Abstract
We demonstrated previously that CD45RA+ CD4+ T cells are infected primarily by syncytium-inducing (SI) HIV-1 variants, whereas CD45RO+ CD4+ T cells harbor both non-SI (NSI) and SI HIV-1 variants. Here, we studied evolution of tropism for CD45RA+ and CD45RO+ CD4+ cells, coreceptor usage, and molecular phylogeny of coexisting NSI and SI HIV-1 clones that were isolated from four patients in the period spanning SI conversion. NSI variants were CCR5-restricted and could be isolated throughout infection from CD45RO+ CD4+ cells. SI variants seemed to evolve in CD45RO+ CD4+ cells, but, in time, SI HIV-1 infection of CD45RA+ CD4+ cells equaled infection of CD45RO+ CD4+ cells. In parallel with this shift, SI HIV-1 variants first used both coreceptors CCR5 and CXCR4, but eventually lost the ability to use CCR5. Phylogenetically, NSI and SI HIV-1 populations diverged over time. We observed a differential expression of HIV-1 coreceptors within CD45RA+ and CD45RO+ cells, which allowed us to isolate virus from purified CCR5+ CXCR4– and CCR5– CXCR4+ CD4+ cells. The CCR5+ subset was exclusively infected by CCR5-dependent HIV-1 clones, whereas SI clones were preferentially isolated from the CXCR4+ subset. The differential expression of HIV-1 coreceptors provides distinct cellular niches for NSI and SI HIV-1, contributing to their coexistence and independent evolutionary pathways.
Introduction
In HIV type 1 (HIV-1) infection, an estimated number of viral particles, between 108 and 1010, are produced and eliminated from the human body per day (1–3). The error-prone viral reverse-transcriptase enzyme and lack of proofreading mechanisms during reverse transcription of the viral RNA results in the frequent incorporation of mutations in the viral genome. The large turnover of virus in combination with the high mutation rate of HIV-1 results in a mixed population of related but distinct virus variants, also termed viral quasispecies (4). Viral quasispecies are defined by a dominant sequence and a spectrum of mutants, which may be relatively stable for many generations (population equilibrium). Variants within the viral quasispecies are continuously competing, and the dominant sequence reflects the most fit variant at that time point. Due to the introduction of beneficial mutations in the viral genome or changing environmental factors, such as the introduction of antiretroviral agents, a previously minor population may become the dominant sequence and thus an altered population equilibrium is established.
Altered biological characteristics such as replication rate, cytopathicity, and cellular tropism may influence viral fitness and therefore HIV-1 quasispecies behavior. Indeed, viral biological characteristics continue to evolve during the course of HIV-1 infection. Early in infection a homogeneous population of generally macrophage-tropic, non–syncytium-inducing (NSI) virus variants is present (5, 6). However, during the course of infection, virus variants become predominantly T-cell tropic (7), and in 50% of HIV-1–infected patients syncytium-inducing (SI) HIV-1 variants appear (8). SI HIV-1 variants can be discriminated from NSI variants by the ability to replicate and induce syncytia in T-cell lines (9). The emergence of SI HIV-1 variants is associated with a subsequent, more-rapid decline of CD4+ T cells, an accelerated disease progression, and a poor prognosis for survival (8).
CC chemokine-receptor 5 (CCR5) has been identified as the coreceptor for entry of NSI HIV-1 into CD4+ T cells. SI variants may, in addition, or alternatively, use CXC chemokine-receptor 4 (CXCR4) as their main coreceptor (10–15). Other chemokine receptors (such as CCR2, CCR3, CCR8, CCR9, CX3CR1) or structurally related molecules (the orphan receptors BOB/gpr15, BONZO/strl33, apj, Chem23) have been identified as coreceptors for entry of HIV-1, as well (14–23). Still, the majority of primary NSI HIV-1 isolates were shown to be restricted to CCR5 usage exclusively (24–26). Expanded coreceptor usage, including usage of CXCR4 and possibly additional coreceptors, was associated with more rapid disease progression (27–29), but is not a prerequisite for a progressive disease course (26).
The biological significance of coreceptors other than CCR5 and CXCR4 remains to be established. These molecules seem to mediate entry of only a minority of HIV-1 variants in vitro, and usage of these coreceptors was always in addition to usage of either CCR5 or CXCR4. In an ex vivo lymphoid tissue model, in vitro specificity for CCR3, CCR8, BOB, and BONZO, in addition to CXCR4 usage, did not correlate with additional cytopathicity or reduced sensitivity to CXCR4 antagonists (30), suggestive of a limited role of minor coreceptors other than CXCR4 for SI HIV-1.
SI phenotype is determined mainly by the second and third variable loop (V2 and V3) of gp120 (31, 32). Specifically, the presence of a positively charged amino acid at either one or both of two fixed positions of the V3 loop (positions 11 and 25) was highly associated with SI phenotype of primary isolates (33, 34), which suggests that these amino acids play a crucial role in the interaction of gp120 with CXCR4. Indeed, it has been shown the V1/V2 and V3 regions are involved in the interaction with CCR5, CCR3, and CXCR4 (35–40). This fits insights from structural studies in which V3 and a conserved coreceptor-binding site are involved in coreceptor binding. In a trimeric model of the gp41-gp120 complex, V2 juxtaposes V3 and may thus interfere with coreceptor binding (41–44).
Though only a limited number of amino acid substitutions in V3 can confer SI phenotype to NSI isolates in vitro (45), SI conversion occurs in only 50% of HIV-1–infected individuals in the course of HIV-1 infection (9). This is puzzling in light of the apparent beneficial effect of SI conversion for the viral population, since, theoretically, the potential target-cell population is greatly enlarged through acquisition of the ability to enter cells via CXCR4. The finding that SI conversion seems to occur only once during HIV-1 infection (46), the rarity of virus variants with intermediate genotypes, and the nongradual character of the evolution from NSI to SI HIV-1 (47) might suggest that the virus evolves to the SI phenotype through less-fit intermediate stages. The relatively low incidence of the SI phenotype, despite the high mutation frequency of HIV-1, might then be explained by an incompetence of these intermediate variants to compete with the well-established NSI virus population.
Once established, SI variants appear to be more replication competent than NSI variants, given the broader target-cell range and the generally higher replication kinetics of SI HIV-1 in vitro (46, 48, 49). In this light it seems paradoxical that NSI variants persist and may even expand in vivo after the emergence of SI HIV-1 (50). A possible explanation of coexistence of NSI and SI HIV-1 would be that both phenotypic variants infect distinct target cells within an infected patient. Indeed, recently we showed that NSI variants are isolated mainly from memory CD45RO+ CD4+ T cells, whereas SI variants are isolated from both CD45RA+ and CD45RO+ CD4+ T cells (51).
To explore the concept that distinct T-cell subsets provide niches for NSI and SI HIV-1 variants, we longitudinally studied in vivo tropism for CD45RA+ and CD45RO+ CD4+ T cells during NSI to SI evolution. Changes in in vivo tropism of biological virus clones were related to in vitro coreceptor usage, phenotype-associated mutations in gp120 V3, and molecular phylogeny. Based on differential expression of NSI and SI coreceptors on CD45RA+ and CD45RO+ CD4+ T cells, three major potential HIV-1 target-cell populations can be distinguished: CD45RA+ CXCR4+ CCR5–; CD45RO+ CXCR4+ CCR5–; and CD45RO+ CXCR4– CCR5+ CD4+ T cells. From these purified subsets we isolated biological virus clones and determined their NSI or SI phenotype. We showed that the differentially expressed coreceptors indeed provide the cellular compartments that allow independent evolutionary pathways and coexistence of NSI and SI HIV-1. Finally, we propose a model of NSI to SI HIV-1 evolution and discuss the biological relevance of niche differentiation for HIV-1 quasispecies behavior.
Methods
Patients.
Four patients, ACH039, ACH171, ACH208, ACH490, all of whom developed SI HIV-1 variants during a progressive disease course, were selected (Figure 1). All patients were male homosexual participants of the Amsterdam Cohort studies on HIV-1 and AIDS. In this cohort, the presence of SI HIV-1 variants in peripheral blood is prospectively determined at every visit (in general every 3 months) by cocultivation of 1.106 patient PBMCs with 1.106 MT2 cells, as described previously (9). The moment of SI conversion was calculated as the midpoint between the last MT2-negative and the first MT2-positive visit. It is important to note that SI conversion is, by definition, the moment of first detection in the MT2 assay performed on total patient PBMCs, which is not necessarily the moment of first appearance of SI variants in vivo, due to detection limits of the assay.
