Abstract
Nuclear factor I (NFI)-A is a member of the NFI family of transcription factors implicated in regulation of granulocyte differentiation. However, its role in the lymphoid lineage is not known. NFI-A deficiency results in perinatal lethality, thus precluding analysis of the role of NFI-A in lymphocyte development and function. Using recombination activation gene-2-deficient (RAG-2−/−) blastocysts and embryonic stem cells with homozygous NFI-A gene deletion, we show an essential role for NFI-A in T-cell activation. NFI-A−/−→RAG-2−/− chimeric mice had normal distributions of CD4−CD8− double negative, CD4+CD8+ double positive, CD4+CD8− and CD4−CD8+-single positive cells in the thymus and CD4+CD8− and CD4−CD8+ cells in spleen and lymph nodes. However, NFI-A−/−→RAG-2−/− mice had severely reduced thymus size and hypocellularity. The decrease in thymocytes and peripheral T cells in NFI-A−/−→RAG-2−/− chimeric mice is attributed to proliferative defects associated with decreased blast transformation, CD69 expression and DNA synthesis in response to T antigen receptor stimulation. Interestingly, NFI-A-null T cells showed increased levels of c-myc transcription that is inhibited in response to antigen receptor-mediated activation. These studies demonstrate for the first time a requirement for the NFI-A transcription factor in antigen receptor-induced T-cell activation events.
Keywords: activation, lymphocyte, thymocyte development, transcription factor
Introduction
Nuclear factor I (NFI)-A is a member of the NFI family of transcription factors that bind to a conserved TTGGCN5GCCAA DNA motif through a putative α-helical segment (1). NFI-A proteins mediate initiation of adenoviral replication in vitro and transcription of many cellular and viral genes (1–6). The NFI gene family is highly conserved from chickens to humans and is composed of Nfi-a, Nfi-b, Nfi-c and Nfi-x (7, 8). The proteins encoded by these genes, sometimes referred to as CCAAT-binding transcription factors or CTF, contain a highly conserved 220 amino acid N-terminal region that mediates DNA binding and dimerization (8, 9). Different NFI proteins exhibit variation within their COOH terminal domains that results in distinct transcriptional modulatory functions (8, 9). Gene diversity, alternative splicing and unrestricted dimerization add multiple levels of diversity to regulation of gene expression by NFI family of proteins. The occurrence of multiple NFI genes in vertebrates, their differential expression during mouse development and the expression of NFI-dependent genes in multiple organs including brain, liver, muscle and other terminally differentiated tissues, suggested an important role for NFI proteins in transcription during development (7, 10–15).
Recently, interplay among microRNA-223, NFI-A and C/EBPα has been demonstrated in human granulopoiesis (16). The role of NFI-A in the lymphoid lineage has not been evaluated. Targeted deletion of NFI-A protein in mice resulted in lack of corpus callosum associated with ventricular dilation indicating early hydrocephalus with defective development of the CNS and perinatal lethality (3). Expression of NFI-A in spleen and T cells provided a rationale to develop the RAG-2−/− blastocyst complementation model to delineate the role of NFI-A in cell proliferation in general and lymphoid lineage in particular (7). Toward this goal, we generated chimeric mice by injecting NFI-A−/− embryonic stem (ES) cells into blastocyst derived from RAG-2−/− mice. Analysis of this genetically modified mouse model has identified a selective requirement of NFI-A in antigen receptor-induced T lymphocyte, activation, proliferation and cell cycle progression.
Methods
Generation of chimeric mice
RAG-2−\− mice used in these studies were backcrossed 10 generations onto the C57Bl/6 background. The targeting construct used in the generation of targeted ES cells is described elsewhere (3). The NFI-A+/− heterozygous ES cells were subjected to high G418 selection to obtain the NFI-A−/− ES cell clones as described by us previously (17). The NFI-A−/− ES cell or wild-type NFI-A+/+ ES cells were injected into 3.5-day post coitus blastocysts obtained from RAG-2−/− mice to generate the NFI-A−/−→RAG-2−/− chimeric mice. All animal experiments were approved by the Institutional Animal Care and Use Committee.
Southern blot analysis
Genomic DNA samples, prepared from the ES cells or indicated tissues from the RAG-2, NFI-A+/+→RAG-2−/− or NFI-A−/−→RAG-2−/− chimeric mice, were digested with HindIII, fractionated on a 0.8% non-denaturing agarose gel, transferred to nylon membranes and probed with an NFI-A-specific probe as described previously (3). The relative levels of the 7.0-kb wild-type and 5.0-kb mutant NFI-A alleles were quantified by phosphorimaging (Molecular Dynamics).
