ubiquitinated IκBα to VCP, a 26S proteasome–associated protein, is necessary, but not sufficient, for subsequent IκBα degradation (24). Western blotting with the anti-VCP Ab demonstrated the presence of this protein in the anti-PR39 Ab immunoprecipitate of the ECV-PR39 cells (not shown). Western blot analysis of anti-IκBα Ab immunoprecipitated material from ECV304 cells demonstrated the presence of VCP in accord with the study reported previously (24). Pretreatment with PR39 increased the amount of VCP in the IκBα immunoprecipitate (Figure 5d), as would be expected given increased amount of ubiquitinated forms of IκBα in the PR39-treated cells. We conclude from these studies that PR39 does not interfere with IκBα-VCP binding.
Discussion
In this study we have presented several types of evidence that the naturally occurring antibacterial 39–amino acid peptide, PR39, inhibits IκBα degradation in cultured cells and in two different mice models and that functional effects of PR39 treatment is mediated by inhibition of NF-κB–dependent gene expression. Several lines of evidence support this conclusion. In cell culture the peptide inhibited TNF-α–induced activation of NF-κB-TK–promoter expression and induction of ICAM-1 and VCAM-1 expression in HUVEC cells. In mice studies, the peptide blocked NF-κB–dependent transcription in the setting of acute pancreatitis and prevented increase in ICAM-1 and VCAM-1 expression after induction of acute myocardial infarction.
The later model was used to assess the functional significance of PR39 activity. Recent studies have documented that the ischemia-reperfusion is associated with TNF-α release from the heart (44, 45), which is associated with early induction of NF-κB gene expression (39). This, in turn, is associated with increased expression of adhesion molecules including ICAM and VCAM (46, 47), which can be blocked by proteasome inhibitors (47) or other compounds capable of inhibiting NF-κB activity (40). Furthermore, inhibition of proteasome-mediated IκBα degradation (48) or adhesion molecule expression has been shown to reduce significantly ischemia-reperfusion injury (49–51).
The link between PR39, inhibition of IκBα degradation, inhibition of NF-κB–dependent transcription, fall in ICAM-1 and VCAM-1 gene expression, and reduction in the size of myocardial infarction does not rule out that PR39 may have other biological effects. Indeed, we have shown that the peptide increases HIF-1α protein levels (32), and others have reported PR39 interactions with p47phox (27) and p130Cas (28).
Whereas the ability to inhibit IκBα degradation may suggest that PR39 acts as a proteasome inhibitor, several observations clearly set it apart from the small-molecule proteasome inhibitors described to date. Most important in this regard is the relatively selective nature of the PR39 effect. This selectivity is indicated by the lack of any effect on overall proteasome-dependent protein degradation in cells as well as degradation of two cell-cycle genes known to be regulated by proteasome-dependent degradation — the cell cycle inhibitor p21Cip1/Waf1 (52) and a transcription factor c-fos (53). Additional evidence in favor of selective inhibition of proteasome activity by PR39 includes the absence of induction of heat shock proteins such as HSP-70 and the lack of toxicity after a long-term exposure in cell culture or transgenic expression in mice.
In contrast, exposure of cultured cells to proteasome inhibitors MG132 and lactacystin leads to a general reduction in intracellular proteolysis, a rapid increase in HSP-70 expression, and cell death (37, 41, 42). In this regard it is interesting to note that the lack of PR39 effect on cell proliferation in culture is consistent with the lack of increase in p21Cip1/Waf1 expression seen with proteasome inhibitors such as MG132, which leads to inhibition of cell growth (54).
Whereas known inhibitors of 20S proteasome such as lactacystin and MG132 affect its active sites, and therefore the inhibition of proteolysis is not specific to any particular protein or class of proteins, selective inhibition of proteasome-mediated degradation potentially can be achieved by blocking substrate-specific steps in the degradative pathway. We therefore carried out experiments to determine whether the inhibition of IκBα degradation by PR39 is due to inhibition of known steps required for its proteasome-mediated degradation, including phosphorylation, ubiquitination, and VCP binding.
