Abstract
SpoIIGA is a novel type of membrane-associated aspartic protease that responds to a signal from the forespore by cleaving Pro-σE in the mother cell during sporulation of Bacillus subtilis. Very little is known about how SpoIIGA recognizes Pro-σE. By co-expressing proteins in Escherichia coli, it was shown that charge reversal substitutions for acidic residues 24 and 25 of Pro-σE, and for basic residues 245 and 284 of SpoIIGA, impaired cleavage. These results are consistent with a model predicting possible electrostatic interactions between these residues; however, no charge reversal substitution for residue 245 or residue 284 of SpoIIGA restored cleavage of Pro-σE with a charge reversal substitution for residue 24 or residue 25. Bacillus subtilis SpoIIGA cleaved Pro-σE orthologs from Bacillus licheniformis and Bacillus halodurans, but not from Bacillus cereus. A triple substitution in the pro-sequence of B. cereus Pro-σE allowed cleavage by B. subtilis SpoIIGA, indicating that residues distal from the cleavage site contribute to substrate specificity. Co-expression of SpoIIGA and Pro-σE orthologs in different combinations suggested that B. licheniformis SpoIIGA has a relatively narrow substrate specificity as compared with B. subtilis SpoIIGA, whereas B. cereus SpoIIGA and B. halodurans SpoIIGA appear to have broader substrate specificity.
Keywords: Aspartic protease, bacillus subtilis, sigma factor, signal transduction, sporulation
The Gram-positive bacterium Bacillus subtilis undergoes sporulation in response to starvation. Sporulation involves a highly ordered program of gene expression and morphological change (1). The first morphological change of sporulation is the appearance of an asymmetrically positioned septum that divides the cell into a larger mother cell and a smaller forespore compartment (Fig. 1A, left). The first compartment-specific transcription factor to become active is σF in the forespore. This subunit of RNA polymerase (RNAP) directs transcription of at least 47 genes (2, 3), including spoIIR, whose product is believed to be secreted from the forespore into the space between the two membranes of the polar septum, and signals activation of σE in the mother cell (4–6) (Fig. 1B, middle). The σE is synthesized as an inactive precursor, Pro-σE, and is activated by proteolytic cleavage that removes its N-terminal 27 residues (7, 8). The gene (spoIIGB or sigE) encoding Pro-σE is co-transcribed in an operon with spoIIGA (9), whose product is necessary for the processing of Pro-σE to σE. The σE RNAP transcribes over 270 genes (2, 10–12). SpoIIGA and Pro-σE are expressed predominantly in the mother cell (13) (Fig. 1). SpoIIGA is predicted to have an N-terminal domain with five transmembrane segments embedded in the mother cell membrane (14) and a C-terminal aspartic protease domain in the mother cell cytoplasm (15). SpoIIGA dimerizes to form the catalytic site of the aspartic protease (see Fig. 3 which appears later). Pro-σE associates with the mother cell membrane via its N-terminal pro-sequence (16, 17) and is cleaved (Fig. 1, arrowhead) by SpoIIGA, releasing active σE into the mother cell. The products of certain genes under σF control in the forespore and other genes under σE control in the mother cell bring about further morphological change, resulting in the formation of an endospore (1).
Fig. 1.
Model of Pro-σE processing during sporulation of B. subtilis and in E. coli. (Left) Asymmetric septation during B. subtilis sporulation generates a smaller forespore compartment and a larger mother cell compartment. (Middle) An expanded view of the septum depicts proteins involved in Pro-σE processing. SpoIIR is produced in the forespore and is believed to cross the forespore membrane and interact with SpoIIGA, activating Pro-σE processing (4–6). (Right) SpoIIR, SpoIIGA and Pro-σE are produced under T7 RNAP control in E. coli. (Middle) An expanded view of the E. coli outer membrane, periplasm and inner membrane.
Fig. 3.
Model of SpoIIGA in complex with a short segment of Pro-σE. Only the C-terminal domain of SpoIIGA is shown, with the two chains of the putative dimer coloured black and white (arrows and helices indicate β-sheet and α-helix secondary structures, respectively), except van der Waals surfaces are shown for the putative catalytic aspartates (D183) and residues hypothesized to play a role in substrate recognition (R245 and K284) or flap function (P259) for each chain. Only a short segment of Pro-σE is shown (residues 22–33), in stick representation coloured yellow, except van der Waals surfaces are shown for K22, D24 and E25. The left part shows a ‘side view’ and the right part shows a ‘top view’ of the complex. The ‘side view’ is the orientation typically shown for HIV-1 protease. Note that both the N-terminus (i.e. the point of attachment to the N-terminal membrane domain) and C-terminus of each SpoIIGA chain is at the bottom in the ‘side view’, which is upside down relative to the depiction in Fig. 1. This image was made with Visual Molecular Dynamics (VMD) software support. VMD is developed with NIH support by the Theoretical and Computational Biophysics group at the Beckman Institute, University of Illinois at Urbana-Champaign.