Expression analysis of NFI-A, NFI-B, NFI-C and NFI-X
Real-time quantitative PCR was performed using primers specific for each NFI gene with β2 microglobulin levels used for normalization. The following primers were used:
mβ2microglobulin—B2MM AGACTGATACATACGCCTGCAG 119 bp product and B2MM C GCAGGTTCAAATGAATCTTCAG; mA total mNFI-AE23 TGGCATACTTTGTACA TGCAGC 128 bp product and mNFI-AE4C2 ACCTGATGTGACAAAGCTGTCC; mB total mNFI-BE2 GTTTTTGGCATACTACGTGCAGG 133 bp product and mNFI-BE3C CTCTGATA CATTGAAGACTCCG; mC total mNFI-CE22 GACCTGTACCT GGCCTACTTTG 147 bp product and mNFI-CE4C CACACCT GACGTGACAAAGCTC and mX total mNFI-XE2 CTGGCTT ACTTTGTCCACACTC 148 bp product and mNFI-XE4C CCAGCTCTGTCACATTCCAGAC.
Total RNA was isolated using TRIzol Reagent (GIBCO/BRL) following the manufacturer’s instructions. cDNA was generated from 2 to 5 μg of RNA using Superscript (GIBCO/BRL) following the manufacturer’s random-prime protocol. Nfi-a, Nfi-b, Nfi-c and Nfi-x transcript levels were normalized to β2 microglobulin levels by real-time quantitative PCR using a Bio-Rad real-time thermocycler with SYBRgreen detection and the deltadelta Ct method as recommended by the manufacturer.
Reagents
Hamster anti-mouse CD3 antibody (2C11.2) was purchased from BD Pharmingen. Phorbol myristate acetate (PMA) and calcium ionophore (ionomycin) were obtained from Calbiochem (La Jolla, CA, USA). Propidium iodide was purchased from Sigma Chemical Co (St Louis, MO, USA). Fluorochrome-labeled anti-CD4 (PE), anti-CD8 (FITC), anti-CD3-(FITC), anti-TCRαβ (PE), anti-TCRγδ (FITC), anti-IgM (FITC), anti-B220 (PE) and anti-CD69 (FITC) were purchased from BD Pharmingen.
Culture conditions and assay for DNA synthesis
Thymocytes and splenic T cells were prepared as described previously, using commercially available T-cell enrichment columns (R&D system) (17). Thymocytes (4.0 × 105) or splenic T cells were stimulated with plate immobilized anti-CD3 antibody (15 μg ml−1) or PMA (100 ng ml−1) ± ionomycin (0.5 mg ml−1) in 96-well tissue culture plates, in 200 μl volumes. DNA synthesis was measured by incorporation of [3H]thymidine, for 18 h following 40 h of stimulation. Results are presented as the geometric mean of counts per minute ± standard error from triplicate cultures.
Flow cytometry
Single cell suspensions (0.5 to 1 × 106) of thymus, spleen, bone marrow or lymph node cells from 8-week-old mice (n = 5) were stained with fluorochrome-labeled anti-CD4 (PE), anti-CD8 (FITC), anti-CD3-(FITC), anti-TCRαβ (PE), anti-TCRγδ (FITC), anti-IgM (FITC) and anti-B220 (PE) or anti-CD69 (FITC) as described previously (18). All samples were analyzed on an EPICS ELITE ESP flow cytometer (Hialeah, FL, USA). At least 20 000 events gated for live lymphocytes based on forward and side scatter were collected for each sample.
