Abstract
Activating B-RAF(V600E) (also known as BRAF) kinase mutations occur in ~7% of human malignancies and ~60% of melanomas1. Early clinical experience with a novel class I RAF-selective inhibitor, PLX4032, demonstrated an unprecedented 80% anti-tumour response rate among patients with B-RAF(V600E)-positive melanomas, but acquired drug resistance frequently develops after initial responses2. Hypotheses for mechanisms of acquired resistance to B-RAF inhibition include secondary mutations in B-RAF(V600E), MAPK reactivation, and activation of alternative survival pathways3–5. Here we show that acquired resistance to PLX4032 develops by mutually exclusive PDGFRβ (also known as PDGFRB) upregulation or N-RAS (also known as NRAS) mutations but not through secondary mutations in B-RAF(V600E). We used PLX4032-resistant sub-lines artificially derived from B-RAF(V600E)-positive melanoma cell lines and validated key findings in PLX4032-resistant tumours and tumour-matched, short-term cultures from clinical trial patients. Induction of PDGFRβ RNA, protein and tyrosine phosphorylation emerged as a dominant feature of acquired PLX4032 resistance in a subset of melanoma sub-lines, patient-derived biopsies and short-term cultures. PDGFRβ-upregulated tumour cells have low activated RAS levels and, when treated with PLX4032, do not reactivate the MAPK pathway significantly. In another subset, high levels of activated N-RAS resulting from mutations lead to significant MAPK pathway reactivation upon PLX4032 treatment. Knockdown of PDGFRβ or N-RAS reduced growth of the respective PLX4032-resistantsubsets. Overexpression of PDGFRβ or N-RAS(Q61K) conferred PLX4032 resistance to PLX4032-sensitive parental cell lines. Importantly, MAPK reactivation predicts MEK inhibitor sensitivity. Thus, melanomas escape B-RAF(V600E) targeting not through secondary B-RAF(V600E) mutations but via receptor tyrosine kinase (RTK)-mediated activation of alternative survival pathway(s) or activated RAS-mediated reactivation of the MAPK pathway, suggesting additional therapeutic strategies.
We selected three B-RAF(V600E)-positive parental (P) cell lines, M229, M238 and M249, exquisitely sensitive to PLX4032-mediated growth inhibition in vitro and in vivo6, and derived PLX4032-resistant (R) sub-lines by chronic PLX4032 exposure. In cell survival assays, M229 R, M238 R and M249 R sub-lines displayed strong resistance to PLX4032 (GI50, the concentration of drug that inhibits growth of cells by 50%, not reached up to 10 μM) and paradoxically enhanced growth at low PLX4032 concentrations, in contrast to parental cells (Supplementary Fig. 1a). Morphologically, both M229 R and M238 R sub-lines appear flatter and more fibroblast-like compared to their parental counterparts, but this morphologic switch was not seen in the M249 P versus M249 R4 pair (Supplementary Fig. 2a).
There were no secondary mutations in the drug target B-RAF (V600E) observed on bi-directional Sanger sequencing of all 18 B-RAF exons in 15 M229 R (R1–R15), two M238 R (R1 and R2), and one M249 R (R4) acquired resistant sub-lines (Supplementary Table 1 and Supplementary Fig. 3a, left column). Based on Sanger sequencing, this lack of secondary B-RAF (V600E) mutation along with retention of the original B-RAF(V600E) mutation was confirmed in 16/16 melanoma tumour biopsies (from 12 patients) with clinically acquired resistance to PLX4032 (that is, initial >30% tumour size decrease or partial response, as defined by RECIST (response evaluation criteria in solid tumours) and subsequent progression on PLX4032 dosing; see examples in Supplementary Fig. 4) and 5/5 short-term melanoma cultures established from 5 resistant tumours obtained from 4 patients (Supplementary Table 2). Given recent reports of B-RAF-selective inhibitors having a growth-promoting effect on B-RAF wild-type tumour cells7–9, retention of the original B-RAF alleles in PLX4032-resistant sub-lines, tissues and cultures indicates that PLX4032 chronic treatment did not select for the outgrowth of a pre-existing, minor B-RAF wild-type sub-population. Furthermore, immunoprecipitated B-RAF kinase activities from resistant sub-lines and short-term cultures were similarly sensitive to PLX4032 as B-RAF kinase activities immunoprecipitated from parental cell lines (Supplementary Fig. 3b; Pt48 R and Pt55 R resistance to PLX4032 (ref. 10) and the pre-clinical analogue PLX4720 (ref. 11) shown in Supplementary Fig. 5a and b, respectively; Pt, patient). These results demonstrate that, in all tested acquired resistant cell lines and cultures, the mutated B-RAF(V600E) kinase lacks secondary mutations and hence retains its ability to respond to PLX4032.