Since in vitro processing of Pro-σE has not been achieved yet, it remained possible that SpoIIGA and SpoIIR modify the activity of an unidentified protein that directly cleaves Pro-σE. Recently, we reconstituted Pro-σE processing in Escherichia coli (Fig. 1C, right) and provided evidence that SpoIIR and SpoIIGA are necessary and sufficient for accurate processing of Pro-σE in a heterologous host, and that SpoIIGA is a novel type of signal-transducing aspartic protease that cleaves Pro-σE to σE (15). This E. coli system is also a powerful tool for mutagenesis to analyse Pro-σE processing. Pro-σE was co-expressed in E. coli with C-terminally double-FLAG-tagged SpoIIR (SpoIIR-F2) and with C-terminally tagged SpoIIGA (SpoIIGA-GFP-F2). SpoIIR is expected to be secreted to the periplasm where it could interact with SpoIIGA's inner membrane-embedded N-terminal domain, stimulating SpoIIGA's C-terminal domain to cleave inner membrane-associated Pro-σE, releasing σE to the cytoplasm (15) (Fig. 1). Expression of both SpoIIR-F2 and SpoIIGA-GFP-F2 was necessary and sufficient for cleavage of Pro-σE to σE in E. coli. Modelling and mutational analysis of the C-terminal half of SpoIIGA provided evidence that it forms a dimer similar to the human immunodeficiency virus type 1 (HIV-1) protease and related retroviral aspartic proteases. However, unlike the retroviral proteases, SpoIIGA has an N-terminal, membrane-embedded domain (14, 18). SpoIIGA-GFP-F2 expressed in E. coli was membrane associated and appeared to form inactive dimers or oligomers, that upon interaction between their N-terminal domains and SpoIIR-F2 on the periplasmic side of the inner membrane was proposed to cause a conformational change in the cytoplasmic C-terminal domains of SpoIIGA-GFP-F2, allowing formation of active aspartic protease dimer capable of cleaving membrane-associated Pro-σE (15). According to this model, SpoIIGA is unique since previously described membrane-associated aspartic proteases have two catalytic aspartate residues in a single-polypeptide chain, and they have not been proposed to transduce a signal across a membrane. Very little is known about how SpoIIGA recognizes its substrate, Pro-σE. A previous study showed that E25 of Pro-σE is important for processing in B. subtilis (19). Also, the N-terminal 28 residues of Pro-σE were shown to be sufficient for processing of a ProσE–σK fusion protein (20), and the N-terminal 52–55 amino acids of Pro-σE were sufficient for processing when fused to proteins unrelated to σ-factors (16). These results suggested that some, if not all, of the information necessary for recognition by the SpoIIGA protease resides in the pro-sequence of Pro-σE (residues 1–27). Here we investigated substrate specificity of SpoIIGA in Bacilli.
Experimental Procedures
Plasmids
Plasmids used in this study are described in Supplementary Table SI, and primers used in their construction are listed in Supplementary Table SII. All cloned PCR products and all genes subjected to mutagenesis (QuikChange kit, Stratagene) were sequenced to confirm that no undesired mutations were present.
Co-transformation
Plasmids bearing different antibiotic-resistance genes were co-transformed into E. coli strain BL21(DE3) (Novagen) as described previously (15).
Induction of gene expression
Escherichia coli BL21(DE3) can be induced to synthesize T7 RNAP by addition of isopropyl β-d-thiogalactopyranoside (IPTG). This strain, bearing plasmids with genes fused to the T7 RNAP promoter, was grown in Luria–Bertani medium containing 50 µg/ml kanamycin sulphate and/or 50 µg/ml ampicillin overnight at 37°C with shaking. A 50 µl of culture was transferred to 5 ml of fresh Luria–Bertani medium with antibiotics, and incubation was continued at 37°C with shaking at 140 r.p.m. until the culture reached 0.5 of OD660. To induce gene expression, IPTG (0.5 mM) was added, and incubation with shaking was continued for 2 h since cleavage was shown previously to reach a maximum at this time (15).
Western blot analysis
Escherichia coli cells from cultures (1 ml) induced as described above were collected by centrifugation (12,000g). Samples were prepared by resuspending cells in 100 µl of sample buffer [50 mM Tris–HCl, pH 6.8, 2% SDS, 10% (v/v) glycerol and 0.015% bromphenol blue] and boiling for 3 min. Samples were separated on SDS–12% polyacrylamide gels with Tris–Tricine electrode buffer (0.1 M Tris, 0.1 M Tricine and 0.1% SDS) and electroblotted to Immobilon-P membranes (Millipore). Membranes were incubated in TBS [20 mM Tris–HCl (pH 7.5), 0.5 M NaCl] with 5% non-fat dry milk for 1 h at room temperature with shaking to block non-specific antibody binding and then incubated overnight at room temperature with shaking in monoclonal antibody against σE (a gift from W. Haldenwang) at 1 : 1,000 dilution into TBS with 2% non-fat dry milk to detect pro-σE and σE. Horseradish peroxidase-conjugated goat anti-mouse IgG (Promega) were used at 1 : 1,000 dilution for 1 h at room temperature with shaking serving as the secondary antibodies. Horseradish peroxidase-conjugated antibodies to FLAG (Sigma) were used at 1 : 5,000 dilution for 1 h at room temperature with shaking to detect SpoIIR-F2 and SpoIIGA-GFP-F2. Chemiluminescent detection was performed according to the manufacturer's instructions (ECL kit; Amersham). Signals were captured using a charge-coupled-device camera (ATTO CoolSaver AE-6955; ATTO Corporation, Tokyo, Japan).