Results
Generation of NFI-A−/−→RAG-2−/− chimeric mice
The NFI-A wild-type genomic locus, the targeting vector used to mutate the NFI-A locus and the disrupted NFI-A locus are schematically shown in Fig. 1(a). The targeting vector replaces part of exon 2 resulting in disruption of the NFI-A locus (3). HindIII digestion of the wild type and the targeted loci resulted in 7- and 5-kb fragments, respectively, identified with the probe shown in Fig. 1(a). Southern blot analysis of DNA from NFI-A+\+ wild type, NFI-A+/− heterozygous and NFI-A−/− homozygous ES cells generated by high G418 selection revealed the expected 7- and 5-kb-targeted fragments (Fig. 1b). The generation of null NFI-A mutation was confirmed by reverse transcription–PCR analysis of the cDNA prepared using RNA isolated from the NFI-A+/+ and NFI-A−/− ES cells, using primers specific for the àNfi-a, Nfi-c and Nfi-x and β2 microglobulin as described previously (3). The expression of NFI-A, NFI-C and NFI-X in relation to the levels of β2 microglobulin is shown in Fig. 1(c). No detectable levels of NFI-A transcripts were found in NFI-A−/− ES cells, indicating the lack of expression of NFI-A in the homozygous mutant ES cells. Comparable levels of expression of NFI-B, NFI-C and NFI-X transcripts were observed in NFI-A+/+ and NFI-A−/− ES cells indicating the absence of compensatory modulation of other NFI family members. The NFI-A−/− and NFI-A+/+ ES cells were used to reconstitute RAG-2−/− blastocysts, to generate NFI-A−/−→RAG-2−/− and NFI-A+/+→RAG-2−/− chimeric mice, respectively. Southern blot analysis of genomic DNA prepared from kidney, lung, heart and intestine from NFI-A+/+ or NF-1A−/−→RAG-2−/− chimeric mice and control RAG-2−/− mice was performed to determine the ES cell contribution to the chimeric mice. The probe derived from a genomic region outside of the targeting vector detected expected 7- and 5-kb HindIII fragments in tissues derived from NFI-A−/−→RAG-2−/− chimeric mice. The tissues from wild-type ES cell injected chimeras and RAG-2−/− mice revealed the expected 7-kb wild-type band (Fig. 2a). Figure 2(b) shows contribution of injected mutant ES cell to 40–62% chimerism in the kidney, lung, heart and intestine as determined by the relative levels of the targeted and wild-type alleles detected by Southern blot analysis as described by us previously (17).
Fig. 1.
(a) Targeted disruption of NFI-A locus. Schematic representations of the NFI-A wild-type genomic locus, the targeting vector used to mutate the NFI-A gene and the disrupted NFI-A locus are shown. The expected 7-kb wild-type and the 5-kb-targeted HindIII fragments detected with the probe outside of the region of homology are shown. The Neomycin resistant gene (Neo) was used for positive selection of targeted clones. Herpes Simplex Virus-thymidine kinase (HSV-tk) gene flanking the 5′end of the targeting vector is used for negative selection of random integrants using gancyclovir. (b) Southern blot analysis of NFI-A-deficient ES cells. Genomic DNA from wild type (+/+) and targeted (+/−) ES cells were restricted with HindIII enzyme and analyzed by Southern blot analysis using the p32 labeled probe shown in Fig. 1(a). The 7-kb wild-type (wt) and the 5-kb-targeted (t) fragments identified are shown. The homozygous (−/−) ES cells were generated by high G418 selection of heterozygous NFI-A+/− ES cells. The single 5-kb-targeted fragment in two independent homozygous NFI-A−/− ES cells is shown in the right panel. (c) Analysis of NFI-A, NFI-C and NFI-X transcripts in NFI-A+/+ and NFI-A−/− ES cells. NFI-A, NFI-C, NFI-X and β2 microglobulin transcript levels were measured in cDNA prepared from RNA isolated from NFI-A+/+ and NFI-A−/− ES cells using transcript-specific primers. The NFI transcripts were normalized with β2 microglobulin transcripts and the data are represented as moles of NFI gene transcripts per mole of β2 microglobulin.
Fig. 2.
Southern blot analysis of NFI-A+/+→RAG-2−/−, NFI-A−/−→RAG-2−/− and RAG-2−/−mice. NFI-A+/+ or NFI-A−/− ES cells were injected into RAG-2−/− blastocyst to generate NFI-A+/+→RAG-2−/− and NFI-A−/−→RAG-2−/− chimeric mice, respectively. Southern blot analysis of genomic DNA prepared from various tissues from NFI-A+/+- or NFI-A−/− ES cell-reconstituted RAG-2−/− chimeric mice and RAG-2−/− mice are shown in panel A. The probe shown in Fig. 1(a) detected a 7-kb HindIII fragment from the wild-type allele and a 5-kb fragment from the targeted allele. The contribution of injected mutant ES cells to the kidney (K), lung (L), heart (H) and intestine (I) in the NFI-A−/−→RAG-2−/− chimeric mice is shown in panel B. The % of ES cell contribution was determined by the quantification of the intensity of wild type and targeted bands by phosphorimager analysis.
