Abstract
Human embryonic stem (ES) cells are pluripotent and are believed to be able to generate all cell types in the body. As such, they have potential applications in regenerative therapy for kidney disease. However, before this can be achieved, a protocol to differentiate human ES cells to mesodermal renal progenitor lineages is required. Reduction of serum concentration and feeder layer density reduction cultures were used to differentiate human ES cells for 14 days. Differentiated ES cells were then fractionated by flow cytometry based on expression of the markers CD24, podocalyxin, and GCTM2 to isolate putative renal cells. These cells up-regulated the expression of the renal transcription factors PAX2, LHX1, and WT1 when compared with unfractionated human ES cells. Immunohistochemical assays confirmed that a subset of cells within this fraction co-expressed nuclear WT1 and PAX2 proteins. Transcriptome profiling also showed that the most differentially up-regulated genes in this fraction preferentially associated with kidney development in comparison with any other lineage. When compared with a transcriptome profile database of urogenital development (GUDMAP), the top 200 differentially up-regulated genes in this fraction strongly clustered into a group of genes associated with the metanephric mesenchyme at E11.5 and the corticonephrogenic interstitium at E15.5 of murine kidney development. Hence, this approach confirms an ability to direct human ES cells toward a renal progenitor state.
Introduction
Embryonic stem (ES) cells are pluripotent and have unlimited self-renewal properties. They represent an inexhaustible source of cells for the study of physiological diseases and are an excellent model system for the study of development. However, it is arguably their potential as a renewable source of specialized cells for cell-based therapies that has stimulated much research in the ES cell field. Indeed, human ES cells have been successfully directed to differentiate in vitro using a variety of methods into many cell types including neural progenitors and their differentiated progeny [1,2], endothelial cells [3,4], osteogenic cells [5–7], cardiomyocytes [8,9], insulin-producing cells [10,11], hepatocytes [12,13], keratinocytes [14,15], germ cells [16,17], and trophoblast cells [18,19]. However, differentiation to the renal lineage has not yet been examined in detail. Murine ES cells have been shown to be able to integrate with E12–E13 metanephros [20] and have also been demonstrated to have the capacity to differentiate into renal epithelial cells that can integrate into developing kidneys [21, 22]. In the case of human ES cells, kidney-like structures have been observed in teratomas formed from human ES cells [23–25] and reverse transcription-polymerase chain reaction (RT-PCR) analyses of heterogeneous human ES cell-derived populations in embryoid bodies have indicated the presence of transcripts associated with kidney development [26]. In addition, in a recent report by Batchelder and colleagues, genes associated with the intermediate mesoderm and developing kidney were shown to be expressed in human ES cell colonies cultured in the presence of retinoic acid, activin A, and BMP7 on different substrates [27]. We sought to provide further evidence that human ES cells have the capacity to differentiate along this lineage so as to assess the potential utility of human ES cells in renal research and regenerative medicine.
The permanent postnatal kidney, the metanephros, is derived from 2 intermediate mesoderm-derived structures: the ureteric bud (UB) and the metanephric mesenchyme (MM) [28,29]. While the UB is a critical inducer tissue for the formation and patterning of the functional units of the kidney, the nephrons, this tissue ultimately only gives rise to the calyceal system of the kidney, collecting ducts, and the ureter. It is the MM that is regarded as the renal progenitor population as this tissue gives rise to all remaining segments of the nephrons including the glomeruli, as well as contributing extensively to the interstitial elements of the kidney, including portions of the vasculature. For this reason, the generation and competent culture of MM is a major target for renal regeneration and bioengineering. The paradigm for directed differentiation of human ES cells suggests that the cells will need to recapitulate the normal steps of embryology to generate a specific cell type. However, as differentiation of human ES cells occurs in the absence of the intrinsic structural architecture present in a developing embryo, combinations of specific antigenic and molecular markers identified from mammalian embryological studies are required to categorically identify the target cell population within the mixture of other cell types also present in cells differentiating from human ES cells. In the case of kidney differentiation, specific markers of MM are required. Microarray investigations coupled with in situ hybridization analyses conducted by Challen and colleagues [30] revealed that the cell surface marker CD24 was present in the mouse MM from E10.5 while podocalyxin is expressed in both MM and the surrounding intermediate mesoderm. Gene-targeting studies have identified the transcription factors Pax2, Lim1 (LHX1), and Wt1 as critically important for kidney development [31–33]. In this study, to assess potential renal differentiation we employed a differentiation regime of low serum concentration that has previously been shown to permit cardiomyocyte and definitive endoderm differentiation [34–36], in combination with a reduction in the density of the mouse embryonic fibroblast (MEF) feeder layer to induce human ES cell differentiation. Coupled with this differentiation regime, we have utilized combinations of the markers listed above to prospectively isolate cell populations displaying a similar phenotype to MM from a heterogeneous population of differentiated human ES cells. Quantitative PCR, immunofluorescence staining, and gene microarray analyses are also presented to demonstrate the identification of a specific fraction enriched for cells expressing genes associated with the developing kidney.
Materials and Methods
Human ES cell culture and differentiation
Human ES cell lines HES-2, HES-3, and HES-4 [37] were maintained as previously described [24] in the presence of 20% fetal calf serum on a layer of MEFs at a density of 6 × 104 cells/cm2. For differentiation experiments, human ES cells were cultured on a reduced density (2 × 104 cells/cm2) of feeder cells in 20% serum concentration for 2 days and 5% serum concentration for 12 more days with a media change every second day.
Human embryos
Histological sections of first trimester human embryos were obtained from archival material donated with informed consent by patients undergoing termination of pregnancy at the John Radcliffe Hospital, Oxford, UK. The tissue was fixed in absolute alcohol, embedded in paraffin, and sectioned at 5-μm thickness. All tissue was obtained following local ethical committee approval.
Teratoma assays
Teratomas were formed as described previously [38]. In brief, ∼50,000 human ES cells were injected beneath the testis capsule of SCID mice between 5 and 6 weeks of age. The animals were monitored weekly beginning at around 4 weeks for tumor development. Lesions usually became apparent as swellings in about 5 weeks and teratomas removed at ∼6 weeks. The tumors were removed, fixed in formalin, and sent for routine histological processing. Every 10th section was visually examined for kidney-like areas after hematoxylin and eosin staining.
Immunohistochemistry
Alkaline phosphatase-based immunohistochemistry was performed on the human embryo and teratoma sections using the Vectastain ABC kit (Vector Laboratories, Burlingame, CA) as per the manufacturer's instructions. Embryo sections were subjected to antigen retrieval in 10 mM citric acid buffer (pH 6.4) prior to immunohistochemistry. Primary antibodies used were mouse anti-cadherin 16 and mouse anti-cadherin 11 (Zymed Laboratories, San Francisco, CA), mouse anti-human WT1 (Dako, Carpinteria, CA), mouse anti-podocalyxin (PHM5, kind gift from Dr. David Nikolic-Paterson, Department of Nephrology, Monash Medical Centre), and mouse anti-CD24 (BD Pharmingen, San Diego, CA). Isotype-matched negative control antibodies (Dako, CA) were also employed to ensure specific staining.