Figure 1.
The clinical course of HIV-1 infection by CD4+ T-cell number (filled circles) and serum RNA load (open triangles). Filled triangles at the x axis indicate time points selected for isolation and analysis of virus. Open triangles at the x axis indicate time points of virus isolation after cell sort based on HIV-1–coreceptor expression. SI conversion occurred 16, 62, 14, and 75 months after seroconversion in patient ACH039, ACH171, ACH208, and ACH490, respectively.
FACS analysis of cell-surface markers and cell sorting.
Cryopreserved patient PBMCs were incubated with mAb’s against CD45 RA (phycoerythrin [PE] conjugated), CD45 RO (FITC), and CD4 (Tricolor; all Ab’s were obtained from Caltag Laboratories, Burlingame, California, USA), and sorted on a FACStar (Becton Dickinson, San Jose, California, USA), as described (51). Cells were separated in CD45RO+ CD45RA– and CD45RO– CD45RA+ CD4+ T-cell populations. CD45RAdull and CD45ROdull CD4+ cells were not sorted or analyzed.
Expression of CCR5 and CXCR4 on the CD45RO+ and CD45RO– CD4+ T-cell subsets was determined by four-color flow cytometry. Cryopreserved PBMCs of patients and eight healthy donors were incubated with mAb’s against CCR5 (clone 2D7, FITC conjugated; PharMingen, San Diego, California, USA), CXCR4 (clone 12G5, PE; PharMingen), CD4 (PerCP; Becton Dickinson), and CD45RO (APC; Becton Dickinson) and analyzed on a FACScalibur flow cytometer (Becton Dickinson). Isotype-matched control Ab’s were included and used to set the quadrant location. Patient PBMCs obtained 19.8, 10.5, 11.4, and 7.3 months after SI conversion (ACH039, ACH171, ACH208, and ACH490) were sorted in CD45RO– CXCR4+ CCR5–; CD45RO+ CXCR4+ CCR5–; and CD45RO+ CXCR4– CCR5+ CD4+ T cells on a MOFLO cell sorter (Cytomation Inc., Ft. Collins, Colorado, USA). After cell sorting, virus was isolated from these cells as described below.
Clonal isolation of virus.
Biological virus clones were obtained by cocultivation of serial dilutions of patient PBMCs, purified CD45RA+ or CD45RO+ CD4+ T cells, or purified CCR5+ CXCR4– and CCR5– CXCR4+ CD4+ T cells with 2–3-day phytohemagglutinin-stimulated PBMCs (PHA-PBMCs) of at least two healthy donors. The number of patient cells per well was either 10,000–40,000 PBMCs, 100–8,000 CD45RA+ or CD45RO+ CD4+ T cells, or 50–10,000 CCR5+ or CXCR4+ CD4+ T cells. The cells were cocultivated with 105 PHA-PBMCs in a final volume of 200 μl rIL-2–supplemented (proleukin; Chiron Benelux BV, Amsterdam, The Netherlands) Iscove’s medium for 28 days in a 96-well flat-bottom plate. For each cell dilution, multiple cocultures (24–48 wells) were performed. Weekly, one third of the culture supernatant was harvested for analysis of p24 production by an in-house ELISA. Cells were resuspended, and half of the cells were transferred to fresh PHA-PBMCs from healthy donors. To determine SI phenotype of the viral clones, 25,000 MT2 cells per well were added to the remainder of the culture, and these were scored by visual inspection for the presence of syncytia from day 3 to day 7. From positive wells in the PBMC cultures, viral stocks were grown, cell-free supernatants were stored, and infected PBMCs were used to confirm SI phenotype by cocultivation with 1.106 MT2 cells in a final volume of 3 ml for 10 days. In cases where MT2 infection did not correspond with the U87-CXCR4 infection, the MT2 assay was repeated, and infection was determined by visual inspection of syncytia formation and by p24 ELISA.
The frequency of productively infected cells (F) was calculated assuming a Poisson distribution with the formula F = –ln(F0), in which F0 is the fraction of negative cultures using the jackknife resampling technique (52). The frequency of infected cells is expressed as tissue-culture infectious dose (TCID) per 106 cells. If less than one third of the wells were positive within a dilution step, viruses were considered to be clonal, since it is unlikely that more than two infected patient cells were present in a single well. Furthermore, no evidence of mixed viral populations was obtained in sequence analyses of tested viral isolates, which underlines the clonality of the isolates obtained by this clonal isolation method.
Determination of coreceptor usage.
Coreceptor usage was tested using human astroglioma U87 cells, stably expressing CD4 and one of the HIV-1 coreceptors CCR1, CCR2, CCR3, CCR5, CXCR4, BOB/gpr15, or BONZO/strl33. All coreceptor-expressing cell lines were maintained in Iscove’s medium supplemented with 10% FCS, 5 μg/ml polybrene, 100 U/ml penicillin, 100 μg/ml streptomycin, and 1 μg/ml puromycin, and regularly selected for CD4 expression with 200 μg/ml g418. A control U87 cell line expressing CD4 but no HIV-1 coreceptor (U87-CD4) was maintained in the same medium in the absence of puromycin.
CD4 expression on each cell line and CCR1, CCR2, CCR3, CCR5, and CXCR4 expression on the corresponding U87 cell lines was confirmed by flow cytometry or RT-PCR (26). After finishing the experiments, expression of CD4 and coreceptors on U87-CCR3, CCR5, and CXCR4 was reconfirmed by flow cytometry (data not shown). U87-BOB and U87-BONZO were obtained through retroviral transfer of the U87-CD4 cell line. The transfected cells were maintained in selection medium, and expression of BOB and BONZO was confirmed by RT-PCR (data not shown).
Twenty-four hours before virus inoculation, 104 cells per well of each cell line were seeded in 96-well plates. The cells were inoculated in a cell-free manner with a minimum of 102 TCID50 per clone in a total volume of 200 μl. After 24 hours, the cells were washed twice with PBS to remove unbound virus and cultured in 200 μl of medium. At day 7, the cells were transferred to 24-well plates and maintained in 1 ml of fresh medium. Supernatant was collected at 7, 14, 21, and 28 days after infection and analyzed by p24 ELISA. Clones from all patients were tested for the ability to use CCR3, CCR5, and CXCR4. Clones from ACH039 and ACH208 were additionally tested for the ability to use CCR1 and CCR2 and in a random subset (n = 19 and n = 29) for the ability to use BOB and BONZO. In all experiments, the U87-CD4 cell line not expressing an HIV-1 coreceptor was included as a control.
Viral clones were tested for the ability to infect PHA-PBMCs from a healthy donor who is homozygous for a 32-bp deletion in CCR5 (CCR5 Δ32/Δ32) by inoculation of 105 cells per well PHA-PBMCs in a cell-free manner in a 96-well plate and cultured for 21 days. Weekly, half of the cultures were transferred to fresh PHA-PBMCs, and culture supernatant was tested for virus replication using an in-house p24 ELISA.
Sequencing.
Viral gp120 V3 sequences were amplified by PCR from DNA isolated from infected PBMC, as described (46). PCR products were purified and sequenced with the ABI prism BigDye Terminator sequencing kit (Perkin Elmer, Foster City, California, USA) according to the instructions from the manufacturer using primers seq 5 and seq 6 (46, 53) or primer d (54). Sequences were analyzed on an ABI prism 377 DNA sequencer. A number of sequences were available from previous studies (46, 53). For ACH039, accession numbers are: AF022258, AF022262, AF022263, AF022267, AF022271, AF022273, AF022274, AF022277, AF022278, AF022280-AF022283, AF022287-AF022291, AF022293, AF022294, and AF022299-AF022301. For ACH208, accession numbers are: AF021477, AF021494, AF021499, AF021500, AF021502, AF021503, AF021505, AF021510, AF021514, AF021518, AF021523, AF021524, AF021532, AF021533, AF021536, AF021607, AF021608, AF021612-AF021614, AF021616-AF021618, AF021620, AF021622, AF021627-AF021630, AF021639, AF021647, AF021650, AF021651, and AF021668-AF021670. Newly generated sequences are deposited in Genbank under accession numbers AF180907 to AF180916 (ACH039); AF259011 to AF259052 (ACH171); AF180898 to AF18096 (ACH208); and AF258957 to 259010 (ACH490).