Developmental analysis of NFI-A−/− lymphocytes in NFI-A−/−→RAG-2−/− chimeric mice
Analysis of NFI-A in various tissues has revealed NFI-A-binding activity in spleen and T-cell lines, suggesting a role for NFI-A in T-lymphocyte development and/or function (ref. 19 and data not shown). In order to directly test the role of NFI-A in lymphocyte development, thymocytes from RAG-2−/−, NFI-A+/+→RAG-2−/− and NFI-A−/−→RAG-2 chimeric mice were stained with fluorochrome-labeled anti-CD4, anti-CD8, anti-TCRαβ or CD3 antibodies. As previously reported, the RAG-2−/− mice exhibited developmental arrest in CD4−/CD8− double negative stage resulting in the absence of CD4+/CD8+ double positive (DP) or CD4+/CD8− single positive (SP) or CD4−CD8+ SP T cells (Fig. 3) (17). In contrast to RAG-2−/− mice, NFI-A−/−→RAG-2−/− chimeric mice revealed a normal distribution of CD4+/CD8+ DP, CD4+/CD8- SP or CD4-/CD8+ SP T cells comparable to NFI-A+/+→RAG-2−/− chimeric mice (Fig. 3). The ratio of CD4+CD8− SP and CD4−CD8+ SP cells were comparable in both the NFI-A+/+→RAG-2−/− and the NFI-A−/−→RAG-2−/− chimeric mice. Consistent with the normal development in the thymus, flow cytometric analysis of splenocytes and lymph node cells revealed normal profiles and distribution of CD4 and CD8 SP cells (Fig. 3 and data not shown). Further, staining for the surface IgM and the B220 molecules showed comparable development of mature B cells in the spleen and bone marrow in NFI-A+/+→RAG-2−/− and NFI-A−/−→RAG-2−/− chimeric mice (Fig. 3 and data not shown).
Fig. 3.
Developmental analysis of lymphocytes in RAG-2−/−, NFI-A+/+→RAG-2−/− and NFI-A−/−→RAG-2−/− chimeric mice. One million thymocytes or splenocytes from RAG-2−/−, NFI-A+/+→RAG-2−/− and NFI-A−/− RAG-2 chimeric mice were stained with PE-labeled anti-CD4 and FITC-labeled anti-CD8 or FITC-labeled anti-IgM and PE-labeled anti-B220 antibodies. Twenty thousand events were collected by list mode using FACS. The numbers in each of the quadrants indicate the percentages of live cells within those quadrants.
T-cell antigen receptor-induced proliferative defect in NFI-A−/−→ RAG-2−/− chimeric mice
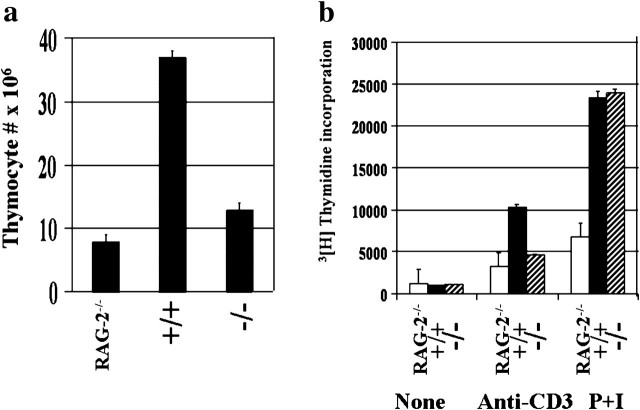
In contrast to the observed normal distribution of CD4 SP, CD8 SP cells and CD4+CD8+ DP cells, the thymi and spleen from NFI-A−/−→RAG-2−/− chimeric mice were consistently smaller in size by visual appearance and ∼60% loss in weight compared with the equally reconstituted NFI-A+/+→RAG-2−/− chimeric mice. Consistent with the decreased size and weight, the total thymocytes in NFI-A−/−→RAG-2−/− chimeric mice were significantly reduced in numbers by >80% (Fig. 4a). This is further reflected by the corresponding decrease in the absolute numbers of CD4+CD8− and CD4−CD8+ SP and CD4+CD8+ DP cells in the thymus. To directly test if the decreased in cell number is due to limited expansion of T cells, the proliferative potential of NFI-A−/−→RAG-2−/− and NFI-A+/+→RAG-2−/− thymocytes was determined in response to stimulation with T-cell antigen receptor using plate immobilized anti-CD3 antibody or PMA and ionomycin. Activation through the TCR resulted in a 60–80% decrease in the proliferation of NFI-A−/− T cells compared with NFI-A+/+ T