Flow cytometric analyses of differentiated human ES cells
For flow cytometric analyses with CD24 (BD Pharmingen, CA), podocalyxin, and GCTM2, colonies were incubated with Dispase (Gibco, Carlsbad, CA) at a concentration of 10 mg/mL in human ES media [24], lifted intact by nudging gently with a 1,000-μL pipette tip and transferred to a 15-mL tube containing 5 mL phosphate-buffered saline (PBS). Cells were then dissociated into single cells by incubation in TrypLE™ Express (Gibco, Carlsbad, CA) at 37°C for 3 min after which the reaction was inactivated by the addition of an equal volume of serum containing human ES cell media. Harvested cells were blocked in 3 mL 1% normal goat serum in PBS and stained with CD24, podocalyxin, and GCTM2 antibodies diluted in 1% normal goat serum in PBS. A negative control tube was included comprising cells labeled with secondary antibody only. Just prior to sorting, cells were transferred to a 5-mL polypropylene tube and the viability dye propidium iodide (Sigma, St. Louis, MO) was added to the cells at a concentration of 1 μg/mL to distinguish between live and dead cells using the BD FACSVantage DiVA cell sorter (Becton Dickinson, Franklin Lakes, NJ). Sorting was performed with a nozzle size of 80 μm and a sheath pressure of 30 psi into 5-mL collecting tubes containing 1 mL human ES cell media. Cells were initially gated based on forward and side scatter to exclude cellular debris and nonviable cells were excluded by their uptake of the viability marker, propidium iodide. Subsequently, gates for each marker were set using negative controls.
Total RNA isolation and quantitative PCR
Total RNA was isolated using the RNAqueous®-Micro RNA Isolation Kit (Ambion, Foster City, CA) and reverse-transcribed using Superscript™ III Reverse Transcriptase (Invitrogen, Carlsbad, CA) according to manufacturer's instructions. Real-time quantitative gene expression analyses were performed using the Taqman® gene expression assays (Applied Biosystems, Foster City, CA) using predesigned Taqman® probes for the genes of interest ordered from the manufacturer according to supplied instructions (Table 1).For each reaction, 0.5 μL of cDNA template was incubated with 5 μL Universal Master Mix, 0.5 μL 20× predesigned probe, and 4 μL water in one well of a MicroAmp™ Optical 96-well Reaction Plate (Applied Biosystems). The thermal cycling conditions were as follows: 50°C for 2 min, 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 1 min. Gene expression levels were normalized to the housekeeping gene 18S rRNA and transcript abundance of each fraction was compared with those in the unfractionated global population. For validation of microarrays, gene expression levels normalized to the housekeeping gene 18S rRNA were calibrated against a large stock of cDNA of an in-house undifferentiated human ES cell line [39].
Table 1.
List of Predesigned Taqman® Probes for QPCR (Applied Biosystems)
| Catalog Number | Gene Symbol | Description |
|---|---|---|
| Hs00742896_s1 | POU5F1 | Homo sapiens POU domain, class 5, transcription factor 1 transcript variant 1, mRNA |
| Hs00220998_m1 | GDF3 | Homo sapiens growth differentiation factor 3, mRNA |
| Hs00232144_m1 | LHX1 | Homo sapiens LIM homeobox 1, mRNA |
| Hs00240858_m1 | PAX2 | Homo sapiens paired box 2, mRNA |
| Hs00240913_m1 | WT1 | Homo sapiens Wilms' tumor 1, mRNA |
| Hs00377071_m1 | OSR1 | Homo sapiens odd skipped related 1 (Drosophila), mRNA |
| Hs00244943_m1 | MEOX1 | Homo sapiens mesenchyme homeobox 1, mRNA |
| Hs00361186_m1 | TWIST1 | Homo sapiens twist homolog 1, mRNA |
| Hs00156373_m1 | CD34 | Homo sapiens CD34 molecule, mRNA |
| Hs00171403_m1 | GATA4 | Homo sapiens GATA-binding protein 4, mRNA |
| Hs00172991_m1 | IKZF1 | Homo sapiens IKAROS family zinc finger 1 (Ikaros), mRNA |
| Hs00230962_m1 | FOXF1 | Homo sapiens forkhead box F1, mRNA |
| Hs00174344_m1 | CDH5 | Homo sapiens cadherin 5, type 2, (vascular endothelium), mRNA |
| Hs00246256_m1 | FST | Homo sapiens follistatin, mRNA |
| Hs00365539_m1 | TBX6 | Homo sapiens T-box 6, mRNA |
| Hs00231821_m1 | TCF15 | Homo sapiens transcription factor 15 (basic helix-loop-helix), mRNA |
RNA amplification and microarray analyses
Total RNA preparations (4 biological replicates of separate passage numbers for HES-4) with a minimum concentration of 20 ng/μL and an A260/A280 ratio of >1.8 were sent to the SRC Microarray facility at the University of Queensland for RNA amplification and microarray analyses. RNA samples were analyzed for integrity using the Agilent Bioanalyzer 2100 and an RNA Integrity Number (RIN) was obtained using the Agilent Expert 2100 software vB.02.02. Samples with a RIN score of above 8.5 were deemed suitable for array analyses. Subsequently, 200 ng of total RNA from each sample was amplified using the Illumina® TotalPrep™ RNA Amplification Kit (Ambion, Foster City, CA) according to the manufacturer's guidelines with a 14-h in vitro transcription step. The 1.5 μg of amplified cRNA per array was hybridized to Human-6 v2 BeadChip arrays according to the manufacturer's instructions and detected using Fluorolink Streptavidin-Cy3 (GE Healthcare Biosciences, Freiburg, Germany). BeadChip arrays were scanned using the BeadStation array scanner (Illumina, San Diego, CA) and raw image intensity values were compiled using the Illumina BeadStudio software v2.3.41. The raw array data was normalized in R, a language and environment for statistical computing and graphics (www.r-project.org) before being imported into GeneSpring GX v7.3.1 (Agilent Technologies, Santa Clara, CA) for visualization.
Cytospinning FACS-fractionated cells and immunofluorescent labeling
Fluorescence-activated cell sorter (FACS)-sorted cells were washed twice in PBS+ and transferred in a small volume of PBS+ to V-bottomed tubes containing 3 mL of 4% paraformaldehyde and incubated for 10 min at room temperature. Cells were then washed and cytospun onto poly-l-lysine-coated slides in the Shandon Cytospin® 4 cytocentrifuge at 7g for 10 min at low acceleration. Cytospun slides were then washed in a Coplin jar containing Milli-Q® H2O with shaking for 5 min before permeabilization with 0.1% Triton X-100 in PBS for 10 min at room temperature. Slides were then marked with a wax pen and blocked with 10% normal goat serum in PBS+ at room temperature for 30 min. WT1 (Dako, Carpinteria, CA) and PAX2 (Zymed Laboratories, San Francisco, CA) antibodies diluted with blocking buffer were added to the slides and incubated for 1 h at room temperature or at 4°C overnight. Following 2 washes in PBS+, isotype-matched secondary antibodies (Invitrogen, Carlsbad, CA) diluted in blocking buffer were added and allowed to incubate for 30 min at room temperature. Slides were protected from the light from this step onward to prevent photobleaching. After incubation, five 2-min washes were performed as above and cells were labeled with DAPI (Invitrogen, CA) diluted in PBS+ and mounted in Vectashield® (Vector Laboratories, Burlingame, CA).