Phylogenetic analyses.
Envelope V3 region sequences were aligned using ClustalW (version 1.7) (55) and checked manually. Alignments were displayed and consensus sequences were calculated using PRETTY (GCG package version 8; Genetics Computer Group, Madison, Wisconsin, USA). Phylogenetic analyses were produced using the neighbor joining method (NEIGHBOR) as implemented in the Phylip package (version 3.5c) (56). Endgaps were excluded from analyses, leaving 348 nt (ACH039), 297 nt (ACH171), 160 nt (ACH208), and 339 nt (ACH490) of sequence data for analyses. Distance matrices were produced in DNADIST using the maximum-likelihood model for nucleotide substitutions, which corrects for differences in nucleotide frequencies and different transition and transversion ratios (Ti/Tv ratio). PUZZLE (version 4.0.2) (57) was used to estimate Ti/Tv ratios from the data sets, which were used as input for DNADIST (Ti/Tv ratio of 1:72; 2:54; 1:27, and 2:71 for sequences from ACH039, ACH171, ACH208, and ACH490, respectively). For bootstrapping, SEQBOOT, DNADIST, NEIGHBOR, and CONSENSE (Phylip package) were used. Phylip’s DRAWTREE was used to prepare the plots. A phylogenetic tree of all sequences from all patients was constructed. Within-patient sequences clustered together and apart from sequences from other patients with high bootstrap values (over 98% of 50 bootstrap analyses), indicating that no mix-up had occurred during handling and processing of samples and sequence data (data not shown). Pairwise hamming distances, the number of different nucleotides between each pair of sequences, were calculated using DISTANCES (GCG package).
Results
NSI to SI conversion during the course of HIV-1 infection.
To analyze evolution of cellular tropism and coreceptor usage during transition from NSI to SI HIV-1, four patients who developed SI variants during follow-up were selected from the Amsterdam Cohort Studies on HIV-1 infection and AIDS. The course of infection as reflected by changes in CD4+ T-cell numbers and serum viral RNA load is depicted in Figure 1. In two of the patients SI variants were first detected within 1.5 years after seroconversion (16 and 14 months, ACH039 and ACH208, respectively). In the other patients, SI conversion occurred 5 (ACH171) and 6 (ACH490) years after seroconversion. To isolate SI variants as early as possible after their first appearance, biological clones were isolated from several time points encompassing the estimated moment of SI conversion. In addition, time points were selected well before and after the moment of SI conversion.
Tropism of SI HIV-1 for both CD45RO+ and CD45RA+ CD4+ T cells.
Previously, we demonstrated in a cross-sectional study that NSI HIV-1 variants are mainly isolated from CD45RO+ CD4+ T cells, whereas SI HIV-1 variants can be isolated from both CD45RA+ and CD45RO+ CD4+ T cells. Here, we longitudinally study in vivo tropism for these T-cell subsets in two patients (ACH171 and ACH490) in relation to SI conversion. Clonal virus isolation was performed on total PBMCs and purified CD45RA+ and CD45RO+ CD4+ T cells from six sequential PBMC samples, obtained during a 40-month period spanning the moment of SI conversion.
The frequency of cells infected with either NSI or SI HIV-1 (hereafter, NSI or SI load) in total PBMCs and purified CD45RA+ and CD45RO+ CD4+ T cells is depicted in Figure 2a. Throughout infection, NSI variants were predominantly isolated from CD45RO+ CD4+ T cells, though NSI variants could be isolated from CD45RA+ CD4+ T cells, as well (Figure 2a). NSI load in CD45RO+ T cells is about 100 times and 40 times higher than NSI load in CD45RA+ T cells in ACH171 and ACH490, respectively (Figure 2b). Up to 20 months after SI conversion, SI variants were also isolated predominantly from the CD45RO+ CD4+ T cells. However, at 2 years after SI conversion, SI load was slightly higher in CD45RA+ than in CD45RO+ CD4+ T cells (approximately 1.5 times in both patients, Figure 2b), indicating that especially late-stage SI variants differ from NSI variants with respect to tropism for CD45RA+ and CD45RO+ cells. At this time point, CD45RA+ CD4+ T cells were exclusively infected by SI clones, whereas CD45RO+ cells were infected by both NSI and SI HIV-1 (Figure 2c).
Figure 2.
Tropism for CD45RA+ and CD45RO+ CD4+ T cells during NSI to SI HIV-1 conversion. Patient PBMCs were FACS sorted in CD45RA+ CD45RO– CD4+ and CD45RA– CD45RO+ CD4+ T cells, and clonal virus isolation was performed on these T-cell subsets. (a) The frequency of infected cells with either NSI or SI HIV-1 variants (NSI or SI load) in total (top), CD45RO+ (middle), and CD45RA+ (bottom) CD4+ T cells relative to SI conversion in patient ACH171 (left panels) and 490 (right panels). Filled symbols indicate SI clones, and open symbols indicate NSI clones. In the top panel, serum RNA load is indicated by triangles. (b) Ratio of NSI load in CD45RO+ to NSI load in CD45RA+ CD4+ T cells (left) and ratio of SI load in CD45RA+ to SI load in CD45RO+ CD4+ T cells (right) of patient ACH171 (filled circles) and ACH490 (open circles). (c) Prevalence of SI phenotype among HIV-1 clones isolated from CD45RA+ (circles) or CD45RO+ (squares) CD4+ T-cell subset in ACH171 and ACH490.
Evolution of coreceptor usage of NSI and SI HIV-1 variants.
We analyzed whether the gradual evolution of SI variants from preferential tropism for CD45RO+ CD4+ T cells to preferential tropism for CD45RA+ CD4+ T cells is reflected by evolution of coreceptor usage after SI conversion. In addition to clones obtained from CD45RA+ and CD45RO+ CD4+ T cells from ACH171 and ACH490, we analyzed clones from ACH039 and ACH208, available from a previous study (46). Coreceptor usage was analyzed by the capacity to productively infect U87 cell lines stably expressing CD4 in combination with either CCR3, CCR5, or CXCR4. Clones from ACH039 and ACH208 were additionally tested for the ability to infect U87-CCR1, U87-CCR2, U87-BOB, and U87-BONZO. None of the tested clones were able to infect these cell lines (data not shown). A U87 cell line expressing only CD4 but no coreceptor was included, but remained uninfected in all experiments (data not shown).
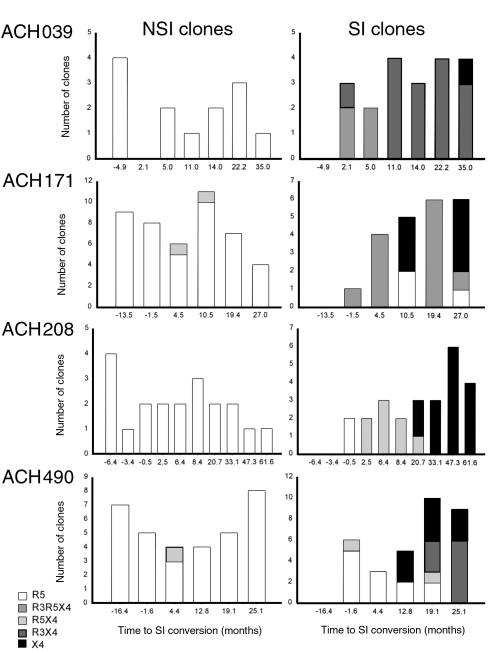
In all patients, the majority of NSI variants were restricted to CCR5 usage throughout infection (108 of 111 clones: 97% (Figure 3a). In line with their tropism for MT2 cells, the majority of SI HIV-1 variants were able to infect the U87-CXCR4 cell line (83 of 100 clones: 83%). CCR3 usage was observed only in addition to CXCR4 usage in three of the patients (ACH039, ACH171, ACH490). Interestingly, in three out of four patients, early SI variants were able to use both CCR5 and CXCR4 (R5X4 or R3R5X4), whereas at the last time point analyzed the majority (67%, ACH171) or all (ACH039, 208, 490) of the SI variants were restricted to usage of CXCR4, either in combination with or not in combination with CCR3 (X4 or R3X4 variants). Thus, the capacity to use both CCR5 and CXCR4 seems to be an early stage in the evolution of SI HIV-1, whereas late-stage SI variants lost the ability to infect cells via CCR5.
Figure 3.