Results and discussion
Importance of conserved, charged residues close to the cleavage site in Pro-σE
An alignment of B. subtilis Pro-σE and its orthologs revealed that the pro-sequence is not as conserved as the downstream sigma factor domain (Supplementary Fig. S1). However, E25 in the pro-sequence of B. subtilis Pro-σE appears to be conserved in Bacilli and some other species, although not in Clostridia. Likewise, D24 and K22 in the pro-sequence of B. subtilis Pro-σE appear to be highly conserved in Bacilli. In the HIV-1 protease, it is proposed that the opening of flaps exposes the electronegative active site, then, a neutral or positively charged substrate could potentially be guided into a conformation optimal for binding (21). We reasoned that one or more of the conserved, charged residues in pro-sequence of Pro-σE might interact electrostatically with a residue of opposite charge in SpoIIGA to help position the substrate for cleavage. To test this hypothesis, we engineered substitutions in B. subtilis Pro-σE and in SpoIIGA-GFP-F2, and we co-expressed mutant forms of these proteins together with SpoIIR-F2 in E. coli as described previously (15). Expression of SpoIIR-F2 from pID50 and its derivatives was normal (Supplementary Fig. S2). As noted previously (15), pID50 expresses Pro-σE with its methionine at residue 17 changed to isoleucine (M17I) to avoid initiation of translation at this position, which would interfere with the analysis of cleavage by SpoIIGA-GFP-F2 (expressed from pID48) because the species produced from the alternative start codon co-migrates with σE. Also, the Pro-σE is C-terminally tagged with 6-histidine residues (H6). Neither the H6 tag nor the M17I substitution impairs cleavage (15), and we refer to pro-σE(M17I)-H6 as the wild-type substrate here.
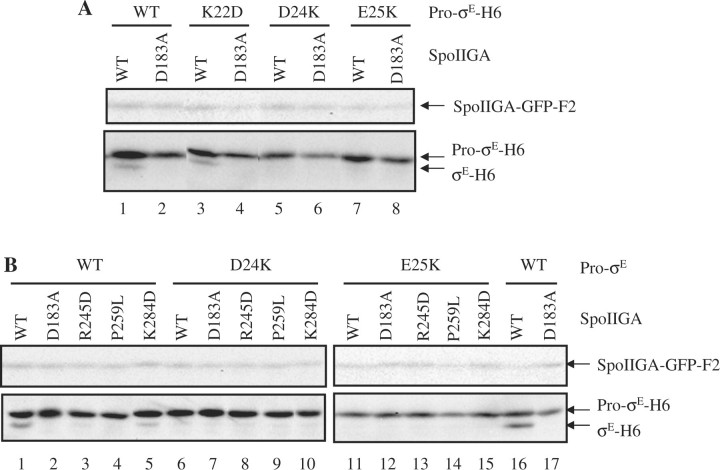
We engineered three charge reversals in Pro-σE (K22D, D24K and E25K) by subjecting pID50 to site-directed mutagenesis, and the sigE gene was sequenced to ensure that only the desired mutation was created. We chose to make charge reversal substitutions because we hoped to make compensatory charge reversals in SpoIIGA (see below). As expected, the wild-type substrate was cleaved to σE-H6 by SpoIIGA-GFP-F2 (Fig. 2A, lane 1) but not by the D183A mutant form (Fig. 2A, lane 2) (15). D183 is the putative catalytic aspartate residue of SpoIIGA and was used as a negative control for SpoIIGA activity in this study. The K22D substitution in Pro-σE-H6 had little or no effect on cleavage, indicating that charge reversal at residue 22 is tolerated (Fig. 2A, lanes 3 and 4). However, the D24K and E25K substitutions in Pro-σE-H6 prevented cleavage (Fig. 2A, lanes 5–8). The failure of the E25K mutant form of Pro-σE to be cleaved in the E. coli system is consistent with its failure to be cleaved by wild-type SpoIIGA during B. subtilis sporulation (19). The inability of SpoIIGA to cleave the D24K and E25K mutant forms of Pro-σE shows that charge reversals are not tolerated at these positions. These results are consistent with a model predicting that D24 and/or E25 might interact electrostatically with a residue of opposite charge in SpoIIGA (see below).
Fig. 2.