Statistical analyses
Statistical analyses were performed using the SigmaStat 3.1 Software. The differences between 2 means were nonparametrically analyzed using the Mann–Whitney rank sum test. Values were considered to be significantly different when the achieved significance level (P value) was ≤0.05 (denoted by *).
Results
Human ES cell-derived renal structures form in teratomas
Identifying renal cells from a mixture of other cell types in a human ES cell-derived teratoma requires the use of markers indicative of cell types of the kidney. To this aim, we initially carried out immunological studies in first trimester human embryo sections. Immunoreactivity for WT1, cadherin 16, and podocalyxin was observed in derivatives of both the UB and the MM (Fig. 1A–1F) although of these WT1 and podocalyxin were also found in other tissues both in the developing embryo and the adult. Cadherin 16 expression, although specific to the adult kidney, has been reported in the mouse sex duct and lung [40]. However, we did not observe any immunoreactivity to cadherin 16 in human embryonic sex ducts or lungs, suggesting that in the human, cadherin 16 expression may be restricted to cells of the kidney.
FIG. 1.
Immunohistochemical analyses of human embryo sections (A–F) and kidney-like structures within teratomas (G–J) formed from hESC injected into SCID mice. a. Staining of 8-week embryonic kidney for WT1 revealed cytoplasmic staining of podocytes (arrow). (B) Isotype-matched negative control for A. (C and D) Staining of 7-week embryonic kidney with cadherin 16 showed immunoreactivity within nephric duct (black arrow) and MM-derived (white arrow) structures. (E) Isotype-matched control for C and D. (F) Podocalyxin was localized to nephric duct (black arrow) and MM-derived (white arrow) structures within the 9.5-week human embryonic kidney. (G) Cadherin 16 staining of a duct-like structure within a teratoma. (H) WT1 immunoreactivity on rare cells within a teratoma. (I) Podocalyxin staining of a glomerular-like structure found in a teratoma and the epithelial tubule, possibly a proximal tubule (white arrow). (J) Isotype-matched negative control for I. Scale bar = 100 μm.
These markers were then used to investigate the ability of human ES cells to differentiate to cells of the nephric lineage in teratomas following ES cell injection into the testis capsule of SCID mice. Teratomas derived from human ES cells typically contain a variety of tissues such as muscle, neural tissue, and various forms of epithelia, with some showing a high degree of tissue-specific organization [23,24]. However, such structures are usually immature and therefore definitively identifying the tissues present in such lesions can be difficult. In order to identify areas of potential renal tissue, teratomas were examined microscopically for structures indicative of the kidney. After identification of the rare putative regions of renal tissue, serial sections were then stained with epithelial or renal markers (Fig. 1G—1J). Structures reminiscent of renal tubules and collecting ducts marked by cadherin 16 were observed (Fig. 1G). WT1 was also localized to nonorganized regions of the teratoma (Fig. 1H). Glomerular-like structures were observed in the teratoma and displayed a remarkable degree of organization as evidenced by Figure 1I and 1J. This glomerular-like structure stained positively for podocalyxin and demonstrated a structure resembling a renal corpuscle with a glomerulus (with podocytes), urinary space, Bowman's parietal epithelium, and a urinary pole extending into the proximal tubule (white arrow). The formation of glomerular-like structures immunoreactive for the podocyte marker (in this context) podocalyxin alongside the cadherin 16 immunoreactive tubule-like structure provides strong evidence that human ES cells in a teratoma are capable of forming organized kidney-like structures. However, these structures were exceedingly rare and their formation was time-consuming (2–3 months duration). Therefore, in vitro strategies for the production of renal cells were explored.
Enrichment of Human ES-derived renal cells by FACS selection and characterization by gene expression analysis
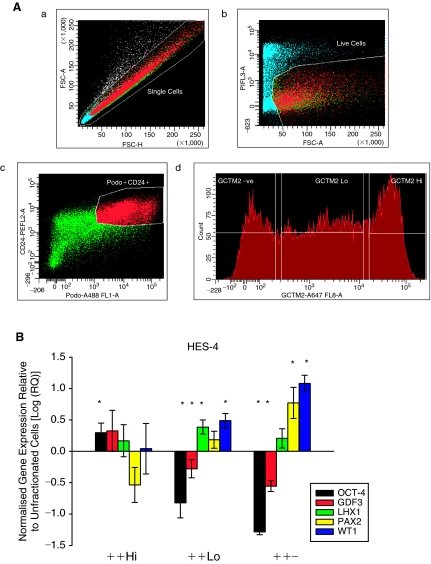
The majority of the 26 or more cell types of the adult kidney arise primarily from the MM [28]. Therefore, the differentiation strategy employed aimed to produce cells of this phenotype. Reduction of serum concentration was selected as it has previously been used to induce human ES cell differentiation toward the pancreatic and cardiomyocyte lineages [34,36], suggesting selection for mesendodermal tissues. Furthermore, we consistently observe more marked differentiation in colonies of human ES cells cultured on a reduced density of feeder cells (unpublished). However, inducing human ES cells to differentiate under these culture conditions results in a heterogeneous population of cells comprised of cells of interest interspersed with a myriad of other cell types. We therefore sought to enrich for renal cells using FACS selection. Two cell surface markers, CD24 and podocalyxin, were selected for FACS based on microarray and in situ hybridization assays performed by Challen and colleagues [30]. In the latter study, CD24 was strongly up-regulated in the uninduced murine MM at E10.5 while podocalyxin was highly expressed in MM and the surrounding intermediate mesoderm, which may play a crucial role in directing nephrogenesis of the uninduced MM. These markers are not renal-specific and CD24 and podocalyxin transcripts were also detectable in highly purified human ES cell populations [41,42] and some differentiated cell types. Therefore, a well-characterized marker of pluripotency, GCTM2 [24], was included to discriminate between undifferentiated (GCTM2+) and differentiated (GCTM2Low, GCTM2−) cells within the global population. While podocalyxin and GCTM2 have previously been reported to be identical [43], we do not believe this to be the case [41]. Live cells showing double-positive immunoreactivity to CD24 and podocalyxin were fractionated into populations according to expression level of GCTM2 and denoted ++Hi, ++Low, and ++−, respectively (Fig. 2A). These fractions were then analyzed for gene expression by RT-quantitative PCR.
FIG. 2.
Cell-sorting strategy. (A) Representative FACS plots showing viable, differentiated human embryonic stem (ES) cells fractionated based on co-expression of CD24, podocalyxin, and varying levels of GCTM2 (denoted ++Hi, ++Low, and ++−). (a) Cells were first distinguished based on forward and side scatter and (b) dead cells were excluded based on the uptake of the viability marker propidium iodide. (d) Cells that co-expressed CD24 and podocalyxin were further gated based on GCTM2 expression resulting in 3 populations. (B) Quantitative gene expression analysis indicates that when compared with unfractionated cells, the ++Low and ++− fractions showed a trend of increased expression of kidney development genes LHX1, PAX2, and WT1 and reduced expression of pluripotent genes OCT4 and GDF3. * indicates a statistically significant difference. Values are presented as means ± standard error of the mean (SEM), n = 5.