Given that minority PLX4032-resistant sub-populations in tissues may acquire B-RAF(V600E) secondary mutations not detectable by Sanger sequencing, we analysed “ultra-deep” (Supplementary Fig. 6) and deep (Supplementary Fig. 7) sequences of B-RAF (exons 2–18) using the Illumina platform for 9/11 acquired resistant tumour samples without tumour-matched short-term cultures (one sample, Pt111-010 DP2, intentionally analysed by both methods; DP, disease progression). Ultradeep B-RAF sequencing of five PLX4032-resistant melanoma tissues resulted in every base of exons 2–18 being sequenced at a median coverage of 127× (27×–128×) (Supplementary Fig. 6a and b). The known variant, V600E, was detected in all five samples with significantly high non-reference allele frequencies (NAF) (Supplementary Fig. 6c). In all five tissues, exon 13, where the T529 gatekeeper residue12 is located, was independently amplified and uniquely bar-coded twice. Rare variants (none at the T529 codon; Supplementary Fig. 6d) detected in these independent exon 13 analyses do not overlap and helped define the true, signal NAF at >4.81% (Supplementary Methods). Furthermore, deep B-RAF (exons 2–18) sequence analysis of PLX4032-resistant melanoma tissues from a whole exome sequencing project resulted in 2,396 base pairs of B-RAF coding regions having coverage ≥10× (average coverage per exon in each tissue shown in Supplementary Fig. 7a). After filtering, no position harboured a variant with a NAF >4.81%, except for the known V600E mutation in all five resistant samples. Together, these data strongly corroborate the lack of B-RAF(V600E) secondary mutations during the evolution of PLX4032 acquired resistance in the majority of patients and their tumours.
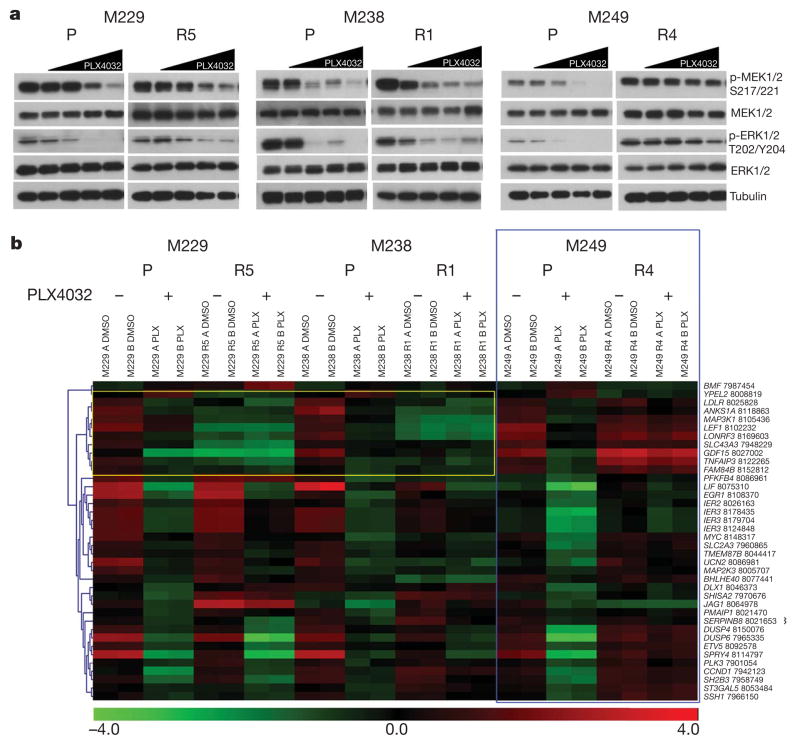
To begin to understand PLX4032-resistance in vitro, we used phospho-specific antibodies to probe the activation status of the RAF downstream effectors, MEK1/2 and ERK1/2 (also know as MAP2K1/2 and MAPK3/1, respectively), in parental versus resistant sub-lines, with and without PLX4032 (Fig. 1a). As expected, PLX4032 induced dose-dependent decreases in p-MEK1/2 and p-ERK1/2 in all parental cells. However, the pattern of MEK-ERK sensitivity to PLX4032 varied among resistant sub-lines, suggesting distinct mechanisms. In contrast to M249 R4, which showed strong resistance to PLX4032-induced MEK/ERK inhibition (suggesting MAPK reactivation), M229 R5 and M238 R1 were both similarly sensitive to PLX4032-induced decreases in the levels of p-MEK1/2 and p-ERK1/2. Gene expression profiling (Fig. 1b) further supported distinct PLX4032 acquired resistant mechanisms represented by M229 R5/M238 R1 versus M249 R4. We first used the gene expression alterations responsive to PLX4032 in parental cells to define a B-RAF(V600E)-responsive