Effects of substitutions in Pro-σE and in SpoIIGA on cleavage. (A) Effects of charge reversals in conserved residues in the pro-sequence of Pro-σE. Escherichia coli bearing pID50 to produce SpoIIR-F2 and Pro-σE-H6 (WT), or a derivative of pID50 to produce SpoIIR-F2 and Pro-σE-H6 with the indicated substitution, and pID48 to produce active SpoIIGA-GFP-F2 (WT), or pID53 to produce inactive SpoIIGA(D183A)-GFP-F2, were induced with IPTG for 2 h. Intervening lanes were deleted from the image. (B) Effects of substitutions in SpoIIGA on cleavage of wild-type and mutant Pro-σE. As described for panel A, E. coli bearing pID50 or a derivative, and pID48 or a derivative, were induced. For both panels, samples (10 µl) were subjected to western blot analysis with antibodies against FLAG (upper blot) or σE (lower blot).
Effects of changes in SpoIIGA on cleavage of Pro-σE
Based on a structural model of SpoIIGA in complex with a short segment of Pro-σE (15), we predicted that R245 of SpoIIGA might interact with D24 and/or E25 of Pro-σE because the residues are in close proximity (Fig. 3) and of opposite charge. R245 is predicted to be located in a flap in each subunit of the putative SpoIIGA dimer. The corresponding flaps in the HIV-1 protease dimer (the template for the structural model) control access of the substrate to the active site. Alignment of B. subtilis SpoIIGA with its orthologs showed that R245 appears to be highly conserved in Bacilli, although not in other species (Supplementary Fig. S3). We engineered an R245D substitution in B. subtilis SpoIIGA-GFP-F2 and co-expressed it with SpoIIR-F2 and wild-type or mutant forms of Pro-σE-H6 in E. coli. The R245D substitution abolished the ability of SpoIIGA-GFP-F2 to cleave wild-type Pro-σE-H6 (Fig. 2B, lane 3), consistent with the idea that R245 interacts with the substrate. However, SpoIIGA(R245D)-GFP-F2 did not cleave the D24K or E25K mutant form of Pro-σE-H6 (Fig. 2B, lanes 8 and 13). Perhaps R245 of SpoIIGA interacts with another residue in Pro-σE or the particular substitutions we chose are incompatible with the predicted interaction. In addition to possibly interacting with Pro-σE, R245 of SpoIIGA is predicted to form a salt bridge with D190 that might be important for structural stability in the substrate binding cavity (15), so the R245D substitution might disrupt that interaction, although the altered protein accumulates normally (Fig. 2B, lanes 3, 8 and 13).
A K284D substitution in SpoIIGA-GFP-F2 was tested in the same way as the R245D substitution. Based on our structural model (15), K284 is predicted to be in a loop near the substrate, so it might interact with D24 and/or E25 (Fig. 3). However, this seemed less likely than an interaction between D24 and/or E25 of Pro-σE and R245 of SpoIIGA, since K284 is not conserved among orthologs of SpoIIGA in Bacilli (Supplementary Fig. S3). The K284D substitution in SpoIIGA-GFP-F2 reduced its activity towards wild-type Pro-σE-H6 (Fig. 2B, lane 5) and did not permit cleavage of the D24K and E25K mutant forms of Pro-σE-H6 (Fig. 2B, lanes 10 and 15).
The effects of the R245D and K284D substitutions in SpoIIGA on cleavage of wild-type Pro-σE provide the first evidence that R245 and K284 are important for SpoIIGA function. Although our results do not provide evidence that R245 or K284 of SpoIIGA interact with D24 or E25 of Pro-σE, neither do they rule out such interactions.
Since SpoIIGA(P259L) had been identified as a suppressor of Pro-σE(E25K) previously in sporulating B. subtilis (22), we also tested the effects of a P259L substitution in SpoIIGA-GFP-F2 using our E. coli system. In B. subtilis during sporulation, SpoIIGA(P259L) cleaves both wild-type Pro-σE and the E25K mutant form (22) [Pro-σE(D24K) was not tested] suggesting that SpoIIGA(P259L) has broadened substrate specificity. It is conceivable that the broadened substrate specificity of SpoIIGA(P259L) is due to increased mobility of flaps that in the HIV-1 protease control access of the substrate to the active site because P259 is located at the base of the flap (Fig. 3). Surprisingly, the P259L substitution in SpoIIGA-GFP-F2 abolished its activity on wild-type Pro-σE (Fig. 2B, lane 4) and failed to permit cleavage of the D24K and E25K mutant forms of Pro-σE (Fig. 2B, lanes 9 and 14). There are many differences between sporulating B. subtilis and the E. coli system that might account for the different results (Fig. 1), including the composition of the membrane into which SpoIIGA is inserted. We speculate that in E. coli, SpoIIGA(P259L) cannot close the flaps tightly enough to bind the substrate and permit cleavage. We note that P259 appears to be highly conserved in SpoIIGA orthologs (Supplementary Fig. S3).