Longitudinal analysis of coreceptor usage of NSI and SI HIV-1 clones. Based on the ability to infect MT2 cells, viral clones were subdivided in NSI (left panels) and in SI (right panels) phenotypes. Coreceptor usage was tested by the ability to productively infect U87 cells transfected with CD4 and one of the HIV-1 coreceptors CCR3, CCR5, or CXCR4 after cell-free infection of these cells. Bars indicate in vitro specificity for HIV-1 coreceptors: CCR5 (R5), open bars; CCR5 and CXCR4 (R5X4), gray bars; CCR3, CCR5, and CXCR4 (R3R5X4), light gray bars; CCR3 and CXCR4 (R3X4), dark gray bars; and CXCR4 (X4), filled bars.
Unexpectedly, MT2 tropism did not correlate absolutely with the capacity to productively infect U87-CXCR4 cells. A minority of non–MT2-tropic NSI variants were able to infect the U87-CXCR4 cell line (3%), whereas, especially in ACH490, MT2-tropic variants were detected that were unable to infect the U87-CXCR4 cell line (Figure 3a). We excluded the possibility that low contaminating amounts of SI HIV-1 in NSI isolates explained low-level infection of MT2 cells by reisolation and sequencing of virus after infection of MT2 cells with these isolates. Passage through MT2 cells would rapidly select for SI sequences if contaminating SI HIV-1 were present; however, SI sequences were not observed in any of the reisolated viruses (data not shown). MT2 cells do not express CCR5, as assessed by FACS analyses or RT-PCR (data not shown); therefore, it is unlikely that entry into MT2 cells was mediated by low-level expression of CCR5. Alternatively, the discordance between infection of MT2 and U87-CXCR4 cells might indicate low affinity of the viral clones for CXCR4.
To further confirm data from MT2 and U87 cells, we determined the capacity of the viral clones to infect PHA-PBMCs from a healthy donor homozygous for the CCR5 Δ32 deletion (CCR5 Δ32/Δ32). These cells do not express CCR5 on their cell surface, therefore entry must be independent from CCR5. The vast majority of X4 SI clones (98%) were able to infect the CCR5 Δ32/Δ32 PBMCs, whereas none of the R5 NSI variants tested (n = 80) were able to infect CCR5 Δ32/Δ32 PBMCs (Table 1). Viral clones with a discordant phenotype (i.e., the R5X4 non–MT2-tropic and R5 MT2-tropic variants) were unable to infect CCR5 Δ32/Δ32 PBMCs (n = 20), supporting the idea that CXCR4 usage of these variants is highly inefficient and may depend on the cell type used. These clones may represent evolutionary intermediates in NSI to SI conversion.
Table 1.
Infection of CCR5 Δ32/Δ32 PHA-PBMCs by MT2 and U87 tropismA
Mutations in V3 loop of gp120 associated with CXCR4 usage.
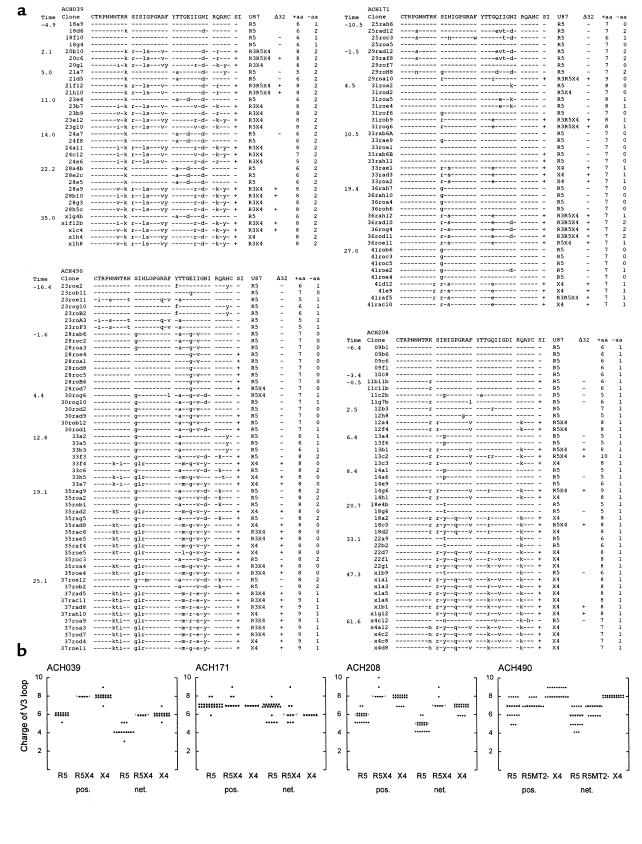
Since we observed a gradual evolution of tropism for CD45RO+ to CD45RA+ cells and a concurrent evolution of coreceptor preference after SI conversion, we studied evolution of the third variable (V3) loop-spanning region of gp120. The deduced amino acid sequences of the V3 loops and viral phenotype of the clones are depicted in Figure 4a.
Figure 4.
Envelope gp120 V3 sequence analyses of NSI and SI HIV-1. (a) Sequence alignment of the deduced amino acid sequences of the gp120 V3 loop. Sequences were aligned with the consensus sequence of the first time point from each patient. SI capacity in MT2 cells, coreceptor usage in U87 cells, the ability to infect CCR5 Δ32/Δ32 PBMCs, and the absolute number of positively charged and negatively charged amino acids in the V3 loop are indicated. Although more sequence data are available, the V3 loop is depicted for clarity only. (b) Number of positively charged amino acids (pos.) and net positive charge (net.) of the gp120 V3 loop of biological clones by coreceptor usage. Horizontal bars reflect median values. R3R5X4 clones are combined with R5X4 clones and R3X4 clones with X4-restricted HIV-1 clones. For patient ACH490, clones with a discordant phenotype (MT2 tropic, but unable to infect CCR5 Δ32/Δ32 PBMCs [R5MT2+]) are also shown.
In agreement with earlier findings (34), all clones from ACH171, ACH208, and ACH490 that were able to use CXCR4 in MT2 cells, U87-CXCR4 cells, and PBMCs carried a positively charged amino acid at either position 11 or 25 of the V3 loop. Early X4 clones from ACH490 were atypical in this respect, since no positively charged amino acid was observed at these positions. Late-stage X4 clones from this patient did display a positively charged arginine at position 25.
A major role has been attributed to the charge of the V3 loop for interaction with CXCR4. The absolute number of positively charged amino acids and the net positive charge of the gp120 V3 loop by viral phenotype are depicted in Figure 4b. V3 loops of X4 or R5X4 clones have a higher net charge or absolute positive charge than R5 clones from the same patient in three of the four patients. Unexpectedly, no association between charge and CXCR4 usage was observed among clones from ACH171, indicating that not merely the number but also the position of positively charged amino acids is important for the interaction with CXCR4. Furthermore, a highly positively charged V3 loop does not warrant X4 usage, since clones with a net charge of +7 to +8 were isolated (ACH171 and ACH490) that nevertheless were unable to use CXCR4 in MT2 or U87-CXCR4 cells.
We observed an evolution from the capacity to use both CCR5 and CXCR4 to a CXCR4-restricted phenotype among SI clones. In all patients, the charge of the V3 loop of R5X4 variants and X4-restricted variants were similar (Figure 4b). Furthermore, the presence of R5X4 clones with a net V3 charge up to +9 indicates that a highly positive charge does not exclude CCR5 usage per se. Therefore, loss of CCR5 usage was not due to a further rise of V3 charge.
Continuous evolutionary divergence of NSI and SI HIV-1.
The molecular evolution of the V3-spanning region of gp120 of HIV-1 clones with different coreceptor usage is shown in unrooted neighbor joining trees in Figure 5a. In general, a large evolutionary distance between (X4 and R5X4) SI clones and R5 NSI clones can be observed. This divergence between R5 and X4 HIV-1 population is stable in bootstrap analyses, with values ranging from 77% to 89% of 100 replicate bootstrap analyses. Although, in patient ACH171, the majority of the R5X4 and X4 clones also clustered apart from R5 clones with high bootstrap value (96%), two early R5X4 clones clustered with R5 clones. Once CXCR4 usage was established, the V3 regions of both NSI and SI populations continued to evolve, as reflected by an ever-increasing distance between NSI and coexisting SI clones.
Figure 5.