Studies in the cell-free system failed to show any effect of PR39 treatment on ubiquitination of IκBα, whereas cell-culture experiments demonstrated an accumulation of phosphorylated and ubiquitinated forms of IκBα in the PR39-treated cells. Recently, it was shown that proteasome-dependent degradation of IκBα requires its binding to a cytoplasmic protein, VCP, that may provide a physical link between the ubiquitinated IκBα and the 26S proteasome (24). In our experiments, exposure of cells to PR39 did not interfere with IκBα-VCP binding, as demonstrated by the presence of the VCP band on the immunoprecipitate of whole-cell lysate with anti- IκBα Ab in the presence or absence of PR39. Thus, PR39 did not interfere with any of the known steps involved in proteasome-mediated degradation of IκBα.
Several experimental results in this study are consistent with direct interaction of PR39 with the 20S particle. The peptide is able to bind with high affinity directly to the α7 subunit of the proteasome as shown in the yeast two-hybrid assay, as well as by coimmunoprecipitation with the anti-PR39 and anti-α7 Ab’s. Furthermore, as a result of binding to α7, PR39 is able to bring down at least one other (α2) 20S subunit, and probably other 20S subunits as well, suggesting that the peptide binds to the whole 20S particle without causing its dissociation. Whereas the site of interaction on the α7 subunit or on PR39 has not been established, one possibility is the high negative charge of the COOH-terminal end of the α7 subunit, which makes it a likely site for binding of the positively charged PR39 molecule. The α7 COOH-terminal sequence is actually the most negatively charged sequence of any of the proteasomal α subunits, and it has the ability to assemble into heptameric ring structures by itself, as well as to induce ring formation of other α subunits (55).
However, it is unclear how PR39 interaction with the 20S proteasome can lead to selective inhibition of the degradation of phosphorylated and ubiquitinated IκBα. One possible mechanism would include interference with binding of ubiquitinated IκBα-VCP complex to the 26S proteasome. The observed inhibition of the NF-κB–dependent transcription in PR39-treated cells or animals is consistent with this possibility, since inhibition of VCP-IκBα binding to the proteasome would leave the ubiquitinated IκBα in complex with NF-κB. Alternatively, PR39 may interfere with another, yet unknown, step involved in IκBα degradation by the proteasome. Finally, PR39 binding to the α7 subunit may alter the three-dimensional proteasome architecture or interaction of the 20S particle and the 19S regulatory subunit that would affect the entry of certain protein substrates into the 20S cylinder. All of these possibilities are consistent with the observation that PR39 inhibition of IκBα degradation is dose dependent and reversible, apparently requiring continuous presence of the peptide. It should be noted that this activity is not strictly limited to IκBα, since we have found that this peptide also inhibits proteasome-dependent degradation of HIF-1α (32). This could imply that the peptide affects a step common to degradation of these two proteins.
Several proteins interacting with subunits of 20S proteasome have been identified to date, including viral proteins Hbx (56), Tat (57), and Tax (58). For example, Tax binding to HC9 has been reported to facilitate binding of p105 NF-κB to the proteasome and accelerates its proteolytic maturation (58). Thus, PR39 is yet another example of α subunit–binding protein that may have a unique regulatory activity of the 26S complex.
In conclusion, the ability of PR39 peptide to block stimulation of NF-κB–dependent gene expression relatively selectively in animal models of pancreatitis and acute myocardial infarction may have important implications as a potential therapeutic approach to management of a number of disease states, including acute inflammation, immune and autoimmune responses, and ischemia-reperfusion injury.
Acknowledgments
We thank G. Walz for HA-ubiquitin plasmid construct MT123 and C.C. Li (NIH) for antiVCP-3 antibody.
This work was supported, in part, by the NIH grants HL-53793 and HL-56993 (M. Simons), DK-31396 (M.L. Steer), GM-51923 and GM-46147 (A.L. Goldberg), F32 HL-10013 (R. Volk); American Heart Association Established Investigator Award 9940074 (M. Simons); and a grant from Chiron Corporation.