Ability of B. subtilis SpoIIGA to cleave Pro-σE orthologs
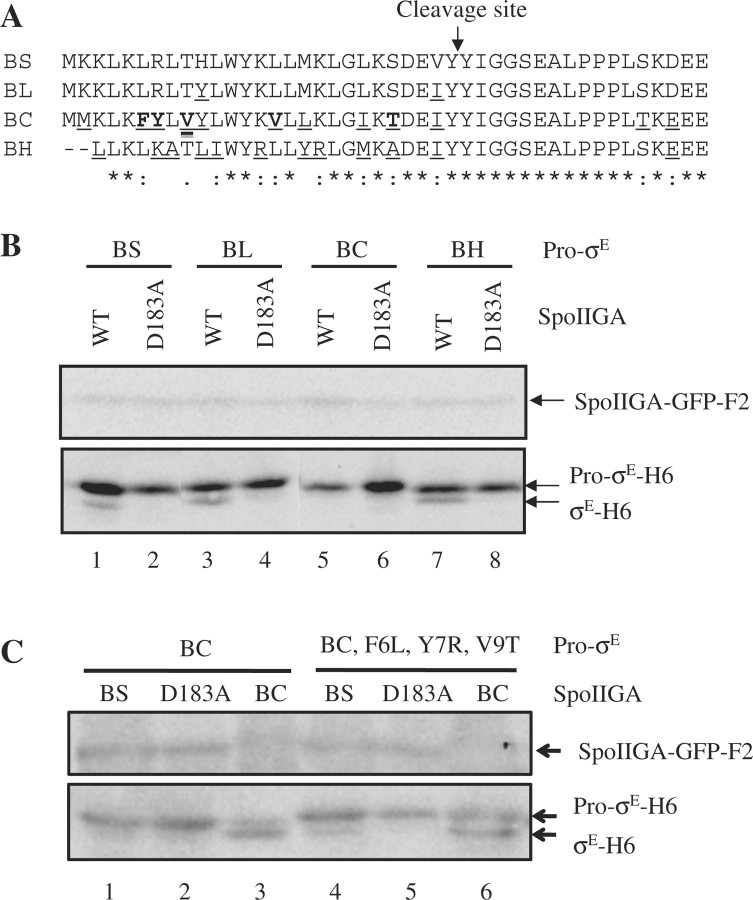
To further investigate how SpoIIGA recognizes Pro-σE, we took advantage of the natural variation in the pro-sequence among Pro-σE orthologs (Supplementary Fig. S1). We cloned the sigE genes that code for Pro-σE orthologs from Bacillus licheniformis, Bacillus cereus and Bacillus halodurans, which differ in their pro-sequence at 2, 10 and 13 positions, respectively, from B. subtilis Pro-σE (Fig. 4A). Each sigE gene was cloned downstream of a T7 RNAP promoter in a plasmid that also expresses SpoIIR-F2 from a T7 RNAP promoter. Expression of Pro-σE-H6 from B. licheniformis, Pro-σE(BL)-H6, in E. coli generated a smaller species (data not shown), presumably because methionine at position 17 served as an alternative translation initiation codon (Fig. 4A), as observed previously for B. subtilis Pro-σE-H6 (15). Therefore, an M17I substitution was made in Pro-σE(BL)-H6 (analogous to the one made in B. subtilis Pro-σE-H6). Expression of Pro-σE-H6 from B. cereus, Pro-σE(BC)-H6, or B. halodurans, Pro-σE(BH)-H6, in E. coli did not produce a smaller species (Fig. 4B, lanes 6 and 8). Interestingly, upon co-expression in E. coli, B. subtilis SpoIIGA-GFP-F2 cleaved Pro-σE(BL)-H6 and Pro-σE(BH)-H6, but not Pro-σE(BC)-H6 (Fig. 4B). These results suggest that the Pro-σE(BC) pro-sequence is not recognized by B. subtilis SpoIIGA.
Fig. 4.
Cleavage of Pro-σE orthologs by B. subtilis SpoIIGA. (A) Alignment of the N-terminal region of Pro-σE orthologs tested for cleavage. The N-terminal sequences of Pro-σE from B. subtilis (BS), B. licheniformis (BL), B. cereus (BC) and B. halodurans (BH) were aligned using ClustalW (23). An arrow indicates the cleavage site in B. subtilis Pro-σE. Underlining indicates differences from the B. subtilis sequence. Bold case letters indicate residues subjected to substitutions in the BC pro-sequence to corresponding BS residues in Supplementary Fig. S4B. At the bottom, asterisks, double dots and dots indicate identical, conserved and semi-conserved residues, respectively. (B) Cleavage of Pro-σE orthologs by B. subtilis SpoIIGA. Escherichia coli bearing pID50, pID129, pID121 or pID122 to produce B. subtilis SpoIIR-F2 and Pro-σE-H6 from B. subtilis (BS), B. licheniformis (BL), B. cereus (BC) or B. halodurans (BH), respectively, and pID48 to produce active B. subtilis SpoIIGA-GFP-F2, or pID53 to produce inactive B. subtilis SpoIIGA(D183A)-GFP-F2, were induced with IPTG for 2 h. Intervening lanes were deleted from the image. (C) Cleavage of triply substituted B. cereus Pro-σE by B. subtilis SpoIIGA. E. coli bearing pID121 or pID176 to produce B. subtilis SpoIIR-F2 and Pro-σE-H6 from B. cereus [Pro-σE(BC)] or its derivative [Pro-σE(BC, F6L, Y7R, V9T)], respectively, and pID48 to produce active B. subtilis SpoIIGA-GFP-F2, pID53 to produce inactive B. subtilis SpoIIGA(D183A)-GFP-F2, or pID126 to produce SpoIIGA(BC)-GFP-F2 were induced with IPTG for 2 h. For (B and C), samples (10 µl) were subjected to western blot analysis with antibodies against FLAG (upper blot) or σE (lower blot).