Molecular phylogeny of NSI and SI HIV-1 based on gp120 V3-spanning region. (a) Neighbor joining phylogenetic trees of gp120 V3 regions. Bootstrap values, based on 100 bootstrap analyses, indicate in how many replicate trees the depicted clustering is observed. Coreceptor usage of the clones is indicated by the color of the symbols: R5, open symbols; R3R5X4 or R5X4, light-gray symbols; R3X4, dark-gray symbols; and X4-restricted clones, filled symbols. Asterisks indicate HIV-1 clones that were MT2 tropic but were unable to infect U87-CXCR4 cells or CCR5 Δ32/Δ32 PBMCs. (b) Pairwise hamming distances between R5 clones to R5 clones (R5), between R5 clones to R5X4 clones (R5X4), and between R5 clones to X4 clones (X4). Uncorrected hamming distances between each pair of clones were displayed in a box plot, depicting tenth, 25th, 50th, 75th, and 90th percentiles. Circles depict fifth and 95th percentiles.
To analyze the evolutionary divergence from R5 clones to R5X4 and X4 clones, we calculated pairwise hamming distances between the viral clones. The evolutionary distance between R5 and R5X4 clones was lower than the distance between R5- and X4-restricted clones (in ACH208 and ACH039), underlining the intermediate character of R5X4 clones in the evolution of SI HIV-1. In this respect, ACH171 is exceptional, since R5X4 clones cluster with X4 clones in an evolutionary tree (Figure 5a) and divergence from R5 to X4 clones is similar to R5 to R5X4 clone (Figure 5b), indicating that loss of CCR5 usage may not be associated with mutations in the V3 region, at least in this patient.
Viral clones with discordant phenotypes that were able to use CXCR4 in either MT2 or U87-CXCR4 cells, but unable to infect CCR5 Δ32/Δ32 PBMCs, clustered in all patients with R5 clones (Figure 5a). If these variants indeed represent early intermediates in the evolution to efficient CXCR4 usage, this finding would indicate that regions outside the V3 region, or only minor mutations in gp120 V3 region, might confer low affinity for CXCR4.
SI HIV-1 variants isolated from CD45RA+ and CD45RO+ CD4+ T cells are phenotypically and evolutionarily indistinguishable.
We observed that SI HIV-1 variants can be isolated from both CD45RA+ and CD45RO+ CD4+ T cells. For ACH171 and ACH490, we analyzed whether SI HIV-1 clones isolated from these T-cell subsets differed with respect to coreceptor usage or V3 sequence. With respect to coreceptor usage, R3X4, R5X4, R3R5X4, and X4 HIV-1 variants could be isolated from both subsets in similar proportions, suggesting that coreceptor preference other than CXCR4 usage does not determine differential tropism for CD45RA+ and CD45RO+ CD4+ T cells of SI HIV-1 (Figure 6a).
Figure 6.
SI HIV-1 clones isolated from CD45RA+ and CD45RO+ CD4+ T cells are phenotypically and evolutionarily indistinguishable. (a) Coreceptor usage of SI clones isolated from CD45RA+ and CD45RO+ CD4+ T cells from ACH171 and ACH490. Virus clones were pooled from the time points in which SI clones were obtained from both subsets (10.5, 19.5, and 27 months for ACH171 and 4.4, 19.2, and 25.1 months for ACH490). Bar descriptions are as in Figure 3. (b) Pairwise hamming distances were calculated between clones obtained from CD45RA+ CD4+ T cells (RA vs. RA), between clones obtained from CD45RO+ CD4+ T cells (RO vs. RO), and between clones isolated from CD45RA+ to clones from CD45RO+ CD4+ T cells (RA vs. RO). Uncorrected hamming distances between each pair of sequences were calculated and are depicted in a box plot as described in Figure 5.
Pairwise hamming distances between CXCR4 using clones isolated from CD45RA+ to clones from CD45RO+ CD4+ T cells were compared to the hamming distances of clones within CD45RA+ and CD45RO+ cells (Figure 6b). Divergence between clones from CD45RA+ to clones from CD45RO+ CD4+ cells did not exceed diversity of clones within the CD45RO+ and within the CD45RA+ CD4+ T-cell subset. Thus, clones from CD45RA cells and CD45RO cells are phenotypically and phylogenetically indistinguishable, suggesting a dynamic exchange of R5X4 and X4 clones between those T-cell subsets.
Differential HIV-1 coreceptor expression on CD45RO– and CD45RO+ CD4+ T cells.
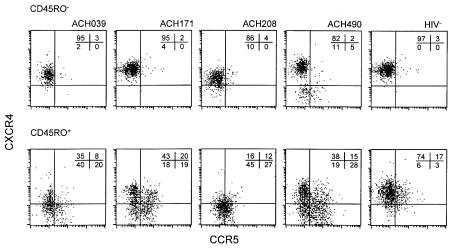
The divergence of NSI and SI variants with respect to V3 sequence and coreceptor usage is compatible with the idea that both variants infect different target cells within the CD4+ T-cell population. However, within the CD45RO+ CD4+ T-cell subset, NSI and SI variants persistently coexisted. To analyze whether distinct NSI and SI target-cell populations can be distinguished within the CD45RO+ CD4+ T-cell population, we determined the expression of CCR5 and CXCR4 on CD45RO+ and CD45RO– CD4+ T cells by four-color flow cytometry on cryopreserved PBMCs obtained at 7 to 20 months after SI conversion and among eight healthy donors.
Within the CD45RO– CD4+ T-cell subset, the majority of the cells were CCR5– CXCR4+, both in healthy donors (average 96% of eight donors, range 94–97%) and HIV+ patients (82%–95%, Figure 7). CCR5 expression in this subset was low, ranging from 2% to 5%. However, among CD45RO+ CD4+ cells, a divergent expression pattern was observed in HIV-1–infected patients as compared with healthy donors. In healthy donors, the majority of the CD45RO+ cells were CXCR4+ CCR5– (on average 71%, range 59–78%) or CCR5+ CXCR4+ (17%, range 10–21%). In contrast, a differential expression of CCR5 and CXCR4 was observed in CD45RO+ CD4+ T cells in HIV-1–infected patients. In this T-cell subset, major populations of CXCR4+ CCR5– (16–43%) and CXCR4– CCR5+ (19–28%) cells and a population negative for both HIV-1 coreceptors (18–45%) can be distinguished. The aberrant expression of coreceptors in HIV-1–infected patients is not due to the presence of SI HIV-1 variants, since similar results were obtained from samples obtained well before SI conversion (25 and 10 months, ACH171 and ACH490) and could also be observed in patients infected with only NSI variants (data not shown).
Figure 7.
Expression of CCR5 and CXCR4 on CD45RO– and CD45RO+ CD4+ T cells. Cryopreserved PBMCs from HIV-1–infected patients and a healthy donor were incubated with mAb’s against CD4, CD45RO, CCR5, and CXCR4. The CD45RO– (top) and CD45RO+ CD4+ T cells (bottom) were gated. Within these subsets, CCR5 and CXCR4 expression was determined. Numbers in the figures depict percentage of gated cells in the corresponding quadrants. A representative healthy donor from eight unrelated donors is shown.
CD45RO+ CD4+ T-cell subsets with distinct HIV-1–coreceptor expression provide distinct niches for NSI and SI HIV-1.
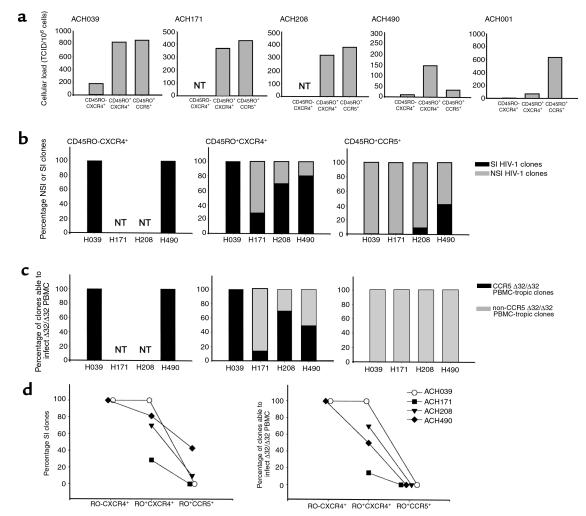
To analyze whether the differential expression of CCR5 and CXR4 among CD45RO+ CD4+ T cells provide distinct target cells for NSI and SI HIV-1 variants, we sorted patient PBMCs based on their coreceptor expression and isolated HIV-1 clones from the sorted cells. Patient PBMCs were sorted in CD45RO– CXCR4+ CCR5–; CD45RO+ CXCR4+ CCR5–, and CD45RO+ CXCR4– CCR5+ CD4+ T cells. The cellular infectious viral load in each subset is shown in Figure 8a. In all SI HIV-1–infected patients, virus could be isolated from both CXCR4+ CCR5+ and CXCR4– CCR5+ CD45RO+ CD4+ T cells. To exclude that the presence of HIV-1 in CXCR4+ CCR5– cells was a late-stage effect of HIV-1 infection rather than a reflection of SI HIV-1 infection, we also included a patient (ACH001, 7 years after seroconversion) who developed AIDS in the absence of SI HIV-1 variants. In this patient virus was isolated mainly from the CCR5+ CD45RO+ CD4+ cells (Figure 8a).