gene signature, which is similar to gene sets defined by a MEK1 inhibitor (PD325901)13 and by PLX4720 (ref. 14; Supplementary Fig. 8). Concordant with the western blot results (Fig. 1a), M249 R4 demonstrated striking resistance to PLX4032 treatment with a gene signature of persistent MEK-ERK activation, whereas both M229 R5 and M238 R1 retained a PLX4032-sensitive gene signature (Fig. 1b). These data confirm that M229 R5 and M238 R1 share key characteristics of resistance, which are in line with unsupervised clustering of these two resistant sub-lines in genome-wide, differential expression patterns (Supplementary Fig. 9).
Figure 1. In vitro models of PLX4032 acquired resistance display differential MAPK reactivation.
a, Parental and PLX4032-resistant sub-lines were treated with increasing PLX4032 concentration (0, 0.01, 0.1, 1 and 10 μM), and the effects on MAPK signalling were determined by immunoblotting for p-MEK1/2 and p-ERK1/2 levels. Total MEK1/2, ERK1/2 and tubulin levels, loading controls. b, Heat map for B-RAF(V600E) signature genes in each of the cell lines treated with DMSO or PLX4032. Colour scale, log2-transformed expression (red, high; green, low) for each gene (row) normalized by the mean of all samples. Blue box showing M249 R4 MAPK reactivation. Yellow box showing diminished, baseline expression of B-RAF(V600E) signature genes in M229 and M238 resistant sub-lines (FDR <0.05). The probeset number is shown after each gene.
Gene set enrichment analysis demonstrated an enrichment of RTK-controlled signalling in M229 R5 and M238 R1 but exclusive of M249 R4 (Supplementary Table 3). Unsupervised clustering of the receptor tyrosine kinome gene expression profiles showed that M229 R5 and M238 R1 clustered away from M229 and M238 parental cell lines largely based on higher expression levels of KIT, MET, EGFR and PDGFRβ (Supplementary Fig. 10a, yellow highlight). RNA upregulation of these four RTKs was consistently not associated with genomic DNA (gDNA) copy number gain (Supplementary Fig. 10b). Of these four candidate RTKs, EGFR and PDGFRβ protein levels were overexpressed (Fig. 2a, left; Fig. 3b; Supplementary Fig. 10c), but only PDGFRβ displayed elevated activation-associated tyrosine phosphorylation in a phospho-RTK array (Fig. 2a, right). PDGFRβ RNA upregulation was a common feature among additional M229 R and M238 R sub-lines (Supplementary Fig. 11a) but could not be observed in any of ten randomly selected parental melanoma cell lines (Supplementary Fig. 11b). Interestingly, tyrosine phosphorylation of PDGFRβ correlated with an upregulation of a gene signature unique to PDGFRβ (ref. 15; Supplementary Fig. 12) but is not due to mutational activation, as PDGFRβ cDNAs derived from M229 R5, M238 R1 and Pt48 R are wild type (Supplementary Table 1).