To demonstrate that Pro-σE(BC) could be cleaved by its cognate enzyme, we cloned the gene encoding, the SpoIIGA ortholog from B. cereus, SpoIIGA(BC), and engineered expression in E. coli. Although SpoIIGA(BC)-GFP-F2 was not expressed as highly as B. subtilis SpoIIGA-GFP-F2 in E. coli, Pro-σE(BC)-H6 was readily cleaved by SpoIIGA(BC)-GFP-F2 (Fig. 5, lane 11). As shown in Supplementary Fig. S4A, this cleavage depended on co-expression of B. subtilis SpoIIR-F2, indicating that the B. subtilis signalling protein stimulates SpoIIGA(BC)-GFP-F2.
Fig. 5.
Combinations of SpoIIGA and Pro-σE orthologs. Escherichia coli bearing pID50 (lanes 1–4), pID129 (lanes 5–8), pID121 (lanes 9–12) or pID122 (lanes 13–16) to produce B. subtilis SpoIIR-F2 and Pro-σE-H6 orthologs, and pID48 (lanes 1, 5, 9 and 13) to produce SpoIIGA(BS)-GFP-F2, pID125 (lanes 2, 6, 10 and 14) to produce SpoIIGA(BL)-GFP-F2, pID126 (lanes 3, 7, 11 and 15) to produce SpoIIGA(BC)-GFP-F2, or pID173 (lanes 4, 8, 12 and 16) to produce SpoIIGA(BH) were induced with IPTG for 2 h. Samples (10 µl) were subjected to western blot analysis with antibodies against FLAG (upper blot) or σE (lower blot).
To investigate the inability of B. subtilis SpoIIGA to cleave Pro-σE(BC), we compared the pro-sequences of the Pro-σE orthologs (Supplementary Fig. S1). We considered F6, Y7, V9, V15 and T23 of Pro-σE(BC) to be the most likely residues to explain its inability to be cleaved by B. subtilis SpoIIGA (Fig. 4A). The five residues of Pro-σE(BC)-H6 were changed individually to those of B. subtilis, creating F6L, Y7R, V9T, V15L and T23S mutant forms of Pro-σE(BC)-H6, which were tested for cleavage by B. subtilis SpoIIGA-GFP-F2 and by SpoIIGA(BC)-GFP-F2 (Supplementary Fig. S4B). None of the individual substitutions enabled Pro-σE(BC)-H6 to be cleaved by B. subtilis SpoIIGA-GFP-F2. We considered the possibility that several residues of Pro-σE(BC) need to be changed simultaneously in order to allow cleavage by B. subtilis SpoIIGA. Noting that F6, Y7 and V9 are highly diverged from the corresponding residues in B. subtilis Pro-σE, we hypothesized that these residues are incompatible with cleavage by B. subtilis SpoIIGA, because this enzyme had been shown to cleave B. subtilis Pro-σE lacking residues 2–11 (19), so this region is not required for substrate recognition. Therefore, we constructed triply substituted Pro-σE(BC)-H6 in which F6, Y7 and V9 were changed to the corresponding residues in B. subtilis Pro-σE. The resulting Pro-σE(BC, F6L, Y7R, V9T)-H6 was reproducibly cleaved by B. subtilis SpoIIGA-GFP-F2 (Fig. 4C, lane 4), although not as efficiently as by SpoIIGA(BC)-GFP-F2 (Fig. 4C, lane 6). Probably, more residues of Pro-σE(BC) would need to be changed to allow full cleavage by B. subtilis SpoIIGA. In any case, the result with triply-substituted Pro-σE(BC)-H6 indicates that residues distal from the cleavage site can inhibit cleavage by SpoIIGA and therefore contribute to the specificity of the cleavage reaction.