Figure 8.
CD4+ T-cell subsets with distinct HIV-1–coreceptor expression provide distinct niches for NSI and SI HIV-1. Cryopreserved PBMCs were sorted based on expression of CD45RO and HIV-1 coreceptors CCR5 and CXCR4, and virus was isolated by cocultivation with PHA-PBMCs under limiting dilution conditions. In addition to SI-infected patients ACH039, ACH171, ACH208, and ACH490, one patient (ACH001) who was infected solely with NSI HIV-1 variants, was studied. (a) Frequency of CD45RO– CXCR4+ CCR5–; CD45RO+ CXCR4+ CCR5–; and CD45RO+ CXCR4– CCR5+ CD4+ T cells productively infected with HIV-1. NT, not tested. (b and c) Phenotype of virus clones from CD45RO– CXCR4+ (left panel), CD45RO+ CXCR4+ (middle), and CD45RO+ CCR5+ (right) cells as defined by the ability to infect MT2 cells (b) or CCR5Δ32/Δ32 PBMCs (c). Filled bars reflect the percentage of clones able to infect the indicator cells, whereas gray bars reflect clones unable to infect these cells. (d) The difference in the proportion of clones with SI phenotype (left panel) and clones able to infect CCR5 Δ32/Δ32 PBMCs (right panel) isolated from CD45RO+ CXCR4+ and CD45RO+ CCR5+ cells were compared pairwise by Wilcoxon signed rank test (P = 0.07, in both panels).
The phenotype of the viral clones obtained from these subsets as defined by the ability to infect MT2 cells and CCR5 Δ32/Δ32 PBMCs is depicted in Figure 8, b and c. From the CD45RO– cells we isolated only SI HIV-1 clones that were able to infect CCR5 Δ32/Δ32 PBMCs. The majority of the clones isolated from CCR5+ CD45RO+ cells had an NSI phenotype (86%), and none of these clones were able to infect PBMCs lacking CCR5 expression. The majority of the clones isolated from CXCR4+ CD45RO+ cells had an SI phenotype (75%) and was able to infect CCR5 Δ32/Δ32 PBMCs (57%). However, NSI R5–dependent clones were isolated from these cells as well in three of the patients. Interestingly, in these patients a relatively high number of CD45RO+ cells expressed both CCR5 and CXCR4 (Figure 7). Chemokine receptors can be upregulated and downregulated rapidly in response to the proper stimuli (58). Therefore, it is likely that in these patients cells expressed both CCR5 and CXCR4 at the moment of infection, thus allowing infection with R5-dependent clones, after which CCR5 expression was downregulated. The prevalence of SI, R5-independent HIV-1 clones among CXCR4+ CD45RO+ is higher than the prevalence in CCR5+ CD45RO+ cells (Wilcoxon signed rank test, P = 0.07; Figure 8d). Taken together, these data show that based on coreceptor expression two distinct target-cell populations can be distinguished within the CD45RO+ CD4+ T-cell subset, providing cellular niches for NSI and SI HIV-1.
Discussion
We studied changes in HIV-1 coreceptor usage, viral tropism for CD45RA+ and CD45RO+ memory CD4+ T cells, and molecular phylogeny of biological cloned virus in four patients during NSI to SI HIV-1 evolution. NSI HIV-1 variants were always isolated predominantly from the CD45RO+ T cells, independent of the presence of coexisting SI variants. Just after their emergence, SI variants were also isolated mainly from the CD45RO+ cells, but with time an equal distribution over CD45RO+ and CD45RA+ CD4+ T-cell subsets was established, in agreement with previous observations from a cross-sectional study (51).
CD45RA+ CD4+ T cells may also include non-naive CD4 cells, especially in HIV-1 infection (59). Since we did not specifically stain for truly naive cells, we cannot formally exclude that sorted CD45RA+ cells also compromise non-naive cells. However, we observed that non-naive CD45RA+ CD4 cells express strongly reduced levels of CXCR4 and a higher percentage of CCR5 when compared with truly naive CD4+ T cells (data not shown), which would not fit the predominant isolation of SI HIV-1 from CD45RA+ CD4+ T cells. Therefore, we conclude that virus clones isolated from CD45RA+ CD4+ T cells are derived from truly naive cells.
Productive infection of naive CD45RA+ CD4+ T cells, as observed in vivo in this and other studies (refs. 51 and 60 and our unpublished observations), is in contrast to in vitro resistance of naive CD4+ T cells to HIV-1 replication (61–63). Productive infection of HIV-1 is dependent on the activation state of the cells and may therefore not occur in quiescent lymphocytes (64, 65). Although naive CD4 T cells are considered to be inactivated and nonproliferating, it has been shown that resting naive CD4 cells can be induced to proliferate by a combination of cytokines without a switch to the memory phenotype (66). Moreover, it was published recently that the cell-proliferation marker Ki67 is expressed among truly naive CD4 cells in HIV-1–infected patients at higher levels than in healthy donors (67). The enhanced activation state of naive cells in HIV-1 infection may provide the necessary intracellular requirements for replication of HIV-1 and explain discrepancies between in vivo and in vitro infection of naive cells.
In line with their restricted tropism for CD45RO+ CD4+ T cells, the vast majority of NSI isolates were restricted to CCR5 usage in U87 cells. In agreement with a dependence on CCR5 usage, NSI clones were unable to infect PBMCs from a donor with a CCR5 Δ32/Δ32 genotype. In three patients, early SI variants could use both CCR5 and CXCR4, whereas late-stage SI clones lost the ability to use CCR5. CCR3 usage was observed in three of the patients only in addition to CXCR4 usage of both early and late-stage SI HIV-1 variants. In phylogenetic trees, R5X4 clones, in general, clustered between R5 clones and late-stage X4 clones, indicating that dual tropism for CCR5 and CXCR4 is an intermediate stage in evolution of SI HIV-1. The continuous divergence between CXCR4-using clones and R5 clones in phylogenetic trees reflects a continuous evolution of V3 gp120 from the acquisition of CXCR4 usage onward. This may implicate that the structure of gp120 continues to evolve to optimize interactions with CXCR4.
It has been proposed that interaction of gp120 with CXCR4 is due mainly to electrostatic interactions between the positively charged conserved coreceptor-binding site and V3 loop in gp120 and negatively charged extracellular domains of CXCR4 (43, 44). In agreement with this, in three of the patients the V3 loop of R5X4 and X4 clones in general had a higher positive charge than the V3 loop of R5 clones. However, in one patient, the acquisition of CXCR4 usage was not accompanied with an increase in V3 charge, despite the concurrent appearance of a SI phenotype–associated positively charged amino acid at position 11. No further increase in charge of the V3 loop of CXCR4-using clones was observed upon the eventual loss of R5 usage. Taken together, these results show that a highly positively charged V3 loop does not exclude CCR5 usage per se, and, importantly, a highly positive charge does not warrant CXCR4 usage, suggesting a complex interaction of gp120 with CXCR4 in which both charge and structure of V3 play an major role.
The divergence of NSI and SI variants with respect to V3 sequence and coreceptor usage and their coexistence throughout infection are compatible with the idea that they occupy different niches within the CD4+ T-cell population. Since in a viral quasispecies virus variants are constantly competing for the available target cells, one would expect that a variant with a slight growth advantage would outcompete the less-fit virus variants if the same niche is occupied (principle of competitive exclusion; ref. 4). Although SI HIV-1 variants are isolated mainly from CD45RA+ CD4+ T cells, NSI and SI HIV-1 populations persistently coexist within the memory CD45RO+ CD4+ T cells.