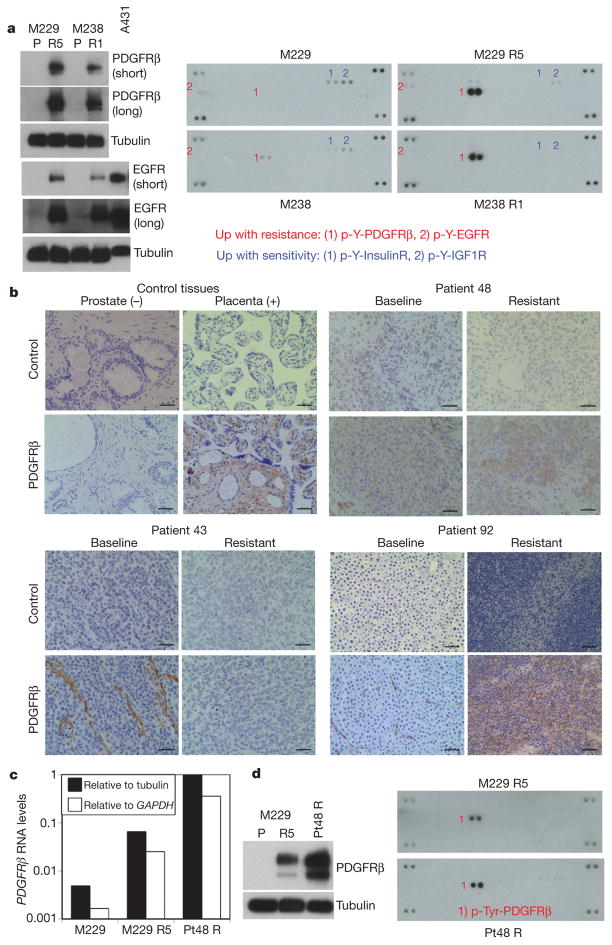
Figure 2. PDGFRβ upregulation is strongly correlated with PLX4032 acquired resistance.
a, Left, total levels of PDGFRβ and EGFR. A431, an EGFR-amplified cell line. Tubulin levels, loading control. Right, whole-cell extracts were incubated on the RTK antibody arrays, and phosphorylation status was determined by subsequent incubation with anti-phosphotyrosine horseradish peroxidase (each RTK spotted in duplicate, positive controls in corners, gene identity below). b, Anti-PDGFRβ immunohistochemistry of formalin-fixed, paraffin-embedded tissues. Prostate, negative control; placenta, positive control. Black bar, 50 μm. c, Relative RNA levels of PDGFRβ in M229 P/R5 and Pt48 R as determined by real-time, quantitative PCR (average of duplicates). d, Total PDGFRβ (left) and p-RTK (right) levels in Pt48 R versus M229 R5.
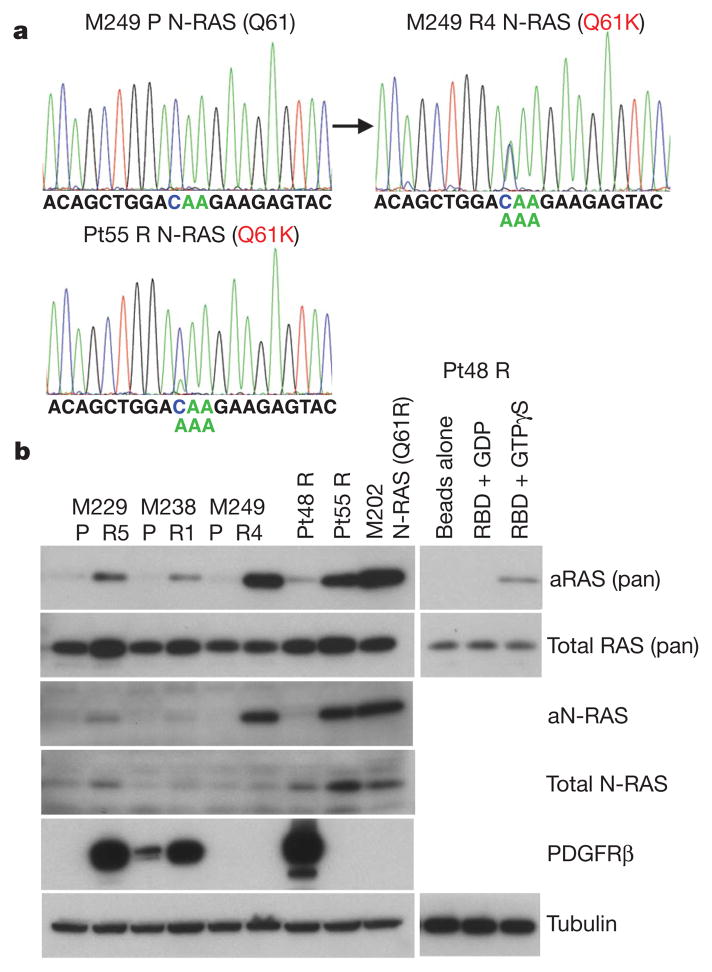
Figure 3. N-RAS upregulation correlates with a distinct subset of PLX4032 acquired resistance.