Specificity of SpoIIGA orthologs
To explore the specificity of SpoIIGA orthologs further, we cloned the putative proteases from B. halodurans and B. licheniformis. We tested the cleavage of all combinations of SpoIIGA and Pro-σE from species used in this study, using co-expression in E. coli, and in each case B. subtilis SpoIIR-F2 was also co-expressed (Fig. 5). The B. halodurans ortholog, SpoIIGA(BH), was not tagged with GFP-F2 because SpoIIGA(BH)-GFP-F2 was inactive (data not shown), so we were not able to verify expression of SpoIIGA(BH). However, all four Pro-σE substrates were cleaved in E. coli engineered to co-express SpoIIGA(BH) (Fig. 5, lanes 4, 8, 12 and 16). Likewise, SpoIIGA(BC)-GFP-F2 cleaved all four substrates (Fig. 5, lanes 3, 7, 11 and 15). As noted above, B. subtilis SpoIIGA(BS)-GFP-F2 cleaved all except Pro-σE(BC)-H6 (Fig. 5, lanes 1, 5, 9 and 13). Interestingly, SpoIIGA(BL)-GFP-F2 cleaved only its cognate substrate (Fig. 5, lanes 2, 6, 10 and 14). These results suggest that SpoIIGA(BL) has a relatively narrow substrate specificity as compared with SpoIIGA(BS), whereas SpoIIGA(BC) and SpoIIGA(BH) appear to have broader substrate specificity. The four enzymes are nearly identical in the region around the catalytic aspartate [D183 of SpoIIGA(BS)] and all have isoleucine or valine at critical positions [I294 and I295 of SpoIIGA(BS)] in a β-sheet secondary structure predicted to position the catalytic aspartate (Supplementary Fig. S3), so it is unlikely that these regions account for differences in substrate specificity. On the other hand, SpoIIGA(BS) and SpoIIGA(BL) differ at a few positions, and SpoIIGA(BC) and SpoIIGA(BH) differ greatly in three regions predicted to interact with substrate; the flap, loop and C-terminal regions corresponding to residues 242–257, 281–292 and 302–309, respectively, of SpoIIGA(BS) (Supplementary Fig. S3). These differences might account for the observed differences in substrate specificity.
Our results shed new light on residues in Pro-σE and in SpoIIGA that are important for cleavage, and demonstrate that SpoIIGA orthologs exhibit different ranges of substrate specificity. Our results show for the first time that residues in the N-terminal one-third of the pro-sequence of Pro-σE contribute to substrate specificity. The experimental determination of the 3D structure of a SpoIIGA·Pro-σE complex and further mutational analysis will be required to clarify the mechanism of substrate recognition by SpoIIGA and its orthologs. As shown in Supplementary Fig. S4A, the cleavage of Pro-σE(BC)-H6 by SpoIIGA(BC)-GFP-F2 depended on co-expression of B. subtilis SpoIIR-F2, indicating that the B. subtilis signalling protein stimulates SpoIIGA(BC)-GFP-F2, presumably by direct interaction, as B. subtilis SpoIIR-F2 was shown to interact directly with B. subtilis SpoIIGA-GFP-H6 when the proteins were co-expressed in E. coli (15). Our finding that B. subtilis SpoIIR stimulates SpoIIGA (BC), and likely SpoIIGA(BL) and SpoIIGA(BH), may not be surprising since an alignment of SpoIIR orthologs (Supplementary Fig. S5) shows higher similarity and identity than for SpoIIGA orthologs (Supplementary Fig. S3). In particular, SpoIIR orthologs are highly similar in a region near their C-termini, and all contain the identical sequence NWWCV (Supplementary Fig. S5), which might be important for interaction with SpoIIGA.
Supplementary Data
Supplementary Data are available at JB online.
Funding
The Science Research Promotion Fund; Grant-in-Aid for Scientific Research (C) (20580089); Grant-in-Aid for Young Scientists (B) (19780066) from the Japanese Society for the Promotion of Science; The Michigan Agricultural Experiment Station; National Science Foundation (0447799 to M.F.); The Alfred P. Sloan Foundation (to M.F.); and National Institutes of Health (GM43585 to L.K.).
Conflict of interest
None declared.
Supplementary Material
Acknowledgements
We are grateful for the gift of monoclonal antibody against σE from Dr William G. Haldenwang (University of Texas Health Science Center, San Antonio, TX, USA).