References
- 1.Goldberg AL, Akopian TN, Kisselev AF, Lee DH, Rohrwild M. New insights into the mechanisms and importance of the proteasome in intracellular protein degradation. Biol Chem. 1997;378:131–140. [PubMed] [Google Scholar]
- 2.Tanaka K. Molecular biology of the proteasome. Biochem Biophys Res Commun. 1998;247:537–541. doi: 10.1006/bbrc.1998.8617. [DOI] [PubMed] [Google Scholar]
- 3.Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 4.Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 5.DeMartino GN, Slaughter CA. The proteasome, a novel protease regulated by multiple mechanisms. J Biol Chem. 1999;274:22123–22126. doi: 10.1074/jbc.274.32.22123. [DOI] [PubMed] [Google Scholar]
- 6.Whiteside ST, Israel A. I kappa B proteins: structure, function and regulation. Semin Cancer Biol. 1997;8:75–82. doi: 10.1006/scbi.1997.0058. [DOI] [PubMed] [Google Scholar]
- 7.Srinivas V, Zhu X, Salceda S, Nakamura R, Caro J. Hypoxia-inducible factor 1 (HIF-1) is a non-heme iron protein. Implications for oxygen sensing. J Biol Chem. 1998;273:18019–18022. doi: 10.1074/jbc.273.29.18019. [DOI] [PubMed] [Google Scholar]
- 8.Salceda S, Caro J. Hypoxia-inducible factor 1alpha (HIF-1alpha) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J Biol Chem. 1997;272:22642–22647. doi: 10.1074/jbc.272.36.22642. [DOI] [PubMed] [Google Scholar]
- 9.Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He H, Qi XM, Grossmann J, Distelhorst CW. c-Fos degradation by the proteasome. An early, bcl-2-regulated step in apoptosis. J Biol Chem. 1998;273:25015–25019. doi: 10.1074/jbc.273.39.25015. [DOI] [PubMed] [Google Scholar]
- 11.Pahl HL, Baeuerle PA. Control of gene expression by proteolysis. Curr Opin Cell Biol. 1996;8:340–347. doi: 10.1016/s0955-0674(96)80007-x. [DOI] [PubMed] [Google Scholar]
- 12.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 13.Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 14.Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 15.Baeuerle PA, Baltimore D. NF-kappa B: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 16.Beg AA, Baltimore D. An essential role for NF-kappa B in preventing TNF-alpha-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 17.Van Antwerp DJ, Martin SJ, Kafri T, Green DR, Verma IM. Suppression of TNF-alpha-induced apoptosis by NF-kappa B. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 18.Chen ZJ, Parent L, Maniatis T. Site-specific phosphorylation of IkappaBalpha by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki H, et al. In vivo and in vitro recruitment of an IkappaBalpha-ubiquitin ligase to IkappaBalpha phosphorylated by IKK, leading to ubiquitination. Biochem Biophys Res Commun. 1999;256:121–126. doi: 10.1006/bbrc.1999.0296. [DOI] [PubMed] [Google Scholar]
- 20.Spencer E, Jiang J, Chen ZJ. Signal-induced ubiquitination of IkappaBalpha by the F-box protein Slimb/beta-TrCP. Genes Dev. 1999;13:284–294. doi: 10.1101/gad.13.3.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kroll M, et al. Inducible degradation of IkappaBalpha by the proteasome requires interaction with the F-box protein h-betaTrCP. J Biol Chem. 1999;274:7941–7945. doi: 10.1074/jbc.274.12.7941. [DOI] [PubMed] [Google Scholar]
- 22.Yaron A, et al. Identification of the receptor component of the IkappaBalpha-ubiquitin ligase. Nature. 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- 23.Gonen H, et al. Identification of the ubiquitin carrier proteins, E2s, involved in signal-induced conjugation and subsequent degradation of IkappaBalpha. J Biol Chem. 1999;274:14823–14830. doi: 10.1074/jbc.274.21.14823. [DOI] [PubMed] [Google Scholar]
- 24.Dai RM, Chen E, Longo DL, Gorbea CM, Li CC. Involvement of valosin-containing protein, an ATPase co-purified with IkappaBalpha and 26 S proteasome, in ubiquitin-proteasome-mediated degradation of IkappaBalpha. J Biol Chem. 1998;273:3562–3573. doi: 10.1074/jbc.273.6.3562. [DOI] [PubMed] [Google Scholar]