Figure 2B shows that in differentiated cultures of the cell line HES-4, expression of the pluripotency genes OCT4 and GDF3 were down-regulated in both the ++Low and ++− fractions compared with the unfractionated population. Conversely, transcripts of PAX2 and WT1 were up-regulated (P ≤ 0.05) in the ++− fraction. Expression of LHX1 was significantly increased in the ++Low but not in the ++− fraction. Similar trends in gene expression were also observed when human ES cell populations were fractionated postinduction from 2 other human ES cell lines tested, HES-2 and HES-3 (Supplementary Fig. 1; Supplementary materials are available online at www.liebertonline.com/scd).
Microarray analysis of FACS-separated fractions show up-regulation of kidney-associated genes in the differentiated populations
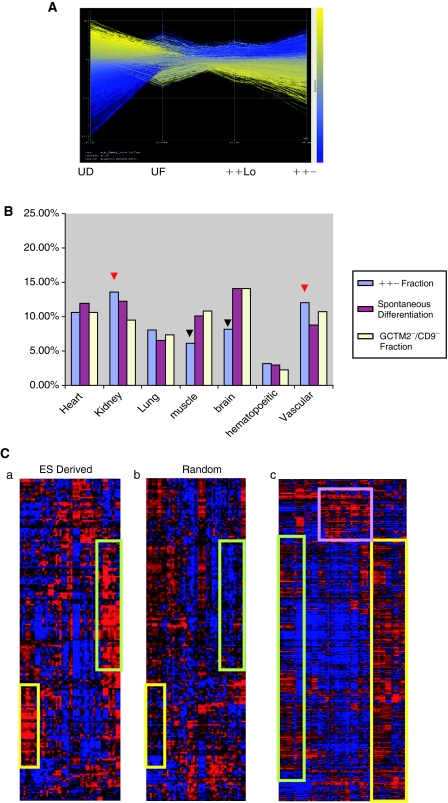
While we detected an up-regulation in the levels of PAX2, WT1, and LHX1 in the differentiated fractions, we recognize that these markers individually are not specific for renal cells. As such, to further confirm the presence of potential renal progenitors in our differentiated human ES cell population, we utilized microarray analysis to examine the global gene expression profile of the FACS-separated fractions. RNA from 4 biological replicates of each fraction was isolated and analyzed by Illumina microarray from the following fractions of HES-4 cells (n = 4 passages); unfractionated, ++Low, ++−, and undifferentiated ES cells of the same passage. The gene expression patterns determined by array analysis were then clustered according to their involvement in pluripotency (yellow) or differentiation and development (blue) and represented on a heat map (Fig. 3A). The heat map shows that genes involved in development are clearly up-regulated as the fractions move from undifferentiated to unfractionated, ++Low, and ++− fractions, while the reverse is true for stem cell genes. In addition, results were subjected to gene ontology and cluster analysis to characterize the fractions (full data sets are available at GEO accession number GSE15257). Initially, the top 1,000 up-regulated genes in the ++− fraction relative to the undifferentiated ES cells of the same passage were compared with markers of development obtained from GO (www.geneontology.org). To ensure that the bioinformatic readout obtained was representative of the differentiation and isolation regime, ontology analysis on the populations isolated here was carried out in parallel with analysis of previously obtained microarray results from spontaneously differentiated human ES cell-derived populations [42]. The results of the gene ontology analyses indicate a higher percentage of renal development genes in the ++− fraction compared with any other lineage investigated (Fig. 3B). While this increase does not appear significant when compared with the spontaneously differentiated or GCTM2−/CD9− fractions, it is interesting to note that genes associated with muscular and neural development appear to be present at a lesser proportion in the ++− fraction than either of the other 2 analyzed populations.
FIG. 3.
Microarray analyses of undifferentiated, unfractionated, and sorted fractions. (A) Heat maps of undifferentiated, unfractionated, and sorted fractions. Combined heat map of 3 biological replicates shows down-regulation of stem cell genes (yellow) and up-regulation of developmental genes (blue) across the undifferentiated (UD), unfractionated (UF), +Low, and ++− fractions. All 3 replicates show similar expression patterns of pluripotent and developmental genes confirming the reproducibility of the array results. (B) Gene ontology analyses performed on the top 1,000 up-regulated genes (B-stat > 0) in the ++− compared with the undifferentiated fraction show a slight increase in percentage of kidney and vascular development-associated genes in the ++− fraction relative to either spontaneously differentiated embryonic stem (ES) cells or cell populations derived from varying stem cell marker expression-based fractionation (red arrowheads). Ontology analysis also indicated a reduced proportion of genes involved in neural and skeletal muscle development in the ++− cell fraction (black arrowheads). (C) Cluster analyses. (a) Cluster analysis of the top 200 up-regulated genes in the ++− fraction compared with undifferentiated ES cells cross-matched with a genitourinary development-specific gene expression database (GUDMAP) shows an overrepresentation of transcripts associated with E11.5 murine metanephric interstitium (green box) and E15.5 nephrogenic and cortical interstitium (yellow box). (b) This overrepresentation was not observed when a randomly generated list of genes was subjected to the same cross-match. (c) Cluster analysis performed on all genes up-regulated only in ++− compared with undifferentiated, spontaneously differentiated, and ++Low fractions shows these genes are associated with the MM at E11.5 (green box), podocytes at E13.5 and E15.5 (yellow box), and the collecting duct and proximal tubules (purple box) when cross-matched with the GUDMAP database.
In order to determine if up-regulated transcripts in the ++− fraction are associated with a particular compartment of the developing kidney or a specific time point of kidney development, a bioinformatic cluster analysis was performed. Expression levels of 200 of the most differentially up-regulated genes in the ++− fraction relative to the undifferentiated ES cells of the same passage were compared with a transcriptome profiling database specific to urogenital development obtained from the Genitourinary Development Molecular Anatomy Project (GUDMAP) [44]. Genes were clustered based on spatial and temporal expression within the kidney. Figure 3C shows that these up-regulated genes cluster into a group of genes associated with the MM at E11.5 (prior to nephron formation) and the cortical and nephrogenic interstitium at E15.5 of murine renal development. This suggests selection for a MM-like population of cells. Conversely, this clustering pattern was not observed when a randomly selected list of genes was compared with the same gene expression database. As this comparison was performed with cells of the ++− fraction and undifferentiated ES cells, it is possible that this may be due to spontaneous differentiation and not the result of enrichment by our sorting strategy. Therefore, we further identified genes selectively up-regulated in the ++− fraction compared with the ++Low, spontaneously differentiated (GCTM2−/CD9−), and undifferentiated ES fractions. This list of genes was then cross-matched again with those from GUDMAP [44] (Supplementary Table 1; Supplementary materials are available online at www.liebertonline.com/scd). Results from this second comparison show that genes that are up-regulated only in the ++− fraction can indeed be clustered into a group associated with the MM at E11.5 (Fig. 3C). It is likely that this finding was not observed in our initial gene ontology analysis as GO utilizes general markers of kidney development and provides an overview compared with GUDMAP that focuses specifically on discrete stages of kidney development. This result both validates the sorting and differentiation strategy and corroborates the predicted markers [30]. Furthermore, there appears to be a cluster of genes associated with podocytes at E13.5 and E15.5, the collecting duct, and proximal tubules (Fig. 3C).