a, Detection of a N-RAS(Q61K) allele in M249 R4 and Pt55 R. b, The levels of activated RAS (aRAS) and N-RAS (aN-RAS) eluted after pull-down using the RAS-binding domain (RBD) of RAF-1. The total levels of RAS, N-RAS, PDGFRβ and tubulin (loading control) from the whole-cell lysates are shown by immunoblotting. Effects of GDP and GTPγS pre-incubation on RBD pull-down and beads without RBD pull-down from Pt48 R lysates are shown as controls.
We then validated our in vitro finding in vivo by studying clinical trial patient-derived samples (Supplementary Table 2; Fig. 2b) and tumour-matched short-term cultures (Fig. 2c and d). In 4/11 available, paired biopsy specimens, the resistant tumours showed a tumour-associated overexpression of PDGFRβ compared to the baseline tumour in the same patients (Fig. 2b; Supplementary Table 2 and Supplementary Fig. 13). PDGFRβ-positive areas of tissue sections were consistently strongly positive for S100 or MART1 (melanoma markers; MART1 is also known as MLANA) but lacked CD31 (an endothelial, platelet, macrophage marker, also known as PECAM1) staining (data not shown). We were able to validate this finding further in an available short-term culture (Pt48 R) derived from a PLX4032-resistant, PDGFRβ-positive tumour. Pt48 R was established from an intracardiac mass progressing 6 months after initiating treatment with PLX4032. The Pt48 R short-term culture demonstrated clear overexpression of PDGFRβ RNA (Fig. 2c), protein and p-Tyr levels (Fig. 2d).
In M249 R4, we sequenced all exons of N-RAS, K-RAS (also known as KRAS) or H-RAS (also known as HRAS) (to include codons 12, 13, and 61 as well as mutational hotspots of emerging significance16) and MEK1 (ref. 17; Supplementary Table 1 and data not shown) because we proposed a resistance mechanism reactivating MAPK despite not having a secondary B-RAF mutation. Interestingly, M249 R4 harbours a N-RAS(Q61K) activating mutation not present in the parental M249 cell line (Fig. 3a). We found N-RAS mutations in 2/16 acquired resistant biopsy samples (note that both came from Pt55; Supplementary Table 2 and Supplementary Fig. 14). A N-RAS(Q61K) mutated sample, Pt55 DP1 (for disease progression 1) was obtained from a biopsy taken from an isolated, nodal metastasis that partially regressed on PLX4032 but increased in size 10 months after starting on therapy with PLX4032 (Supplementary Fig. 4a). This patient continued on therapy with PLX4032 until 6 months later, when several other nodal metastases developed (Supplementary Fig. 14a, b). Analysis of a biopsy taken at a second progression site (Pt55 DP2) demonstrated a different mutation in N-RAS, N-RAS(Q61R) (Supplementary Fig. 14b). Both Pt55 DP1 and DP2 tissue N-RAS mutations were confirmed in their respective short-term cultures, Pt55 R and Pt55 R2 (Fig. 3a and Supplementary Fig. 14b). Also, both DP1 and DP2 (and their respective cultures) harboured increased N-RAS gDNA copy numbers (Supplementary Fig. 14c and d). Both Pt55 R and Pt55 R2 also showed increased N-RAS RNA (Supplementary Fig. 14e) and protein levels (Fig. 3b). In addition, N-RAS(Q61K) mutation in M249 R4 and Pt55 R correlated with a marked increase in activated N-RAS levels (Fig. 3b). Of note, the N-RAS mutations were mutually exclusive with PDGFRβ overexpression in all samples (Supplementary Table 2).