Here we showed that within the CD45RO+ T-cell subset, HIV-1 coreceptors CCR5 and CXCR4 are differentially expressed. In vivo NSI and SI HIV-1 variants were distributed over these subsets according to their coreceptor preference. From CCR5+ CXCR4– CD45RO+ CD4+ T cells, exclusively CCR5-dependent virus strains were isolated, whereas the majority of the viral clones obtained from CCR5– CXCR4+ CD45RO+ CD4+ T cells were MT2 tropic and able to use CXCR4 in PBMCs. Therefore, we conclude that within the CD45RO+ CD4+ T-cell subset, CCR5+ CXCR4– and CCR5– CXCR4+ cells are the distinct cell populations that function as distinct cellular niches for NSI and SI HIV-1 variants. Furthermore, we showed that SI variants isolated from CD45RA+ and CD45RO+ CD4+ T cells were phylogenetically and phenotypically indistinguishable, indicating that CD45RA+ and CXCR4+ CCR5– CD45RO+ CD4+ cells represent one single niche for SI HIV-1 variants.
Besides explaining coexistence, circumvention of competition and independent evolutionary pathways of NSI and SI HIV-1, niche adaptation may result in a differential encounter of environmental and intracellular factors. This is in agreement with previous findings showing a differential effect of NSI and SI HIV-1 in response to monotherapy with AZT and ddI (68), nucleoside analogues that depend on phosphorylation by cellular kinases for their activity.
Based on the results described in this study, we propose a model of continuous virus evolution during and after the conversion to SI HIV-1. NSI clones are CCR5 restricted and therefore able to infect CCR5+ CD45RO+ memory CD4+ T cells throughout infection. NSI infection of CD45RA+-naive CD4+ T-cell infection also occurs, though at low frequency due to low numbers of CCR5-expressing CD45RA+ CD4+ T cells. Mutations introduced in gp120 in the proper structural context may result in a low affinity for CXCR4. Still, these variants preferentially use CCR5 in vivo and therefore have to compete with the coexisting and well-adapted NSI population for the same CCR5-expressing target-cell population. Only upon the acquisition of a combination of additional mutations, generally including the positively charged amino acid at either position 11 or 25 in the V3 loop, efficient CXCR4 usage is obtained. The acquisition of high affinity for CXCR4 might occur initially at the expense of viral fitness, which would then explain the large evolutionary gap between NSI and SI HIV-1 and why SI variants do not occur in all HIV-1–infected individuals. After acquisition of efficient CXCR4 usage, the target-cell population of the SI HIV-1 variants is extended to include the CXCR4+ CCR5– (CD45RA+ and CD45RO+) CD4+ T cells. Infection of these cells offers the opportunity to avoid competition with the NSI virus population, which may be highly replication competent, especially late in infection (69). The existence of distinct cellular niches for NSI and SI HIV-1 allows SI variants to expand in an environment where they are outnumbered initially by the NSI population and eventually reach equal levels as NSI variants. Further adaptations in gp120 may exclude the capacity to use CCR5 because of structural constraints, explaining the eventual loss of the ability to use CCR5 by late-stage SI variants. Eventually, two independent virus populations are established with two distinct target-cell populations: a CCR5-dependent NSI HIV-1 population that, within the T-cell pool, infects CCR5+ CXCR4– memory CD4+ cells and a CXCR4-restricted SI HIV-1 population that infects naive and CXCR4+ CCR5– memory CD4+ T cells.
Acknowledgments
This study was performed as part of the Amsterdam Cohort Studies on HIV infection and AIDS, a collaboration between the Municipal Health Service, the Academic Medical Center, and CLB Sanquin, Amsterdam, The Netherlands. We are greatly indebted to the cohort participants for their continuous participation. Proleukin (rIl2) was kindly provided by R. Rombouts, Chiron Benelux BV. D. Littman kindly provided the U87 cell lines. Expression vectors pBABE-BOB and pBABE-BONZO were obtained from N. Landau through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. The authors wish to thank Berend Hooibrink and Bert Hovenkamp for FACS sorting, Angelique van ’t Wout for advice on phylogenetic analyses, and Frank Miedema for critically reading the manuscript. This study was financially supported by the Netherlands Foundation for Preventive Medicine (grant 28-2547), the Netherlands Ministry of Public Health, and the Dutch AIDS Fund (grant 1305).
Footnotes
Hetty Blaak’s present address is: Institute of Virology, Erasmus University, Rotterdam, The Netherlands.
Margreet Brouwer’s present address is: F.A. Janssens Laboratory of Genetics, Catholic University Leuven, Leuven, Belgium.Ana-Maria de Roda Husman’s present address is: Microbiological Laboratory for Health Protection, National Institute of Public Health and the Environment, Bilthoven, The Netherlands.
References
- 1.Ho DD, et al. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 2.Wei X, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 3.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 4.Domingo E, et al. Basic concepts in RNA virus evolution. FASEB J. 1996;10:859–864. doi: 10.1096/fasebj.10.8.8666162. [DOI] [PubMed] [Google Scholar]
- 5.Zhu T, et al. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 6.Van ‘t Wout AB, et al. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral and vertical transmission. J Clin Invest. 1994;94:2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuitemaker H, et al. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koot M, et al. Prognostic value of human immunodeficiency virus type 1 biological phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993;118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 9.Koot M, et al. HIV-1 biological phenotype in long term infected individuals, evaluated with an MT-2 cocultivation assay. AIDS. 1992;6:49–54. doi: 10.1097/00002030-199201000-00006. [DOI] [PubMed] [Google Scholar]
- 10.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 11.Deng HK, et al. Identification of the major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 12.Dragic T, et al. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 13.Alkhatib G, et al. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 14.Choe H, et al. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 15.Doranz BJ, et al. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3 and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 16.Combadiere C, et al. Identification of CX3CR1, a chemotactic receptor for the human CX3C chemokine fractalkine and a fusion coreceptor for HIV-1. J Biol Chem. 1998;273:23799–23804. doi: 10.1074/jbc.273.37.23799. [DOI] [PubMed] [Google Scholar]
- 17.Owman C, et al. The leucotriene B4 receptor functions as a novel type of coreceptor mediating entry of primary HIV-1 isolates into CD4-positive cells. Proc Natl Acad Sci USA. 1998;95:9530–9534. doi: 10.1073/pnas.95.16.9530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choe H, et al. The orphan seven-transmembrane receptor Apj supports the entry of primary T-cell-line-tropic and dualtropic human immunodeficiency virus type 1. J Virol. 1998;72:6113–6118. doi: 10.1128/jvi.72.7.6113-6118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farzan M, et al. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J Exp Med. 1997;186:405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng HK, Unutmaz D, KewalRamani VN, Littman DR. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 21.Rucker J, et al. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edinger AL, et al. An orphan seven-transmembrane domain receptor expressed widely in the brain functions as a coreceptor for human immunodeficiency virus type 1 and simian immunodeficiency virus. J Virol. 1998;72:7934–7940. doi: 10.1128/jvi.72.10.7934-7940.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samson M, et al. ChemR23, a putative chemoattractant receptor, is expressed in monocyte-derived dendritic cells and macrophages and is a coreceptor for SIV and some primary HIV-1 strains. Eur J Immunol. 2000;28:1689–1700. doi: 10.1002/(SICI)1521-4141(199805)28:05<1689::AID-IMMU1689>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y-J, et al. Use of coreceptors other than CCR5 by non-syncytium-inducing adult and pediatric isolates of human immunodeficiency virus type 1 is rare in vitro. J Virol. 1998;72:9337–9344. doi: 10.1128/jvi.72.11.9337-9344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, et al. Chemokine coreceptor usage by diverse primary isolates of human immunodeficiency virus type 1. J Virol. 1998;72:9307–9312. doi: 10.1128/jvi.72.11.9307-9312.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Roda Husman AM, van Rij RP, Blaak H, Schuitemaker H. Adaptation to promiscuous usage of chemokine receptors is not a prerequisite for HIV-1 disease progression. J Infect Dis. 1999;180:1106–1115. doi: 10.1086/314987. [DOI] [PubMed] [Google Scholar]