Validation of microarrays by RT-QPCR
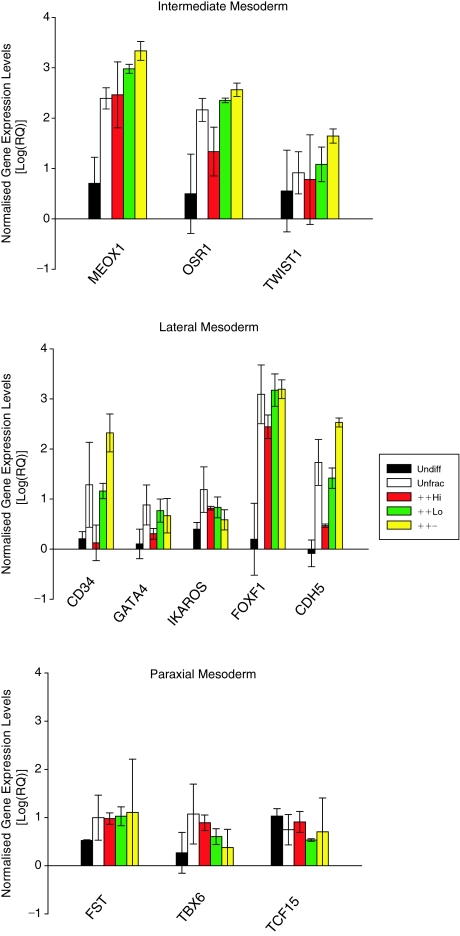
RNA from the same samples used in the microarray analyses were reverse-transcribed and analyzed by RT-QPCR for the expression of genes involved in development of various mesoderm compartments. Results of the quantitative PCR are expressed on a logarithmic scale and show that genes expressed in all 3 mesoderm compartments are up-regulated in the 2-week differentiated cells compared with the calibrator human ES cell line. Genes associated with the intermediate mesoderm were also slightly up-regulated in the ++Low and ++− fractions compared with both the unfractionated and ++Hi populations (Fig. 4). Notably, this pattern of increased expression in the ++Low and ++− fractions was also observed for genes involved in lateral mesoderm development to a more pronounced extent (Fig. 4). Conversely, transcript levels of paraxial mesoderm genes were detected at lower levels than those of the other mesoderm compartments and furthermore did not show much variation in expression between the 4 analyzed fractions (Fig. 4).
FIG. 4.
Validation of microarray results by quantitative polymerase chain reaction (PCR). Results of the microarray were validated by quantitative PCR screen of genes involved in the development of each compartment of the mesoderm where expression levels were first normalized against 18S rRNA and calibrated using a CD30-expressing human embryonic stem (ES) cell line as an internal standard. The QPCR results demonstrated a slight up-regulation in transcript levels of all 3 intermediate mesoderm and 2 lateral mesoderm genes, CD34 and CDH5, in the ++− fraction compared with the other 3 fractions, indicating that cells expressing these transcripts are enriched from the unfractionated cell population by sorting based on CD24+/Podo+/GCTM2−. Conversely, an overall lower level of gene expression was observed for paraxial mesoderm genes, which also showed little variation in transcript abundance across the cellular fractions, indicating that the cells that express these genes are not selectively isolated by sorting with the above markers. Values are presented as means ± SEM, n = 3.
Detection of WT1 and PAX2 co-expressing cells by immunolocalization confirms the presence of renal cells in FACS-fractionated cells
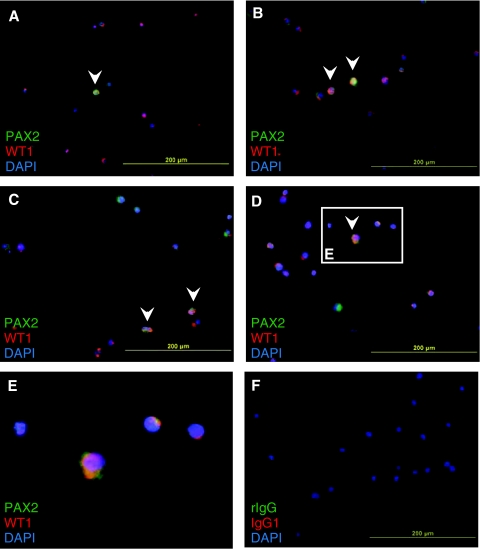
While RT-QPCR and transcriptome analyses provided evidence of the presence of MM-like cells in the sorted fractions, these were performed on heterogeneous populations of cells. In order to exclude the possibility that the observed up-regulation of kidney-related genes is due to increased gene expression in distinct cell populations contained within the FACS-separated cell populations, we performed indirect immunofluorescent analysis of WT1 and PAX2 expression on FACS-separated 2-week differentiated human ES cells. To avoid cross-reactivity of the secondary antibodies to monoclonal antibodies of identical isotypes, differentiated human ES cells were sorted using GCTM2 alone instead of in combination with CD24 and podocalyxin to yield GCTM2− cells that contain the ++− cells of interest before being labeled with WT1 and PAX2. A subset of GCTM2−/PAX2+/WT1+ cells was detectable (Fig. 5). In addition, GCTM2− cells that labeled positive for either PAX2 or WT1 alone could also be seen in these fractionated cells.
FIG. 5.
Detection of a population of WT1+/PAX2+ cells. (A–D) Four fields of view showing the presence of a rare population of cells that co-localize WT1 and PAX2 in GCTM2− cell fractions (arrow). (E) Magnified image of a cell co-expressing WT1 and PAX2. (F) Corresponding nuclear stain for the isotype control.
Discussion
In this study, we demonstrate that it is possible to generate renal cells from human ES cells in vitro. Initially, we established that the markers WT1, cadherin 16, and podocalyxin recognize structures derived from the MM and UB in the developing human embryo. While the pluripotent nature of human ES cells is well accepted, to date there have been 2 reports of the formation of kidney-like structures from human ES cell-derived teratomas [23, 25]. Only one of these reports presented immunohistochemistry for markers of renal structures (WT1 and NCAM) [25]. We have confirmed and extended these findings using the renal markers WT1, cadherin 16, and podocalyxin and show that human ES cells indeed have the ability to differentiate into structures resembling fetal glomeruli. In addition, we also extend these results to validate that the structures observed in the teratomas are of the renal lineage.