cells. Interestingly, NFI-A−/− T cells showed no difference, in response to stimulation with PMA and ionomycin, when compared with NF-1A+/+ T cells (Fig. 4b). The decreased proliferation of the NFI-A−/− T cells is not due to differences in the levels of CD3 molecules in NFI-A+/+ and NFI-A−/− T cells as evidenced by comparable levels of surface expression of CD3 [NFI-A+/+ mean fluorescence intensity (MFI) = 149 ± 12 versus NFI-A−/− MFI = 155 ± 8, n = 3)]. Further the antigen receptor-induced defect in DNA synthesis is not restricted to thymocytes as activation of purified splenic T cells from NFI-A−/− mice also resulted in decreased DNA synthesis (Fig. 5c). This appears to be due to an effect on the early stage of T-cell activation as the NFI-A−/− T cells exhibited a significant decrease in anti-CD3-induced blast transformation (∼88% compared with NFI-A+/+ cells) as detected by forward scatter profiles (Fig. 5a). Further, consistent with the TCR-induced activation defect in NFI-A-deficient T cells, decreased expression of activation antigen CD69 was observed in NFI-A−/− T cells compared with NFI-A+/+ T cells, in response to anti-CD3 stimulation (Fig. 5b). Consistent with the decreased activation, supernatants obtained from anti-CD3-stimulated NFI-A−/− T cells exhibited ∼80% reduction in IL-2 levels compared with NFA-1+/+ T cells (data not shown). The decreased proliferation observed in NFI-A−/− T cells is not due to either endogenous or CD3-induced modulation of CD3 signaling molecules such as GRB-2, LAT, LCK, ZAP-70 or PKB as both NFI-A+/+ and NFI-A−/− T cells showed comparable expression of these gene transcripts (Fig. 6a). In contrast to the proximal signaling molecules, the NFI-A−/− T cells exhibited higher levels of c-Myc expression compared with the NFI-A+/+ T cells. More importantly, stimulation of T cells through the antigen receptor using immobilized anti-CD3 antibody resulted in up-regulation of c-Myc in normal T cells, while down-modulating the c-Myc expression in NFI-A−/− T cells suggesting a potential role for NFI-A in regulation of c-Myc expression in resting and activated T cells (Fig. 6b).
Fig. 4.
Panel (a) decreased thymocyte number in NFI-A−/−→RAG-2−/− chimeric mice. The total thymocytes from RAG-2−/−, NFI-A+/+→RAG-2−/− and NFI-A−/−→RAG-2−/− chimeric mice (8 weeks of age) were enumerated by trypan blue exclusion method. The results are the mean of three independent experiments constituting a total of six to nine animals per group. Panel (b) defective proliferation of NF-1A−/−. Thymocytes in response to anti-CD3 but not PMA + ionomycin stimulation. Thymocytes (4.0 × 105) from RAG-2−/−, NFI-A+/+→RAG-2−/− and NFI-A−/−→RAG-2−/− chimeric mice were stimulated through the T-cell antigen receptor using plate immobilized anti-CD3 antibody (15 μg ml−1) or PMA (P) plus ionomycin (I) as described in the Methods. The cells were cultured at 37° in a 5% CO2 incubator. The cells were pulsed for 18 h with 1 μCi 3[H]thymidine of following 48 h after initiation of culture. The results are represented as counts per minute. The error bars indicate the mean and standard deviation of triplicate cultures. The results are representative of at least three independent experiments.
Fig. 5.