- 25.Agerberth B, et al. Amino acid sequence of PR-39. Isolation from pig intestine of a new member of the family of proline-arginine-rich antibacterial peptides. Eur J Biochem. 1991;202:849–854. doi: 10.1111/j.1432-1033.1991.tb16442.x. [DOI] [PubMed] [Google Scholar]
- 26.Gudmundsson GH, et al. Structure of the gene for porcine peptide antibiotic PR-39, a cathelin gene family member: comparative mapping of the locus for the human peptide antibiotic FALL-39. Proc Natl Acad Sci USA. 1995;92:7085–7089. doi: 10.1073/pnas.92.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi J, Ross CR, Leto TL, Blecha F. PR-39, a proline-rich antibacterial peptide that inhibits phagocyte NADPH oxidase activity by binding to Src homology 3 domains of p47 phox. Proc Natl Acad Sci USA. 1996;93:6014–6018. doi: 10.1073/pnas.93.12.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan YR, Gallo RL. PR-39, a syndecan-inducing antimicrobial peptide, binds and affects p130 (Cas) J Biol Chem. 1998;273:28978–28985. doi: 10.1074/jbc.273.44.28978. [DOI] [PubMed] [Google Scholar]
- 29.Storici P, Zanetti M. A cDNA derived from pig bone marrow cells predicts a sequence identical to the intestinal antibacterial peptide PR-39. Biochem Biophys Res Commun. 1993;196:1058–1065. doi: 10.1006/bbrc.1993.2358. [DOI] [PubMed] [Google Scholar]
- 30.Li J, Brown LF, Laham RJ, Volk R, Simons M. Macrophage-dependent regulation of syndecan gene expression. Circ Res. 1997;81:785–796. doi: 10.1161/01.res.81.5.785. [DOI] [PubMed] [Google Scholar]
- 31.Gallo RL, et al. Syndecans, cell surface heparan sulfate proteoglycans, are induced by a proline-rich antimicrobial peptide from wounds. Proc Natl Acad Sci USA. 1994;91:11035–11039. doi: 10.1073/pnas.91.23.11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, et al. PR39, a peptide regulator of angiogenesis. Nat Med. 2000;6:49–55. doi: 10.1038/71527. [DOI] [PubMed] [Google Scholar]
- 33.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Pasparakis M, Kollias G, Simons M. Myocyte-dependent regulation of endothelial cell syndecan-4 expression. Role of TNF-alpha. J Biol Chem. 1999;274:14786–14790. doi: 10.1074/jbc.274.21.14786. [DOI] [PubMed] [Google Scholar]
- 35.Dyer RB, Herzog NK. Isolation of intact nuclei for nuclear extract preparation from a fragile B-lymphocyte cell line. Biotechniques. 1995;19:192–195. [PubMed] [Google Scholar]
- 36.Frossard JL, et al. The role of intercellular adhesion molecule 1 and neutrophils in acute pancreatitis and pancreatitis-associated lung injury. Gastroenterology. 1999;116:694–701. doi: 10.1016/s0016-5085(99)70192-7. [DOI] [PubMed] [Google Scholar]
- 37.Meriin AB, Gabai VL, Yaglom J, Shifrin VI, Sherman MY. Proteasome inhibitors activate stress kinases and induce Hsp72. Diverse effects on apoptosis. J Biol Chem. 1998;273:6373–6379. doi: 10.1074/jbc.273.11.6373. [DOI] [PubMed] [Google Scholar]
- 38.Marlor CW, Webb DL, Bombara MP, Greve JM, Blue ML. Expression of vascular cell adhesion molecule-1 in fibroblastlike synoviocytes after stimulation with tumor necrosis factor. Am J Pathol. 1992;140:1055–1060. [PMC free article] [PubMed] [Google Scholar]
- 39.Li C, Browder W, Kao RL. Early activation of transcription factor NF-kappaB during ischemia in perfused rat heart. Am J Physiol. 1999;276:H543–H552. doi: 10.1152/ajpheart.1999.276.2.H543. [DOI] [PubMed] [Google Scholar]
- 40.Ferran C, et al. Inhibition of NF-kappa B by pyrrolidine dithiocarbamate blocks endothelial cell activation. Biochem Biophys Res Commun. 1995;214:212–223. doi: 10.1006/bbrc.1995.2277. [DOI] [PubMed] [Google Scholar]
- 41.Bush KT, Goldberg AL, Nigam SK. Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. J Biol Chem. 1997;272:9086–9092. doi: 10.1074/jbc.272.14.9086. [DOI] [PubMed] [Google Scholar]