In order to achieve the long-term goal of utilizing human ES cells in cell replacement therapy for renal disease, methods for obtaining renal cells from human ES cells in a controlled and reproducible manner are required. To date, there has been only one study involving the directed differentiation of human ES cells toward the renal lineage, which indicated that it is possible to detect an up-regulation of MM-related transcripts (OSR1, PAX2, SIX2, and WT1) and genes associated with kidney precursors (EYA1, LIM1, and CD24) in human ES cell colonies cultured in the presence of retinoic acid, activin A, and BMP4 or BMP7 on laminin or gelatin substrates [27]. In the present study, we employed a strategy of serum concentration reduction and feeder depletion to induce differentiation of human ES cells. As noted, serum reduction in combination with growth factors such as activin A [35,36] or in a co-culture system [34] has previously been used to differentiate human ES cells toward the cardiomyocyte and pancreatic lineages, respectively. In the case of endoderm differentiation, this effect at least partially relies on the reduction of insulin-like growth factor (IGF), usually present in the serum, which is thought to antagonize differentiation of human ES cells via the activation of the phosphotidylinositol-3-kinase (PI3K) signaling pathway [45]. While the effect of this pathway on differentiation toward other lineages has not been investigated, we speculate that similar factors are present in serum and either have an inhibitory effect on differentiation, a stimulatory effect on the maintenance of pluripotency, or both. As with most protocols involving human ES cell differentiation, the resulting cell population represented a heterogeneous mixture of cell types. The markers CD24, podocalyxin, and GCTM2 were selected for use in FACS to isolate renal cells from this mixture of cells. Individually, neither CD24 nor podocalyxin are restricted in their expression to the developing kidney. However, using them in combination with GCTM2 exclusion enables a subset of differentiated cells containing the renal precursor cells of interest to be isolated from a global heterogeneous population of differentiated and pluripotent cells. We therefore isolated 3 fractions of cells CD24+/podocalyxin+/GCTM2Hi (++Hi), CD24+/podocalyxin+/GCTM2Low (++Low), and CD24+/podocalyxin+/GCTM2-(++−). While each of these fractions are still heterogeneous, we hypothesized that the ++Hi fraction would contain the highest proportion of pluripotent cells, the ++Low fraction contains cells that have begun to commit to a mesoderm fate, while the ++− fraction contains the renal cells of interest. We confirmed this hypothesis using quantitative gene expression analysis and showed that the ++Low and ++− fractions have indeed proceeded along the pathway of differentiation based on their down-regulation of the stemness genes, OCT4 and GDF3. With particular regards to HES-4, the observed up-regulation of LHX1 transcripts in the ++Low but not the ++− fraction lends further weight to the hypothesis that this fraction contains less differentiated cells than ++− since Lim1 is critical for intermediate mesoderm formation in the mouse. While the quantitative PCR readout indicated that HES-4 is the cell line that is most amenable to the differentiation regime, similar trends in gene expression were observed in the other 2 human ES cell lines (Supplementary Table 1).This variation in responsiveness of different cell lines is not uncommon and has been reported with differentiation protocols aimed at obtaining pancreatic endocrine cells from human ES cells [46]. Notably, these gene expression results are in line with reports by D'Amour and colleagues (2005) who showed that culture of human ES cells for 4 days in the presence of low serum results in high expression of mesoderm genes.
As organogenesis is a complex process that involves the expression of a multitude of genes, evidence of lineage commitment requires larger-scale characterization via transcriptome profiling. Our microarrays provided strong evidence of a MM-like population within the FACS-isolated fractions of differentiated human ES cells. From our gene ontology results, the observed up-regulation of kidney-associated genes and the significant decrease in transcripts associated with neural and muscular development in the ++− fraction relative to spontaneously differentiated human ES cells is particularly encouraging, as the induction of neural differentiation is a default pathway that occurs autonomously when inhibitory signals are removed [47]. With respect to muscular lineages, while it appears that the sorting strategy may also be selecting against cells that have differentiated along muscular lineages, the ontology analysis for markers of muscle development includes genes associated with skeletal, cardiac, visceral, and smooth muscles based on cluster and gene ontology analyses. The efficacy of our sorting strategy is also promising based on results of our cluster analysis, which show that the ++− fraction contains cells that have an increased expression of genes associated with specific compartments of the kidney during development.
Overall, our results suggest the selection of a population enriched in cells adopting a phenotype derived from intermediate mesoderm, the source tissue for the kidney. In addition, this fraction showed differential expression of markers previously associated with the MM. As noted previously, this tissue gives rise to both the nephrons and components of the renal interstitium. The former target is the most critical for any potential renal regenerative option. There are many known transcription factors that differentiate between the MM fated to become nephron versus interstitium and while this differentiation and enrichment strategy showed up-regulation of MM markers, this did not include those specifically marking the nephron progenitors. For the MM to undergo a mesenchymal to epithelial transition to form a nephron, the cells must co-express WT1 and PAX2. While expression of each of these markers individually have been reported in a variety of nonrenal cell types, their co-expression during embryonic development is a strong indicator of nephron progenitor activity [22]. We identified a small population of WT1+PAX2+ cells within the ++− fraction suggesting that such progenitors were being induced at a low rate. What will be critical now is to optimize further selective strategies for the enrichment or selective culture of that nephron progenitor subfraction. Such a population would be invaluable as a source for the terminal differentiation of mature renal cells from human ES cells for toxicology screening and potentially for renal therapy.
Supplementary Material
Acknowledgments