Defective activation of NFI-A−/− T cells in response to antigen receptor stimulation. Purified splenic T cells (4.0 × 106 ml−1) from NFI-A+/+→RAG-2−/− and NFI-A−/−→RAG-2−/− chimeric mice were stimulated through the T-cell antigen receptor using plate immobilized anti-CD3 antibody. The cells were harvested at 36 h after stimulation and analyzed by FACS. Panel (a) shows the forward (FSC) and side scatter (SSC) profiles of the activated cells. The numbers in each panel shows the percentage of cells undergoing blast transformation. Panel (b) shows the expression of the activation marker CD69, in anti-CD3-activated NFI-A+/+ and NFI-A−/− T cells. The numbers indicate the mean fluorescence intensity (MFI) of CD69+ cells in the total activated T cells. Panel (c) shows the DNA synthesis in NFI-A+/+ or NFI-A−/− T cells stimulated with the anti-CD3 as measured by 3[H]thymidine incorporation. The cells were cultured 48 h and then pulsed with 1 μCi 3[H]thymidine for 18 h. The error bars indicate the mean and standard deviation of triplicate cultures. The results are representative of two independent experiments.
Fig. 6.
NFI-A-null T cells show increased c-Myc transcription that is inhibited in response to antigen receptor-mediated activation. Purified T cells from NFI-A+/+→Rag-2 or NFI-A-/→Rag-2−/− mice were stimulated with media or immobilized anti-CD3 (10 μg ml−1). Total RNA was isolated 24 h following stimulation and the expression of GRB-2, LAT, LCK, ZAP70, CD3ζ, PKB and HPRT transcripts were analyzed by semiquantitative real-time PCR as described previously (18). HPRT was used as an internal control. Panel b shows increased c-Myc transcription that is inhibited in response to antigen receptor-mediated activation in NFI-A−/−. This is in contrast to induction of c-Myc in NFI-A+/+ T cells. Results are representative of three independent experiments.
Discussion
NFI-A has been implicated in the expression of many cellular and viral genes. Recently, its role in human granulopoiesis has been established through interplay between C/EBPa and mir 223 (16). The role of NFI-A in lymphocyte development or proliferation has not been previously analyzed. The RAG-2−/− blastocyst complementation model described in this report describes a role for NFI-A in T-cell proliferation and activation functions. Consistent with the previous observations, we observed >90% contribution of the injected ES cells to the thymus in the chimeric mice (20). Normal development of T cells in thymus, spleen and lymph node and B cells in the bone marrow, spleen and lymph nodes indicated that NFI-A is not required for normal T- and B-cell development. It is formally possible that the binding of other NFI family members such as NFI-B, NFI-C or NFI-X could functionally compensate for NFI-A in the development of lymphocytes. This possibility remains to be tested using the combinatorial mutants of the NFI family members when these mice become available. Absence of NFI-A does not alter the expression of other NFI family members such as NFI-B, NFI-C and NFI-X. However, since each of these gene products share similar DNA binding and dimerization domains, the effects of NFI-A deficiency on homo- and heterodimerization of NFI family members or regulation of genes involved in T-cell activation remain to be determined. The observed defects in T-cell activation could be directly due to the deregulation of NFI-A target genes or indirectly by allowing novel associations of other NFI family of proteins in the absence of NFI-A resulting in altered gene expression. The finding that the loss of NFI-A dramatically impairs proliferative responses to antigen receptor stimulation, points to NFI-A being a key element in TCR-induced signal transduction pathways. The comparable levels of transcription of CD3-associated downstream signalosomes including GRB-2, LAT, LCK, ZAP-70 and PKB in NFI-A+/+ and NFI-A−/− T cells suggests that the decreased proliferation observed in NFI-A−/− T cells is not due to either endogenous or CD3-induced modulation of these molecules. In contrast, these differences could be attributed to the differential regulation of endogenous and anti-CD3-induced c-Myc expression in NFI-A−/− and NFI-A+/+ T cells. Understanding the molecular mechanisms and role of NFI-A in regulation of c-Myc will provide insights into the role of NFI-A in antigen receptor-induced T-cell activation and functional differentiation.
Funding
This work was supported in part by American Cancer Society to N.M and National Institutes of Health (HD34908 to R.M.G.).