- 27.Scarlatti G, et al. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 28.Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao L, Rudolph DL, Owen SM, Spira TJ, Lal RB. Adaptation to promiscuous usage of CC and CXC-chemokine coreceptors in vivo correlates with HIV-1 disease progression. AIDS. 1998;12:F137–F143. doi: 10.1097/00002030-199813000-00001. [DOI] [PubMed] [Google Scholar]
- 30.Schramm B, et al. Viral entry through CXCR4 is a pathogenic factor and therapeutic target in human immunodeficiency virus type 1 disease. J Virol. 2000;74:184–192. doi: 10.1128/jvi.74.1.184-192.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Groenink M, et al. Relation of phenotype evolution of HIV-1 to envelope V2 configuration. Science. 1993;260:1513–1516. doi: 10.1126/science.8502996. [DOI] [PubMed] [Google Scholar]
- 32.De Jong JJ, et al. Human immunodeficiency virus type-1 clones chimeric for the envelope V3 domain differ in syncytium formation and replication capacity. J Virol. 1992;66:757–765. doi: 10.1128/jvi.66.2.757-765.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fouchier RAM, Brouwer M, Broersen SM, Schuitemaker H. Simple determination of human immunodeficiency virus type 1 syncytium inducing genotype by PCR. J Clin Microbiol. 1995;33:906–911. doi: 10.1128/jcm.33.4.906-911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fouchier RAM, et al. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho MW, et al. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J Virol. 1998;72:2509–2515. doi: 10.1128/jvi.72.3.2509-2515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ross TM, Cullen BR. The ability of HIV-1 to use CCR-3 as a co-receptor is controlled by envelope V1/V2 sequences acting in conjunction with a CCR-5 tropic V3 loop. Proc Natl Acad Sci USA. 1998;95:7682–7686. doi: 10.1073/pnas.95.13.7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffman TL, Stephens EB, Narayan O, Doms RW. HIV type 1 envelope determinants for use of the CCR2b, CCR3, STRL33, and APJ coreceptor. Proc Natl Acad Sci USA. 1998;95:11360–11365. doi: 10.1073/pnas.95.19.11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee MK, Heaton J, Cho MW. Identification of determinants of interaction between CXCR4 and gp120 of a dual tropic HIV-1DH12 isolate. Virology. 1999;257:290–296. doi: 10.1006/viro.1999.9686. [DOI] [PubMed] [Google Scholar]
- 39.Smyth RJ, Yi Y, Singh A, Collman RG. Determinants of entry cofactor utilization and tropism in a dualtropic human immunodeficiency virus type 1 primary isolate. J Virol. 1998;72:4478–4484. doi: 10.1128/jvi.72.5.4478-4484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohagen A, et al. Apoptosis induced by infection of primary brain cultures with diverse human immunodeficiency virus type 1 isolates: evidence for a role of the envelope. J Virol. 1999;73:897–906. doi: 10.1128/jvi.73.2.897-906.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rizzuto CD, et al. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 42.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwong PD, et al. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J Virol. 2000;74:1961–1972. doi: 10.1128/jvi.74.4.1961-1972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moulard M, et al. Selective interactions of polyanions with basic surfaces on human immunodeficiency virus type 1 gp120. J Virol. 2000;74:1948–1960. doi: 10.1128/jvi.74.4.1948-1960.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Jong JJ, De Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van ‘t Wout AB, et al. Evolution of syncytium inducing and non-syncytium inducing biological virus clones in relation to replication kinetics during the course of HIV-1 infection. J Virol. 1998;72:5099–5107. doi: 10.1128/jvi.72.6.5099-5107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuiken CL, et al. Evolution of the V3 envelope domain in proviral sequences and isolates of human immunodeficiency virus type 1 during transition of the viral biological phenotype. J Virol. 1992;66:4622–4627. doi: 10.1128/jvi.66.7.4622-4627.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Asjo B, et al. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet. 1986;2:660–662. [PubMed] [Google Scholar]
- 49.Connor RI, Mohri H, Cao Y, Ho DD. Increased viral burden and cytopathicity correlate temporally with CD4+ T-lymphocyte decline and clinical progression in human immunodeficiency virus type 1 infected individuals. J Virol. 1993;67:1772–1777. doi: 10.1128/jvi.67.4.1772-1777.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koot M, et al. Relation between changes in cellular load, evolution of viral phenotype, and the clonal composition of virus populations in the course of human immunodeficiency virus type 1 infection. J Infect Dis. 1996;173:349–354. doi: 10.1093/infdis/173.2.349. [DOI] [PubMed] [Google Scholar]
- 51.Blaak H, et al. In vivo HIV-1 infection of CD45RA+CD4+ T cells is established primarily by syncytium-inducing variants and correlates with the rate of CD4+ T cell decline. Proc Natl Acad Sci USA. 2000;97:1269–1274. doi: 10.1073/pnas.97.3.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strijbosch LWG, Does RJMM, Buurman WA. Computer aided design and evaluation of limiting dilution and serial dilution experiments. Int J Biomed Comput. 1988;23:279–290. doi: 10.1016/0020-7101(88)90020-7. [DOI] [PubMed] [Google Scholar]
- 53.Van’t Wout, et al. Analysis of the temporal relationship between human immunodeficiency virus type 1 quasispecies in sequential blood samples and various organs obtained at autopsy. J Virol. 1998;72:488–496. doi: 10.1128/jvi.72.1.488-496.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simmonds P, Balfe P, Ludlam CA, Bishop JO, Brown AJL. Analysis of sequence diversity in hypervariable regions of the external glycoprotein of human immunodeficiency virus type 1. J Virol. 1990;64:5840–5850. doi: 10.1128/jvi.64.12.5840-5850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson JD, Higgins DG, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Felsenstein J. PHYLIP-phylogeny package. Cladistics. 1989;5:164–166. [Google Scholar]
- 57.Strimmer K, Von Haeseler A. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 58.Loetscher P, Seitz M, Baggiolini M, Moser B. Interleukin-2 regulates CC chemokine receptor expression and chemotactic responsiveness in T lymphocytes. J Exp Med. 1996;184:569–577. doi: 10.1084/jem.184.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roederer M, et al. CD8 naive T cell counts decrease progressively in HIV-infected adults. J Clin Invest. 1995;95:2061–2066. doi: 10.1172/JCI117892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ostrowski MA, et al. Both memory and CD45RA+/CD62L+ naive CD4+ T cells are infected in human immunodeficiency virus type 1-infected individuals. J Virol. 1999;73:6430–6435. doi: 10.1128/jvi.73.8.6430-6435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spina CA, Prince HE, Richman DD. Preferential replication of HIV-1 in the CD45RO memory cell subset of primary CD4 lymphocytes in vitro. J Clin Invest. 1997;99:1774–1785. doi: 10.1172/JCI119342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roederer M, Raju PA, Mitra DK, Herzenberg LA, Herzenberg L. HIV does not replicate in naive CD4 T cells stimulated with CD3/CD28. J Clin Invest. 1997;99:1555–1564. doi: 10.1172/JCI119318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woods TC, Roberts BD, Butera ST, Folks TM. Loss of inducible virus in CD45RA naive cells after human immunodeficiency virus-1 entry accounts for preferential viral replication in CD45RO memory cells. Blood. 1997;89:1635–1641. [PubMed] [Google Scholar]
- 64.Zack JA, Haislip AM, Krogstad P, Chen ISY. Incompletely reverse-transcribed human immunodeficiency virus type 1 genomes in quiescent cells can function as intermediates in the retroviral life cycle. J Virol. 1992;66:1717–1725. doi: 10.1128/jvi.66.3.1717-1725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bukrinsky MI, Stanwick TL, Dempsey MP, Stevenson M. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science. 1991;254:423–427. doi: 10.1126/science.1925601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Unutmaz D, Pileri P, Abrignani S. Antigen-independent activation of naive and memory resting T cells by a cytokine combination. J Exp Med. 1994;180:1159–1164. doi: 10.1084/jem.180.3.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hazenberg MD, et al. T cell division in human immunodeficiency virus (HIV-1)-infection is mainly due to immune activation: a longitudinal analysis in patients before and during highly active anti-retroviral therapy. Blood. 2000;95:249–255. [PubMed] [Google Scholar]
- 68.Van’t Wout AB, et al. Selective inhibition of syncytium-inducing and nonsyncytium-inducing HIV-1 variants in individuals receiving didanosine or zidovudine respectively. J Clin Invest. 1997;100:2325–2332. doi: 10.1172/JCI119771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blaak H, Brouwer M, Ran LJ, De Wolf F, Schuitemaker H. In vitro replication kinetics of HIV-1 variants in relation to viral load in long-term survivors of HIV-1 infection. J Infect Dis. 1998;177:600–610. doi: 10.1086/514219. [DOI] [PubMed] [Google Scholar]