The authors thank Dr. David Nikolic-Paterson for providing the podocalyxin antibody (PHM5), Genevieve Browne and Stephanie Wood for technical assistance with immunohistochemistry, the Human Embryonic Stem Cell Core Facility at the Australian Stem Cell Centre (ASCC) for provision of cells, and FlowCore (a collaborative initiative between Monash University, the ASCC, and the Australian Regenerative Medicine Institute [ARMI]) for assistance with FACS. This work was supported by grants from the Australian National Health & Medical Research Committee (ALL), Kidney Health Australia (ALL) and from the National Institute for Diabetes, Digestion and Kidney Disease, National Institutes of Health (DK63400 to SG, MHL, MFP, SDR, and JFB) as part of the Stem Cell Genome Anatomy Project (www.scgap.org). SAL was supported by a postgraduate scholarship from Kidney Health Australia.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Erceg S. Laínez S. Ronaghi M. Stojkovic P. Pérez-Aragó MA. Moreno-Manzano V. Moreno-Palanques R. Planells-Cases R. Stojkovic M. Differentiation of human embryonic stem cells to regional specific neural precursors in chemically defined medium conditions. PLoS ONE. 2008;3:e2122. doi: 10.1371/journal.pone.0002122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen MA. Itsykson P. Reubinoff BE. Neural differentiation of human ES cells. Curr Protoc Cell Biol. 2007;36:23.7.1–23.7.20. doi: 10.1002/0471143030.cb2307s36. [DOI] [PubMed] [Google Scholar]
- 3.Ferreira LS. Gerecht S. Shieh HF. Watson N. Rupnick MA. Dallabrida SM. Vunjak-Novakovic G. Langer R. Vascular progenitor cells isolated from human embryonic stem cells give rise to endothelial and smooth muscle like cells and form vascular networks in vivo. Circ Res. 2007;101:286–294. doi: 10.1161/CIRCRESAHA.107.150201. [DOI] [PubMed] [Google Scholar]
- 4.Nakagami H. Nakagawa N. Takeya Y. Kashiwagi K. Ishida C. Hayashi S. Aoki M. Matsumoto K. Nakamura T. Ogihara T. Morishita R. Model of vasculogenesis from embryonic stem cells for vascular research and regenerative medicine. Hypertension. 2006;48:112–119. doi: 10.1161/01.HYP.0000225426.12101.15. [DOI] [PubMed] [Google Scholar]
- 5.Karner E. Bäckesjö CM. Cedervall J. Sugars RV. Ahrlund-Richter L. Wendel M. Dynamics of gene expression during bone matrix formation in osteogenic cultures derived from human embryonic stem cells in vitro. Biochim Biophys Acta. 2009;1790:110–118. doi: 10.1016/j.bbagen.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Kärner E. Linger C. Sloan AJ. Ahrlund-Richter L. Sugars RV. Wendel M. Bone matrix formation in osteogenic cultures derived from human embryonic stem cells in vitro. Stem Cells Dev. 2007;16:39–52. doi: 10.1089/scd.2006.0010. [DOI] [PubMed] [Google Scholar]
- 7.Lee KW. Yook JY. Son MY. Kim MJ. Koo DB. Han YM. Cho YS. Rapamycin promotes the osteoblastic differentiation of human embryonic stem cells by blocking the mTOR pathway and stimulating the BMP/Smad pathway. Stem Cells Dev. 2009;19:557–568. doi: 10.1089/scd.2009.0147. [DOI] [PubMed] [Google Scholar]
- 8.Bu L. Jiang X. Martin-Puig S. Caron L. Zhu S. Shao Y. Roberts DJ. Huang PL. Domian IJ. Chien KR. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460:113–117. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- 9.Tran TH. Wang X. Browne C. Zhang Y. Schinke M. Izumo S. Burcin M. Wnt3a-induced mesoderm formation and cardiomyogenesis in human embryonic stem cells. Stem Cells. 2009;27:1869–1878. doi: 10.1002/stem.95. [DOI] [PubMed] [Google Scholar]
- 10.Borowiak M. Maehr R. Chen S. Chen AE. Tang W. Fox JL. Schreiber SL. Melton DA. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell. 2009;4:348–358. doi: 10.1016/j.stem.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Hoof D. D'Amour KA. German MS. Derivation of insulin-producing cells from human embryonic stem cells. Stem Cell Res. 2009;3:73–87. doi: 10.1016/j.scr.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Sasaki K. Ichikawa H. Takei S. No HS. Tomotsune D. Kano Y. Yokoyama T. Sirasawa S. Mogi A. Yoshie S. Sasaki S. Yamada S. Matsumoto K. Mizuguchi M. Yue F. Tanaka Y. Hepatocyte differentiation from human ES cells using the simple embryoid body formation method and the staged-additional cocktail. Scientific World Journal. 2009;9:884–890. doi: 10.1100/tsw.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Synnergren J. Heins N. Brolen G. Eriksson G. Lindahl A. Hyllner J. Olsson B. Sartipy P. Bjorquist P. Transcriptional profiling of human embryonic stem cells differentiating to definitive and primitive endoderm and further towards the hepatic lineage. Stem Cells Dev. 2009 doi: 10.1089/scd.2009.0220. [DOI] [PubMed] [Google Scholar]
- 14.Ji L. Allen-Hoffmann BL. de Pablo JJ. Palecek SP. Generation and differentiation of human embryonic stem cell-derived keratinocyte precursors. Tissue Eng. 2006;12:665–679. doi: 10.1089/ten.2006.12.665. [DOI] [PubMed] [Google Scholar]
- 15.Metallo CM. Ji L. de Pablo JJ. Palecek SP. Retinoic acid and bone morphogenetic protein signaling synergize to efficiently direct epithelial differentiation of human embryonic stem cells. Stem Cells. 2008;26:372–380. doi: 10.1634/stemcells.2007-0501. [DOI] [PubMed] [Google Scholar]
- 16.Clark AT. Bodnar MS. Fox M. Rodriquez RT. Abeyta MJ. Firpo MT. Pera RA. Spontaneous differentiation of germ cells from human embryonic stem cells in vitro. Hum Mol Genet. 2004;13:727–739. doi: 10.1093/hmg/ddh088. [DOI] [PubMed] [Google Scholar]
- 17.Park TS. Galic Z. Conway AE. Lindgren A. van Handel BJ. Magnusson M. Richter L. Teitell MA. Mikkola HK. Lowry WE. Plath K. Clark AT. Derivation of primordial germ cells from human embryonic and induced pluripotent stem cells is significantly improved by coculture with human fetal gonadal cells. Stem Cells. 2009;27:783–795. doi: 10.1002/stem.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu RH. Chen X. Li DS. Li R. Addicks GC. Glennon C. Zwaka TP. Thomson JA. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20:1261–1264. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- 19.Xu RH. In vitro induction of trophoblast from human embryonic stem cells. Methods Mol Med. 2006;121:189–202. doi: 10.1385/1-59259-983-4:187. [DOI] [PubMed] [Google Scholar]
- 20.Steenhard BM. Isom KS. Cazcarro P. Dunmore JH. Godwin AR. St John PL. Abrahamson DR. Integration of embryonic stem cells in metanephric kidney organ culture. J Am Soc Nephrol. 2005;16:1623–1631. doi: 10.1681/ASN.2004070584. [DOI] [PubMed] [Google Scholar]
- 21.Kim D. Dressler GR. Nephrogenic factors promote differentiation of mouse embryonic stem cells into renal epithelia. J Am Soc Nephrol. 2005;16:3527–3534. doi: 10.1681/ASN.2005050544. [DOI] [PubMed] [Google Scholar]
- 22.Vigneau C. Polgar K. Striker G. Elliott J. Hyink D. Weber O. Fehling HJ. Keller G. Burrow C. Wilson P. Mouse embryonic stem cell-derived embryoid bodies generate progenitors that integrate long term into renal proximal tubules in vivo. J Am Soc Nephrol. 2007;18:1709–1720. doi: 10.1681/ASN.2006101078. [DOI] [PubMed] [Google Scholar]