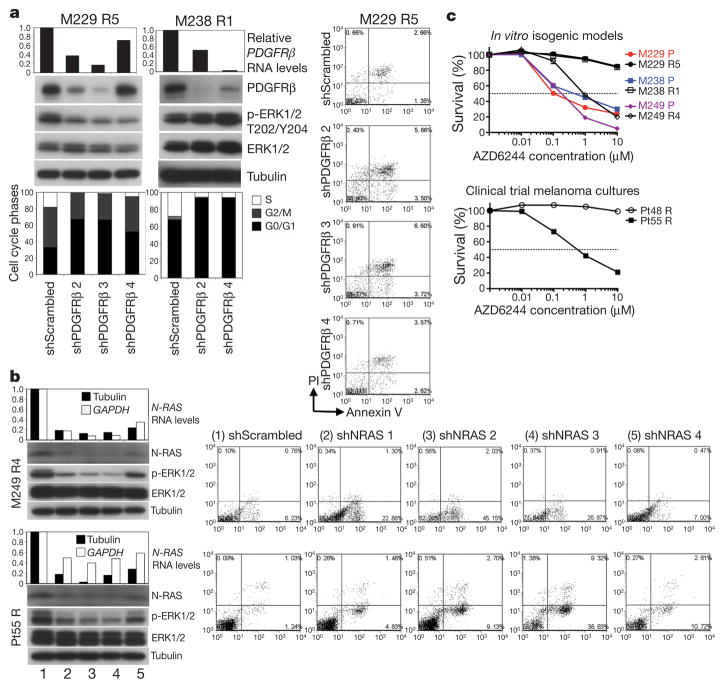
Knockdown of PDGFRβ or N-RAS using small interfering RNA (siRNA) pools preferentially growth-inhibited melanoma cells with upregulated PDGFRβ or N-RAS, respectively (Supplementary Fig. 15a, b and Supplementary Table 4). We then selected two resistant sub-lines or cultures to test the effects of individual PDGFRβ and N-RAS short hairpin RNAs (shRNAs; Fig. 4a and b, respectively). Stable knockdown of PDGFRβ caused an admixture of G0/G1 cell cycle arrest (in a MEK inhibitor-dependent manner due to compensatory signalling; Supplementary Fig. 16a and data not shown) and apoptosis in M229 R5 and a G0/G1 cell cycle arrest in M238 R1. This effect was specific, as stable PDGFRβ knockdown in M249 R4 and Pt55 R did not result in G0/G1 cell cycle arrest (Supplementary Fig. 17a). In contrast, stable N-RAS knockdown resulted in a predominantly apoptotic response in M249 R4 and Pt55 R (Fig. 4b) but not in M229 R5, M238 R1 or Pt48 R (Supplementary Fig. 17b). Moreover, stable N-RAS knock down markedly conferred PLX4032 sensitivity to M249 R4 and Pt55 R but had no effect on M229 R5 PLX4032 resistance (Supplementary Fig. 18a). Flag–N-RAS(Q61K) stable overexpression conferred PLX4032 resistance in the M249 parental cell line (Supplementary Fig. 18b), whereas stable PDGFRβ–MYC overexpression conferred reduced PLX4032 sensitivity in both M229 and M238 parental cell lines (Supplementary Fig. 19).
Figure 4. PDGFRβ- and N-RAS-mediated growth and survival pathways differentially predict MEK inhibitor sensitivity.
a, Transduction of PDGFRβ shRNAs in M229 R5 and M238 R1 (1 μM PLX4032), RNA (relative to GAPDH) and protein knockdown, effects on p-ERK levels, cell cycle distribution, and apoptosis (when applicable). M229 R5 was also treated with 0.5 μM AZD6244. PI, propidium iodide.
b, Transduction of N-RAS shRNAs in M249 R4 and Pt55 R (1 μM PLX4032), RNA and protein knockdown, effects on p-ERK levels and apoptosis.
c, Survival curves for isogenic cell line pairs and melanoma cultures treated with the indicated AZD6244 concentration for 72 h (relative to DMSO-treated controls; mean ± s.e.m., n = 5). PLX4032-resistant cells were grown with PLX4032. Dashed line, 50% cell killing.
We then asked whether N-RAS-dependent growth and reactivation of the MAPK pathway (Fig. 1a and Supplementary Fig. 20) would selectively sensitize M249 R4 and Pt55 R to MEK inhibition. Indeed, whereas the growth of M229 R5, M238 R1 and Pt48 R was uniformly highly resistant to the MEK inhibitor AZD6244 (and U0126, Supplementary Fig. 21), the growth of M249 R4 and Pt55 R was sensitive to MEK inhibition in the presence of PLX4032 (Fig. 4c) or absence of PLX4032 (Supplementary Fig. 22). It is known that activated N-RAS in melanoma cells uses C-RAF (also known as RAF1) over B-RAF to signal to MEK-ERK18. Thus, N-RAS activation would be capable of bypassing PLX4032-inhibited B-RAF, reactivating the MAPK pathway. It is worth noting that PDGFRβ-upregulated, PLX4032-resistant melanoma sub-lines (M229 R5 and M238 R1) and culture (Pt48 R) are resistant not only to AZD6244 but also to imatinib, which is at least partially due to rebound, compensatory survival signalling (Supplementary Fig. 23 and unpublished observations, H.S. and R.S.L.).