- 42.Lee DH, Goldberg AL. Proteasome inhibitors cause induction of heat shock proteins and trehalose, which together confer thermotolerance in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:30–38. doi: 10.1128/mcb.18.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fenteany G, Schreiber SL. Lactacystin, proteasome function, and cell fate. J Biol Chem. 1998;273:8545–8548. doi: 10.1074/jbc.273.15.8545. [DOI] [PubMed] [Google Scholar]
- 44.Gurevitch J, et al. Tumor necrosis factor-alpha is released from the isolated heart undergoing ischemia and reperfusion. J Am Coll Cardiol. 1996;28:247–252. doi: 10.1016/0735-1097(96)00105-2. [DOI] [PubMed] [Google Scholar]
- 45.Meldrum DR. Tumor necrosis factor in the heart. Am J Physiol. 1998;274:R577–R595. doi: 10.1152/ajpregu.1998.274.3.R577. [DOI] [PubMed] [Google Scholar]
- 46.Collins T, et al. Transcriptional regulation of endothelial cell adhesion molecules: NF-kappa B and cytokine-inducible enhancers. FASEB J. 1995;9:899–909. [PubMed] [Google Scholar]
- 47.Cobb RR, Felts KA, Parry GC, Mackman N. Proteasome inhibitors block VCAM-1 and ICAM-1 gene expression in endothelial cells without affecting nuclear translocation of nuclear factor-kappa B. Eur J Immunol. 1996;26:839–845. doi: 10.1002/eji.1830260417. [DOI] [PubMed] [Google Scholar]
- 48.Campbell B, Adams J, Shin YK, Lefer AM. Cardioprotective effects of a novel proteasome inhibitor following ischemia and reperfusion in the isolated perfused rat heart. J Mol Cell Cardiol. 1999;31:467–476. doi: 10.1006/jmcc.1998.0880. [DOI] [PubMed] [Google Scholar]
- 49.Palazzo AJ, et al. Myocardial ischemia-reperfusion injury in CD18- and ICAM-1-deficient mice. Am J Physiol. 1998;275:H2300–H2307. doi: 10.1152/ajpheart.1998.275.6.H2300. [DOI] [PubMed] [Google Scholar]
- 50.Kusterer K, et al. Soluble ICAM-1 reduces leukocyte adhesion to vascular endothelium in ischemia-reperfusion injury in mice. Am J Physiol. 1998;275:G377–G380. doi: 10.1152/ajpgi.1998.275.2.G377. [DOI] [PubMed] [Google Scholar]
- 51.Zhao ZQ, et al. Monoclonal antibody to ICAM-1 preserves postischemic blood flow and reduces infarct size after ischemia-reperfusion in rabbit. J Leukoc Biol. 1997;62:292–300. doi: 10.1002/jlb.62.3.292. [DOI] [PubMed] [Google Scholar]
- 52.Cayrol C, Ducommun B. Interaction with cyclin-dependent kinases and PCNA modulates proteasome-dependent degradation of p21. Oncogene. 1998;17:2437–2444. doi: 10.1038/sj.onc.1202189. [DOI] [PubMed] [Google Scholar]
- 53.He H, Qi XM, Grossmann J, Distelhorst CW. c-Fos degradation by the proteasome. An early, Bcl-2-regulated step in apoptosis. J Biol Chem. 1998;273:25015–25019. doi: 10.1074/jbc.273.39.25015. [DOI] [PubMed] [Google Scholar]
- 54.Adams J, et al. Proteasome inhibitors: a novel class of potent and effective antitumor agents. Cancer Res. 1999;59:2615–2622. [PubMed] [Google Scholar]
- 55.Gerards WL, de Jong WW, Bloemendal H, Boelens W. The human proteasomal subunit HsC8 induces ring formation of other alpha-type subunits. J Mol Biol. 1998;275:113–21. doi: 10.1006/jmbi.1997.1429. [DOI] [PubMed] [Google Scholar]
- 56.Fischer M, Runkel L, Schaller H. HBx protein of hepatitis B virus interacts with the C-terminal portion of a novel human proteasome alpha-subunit. Virus Genes. 1995;10:99–102. doi: 10.1007/BF01724303. [DOI] [PubMed] [Google Scholar]
- 57.Seeger M, Ferrell K, Frank R, Dubiel W. HIV-1 tat inhibits the 20 S proteasome and its 11 S regulator-mediated activation. J Biol Chem. 1997;272:8145–8148. doi: 10.1074/jbc.272.13.8145. [DOI] [PubMed] [Google Scholar]
- 58.Rousset R, Desbois C, Bantignies F, Jalinot P. Effects on NF-kappa B1/p105 processing of the interaction between the HTLV-1 transactivator Tax and the proteasome. Nature. 1996;381:328–331. doi: 10.1038/381328a0. [DOI] [PubMed] [Google Scholar]