- 23.Thomson JA. Itskovitz-Eldor J. Shapiro SS. Waknitz MA. Swiergiel JJ. Marshall VS. Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 24.Reubinoff BE. Pera MF. Fong CY. Trounson A. Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat B iotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- 25.Gertow K. Wolbank S. Rozell B. Sugars R. Andäng M. Parish CL. Imreh MP. Wendel M. Ahrlund-Richter L. Organized development from human embryonic stem cells after injection into immunodeficient mice. Stem Cells Dev. 2004;13:421–435. doi: 10.1089/scd.2004.13.421. [DOI] [PubMed] [Google Scholar]
- 26.Schuldiner M. Yanuka O. Itskovitz-Eldor J. Melton DA. Benvenisty N. Effects of eight growth factors on the differentiation of cells derived from human embryonic stem cells. Proc Natl Acad Sci USA. 2000;97:11307–11312. doi: 10.1073/pnas.97.21.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Batchelder CA. Lee CC. Matsell DG. Yoder MC. Tarantal AF. Renal ontogeny in the rhesus monkey (Macaca mulatta) and directed differentiation of human embryonic stem cells towards kidney precursors. Differentiation. 2009;78:45–56. doi: 10.1016/j.diff.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saxen L. Organogenesis of the Kidney. Cambridge University Press; Cambridge: 1987. [Google Scholar]
- 29.Rumballe B. Georgas K. Wilkinson L. Little M. Molecular anatomy of the kidney: what have we learned from gene expression and functional genomics? Pediatr Nephrol. 2010;25:1005–1016. doi: 10.1007/s00467-009-1392-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Challen GA. Martinez G. Davis MJ. Taylor DF. Crowe M. Teasdale RD. Grimmond SM. Little MH. Identifying the molecular phenotype of renal progenitor cells. J Am Soc Nephrol. 2004;15:2344–2357. doi: 10.1097/01.ASN.0000136779.17837.8F. [DOI] [PubMed] [Google Scholar]
- 31.Torres M. Gómez-Pardo E. Dressler GR. Gruss P. Pax-2 controls multiple steps of urogenital development. Development. 1995;121:4057–4065. doi: 10.1242/dev.121.12.4057. [DOI] [PubMed] [Google Scholar]
- 32.Kreidberg JA. Sariola H. Loring JM. Maeda M. Pelletier J. Housman D. Jaenisch R. WT-1 is required for early kidney development. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- 33.Carroll TJ, editor; McMahon AP, editor. Overview: The Molecular Basis of Kidney Development. Academic Press; London: 2003. [Google Scholar]
- 34.Passier R. Oostwaard DW. Snapper J. Kloots J. Hassink RJ. Kuijk E. Roelen B. de la Riviere AB. Mummery C. Increased cardiomyocyte differentiation from human embryonic stem cells in serum-free cultures. Stem Cells. 2005;23:772–780. doi: 10.1634/stemcells.2004-0184. [DOI] [PubMed] [Google Scholar]
- 35.D'Amour KA. Agulnick AD. Eliazer S. Kelly OG. Kroon E. Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 36.D'Amour KA. Bang AG. Eliazer S. Kelly OG. Agulnick AD. Smart NG. Moorman MA. Kroon E. Carpenter MK. Baetge EE. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 37.Adewumi O. Aflatoonian B. Ahrlund-Richter L. Amit M. Andrews PW. Beighton G. Bello PA. Benvenisty N. Berry LS. Bevan S. Blum B. Brooking J. Chen KG. Choo AB. Churchill GA. Corbel M. Damjanov I. Draper JS. Dvorak P. Emanuelsson K. Fleck RA. Ford A. Gertow K. Gertsenstein M. Gokhale PJ. Hamilton RS. Hampl A. Healy LE. Hovatta O. Hyllner J. Imreh MP. Itskovitz-Eldor J. Jackson J. Johnson JL. Jones M. Kee K. King BL. Knowles BB. Lako M. Lebrin F. Mallon BS. Manning D. Mayshar Y. McKay RD. Michalska AE. Mikkola M. Mileikovsky M. Minger SL. Moore HD. Mummery CL. Nagy A. Nakatsuji N. O'Brien CM. Oh SK. Olsson C. Otonkoski T. Park KY. Passier R. Patel H. Patel M. Pedersen R. Pera MF. Piekarczyk MS. Pera RA. Reubinoff BE. Robins AJ. Rossant J. Rugg-Gunn P. Schulz TC. Semb H. Sherrer ES. Siemen H. Stacey GN. Stojkovic M. Suemori H. Szatkiewicz J. Turetsky T. Tuuri T. van den Brink S. Vintersten K. Vuoristo S. Ward D. Weaver TA. Young LA. Zhang W International Stem Cell Initiative. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- 38.Pera MF. Filipczyk AA. Hawes SM. Laslett AL. Isolation, characterization, and differentiation of human embryonic stem cells. Meth Enzymol. 2003;365:429–446. doi: 10.1016/s0076-6879(03)65030-5. [DOI] [PubMed] [Google Scholar]
- 39.Herszfeld D. Wolvetang E. Langton-Bunker E. Chung TL. Filipczyk AA. Houssami S. Jamshidi P. Koh K. Laslett AL. Michalska A. Nguyen L. Reubinoff BE. Tellis I. Auerbach JM. Ording CJ. Looijenga LH. Pera MF. CD30 is a survival factor and a biomarker for transformed human pluripotent stem cells. Nat Biotechnol. 2006;24:351–357. doi: 10.1038/nbt1197. [DOI] [PubMed] [Google Scholar]
- 40.Wertz K. Herrmann BG. Kidney-specific cadherin (cdh16) is expressed in embryonic kidney, lung, and sex ducts. Mech Dev. 1999;84:185–188. doi: 10.1016/s0925-4773(99)00074-x. [DOI] [PubMed] [Google Scholar]
- 41.Laslett AL. Grimmond S. Gardiner B. Stamp L. Lin A. Hawes SM. Wormald S. Nikolic-Paterson D. Haylock D. Pera MF. Transcriptional analysis of early lineage commitment in human embryonic stem cells. BMC Dev Biol. 2007;7:12. doi: 10.1186/1471-213X-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolle G. Ho M. Zhou Q. Chy HS. Krishnan K. Cloonan N. Bertoncello I. Laslett AL. Grimmond SM. Identification of human embryonic stem cell surface markers by combined membrane-polysome translation state array analysis and immunotranscriptional profiling. Stem Cells. 2009;27:2446–2456. doi: 10.1002/stem.182. [DOI] [PubMed] [Google Scholar]
- 43.Schopperle WM. Kershaw DB. DeWolf WC. Human embryonal carcinoma tumor antigen, Gp200/GCTM-2, is podocalyxin. Biochem Biophys Res Commun. 2003;300:285–290. doi: 10.1016/s0006-291x(02)02844-9. [DOI] [PubMed] [Google Scholar]
- 44.Brunskill EW. Aronow BJ. Georgas K. Rumballe B. Valerius MT. Aronow J. Kaimal V. Jegga AG. Yu J. Grimmond S. McMahon AP. Patterson LT. Little MH. Potter SS. Atlas of gene expression in the developing kidney at microanatomic resolution. Dev Cell. 2008;15:781–791. doi: 10.1016/j.devcel.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McLean AB. D'Amour KA. Jones KL. Krishnamoorthy M. Kulik MJ. Reynolds DM. Sheppard AM. Liu H. Xu Y. Baetge EE. Dalton S. Activin a efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells. 2007;25:29–38. doi: 10.1634/stemcells.2006-0219. [DOI] [PubMed] [Google Scholar]
- 46.Phillips BW. Hentze H. Rust WL. Chen QP. Chipperfield H. Tan EK. Abraham S. Sadasivam A. Soong PL. Wang ST. Lim R. Sun W. Colman A. Dunn NR. Directed differentiation of human embryonic stem cells into the pancreatic endocrine lineage. Stem Cells Dev. 2007;16:561–578. doi: 10.1089/scd.2007.0029. [DOI] [PubMed] [Google Scholar]
- 47.Tropepe V. Hitoshi S. Sirard C. Mak TW. Rossant J. van der Kooy D. Direct neural fate specification from embryonic stem cells: a primitive mammalian neural stem cell stage acquired through a default mechanism. Neuron. 2001;30:65–78. doi: 10.1016/s0896-6273(01)00263-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.