We propose (Supplementary Fig. 1b) that B-RAF(V600E)-positive melanomas, instead of accumulating B-RAF(V600E) secondary mutations, can acquire PLX4032 resistance by (1) activating an RTK (PDGFRβ)-dependent survival pathway in addition to MAPK, or (2) reactivating the MAPK pathway via N-RAS upregulation. These two mechanisms account for acquired PLX4032 resistance in 5/12 patients in our study cohort, and additional mechanisms await future discovery. Some patients who relapse on PLX4032 are already being enrolled in a phase II MEK inhibitor trial (ClinicalTrials.gov identifier NCT01037127) based on the assumption of MAPK reactivation. Our findings directly imply a strategy to stratify patients who relapse on PLX4032 and should prompt a search for rational combinations of targeting agents most optimal for distinct mechanisms of acquired resistance to PLX4032 as well as other B-RAF inhibitors (for example, GSK2118436) in clinical development.
METHODS SUMMARY
Cell culture, infections and compounds
Cells were maintained in Dulbecco’s modified Eagle medium (DMEM) with 10 or 20% fetal bovine serum and glutamine. shRNAs were sub-cloned into the lentiviral vector pLL3.7 and infections carried out with protamine sulphate. Stocks of PLX4032 (Plexxikon) and AZD6244 (commercially available) were made in DMSO. Cells were quantified using CellTiter-GLO Luminescence (Promega).
Protein detection
Western blots were probed with antibodies against p-MEK1/2 (S217/221), MEK1/2, p-ERK1/2 (T202/Y204), ERK1/2, PDGFRβ, and EGFR (Cell Signaling Technologies), and N-RAS (Santa Cruz Biotechnology), pan-RAS (Thermo Scientific) and tubulin (Sigma). p-RTK arrays were performed according to the manufacturer’s recommendations (Human Phospho-RTK Array Kit, R&D Systems). For PDGFRβ immunohistochemistry, paraffin-embedded formalin-fixed tissue sections were antigen-retrieved, incubated with a PDGFRβ antibody followed by horseradish peroxidase-conjugated secondary antibody (Envision System, DakoCytomation). Immunocomplexes were visualized using the DAB (3,3′-diaminobenzidine) peroxidase method and nuclei haematoxylin-counterstained. For activated RAS pull-down, lysates were incubated with beads coupled to glutathione-S-transferase (GST)–RAF-1–RAS-binding domain of RAF1 (RBD) (Thermo) for 1 h at 4 °C.
RNA quantifications
For real-time quantitative PCR, total RNA was extracted and cDNA quantified. Data were normalized to tubulin and GAPDH levels. Relative expression is calculated using the delta-Ct method. For RNA expression profiling, total RNAs were extracted, and generated cDNAs were fragmented, labelled and hybridized to the GeneChip Human Gene 1.0 ST Arrays (Affymetrix). Expression data were normalized, background-corrected, and log2-transformed for parametric analysis. Differentially expressed genes were identified using significance analysis of microarrays (SAM) with the R package ‘samr’ (false discovery rate (FDR) <0.05; fold change >2).
Cell cycle and apoptosis
For cell cycle analysis, cells were fixed, permeabilized and stained with propidium iodide (BD Pharmingen). Cell cycle distribution was analysed by Cell Quest Pro and ModiFit software. For apoptosis, cells were co-stained with Annexin V-V450 and propidium iodide (BD Pharmingen). Data were analysed with the FACS Express V2 software.