Glossary
Abbreviations
- BC
Bacillus cereus
- BH
Bacillus halodurans
- BL
Bacillus licheniformis
- BS
Bacillus subtilis
- F2
double FLAG tag
- GFP
green fluorescent protein
- H6
6 histidine tag
- HIV-1
human immunodeficiency virus type 1
- IPTG
isopropyl β-D-thiogalactopyranoside
- RNAP
RNA polymerase
- VMD
Visual Molecular Dynamics
- WT
wild type
References
- 1.Kroos L. The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Annu. Rev. Genet. 2007;41:13–39. doi: 10.1146/annurev.genet.41.110306.130400. [DOI] [PubMed] [Google Scholar]
- 2.Steil L, Serrano M, Henriques AO, Völker U. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology. 2005;151:399–420. doi: 10.1099/mic.0.27493-0. [DOI] [PubMed] [Google Scholar]
- 3.Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, Sato T, Setlow P, Losick R, Eichenberger P. The forespore line of gene expression in Bacillus subtilis. J. Mol. Biol. 2006;358:16–37. doi: 10.1016/j.jmb.2006.01.059. [DOI] [PubMed] [Google Scholar]
- 4.Hofmeister AE, Londoño-Vallejo A, Harry E, Stragier P, Losick R. Extracellular signal protein triggering the proteolytic activation of a developmental transcription factor in B. subtilis. Cell. 1995;83:219–226. doi: 10.1016/0092-8674(95)90163-9. [DOI] [PubMed] [Google Scholar]
- 5.Karow ML, Glaser P, Piggot PJ. Identification of a gene, spoIIR, that links the activation of σE to the transcriptional activity of σF during sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA. 1995;92:2012–2016. doi: 10.1073/pnas.92.6.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Londoño-Vallejo JA, Stragier P. Cell-cell signaling pathway activating a developmental transcription factor in Bacillus subtilis. Genes Dev. 1995;9:503–508. doi: 10.1101/gad.9.4.503. [DOI] [PubMed] [Google Scholar]
- 7.LaBell TL, Trempy JE, Haldenwang WG. Sporulation-specific σ factor σ29 of Bacillus subtilis is synthesized from a precursor protein, P31. Proc. Natl. Acad. Sci. USA. 1987;84:1784–1788. doi: 10.1073/pnas.84.7.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyao A, Theeragool G, Takeuchi M, Kobayashi Y. Bacillus subtilis spoVE gene is transcribed by σE-associated RNA polymerase. J. Bacteriol. 1993;175:4081–4086. doi: 10.1128/jb.175.13.4081-4086.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kenney TJ, Moran CP., Jr. Organization and regulation of an operon that encodes a sporulation-essential sigma factor in Bacillus subtilis. J. Bacteriol. 1987;169:3329–3339. doi: 10.1128/jb.169.7.3329-3339.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eichenberger P, Fujita M, Jensen ST, Conlon EM, Rudner DZ, Wang ST, Ferguson C, Haga K, Sato T, Liu JS, Losick R. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2004;2:e328. doi: 10.1371/journal.pbio.0020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eichenberger P, Jensen ST, Conlon EM, van Ooij C, Silvaggi J, González-Pastor JE, Fujita M, Ben-Yehuda S, Stragier P, Liu JS, Losick R. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 2003;327:945–972. doi: 10.1016/s0022-2836(03)00205-5. [DOI] [PubMed] [Google Scholar]
- 12.Feucht A, Evans L, Errington J. Identification of sporulation genes by genome-wide analysis of the σE regulon of Bacillus subtilis. Microbiology. 2003;149:3023–3034. doi: 10.1099/mic.0.26413-0. [DOI] [PubMed] [Google Scholar]
- 13.Fujita M, Losick R. An investigation into the compartmentalization of the sporulation transcription factor σE in Bacillus subtilis. Mol. Microbiol. 2002;43:27–38. doi: 10.1046/j.1365-2958.2002.02732.x. [DOI] [PubMed] [Google Scholar]
- 14.Stragier P, Bonamy C, Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988;52:697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- 15.Imamura D, Zhou R, Feig M, Kroos L. Evidence that the Bacillus subtilis SpoIIGA protein is a novel type of signal-transducing aspartic protease. J. Biol. Chem. 2008;283:15287–15299. doi: 10.1074/jbc.M708962200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ju J, Luo T, Haldenwang WG. Bacillus subtilis Pro-σE fusion protein localizes to the forespore septum and fails to be processed when synthesized in the forespore. J. Bacteriol. 1997;179:4888–4893. doi: 10.1128/jb.179.15.4888-4893.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ju J, Haldenwang WG. Tethering of the Bacillus subtilis σE proprotein to the cell membrane is necessary for its processing but insufficient for its stabilization. J. Bacteriol. 2003;185:5897–5900. doi: 10.1128/JB.185.19.5897-5900.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Londono-Vallejo JA. Mutational analysis of the early forespore/mother-cell signalling pathway in Bacillus subtilis. Microbiology. 1997;143:2753–2761. doi: 10.1099/00221287-143-8-2753. [DOI] [PubMed] [Google Scholar]
- 19.Peters HK, 3rd, Carlson HC, Haldenwang WG. Mutational analysis of the precursor-specific region of Bacillus subtilis σE. J. Bacteriol. 1992;174:4629–4637. doi: 10.1128/jb.174.14.4629-4637.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlson HC, Lu S, Kroos L, Haldenwang WG. Exchange of precursor-specific elements between Pro-σE and Pro-σK of Bacillus subtilis. J. Bacteriol. 1996;178:546–549. doi: 10.1128/jb.178.2.546-549.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott WR, Schiffer CA. Curling of flap tips in HIV-1 protease as a mechanism for substrate entry and tolerance of drug resistance. Structure. 2000;8:1259–1265. doi: 10.1016/s0969-2126(00)00537-2. [DOI] [PubMed] [Google Scholar]
- 22.Peters HK, III, Haldenwang WG. Isolation of a Bacillus subtilis spoIIGA allele that suppresses processing-negative mutations in the Pro-σE gene (sigE) J. Bacteriol. 1994;176:7763–7766. doi: 10.1128/jb.176.24.7763-7766.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.