References
- 1.Leegwater PAJ, van Driel W, van der Vilet PC. Recognition site of nuclear factor I, a sequence-specific DNA-binding protein from HeLa cells that stimulates adenovirus DNA replication. EMBO J. 1985;6:1515. doi: 10.1002/j.1460-2075.1985.tb03811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagata K, Guggenheimer R, Enomoto T, Lichy J, Hurwitz J. Adenovirus DNA replication in vitro: identification of a host factor that stimulates synthesis of the preterminal protein-dCMP complex. Proc. Natl Acad. Sci. USA. 1982;79:6438. doi: 10.1073/pnas.79.21.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Das Neves L, Duchala CS, Godinho F, et al. Disruption of the murine nuclear factor I-A gene (NFI-A) results in perinatal lethality, hydrocephalus, and agenesis of the corpus callosum. Proc. Natl Acad. Sci. USA. 1999;96:1. doi: 10.1073/pnas.96.21.11946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mermod N, O'Neill EA, Kelly TJ, Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989;58:741. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- 5.Gounari F, De Francesco R, Schmitt J, van der Vliet P, Cortese R, Stunnenberg H. Amino-terminal domain of NF1 binds to DNA as a dimer and activates adenovirus DNA replication. EMBO J. 1990;9:559. doi: 10.1002/j.1460-2075.1990.tb08143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puzianowska-Kuznicka M, Shi YB. Nuclear factor I as a potential regulator during postembryonic organ development. J. Biol. Chem. 1996;271:6273. doi: 10.1074/jbc.271.11.6273. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhry AZ, Lyons CE, Gronostajski RM. Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development. Dev. Dyn. 1997;208:313. doi: 10.1002/(SICI)1097-0177(199703)208:3<313::AID-AJA3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 8.Santoro C, Mermod N, Andrews PC, Tjian R. A family of human CCAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature. 1988;334:218. doi: 10.1038/334218a0. [DOI] [PubMed] [Google Scholar]
- 9.Kruse U, Sippel AE. Transcription factor nuclear factor I proteins form stable homo- and heterodimers. FEBS Lett. 1994;348:46. doi: 10.1016/0014-5793(94)00585-0. [DOI] [PubMed] [Google Scholar]
- 10.Aoyama A, Tamura TA, Mikoshiba K. Regulation of brain-specific transcription of the mouse myelin basic protein gene: function of the NFI-binding site in the distal promoter. Biochem. Biophys. Res. Commun. 1990;167:648. doi: 10.1016/0006-291x(90)92074-a. [DOI] [PubMed] [Google Scholar]
- 11.Amemiya K, Traub R, Durham L, Major EO. Adjacent nuclear factor-1 and activator protein binding sites in the enhancer of the neurotropic JC virus. A common characteristic of many brain-specific genes. J. Biol. Chem. 1992;267:14204. [PubMed] [Google Scholar]
- 12.Cereghini S, Raymondjean M, Carranca AG, Herbomel P, Yaniv M. Factors involved in control of tissue-specific expression of albumin gene. Cell. 1987;50:627. doi: 10.1016/0092-8674(87)90036-5. [DOI] [PubMed] [Google Scholar]
- 13.Darville MI, Antoine IV, Rousseau GG. Characterization of an enhancer upstream from the muscle-type promoter of a gene encoding 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase. Nucleic Acids Res. 1992;20:3575. doi: 10.1093/nar/20.14.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edmondson DG, Cheng TC, Cserjesi P, Chakraborty T, Olson EN. Analysis of the myogenin promoter reveals an indirect pathway for positive autoregulation mediated by the muscle-specific enhancer factor MEF-2. Mol. Cell. Biol. 1992;12:3665. doi: 10.1128/mcb.12.9.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graves RA, Tontonoz P, Ross SR, Spiegelman BM. Identification of a potent adipocyte-specific enhancer: involvement of an NF-1-like factor. Genes Dev. 1991;5:428. doi: 10.1101/gad.5.3.428. [DOI] [PubMed] [Google Scholar]
- 16.Fazi F, Rosa A, Fatica A, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 17.Muthusamy N, Barton K, Leiden JM. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature. 1995;377:639. doi: 10.1038/377639a0. [DOI] [PubMed] [Google Scholar]
- 18.Chen H-C, Byrd JC, Muthusamy N. Transgenic mice over-expressing a dominant negative cyclic AMP responsive element binding protein reveal a differential role for CREB-1 transcription factor in multiple stages of B cell development, maturation and survival. J. Immunol. 2006;176:2208. doi: 10.4049/jimmunol.176.4.2208. [DOI] [PubMed] [Google Scholar]
- 19.Safford M, Collins S, Lutz MA, et al. Egr-2 and Egr-3 are negative regulators of T cell activation. Nat. Immunol. 2005;6:472. doi: 10.1038/ni1193. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Lansford R, Stewart V, Young F, Alt FW. RAG-2-deficient blastocyst complementation: an assay of gene function in lymphocyte development. Proc. Natl Acad. Sci. USA. 1993;90:4528. doi: 10.1073/pnas.90.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]