Supplementary Material
Acknowledgments
We are grateful to G. Bollag and P. Lin (Plexxikon) for providing PLX4032, J. S. Economou for biopsies, B. Comin-Anduix for FACS assistance, S. Mok for assistance with virus production, C. Ng for tissue acquisition and culture establishment, R. Huang for patient tissue processing, N. Doan for immunohistochemistry, P. Mischel for discussion, B. Chmielowski for coordinated patient care, T.L. Toy for technical help with library generation for deep sequencing, and B. Harry for help with analysis of deep sequence data. R.S.L. acknowledges funding from the following: Dermatology Foundation, Burroughs Wellcome Fund, STOP CANCER Foundation, Margaret E. Early Medical Trust, Ian Copeland Memorial Melanoma Fund, V Foundation for Cancer Research, Melanoma Research Foundation, American Skin Association, Caltech-UCLA Joint Center for Translational Medicine, Wesley Coyle Memorial Fund, and Melanoma Research Alliance. R.N. is supported by a post-doctoral fellowship from the T32 Tumor Immunology Training Grant (S. Dubinett). A.R. is supported by the California Institute for Regenerative Medicine (CIRM), the Jonsson Cancer Center Foundation (JCCF), and Caltech-UCLA Joint Center for Translational Medicine. Array and sequence work were performed within the Jonsson Comprehensive Cancer Center Gene Expression Shared Resource. Patient-informed consent was obtained for the research performed in this study. We would like to thank all the patients that participated in this study.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature. . A figure summarizing the main result of this paper is also included as SI.
Author Contributions R.N., H.S., Q.W., X.K., H.L., Z.C. designed and performed experiments and analysed data. M.L. helped analyse data. R.C.K., N.A., H.S., T.C., G.M., J.A.S., and A.R. provided reagents. S.F.N. helped design experiments and interpreted data. A.R. and R.S.L. designed research aims. H.L., S.F.N., G.M., J.A.S. and A.R. helped write the paper. R.S.L. designed and performed experiments, analysed data, provided reagents, and wrote the paper.
Author Information Gene expression and copy number data are deposited at Gene Expression Omnibus under accession numbers GSE24862 and GSE24890, respectively. Reprints and permissions information is available at www.nature.com/reprints. The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Flaherty KT, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jänne PA, Gray N, Settleman J. Factors underlying sensitivity of cancers to small-molecule kinase inhibitors. Nature Rev Drug Discov. 2009;8:709–723. doi: 10.1038/nrd2871. [DOI] [PubMed] [Google Scholar]
- 4.Montagut C, et al. Elevated CRAF as a potential mechanism of acquired resistance to BRAF inhibition in melanoma. Cancer Res. 2008;68:4853–4861. doi: 10.1158/0008-5472.CAN-07-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Søndergaard JN, et al. Differential sensitivity of melanoma cell lines with BRAFV600E mutation to the specific Raf inhibitor PLX4032. J Transl Med. 2010;8:39–50. doi: 10.1186/1479-5876-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halaban R, et al. PLX4032, a selective BRAFV600E kinase inhibitor, activates the ERK pathway and enhances cell migration and proliferation of BRAFWT melanoma cells. Pigment Cell Melanoma Res. 2010;23:190–200. doi: 10.1111/j.1755-148X.2010.00685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatzivassiliou G, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 9.Heidorn SJ, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bollag G, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai J, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci USA. 2008;105:3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whittaker S, et al. Gatekeeper mutations mediate resistance to BRAF-targeted therapies. Sci Transl Med. 2010;2:35ra41. doi: 10.1126/scitranslmed.3000758. [DOI] [PubMed] [Google Scholar]
- 13.Pratilas CA, et al. V600EBRAF is associated with disabled feedback inhibition of RAF–MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci USA. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Packer LM, East P, Reis-Filho JS, Marais R. Identification of direct transcriptional targets of BRAF/MEK signalling in melanoma. Pigment Cell Melanoma Res. 2009;22:785–798. doi: 10.1111/j.1755-148X.2009.00618.x. [DOI] [PubMed] [Google Scholar]
- 15.Wu E, et al. Comprehensive dissection of PDGF-PDGFR signaling pathways in PDGFR genetically defined cells. PLoS ONE. 2008;3:e3794. doi: 10.1371/journal.pone.0003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith G, et al. Activating K-Ras mutations without ‘hotspot’ codons in sporadic colorectal tumours - implications for personalised cancer medicine. Br J Cancer. 2010;102:693–703. doi: 10.1038/sj.bjc.6605534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emery CM, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci USA. 2009;106:20411–20416. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dumaz N, et al. In melanoma, RAS mutations are accompanied by switching signaling from BRAF to CRAF and disrupted cyclic AMP signaling. Cancer Res. 2006;66:9483–9491. doi: 10.1158/0008-5472.CAN-05-4227. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.